Abstract
Anaerobic enrichment cultures with elemental sulfur as electron acceptor and either acetate or propionate as electron donor and carbon source at pH 10 and moderate salinity inoculated with sediments from soda lakes in Kulunda Steppe (Altai, Russia) resulted in the isolation of two novel members of the bacterial phylum Chrysiogenetes. The isolates, AHT11 and AHT19, represent the first specialized obligate anaerobic dissimilatory sulfur respirers from soda lakes. They use either elemental sulfur/polysulfide or arsenate as electron acceptor and a few simple organic compounds as electron donor and carbon source. Elemental sulfur is reduced to sulfide through intermediate polysulfide, while arsenate is reduced to arsenite. The bacteria belong to the obligate haloalkaliphiles, with a pH growth optimum from 10 to 10.2 and a salt range from 0.2 to 3.0 M Na+ (optimum 0.4–0.6 M). According to the phylogenetic analysis, the two strains were close to each other, but distinct from the nearest relative, the haloalkaliphilic sulfur-reducing bacterium Desulfurispirillum alkaliphilum, which was isolated from a bioreactor. On the basis of distinct phenotype and phylogeny, the soda lake isolates are proposed as a new genus and species, Desulfurispira natronophila (type strain AHT11T = DSM22071T = UNIQEM U758T).
Electronic supplementary material
The online version of this article (doi:10.1007/s00792-010-0314-7) contains supplementary material, which is available to authorized users.
Keywords: Desulfurispirillumalkaliphilum, Haloalkaliphilic, Soda lakes, Sulfur-reducing bacteria (SuRB)
Introduction
The first haloalkaliphilic sulfur-respiring bacterium described as Desulfurispirillum alkaliphilum has recently been isolated from a full-scale bioreactor removing sulfide from biogas (Sorokin et al. 2007). In this anthropogenic system a short sulfur cycle at moderately haloalkaline conditions apparently takes place, i.e., sulfide oxidation to elemental sulfur at oxygen limitation as the main process and sulfur reduction at the expense of organic carbon excreted by the sulfide oxidizers as a minor unwanted reaction (Janssen et al. 2009).
Investigation of the reductive branch of the sulfur cycle (i.e., sulfidogenesis) at extremely haloalkaline conditions has so far been limited by dissimilatory sulfate reduction, which demonstrated that this process is very active in soda lake habitats and that novel lineages within the Deltaproteobacteria are responsible for this activity (Gorlenko et al. 1999; Sorokin et al. 2004; Scholten et al. 2005; Kulp et al. 2006; 2007; Foti et al. 2007; Zhilina 2007; Sorokin et al. 2008a). Until now, only a single report has been published on the presence of non sulfate-reducing sulfidogenic haloalkaliphiles in soda lakes (Sorokin et al. 2008b). Meanwhile, the existence of short sulfur cycling similar to the example mentioned above might be predicted for the conditions of shallow hypersaline soda lakes, where sediments have high sulfide content at the top and low oxygen solubility in near bottom brines. One of the most interesting features of sulfur speciation at high pH is the appearance of linear polysulfide (−S–Sx –S−) as a stable intermediate, which, in contrast to cyclic elemental sulfur, is soluble, and, therefore, much more reactive. Polysulfides are the products of spontaneous reaction of sulfide and elemental sulfur. This makes sulfur cycling at high pH and, particularly, dissimilatory sulfur reduction where polysulfide is the actual electron acceptor (Hedderich et al. 1999), substantially different from those in the pH-neutral marine habitats.
Here we describe the properties of two novel obligately anaerobic bacterial isolates from hypersaline soda lakes in south-west Siberia, which represent the first highly specialized haloalkaliphilic dissimilatory sulfur/polysulfide reducers. The most important function of these bacteria is the potential to utilize acetate as electron donor with sulfur as electron acceptor at extremely haloalkaline anoxic conditions.
