Abstract
A low-molecular-weight human immunodeficiency virus type 1 (HIV-1) inhibitor, PF-68742 (molecular weight, 573), has been identified in a high-throughput screen for compounds that block HIV-1 envelope glycoprotein (Env)-mediated fusion. The compound is shown to be potent against R5 and X4 isolates in both cell-cell fusion and antiviral assays (50% effective concentrations of ∼0.1 to 1 μM). Postfusion and HIV-1 pseudotyping control experiments confirm that PF-68742 is an entry inhibitor with Env as the specific target for antiviral action. PF-68742 was not able to block binding of monomeric gp120 to soluble CD4 or the binding of gp120:CD4 complexes to cell-associated CCR5, thus distinguishing PF-68742 from described gp120 antagonists and coreceptor binders. Escape variants of HIV-1NL4-3 were selected, and all resistant viruses were found to contain a common G514R (HxB2 numbering) mutation in Env, located proximal to the furin cleavage site in the fusion peptide of gp41. When introduced into wild-type NL4-3 gp41, G514R conferred resistance to PF-68742. Resistance via G514R is shown to be associated with enhancement of virion infectivity by PF-68742 that may result from altered properties of inhibitor-bound Env, rather than from a loss of compound binding. Wild-type viruses and those with substitutions in the disulfide loop (DSL) region of gp41 were also examined for PF-68742 sensitivity. Here, complete resistance to PF-68742 was found to occur through changes outside of position 514, including in the gp41 DSL region. The results highlight PF-68742 as a starting point for novel therapies against HIV-1 and provide new insights into models of Env-mediated fusion.
Around 33.2 million people are currently infected with human immunodeficiency virus type 1 (HIV-1) worldwide, and there were 2.1 million deaths as well as 2.5 million newly infected individuals in 2007 alone (62). Current HAART (for highly active antiretroviral therapy) regimes combine different classes of antiviral drugs to suppress viral replication, limit the breakthrough of drug-resistant virus, and maintain immune system function (48). However, as resistance to these agents becomes more widespread, there is a corresponding need for novel agents that are active against emerging resistant strains. Moreover, anti-HIV-1 agents with improved tolerability and convenience are highly desirable, since noncompliance is a key factor in regimen failure and the consequent development of resistant virus. Established classes of antiviral drugs include protease inhibitors (PI), nucleoside reverse transcriptase inhibitors (NRTI), and non-nucleoside inhibitors (NNRTI). More recently, this armamentarium has been complemented with integrase inhibitors, exemplified by raltegravir (59), and with entry inhibitors. Entry (or fusion) inhibitors are unique in that they prevent the virus from entering the target cell, instead of acting on subsequent steps of viral replication. The first fusion inhibitor to be made available to patients was enfuvirtide/T20 (41), an injectable peptide that targets the transmembrane glycoprotein of HIV-1, gp41. The more recently approved entry inhibitor maraviroc targets CCR5 coreceptors on the host cell and is the first small molecule entry inhibitor that can be taken orally (18).
The mechanism by which HIV-1 is able to enter host cells has been extensively studied (see references 20 and 26 for reviews). HIV-1 envelope glycoprotein (Env), in its native and functional state, is presented on the surface of virions and infected cells as a complex of three gp120 outer subunits noncovalently associated with three gp41 inner subunits that are anchored in the membrane. Each gp120 subunit contains the binding sites for the primary host cell receptor, CD4, and the coreceptor (i.e., CCR5 or CXCR4). The gp41 molecule consists of several distinct regions, including (from the N to the C terminus): the fusion peptide (FP), the N-terminal leucine zipper heptad repeat (HR1), a central disulfide loop-containing region (DSL), the C-terminal heptad repeat (HR2), the membrane-proximal external region (MPER), the transmembrane domain (TM), and a long cytoplasmic tail (CT). The fusion process is triggered by the binding of the gp120 subunit of Env to host cell CD4, resulting in a conformational change that enables a further interaction between the gp120:CD4 complex and a coreceptor, either CCR5 (R5) or CXCR4 (X4). Coreceptor binding appears to trigger the fusion process. This process also involves the extrusion from the Env trimer of the N-terminal FP of gp41, resulting in the formation of a prefusion intermediate. Here, the FP and TM are engaged in the host membrane and the viral envelope, respectively; a physical link (protein bridge) between the two lipid bilayers is thus generated. This metastable gp41 extended conformation exposes both the trimeric HR1 coiled-coil (35) and the HR2 (15, 37) regions to potential inhibitors of the fusion process over a period of many minutes (17). The HR1 and the HR2 regions then form a very stable six-helical bundle (6HB) structure that appears to bring the viral envelope and plasma membrane together, leading to lipid bilayer mixing and viral entry (26).
