Abstract
VjbR is a LuxR-type quorum-sensing (QS) regulator that plays an essential role in the virulence of the intracellular facultative pathogen Brucella, the causative agent of brucellosis. It was previously described that VjbR regulates a diverse group of genes, including the virB operon. The latter codes for a type IV secretion system (T4SS) that is central for the pathogenesis of Brucella. Although the regulatory role of VjbR on the virB promoter (PvirB) was extensively studied by different groups, the VjbR-binding site had not been identified so far. Here, we identified the target DNA sequence of VjbR in PvirB by DNase I footprinting analyses. Surprisingly, we observed that VjbR specifically recognizes a sequence that is identical to a half-binding site of the QS-related regulator MrtR of Mesorhizobium tianshanense. As shown by DNase I footprinting and electrophoretic mobility shift assays, generation of a palindromic MrtR-like-binding site in PvirB increased both the affinity and the stability of the VjbR-DNA complex, which confirmed that the QS regulator of Brucella is highly related to that of M. tianshanense. The addition of N-dodecanoyl homoserine lactone dissociated VjbR from the promoter, which confirmed previous reports that indicated a negative effect of this signal on the VjbR-mediated activation of PvirB. Our results provide new molecular evidence for the structure of the virB promoter and reveal unusual features of the QS target DNA sequence of the main regulator of virulence in Brucella.
Quorum sensing (QS) is a widespread mechanism of gene regulation that mediates bacterial cell-to-cell communication. In Gram-negative bacteria, most of the identified QS circuits consist of components that resemble those of the canonical LuxI/LuxR system of Vibrio fischeri (17). LuxI is the enzyme responsible for the synthesis of an acylated homoserine lactone signaling molecule (AHL) known as an autoinducer whose concentration is proportional to the bacterial population density. As bacterial cell density increases, the extracellular concentration of the autoinducer reaches a threshold value that activates the DNA-binding protein LuxR to control specific gene transcription.
In addition to the bioluminescence of Vibrio fischeri, QS-related systems have been shown to participate in the regulation of many bacterial physiological functions, including biofilm formation and the expression of virulence factors (7, 11, 16). In Brucella, it was also found that a LuxR-type regulator is directly involved in the control of transcription of important virulence determinants of this facultative intracellular bacterium (9).
Brucella is a genus of Gram-negative bacteria that cause brucellosis, a debilitating zoonotic disease that affects different species of domestic mammals. The Brucella species differ in their host specificities. In addition to their animal host, Brucella abortus, Brucella melitensis, and Brucella suis are also able to infect humans. The virulence of Brucella is determined by its ability to survive and replicate within macrophages and nonprofessional phagocytes. To achieve this, Brucella displays mechanisms that allow the bacterium to actively control its intracellular trafficking. After internalization into the host cells, Brucella is located in a vacuole that transiently interacts with endoplasmic reticulum (ER)-derived membranes and lysosomes (4, 24). Subsequently, the bacterium promotes the formation of the replicative compartment, which has the structural and functional properties of an ER. The virB operon of Brucella codes for a type IV secretion system (T4SS) that plays an essential role in the establishment of the replicative niche. It was observed that virB mutants of Brucella are not able to survive within the host cells and undergo lysosomal degradation (6, 18, 23). As in other pathogenic bacteria, the T4SS of Brucella is thought to act as a translocator of effector proteins into the host cell, which subvert eukaryotic cellular functions and allow the bacterium to overcome the host defenses (2, 8).
Analyses of regulation of the T4SS of Brucella showed that expression of the virB genes is rapidly induced after internalization into the host cells (3, 22). Studies carried out with bacteria cultured in vitro showed that the virB operon is expressed in nutrient-poor media at pH 4.5, which are conditions similar to those encountered by Brucella within the intracellular environment (3, 13). On the other hand, it was recently found that expression of the virB genes is linked to the histidine utilization pathway, both within the host cell and in cultured bacteria (21).
To date, the transcriptional regulators that were found to regulate virB expression through binding to the virB promoter (PvirB) are IHF, HutC, and VjbR (8, 21, 22). VjbR, together with BlxR, is one of the two QS-related LuxR-type factors of Brucella that regulate the expression of each other and control the transcription of an overlapping set of targets (19). In addition to the virB genes, VjbR also controls, either directly or indirectly, the expression a tetR-like regulator, flagellar components, outer membrane proteins, and genes coding for the recently identified VirB-translocated effectors VceA and VceC, among others (8, 9, 15, 27). Deletion of vjbR abrogates both virB expression and intracellular replication within infected cell lines, which demonstrates that this QS-related regulator plays a central role in the regulation of virulence of Brucella.
In B. melitensis, an N-dodecanoyl-AHL (C12-HSL) autoinducer molecule, was isolated from highly concentrated bacterial culture supernatants (26). It was observed that the exogenous addition of C12-HSL suppresses the expression of the virB operon in cultured bacteria and reduces the intracellular multiplication of B. melitensis in macrophages (9, 27). These observations suggested that a C12-HSL-mediated mechanism negatively modulates the regulatory activity of VjbR, which may play a role in the regulation of virB expression in vivo.
