ADARs are a family of enzymes, present in all animals, that convert adenosines to inosines within double-stranded RNA. Inosine has different base-pairing properties from adenosine, and thus, editing alters RNA structure, coding potential and splicing patterns. The first identified ADAR substrates were edited in codons, and ADARs were presumed to function primarily in proteome diversification. Although this is an important ADAR function, especially in the nervous system, editing in coding sequences is rare compared to editing in non-coding sequences. Introns and untranslated regions of mRNA are the primary non-coding targets, but editing also occurs in small RNAs, such as miRNAs. Although the role of editing in non-coding sequences remains unclear, ongoing research suggests functions in the regulation of a variety of post-transcriptional processes.
Basics of RNA editing by ADARs
RNA editing is the alteration of RNA by nucleotide modification, insertion or deletion (reviewed in [1]). In metazoans, a common type of RNA editing is a nucleotide modification involving the hydrolytic deamination of adenosine (A) to inosine (I). This type of editing is catalyzed by the Adenosine Deaminases that act on RNA (ADARs) family [2–4], which target RNA that is completely, or largely, double-stranded. ADARs are likely present in all animals and have been studied in squids, worms, flies and mammals. A single organism encodes between 1–3 ADARs, and all family members contain a highly conserved catalytic domain at their C-terminus, and variable numbers of N-terminal dsRNA binding motifs (dsRBMs). Whereas ADARs are essential in mammals, invertebrates that lack ADARs are viable, but exhibit behavioral defects [5–8].
Inosine is recognized as guanosine (G) by the translation and splicing machineries, and thus, ADARs can alter the protein-coding information of an mRNA. In addition, because inosine prefers to pair with cytidine (C), ADARs destabilize dsRNA by changing AU base-pairs to IU mismatches [9,10], or increase its stability by changing AC mismatches to IC base-pairs [11]; while other changes to RNA structure have not been documented, conceivably, ADARs could promote more complex structural rearrangements as well. In choosing which adenosines to target, ADARs show slight sequence preferences [12, 13], and in addition, the number of adenosines targeted in a dsRNA is dependent on its length and structure. For example, in vitro studies show that duplexes of about 15–40 bp are edited selectively, at very few sites, whereas those greater than 50 base-pairs are extensively, or non-selectively deaminated (50–60% of adenosines) [12, 14]. Selective deamination is also observed in dsRNA that is interrupted by bulges, loops and mismatches [15, 16].
The two modes of editing, selective and non-selective, are observed in endogenous ADAR substrates. Coding regions of mRNAs typically undergo selective deamination (e.g. see [17–20]), which is not surprising, given that non-selective editing would lead to multiple amino acid changes, and likely, a nonfunctional protein. Non-coding regions of mRNA such as introns and untranslated regions (UTRs) are also targeted by ADARs, usually within dsRNA formed by pairing of inverted repetitive elements [2, 21, 22]. These structures are long and almost completely base-paired, and correspondingly, are subject to non-selective deamination.
Although the first non-coding substrates of ADARs were identified a decade ago [23], and five years have passed since the identification of over 10,000 non-coding editing sites in the human genome [11, 24–26], the function of these editing events remains intriguing, but unclear. Indeed, studies point to a wide-range of effects on gene expression. Many provocative "proof of principle” and in vitro studies have been performed, and although some of these will be discussed, this review will focus on editing events identified in endogenous RNA.
Do edited 3’ UTR structures affect cellular localization?
Based on analyses of synthetic RNAs, an early and attractive hypothesis posited that inosines within an RNA caused nuclear retention by promoting binding to the multifunctional RNA binding protein, p54nrb [27, 28]. mRNAs selectively deaminated in codons are clearly translated in the cytoplasm [29, 30], so nuclear retention was proposed to require the non-selective deamination found in non-coding sequences (e.g., UTRs). However, endogenous mRNAs with multiple inosines in their 3' UTRs have now been found in the cytoplasm, in both mammalian cells and Caenorhabditis elegans, emphasizing that nuclear retention is not a general phenomenon (Figure 1) [31–33]. Further, inosine is not required for in vivo binding of p54nrb to RNA, as an interaction between p54nrb and NEAT1, a nuclear enriched non-coding RNA that is not edited, was recently detected [34–36]. Interestingly, in earlier studies, cross-linking of p54nrb to inosine-containing RNAs was efficiently competed by inosines in the context of the 4 Watson-Crick nucleotides, but poorly by polyinosine; the latter emphasizes that p54nrb binding to RNA involves more than recognition of inosine [27].
