Abstract
Streptococcus uberis, strain 0140J, contains a single copy sortase A (srtA), encoding a transamidase capable of covalently anchoring specific proteins to peptidoglycan. Unlike the wild-type, an isogenic mutant carrying an inactivating ISS1 insertion within srtA was only able to infect the bovine mammary gland in a transient fashion. For the first 24 h post challenge, the srtA mutant colonised at a similar rate and number to the wild type strain, but unlike the wild type did not subsequently colonise in higher numbers. Similar levels of host cell infiltration were detected in response to infection with both strains, but only in those mammary quarters infected with the wild type strain were clinical signs of disease evident. Mutants that failed to express individual sortase substrate proteins (sub0135, sub0145, sub0207, sub0241, sub0826, sub0888, sub1095, sub1154, sub1370, and sub1730) were isolated and their virulence determined in the same challenge model. This revealed that mutants lacking sub0145, sub1095 and sub1154 were attenuated in cattle. These data demonstrate that a number of sortase anchored proteins each play a distinct, non-redundant and important role in pathogenesis of S. uberis infection within the lactating bovine mammary gland.
Keywords: mastitis, S. uberis, virulence, sortase, vaccine
1. INTRODUCTION
Streptococcus uberis is one of the most common pathogens isolated from cases of bovine mastitis [7]. It impacts negatively on animal health, welfare and the economics of milk production. S. uberis has been shown in experimental infection models within the dairy cow to colonise the mammary gland rapidly, induce neutrophil diapedesis and cause an acute local inflammation during which milk becomes denatured and the mammary gland distended, swollen and painful. S. uberis persists within the mammary gland in the presence of neutrophils and other host defences, which seem to offer little control. However, the infection remains localised to the mammary gland and the bacterium is found largely in the secretion present within the lumen of the gland. Although a common mammary pathogen, S. uberis is often found as part of the normal, commensal microflora at other body sites and is not commonly associated with other disease states.
The interaction of S. uberis with its immediate environment in the host is likely to be dependent on components that are at the bacterial surface or are released into the surrounding medium during the infection process. The surface of Gram-positive bacteria is decorated with many different macromolecules, including carbohydrates, teichoic acids, polysaccharides and proteins. In many species a subset of surface proteins have been reported to be covalently linked to the peptidoglycan matrix of the cell wall [22, 30, 39]. In almost all studies to date, covalent attachment of proteins relies on the transamidase, sortase [12]. This activity was first described in Staphylococcus aureus [27] and since then five different classes of sortase enzyme have been identified in Gram positive bacteria [13, 33].
Proteins destined for surface location in Gram-positive bacteria are targeted for transport across the cytoplasmic membrane due to the presence of a hydrophobic N-terminal signal peptide. Following transport through the membrane and removal of the signal peptide, the polypeptide is cleaved within a specific sorting motif located towards the C-terminus of the protein. Sortase subsequently reforms an amide bond between the carboxyl group of the new C-terminal amino acid and the amino group of peptides that cross bridge the peptidoglycan of the cell wall. In the absence of sortase, proteins are not anchored and may be released into the surrounding medium with their C-termini intact [25, 46].
Each class of sortase has been reported to anchor a specific and discrete subset of proteins by recognition of a specific amino acid sequence motif contained within the substrate protein. A recent investigation in S. uberis revealed two sequences (LPXXG and LPXXXD) that appear to function as cell wall attachment motifs in this and other streptococci [14].
The published sequence of S. uberis [44] contains a single coding sequence (sub0881) with homology to sortase (SrtA) and 10 CDS (sub0135, sub0145, sub0207, sub0241, sub0826, sub0888, sub1095, sub1154 and sub1370, sub1730) containing the LPXXG or LPXXXD motifs along with other features typically required for cell wall anchoring [14]. Of these, the presence of nine was confirmed by comparative proteomic analysis of the wild type and SrtA mutant [14]. The sortase anchored proteins identified in S. uberis include a number of putative virulence determinants identified in other streptococci [14, 44].
In this communication, we report the ability of S. uberis to infect the bovine mammary gland and induce mastitis in the absence of SrtA or individual SrtA-anchored proteins.
2. MATERIALS AND METHODS
2.1. Bacterial strains and reagents
S. uberis strain 0140J and an isogenic insertional mutant lacking SrtA, as described previously [14] were used throughout this study. Bacterial cultures were routinely grown in Todd Hewitt broth and plated on sheep blood agar containing 1% (w/v) aesculin (ABA). Other bacterial strains and reagents were used as described in the text. Oligonucleotide primers for PCR amplification are referred to in the text by their specific reference numbers (PXXX) and the sequence of each is shown in Table I.
Table I.
Oligonucleotide primer sequences used in this study.
| Designation | Sequence (5′–3′). Lower case indicates base changes to introduce restriction sites | Application | Template | Annealing temp (°C) |
|---|---|---|---|---|
| p247 ISS1 fwd | GCTCTTCGGATTTTCGGTATC | ISS1 probe | pGh9::ISS1 | 58 |
| p250 ISS1 rev | CATTTTCCACGAATAGAAGGACTGTC | ISS1 probe | pGh9::ISS1 | 61 |
| p261 | TGGTTGAAGCAGAAGCTGAA | Screening for ISS1 within srtA ORF vs p247 | pGh9::ISS1 | 55 |
| p616 | CTAAGAAAATCACAGTTGACATGC | Screening for ISS1 within sub0826 ORF vs p247/p250 | pGh9::ISS1 | 59 |
| p621 | CAACGAATCAACAAACTGAAAGC | Screening for ISS1 within sub1370 ORF vs p247/p250 | pGh9::ISS1 | 59 |
| p628 | CATGTCTTTCGGTACCGTGGTTTTCTCG | Amplification of srtA ORF | S. uberis 0140J genomic DNA | 62.5 |
| p633 | ACAGTATTAAAGGCACAAATGGAAG | Amplification of srtA ORF | S. uberis 0140J genomic DNA | 60 |
| p641 | CAAATGCTAAAGTTGGCTTAAAAAC | Screening for ISS1 within sub0135 ORF vs p247/p250 | pGh9::ISS1 | 60 |
| p642 | CAATAATCAACAAATAAAGCGCAAT | Screening for ISS1 within sub0135 ORF vs p247/p250 | pGh9::ISS1 | 60 |