Such a complex multistep process is vulnerable to several types of inhibitor including CD4 blockers (14), gp120 binders (28, 58), coreceptor inhibitors (18), and gp41-targeted peptide inhibitors. The latter peptides typically bear sequences that correspond to either HR1 (4, 45) or HR2 (e.g., T20/enfuvirtide and T-1249) (19-21, 32, 35, 41). A wide range of low-molecular-weight antagonists have also been reported that target gp41, with much of the research devoted to 6HB disruptors (56). Accordingly, most described gp41 binding compounds are 6HB disruptors, such as low-molecular-weight pyrrole derivatives (33), the hydroxytyrosols from olive leaf extracts (43), the 4-benzamidobenzoic peptidomimetics (9), the thiazolidines (12), trifluoromethylnaphthyridine analogs (24), and an aminophenol series (63). These molecules have various levels of antiviral activity, usually in the high micromolar range. A more active chemical series with unspecified structures has been reported that can block exposure of 6HB epitopes in activated Env (23). Other independent research efforts have identified the betulinic acid derivatives RPR103611, and its stereoisomer IC9564, which target a postattachment, pre-entry step (38, 49). Viruses resistant to RPR103611 have implicated gp41 as a target, whereas resistance to IC9564 appears to emerge from changes to V3 of gp120 (29, 30, 38-40, 49). Low-molecular-weight inhibitors of fusion are widely regarded as desirable for therapeutic and/or prophylactic intervention of HIV-1 infection due to the reduced cost of production and superior pharmacological properties compared to peptides and peptidomimetics (22, 44, 66). We now report on a low-molecular-weight viral entry inhibitor, a benzene sulfonamide, PF-68742, which targets distinct sequences on gp41.
MATERIALS AND METHODS
Cell lines and virus strains.
HeLa P4 and CHO-tat10 (expressing JR-FL gp160) have been described previously (7). HEK Freestyle cells (Invitrogen) were cultured in Freestyle Expression medium (Invitrogen). All laboratory adapted HIV-1 strains and primary isolates were obtained from the AIDS Reagent Project, National Institute of Biological Standards and Control, Potters Bar, Hertfordshire, United Kingdom. CHO cells stably transfected with the His6-tagged gp120 expression plasmid pEE14.1 (BaL strain; Lonza Biologics) were treated as previously described (18). MIP34.10 cells were cultured in Dulbecco modified Eagle medium (DMEM), supplemented with 10% fetal calf serum (FCS), 100 U of penicillin/ml, 0.1 mg of streptomycin/ml, and 0.5 mg of Geneticin/ml. Pseudotyped virions were produced by transfection of HEK 293T cells. U87.CD4.CCR5 (6) or TZM-bl (64) cells were used as target reporter cells for infection and were obtained through the National Institutes of Health (NIH) AIDS Research and Reference Reagent Program. The T-cell leukemia cell line MT2 was maintained in RPMI 1640 medium and the human epithelioid β-galactosidase reporter cell line JC53BL (1) was maintained in DMEM lacking l-glutamine. All cells were maintained at 37°C and 5% CO2 in medium containing 10% (vol/vol) fetal calf serum, 2 mM l-glutamine, 1 U of penicillin/ml, and 0.1 mg of streptomycin/ml. The HIV-1 CXCR4-tropic laboratory-adapted strain NL4-3 was used to generate the PF-68742-resistant viruses and viruses resistant to PF-4087738, a close structural analog to PF-68742.
Plasmids (Env-only and molecular clones).
Env genes from NL4-3, LAI, AD8, TH93074, UG92001; BR92014, SE12808, ZA97012, IN98017, IN98022, and BR92017 were PCR amplified and then subcloned into the BstX1 site in expression plasmid pcDNA3.1/V5HisTOPO (Invitrogen). All constructs were sequenced (LARK) prior to transient transfection. Plasmid pSVIIIexE7pA− (27), a gift from J. Sodroski (Harvard), was used to express full-length gp160JR-FL, and pCAGGS (50) was used to express gp140JR-FL (CT-truncated, ΔCT) and SIVmac239 gp160ΔCT on pseudotyped virions by cotransfection with plasmid pNL4-3.Luc.R-E (11), obtained from the NIH ARRRP. A plasmid expressing vesicular stomatitis virus G protein (pVSV-G) was a gift from T. Friedmann (University of California at San Diego) and has been described previously (11, 47). Mutations in the vicinity of the DSL of gp41 were introduced into the pLAI-JRFL plasmid, a derivative of pLAI.2 (11, 47). Plasmid pLAI-JRFL was generated by subcloning of the ectodomain of JR-FL env (from residues 25 to 690, HxB2 numbering) in place of the corresponding LAI sequence in the pLAI.2 molecular clone. Sequences are available upon request.
CD4-gp120 binding assay.
The commercially available AlphaScreen (Perkin-Elmer) was used to measure binding of gp120 to soluble CD4. Purified, C-terminal His6-tagged BaL gp120 (0.1 μg/μl) was immobilized onto AlphaScreen nickel chelate acceptor beads (100 ng/μl) in assay buffer (25 mM HEPES [pH 7.4], 100 mM NaCl, 0.03% Tween 20, 0.3% bovine serum albumin). Soluble biotinylated CD4 (100 nM; AutogenBioClear) was immobilized onto AlphaScreen streptavidin-coated donor beads (100 ng/μl) in assay buffer. After incubation at room temperature for 30 min, equal volumes of coated acceptor beads and donor beads were added to a 0.5 volume of test compound, serially diluted with 5% dimethyl sulfoxide-phosphate-buffered saline (PBS). The plate was incubated in the dark for 2 h, and the chemiluminescence was measured by using a Fusion FT plate reader.