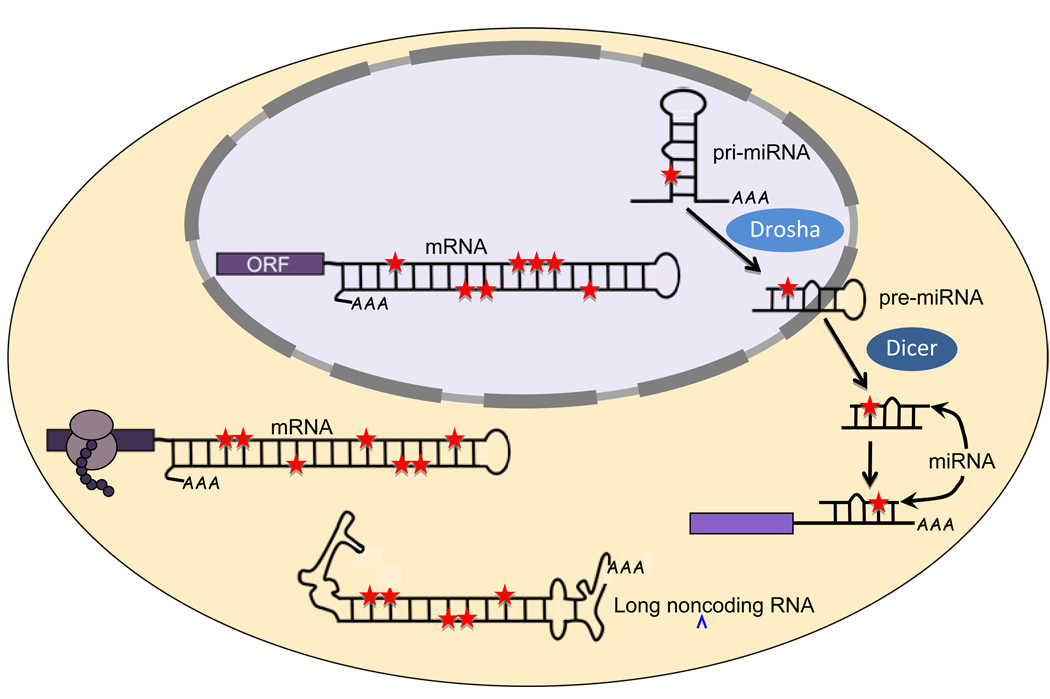
Figure 1.
Endogenous RNAs edited by ADARs in non-coding sequences. The various types of endogenous RNA targeted by ADARs in base-paired, non-coding sequences are depicted in the subcellular compartment where they are observed (nucleus, purple oval; cytoplasm, beige oval). Colored rectangles show open-reading frames (ORF), with twin circles representing ribosomes translating an mRNA. In the cartoon, the exact sites of inosines (red stars) are not meaningful, but selective deamination is denoted by a single star in an RNA and non-selective deamination by multiple stars in an RNA. Small RNAs that are edited, including many pri-miRNAs and at least one pre- and one mature miRNA, have been identified in human cell lines [42, 54–56, 58]. In addition, a large (~800 nucleotides), polyadenylated, non-coding RNA, rncs-1 (depicted with branched terminal structures), with multiple editing sites is present in C. elegans and antagonizes Dicer function [71]. Although some mammalian mRNAs with edited 3’ UTRs have been detected in the nucleus [32, 37], in both worms and human cell lines, mRNAs with edited 3’ UTRs are also present on translating ribosomes [31].
Although editing within a 3’ UTR clearly does not preclude an mRNA from reaching the cytoplasm, it remains possible that some inosine-containing RNAs are retained in the nucleus, and in this regard, data for mouse Cat2 Transcribed Nuclear-RNA (CTN-RNA) are compelling. CTN-RNA was the first endogenous inosine-containing RNA found to be associated with p54nrb [37]. It is produced using an alternative polyadenylation site that results in an extended 3’ UTR containing a large double-stranded region that is edited at multiple sites. Although the extended 3’ UTR is essential for CTN-RNA nuclear enrichment, whether the inosines are important for retention has not been tested.
All mRNAs with inosine-containing 3’ UTRs studied to date also contain double-stranded structures within their 3’ UTRs, and some of these are cytoplasmic [31–33]. Thus, the mere presence of a double-stranded structure in a 3' UTR does not preclude nuclear export of the mRNA. Interestingly, both endogenous mammalian inosine-containing RNAs reported to be retained in the nucleus, CTN-RNA and NICN-1, utilize alternative polyadenylation [32, 37]. These transcripts have identical open-reading frames as other transcripts from the same gene locus (mCAT2 for CTN-RNA; AJ299740 for NICN-1), but due to the use of a distal polyadenylation site, contain extended 3’ UTRs. Similarly, at least one other nuclear enriched mRNA, which lacks inverted repeats and inosines, is a product of alternative polyadenylation [38]. It is intriguing to consider that the common feature, alternative polyadenylation, is important for nuclear retention. Possibly, the element or protein important for skipping the proximal polyadenylation site confers nuclear retention. Interestingly, consistent with its ability to recognize nucleotides other than inosine, p54nrb is important for binding and regulating alternative polyadenylation of mRNAs [39]. Could the interaction of p54nrb with CTN-RNA and NICN-1 be driven by elements important for alternative polyadenylation and not inosine?
Although the elements within an RNA that cause nuclear retention are unclear, a discrete territory within the nucleus, the paraspeckle, is critical for nuclear retention of certain RNAs [40]. Correspondingly, in human embryonic stem cells, which do not contain paraspeckles, at least one mRNA (PAICS1) found in the nucleus of other cell types is present in the cytoplasm and translated [33]. The importance of paraspeckles to nuclear retention is not unexpected, as the nuclear-enriched CTN-RNA co-purifies with these structures [37]. Given that multiple studies point to the importance of paraspeckles in nuclear retention of RNA, analysis of other mRNAs associated with these structures seems critical in determining the elements required for nuclear retention, and what role, if any, inosines play in this process.
Do edited 3’ UTR structures affect translation?
Obviously, one of the best ways to evaluate effects of inosine on gene expression is to examine mRNAs in animals that lack editing. C. elegans have been used for such studies as ADARs are not essential for viability in this species [8]. Comparisons of endogenous and reporter mRNAs with structured 3’ UTRs show no differences in mRNA translation between wildtype animals and mutants that lack editing [31]. However, these studies revealed that, in both wildtype C. elegans and mutants that lack editing, mRNAs with double-stranded 3’ UTR structures associate with fewer ribosomes than mRNAs without these structures. Thus, in at least some cases, double-stranded 3’ UTRs affect translational efficiency independent of editing.