| p645 | ATCAAACTATTGTACCCTTTGGTGA | Screening for ISS1 within sub1095 ORF vs p247/p250 | pGh9::ISS1 | 60 |
| p646 | TGTAATGAATCTAGGAGGGGTAGGT | Screening for ISS1 within sub1095 ORF vs p247/p250 | pGh9::ISS1 | 60 |
| p649 | ATTAAAGAAGGTGATGTGGTTCAAA | Screening for ISS1 within sub1730 ORF vs p247/p250 | pGh9::ISS1 | 60 |
| p650 | GTCATTTAGCTTCTCCCATGTAAAA | Screening for ISS1 within sub1730 ORF vs p247/p250 | pGh9::ISS1 | 60 |
| p653 | CTTTATGAAAATAGCCAAGCTGAAA | Screening for ISS1 within sub0888 ORF vs p247/p250 | pGh9::ISS1 | 60 |
| p654 | CTTAATTGTTGCTGGTCTTTCATTT | Screening for ISS1 within sub0888 ORF vs p247/p250 | pGh9::ISS1 | 60 |
| p657 | TTTTCTTCTTAATTGCACACCTGA | Screening for ISS1 within sub0207 ORF vs p247/p250 | pGh9::ISS1 | 60 |
| p658 | AATCCAAGCTGTTGAACTATCCTC | Screening for ISS1 within sub0207 ORF vs p247/p250 | pGh9::ISS1 | 60 |
| p667 | TGATCTTGTGCATATCAATCTGG | Screening for ISS1 within sub0145 ORF vs p247/p250 | pGh9::ISS1 | 59 |
| p668 | AAGGAACCTGAAAGAACCATCTC | Screening for ISS1 within sub0145 ORF vs p247/p250 | pGh9::ISS1 | 61 |
| p675 | ACGTTCCCTACAAAAGAAAAAGG | Screening for ISS1 within sub0241 ORF vs p247/p250 | pGh9::ISS1 | 59 |
| p676 | TTGCCACTTAGGCTGATAAAAAC | Screening for ISS1 within sub0241 ORF vs p247/p250 | pGh9::ISS1 | 59 |
| p689 | GTGATTAAGCgAAtTCAATAAGGAAATAATGG | Amplification of sub1154 gene fragment and upstream flanking sequence. Incorporating an EcoRI restriction site. | S. uberis 0140J genomic DNA | 62 |
| p690 | CTTGTCGTCGTCATCCTTGTAGTCATCCGACTGCTTTTCATTTTGCAA | Amplification of sub1154 gene fragment, incorporating FLAG-TAG | S. uberis 0140J genomic DNA | > 60 |
| p691 | GACTACAAGGATGACGACGACAAGTATCCGCAGTCAATAAGCCAAA | Amplification of sub1154 gene fragment, incorporating FLAG-TAG | S. uberis 0140J genomic DNA | > 60 |
| p692 | ACTTACTTGAAAGCtTCACAAGTGGTAAAGT | Amplification of sub1154 gene fragment and downstream flanking sequence. Incorporating a HindIII restriction site. | S. uberis 0140J genomic DNA | 63 |
Table II.
Manifestation of clinical response to infection with Streptococcus uberis. All quarters and milk samples were analyzed against these criteria at each milking following challenge.
| Score | Appearance of quarter | Score | Appearance of milk |
|---|---|---|---|
| 1 | Normal | 1 | Normal |
| 2 | Minor changes (e.g. hardness) | 2 | Minor changes (e.g. a few flakes) |
| 3 | Moderate signs (e.g. heat, tenderness) | 3 | Moderate signs (e.g. clots and/or clumps) |
| 4 | Severe signs (e.g. distended and discomfort on palpation) | 4 | Severe signs (e.g. changes in colour and/or composition) |
2.2. Production and isolation of mutant strains of S. uberis
Insertionally inactivated mutants were located within a random insertional mutant bank of S. uberis strain 0140J by PCR as previously described [43]. Primers specific for each locus of interest were used in conjunction with those specific to ISS1 (Tab. I).
Attempts to isolate an insertion mutant with ISS1 located near to the start of the sub1154 coding sequence proved unsuccessful. Subsequently, a targeted deletion strategy was used to ablate production of the sub1154 gene product. Briefly, two fragments located at either end of the 3.4 kb open reading frame were amplified from genomic DNA using primer pairs P689/P690 and P691/P692 (Tab. I). The two fragments were purified and used in equal proportion as template in a further amplification reaction with primers P689 and P692 to generate a single ∆1154 product lacking 3169 bp of the sub1154 coding sequence. This amplicon was subcloned into the multiple cloning site of the low copy pG+h9 temperature sensitive plasmid using restriction sites introduced during amplification with P689 and P692. The plasmid construct was transformed into E. coli TG1 RepA and transformants cultured on LB agar/broth (200 μg/μL erythromycin) at 37.5 °C. The re-isolated purified plasmid (10 ng) was used to transform S. uberis 0140J and transformants selected on THA (1 μg/mL erythromycin) at 28 °C. S. uberis 0140J / pG+h9::∆1154 transformants were grown to OD550nm 0.5 in Todd Hewitt broth at 28 °C, the growth temperature was then raised to 37.5 °C to force single cross-over chromosomal integration. Integrant colonies were selected on THA ery (1 μg/mL) at 37 °C and subsequently grown in THB lacking antibiotic at 28 °C to promote excision of the pG+h9 replicon by a second cross-over event. Resulting erythromycin sensitive colonies were picked from THA plates, grown overnight at 37 °C and deletion of the sub1154 locus was determined by PCR amplification. Southern analysis and DNA sequencing were employed to confirm the exact nature of the deletion.
2.3. Extraction of chromosomal DNA from S. uberis
Genomic DNA was prepared using a variation of the method of Hill and Leigh as described previously [19]. Briefly, 1.5 mL of an overnight culture was centrifuged at 10 000 × g for 2 min and the cell pellet washed with 500 μL of 10 mM Tris-Cl, 5 mM EDTA, pH 7.8. Bacterial cell walls were disrupted by resuspension in 375 μL of 10 mM Tris-Cl, 5 mM EDTA pH 7.8 containing 30 units/mL mutanolysin and 10 mg/mL lysozyme (both from Sigma, Poole, UK) and subsequent incubation at 37 °C for 30 min. Total cell lysis was achieved by addition of 20 μL of SDS solution (20% w/v in 50 mM Tris-Cl, 20 mM EDTA, pH 7.8) and proteinase K (Sigma) to a final concentration of 150 μg/mL and a further incubation at 37 °C for 1 h. Cell wall material was removed by precipitation following the addition of 200 μL of saturated NaCl and subsequent centrifugation at 12 000 × g for 10 min. The supernatant was extracted with phenol chloroform and DNA precipitated by addition of 2 volumes of absolute ethanol. DNA pellets were washed with 70% aqueous ethanol and air-dried prior to resuspension in TE buffer containing 20 μg/mL RNaseA (Sigma).
2.4. Challenge of lactating dairy cows with S. uberis
The requirement by S. uberis strain 0140J for SrtA and/or individual SrtA-substrates (Tab. III) for virulence was determined by experimental challenge in a well established intramammary infection model in the dairy cow. Bacteria were grown for 18 h at 37 °C in Todd Hewitt broth. Cells were recovered by centrifugation (10 000 × g, 10 min), suspended in pyrogen-free saline (Sigma) and diluted in the same to the appropriate cell density. Suspensions of each strain were held on ice prior to being used to challenge animals. The actual number of viable bacteria in identical aliquots of each suspension was enumerated both prior to and following challenge.