CD4:gp120 complex and coreceptor binding studies.
These experiments were performed essentially as previously described (18). Briefly, a time-resolved fluorescence immunoassay was used to determine soluble CD4:gp120 complex binding to CCR5 coreceptor on MIP34.10 cells (expressing high levels of CCR5) in a 96-well plate format. Ten thousand MIP34.10 cells were seeded in 96-well plates coated with poly-d-lysine and then incubated overnight at 37°C in 5% CO2. CD4:gp120 complexes were generated by incubating 36 nM soluble CD4 (Immunodiagnostics) with 60 nM purified His6-tagged BaL gp120 in assay buffer (DMEM-10% FCS) on ice for 15 min. After the MIP34.10 monolayer was rinsed, 20 μl of serially diluted test compounds was added to each well, followed by 40 μl of CD4:gp120 complex. Background wells containing only gp120 (20 μl) and assay buffer (40 μl) were included to control for nonspecific effects; positive control wells for coreceptor blocking included 20 μl of MIP-1 plus 40 μl of CD4:gp120 complex, and compound control wells included 20 μl of assay buffer plus 40 μl of CD4:gp120 complex. The plates were incubated for 1 h at 37°C with rocking, after which the wells were washed with assay buffer. A total of 100 μl of sheep anti-gp120 antibody specific for the C terminus (1:500 in assay buffer Aalto Bio [D7324]) was added to the wells, followed by incubation for 90 min at room temperature. The wells were washed with assay buffer and probed using 100 μl of Eu3+ labeled donkey anti-sheep IgG antibody (Sigma) for 15 min at room temperature with rocking. After washing with wash buffer (EG&G Wallac, catalog no. 1380-0865/R) and rinsing with PBS, the wells were developed by adding 200 μl of Enhancement solution (EG&G Wallac catalog no. 1244-104) and vortexing for 2 to 3 min. The plate was read in a Delfia 1234 fluorimeter after 15 to 60 min.
Cell fusion and cytotoxicity assays.
The assay is essentially as previously described (7). Briefly, 6 × 105 HeLa P4 (containing LTR- β-galactosidase) cells were coincubated with 6 × 105 CHO-tat 10 cells (for CCR5-gp160 fusion) in the presence of serially diluted test compounds. For CXCR4-gp160 fusion assays, 6 × 105 HEK Freestyle cells, transiently transfected with a NL4-3 gp160 pcDNA3.1 expression construct, were used instead of CHO-tat 10 cells. After overnight incubation, the cocultures were lysed and developed for β-galactosidase expression, as previously described (18). In the postattachment inhibition experiments, the compounds were added after coculture at the indicated times. Cytotoxicity assays were set up in parallel using HelaP4 cells and HEK Freestyle cells plated separately and developed using CellTitreGlo after 3 days.
HIV-1 virus entry inhibition assays. (i) HIV-1 (NL4-3 or BaL) against HeLa P4 cells.
HeLaP4 cells, maintained as described above, were detached from the culture flask by using cell dissociation fluid. Cells were resuspended in culture medium, and the cell count and viability were determined by trypan blue dye exclusion. Only cells >95% viable cells were used in the assay. HeLaP4 cells were infected with various multiplicities of infection (MOIs) of wild-type virus (NL4-3 or BaL) based on the 50% tissue culture infective dose of the virus stock. After 4 days of culture, the volume of virus that gave a β-galactosidase signal within the linear range of the β-galactosidase standard curve and having a window >10-fold that of the uninfected cell control was used as the assay input. These volumes of virus were then used to infect the cells in the antiviral assay (final cell density, 105 cells/ml). The infected cells were mixed thoroughly, and 90 μl was added per well of the assay plate. Similarly, 90 μl of uninfected cells at the same cell density (105 cells/ml) was added to the “negative” control wells. Assay plates were incubated for 5 days at 37°C in a CO2 incubator. Virus was quantified in cultures by using a FluorAce β-galactosidase reporter assay kit (Bio-Rad) according to the manufacturer's guidelines. Briefly, a 10-point standard curve ranging from 0 to ∼2,370 U of β-galactosidase (the exact concentration range being dependent on the β-galactosidase provided in the kit) was prepared in culture medium, and 100 μl was transferred to the wells of a 96-well, flat-bottom black plate with a clear base. Assay plates were developed in 250 mM Tris (pH 7.5)-1% Triton X-100-10% glycerol-500 μM MUG-10 mM β-mercaptoethanol, and the plates were incubated for 2 h at 37°C in a CO2 incubator. The reaction was stopped by the addition of 20 μl of 10× stop buffer (Bio-Rad), and the relative fluorescence unit value was determined within 4 h on a fluorimeter (excitation, 355 to 360 nm; emission, 460 nm).
(ii) Pseudotyped HIV-1, pNL4-3, and pLAI-JRFL molecular clones using U87.CD4.CCR5 or TZM-bl target cells.
