Abstract
We show that natural products target proteins with a high number of protein−protein functional interactions (high biological network connectivity) and that these protein targets have higher network connectivity than disease genes. This feature may facilitate disruption of essential biological pathways, resulting in competitor death. This result also suggests that additional sources of small molecules will be required to discover drugs targeting the root causes of human disease in the future.
Naturally occurring small molecules (“natural products”) or their derivatives comprise a substantial fraction of the current pharmacopeia.1,2 We wished to understand the relationship of protein targets of natural products to heritable disease genes by comparing the biological network connectivities (functional connections between genes and gene products, e.g., protein/protein interactions) of these targets and genes. Understanding such relationships might facilitate future drug discovery, e.g., by determining whether natural products are intrinsically suited for targeting disease genes and whether their enrichment among current drugs reflects a historical focus or special properties intrinsic to these molecules. We seek to learn, in a data-driven way, whether or not the propensity of natural products for interaction with biological targets is an advantage for probe or drug discovery directed at the genes determined to be causal for human disease.
Discussions about the past and future roles of natural products in drug discovery continue while large-scale screening of pure synthetic compounds has come to dominate the drug-discovery landscape.1,3 In many cases, such discussions focus on the chemical structures of natural products and how they are similar to or different from synthetic compounds,(2) particularly in light of “rules” to guide drug-discovery efforts.4,5 Other perspectives have focused on the accessibility of these molecules, their relative difficulty of synthetic modification, and whether they are suited to prevailing screening methods within the pharmaceutical industry.1,3 Here, we investigate natural products from a different perspective, bioinformatic analysis of natural product targets.
Earlier studies show that disease genes (genes having polymorphisms within the human population that correlate with the frequency of disease; e.g., the CFTR gene and cystic fibrosis) have intermediate connectivities in biological networks.6,7 Both highly connected and less connected genes are less likely to be associated with disease phenotypes. The network connectivity of natural product targets is presumed to be high; however, we believe this is the first computational study of network connectivity of natural product targets compared with disease genes. We used the publicly available STRING database of protein/protein associations(8) as a foundation for analyzing network connectivities.9,10 In our analysis, we included only proteins with at least one connection and only connections with very high confidence scores assigned by STRING (≥0.9).
To identify natural product targets, we evaluated a commercial database of natural products and targets from GVKBio.(11) We standardized 5581 target names and species to human proteins, as either direct natural product targets or orthologous human target proteins, and mapped these targets to 946 human proteins with connections in STRING. For human disease genes, we combined 3655 genes contained in the OMIM Morbid Map(12) with 1580 genes from a genome-wide association study SNP database(13) and mapped these to 2681 human proteins with connections in STRING.
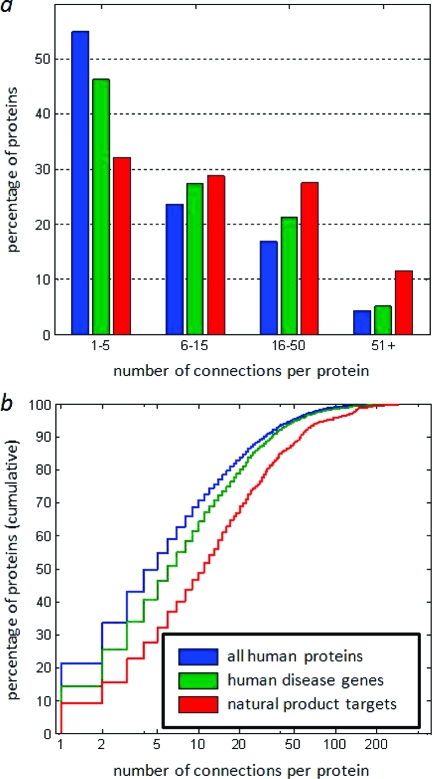
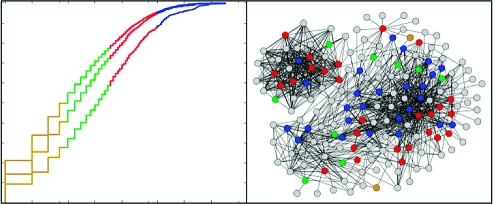
We assessed the distributions of protein connectivities among all STRING proteins with at least one connection, natural product targets mapped to STRING proteins, and heritable disease genes mapped to STRING proteins (Figure 1a). Our analysis confirms that STRING proteins mapped from disease genes display intermediate connectivity in biological networks. In contrast, STRING proteins mapped from natural product targets are enriched for more highly connected proteins compared to both STRING proteins mapped from disease genes and all STRING proteins. We used a Kolmogorov−Smirnov goodness-of-fit test(14) between cumulative connectivity distributions (Figure 1b) to assess the statistical significance of this finding. Differences in distributions of network connectivities for natural product targets vs disease genes (p = 2.6 × 10−15), natural product targets vs all STRING proteins (p = 1.9 × 10−39), and disease genes vs all STRING proteins (p = 2.8 × 10−15) were all statistically significant.