Table III.
Sortase-anchored gene products identified by bioinformatic and proteomic analysis and description of their mutation in S. uberis 0140J.
| Gene/Producta | Protein accession numberb | Mutation | ISS1 orientation relative to disrupted CDS | Location of ISS1 insertion/deletion from ATG start codon | Predicted translation of disrupted mature protein (%) |
|---|---|---|---|---|---|
| sub0135 | YP_002561505 | Insertion | Forward | 389 bp | 10 |
| sub0145 | YP_002561515 | Insertion | Reverse | 485 bp | 24 |
| sub0207 | YP_002561573 | Insertion | Forward | 682 bp | 42 |
| sub0241 | YP_002561606 | Insertion | Forward | 896 bp | 34 |
| sub0826 | YP_002562161 | Insertion | Reverse | 664 bp | 13 |
| sub0888 | YP_002562216 | Insertion | Forward | 205 bp | 12 |
| sub1095 | YP_002562412 | Insertion | Reverse | 312 bp | 14 |
| sub1154 | YP_002562469 | Deletion | Disruption extends from 118 bp | 0.6 | |
| sub1370 | YP_002562668 | Insertion | Forward | 280 bp | 5 |
| sub1730 | YP_002563003 | Insertion | Reverse | 423 bp | 32 |
Gene annotation according to the genomic sequence of S. uberis 0140J [40].
Protein accession number according to NCBI protein database.
Dairy cows (Holstein, Friesian), 2–10 weeks into their first lactation, were selected for challenge. Criteria for selection were: absence of signs of mastitis, absence of bacteria in milk samples taken 24–48 h prior to challenge, no history of mastitis during the current lactation and no evidence of intramammary infection in milk samples taken at 7 and 14 days after parturition. Animals were challenged in mammary quarters after milking by infusion of 1 mL of pyrogen-free saline (Sigma) containing S. uberis.
To determine the role of SrtA, two animals were challenged in a total of four quarters with 6.0 × 102 cfu of strain 0140J and a further four animals, were challenged in a total of eight quarters with a similar dose of the srtA mutant. To subsequently determine the requirement for individual SrtA-anchored proteins, mutants lacking each protein were used to challenge two mammary quarters on the same animal with between 5.0 × 102–1.5 × 103 cfu. Any mutant that failed to induce overt signs of mastitis and that failed to colonise to high levels was subsequently rescreened in a similar challenge of a further two mammary quarters in another dairy cow.
Following challenge, animals were milked and inspected twice daily (07:00 h and 15:30 h) and those in which predetermined criteria (Tab. II) for clinical end points (clotted and discoloured milk and/or udder quarter swollen or causing discomfort on palpation) had been reached were treated with proprietary branded antibiotics. Milk samples were taken at each milking and analysed for bacteria and somatic cells, as described below.
2.5. Analysis of milk samples
The number of viable bacteria present was estimated by direct plating of up to 1 mL of each milk sample on to ABA. Samples were also diluted in saline and 50 μL of each dilution plated directly onto ABA. In each case, the presence and/or number of S. uberis was determined and the genotype of the recovered isolates was determined by PCR amplification of the altered locus. The number of somatic cells present in milk samples was determined using a DeLaval portable cell counter in line with the manufacturer’s instructions.
2.6. Generation of recombinant SrtAΔ65 protein
A clone expressing a truncated version of the recombinant sortase protein (SrtAΔ65) was generated by PCR amplification from S. uberis 0140J template DNA using the sense primer p633 (located 195 bp after the srtA gene (SUB0881) start codon) and the antisense primer p628 (located 174 bp downstream of srtA gene stop codon), incorporating a KpnI site for cloning into the pQE-1 expression plasmid (Qiagen, Hilden, Germany). The 738 bp fragment was generated using Phusion™ high fidelity polymerase (New England Biolabs, Ipswich, USA), purified using a MinElute PCR Purification Kit (Qiagen) and digested with KpnI (New England Biolabs). Fragments were cloned into pQE1 vector pre-treated with PvuII, KpnI and Antarctic phosphatase (all New England Biolabs) using T4 DNA Ligase (New England Biolabs) at 14 °C according to the manufacturer’s instructions. Twenty microlitres of the ligation mixture was desalted [2] and approximately 10 ng of the desalted ligation mixture was transformed into Escherichia coli M15 pREP4 (Qiagen). Transformants were selected on LB agar plates containing 50 μg/mL ampicillin (Amp) and 25 μg/mL kanamycin (Kan). Recombinant (6 × His-tagged) SrtAΔ65 protein was generated from overnight cultures inoculated at a 1/50 dilution into LB broth containing 50 μg/mL of Amp and 25 μg/mL of Kan. Cultures were grown at 37 °C with aeration (shaking at 200 rpm) and protein expression induced, once cultures reached an OD600nm of 0.8, by addition of 0.2 mM IPTG. Following further incubation (2.5 h), cells were harvested by centrifugation (16 000 × g, 10 min) and the bacterial pellets lysed using CelLytic in the presence of 2 μ/mL RNase, 2 μ/mL DNase (all from Sigma) and Complete-EDTA-free protease inhibitors (Roche, Mannheim, Germany). Approximately 3 mg of soluble SrtAΔ65 recombinant protein was purified using HIS-Select Cartridges (Sigma) and dialysed overnight against MilliQ water using Slide-A-Lyzer dialysis cassettes (Pierce, Rockford, USA) according to the manufacturer’s instructions.
2.7. Preparation of cellular extracts of S. uberis
Overnight cultures (400 mL) of S. uberis 0140J and SrtA mutant were grown in THB overnight and bacteria were harvested by centrifugation (16 000 × g, 10 min). Complete protease inhibitors (Roche) were added to all cellular extracts at a 1× concentration. The bacterial growth medium was filter sterilised through a 0.22 μm filter (Millipore, Bedford, USA) and concentrated approximately 100-fold using Amicon centrifugal filter devices (Millipore) with 10 kDa molecular weight cut off. To precipitate proteins, 4 volumes of methanol and chloroform (3:1) was added to 1 volume of concentrated bacterial growth media. The preparation was vortexed and 2 volumes of MilliQ water were added prior to centrifugation (16 000 × g, 2 min). The upper phase was carefully removed and discarded. Proteins were precipitated with 3 volumes of methanol, and collected by centrifugation (16 000 × g, 2 min). The supernatant was discarded and the remaining pellet air-dried before resuspension in SDS-PAGE-loading buffer.