Methods
Samples
Samples of the top 5 cm sediments from soda lakes Tanatar-5 (pH = 10.15; salinity = 80 g l−1; total alkalinity = 0.98 M; HS− = 2.0 mM; July 2008) and Cock Soda Lake (pH = 10.3; salinity = 120 g l−1; total alkalinity = 0.82 M; HS− = 0.50 mM; July 2009) were used to enrich for sulfur-respiring haloalkaliphiles.
Enrichment and cultivation of SuRB
Enrichment and routine cultivation of haloalkaliphilic SuRB was performed at 28°C on a mineral medium containing sodium carbonate buffer (0.5 M Na+) with pH 10, 0.1 M NaCl, and 0.5 g l−1 of K2HPO4. After sterilization in closed bottles, the medium was supplemented with either 20 mM acetate or propionate as carbon and energy source, 20 mg l−1 of yeast extract, 4 mM NH4Cl, 1 mM MgSO4, 1 ml l−1 each of acidic trace metal solution and vitamin mix (Pfennig and Lippert 1966) and 1 ml l−1 of alkaline Se/W solution (Plugge 2005). Elemental sulfur (Fluka) was autoclaved as a thick water paste at 110°C for 40 min in closed bottles and added in excess of approximately 3 g l−1. Other electron acceptors used were KNO3 and Na2S2O3 (20 mM each), KNO2, Na2SO3, sodium selenate and selenite, sodium arsenate, DMSO (5 mM each), sodium fumarate (20 mM), and freshly prepared amorphous ferrihydrite (20 mM). The latter was prepared by mixing 1:1 0.5 M solution of FeCl3 and 1 M NaOH with subsequent washing of the precipitate with distilled water until neutral pH reaction. In all cases, except for the latter, 1 mM HS− was added to the medium as a reductant. Growth at micro-oxic conditions was tested with an oxygen concentration in the gas phase of 1%. Routine cultivation was performed either in 15 ml Hungate tubes with 10 ml medium, or in 50 ml serum bottles with 40 ml medium with argon in the gas phase. The pH dependence was examined at Na+ content of 0.6 M, using the following filter-sterilized mineral medium: for pH 6–8, 0.1 M HEPES and NaCl/NaHCO3; for pH 8.5–11, a mixture of sodium bicarbonate/sodium carbonate containing 0.1 M NaCl. To study the influence of salt concentration on growth, sodium carbonate media with pH 10, containing 0.1 and 3.5 M of total Na+ were mixed in different proportions.
Analytical procedures
Free sulfide and the sulfane moiety of polysulfide were measured colorimetrically (Trüper and Schlegel 1964) after precipitation in 10% (w/v) Zn acetate. The internal sulfur of the polysulfide was separated by acidification of the sample to pH <3 with concentrated HCl, then precipitated by centrifugation, washed with distilled water, dried, extracted from the cell pellet with acetone overnight and assayed by a cyanolytic procedure (Sörbo 1957). Cell protein was determined according to Lowry et al (1951) after removal of sulfide/polysulfide and washing the cell pellet several times with 0.6 M NaCl. Arsenite was detected by anionic chromatography after neutralization of the supernatant using Biotronic chromatograph, anion-exchange column BT11AN, and 1 mM Na2CO3/1.2 mM NaHCO3 as eluent with a flow rate of 1.5 ml min−1. Acetate was analyzed in filtrated supernatant after acidification to pH 4 by anionic chromatography (Biotronic IC-1000, Germany; column BT III OS; conductivity detection; 1 mM HCl as eluent, 0.8 ml min−1). Phase-contrast microphotographs were obtained with a Zeiss Axioplan Imaging 2 microscope (Göttingen, Germany). For electron microscopy, the cells were negatively contrasted with 1% (w/v) neutralized phosphotungstate. Polar lipids were extracted from 50 mg of freeze dried cells with acidic methanol and the fatty acid methyl esters were analyzed by GC–MS according to Zhilina et al. (1997).