Although it is unclear how these UTR structures mediate effects on translation, it is possible that a dsRNA binding protein (dsRBP) interacts with the UTR. As dsRBPs are not sequence-specific, this possibility is consistent with the observation that structured UTRs affect translation regardless of their length and sequence [31]. ADAR editing alters RNA structure, and this hypothesis at first seems inconsistent with the fact that editing in UTRs does not alter translation. However, at present, it is unclear how editing affects the in vivo binding of dsRBPs to dsRNA. In vitro, both human ADAR2 [41] and a complex containing two dsRBPs, Dicer and TRBP [42], bind similarly to unedited dsRNAs and selectively deaminated dsRNAs. Although the structures in 3’ UTRs typically undergo nonselective deamination events, even these may not be frequent enough to alter binding. Further, within non-coding regions of the human transcriptome, ADARs often edit AC mismatches to create IC pairs that stabilize the double-stranded structure, emphasizing that editing does not necessarily abrogate dsRBP binding [11, 25]. Further studies will be critical to determine what proteins bind to structured UTRs in vivo and whether editing plays a role in regulating these interactions.
Do inosines target RNAs for degradation?
Tudor staphylococcal nuclease (Tudor-SN) is a eukaryotic protein associated with diverse processes, including transcription [43, 44, 45], splicing [46], RNA interference (RNAi) [47] and RNA editing [48]. In vitro studies indicate that Tudor-SN specifically binds dsRNA containing runs of IU or UI base-pairs [48, 49]. Whereas some studies indicate that Tudor-SN cleaves inosine-containing RNA [50], others indicate that the nuclease function is provided by another, as yet unidentified, factor [48]. Inosine and guanosine differ only in that the latter has an amino group at N2, and both form wobble-pairs with uridine using an identical hydrogen bonding scheme [3]. However, Tudor-SN does not bind runs of GU or UG base-pairs in dsRNA or inosines in single-stranded RNA [48]. These data suggest that cleavage by the Tudor-SN complex requires a specific three-dimensional structure adopted by runs of IU and UI base-pairs.
ADARs often target adjacent adenosines [12, 13], so the properties of Tudor-SN are ideal for degradation of ADAR products, alone or associated with another factor. However, there are currently no known endogenous substrates of Tudor-SN. In fact, under normal growth conditions, levels of endogenous C. elegans mRNAs with structured 3’ UTRs are unaffected by the presence or absence of inosines, suggesting these mRNAs are not targeted by Tudor-SN [31]. It is possible that endogenous RNA is targeted by Tudor-SN only in response to certain environmental conditions. As Tudor-SN was originally purified from ribosomes [47], it is interesting to speculate that it exerts translational control by promoting cleavage of inosine-containing 3’ UTRs in response to specific stimuli.
Indeed, there is precedence for cleavage of an endogenous inosine-containing mRNA, CTN-RNA, in response to cellular stimuli [37]. After treatment to induce stress, nuclear CTN-RNA is cleaved to remove the structured 3’ UTR, presumably re-polyadenylated, and exported to the cytoplasm for translation. The CTN-RNA stored in the nucleus provides the cell with a rapid means to produce mCAT2 protein in response to stress. As yet, the exact cleavage site in CTN-RNA is unknown, so it remains possible that inosines or UTR structures are important for cleavage. However, as Tudor-SN localizes primarily to the cytoplasm, it is unlikely to be the nuclease [48]. Regardless, these studies emphasize the importance of monitoring effects of inosine under a variety of conditions.
Does editing of endogenous small RNAs affect their biological function?
Inosines have also been observed in endogenous small RNAs [51, 52], primarily those of the microRNA (miRNA) gene silencing pathway [53]. miRNAs are transcribed from long primary transcripts (pri-miRNAs; Figure 1) that form intramolecular stem-loop structures that are processed into smaller precursor miRNAs (pre-miRNAs) by the RNase III enzyme Drosha. Following Drosha processing, pre-miRNAs are exported from the nucleus and processed into mature miRNAs by Dicer. Mature miRNAs bind with partial complementarity to mRNA, typically within the 3’ UTR, to inhibit translation and/or promote mRNA degradation.
The first searches for inosines in endogenous small RNAs focused on specific miRNA precursors. Although editing events were detected in endogenous pri-miRNAs, editing at specific adenosines was detected in only a small fraction of the RNA population (~5%) [50, 54]. Recent systematic approaches have revealed a larger pool of edited pri-miRNAs (greater than 50) and higher levels of editing at specific adenosines (up to 70% of the RNA population) [55, 56]. These studies predict that 6–16% of human pri-miRNAs are edited, raising the possibility that ADAR editing has a large impact on miRNA-mediated gene silencing.
Small RNAs also modulate gene expression by RNAi. In this pathway, siRNAs (~20–30 nucleotides), the products of dsRNA processing by Dicer, direct sequence-specific degradation of mRNA [53]. RNAi refers to a process triggered by exogenous dsRNA, but the detection of endogenous siRNAs (endo-siRNA) in a number of organisms indicates that endogenous dsRNA triggers a similar pathway in vivo [57]. Edited endo-siRNAs have not been reported; however, as this pathway was only recently discovered, it seems likely that inosines will also be detected in endo-siRNAs and their precursors.
The key question is whether inosines in small RNAs or their precursors have a biological function. Inosine has been proposed to affect processing of precursors, as well as allow targeting of an alternate mRNA (Figure 2). Effects on precursors are thought to occur because the conversion of AU base-pairs to IU mismatches prevents binding and/or cleavage by RNase III enzymes. Further, as inosine prefers to pair with cytidine, editing in small RNAs is proposed to allow targeting of an entirely new set of mRNAs.