Figure 1.
(a) Connectivity summary of different protein groups: all human proteins in STRING database (blue: n = 8799; median = 5; mean = 11.7), disease-associated proteins (green: n = 2681; median = 6; mean = 14.0), natural product targets (red: n = 946; median = 11; mean = 22.5). (b) Cumulative connectivity distributions.
The STRING database contains potential targets from multiple species and connects proteins using multiple lines of evidence, including experimental evidence, pathway database information, and connections from literature text mining. We performed several control comparisons to examine the impact of these factors on our findings. First, when clusters of orthologous groups (COGs) of proteins were used as network nodes, these distributions and their relative relationships were essentially unchanged (see Supplementary Figure S1). Second, when we restricted STRING connections to those obtained from experimental evidence, we found that the relative connectivities of the three groups were unchanged and each remained significantly different from the others (see Supplementary Figure S2). Third, when we restricted STRING connections to those obtained by mining pathway databases, we observe much more similar connectivities between all proteins and disease-associated proteins, while natural product targets remain more connected than both (see Supplementary Figure S3).
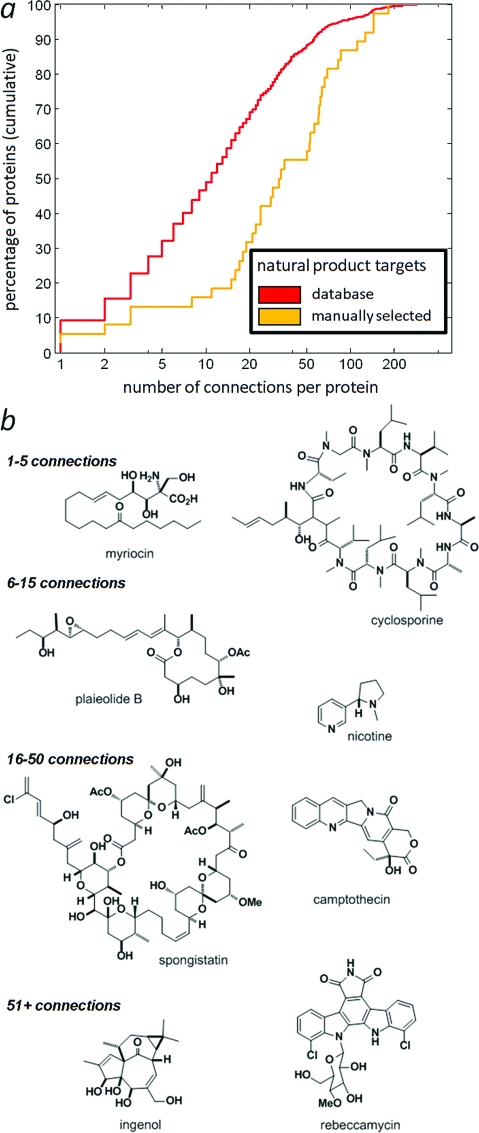
The GVKBio database contains some protein targets that are only weakly or nonselectively inhibited. Therefore, we undertook an annotation of the primary literature by searching for and capturing experimentally confirmed natural product/protein interactions, with a particular focus on highly potent and selective natural products. 76 natural products and their protein targets were captured from the primary literature (see Supporting Information). Protein names were standardized to human genes/proteins or mapped to human orthologues. We analyzed the distribution of protein connectivities of these targets using the STRING database as before and found that these protein targets exhibit even higher connectivities than the larger GVKBio set (Figure 2a), further supporting the notion that natural products target highly connected biological networks to disrupt cellular functions in competing organisms. The natural products that target these highly connected networks display a wide variety of structural features (Figure 2b), suggesting that it is unlikely that there is a single structural feature in natural products that specifically targets highly connected biological nodes. As an alternative method to approach the issue of target “quality” among interactions in the GVKBio database, we analyzed the subset of GVKBio small molecule/protein activities with reported effective concentrations of half-maximal effect (EC50) in the nanomolar range or below (<10−7.5 M). This subset comprises 10.4% of the full GVKBio data set, corresponding to high-potency interactions, and their network connectivities were essentially the same as using the larger GVKBio data set (see Supplementary Figure S4), suggesting that the observation that natural product targets are more highly connected is not skewed by targets that are only weakly or nonselectively inhibited.