To isolate capsule extracts, bacterial pellets from above were washed 3 times with 40 mL of PBS and centrifuged (16 000 × g, 10 min, 4 °C), then resuspended in 500 μL of PBS containing hyaluronidase (100 U/mL, Sigma). Cells were incubated for 2 h at 37 °C and the capsule extract (supernatant) collected following centrifugation (8000 × g, 5 min).
To isolate cell wall fractions, acapsular cells from the preparation above were washed 3 times with 40 mL of PBS and resuspended in 500 μL of 40% sucrose/PBS containing mutanolysin (100 U/mL, Sigma). The cell wall extract (supernatant) was collected following centrifugation (1 500 × g, 15 min). The remaining pellet was washed 3 times with 1 mL of cell membrane buffer (100 mM NaCl, 100 mM Tris-Cl ph 7.5, 10 mM MgCl2) and cycled through 3 periods of freeze thawing (−20 °C to 37 °C) and the cytoplasmic contents (supernatant) were harvested following centrifugation (10 000 × g, 10 min). The resulting pellet was washed 3 times in 1 mL of cell membrane buffer and resuspended in 500 μL of cell membrane buffer containing 1% v/v Triton X-100 and incubated at 37 °C for 1 h. The cell membrane fraction (supernatant) was harvested following centrifugation (10 000 × g, 10 min) and all extracts were stored at −20 °C.
2.8. Production of antiserum in rabbits and immunoblotting
Five aliquots of approximately 50 μg of freeze dried purified recombinant SrtAΔ65 was supplied to Davids Biotechnologie GmbH (Regensburg, Germany) for anti-serum production in rabbits. Anti-serum (50 mL) was supplied filter sterilized and containing 0.02% sodium azide as a preservative. Cellular extracts were separated on 12% or 15% sodium-dodecyl sulphate polyacrylamide (SDS-PAGE) gels and were stained using InstantBlue (Expedeon protein solutions, Cambridge, UK) or transferred onto Amersham Hybond™-C Extra nitrocellulose membranes (GE Healthcare, Little Chalfont, UK) for immuno-detection. Transfer was performed at 170 mA for 1 h in a Transblot apparatus (Biorad, Hemmel Hemstead, UK) in transfer buffer consisting of 25 mM Tris-base, 192 mM glycine and 20% (v/v) methanol, pH 8.1–8.4. Following transfer, membranes were incubated in a solution of 1% (w/v) skim milk (Marvel; Premier International Foods, UK) in PBS at 4 °C overnight. Membranes were washed three times for 5 min in PBS containing 0.1% Tween 20 (PBST) then incubated with SrtA rabbit antiserum at a 1:1 000 dilution in 1% (w/v) skim milk in PBS for 1 h. Membranes were washed three times for 5 min in PBST then incubated with goat anti-rabbit immunoglobulin G conjugated to HRP (Southern Biotech, Alabama, USA) at a 1:1 000 dilution for 1 h. Membranes were washed again as above and HRP conjugate was detected using a solution of 0.5 mg/mL 4-chloronaphthol in PBS containing 16.7% methanol and 0.00015% (v/v) of H2O2. The reaction was incubated in the dark until coloured bands were visible. Subsequently, membranes were washed in PBS, air-dried and stored in the dark.
3. RESULTS
3.1. Sub-cellular localisation of SrtA in S. uberis
A protein band of approximately 26 kDa was detected in the cell membrane-enriched fraction and to a lesser extent in the cytoplasmic-enriched fraction of the wild type by immunoblotting with antiserum raised against rSrtA (Fig. 1). The size of the detected protein was consistent with that of the translated product from sub0881 following the predicted N-terminal cleavage [44]. No such protein was detected in any of the sub-cellular fractions generated from the SrtA mutant (Fig. 1).
Figure 1.

Cellular localisation of sortase A (SrtA) in Streptococcus uberis. Protein extracts were obtained from S. uberis 0140J and srtA mutant from cultures grown overnight in THB media. Fractions were separated on 15% SDS PAGE gels, transferred to nitrocellulose and probed with antiserum to SrtA (size of proteins of known molecular mass are indicated). Extracts were loaded as follows: from S. uberis 0140J capsule (lane 1); mutanolysin-derived cell wall (lane 3); cytoplasmic (lane 5); cell membrane (lane 7) and precipitated media extracts (lane 9) and from the srtA mutant capsule (lane 2); mutanolysin-derived cell wall (lane 4); cytoplasmic (lane 6); cell membrane (lane 8) and precipitated media extracts (lane 10). (A color version of this figure is available online at www.vetres.org.)
3.2. Infectivity and virulence of the wild-type and SrtA mutant following experimental challenge in the lactating bovine mammary gland
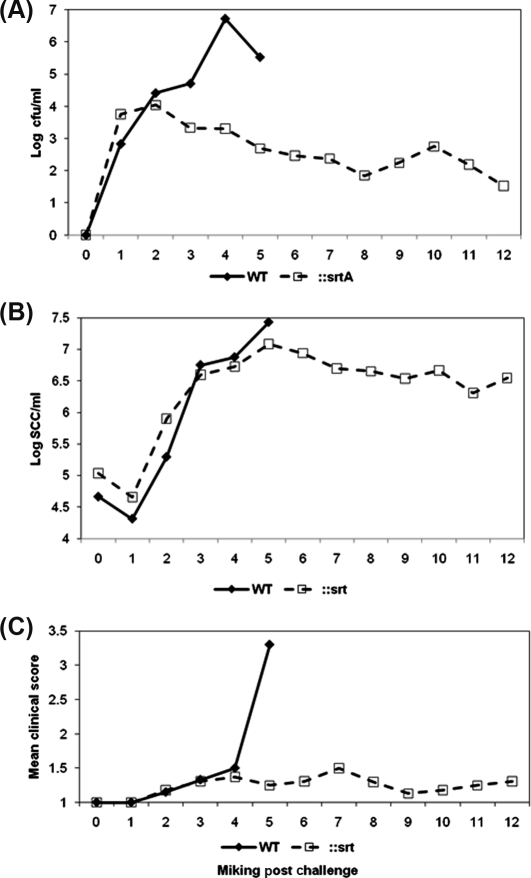
All challenged quarters of animals that received wild type S. uberis became infected and shed bacteria at around 106 cfu/mL by 48–60 h post challenge (Fig. 2A). Following challenge of eight quarters on four animals with the SrtA mutant, all showed evidence of infection and the SrtA mutant was detected in milk at levels similar to those for the wild type for up to 24 h post challenge. However, subsequent bacterial colonization declined, from a typical level of 104 cfu/mL of milk by 24 h post challenge, such that by the end of the experiment (7 days post challenge) the mean bacterial number present was around 10 cfu/mL (Fig. 2B). By this time, only 3 of the 8 quarters continued to shed bacteria the remainder having eliminated the infection (< 1 cfu/mL milk).
Figure 2.