Pseudotyped HIV-1 virions, competent for a single round of infection, were generated by cotransfection of HEK 293T cells, using the PEI transfection reagent (53), the luciferase reporter plasmid pNL4-3.Luc.R-E (11), and an Env complementation plasmid, pSVIIIexE7pA− (27) or pCAGGS (50). Replication-competent HIV-1 molecular clones (i.e., NL4-3, pLAI-JRFL, and their cognate gp41 mutants) were produced by transfection of 293T cells with the molecular clone plasmid DNA. Pseudotyped virus and pLAI-JRFL molecular clones were assayed for viral entry using U87.CD4.CCR5 cells (6) and TZM-bl cells (64), as target cells, respectively. Incremental concentrations of antibody or inhibitor were added (1:1) to HIV-1, and the mixture incubated for 1 h at 37°C prior to transferring (1:1) to the target cells. The assay was developed by using the luciferase assay system (Promega) after a 72-h incubation at 37°C, and the luminescence in relative light units was measured by using an Orion microplate luminometer (Berthold Detection Systems). The extent of viral entry was determined as a percentage reduction of viral infectivity measured against that of an inhibitor-free control. All experiments were performed in triplicate.
Selection and characterization for PF-68742 resistance mutations.
MT2 cells (7 × 106 cells) were infected with the laboratory-adapted strain NL4-3 at an MOI of 10−3 for 1 h at 37°C. Infected cells were subsequently cultured in 12-well plates with 2 × 105 cells/ml containing PF-68742 or PF-4087738 at 1 μM. Virus levels were monitored weekly by observing for the presence of cell syncytia. Whenever syncytia were absent from cultures, virus levels were quantified by infection of the JC53BL reporter cell line. The cultures were split on day 8 by transferring virus supernatant onto fresh cells, at the same cell density, with medium containing fresh compound. Compounds were added at final concentrations two times, four times, and equal to that of presplit concentrations, depending on whether evidence of viral replication was observed. This procedure was continued on a weekly basis. For all experiments with NL4-3, compound-free passages were set up in parallel to control for changes in sensitivity to compound PF-68742 or PF-4087738 upon prolonged culture of virus. For all compound-associated passages, when the virus no longer appeared to be inhibited by the highest compound concentrations feasible in culture, virus stocks were generated for further analysis. The resistant virus strains were characterized as follows. HIV-1 genomic RNA extraction from PF-68742- or PF-4087738-resistant viruses and respective controls were prepared by using a QIAamp viral RNA minikit (Qiagen). Subsequent cDNA synthesis was performed with genomic RNA as a template and a SuperScript III first-strand synthesis system (Invitrogen). A total of 16 pmol of HIV-1 env-specific primer (5′-GCCATCC)/μl was used to produce the 2.56-kb cDNA fragment of the entire env gene. For PCR amplification of cDNA fragments, each sample was mixed with 50 mM MgSO4, 10 mM deoxynucleoside triphosphates, 30 μM (each) HIV-1 env-specific primers (5′-GAGCAGAAGACAGTGGCA and 5′-GACCACTTGCCACCCATC, 10× high-fidelity PCR buffer (Invitrogen), and 2.5 U of Taq DNA high-fidelity polymerase (Invitrogen) in a final volume of 50 μl. The DNA was denatured for 2 min at 94°C and then amplified by 35 PCR cycles of 94°C for 30 s, 55°C for 30 s, and 68°C for 3 min. Sequencing of PCR products was performed by LARK cogenics (Hope End, Takeley, Essex, United Kingdom). Mutations were numbered according to the HxB2 reference sequence.
p24 and gp120 ELISAs.
Enzyme-linked immunosorbent assays (ELISAs) were performed as previously described (42). Briefly, for p24 ELISAs, microtiter wells were coated with 5 μg of sheep anti-p24 (Aalto)/ml and blocked by using 4% nonfat dry milk in PBS. Virions, lysed with 1% Empigen, were transferred to anti-p24-coated wells and incubated for 2 h at 37°C. p24 was probed using alkaline phosphatase-conjugated sheep anti-p24 (Aalto), and the assay was developed by using an AMPAK amplification kit (Argene), according to the manufacturer's directions. gp120 ELISAs were performed similarly except that microtiter wells were coated with Galanthus nivalis lectin (GNL; Sigma) and, after incubation with lysed virions, wells were probed using an anti-gp120 monoclonal antibody (MAb) cocktail (b12 [8], B4e8 [10], and 2G12 [60]; 2 μg/ml each), followed by detection using an horseradish peroxidase-conjugated anti-human Fc (Jackson) secondary reagent. The colorimetric signal was produced using TMB substrate (Pierce).
RESULTS
PF-68742 inhibits HIV-1 Env-mediated fusion in a coreceptor-independent fashion.