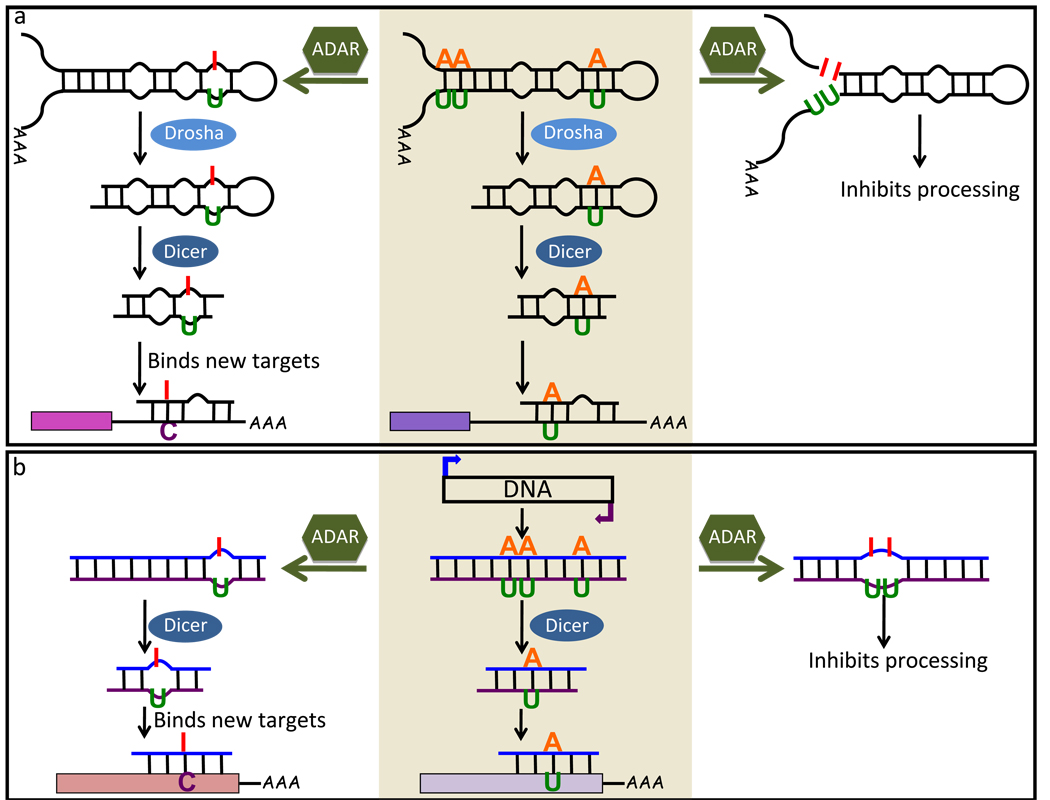
Figure 2.
Models proposed to explain effects of ADARs on dsRNA-mediated gene silencing. miRNA (a) and siRNA (b) pathways are illustrated with precursors, intermediates and mature small RNAs represented as base-paired molecules. Middle pathways (shaded in light brown) show processing by Drosha (light blue oval) and/or Dicer (dark blue oval) in the absence of ADAR (green hexagon), and outer panels show effects of ADARs on processing (right) or re-targeting (left). Representative AU base-pairs are shown being converted to IU mismatches. (a) Drosha processes pri-miRNAs into ~70 nucleotide pre-miRNAs that are then processed into ~22 nucleotide miRNAs by Dicer; as shown, the mature miRNA strand then pairs with the target mRNA [53]. Editing events are detected at a variety of positions in pri-miRNAs, and inosines within both pri- and pre-miRNAs inhibit in vitro cleavage by Drosha and Dicer, respectively [42, 50]. In addition, inosine in at least one endogenous mature miRNA affects binding to mRNA targets [58]. (b) Transgenes that give rise to sense and antisense transcripts produce dsRNA that is edited by ADARs [62]. Inosines within such exogenous dsRNA, or within in vitro transcripts, inhibit processing by Dicer [61]. Although not yet experimentally validated, inosines within siRNAs are predicted to affect binding of siRNAs to target mRNAs. Editing has not been shown to affect the in vivo correlate of this pathway, the endo-siRNA pathway, and there are no reports of endo-siRNAs that contain inosine. However, the characteristics of ADARs make it likely to intersect with the endo-siRNA pathway.
At present, there is only one example of an edited small RNA whose function has been validated in vivo. While searching for edited miRNA precursors, Nishikura and colleagues identified the first example of editing within an endogenous mature miRNA [58]. Although miRNAs do not require complete sequence complementarity to bind to target mRNAs, contiguous base-pairing of 7 nucleotides within their 5’ termini, the seed element, is critical [53]. The edited nucleotide of miR-376a resides in the seed, and using bioinformatics, new target mRNAs that could base-pair with the edited seed were predicted. Using an in vivo reporter assay, the authors demonstrated that unedited and edited versions of miR-376a target distinct genes [58]. With this knowledge in hand, the authors then demonstrated that endogenous expression of PRPS1 mRNA, a target of edited miR-376a, increased in the absence of ADAR2. Further, consistent with a role for PRPS1 in uric acid production, mice lacking ADAR2 had increased uric acid levels.