Genetic and phylogenetic analysis
The isolation of the DNA and determination of the G + C content of the DNA was performed according to Marmur (1961) and Marmur and Doty (1962), respectively. For molecular analysis, the DNA was extracted from the cells using alkaline SDS lysis at 60°C and purified with the Wizard Preps Kit (Promega, USA). The nearly complete 16S rRNA gene was obtained using general bacterial PCR primers 11f and 1492r (Lane 1991). The sequences were aligned with sequences from the GenBank using CLUSTAL W and the phylogenetic tree was reconstructed using neighbor-joining algorithm in the TREECONW program package (van de Peer and de Wachter 1994).
Results and discussion
Enrichment and isolation of pure cultures
Two positive anaerobic enrichment cultures with sulfur as electron acceptor were obtained from soda lake sediments at pH 10 and salinity 0.6 M total Na+: with acetate (lake Tanatar-5) and with propionate (Cock Soda Lake) as electron donor. Both cultures actively dissolved sulfur in the form of polysulfide and were dominated by highly motile spirilla. Several rounds of serial dilutions resulted in the isolation of two pure cultures designated strains AHT11 (acetate) and AHT19 (propionate). The purity of the isolates was verified by homogenous morphology and DGGE analysis (a single band with pure sequence).
Cell morphology and identification
The isolates were spiral shaped highly motile with bipolar tubular flagella (Fig. 1a–c). Colony formation was not observed either inside agar or on the surface of agar plates incubated at anoxic conditions. Liquid cultures at pH 9–10.5 developed in a peculiar structured way: active dissolution of insoluble elemental sulfur (which was at a large excess) occurred right above the flask bottom with a formation of an intense yellow layer of soluble polysulfide, while the upper part of the culture remained colorless or greenish (Supplementary Figure). This can be explained by the following: the bacteria utilized soluble polysulfide as the actual electron acceptor and produced sulfide which, in turn, immediately attacked the insoluble sulfur on the bottom to form polysulfide. Polysulfide molecules diffusing off the reaction site were reduced completely to sulfide which, in turn, rapidly reacted with sulfur. The greenish color is due to a presence of trisulfide (−S–S–S−) formed by an excess of HS−, while the yellow color indicates a presence of longer polysulfides at excess of elemental sulfur.
Fig. 1.
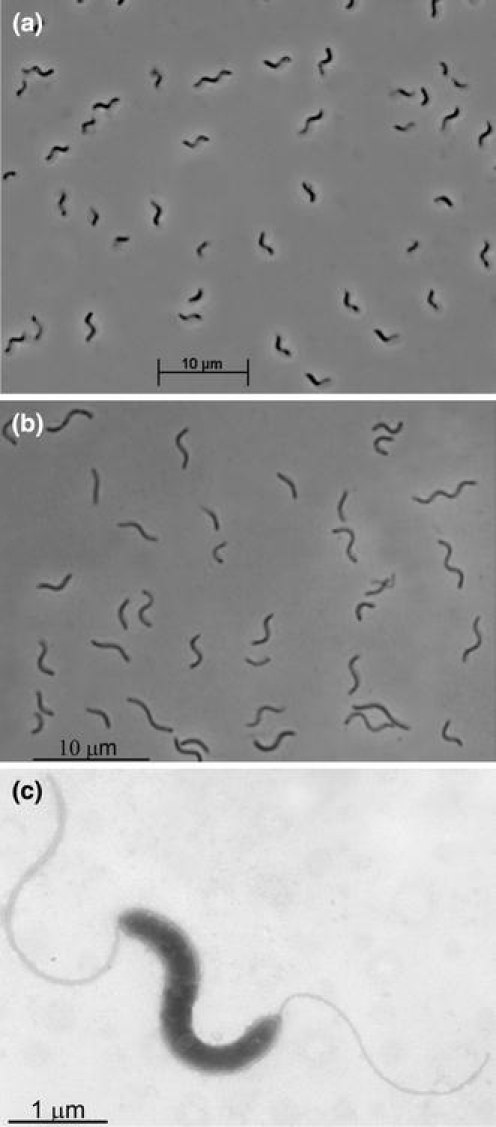
Morphology of sulfur-reducing isolates. a and b phase-contrast microphotograph of cells of AHT11 and AHT19 grown with acetate and sulfur; c electron microphotograph of a positively stained cell of strain AHT11
Phylogenetic analysis (Fig. 2) demonstrated that the isolates are closely related to each other (97% sequence similarity) and belong to the class Chrysiogenetes, a deep bacterial lineage which, at the moment, contains only two valid taxa, Chrysiogenes arsenatis (Macy et al. 1996) and Desulfurispirillum alkaliphilum (Sorokin et al. 2007). The latter is the closest phylogenetic relative of AHT11 and AHT19 (93% sequence similarity) together with two unclassified anaerobic respirers, i.e., the selenate/arsenate-reducing strain S5 from freshwater sediments (Narasingarao and Haggblom 2007) and an iron-reducing isolate from deep subsurface alkaline strata in China (Zhang et al. 2005).