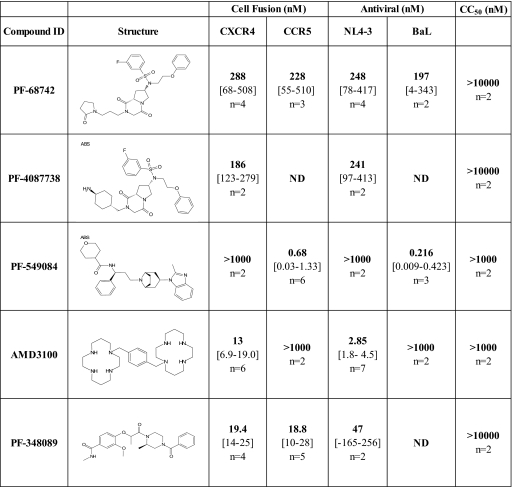
PF-68742 was identified from a high-throughput screen based upon inhibition of cell fusion mediated by the HIV-1JR-FL (R5) envelope (7). Subsequent dose-response experiments generated a 50% effective concentration (EC50) of 0.23 μM (Fig. 1). Active compounds in the cell fusion assay could have a number of different mechanisms of action, including antagonism of CCR5, CD4, or Env, and nonspecific interference with the β-galactosidase reporter activity and/or gene expression. To begin characterizing the mechanism of action for PF-68742, we compared inhibition curves obtained using PF-68742 to those obtained using other well-characterized inhibitors. For this purpose, we picked the specific CXCR4 antagonist AMD3100 (13), a specific CCR5 antagonist PF-549084, an analogue of maraviroc (18), and the gp120 antagonist PF-348089, an analogue of BMS-378806 (28). As expected, AMD3100 and PF-549084 were uniquely active in the CXCR4-gp160- and CCR5-gp160-mediated fusion assays, respectively, whereas the gp120 antagonist PF-348089 was active in both assays (Fig. 1 and 2A). Interestingly, PF-68742 was also active in both assays (Fig. 1 and 2A). Thus, PF-68742 can inhibit HIV-1 Env mediated fusion in a coreceptor independent fashion. PF-4087728, a close structural analog to PF-68742, showed properties similar to those of PF-68742 (Fig. 1).
FIG. 1.
Comparison of PF-68742 with other HIV-1 entry inhibitors in different in vitro assay systems. PF-4087738 is a close structural analog to PF-68742. PF-549084 is a CCR5 antagonist (analog of maraviroc), AMD3100 is a CXCR4 antagonist and PF-348089 is a HIV-1 gp120 antagonist (analog of BMS-378806). The numbers in boldface are the EC50s in the cell fusion assay and antiviral assays, which are described in Materials and Methods. The 50% cytotoxicity concentration (CC50) values represent the concentration of compound able to produce 50% cytotoxicity in HEK cells. The numbers in square brackets are 90% confidence limits for the mean, and “n” is number of independent assays performed (each assay done in replicate). ND, not determined.
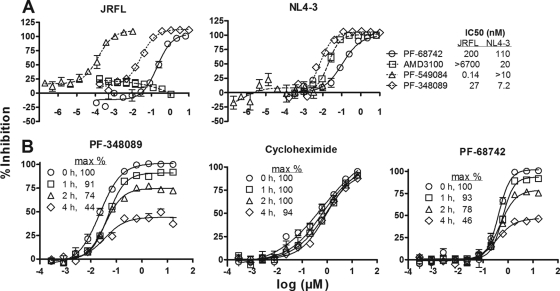
FIG. 2.
Env-mediated cell fusion inhibition by PF-68742. (A) Dose-response curves are shown for PF-68742 (circles) and three standard antiviral compounds with known mechanism of action. AMD3100 (squares) is a CXCR4 antagonist, PF-549084 (triangles) is a CCR5 antagonist, and PF-348089 (diamonds) is a gp120 antagonist. Both EnvJR-FL-CCR5-mediated fusion (left panel) and EnvNL4-3-CXCR4-mediated fusion (right panel) assays are shown. Each curve is derived from at least two independent experiments at n = 2 replicates. (B) To help determine whether PF-68742 is an entry inhibitor, we studied the effect of delayed addition of compound on the maximal (100%) effect in the dose-response curves using the fusion assay. The positive control, gp120 antagonist PF-348089 (left panel), the negative control, cycloheximide (inhibitor of protein biosynthesis; center panel), and PF-68742 (right panel) were added to an EnvNL4-3-CXCR4 fusion assay at time zero (circles), 1 h postfusion (squares), 2 h postfusion (triangles), and 4 h postfusion (diamonds). Each curve is derived from at least two independent experiments at n = 2 replicates.
PF-68742 is an HIV-1 entry inhibitor.
To further delimit the mechanism of action of PF-68742 using the cell fusion assay, control experiments were conducted using various inhibitor types and addition times. If PF-68742 is a genuine viral entry inhibitor, its maximal efficacy should be significantly reduced if added after cell fusion had occurred. In contrast, efficacy should remain unaffected if the compound inhibited a fusion-independent event. We assayed PF-68742 and a gp120 antagonist PF-348089 alongside a nonspecific protein biosynthesis inhibitor (cycloheximide) in the cell fusion assay and showed that delaying the addition of PF-68742 reduced maximum efficacy, as expected for a genuine entry inhibitor (Fig. 2B). Such behavior was also observed with the positive control PF-348089, as expected (Fig. 2B). A completely different inhibitory profile was observed when using the negative control cycloheximide (Fig. 2B). These experiments and other cytotoxicity assays (Fig. 1; data not shown) confirmed that PF-68742 has a specific and nontoxic mechanism of action.