Bacterial isolation, somatic cell count and clinical response following challenge with S. uberis 0140J and SrtA mutant in dairy cattle. (A) Bacterial recovery of S. uberis following challenge. Data are represented as the geometric means of the number of bacteria obtained from the milk of animals challenged with either strain 0140J (n = 4) or the SrtA mutant (n = 8), as indicated on the figure. (B) Cellular influx at each milking following challenge. Data are represented by the geometric means of the number of somatic cells obtained from the milk of animals challenged with either strain 0140J (n = 4) or the SrtA mutant (n = 8), as indicated on the figure. (C) Combined clinical scores from clinical manifestations following S. uberis challenge. Data are represented by the mean of clinical scores given for the appearance of the quarter and appearance of the milk, as outlined in Table II with either strain 0140J (n = 4) or the SrtA mutant (n = 8), as indicated on the figure.
The cellular infiltration into the mammary gland in response to infection was identical in both groups of animals and was not dependent on the challenge strain. In each case, this was similar to that reported previously in this model and reached a maximum of between 106 and 107 cells/mL of milk by 48–60 h post challenge. In animals challenged with the wild type strain this coincided with the appearance of acute clinical signs of mastitis (Tab. II; Fig. 2C) which required administration of antibiotic therapy to eliminate infection and to alleviate signs of disease. In stark contrast, animals that had received the SrtA mutant showed only very minor, if any, signs of mastitis (Tab. II; Fig. 2C).
3.3. Isolation of mutants lacking individual SrtA anchored proteins
Mutants carrying lesions within each of the genes encoding SrtA anchored protein were isolated by genotypic selection from a random mutant bank. Each mutant was cured of the plasmid insertion and shown by Southern blotting to contain one copy of the ISS1 element (data not shown). In one case (sub1154) ISS1 insertion was not identified in sufficiently close proximity to the start codon to ensure loss of function and this gene was subsequently disrupted by allelic exchange mutagenesis. Following the allelic exchange procedures, PCR analysis revealed that 6 of 24 colonies exhibited the deleted sub1154 locus. This was subsequently confirmed by southern analysis (data not shown). One such clone was stored and used throughout this study as the ∆sub1154 mutant strain. Sequence analysis of the deleted locus also identified a single base pair deletion 116 bp beyond the ATG start codon. This additional frameshift event had the effect of prematurely terminating the theoretical deleted translation product to just 55 amino acids, some 22 residues beyond the putative signal peptide cleavage point.
3.4. Screening of mutants lacking SrtA substrates for virulence/attenuation
Each mutant was used to challenge two mammary quarters to determine if the insertional mutation resulted in major attenuation of S. uberis. In all cases, the bacteria recovered post-challenge were genotyped by PCR and in each case showed the presence of the correctly mutated gene. In the case of isolates obtained from contemporaneous challenges with the wild type strain, a similar procedure showed the presence of appropriate wild-type alleles (data not shown).
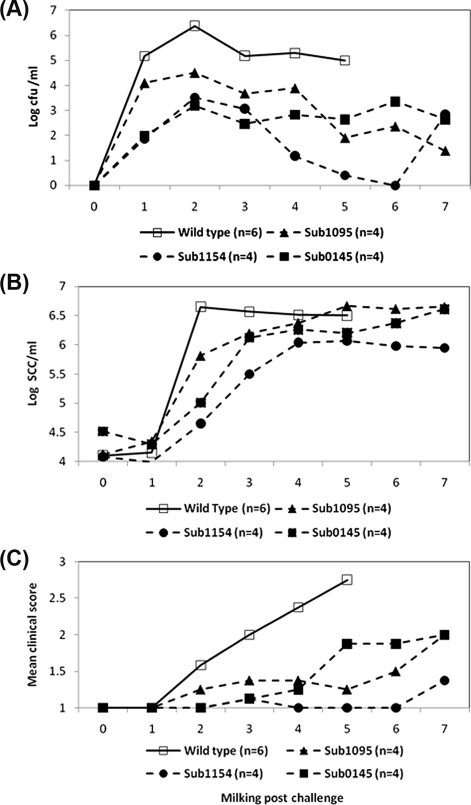
Challenge with mutants carrying insertions within sub0135, sub207, sub0241, sub0826, sub1370 or sub1730 had little if any effect on virulence and each strain induced moderate to severe clinical signs of mastitis (data not shown). In contrast, strains lacking any of sub0145, sub0888, sub1095 or sub1154 showed few, if any, overt signs of infection during the initial screening in two mammary quarters. Subsequent rescreening, in a further two mammary quarters, confirmed the attenuation in strains lacking sub0145, sub1095 or sub1154 (Fig. 3C). However, in this instance the strain lacking sub0888 colonised to high levels. In contemporaneous challenges with the wild type strain all quarters became colonised at high levels (typically 105–107) and moderate to severe signs of mastitis were noted in all six challenged quarters (Fig. 3C). All animals challenged with the wild type strain required intervention with antibiotic therapy by the fifth milking post challenge.
Figure 3.
Bacterial isolation, somatic cell count and clinical response following challenge with S. uberis 0140J and mutants lacking individual SrtA- anchored proteins in dairy cattle. (A) Bacterial recovery of S. uberis at each milking following challenge. Data are represented as the geometric means of the number of bacteria obtained from the milk of animals challenged with either strain 0140J (n = 6) or the individual SrtA-anchored protein mutants (sub1095 n = 4; sub1154 n = 4; sub0145 n = 4), as indicated on the figure. (B) Cellular influx at each milking following challenge. Data are represented by the geometric means of the number of somatic cells obtained from the milk of animals challenged with either strain 0140J (n = 6) or the individual SrtA-anchored protein mutants (sub1095 n = 4; sub1154 n = 4; sub0145 n = 4), as indicated on the figure. (C) Combined clinical scores at each milking following challenge. Data are represented by the mean of the combined clinical scores given for the appearance of the quarter and appearance of the milk, as outlined in Table II with either strain 0140J (n = 6) or the individual SrtA-anchored protein mutants (sub1095 n = 4; sub1154 n = 4; sub0145 n = 4), as indicated on the figure.
The mutant lacking sub1095 appeared to colonise in a manner similar to the wild-type strain for 24 h, after which the numbers of the mutant strain declined (Fig. 3A). In one animal, bacterial colonisation fell below the level of detection in both challenged quarters by 7 milkings post challenge. The mutants lacking either sub0145 or sub1154 showed impaired colonisation compared to the wild type strain (Fig. 3A). In marked contrast to the wild type, colonisation with the mutant lacking sub1154 declined to the point where it was not detected in any challenged quarter by the sixth milking after challenge. However in both animals challenged with this strain, bacteria were subsequently re-isolated from one of the two challenged quarters at milking 7. The mutant lacking sub0145 showed reduced colonisation throughout the experimental period and was consistently re-isolated at levels 100 to 1000-fold less than the wild type strain (Fig. 3A).