In order to investigate the regulatory mechanism of VjbR on the virB operon, we analyzed the interaction of this LuxR-type regulator with the virB promoter region. By DNase I footprinting, we identified the VjbR-binding site and determined the effect of C12-HSL on the DNA-binding activity of VjbR. Our results provide new molecular evidence for the structure of PvirB and reveal similarities between the target DNA sequences of VjbR and the Mesorhizobium QS-related regulator MrtR.
MATERIALS AND METHODS
Bacterial growth conditions.
The Escherichia coli strains were cultured at 28 or 37°C in a rotary shaker at 250 rpm. The media were supplemented with kanamycin (50 μg/ml) or ampicillin (100 μg/ml), as needed.
Construction of plasmids. (i) pGEM-T-vjbR.
A 790-bp DNA fragment that contains sequences corresponding to the vjbR gene (GenBank accession number BAB2_0118) was amplified by PCR using Pfx (Invitrogen), genomic DNA from B. abortus 2308 as the template, and primers rVjbRupBamHI (5′-GGATCCGAGTCTTGATCTCGTTCATTTTC-3′) and rVjbRdownKpnI (5′-GGTACCTCAGACGAGATGCTGTACC-3′). The PCR product was ligated into plasmid GEM-T Easy (Qiagen).
(ii) Expression vector pQE-31-vjbR.
A fragment that contains the vjbR gene was excised from plasmid pGEM-T-vjbR using BamHI and PstI (New England Biolabs) and ligated into plasmid pQE-31 (Qiagen) digested by the same enzymes. The resulting plasmid (pQE-31-vjbR) contains sequences that code for a six-histidine tag fused to the N terminus of the VjbR protein.
Expression and purification of recombinant proteins.
Recombinant HutC and IHF were prepared as described previously (21). Recombinant VjbR was prepared as follows. Plasmid pQE-31-vjbR was transferred into Escherichia coli M15(pREP4) (Qiagen). The bacteria were grown at 37°C in LB medium until exponential phase (optical density at 600 nm = 0.6). Subsequently, the cultures were incubated at 28°C and induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h. The bacteria were harvested, suspended in lysis buffer (20 mM Tris-HCl [pH 7.6], 1 mM phenylmethylsulfonyl fluoride [PMSF]), and disrupted by sonication. After centrifugation, NaCl was added to a final concentration of 0.35 M and the sample was loaded into a Hi-Trap nickel-chelating column (Amersham Biosciences). After a wash with buffer A (20 mM Tris-HCl [pH 7.6], 0.35 M NaCl), the column was eluted with a linear gradient of buffer B (20 mM Tris-HCl [pH 7.6], 03.5 M NaCl, 1 M imidazole). The eluates were analyzed by 12.5% SDS-PAGE, and the fractions containing VjbR (purity, near 95%) were pooled and dialyzed against buffer C (20 mM Tris-HCl [pH 7.6], 0.35 M NaCl, 3 mM β-mercaptoethanol). Samples were stored at −20°C with 5% sucrose.
DNase I footprinting.
Probe PvirB, which contains sequences corresponding to positions −201 to +24, was generated as follows. Primer pvirdownI (22) was 5′ end labeled with 32P by using [γ-32P]ATP and T4 polynucleotide kinase (New England Biolabs). Subsequently, a PCR was performed using Taq (Invitrogen), the 32P-labeled primer pvirdownI, primer pvu229 (22), and the genomic DNA of B. abortus 2308 as the template.
Probe PvirB hM− was generated as follows. Two PCRs were performed using Pfx and primers pvirup (22) and VJBSup (5′-GGGCGCTTGCACTAAATAGATCGGGTGTGTGAATGACGCCCAG-3′) or primers VJBSdown (5′-CCCGATCTATTTAGTGCAAGCGCCCTTGTCCATATATCGGCTTAAAC-3′) and pvirdownII (5′-GTCTGAGGTGCAACAGT-3′). Both products were annealed and used as templates for a PCR performed with Pfx, primer pvirup, and primer pvirdownII. The resulting product, which contains a DNA fragment corresponding to positions −430 to +82 of PvirB with a replacement of the VjbR-binding site by a nonrelated sequence, was used as the template for a PCR performed with Taq, the 32P-labeled primer pvirdownI, and primer pvu229.
Probe PvirB-MrtR-bs was generated as follows. Two PCRs were performed using Pfx and primers pvirup and VJMRup (5′-AGATGGGCCCCCTCAGATGAGGGGGCTATATATTGTG-3′) or primers VJMRdown (5′-TCTGAGGGGGCCCATCTCAAGCATATTTGTCCATATATC-3′) and pvirdownII. Both products were annealed and used as the templates for a PCR performed with Pfx, primer pvirup, and primer pvirdownII. The resulting product, which contains a DNA fragment corresponding to positions −430 to +82 of PvirB with an entire dyad symmetric MrtR-binding site, was used as the template for a PCR performed with Taq, the 32P-labeled primer pvirdownI, and primer pvu229.