Figure 2.
(a) Comparison of cumulative connectivity distributions between targets of 76 manually selected natural products (gold: n = 38; median = 32.5; mean = 48.5) and the database of natural products (red; same as Figure 1b). (b) Representative natural products obtained from literature review, grouped according to connectivity associated with the target.
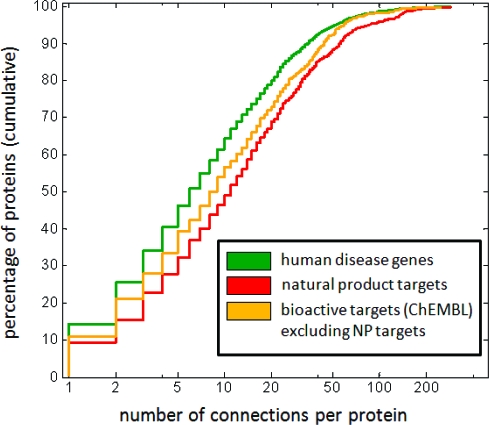
Our basic observation that natural product targets are more connected than disease genes compares a collection of targets of small molecules (natural products) with a collection of proteins obtained by mapping disease-implicated genes onto the proteome. Therefore, we sought to examine whether other small-molecule targets exhibit similarly high connectivities. To test this, we examined protein targets of “bioactive drug-like small molecules” in ChEMBL.(15) In this analysis, we identified the unique human targets in ChEMBL and removed those targets already in our list of natural product targets from GVKBio, giving a set of proteins targeted by only non-natural product small molecules. Consistent with the idea that proteins that are targets of natural products are more highly connected than other proteins, these ChEMBL targets exhibit intermediate connectivity between targets of natural products and proteins encoded by disease genes (Figure 3).
Figure 3.
Comparison of cumulative connectivity distributions between other small-molecule targets from ChEMBL(15) (gold: n = 729; median = 8; mean = 17.4), natural product targets (red; same as Figure 1b), and human disease genes (green; same as Figure 1b).
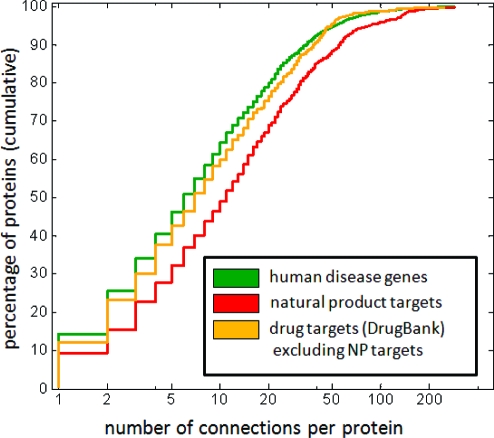
Finally, we examined the targets of approved drugs listed in DrugBank(16) to determine whether their connectivity distribution is more similar to that of targets of natural products than to that of human disease genes. We examined the subset of approved drug targets that are not also natural product targets in GVKBio. We observe that approved drug targets that are not also natural product targets exhibit a connectivity distribution much closer to the case for human disease genes than natural product targets, which remain the most highly connected targets (Figure 4). These results suggest that synthetic compounds that target human proteins are accessing targets with a lower degree of connectivity than natural products. In further support of the idea that disease genes, which encode conceptually and in practice attractive drug targets, exhibit intermediate or low connectivity, we note that G-protein coupled receptors, estimated to be more than 25% of all drug targets,(17) represent one of the least connected subgroups in STRING, with a connectivity distribution much lower than that of STRING as a whole (see Supplementary Figure S5).