One hint that ADARs function to antagonize the endo-siRNA pathway comes from work in C. elegans. Worms lacking ADARs have defects in chemotaxis, the ability to seek out or avoid chemicals [8]. Chemotaxis defects in adr mutant worms are rescued by mutations in genes required for the endo-siRNA pathway, suggesting that ADARs normally “protect” chemotaxis genes from gene silencing [59]. In future studies it will be interesting to determine whether any endo-siRNAs target C. elegans genes involved in chemotaxis.
Do inosines in dsRNA affect its processing by Drosha or Dicer?
Almost as soon as dsRNA-meditated gene silencing was discovered, the question arose as to whether inosine within dsRNA would affect these pathways [60]. As discussed above, in only one case is there a clear biological function for an inosine in a small RNA. However, results of many in vitro studies, and those that monitor expression of exogenous dsRNA in vivo, provide proof-of-principle evidence that ADARs affect gene silencing.
For example, when compared to unedited dsRNA, dsRNA reacted with ADARs in vitro is less effective in silencing a target gene in Drosophila extracts [61]. The loss of efficacy is accompanied by a decrease in the processing of the dsRNA to siRNA, suggesting that the presence of inosines precludes cleavage by Dicer (Figure 2b). A similar situation occurs when repetitive transgenes, which give rise to dsRNA, are expressed in C. elegans. Whereas such transgenes are expressed in wildtype animals, they are silenced in strains lacking the two C. elegans ADARs, adr-1 and adr-2 [62]. Again, the most straightforward interpretation of these experiments is that inosine-containing dsRNA cannot be cleaved by Dicer, and thus does not yield the siRNAs required for gene-silencing. These proof-of-principle experiments raise the possibility that ADARs regulate the in vivo expression of loci that give rise to dsRNA.
Inosines in pri-miRNAs also inhibit their processing in vitro, again presumably because the A to I conversion alters RNA structure (Figure 2a). For example, in vitro editing and processing studies demonstrate that Dicer and Drosha cleavage is inhibited by inosines present within pre-miR-151 and pri-miR-142, respectively [42, 50]. Consistent with an inability of Dicer to process inosine-containing pre-miR-151, all pre-miR-151 detected in human amygdala contains inosine, and edited, mature miR-151 is undetectable [42]. Likewise, consistent with a defect in miRNA processing, both Adar1 and Adar2 null mice have increased levels of mature miR-142 [50]. Both studies showed that editing of two adenosines was sufficient to inhibit precursor processing, and as the editing created mismatches, a change in RNA structure was proposed to cause inhibition.
Do ADARs affect silencing through competition for dsRNA?
Although several in vitro studies show that inosines in dsRNA inhibit its processing to small RNAs, others show that simply by binding dsRNA, ADARs can antagonize gene silencing [51]. The first hint that ADARs could cause effects simply by binding dsRNA came from the study of C. elegans transgene silencing [62]. Of the two ADARs in C. elegans, only ADR-2 has the active site residues important for catalysis, and thus, worms lacking only adr-2, or both adr-1 and adr-2, have no detectable editing [8]. If inosines were solely responsible for antagonism of RNAi, all strains should have identical silencing phenotypes. However, transgene silencing in adr-1;adr-2 animals is more severe than in adr-2 animals, suggesting that binding of ADR-1 to dsRNA contributes to antagonism [62].
The idea that ADARs and RNAi components compete for dsRNA is further supported by studies in mammalian cells and Drosophila melanoagaster [63, 64]. By in vitro gel shift and filter binding assays, a recombinant version of the cytoplasmic form of human ADAR1 (ADAR1-p150) shows a 15–30 fold stronger affinity for siRNAs than the nuclear form of human ADAR1 (ADAR1-p110) or human ADAR2 [63]. Correspondingly, the efficacy of RNAi in Adar1−/− mouse embryonic fibroblasts (MEFs) is three-fold higher than in wild type MEFs. The increased efficacy of RNAi is restored to wildtype levels by expression of ADAR1-p150, but not by a mutant version of ADAR1p-150 lacking the dsRBMs, consistent with a model whereby cytoplasmic ADAR1p-150 binds siRNAs and prevents their ability to silence mRNAs.
Similarly, when human ADARs and an RNAi reporter are co-expressed in the Drosophila eye, ADAR1-p150, but not ADAR1-p110 or ADAR2, antagonizes silencing [64]. An editing inactive ADAR1-p150 mutant also antagonizes RNAi, but to a lesser extent than wildtype, suggesting that both deamination and binding play a role in this process. The high affinity binding of ADAR1-p150 to siRNA, not just the cytoplasmic localization, seems important as a mutant of ADAR2 that localizes to the cytoplasm does not antagonize RNAi. Interestingly, a significant fraction of C. elegans ADR-1 is located in the cytoplasm [31], raising the possibility that this protein could also affect dsRNA-mediated gene silencing by binding to small RNAs.
Do ADARs and inosine-containing RNAs have general effects on gene expression?
Eukaryotes express other dsRBPs in addition to those involved in gene silencing [65], and in theory, ADARs could compete with these as well. In fact, ADARs are known to affect the function of PKR, a dsRBP whose kinase activity is activated by binding to viral dsRNA. Once activated, PKR phosphorylates the translation initiation factor, eIF2α, to shut down all translation [66]. ADAR1 overexpression decreases PKR activation, leading to a general increase in translation of a variety of co-transfected genes [67]. Expression of only ADAR1's dsRBMs is sufficient for increasing translation, indicating that ADAR1 binding to dsRNA is critical, presumably because ADARs compete with PKR for cytoplasmic dsRNA. Although these studies show that overexpression of ADAR1 affects exogenous gene expression, effects on endogenous gene expression were not detected, and the biological significance of these observations remains unclear. However, the increasing number of edited dsRNAs detected in the cytoplasm (Figure 1) raises the question of how these RNAs avoid eliciting a cellular response to dsRNA.