Probe PtetR was generated by PCR using Taq, primer PtetRforward (5′-CTGCGTTCTTTCATTGGCA-3′), 32P-labeled primer PtetRreverse (5′-AGTGTTGCGTGCAGGTTTC-3′), and the genomic DNA of B. abortus 2308 as the template.
Binding reactions were performed by incubating 100,000 cpm of the 32P-labeled probe and different concentrations of recombinant VjbR in binding buffer (15 mM Tris-HCl [pH 8.0], 0,1 mg ml−1 bovine serum albumin [BSA], 1 mM dithiothreitol [DTT], 30 mM KCl) in a final volume of 20 μl. For the analyses of the effect of the autoinducer signal on the DNA-binding activity of VjbR, C12-HSL (Sigma-Aldrich) was dissolved in acetonitrile to a concentration of 400 μM. Different volumes of the C12-HSL solution or acetonitrile were added to the binding reactions at the indicated final concentrations. After incubation of the binding reaction mixtures for 20 min, the MgCl2 and CaCl2 concentrations were adjusted to 1.5 and 0.5 mM, respectively. Subsequently, each reaction mixture was incubated with RQ1 DNase I (Promega) for 1 min at room temperature. The reactions were terminated by the addition of 5 μl of stop solution (25 mM EDTA, 0.6 M sodium acetate). The digested products were extracted with phenol-chloroform, ethanol precipitated, and resuspended in 4 μl of sequencing gel loading buffer. The DNA fragments were separated on a 6% polyacrylamide DNA sequencing gel and were visualized by autoradiography. DNA sequencing reactions carried out with primer pvirdown I or PtetRreverse were used to localize the position of the protected regions.
EMSAs.
The probes were internally labeled by PCR through inclusion of 50 mCi [α-32P]dCTP in the reaction mixture and subsequently purified on native polyacrylamide gels. The control probe, which contains sequences corresponding to 226 bp of virB10 from B. abortus 2308, was constructed as indicated previously (21). Probes PvirB and PvirB-MrtR bs were generated using Taq, primers pvu229 and pvirdownI, and the corresponding templates that were used for the DNase I footprinting probes described above.
The electrophoretic mobility shift assays (EMSAs) were performed in a volume of 20 μl containing DNA-binding buffer [15 mM Tris-HCl (pH 8.0), 0.5 mM EDTA, 50 μg ml−1 BSA, 1 mM DTT, 30 mM KCl, 50 μg ml−1 poly(dI·dC) (Amersham), 6% glycerol], 10,000 cpm of the 32P-labeled probe, and the indicated recombinant proteins. After incubation at room temperature for 30 min, the protein-DNA complexes were separated from the free probes by electrophoresis on 8% nondenaturing polyacrylamide gels at a constant voltage of 220 V. The results of the EMSAs were visualized by exposure of the gels to X-ray films.
RESULTS AND DISCUSSION
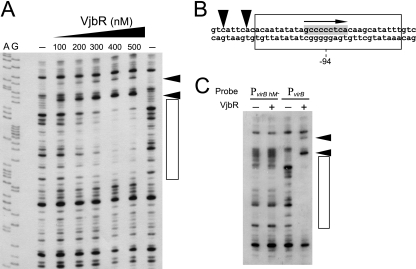
Although the regulatory role of VjbR has been extensively studied by genetic analyses (8, 9, 19, 27), the molecular basis for the observed VjbR-mediated activation of the virB operon is still not fully understood. To identify the VjbR-binding site and gain more insight into the regulatory mechanism of this LuxR-type transcription factor, we performed DNase I footprinting analyses using His-tagged recombinant protein VjbR and a probe that contains sequences corresponding to PvirB of B. abortus. As shown in Fig. 1 A, the binding of VjbR generated two DNase I hypersensitivity sites and a 30-bp protected region centered at position −94. These results also indicated that the affinity of the VjbR-PvirB interaction seemed to be relatively low, since the protected region was observed at concentrations higher than 200 nM VjbR.
FIG. 1.
Identification of the VjbR-binding site in PvirB. (A) DNase I footprinting analysis performed with a probe corresponding to 224 bp of PvirB and increasing concentrations of VjbR, as indicated. The contents of lanes A and G are from DNA sequencing reactions performed by the Sanger method. The VjbR-protected region is indicated by an open rectangle. Arrowheads indicate DNase I hypersensitivity sites. (B) Schematic representation of the VjbR-protected sequence. The protected region and DNase I hypersensitivity sites are as indicated in panel A. Nucleotides that match the MrtR-binding site of Mesorhizobium tianshansense are highlighted in gray. The orientation of the half-dyad symmetric sequence corresponding to the MrtR-binding site is indicated by an arrow. The position relative to the transcription start site is indicated. (C) DNase I footprinting experiment performed with probe PvirB or control probe PvirB hM− in the absence or in the presence of 400 nM VjbR.