Fig. 2.

Phylogenetic position of sulfur-reducing isolates based on 16S rRNA gene sequence analysis. Tree topography and evolutionary distances are obtained by the neighbor-joining method with Jukes and Cantor distances. The scale bar represents 10 nucleotide changes per 100 nucleotides
Comparison of cellular fatty acids in polar lipids demonstrated a general similarity of the profiles in novel isolates and Desulfurispirillum with a domination of C18:1 and C16:0. On the other hand, there was a peculiar difference in the distribution of two isomers of C18:1 in AHT11 and AHT19 (Supplementary Table). Affiliation of the sulfur reducers from soda lakes with Desulfurispirillum alkaliphilum, an acetate-utilizing alkaliphilic sulfur reducer, suggests that such a phenotype is conserved within this phylum. It is also clear that the whole phylum is specialized in ecologically important anaerobic transformations, such as dissimilatory sulfur, nitrate, iron, selenate and arsenate respiration. Direct detection of the members of the Chrysiogenetes in hypersaline soda lakes using a functional phylogenetic marker for arsenate reductase (Hollibaugh et al. 2006; Kulp et al. 2006) indicates that it might be present in significant numbers in alkaline habitats. It is also interesting to note, that the key enzymes responsible for the dissimilatory reduction of arsenate (arr) and polysulfide (psr) are related within the molybdopterin-containing class of enzymes and, therefore, these two modes of anaerobic respiration might have originated from a common ancestor (Duval et al. 2008).
Growth characteristics
Both isolates were obligate anaerobes unable to ferment sugars and pyruvate. They utilized a limited number of simple organic electron donors and were unable to grow autotrophically, although strain AHT11 could use H2 and formate as electron donor in presence of acetate as carbon source (Table 1). During growth with sulfur and acetate, polysulfide was gradually accumulating at extremely high concentrations parallel to sulfur dissolution. As mentioned above, near the solid sulfur surface an intense yellow layer was forming which contained a polysulfide with an average formula S4 2−. After mixing with the bulk culture, the color turned greenish due to rapid formation of a lower polysulfide with an average formula S3 2− due to a presence of excess of free HS−. Further fate of polysulfide depended on initial sulfur concentration. When limited, polysulfide started to disappear after complete sulfur dissolution and sulfide was the final product. Acetate consumption was parallel to the biomass and total reduced sulfur formation (Fig. 3). The amount of sulfane formed (70 mM) after complete utilization of 20 mM acetate is close to theoretical maximum (20 mM acetate × 8 ē = 160 mM ē; 70 mM sulfane × 2 ē = 140 mM ē). Maximal specific growth rate with acetate and sulfur at 0.6 M Na+ and pH 10 was 0.11 and 0.07 h−1 in strains AHT11 and AHT19, respectively.
Table 1.