Finally, an intriguing study indicates that short dsRNA containing multiple IU mismatches actively decreases gene expression in trans [68]. Specifically, transfection of short dsRNAs (~20 nucleotides) which harbor IU mismatches decreased mammalian gene expression by reducing mRNA levels and inhibiting translation. The mere presence of inosine was not sufficient for reducing gene expression, but IU mismatches, and to a lesser extent GU mismatches, produced the effect. Complementing the transfection studies, in cell lysates, IU dsRNA, and to a lesser extent GU dsRNA, interacts with a complex of proteins that are components of stress granules. During times of cellular stress, up to 50% of eukaryotic cellular mRNAs are translationally repressed by sequestration in stress granules [69]. Together these data support a model in which edited dsRNA in the cytoplasm leads to stress granule formation, thus promoting a general decrease in gene expression [68]. Although the model is intriguing, it is important to remember that the studies were performed with synthetic inosine-containing RNAs, and it is unclear which cellular RNAs might serve as regulators of stress granules. Furthermore, the ability of mismatches within dsRNA to promote stress granule formation suggests that it is the structure of hyperedited RNAs, and not specifically the inosines, that is important.
Concluding remarks
ADAR editing of RNA alters information content and structure of cellular RNAs. The first identified endogenous substrates of ADARs revealed that editing in codons is essential for generating diverse protein isoforms. Although codon editing is clearly important, it represents only a small fraction of the editing events in the transcriptome. Editing sites in non-coding regions of RNA are vastly more prevalent, and their identification has exponentially expanded the number of known ADAR substrates. The biological function of editing in non-coding RNA sequences remains mysterious (Box 1). Yet, as proposed previously [2], the non-selective editing that occurs in these substrates likely reflects the primordial function of ADARs.
But what was that primordial function? Our favorite hypothesis is that ADARs evolved to regulate levels of cellular dsRNA. If not kept in check, such molecules are potent triggers of gene-silencing as well as signaling pathways such as the mammalian interferon response. By changing AU base-pairs to IU mismatches, ADARs effectively lower dsRNA concentration, and in-line with this idea, interferon-inducible genes are upregulated in Adar1−/− mice [70]. As new biological functions typically arise from existing pathways, it seems very likely that inosines have additional roles in modern day cells. The numerous editing events now documented in non-coding regions of endogenous RNAs set the stage for defining these functions.
Box 1. Outstanding questions
1. Is there a function for inosine in 3’UTRs?
Only in C. elegans has the fate of edited 3' UTRs been compared in wildtype and mutants that lack ADARs, and similar studies in other organisms would be helpful in answering this question. Further, although inosines in 3’ UTRs had no effects on gene expression in C. elegans grown under normal conditions, a variety of cellular conditions should be monitored.
2. Do paraspeckles serve as reservoirs for edited cellular RNAs?
CTN-RNA provides a striking example of how nuclear retention, specifically in paraspeckles, can regulate gene expression, in this case, in response to stress. However, it is unclear what function inosine has, if any, in promoting the association of cellular RNAs with paraspeckles. Identification of other paraspeckle-associated RNAs, and analysis of how editing affects the cellular localization of known nuclear enriched inosine-containing RNAs, will be key to understanding the function of paraspeckles.
3. Is Tudor-SN involved in the metabolism of endogenous inosine-containing RNAs?
In vitro studies convincingly show that Tudor-SN specifically binds dsRNA containing multiple IU and UI mismatches, but the identification of endogenous substrates is essential to confirm the biological relevance of these studies. Further, whether Tudor-SN itself is a nuclease, or promotes degradation in concert with another factor, is not yet conclusively determined.
4. How many endogenous small RNAs contain inosine?
A genome-wide study to identify inosines in small RNAs will be critical in answering this question, and analyses comparing wildtype and ADAR mutants will be helpful in distinguishing editing from sequencing errors. Such studies will reveal whether inosines are present in endo-siRNAs, and identify additional editing events in miRNAs. Knowledge of the identity of endogenous small RNAs that contain inosine will pave the way for elucidating the effects of editing.