The identified VjbR-binding site exhibited very unusual features. First, instead of having a typical 18- to 20-bp dyad symmetric sequence, such as those observed for the DNA-binding sites of well-studied QS-related regulators (e.g., TraR and LuxR) (10, 32), the VjbR-protected region contains the sequence GCCCCCTCA (Fig. 1B). This motif is reminiscent of the binding site of MrtR, a QS-related transcriptional regulator of Mesorhizobium tianshanense that is involved in the control of nodulation (30). Interestingly, the motif found at the center of the VjbR-protected region is identical to a half site of the dyad symmetric sequence recognized by MrtR in the promoter of the mrtI locus of M. tianshanense (PmrtI), which codes for the AHL synthase MrtI (Fig. 2 A). To determine whether GCCCCCTCA is the sequence recognized by VjbR, we constructed a probe that lacks this 9-bp motif due to a replacement by a nonrelated sequence (probe PvirB hM−). Using DNase I footprinting, we observed that VjbR was completely unable to bind to the mutant probe (Fig. 1C), which demonstrated that this transcriptional regulator specifically binds to the sequence that is related to the cognate DNA-binding site of MrtR. The second unusual characteristic is that the DNase I-hypersensitive sites observed upstream of the VjbR-protected region were generated at 100 nM VjbR, a concentration that was insufficient for generating the DNase I protection (Fig. 1A). This indicates that VjbR is somehow interacting with its binding site before it protects it from cleavage by DNase I. Such an interaction may participate in recognition of structural elements of the VjbR-binding site, probably by a mechanism similar to that of TraR, whose binding to target DNA sequences involves detection of DNA flexibility (29).
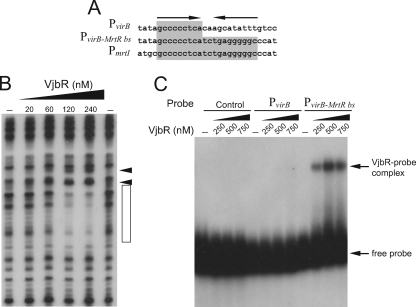
FIG. 2.
Generation of a dyad symmetric MrtR-binding site in PvirB increased the affinity and stability of the VjbR-DNA complex. (A) Schematic representation of sequences corresponding to wild-type probe PvirB, probe PvirB-MrtR bs, or the mrtI promoter (PmrtI) of M. tianshanense. Sequences that match the MrtR-binding site of PmrtI are highlighted in gray. The dyad symmetry of the MrtR-binding site is indicated by arrows. (B) DNase I footprinting analysis performed with probe PvirB-MrtR bs and increasing concentrations of VjbR, as indicated. The VjbR-protected region is indicated by an open rectangle. Arrowheads indicate DNase I hypersensitivity sites. (C) EMSA performed with a control probe, probe PvirB, or probe PvirB-MrtR bs and increasing concentrations of VjbR.
Besides recognizing similar binding sites, the protein VjbR shares more sequence similarity to MrtR than to other QS regulators, such as LuxR from V. fischeri and TraR from Agrobacterium tumefaciens (BLAST E values for similarity to MtrR, LuxR, and TraR, 5E−22, 7E−11, and 5E−07, respectively). On the basis of these findings, we hypothesized that generation of the entire dyad symmetric MrtR-binding site in PvirB may increase the affinity of VjbR to the promoter. To investigate this possibility, we performed DNase I footprinting using increasing amounts of VjbR and the PvirB-MrtR-bs probe, which contains an entire dyad symmetric sequence generated by the insertion of an additional GCCCCCTCA motif in the complementary strand (Fig. 2A). Figure 2B shows that the affinity of VjbR to PvirB-MrtR-bs was higher than to the wild-type probe, since the protected region was detected at 120 nM VjbR. Thus, taken together, these observations support the notion that both the structure and target DNA sequences of VjbR are closely related to those of MrtR.
To further characterize the interaction between VjbR and PvirB, we performed EMSAs using the recombinant protein and the PvirB probe or a control probe. Surprisingly, we did not observe any retarded signal with any of these probes (Fig. 2C). This observation suggests that the relatively weak interaction between VjbR and PvirB observed by DNase I footprinting (Fig. 1A) does not withstand the electrophoretic conditions of EMSA. When PvirB-MrtR-bs was used as the probe, a signal corresponding to a protein-DNA complex was observed by EMSA, which indicates that an increase of the affinity of VjbR for its target DNA sequence contributed to the stability of the VjbR-DNA complex during electrophoresis (Fig. 2C).
Subsequently, we asked whether the binding of other regulators would increase the affinity of VjbR for the promoter by inducing changes in the DNA structure. Using EMSA or DNase I footprinting, no changes in DNA-binding activity were observed for VjbR when it was coincubated with HutC or IHF (data not shown). Therefore, these results suggest that the regulatory mechanism exerted by VjbR on the virB operon does not depend on the positive modulation of its affinity for the promoter sequences.