Phenotypic comparison of soda lake isolates with related bacteria from the class Chrysiogenetes
| Characteristic | AHT11 | AHT19 | Desulfurispirillum alkaliphilum | Chrysiogenes arsenatis |
|---|---|---|---|---|
| Cell morphology | Spirillum with bipolar flagellum | Spirillum with bipolar flagellum | Spirillum with bipolar flagellum | Vibrio, single polar flagellum |
| Autotrophic growth | − | − | − | − |
| Electron donors | ||||
| H2 | + | − | + | − |
| Formate | + | − | − | − |
| Acetate, pyruvate, lactate malate, succinate, fumarate | + | + | + | + |
| Propionate | + | + | + | − |
| Citrate | − | − | + | − |
| Glycerol | + | − | − | − |
| Electron acceptors | ||||
| O2 | − | − | − | − |
| Sulfur (polysulfide) | + | + | + | − |
| NO3 −, NO2 − (to NH3) | − | − | + | + |
| N2O | − | − | − | − |
| Fumarate | − | − | + | − |
| AsO4 2− | + | + | − | + |
| DMSO, SeO4 2−, Fe3+, S2O3 2− | − | − | − | − |
| Oxidation of HS− with nitrate | − | − | + | n.d. |
| pH range (optimum) | 8.5–10.9 (10.2) | 8.2–10.5 (9.8) | 8.0–10.2 (9.0) | Neutrophilic |
| Salt range (M Na+) | 0.2–3.0 | 0.2–3.0 | 0.1–2.5 | Na-independent |
| Habitat | Soda lakes | Soda lakes | Bioreactor | Gold mine |
n.d. not determined
Fig. 3.
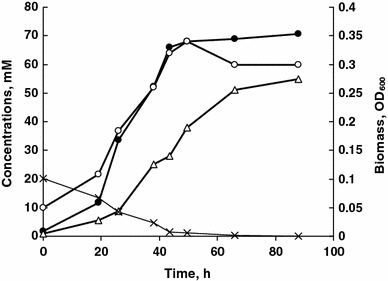
Anaerobic growth of strain AHT11 at pH 10 and 0.6 M Na+ with acetate and sulfur. Open circles biomass, closed circles total sulfane of sulfide/polysulfide, open triangles zero-valent sulfur dissolved in polysulfide, crosses acetate
Apart from sulfur, only arsenate could serve as alternative electron acceptor for both isolates at concentrations up to 20 mM. However, the growth and arsenate reduction were much poorer than with sulfur being limited to 5 mM arsenate consumption with stoichiometric formation of arsenite. Growth inhibition by the latter might be responsible for this effect.
Influence of pH and sodium on the growth and activity of the sulfur reducers
With acetate and sulfur both cultures showed obligate alkaliphily, starting to grow only at a pH above 8.2. On its pH optimum and highest pH limit, however, strain AHT11 was more alkaliphilic than strain AHT19 (Fig. 4a). Sulfidogenic activity of washed cells had very similar strictly alkaliphilic pH profiles similar to the profiles of growing cells except that in strain AHT19 washed cells tolerated much higher pH than a growing culture (Fig. 4b). It is necessary to mention that when the initial pH was above 9 a substantial drop was observed both during growth and during sulfur reduction by washed cells despite a high buffering capacity of the sodium carbonate system. Therefore, the presented profiles are reflecting the final pH values.
Fig. 4.
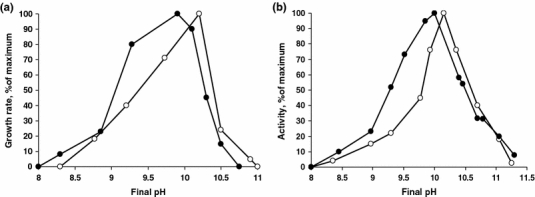
Influence of pH at 0.6 M Na+ on anaerobic growth (a) and sulfidogenic activity of washed cells (b) of strains AHT11 (open circles) and AHT19 (closed circles) with acetate as electron donor and sulfur as electron acceptor. 100% activity = 0.93 and 0.55 μmol HS− mg protein−1 min−1 in AHT11 and AHT19, respectively
When grown at pH 10 in sodium carbonate-based medium, both strains demanded at least 0.2 M total Na+ and grew up to 3.0 M with an optimum at 0.4–0.6 M, which characterizes them as moderately salt-tolerant organisms (Fig. 5a). Washed cells were active at sodium concentrations as low as 0.1 M, (but lysed rapidly), and up to 3–3.5 M (Fig. 5b).