An influx of somatic cells was evident in all mammary quarters challenged with S. uberis; the magnitude of the response was similar in animals challenged with wild type or the mutants lacking either sub0145 or sub1095. However, the speed of the cellular response to infection with the mutant strains was slower than that to the wild-type. The speed and magnitude of the cellular response to challenge with the strain lacking sub1154 was markedly reduced compared to that detected following challenge with the wild type strain (Fig. 3B).
4. DISCUSSION
A homologue of srtA detected within S. uberis 0140J (sub0881) was shown during this investigation to be required for the full expression of virulence by this bacterium. This finding correlates with studies on other streptococci associated with different diseases that have revealed the essential nature of SrtA activity. In the cases of S. pyogenes [5], S. sanguinis [46], S. agalactiae [25] and S. gordonii [6] virulence or virulence related phenotypes were markedly decreased in strains where SrtA was either defective or absent. In some of these instances the altered phenotype, initially attributed to the absence of SrtA, was subsequently shown to be directly linked to one or more sortase anchored proteins [24, 46].
The SrtA isogenic mutant of S. uberis strain 0140J was isolated with an inserted ISS1 element disrupting the gene between 248 bp and 249 bp of the nucleotide sequence, resulting in a theoretically translated product that did not code for any of the predicted active site of the SrtA enzyme [14]. In line with the predicted location for SrtA, this protein was only detected in the cell membrane and cytoplasmic fractions of wild type S. uberis and was not apparent in any fractions from the isogenic mutant (Fig. 1). A previous comparative proteomic analysis revealed the requirement for sortase to anchor proteins bearing the LPXTG or LPXXXD motif at the surface of S. uberis [14].
Although initially able to colonise the bovine mammary gland similarly to the wild-type S. uberis, the mutant lacking SrtA was unable to colonise the gland to high levels; with bacterial numbers remaining approximately 1000-fold lower than those detected in milk from animals challenged with the wild type strain. This corresponded with the failure of the srtA mutant to induce progressive clinical signs of disease. Thus it was postulated that anchoring of proteins by SrtA was required for high level colonisation and/or induction of severe inflammatory reactions associated with clinical disease. As this mutant strain grew at a rate and to a final yield similar to that of the wild type strain in both laboratory media and skimmed milk from healthy, uninfected mammary glands1 interactions by SrtA anchored proteins were considered to be specific to disease pathogenesis.
An increase in the somatic cell count (SCC) is an indicator commonly used as a surrogate of bacterial infection of the mammary gland and mastitis. The inflammatory response to intramammary bacterial infection results in a massive influx of circulating polymorphonuclear neutrophils (PMN) into the mammary tissue and secretion. As in the situation during the current investigation, the SCC can reach in excess of 5 million cells/mL; over 90% of which are PMN [31, 35]. The presence of such high numbers of leukocytes in milk usually coincides with the onset of clinical signs of disease following challenge with a variety of mammary pathogens including E. coli [18], S. aureus [18, 38] and S. uberis [15, 16, 20, 26]. In this study, following challenge with the wild-type, SCC rose to between 106 and 107/mL by 4 milkings (2 days) post challenge and severe clinical signs of disease were detected in all quarters by five milkings post challenge. However, although challenge with the srtA mutant resulted in an increase in the SCC at a similar speed and magnitude in response to challenge with wild-type S. uberis, there was little or no evidence of clinical signs of disease in these animals. These data therefore uncouple the response that stimulates influx of inflammatory cells from the responses that induce the inflammatory changes leading to overt signs of disease and imply that such signs are dependent on the interplay between pathogen and host during the process of infection.
Mutants lacking proteins identified previously to be anchored by SrtA (14) and one further protein (sub0241) that was shown to conform to that expected of a SrtA substrate were isolated. In each case, the position of the insertional element (ISS1) was such that disruption of the function of any residual gene product was predicted. The bank did not contain a mutant with a suitable lesion within sub1154; although a mutant with an insertion near to the end of this CDS was detected. This mutant produced a large C-terminally truncated polypeptide likely to contain the serine-protease active site domain. It is of note that the truncated protein was predicted to lack the C-terminal cell wall anchor (sorting domain) and that this protein, unlike the full length protein in the wild type, was not anchored at the bacterial surface1. Consequently, a mutant carrying a deletion of sub1154 was produced by allelic exchange mutagenesis.
Three of the mutant strains failed to induce clinically apparent mastitis in any of four challenged quarters; this is in contrast to the wild type strain that in contemporary challenges induced mastitis in all six cases. This challenge model using strain 0140J has proven to be very reproducible and has been conducted on numerous occasions by different research groups around the world [4, 21, 29]. In each case, disease was evident in approximately 90% of the quarters challenged and in no instance has there been any report of an animal within a study that has resisted challenge in two mammary quarters. Therefore, the absence of clinical signs in 4 of 4 challenged mammary quarters would strongly indicate that each of sub0145, sub1095 and sub1154 play an important, independent, non-redundant role in the pathogenesis of mastitis due to S. uberis. It is possible that other SrtA-anchored proteins also play a role in pathogenesis but that any effect is masked by the presence of another protein/activity that is sufficient to complement that missing from the mutant.
It is also worthy of note that 2 of the 3 individual mutant strains, lacking sub0145 or sub1154, appeared less able to colonise the bovine mammary gland than that lacking sub0881 (srtA). This probably reflects the complete absence of the target protein in these strains. However, like the SrtA mutant, these strains grew at a rate and to a final yield similar to that of the wild type strain in Todd Hewitt broth and skimmed bovine milk from healthy uninfected mammary glands1.
In the srtA mutant, the proteins are likely to be produced but not anchored to the bacterial cell. Therefore, we can conclude that in addition to the requirement of sub0145, sub1095 and/or sub1154 for full expression of virulence there may also be a requirement for the bacterium to hold any or all of these, and or other SrtA-anchored proteins, in close proximity to the cell.
A number of the putative sortase substrate proteins have been identified as virulence determinants in other streptococcal species. The protein encoded by sub1154 shows 34% identity at the protein level (BLASTP) to the C5a peptidase of S. pyogenes. In Lancefield group A and B streptococci this protein acts as a highly specific serine peptidase and is also found on the bacterial surface [11, 45]. The C5a peptidase cleaves the C5a complement factor, which is involved in the recruitment and activation of phagocytic cells to the site of infection [23]. The equivalent protein in Group B Streptococcus has also been shown to interact with fibronectin [42] and has been shown to have a role during invasion of human epithelial cells in culture [10].