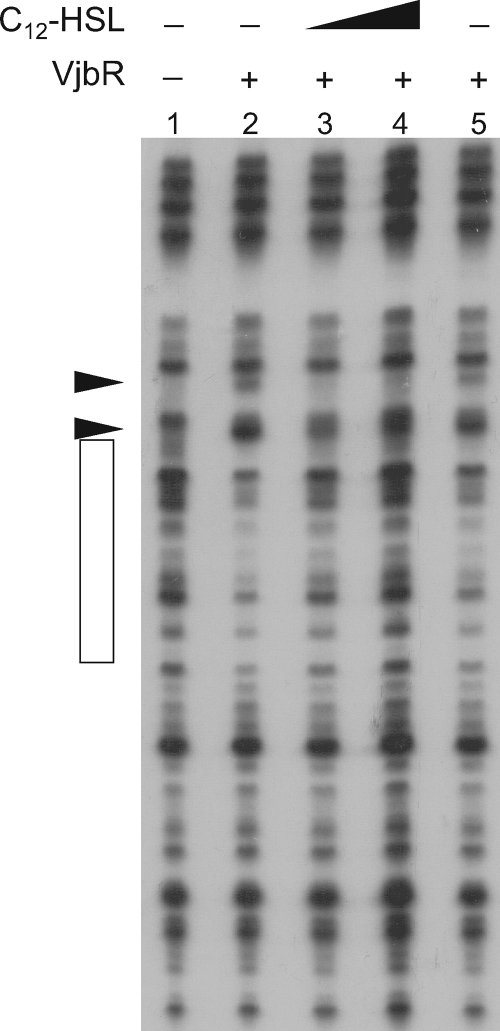
It was previously reported that the exogenous addition of C12-HSL reduces the expression of the virB operon (9). Studies performed with mutations in the putative AHL-binding domain suggested that the autoinducer interacts with VjbR and modulates its regulatory function (27). To determine whether C12-HSL affects the DNA-binding activity of VjbR, DNase I footprinting experiments were performed using VjbR and the PvirB probe in the presence of the autoinducer signal. Figure 3 shows that the addition of 10 μM C12-HSL impaired both DNase I protection and the VjbR-induced DNase I hypersensitivity sites, whereas the acetonitrile used to dissolve the autoinducer signal had no effect. These results are consistent with those in previous reports that showed that C12-HSL reduces the VjbR-mediated activation of virB expression and demonstrate that the autoinducer molecule negatively modulates the DNA-binding activity of this transcriptional regulator.
FIG. 3.
The addition of C12-HSL dissociates VjbR from PvirB. DNase I footprinting analysis was performed with PvirB, VjbR, and different concentrations of C12-HSL. Concentration of VjbR: lane 1, no protein; lanes 2 to 5, 300 nM. The concentrations of C12-HSL were 0 (lanes 1, 2, and 5), 10 μM (lane 3), and 20 μM (lane 4). Lane 5, acetonitrile added to a final concentration equivalent to that in lanes 3 and 4. The VjbR-protected region is indicated by an open rectangle. DNase I hypersensitivity sites are indicated by arrowheads.
Bacterial pathogens have evolved different mechanisms to avoid host defenses and promote the formation of niches permissive for their replication. In many cases, regulation of the virulence determinants involved in such mechanisms is directed by QS circuits. As in other pathogenic bacterial genera, such as Pseudomonas, Erwinia, and Agrobacterium, it was recently found that a LuxR-type regulator is the main activator of essential components for the virulence of Brucella (9, 16). Here, we identified the target DNA sequence of this QS-related transcription factor in PvirB, which exhibited features similar to those of the LuxR-type regulator MrtR. Curiously, VjbR recognizes a sequence that is identical to the half-binding site of MrtR in PmrtI of M. tianshaense. The existence of transcriptional regulators that bind to functional half-binding sites, both in prokaryotes and in eukaryotes, was reported previously (1, 31). PrgX is a regulatory protein involved in the pheromone-inducible conjugation of the Gram-positive bacterium Enterococcus faecalis (5). It was recently found that PrgX binds to a palindromic high-affinity primary PrgX-binding site, as well as to a low-affinity secondary PrgX half-binding site (1). The authors demonstrated that the binding to the low-affinity half-binding site exerts an important physiological role on the autoregulation of PrgX. Similarly, we observed that VjbR regulates the main virulence factor of Brucella through binding to a relatively low-affinity half-binding site. What is intriguing is that, unlike PmrtI of Mesorhizobium, no palindromic GCCCCCTCA-containing MrtR-like binding sites were found by extensive sequence searches in the Brucella genomes (R. Sieira, unpublished results). This observation suggests that Brucella lacks high-affinity binding sites for VjbR. The relatively weak and/or unstable interaction between VjbR and its target DNA sequences probably facilitates dissociation of the complex in response to the autoinducer signal and allows the rapid inactivation of promoter activity, consistent with the fine-tuned regulation of the intracellular virB expression of B. abortus observed (22).