Fig. 5.
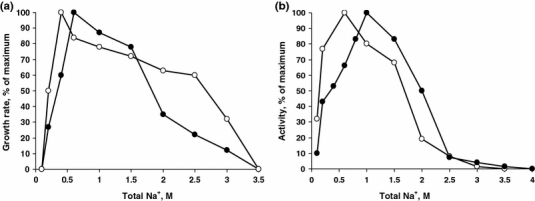
Influence of sodium carbonate at pH 10 on anaerobic growth (a) and sulfidogenic activity of washed cells (b) of strains AHT11 (open circles) and AHT19 (closed circles) with acetate as electron donor and sulfur as electron acceptor
Overall, strains AHT11 and AHT19 isolated from sediments of Siberian soda lakes represent the first example of highly specialized haloalkaliphilic anaerobes, whose most important environmental function can be defined as the acetate and propionate sink in anaerobic sediments. The isolates are obligately alkaliphilic and can function within a very broad salinity range. Their sulfur metabolism needs further investigation, since it must be significantly different from well studied sulfur-reducing enzyme system of neutrophiles. From its phylogenetic relatives, such as Deslfurispirillum alkaliphilum and Chrysiogenes arsenatis, the novel isolates can be distinguished by a number of important phenotypic properties (Table 1). Taken together with distinct phylogeny, the novel sulfur-reducing isolates AHT11 and AHT19 are proposed to be assigned into a new genus and species Desulfurispira natronophila within the family Chrysiogenaceae.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was supported by RFBR (10-04-00152). We are grateful to E. Detkova for the DNA analysis and to G. Osipov for the cellular fatty acid analysis.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Appendix 1: Description of Desulfurispira gen. nov.
Desulfurispira (De.sul.fu.ri.spi’ra. L. pref. de-, from; L.n. sulfur, sulfur; L. fem. n. spira, a coil; N. L. fem. n. Desulfurispira a sulfur-reducing spirillum).
Gram-negative motile spirilla. Obligately anaerobic with respiratory metabolism. Use short-chain fatty acids as electron donors. Do not grow autotrophically. Obligately alkaliphilic and moderately salt-tolerant. Belong to the class Chrysiogenetes. Habitats, soda lakes. The type species is Desulfurispira natronophila.
Appendix 2: Description of Desulfurispiranatronophila sp. nov.
natronophila [na.tro.no.phi’la. N.L. n. natron (arbitrarily derived from the Arabic n. natrun or natron), soda; N.L. pref. natrono-, pertaining to soda; N.L. fem. adj. phila (from Gr. fem. adj. philê), friend, loving; N.L. fem. adj. natronophila soda-loving].
Cells are spiral shaped, 0.3–0.4 × 2–5 μm, motile by single bipolar flagella. Gram-negative. Strictly anaerobic, using elemental sulfur, polysulfide and arsenate as electron acceptors. The final products are sulfide and arsenite. Obligatory heterotrophic. Utilize short-chain fatty acids, including acetate, propionate, lactate, pyruvate and fumarate as electron donor and carbon source. H2 and formate can be used as electron donor by some strains. Obligately alkaliphilic with a pH range for growth between 8.2 and 10.9 and an optimum at pH 9.8–10.2 and moderately salt-tolerant with a range from 0.2 to 3.0 M Na+ (optimum at 0.4–0.6 M). Mesophilic, with a maximum temperature for growth at 42°C and an optimum at 30°C (AHT11T). The predominant fatty acids in the polar membrane lipids include 18:1 and 16:0. The G + C content of the genomic DNA is 49.2 for strain AHT11T and 51.8 for strain AHT19 mol% (Tm). The type strain is AHT11T (DSM22071T = UNIQEM U758T). Isolated from sediments of soda lakes in south-western Siberia. The GenBank 16S rRNA gene sequence accession numbers are GQ922842 (strain AHT11T) and GQ922843 (AHT19).