Figure 4.
Comparison of cumulative connectivity distributions between approved drug targets from DrugBank(16) (gold: n = 731; median = 7; mean = 14.9), natural product targets (red; same as Figure 1b), and human disease genes (green; same as Figure 1b).
Overall, our results indicate that targets of natural products are highly connected, much more so than genes implicated in human disease, which exhibit intermediate connectivity, and more so even than other groups of small-molecule targets. This finding may indicate that natural products tend to target proteins more essential to an organism than are disease genes. This result is logical, as many natural products function as basic defense mechanisms against invaders in the absence of tissue specialization or an advanced immune response. This type of nonspecific defense often results in the death of the invading organism to ensure the producing organism’s survival. Therefore, it is not surprising to find that natural products would target more highly connected proteins, interrupting essential protein activities of the invader. Here, we present computational evidence that this “intuition” is correct. Our results also imply that natural products, as a group, may not display enough versatility to be suitable for treatment of all heritable human diseases. If this is the case, additional sources of small molecules will be required to generate treatments that target the root causes of disease.
Acknowledgments
This work was principally supported by the NIGMS-funded Center of Excellence in Chemical Methodology and Library Development (CMLD) at the Broad Institute (P50-GM069721). V.D. and P.A.C. are supported in part by an NIH grant aimed at small-molecule target identification (RL1-HG004671). S.LS. is an Investigator at the Howard Hughes Medical Institute.
Supporting Information Available
Supplementary analysis, including several figures, detailed statistical methods and tables, and compound information. This material is available free of charge via the Internet at http://pubs.acs.org.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Harvey A. L. Drug Discovery Today 2008, 13, 894–901. [DOI] [PubMed] [Google Scholar]
- Ganesan A. Curr. Opin. Chem. Biol. 2008, 12, 306–17. [DOI] [PubMed] [Google Scholar]
- Li J. W.; Vederas J. C. Science 2009, 325, 161–5. [DOI] [PubMed] [Google Scholar]
- Lipinski C. A.; Lombardo F.; Dominy B. W.; Feeney P. J. Adv. Drug Delivery Rev. 1997, 23, 3–25. [DOI] [PubMed] [Google Scholar]
- Lipinski C. A.; Hopkins A. Nature 2004, 432, 855–861. [DOI] [PubMed] [Google Scholar]
- Feldman I.; Rzhetsky A.; Vitkup D. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 4323–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh K. I.; Cusick M. E.; Valle D.; Childs B.; Vidal M.; Barabasi A. L. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 8685–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STRING v8.2 (http://string.embl.de).
- Jensen L. J.; Kuhn M.; Stark M.; Chaffron S.; Creevey C.; Muller J.; Doerks T.; Julien P.; Roth A.; Simonovic M.; Bork P.; von Mering C. Nucleic Acids Res. 2009, 37, D412–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering C.; Jensen L. J.; Snel B.; Hooper S. D.; Krupp M.; Foglierini M.; Jouffre N.; Huynen M. A.; Bork P. Nucleic Acids Res. 2005, 33, D433–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GVKBio (http://www.gvkbio.com).
- OMIM Morbid Map (ftp://ftp.ncbi.nih.gov/repository/OMIM/morbidmap).
- A Catalog of Published Genome-Wide Association Studies (http://www.genome.gov/26525384).
- Sheshkin D. J.Handbook of Parametric and Nonparametric Statistical Procedures, 2nd ed.; Chapman & Hall/CRC: 2004. [Google Scholar]
- ChEMBL (http://www.ebi.ac.uk/chembldb/).
- DrugBank (http://www.drugbank.ca/).
- Overington J. P.; Al-Lazikani B.; Hopkins A. L. Nat. Rev. Drug Discovery 2006, 5, 993–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.