Sequence analyses revealed that GCCCCCTCA motifs are present in upstream regions of many open reading frames in the Brucella genome. However, DNase I footprinting experiments performed on four of the GCCCCCTCA-containing putative promoter regions failed to identify VjbR-binding sites other than that of PvirB (Sieira, unpublished). This suggests that, besides the 9-bp motif, some other structural component of the VjbR-binding site of PvirB is required for specific recognition. Such hypothetical additional structural components may participate in the VjbR-PvirB interaction as it occurs with c-Maf. This eukaryotic regulator, which recognizes both half and palindromic binding sites, requires an AT-rich 5′-flanking sequence for the recognition of its half-binding site targets, whereas a palindromic dyad symmetric sequence is a sufficient condition for specific high-affinity binding to DNA (31).
Unlike what we have observed in PvirB by DNase I footprinting, VjbR itself was not able to bind to the tetR promoter (PtetR) (data not shown), a regulatory region that was previously shown to be involved in the positive autoregulation of VjbR (8). This is consistent with the fact that the sequence GCCCCCTCA, which was demonstrated here to be necessary for the binding of VjbR to PvirB, was not found upstream of the tetR locus. Furthermore, the 9-bp motif was not found in the upstream regions of other loci that were previously reported to be positively regulated by VjbR. This observation suggests that, except for the virB operon, regulation of the remainder of the identified VjbR-controlled genes is indirect. Therefore, further work will be required to find out additional components of the VjbR-binding site and to identify direct targets of the VjbR regulon. As VjbR affects the expression of many genes involved in diverse functions, it can be speculated that it directly activates the transcription of one or more genes encoding global regulators, which may control the expression of a large number of targets at the transcriptional or posttranscriptional level.
Using EMSA, de Jong et al. (8) recently reported that VjbR interacts directly with PvirB and suggested that the regulator binds to a lux box-like element that is centered at position −37 relative to the transcription start site. However, our results showed that such a putative lux box is not the target DNA sequence of VjbR. It is also worth noting that, as the putative lux box is located between positions −45 and −28 (8), binding of VjbR to this sequence would sterically hinder the access of the RNA polymerase holoenzyme (RNApol) to position −35, with the consequent impairment of promoter activation.
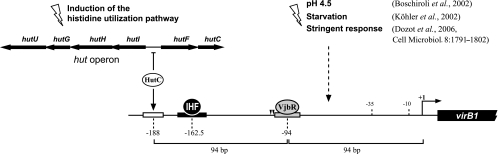
In this work, we provided additional molecular evidence for the structure of the virB promoter of B. abortus. Our results showed that the VjbR-binding site is centered at position −94, far upstream of the transcription start site (Fig. 4). It therefore seems unlikely that VjbR activates virB expression by direct contact with RNApol or with its α-carboxy-terminal domains. However, under certain stress conditions, the bacterial chromosomes undergo modulation of nucleoid organization (25). The nucleoid-associated protein Dps, which participates in nucleoid compaction in starved bacteria, is induced after the internalization of Brucella within host cells (14). One possibility is that the latter, probably in concert with other nucleoid-associated proteins of Brucella, may introduce the compaction of DNA in such a way that allows VjbR to interact with RNApol. On the other hand, it is interesting to note that the VjbR-binding site is positioned halfway between the transcription start site and the region that contains the binding sites for HutC and IHF (Fig. 4), which were previously found to enhance virB expression (21, 22). Thus, an additional possible mechanism is that promoter-bound VjbR induces DNA bending in such an orientation that allows it to bring distant elements closer to RNApol for activation, likely with the involvement of additional factors.
FIG. 4.
Schematic representation of the structure of PvirB and different regulatory inputs that control virB expression. Solid line, sequences corresponding to the virB promoter. The 5′ region of the first gene of the virB operon (virB1) is indicated. Open, black, and gray rectangles, binding sites for HutC, IHF, and VjbR, respectively. The position relative to the transcription start site (+1) is indicated in each case. HutC-mediated activation on PvirB and repression on the hut operon is indicated. Dashed arrow, unidentified pathways that activate virB expression in response to different stimuli.
In addition to EsaR and ExpR, VjbR is one of the few examples of LuxR-type proteins whose regulatory activity is negatively modulated by AHL (20, 28). However, even though it shares the same AHL responsiveness, VjbR exerts a positive regulatory role, which is opposite the regulatory roles of EsaR and ExpR. Thus, VjbR falls into a category that is particularly different from the rest of the LuxR-type members: it activates transcription in the absence of AHL, whereas the addition of exogenous AHL abrogates the VjbR-mediated activation. Here, we demonstrated that C12-HSL dissociates VjbR from the promoter, which is consistent with reports from different studies of a negative effect of AHL on virB expression (9, 26). On the basis of the previous observations, it was suggested that an autoinducer signal is probably involved in the downregulation of virB expression in vivo. However, no homologues for AHL synthases were found in the genome of Brucella, and the pathway for intrinsic production of the autoinducer by B. melitensis remains unidentified. Thus, although this so-called orphan LuxR regulator conserves an AHL-responsive domain, there is a possibility that VjbR mediates interkingdom communication by detection of signaling molecules from its eukaryotic host, as occurs with OryR from Xanthomonas oryzae (12).