Footnotes
References
- Duval S, Ducluzeau A-L, Nitschke W, Schoepp-Cothenet B. Enzyme phylogenies as markers for the oxidation state of the environment: the case of respiratory arsenate reductase and related enzymes. BMC Evol Biol. 2008;8:206. doi: 10.1186/1471-2148-8-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foti M, Sorokin DY, Lomans B, Mussman M, Zakharova EE, Pimenov NV, Kuenen JG, Muyzer G. Diversity, activity and abundance of sulfate-reducing bacteria in saline and hypersaline soda lakes. Appl Environ Microbiol. 2007;73:2093–2100. doi: 10.1128/AEM.02622-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlenko VM, Namsaraev BB, Kulyrova AV, Zavarzina DG, Zhilina TN. Activity of sulfate-reducing bacteria in the sediments of the soda lakes in south-east Transbaikal area. Microbiology (Moscow, English Translation) 1999;68:580–586. [Google Scholar]
- Hedderich R, Klimmek O, Kroëger A, Dirmeier R, Keller M, Stetter KO. Anaerobic respiration with elemental sulfur and with disulfides. FEMS Microbiol Rev. 1999;22:353–381. doi: 10.1111/j.1574-6976.1998.tb00376.x. [DOI] [Google Scholar]
- Hollibaugh JT, Budinoff C, Hollibaugh RA, Ransom B, Bano N. Sulfide oxidation coupled to arsenate reduction by a diverse microbial community in a soda lake. Appl Environ Microbiol. 2006;72:2043–2049. doi: 10.1128/AEM.72.3.2043-2049.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen AJH, Lens P, Stams AJM, Plugge CM, Sorokin DY, Muyzer G, Dijkman H, van Zessen E, Luimes P, Buisman CJN. Application of bacteria involved in the biological sulfur cycle for paper mill effluent purification. Sci Total Environ. 2009;407:1333–1343. doi: 10.1016/j.scitotenv.2008.09.054. [DOI] [PubMed] [Google Scholar]
- Kulp TR, Hoeft SE, Miller LG, Saltikov C, Murphy JN, Han S, Lanoil B, Oremland RS. Dissimilatory arsenate and sulfate reduction in sediments of two hypersaline, arsenic-rich soda lakes: Mono and Searles Lakes, California. Appl Environ Microbiol. 2006;72:6514–6526. doi: 10.1128/AEM.01066-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulp TR, Han S, Saltikov CV, Lanoil BD, Zargar K, Oremland RS. Effects of imposed salinity gradients on dissimilatory arsenate reduction, sulfate reduction, and other microbial processes in sediments from two California soda lakes. Appl Environ Microbiol. 2007;73:5130–5137. doi: 10.1128/AEM.00771-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane DJ. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, UK: John Wiley and Sons; 1991. pp. 115–177. [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Macy JM, Nunan K, Hagen K, Dixon DR, Harbour PJ, Cahill M, Sly LI. Chrysiogenes arsenatis gen. nov., sp. nov., a new arsenate-respiring bacterium isolated from gold mine wastewater. Int J Syst Bacteriol. 1996;46:1153–1157. doi: 10.1099/00207713-46-4-1153. [DOI] [PubMed] [Google Scholar]
- Marmur J. A procedure for isolation of DNA from microorganisms. J Mol Biol. 1961;3:208–214. doi: 10.1016/S0022-2836(61)80047-8. [DOI] [Google Scholar]
- Marmur J, Doty P. Determination of the base composition of deoxyribonucleic acid from microorganisms. J Mol Biol. 1962;5:109–118. doi: 10.1016/S0022-2836(62)80066-7. [DOI] [PubMed] [Google Scholar]