The antiviral target for PF-68742 resides specifically within HIV-1 Env.
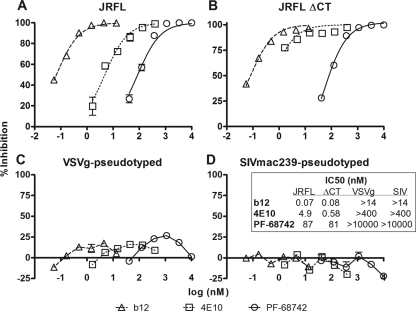
The specificity of PF-68742 was subsequently tested using pseudotyped HIV-1 in a single-round infectivity assay using U87.CD4.CCR5 target cells. HIV-1 pseudotyped with Env from the R5 primary isolate, JR-FL, was compared to that of a CT-truncated JR-FL variant (ΔCT; containing a stop codon at position 709), SIVmac239ΔCT, and VSV-G, each on a common HIV-1 backbone. PF-68742 clearly inhibited HIV-1JR-FL (EC50 = 87 nM), as did the control anti-HIV-1 MAbs b12 (against the CD4 binding site) (8) and 4E10 (against the MPER of gp41) (65) (Fig. 3A). In addition, PF-68742 showed similar potency against JR-FLΔCT, indicating that the CT of gp41 is not targeted (Fig. 3B). However, PF-68742 did not inhibit the infectivity of HIV-1 pseudotyped with either VSV-G or SIVmac239 Env. Thus, PF-68742 is either active or inactive depending upon the Env sequence (Fig. 3). Given that the same target cells were used for VSV-G and SIVmac239 pseudotyped viruses and no entry inhibition was noted, this strongly suggests that CD4, coreceptors, and accessory host factors are not acting as targets for PF-68742. Indeed, these data suggest that PF-68742 inhibits viral entry by specific targeting of HIV-1 Env.
FIG. 3.
Inhibition of HIV-1 entry by PF-68742 in the single round infectivity assay using pseudotyped viruses with different Envs. Single-cycle viral entry assays were performed with PF-68742 and two positive control MAbs, b12 and 4E10, against HIV-1 virions pseudotyped using full-length gp160JR-FL (A), gp145JR-FL ΔCT (B), VSV-G (C), and Env from SIVmac239 (D). Experiments were performed in triplicate.
PF-68742 does not block soluble CD4:gp120 complex formation or subsequent CCR5 engagement.
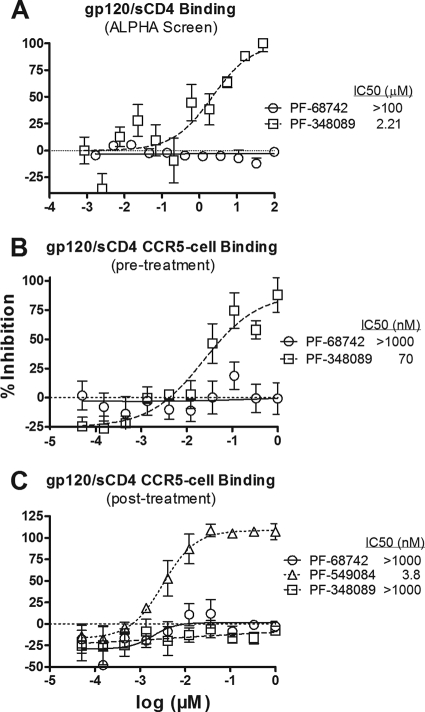
To investigate the effect of PF-68742 on the formation of a CD4:gp120 complex, or the subsequent binding of the CD4:gp120 complex to the coreceptor, we used the AlphaScreen proximity assay (34, 61). A purified recombinant His6-tagged BaL gp120 was bound to nickel chelate acceptor beads, and a biotinylated soluble (s)CD4 was captured on streptavidin donor beads. The gp120 and sCD4 beads were coincubated in the presence or absence of the gp120 antagonist PF-348089 or PF-68742 (Fig. 4A). The data show that the gp120 antagonist clearly blocked sCD4:gp120 complex formation at an IC50 of 1.9 μM, whereas PF-68742 was inactive (IC50 > 100 μM).
FIG. 4.
PF-68742 does not block CD4:gp120 complex formation. (A) AlphaScreen proximity assay. Biotinylated sCD4 bound to streptavidin-coated (donor) beads and His6-tagged gp120 bound to nickel chelate (acceptor) beads were incubated for 2 h in the presence of incremental concentrations of either PF-68742 (circles) or PF-348089 (squares). Chemiluminescence was measured by using a Fusion HP plate reader. (B) PF-68742 does not inhibit label-free sCD4 from complexing with gp120 and interacting with CCR5-bearing cells. HIV-1 gp120 was preincubated with PF-68742 (circles) or the gp120 antagonist PF-348089 (squares) for 1 h. The pretreated gp120 was then incubated with sCD4 for 15 min, and any resulting sCD4:gp120 complexes formed were incubated with CCR5-expressing cells. CCR5-bound sCD4:gp120 was detected by incubation with sheep anti-HIV-1 gp120, followed by Eu-labeled donkey anti-sheep IgG. (C) PF-68742 does not inhibit precomplexed sCD4:gp120 from binding to CCR5 coreceptor. Preformed sCD4:gp120 complexes were incubated with CCR5-expressing cells in the presence of PF-68742 (circles), the CCR5 antagonist PF-549084 (triangles), or the gp120 antagonist PF-348089 (squares). CCR5-bound sCD4:gp120 was incubated with sheep anti-HIV-1 gp120 and detected by Eu-labeled donkey anti-sheep IgG.