To date, several lines of evidence provided by different groups showed that virB expression is under the control of acidic, metabolic, and nutritional stress signals (Fig. 4). Further work will be needed to ascertain the precise regulatory mechanism of the factors that bind to PvirB and to elucidate how the different signal inputs are integrated to modulate expression of the T4SS VirB of Brucella within the host.
Acknowledgments
This work is dedicated to the memory of R.A.U.
We thank Angeles Zorreguieta for critical reading of the manuscript.
This work was supported by grants PICT05-38207 to R.S. and PICT06-651 to R.A.U. from the Agencia Nacional de Promoción Científica y Tecnológica, Buenos Aires, Argentina.
Footnotes
Published ahead of print on 16 April 2010.
REFERENCES
- 1.Bae, T., B. Kozlowicz, and G. M. Dunny. 2002. Two targets in pCF10 DNA for PrgX binding: their role in production of Qa and prgX mRNA and in regulation of pheromone-inducible conjugation. J. Mol. Biol. 315:995-1007. [DOI] [PubMed] [Google Scholar]
- 2.Boschiroli, M. L., S. Ouahrani-Bettache, V. Foulongne, S. Michaux-Charachon, G. Bourg, A. Allardet-Servent, C. Cazevieille, J. P. Lavigne, J. P. Liautard, M. Ramuz, and D. O'Callaghan. 2002. Type IV secretion and Brucella virulence. Vet. Microbiol. 90:341-348. [DOI] [PubMed] [Google Scholar]
- 3.Boschiroli, M. L., S. Ouahrani-Bettache, V. Foulongne, S. Michaux-Charachon, G. Bourg, A. Allardet-Servent, C. Cazevieille, J. P. Liautard, M. Ramuz, and D. O'Callaghan. 2002. The Brucella suis virB operon is induced intracellularly in macrophages. Proc. Natl. Acad. Sci. U. S. A. 99:1544-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Celli, J., C. de Chastellier, D. M. Franchini, J. Pizarro-Cerda, E. Moreno, and J. P. Gorvel. 2003. Brucella evades macrophage killing via VirB-dependent sustained interactions with the endoplasmic reticulum. J. Exp. Med. 198:545-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung, J. W., B. A. Bensing, and G. M. Dunny. 1995. Genetic analysis of a region of the Enterococcus faecalis plasmid pCF10 involved in positive regulation of conjugative transfer functions. J. Bacteriol. 177:2107-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Comerci, D. J., M. J. Martinez-Lorenzo, R. Sieira, J. P. Gorvel, and R. A. Ugalde. 2001. Essential role of the VirB machinery in the maturation of the Brucella abortus-containing vacuole. Cell. Microbiol. 3:159-168. [DOI] [PubMed] [Google Scholar]
- 7.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 8.de Jong, M. F., Y. H. Sun, A. B. den Hartigh, J. M. van Dijl, and R. M. Tsolis. 2008. Identification of VceA and VceC, two members of the VjbR regulon that are translocated into macrophages by the Brucella type IV secretion system. Mol. Microbiol. 70:1378-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delrue, R. M., C. Deschamps, S. Leonard, C. Nijskens, I. Danese, J. M. Schaus, S. Bonnot, J. Ferooz, A. Tibor, X. De Bolle, and J. J. Letesson. 2005. A quorum-sensing regulator controls expression of both the type IV secretion system and the flagellar apparatus of Brucella melitensis. Cell. Microbiol. 7:1151-1161. [DOI] [PubMed] [Google Scholar]
- 10.Egland, K. A., and E. P. Greenberg. 1999. Quorum sensing in Vibrio fischeri: elements of the luxl promoter. Mol. Microbiol. 31:1197-1204. [DOI] [PubMed] [Google Scholar]
- 11.Engebrecht, J., K. Nealson, and M. Silverman. 1983. Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell 32:773-781. [DOI] [PubMed] [Google Scholar]
- 12.Ferluga, S., and V. Venturi. 2009. OryR is a LuxR-family protein involved in interkingdom signaling between pathogenic Xanthomonas oryzae pv. oryzae and rice. J. Bacteriol. 191:890-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Köhler, S., V. Foulongne, S. Ouahrani-Bettache, G. Bourg, J. Teyssier, M. Ramuz, and J. P. Liautard. 2002. The analysis of the intramacrophagic virulome of Brucella suis deciphers the environment encountered by the pathogen inside the macrophage host cell. Proc. Natl. Acad. Sci. U. S. A. 99:15711-15716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lamontagne, J., A. Forest, E. Marazzo, F. Denis, H. Butler, J. F. Michaud, L. Boucher, I. Pedro, A. Villeneuve, D. Sitnikov, K. Trudel, N. Nassif, D. Boudjelti, F. Tomaki, E. Chaves-Olarte, C. Guzman-Verri, S. Brunet, A. Cote-Martin, J. Hunter, E. Moreno, and E. Paramithiotis. 2009. Intracellular adaptation of Brucella abortus. J. Proteome Res. 8:1594-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leonard, S., J. Ferooz, V. Haine, I. Danese, D. Fretin, A. Tibor, S. de Walque, X. De Bolle, and J. J. Letesson. 