- Narasingarao P, Haggblom MM. Identification of anaerobic selenate-respiring bacteria from aquatic sediments. Appl Environ Microbiol. 2007;73:3519–3527. doi: 10.1128/AEM.02737-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfennig N, Lippert KD. Über das Vitamin B12-Bedürfnis phototropher Schwefelbakterien. Arch Mikrobiol. 1966;55:245–256. doi: 10.1007/BF00410246. [DOI] [Google Scholar]
- Plugge CM. Anoxic media design, preparation, and considerations. Methods Enzymol. 2005;397:3–16. doi: 10.1016/S0076-6879(05)97001-8. [DOI] [PubMed] [Google Scholar]
- Scholten JCM, Joye SB, Hollibaugh JT, Murrell JC. Molecular analysis of the sulfate reducing and archaeal community in a meromictic soda lake (Mono Lake, California) by targeting 16S rRNA, mcrA, apsA, and dsrAB genes. Microbial Ecol. 2005;50:29–39. doi: 10.1007/s00248-004-0085-8. [DOI] [PubMed] [Google Scholar]
- Sörbo B. A colorimetric determination of thiosulfate. Biochim Biophys Acta. 1957;23:412–416. doi: 10.1016/0006-3002(57)90346-3. [DOI] [PubMed] [Google Scholar]
- Sorokin DY, Gorlenko VM, Namsaraev BB, Namsaraev ZB, Lysenko AM, Eshinimaev BT, Khmelenina VN, Trotsenko YA, Kuenen JG. Prokaryotic communities of the north-eastern Mongolian soda lakes. Hydrobiologia. 2004;522:235–248. doi: 10.1023/B:HYDR.0000029989.73279.e4. [DOI] [Google Scholar]
- Sorokin DY, Foti M, Tindall BJ, Muyzer G. Desulfurispirillum alkaliphilum gen. nov. sp. nov., a novel obligately anaerobic sulfur- and dissimilatory nitrate-reducing bacterium from a full-scale sulfide-removing bioreactor. Extremophiles. 2007;11:363–370. doi: 10.1007/s00792-006-0048-8. [DOI] [PubMed] [Google Scholar]
- Sorokin DY, Tourova TP, Henstra AM, Stams AJM, Galinski EA, Muyzer G. Sulfidogenesis at extremely haloalkaline conditions by Desulfonatronospira thiodismutans gen. nov., sp. nov., and Desulfonatronospira delicata sp. nov.—a novel lineage of Deltaproteobacteria from hypersaline soda lakes. Microbiology (UK) 2008;154:1444–1453. doi: 10.1099/mic.0.2007/015628-0. [DOI] [PubMed] [Google Scholar]
- Sorokin DY, Tourova TP, Mussmann M, Muyzer G. Dethiobacter alkaliphilus gen. nov. sp. nov., and Desulfurivibrio alkaliphilus gen. nov. sp. nov.—two novel representatives of reductive sulfur cycle from soda lakes. Extremophiles. 2008;12:431–439. doi: 10.1007/s00792-008-0148-8. [DOI] [PubMed] [Google Scholar]
- Trüper HG, Schlegel HG. Sulfur metabolism in Thiorhodaceae. 1. Quantitative measurements on growing cells of Chromatium okenii. Antonie van Leeuwenhoek. 1964;30:225–238. doi: 10.1007/BF02046728. [DOI] [PubMed] [Google Scholar]
- van de Peer Y, de Wachter R. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci. 1994;10:569–570. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]
- Zhang G, Dong H, Xu X, Zhao D, Zhang C. Microbial diversity in ultra-high-pressure rocks and fluids from the Chinese Continental Scientific Drilling project in China. Appl Environ Microbiol. 2005;71:3213–3227. doi: 10.1128/AEM.71.6.3213-3227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhilina TN (2007) Chemotrophic anaerobes of microbial communities of soda lakes. In: Transactions of the Winogradsky Institute of Microbiology, V XIV, Nauka, Moscow, pp 206–216 (in Russian)
- Zhilina TN, Zavarzin GA, Rainey FA, Pikuta EN, Osipov GA, Kostrikina NA. Desulfonatronovibrio hydrogenovorans gen. nov., sp. nov., an alkaliphilic, sulfate-reducing bacterium. Int J Syst Bacteriol. 1997;47:144–149. doi: 10.1099/00207713-47-1-144. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