We next explored PF-68742 binding to gp120 in the solution phase to determine whether the compounds could access soluble gp120 that is free of possible immobilization artifacts. Purified recombinant BaL gp120 was preincubated with up to 10 μM PF-68742 or 1 μM PF-348089, prior to incubation with the sCD4. The sCD4 was added to the mixture of gp120 and inhibitor, and the sCD4:gp120 complexes were added to MIP31.10 cells (CCR5high). Cell-bound sCD4:gp120 complexes were detected using sheep anti-gp120. A MAb against CD4 served as a positive control and produced an EC50 ∼10 nM in this assay (data not shown). PF-68742 was ineffective at micromolar levels despite its submicromolar potency in the fusion assay, whereas the control gp120 antagonist PF-348089 efficiently blocked sCD4:gp120 complex formation (Fig. 4B). These results demonstrate that PF-348089, but not PF-68742, can bind to soluble gp120 and block recognition of CD4. In a second assay format, we investigated whether PF-68742 acted on gp120 sites induced after CD4 binding. Thus, sCD4:gp120 complexes were preformed in the absence of compound and then added to MIP31.10 cells in the presence of either control compounds (i.e., PF-549084, PF-348089, and anti-CD4 MAb) or PF-68742. As expected, the CCR5 antagonist PF-549084 blocked sCD4:gp120 preformed complexes from binding to the coreceptor (Fig. 4C). However, neither PF-68742 nor PF-348089 had a significant effect. We note that the control anti-CD4 MAb was also completely inactive (>10 μg/ml) in this assay format, indicating that the dissociation of preformed sCD4:gp120 complex was very low. Thus, PF-68742 recognizes neither the CD4 binding site nor the CD4-induced coreceptor site on monomeric gp120.
Taken together, the data suggest that PF-68742 is an entry inhibitor that acts as a specific antagonist of Env-mediated fusion but is unable to block the association of gp120 to CD4 or antagonize sCD4:gp120 binding to coreceptors.
HIV-1 strains with selected resistance to PF-68742 and PF-4087738 have a G514R mutation in gp41.
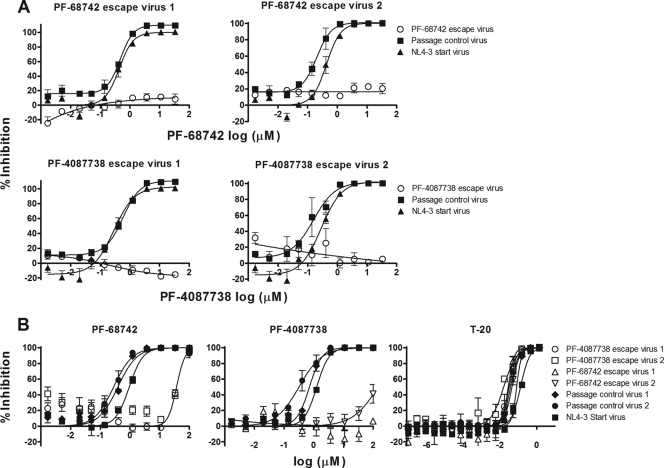
HIV-1 NL4-3 strains resistant to either PF-68742 or PF-4087738 were generated using the appearance of syncytia in host MT2 cells as the endpoint for viral breakthrough and emerging resistance. Untreated NL4-3 passage controls were set up in parallel. Ultimately, after 5 to 6 weeks in culture, two independent viral strains were generated which were >100-fold resistant to either PF-68742 or PF-4087738, (Fig. 5A). The PF-68742 and PF-4087738 escape viruses also exhibited cross-resistance, as may be expected for two closely related structural analogues (Fig. 5B). All of the escape viruses were found to have wild-type level sensitivities to the gp41 fusion inhibitor enfuvirtide (T20; Fig. 5B), suggesting that the cognate Envs adopt a conformation that exposes the T20 binding site similarly to wild-type NL4-3.
FIG. 5.