2007. FtcR is a new master regulator of the flagellar system of Brucella melitensis 16M with homologs in Rhizobiaceae. J. Bacteriol. 189:131-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 17.Nealson, K. H., T. Platt, and J. W. Hastings. 1970. Cellular control of the synthesis and activity of the bacterial luminescent system. J. Bacteriol. 104:313-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Callaghan, D., C. Cazevieille, A. Allardet-Servent, M. L. Boschiroli, G. Bourg, V. Foulongne, P. Frutos, Y. Kulakov, and M. Ramuz. 1999. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol. Microbiol. 33:1210-1220. [DOI] [PubMed] [Google Scholar]
- 19.Rambow-Larsen, A. A., G. Rajashekara, E. Petersen, and G. Splitter. 2008. Putative quorum-sensing regulator BlxR of Brucella melitensis regulates virulence factors including the type IV secretion system and flagella. J. Bacteriol. 190:3274-3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reverchon, S., M. L. Bouillant, G. Salmond, and W. Nasser. 1998. Integration of the quorum-sensing system in the regulatory networks controlling virulence factor synthesis in Erwinia chrysanthemi. Mol. Microbiol. 29:1407-1418. [DOI] [PubMed] [Google Scholar]
- 21.Sieira, R., G. M. Arocena, L. Bukata, D. J. Comerci, and R. A. Ugalde. 2010. Metabolic control of virulence genes in Brucella abortus: HutC coordinates virB expression and the histidine utilization pathway by direct binding to both promoters. J. Bacteriol. 192:217-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sieira, R., D. J. Comerci, L. I. Pietrasanta, and R. A. Ugalde. 2004. Integration host factor is involved in transcriptional regulation of the Brucella abortus virB operon. Mol. Microbiol. 54:808-822. [DOI] [PubMed] [Google Scholar]
- 23.Sieira, R., D. J. Comerci, D. O. Sanchez, and R. A. Ugalde. 2000. A homologue of an operon required for DNA transfer in Agrobacterium is required in Brucella abortus for virulence and intracellular multiplication. J. Bacteriol. 182:4849-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Starr, T., T. W. Ng, T. D. Wehrly, L. A. Knodler, and J. Celli. 2008. Brucella intracellular replication requires trafficking through the late endosomal/lysosomal compartment. Traffic 9:678-694. [DOI] [PubMed] [Google Scholar]
- 25.Stavans, J., and A. Oppenheim. 2006. DNA-protein interactions and bacterial chromosome architecture. Phys. Biol. 3:R1-R10. [DOI] [PubMed] [Google Scholar]
- 26.Taminiau, B., M. Daykin, S. Swift, M. L. Boschiroli, A. Tibor, P. Lestrate, X. De Bolle, D. O'Callaghan, P. Williams, and J. J. Letesson. 2002. Identification of a quorum-sensing signal molecule in the facultative intracellular pathogen Brucella melitensis. Infect. Immun. 70:3004-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uzureau, S., M. Godefroid, C. Deschamps, J. Lemaire, X. De Bolle, and J. J. Letesson. 2007. Mutations of the quorum sensing-dependent regulator VjbR lead to drastic surface modifications in Brucella melitensis. J. Bacteriol. 189:6035-6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.von Bodman, S. B., D. R. Majerczak, and D. L. Coplin. 1998. A negative regulator mediates quorum-sensing control of exopolysaccharide production in Pantoea stewartii subsp. stewartii. Proc. Natl. Acad. Sci. U. S. A. 95:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White, C. E., and S. C. Winans. 2007. The quorum-sensing transcription factor TraR decodes its DNA binding site by direct contacts with DNA bases and by detection of DNA flexibility. Mol. Microbiol. 64:245-256. [DOI] [PubMed] [Google Scholar]
- 30.Yang, M., J. L. Giel, T. Cai, Z. Zhong, and J. Zhu. 2009. The LuxR family quorum-sensing activator MrtR requires its cognate autoinducer for dimerization and activation but not for protein folding. J. Bacteriol. 191:434-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshida, T., T. Ohkumo, S. Ishibashi, and K. Yasuda. 2005. The 5′-AT-rich half-site of Maf recognition element: a functional target for bZIP transcription factor Maf. Nucleic Acids Res. 33:3465-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu, J., and S. C. Winans. 1999. Autoinducer binding by the quorum-sensing regulator TraR increases affinity for target promoters in vitro and decreases TraR turnover rates in whole cells. Proc. Natl. Acad. Sci. U. S. A. 96:4832-4837. [DOI] [PMC free article] [PubMed] [Google Scholar]