The S. uberis protein sub1095 shows 46% protein sequence identity to SclB of S. pyogenes. SclB has been shown to be involved in adhesion to fibroblasts [37] and can bind host Thrombin-activatable fibrinolysis Inhibitor, which is postulated to be utilised by the bacterium to modulate the host response to infection [32]. A collagen-like protein in S. pneumoniae was shown to have a role in invasion of host cells in vitro but was not essential for virulence [34] and in S. pyogenes a collagen like protein was shown to mediate adherence and internalisation via the alpha(2)beta(1) integrin [8]. The requirement for adherence and internalisation during the pathogenesis of bovine mastitis has not been demonstrated in vivo, but evidence put forward from in vitro studies would indicate that adherence and internalisation to a mammary cell line (Mac-T) is largely dependent on Sua, a protein found non-covalently associated with the cell surface [1].
The protein encoded by sub0145 has previously been identified as a possible virulence determinant of S. uberis and at least one allelic variant has been shown to bind bovine lactoferrin [28]. The concentration of this iron-chelating compound increases in the mammary gland during infection [17] however whilst iron limitation is known to have an anti-microbial effect on some bacteria, it appears to have little effect on the growth of S. uberis [9, 36]. However, lactoferrin can also bind other divalent metal ions such as Mn [3, 41]. S. uberis has a requirement for this metal ion in vivo and in the absence of a high affinity Mn uptake system was unable to colonise the bovine mammary gland [40].
In conclusion, the failure of a SrtA deficient strain of S. uberis to cause clinical mastitis in dairy cattle indicates that a number of SrtA-anchored proteins are likely to be involved in the pathogenesis of this bacterium. Further investigation revealed that the list of proteins involved includes sub0145, sub1095 and sub1154, however their precise role in pathogenesis is yet to be established.
Acknowledgments
The authors gratefully acknowledge the financial support of BBSRC, through grants S19514 and BB/E018173.
Footnotes
Leigh J.A., et al., unpublished results.
References
- 1.Almeida R.A., Luther D.A., Park H.M., Oliver S.P., Identification, isolation, and partial characterization of a novel Streptococcus uberis adhesion molecule (SUAM), Vet. Microbiol. (2006) 115:183–191 [DOI] [PubMed] [Google Scholar]
- 2.Atrazhev A.M., Elliott J.F., Simplified desalting of ligation reactions immediately prior to electroporation into E. coli, Biotechniques (1996) 21:1024. [DOI] [PubMed] [Google Scholar]
- 3.Baker E.N., Anderson B.F., Baker H.M., Day C.L., Haridas M., Norris G.E., et al., Three-dimensional structure of lactoferrin in various functional states, Adv. Exp. Med. Biol. (1994) 357:1–12 [DOI] [PubMed] [Google Scholar]
- 4.Bannerman D.D., Paape M.J., Goff J.P., Kimura K., Lippolis J.D., Hope J.C., Innate immune response to intramammary infection with Serratia marcescens and Streptococcus uberis, Vet. Res. (2004) 35:681–700 [DOI] [PubMed] [Google Scholar]
- 5.Barnett T.C., Scott J.R., Differential recognition of surface proteins in Streptococcus pyogenes by two sortase gene homologs, J. Bacteriol. (2002) 184:2181–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolken T.C., Franke C.A., Jones K.F., Zeller G.O., Jones C.H., Dutton E.K., Hruby D.E., Inactivation of the srtA gene in Streptococcus gordonii inhibits cell wall anchoring of surface proteins and decreases in vitro and in vivo adhesion, Infect. Immun. (2001) 69:75–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradley A.J., Leach K.A., Breen J.E., Green L.E., Green M.J., Survey of the incidence and aetiology of mastitis on dairy farms in England and Wales, Vet. Rec. (2007) 160:253–257 [DOI] [PubMed] [Google Scholar]
- 8.Caswell C.C., Lukomska E., Seo N.S., Hook M., Lukomski S., Scl1-dependent internalization of group A Streptococcus via direct interactions with the alpha2beta(1) integrin enhances pathogen survival and re-emergence, Mol. Microbiol. (2007) 64:1319–1331 [DOI] [PubMed] [Google Scholar]
- 9.Chaneton L., Tirante L., Maito J., Chaves J., Bussmann L.E., Relationship between milk lactoferrin and etiological agent in the mastitic bovine mammary gland, J. Dairy Sci. (2008) 91:1865–1873 [DOI] [PubMed] [Google Scholar]
- 10.Cheng Q., Stafslien D., Purushothaman S.S., Cleary P., The group B streptococcal C5a peptidase is both a specific protease and an invasin, Infect. Immun. (2002) 70:2408–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cleary P.P., Prahbu U., Dale J.B., Wexler D.E., Handley J., Streptococcal C5a peptidase is a highly specific endopeptidase, Infect. Immun. (1992) 60:5219–5223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Comfort D., Clubb R.T., A comparative genome analysis identifies distinct sorting pathways in gram-positive bacteria, Infect. Immun. (2004) 72:2710–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dramsi S., Trieu-Cuot P., Bierne H., Sorting sortases: a nomenclature proposal for the various sortases of Gram-positive bacteria, Res. Microbiol. (2005) 156:289–297 [DOI] [PubMed] [Google Scholar]
- 14.Egan S.A., Kurian D., Ward P.N., Hunt L., Leigh J.A., Identification of sortase A (SrtA) substrates in Streptococcus uberis: evidence for an additional hexapeptide (LPXXXD) sorting motif, J. Proteome Res. (2010) 9:1088–1095 [DOI] [PubMed] [Google Scholar]
- 15.Field T.R., Ward P.N., Pedersen L.H., Leigh J.A., The hyaluronic acid capsule of Streptococcus uberis is not required for the development of infection and clinical mastitis, Infect. Immun. (2003) 71:132–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finch J.M., Hill A.W., Field T.R., Leigh J.A., Local vaccination with killed Streptococcus uberis protects the bovine mammary gland against experimental intramammary challenge with the homologous strain, Infect. Immun. (1994) 62:3599–3603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harmon R.J., Schanbacher F.L., Ferguson L.C., Smith K.L., Changes in lactoferrin, immunoglobulin G, bovine serum albumin, and alpha-lactalbumin during acute experimental and natural coliform mastitis in cows, Infect. Immun. (1976) 13:533–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hill A.W., Shears A.L., Hibbitt K.G., The elimination of serum-resistant Escherichia coli from experimentally infected single mammary glands of healthy cows, Res. Vet. Sci. (1978) 25:89–93 [PubMed] [Google Scholar]