5. How many mRNAs and small RNAs do ADARs affect simply by binding?
Evidence from transgenic studies suggests that ADARs affect dsRNA-mediated gene silencing pathways simply by competing for dsRNA. However, the impact of ADAR binding in comparison to ADAR editing is unknown. Comparison of strains expressing wildtype ADARs to those expressing catalytically-inactive versions will be informative in this regard.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gott JM, Emeson RB. Functions and mechanisms of RNA editing. Annu. Rev. Genet. 2000;34:499–531. doi: 10.1146/annurev.genet.34.1.499. [DOI] [PubMed] [Google Scholar]
- 2.Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maydanovych O, Beal PA. Breaking the central dogma by RNA editing. Chem. Rev. 2006;106:3397–3411. doi: 10.1021/cr050314a. [DOI] [PubMed] [Google Scholar]
- 4.Nishikura K. Editor meets silencer: crosstalk between RNA editing and RNA interference. Nat. Rev. Mol. Cell. Biol. 2006;7:919–931. doi: 10.1038/nrm2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Q, et al. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science. 2000;290:1765–1768. doi: 10.1126/science.290.5497.1765. [DOI] [PubMed] [Google Scholar]
- 6.Higuchi M, et al. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 7.Palladino MJ, et al. A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell. 2000;102:437–449. doi: 10.1016/s0092-8674(00)00049-0. [DOI] [PubMed] [Google Scholar]
- 8.Tonkin LA, et al. RNA editing by ADARs is important for normal behavior in Caenorhabditis elegans. EMBO J. 2002;21:6025–6035. doi: 10.1093/emboj/cdf607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bass BL, Weintraub H. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell. 1988;55:1089–1098. doi: 10.1016/0092-8674(88)90253-x. [DOI] [PubMed] [Google Scholar]
- 10.Wagner RW, et al. A double-stranded RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. Proc. Natl. Acad. Sci. U. S. A. 1989;86:2647–2651. doi: 10.1073/pnas.86.8.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levanon EY, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 12.Polson AG, Bass BL. Preferential selection of adenosines for modification by double-stranded RNA adenosine deaminase. EMBO J. 1994;13:5701–5711. doi: 10.1002/j.1460-2075.1994.tb06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lehmann KA, Bass BL. Double-stranded RNA adenosine deaminases ADAR1 and ADAR2 have overlapping specificities. Biochemistry. 2000;39:12875–12884. doi: 10.1021/bi001383g. [DOI] [PubMed] [Google Scholar]
- 14.Nishikura K, et al. Substrate specificity of the dsRNA unwinding/modifying activity. EMBO J. 1991;10:3523–3532. doi: 10.1002/j.1460-2075.1991.tb04916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lehmann KA, Bass BL. The importance of internal loops within RNA substrates of ADAR1. J. Mol. Biol. 1999;291:1–13. doi: 10.1006/jmbi.1999.2914. [DOI] [PubMed] [Google Scholar]
- 16.Dawson TR, et al. Structure and sequence determinants required for the RNA editing of ADAR2 substrates. J. Biol. Chem. 2004;279:4941–4951. doi: 10.1074/jbc.M310068200. [DOI] [PubMed] [Google Scholar]
- 17.Higuchi M, et al. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell. 1993;75:1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- 18.Lomeli H, et al. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science. 1994;266:1709–1713. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- 19.Burns CM, et al. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 20.Hanrahan CJ, et al. RNA editing of the Drosophila para Na(+) channel transcript. Evolutionary conservation and developmental regulation. Genetics. 2000;155:1149–1160. doi: 10.1093/genetics/155.3.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valente L, Nishikura K. ADAR gene family and A-to-I RNA editing: diverse roles in posttranscriptional gene regulation. Prog. Nucleic Acid Res. Mol. Biol. 2005;79:299–338. doi: 10.1016/S0079-6603(04)79006-6. [DOI] [PubMed] [Google Scholar]
- 22.Chen LL, Carmichael GG. Gene regulation by SINES and inosines: biological consequences of A-to-I editing of Alu element inverted repeats. Cell Cycle. 2008;7:3294–3301. doi: 10.4161/cc.7.21.6927. [DOI] [PubMed] [Google Scholar]
- 23.Morse DP, Bass BL. Long RNA hairpins that contain inosine are present in Caenorhabditis elegans poly(A)+ RNA. Proc. Natl. Acad. Sci. U. S. A. 1999;96:6048–6053. doi: 10.1073/pnas.96.11.6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blow M, et al. A survey of RNA editing in human brain. Genome Res. 2004;14:2379–2387. doi: 10.1101/gr.2951204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Athanasiadis A, et al. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim DD, et al. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004;14:1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Z, Carmichael GG. The fate of dsRNA in the nucleus: a p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell. 2001;106:465–475. doi: 10.1016/s0092-8674(01)00466-4. [DOI] [PubMed] [Google Scholar]
- 28.DeCerbo J, Carmichael GG. Retention and repression: fates of hyperedited RNAs in the nucleus. Curr. Opin. Cell Biol. 2005;17:302–308. doi: 10.1016/j.ceb.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 29.Sommer B, et al. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991;67:11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- 30.Morse DP, Bass BL. Detection of inosine in messenger RNA by inosine-specific cleavage. Biochemistry. 1997;36:8429–8434. doi: 10.1021/bi9709607. [DOI] [PubMed] [Google Scholar]
- 31.Hundley HA, et al. C. elegans and H. sapiens mRNAs with edited 3' UTRs are present on polysomes. RNA. 2008;14:2050–2060. doi: 10.1261/rna.1165008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen LL, et al. Alu element-mediated gene silencing. EMBO J. 2008;27:1694–1705. doi: 10.1038/emboj.2008.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear non-coding RNA. Mol. Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clemson CM, et al. An architectural role for a nuclear non-coding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sunwoo H, et al. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sasaki YT, et al. MENepsilon/beta non-coding RNAs are essential for structural integrity of nuclear paraspeckles. Proc. Natl. Acad. Sci. U. S. A. 2009;106:2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prasanth KV, et al. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 38.Kay RA, et al. The expression of migration stimulating factor, a potent oncofetal cytokine, is uniquely controlled by 3'-untranslated region-dependent nuclear sequestration of its precursor messenger RNA. Cancer Res. 2005;65:10742–10749. doi: 10.1158/0008-5472.CAN-05-2038. [DOI] [PubMed] [Google Scholar]