- 19.Hill A.W., Leigh J.A., DNA fingerprinting of Streptococcus uberis: a useful tool for epidemiology of bovine mastitis, Epidemiol. Infect. (1989) 103:165–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill A.W., Finch J.M., Field T.R., Leigh J.A., Immune modification of the pathogenesis of Streptococcus uberis mastitis in the dairy cow, FEMS Immunol. Med. Microbiol. (1994) 8:109–117 [DOI] [PubMed] [Google Scholar]
- 21.Hoeben D., Burvenich C., Eppard P.J., Byatt J.C., Hard D.L., Effect of bovine somatotropin on neutrophil functions and clinical symptoms during Streptococcus uberis mastitis, J. Dairy Sci. (1999) 82:1465–1481 [DOI] [PubMed] [Google Scholar]
- 22.Janulczyk R., Rasmussen M., Improved pattern for genome-based screening identifies novel cell wall-attached proteins in gram-positive bacteria, Infect. Immun. (2001) 69:4019–4026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ji Y., McLandsborough L., Kondagunta A., Cleary P.P., C5a peptidase alters clearance and trafficking of group A streptococci by infected mice, Infect. Immun. (1996) 64:503–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kharat A.S., Tomasz A., Inactivation of the srtA gene affects localization of surface proteins and decreases adhesion of Streptococcus pneumoniae to human pharyngeal cells in vitro, Infect. Immun. (2003) 71:2758–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lalioui L., Pellegrini E., Dramsi S., Baptista M., Bourgeois N., Doucet-Populaire F., et al., The SrtA Sortase of Streptococcus agalactiae is required for cell wall anchoring of proteins containing the LPXTG motif, for adhesion to epithelial cells, and for colonization of the mouse intestine, Infect. Immun. (2005) 73:3342–3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leigh J.A., Finch J.M., Field T.R., Real N.C., Winter A., Walton A.W., Hodgkinson S.M., Vaccination with the plasminogen activator from Streptococcus uberis induces an inhibitory response and protects against experimental infection in the dairy cow, Vaccine (1999) 17:851–857 [DOI] [PubMed] [Google Scholar]
- 27.Mazmanian S.K., Liu G., Ton-That H., Schneewind O., Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall, Science (1999) 285:760–763 [DOI] [PubMed] [Google Scholar]
- 28.Moshynskyy I., Jiang M., Fontaine M.C., Perez-Casal J., Babiuk L.A., Potter A.A., Characterization of a bovine lactoferrin binding protein of Streptococcus uberis, Microb. Pathog. (2003) 35:203–215 [DOI] [PubMed] [Google Scholar]
- 29.Moyes K.M., Drackley J.K., Morin D.E., Bionaz M., Rodriguez-Zas S.L., Everts R.E., et al., Gene network and pathway analysis of bovine mammary tissue challenged with Streptococcus uberis reveals induction of cell proliferation and inhibition of PPARgamma signaling as potential mechanism for the negative relationships between immune response and lipid metabolism, BMC Genomics (2009) 10:542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Navarre W.W., Schneewind O., Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope, Microbiol. Mol. Biol. Rev. (1999) 63:174–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paape M.J., Wergin W.P., Guidry A.J., Schultze W.D., Phagocytic defense of the ruminant mammary gland, Adv. Exp. Med. Biol. (1981) 137:555–578 [PubMed] [Google Scholar]
- 32.Pahlman L.I., Marx P.F., Morgelin M., Lukomski S., Meijers J.C., Herwald H., Thrombin-activatable fibrinolysis inhibitor binds to Streptococcus pyogenes by interacting with collagen-like proteins A, B, J. Biol. Chem. (2007) 282:24873–24881 [DOI] [PubMed] [Google Scholar]
- 33.Pallen M.J., Lam A.C., Antonio M., Dunbar K., An embarrassment of sortases – a richness of substrates?, Trends Microbiol. (2001) 9:97–102 [DOI] [PubMed] [Google Scholar]
- 34.Paterson G.K., Nieminen L., Jefferies J.M., Mitchell T.J., PclA, a pneumococcal collagen-like protein with selected strain distribution, contributes to adherence and invasion of host cells, FEMS Microbiol. Lett. (2008) 285:170–176 [DOI] [PubMed] [Google Scholar]
- 35.Persson K., Larsson I., Hallen Sandgren C., Effects of certain inflammatory mediators on bovine neutrophil migration in vivo and in vitro, Vet. Immunol. Immunopathol. (1993) 37:99–112 [DOI] [PubMed] [Google Scholar]
- 36.Rainard P., Bacteriostatic activity of bovine milk lactoferrin against mastitic bacteria, Vet. Microbiol. (1986) 11:387–392 [DOI] [PubMed] [Google Scholar]
- 37.Rasmussen M., Bjorck L., Unique regulation of SclB – a novel collagen-like surface protein of Streptococcus pyogenes, Mol. Microbiol. (2001) 40:1427–1438 [DOI] [PubMed] [Google Scholar]
- 38.Schalm O.W., Lasmanis J., Jain N.C., Conversion of chronic staphylococcal mastitis to acute gangrenous mastitis after neutropenia in blood and bone marrow produced by an equine anti-bovine leukocyte serum, Am. J. Vet. Res. (1976) 37:885–890 [PubMed] [Google Scholar]
- 39.Scott J.R., Barnett T.C., Surface proteins of gram-positive bacteria and how they get there, Annu. Rev. Microbiol. (2006) 60:397–423 [DOI] [PubMed] [Google Scholar]
- 40.Smith A.J., Ward P.N., Field T.R., Jones C.L., Lincoln R.A., Leigh J.A., MtuA, a lipoprotein receptor antigen from Streptococcus uberis, is responsible for acquisition of manganese during growth in milk and is essential for infection of the lactating bovine mammary gland, Infect. Immun. (2003) 71:4842–4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steijns J.M., van Hooijdonk A.C., Occurrence, structure, biochemical properties and technological characteristics of lactoferrin, Br. J. Nutr. (2000) 84Suppl. 1:S11–S17 [DOI] [PubMed] [Google Scholar]
- 42.Tamura G.S., Hull J.R., Oberg M.D., Castner D.G., High-affinity interaction between fibronectin and the group B streptococcal C5a peptidase is unaffected by a naturally occurring four-amino-acid deletion that eliminates peptidase activity, Infect. Immun. (2006) 74:5739–5746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor D.L., Ward P.N., Rapier C.D., Leigh J.A., Bowler L.D., Identification of a differentially expressed oligopeptide binding protein (OppA2) in Streptococcus uberis by representational difference analysis of cDNA, J. Bacteriol. (2003) 185:5210–5219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ward P.N., Holden M.T., Leigh J.A., Lennard N., Bignell A., Barron A., et al., Evidence for niche adaptation in the genome of the bovine pathogen Streptococcus uberis , BMC Genomics (2009) 10:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wexler D.E., Chenoweth D.E., Cleary P.P., Mechanism of action of the group A streptococcal C5a inactivator, Proc. Natl. Acad. Sci. USA (1985) 82:8144–8148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamaguchi M., Terao Y., Ogawa T., Takahashi T., Hamada S., Kawabata S., Role of Streptococcus sanguinis sortase A in bacterial colonization, Microbes Infect. (2006) 8:2791–2796 [DOI] [PubMed] [Google Scholar]