- 39.Hall-Pogar T, et al. Specific trans-acting proteins interact with auxiliary RNA polyadenylation elements in the COX-2 3'-UTR. RNA. 2007;13:1103–1115. doi: 10.1261/rna.577707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bond CS, Fox AH. Paraspeckles: nuclear bodies built on long non-coding RNA. J. Cell Biol. 2009;186:637–644. doi: 10.1083/jcb.200906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klaue Y, et al. Biochemical analysis and scanning force microscopy reveal productive and nonproductive ADAR2 binding to RNA substrates. RNA. 2003;9:839–846. doi: 10.1261/rna.2167603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawahara Y, et al. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep. 2007;8:763–769. doi: 10.1038/sj.embor.7401011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tong X, et al. The Epstein-Barr virus nuclear protein 2 acidic domain forms a complex with a novel cellular coactivator that can interact with TFIIE. Mol. Cell. Biol. 1995;15:4735–4744. doi: 10.1128/mcb.15.9.4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paukku K, et al. Tudor and nuclease-like domains containing protein p100 function as coactivators for signal transducer and activator of transcription 5. Mol. Endocrinol. 2003;17:1805–1814. doi: 10.1210/me.2002-0256. [DOI] [PubMed] [Google Scholar]
- 45.Yang J, et al. Identification of p100 as a coactivator for STAT6 that bridges STAT6 with RNA polymerase II. EMBO J. 2002;21:4950–4958. doi: 10.1093/emboj/cdf463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang J, et al. Transcriptional co-activator protein p100 interacts with snRNP proteins and facilitates the assembly of the spliceosome. Nucleic Acids Res. 2007;35:4485–4494. doi: 10.1093/nar/gkm470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caudy AA, et al. A micrococcal nuclease homologue in RNAi effector complexes. Nature. 2003;425:411–414. doi: 10.1038/nature01956. [DOI] [PubMed] [Google Scholar]
- 48.Scadden AD. The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nat. Struct. Mol. Biol. 2005;12:489–496. doi: 10.1038/nsmb936. [DOI] [PubMed] [Google Scholar]
- 49.Scadden AD, Smith CW. Specific cleavage of hyper-edited dsRNAs. EMBO J. 2001;20:4243–4252. doi: 10.1093/emboj/20.15.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang W, et al. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bass BL. How does RNA editing affect dsRNA-mediated gene silencing? Cold Spring Harb. Symp. Quant. Biol. 2006;71:285–292. doi: 10.1101/sqb.2006.71.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zinshteyn B, Nishikura K. Adenosine-to-inosine RNA editing. WIREs Systems Biology and Medicine. 2009;1:202–209. doi: 10.1002/wsbm.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luciano DJ, et al. RNA editing of a miRNA precursor. RNA. 2004;10:1174–1177. doi: 10.1261/rna.7350304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blow MJ, et al. RNA editing of human microRNAs. Genome Biol. 2006;7:R27. doi: 10.1186/gb-2006-7-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kawahara Y, et al. Frequency and fate of microRNA editing in human brain. Nucleic Acids Res. 2008;36:5270–5280. doi: 10.1093/nar/gkn479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Golden DE, et al. An inside job for siRNAs. Mol. Cell. 2008;31:309–312. doi: 10.1016/j.molcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kawahara Y, et al. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007;315:1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tonkin LA, Bass BL. Mutations in RNAi rescue aberrant chemotaxis of ADAR mutants. Science. 2003;302:1725. doi: 10.1126/science.1091340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bass BL. Double-stranded RNA as a template for gene silencing. Cell. 2000;101:235–238. doi: 10.1016/s0092-8674(02)71133-1. [DOI] [PubMed] [Google Scholar]
- 61.Scadden AD, Smith CW. RNAi is antagonized by A-->I hyper-editing. EMBO Rep. 2001;2:1107–1111. doi: 10.1093/embo-reports/kve244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Knight SW, Bass BL. The role of RNA editing by ADARs in RNAi. Mol. Cell. 2002;10:809–817. doi: 10.1016/s1097-2765(02)00649-4. [DOI] [PubMed] [Google Scholar]
- 63.Yang W, et al. ADAR1 RNA deaminase limits short interfering RNA efficacy in mammalian cells. J. Biol. Chem. 2005;280:3946–3953. doi: 10.1074/jbc.M407876200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heale BS, et al. Editing independent effects of ADARs on the miRNA/siRNA pathways. EMBO J. 2009;28:3145–3156. doi: 10.1038/emboj.2009.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chang KY, Ramos A. The double-stranded RNA-binding motif, a versatile macromolecular docking platform. FEBS J. 2005;272:2109–2117. doi: 10.1111/j.1742-4658.2005.04652.x. [DOI] [PubMed] [Google Scholar]
- 66.Sadler AJ, Williams BR. Structure and function of the protein kinase R. Curr. Top. Microbiol. Immunol. 2007;316:253–292. doi: 10.1007/978-3-540-71329-6_13. [DOI] [PubMed] [Google Scholar]
- 67.Wang Y, Samuel CE. Adenosine deaminase ADAR1 increases gene expression at the translational level by decreasing protein kinase PKR-dependent eIF-2alpha phosphorylation. J. Mol. Biol. 2009;393:777–787. doi: 10.1016/j.jmb.2009.08.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scadden AD. Inosine-Containing dsRNA Binds a Stress-Granule-like Complex and Downregulates Gene Expression In trans. Mol. Cell. 2007;28:491–500. doi: 10.1016/j.molcel.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Anderson P, Kedersha N. RNA granules. J. Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hartner JC, et al. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat. Immunol. 2009;10:109–115. doi: 10.1038/ni.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hellwig S, Bass BL. A starvation-induced non-coding RNA modulates expression of Dicer-regulated genes. Proc. Natl. Acad. Sci. U. S. A. 2008;105:12897–12902. doi: 10.1073/pnas.0805118105. [DOI] [PMC free article] [PubMed] [Google Scholar]