Characterization of (pooled) escape viruses selected using PF-68742 and PF-4087738. NL4-3 viruses resistant to PF-68742 and PF-4087738 were generated by passaging breakthrough virus in the presence of increasing doses of compound. Two independent resistant populations were generated for each compound. (A) Dose-response curves are shown for each independent, resistant pool after amplification (n = 2) using either PF-68742 (top) or PF-4087738 (bottom). The passage control is NL4-3 virus passaged in parallel but in the absence of added compound and “start” virus is the original NL4-3 virus stock used for the experiment. (B) Cross-resistance study of the PF-68742 resistant (escape) virus and PF-4087738 resistant (escape) virus was performed. In addition, the T-20 sensitivity of breakthrough virus was assayed. NL4-3 viruses resistant to PF-68742 and PF-4087738 were treated with compound and the dose-response curves are shown for each independent, resistant pool after amplification (n = 2) using either PF-68742 (right) or PF-4087738 (center) or T20 (left) with corresponding passage controls and start NL4-3 virus.
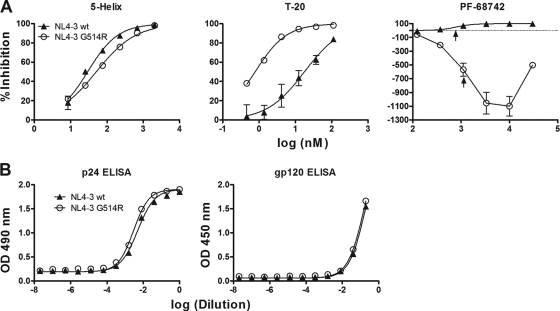
The entire Env sequence from PF-68742-resistant HIV-1 and two independently generated HIV-1 variants resistant to PF-4087738 were isolated by PCR, sequenced, and then compared to Env sequences from two independently generated passage controls (grown in the absence of compound). Only one mutation, G514R (HxB2 numbering) in the FP of gp41, was present in all resistant viruses and absent from all passage controls. The G514R mutation was subcloned into NL4-3 virus and checked for PF-68742 resistance. Whereas wild-type NL4-3 virus was inhibited by PF-68742 as expected, the NL4-3G514R mutant virus was not only resistant to PF-68742 inhibition but also shows marked negative inhibition in the presence of lower concentrations of the compound (Fig. 6A). Interestingly, the EC50s observed for wild-type NL4-3 inhibition and for NL4-3G514R “negative inhibition” are almost identical (consistently within 2-fold of each other), suggesting that PF-68742 is most likely bound to the same site in Env for both NL4-3 and NL4-3G514R. In the absence of PF-68742, we note that the infectivity of the NL4-3G514R mutant virus is ca. 10% that of the wild-type virus. ELISAs for p24 (gag) and gp120 were performed to verify that equivalent levels of both viruses were used in the viral inhibition assays and that the G514R mutation does not affect the relative level of Env in the virion preparations (Fig. 6B). We also tested wild-type and G514R mutant viruses against the gp41 inhibitor, 5-helix (55), as well as MAb b12, and found that the strong resistance phenotype is specific for PF-68742 (Fig. 6A; data not shown). However, interestingly, the G514R mutant is ∼10-fold more sensitive than the wild type to T20 (Fig. 6A), suggesting that G514R may somehow prolong exposure of T20 binding site. This hypersensitivity of NL4-3G514R to T20 contrasts with the original pools of escape virus that had wild-type level sensitivities to T20 (Fig. 5B), perhaps because of additional residue changes outside of position 514 that could restore wild-type-like exposure of the T20 site with the original escape variants. Importantly, our results strongly implicate G514 in the mechanism of action of PF-68742.
FIG. 6.
Sensitivity of HIV-1 NL4-3G514R mutant to PF-68742, 5-helix, and T20. (A) Viral entry assays were performed with the original NL4-3 wild-type molecular clone and an engineered G514R point mutant in the presence of PF-68742, or the control inhibitors, 5-helix and T20. Experiments were performed in triplicate. The positions of the EC50s for the two PF-68742 dose-response curves are indicated by upward arrows. (B) P24 (left) and gp120 (right) ELISAs were performed on NL4-3 wild-type and NL4-3G514R mutant viruses used in panel B to verify that equivalent amounts of virus were used in the viral entry assays and that Env levels were unaffected by the mutation.
Residue changes at positions in gp41 other than G514 confer resistance to PF-68742.
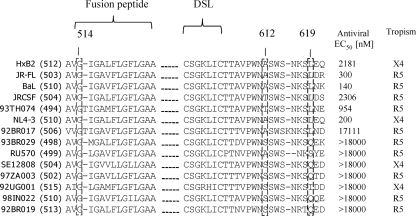
PF-68742 breadth and potency of activity was determined in single-cycle viral entry assays with pseudotyped HIV-1 generated from a panel of Envs from a wide range of lab strains and clinical isolates. The antiviral EC50s for PF-68742 were calculated from dose-response curves using 14 HIV-1 Env variants (Fig. 7). The experiment identified seven Env variants that were susceptible to PF-68742 with a range of sensitivities and seven resistant Envs (EC50 > 18 μM). All of the naturally occurring resistant Envs have the conserved Gly at position 514, demonstrating that resistance to PF-68742 is conferred by residue changes at positions other than 514. Given the small size of PF-68742, it is conceivable that residues conferring resistance would be spatially very close to G514. On the other hand, it is also possible that many seemingly unrelated sequence changes throughout Env could combine to produce a resistance phenotype. To begin to address this issue, we noted relative sequence conservation of gp41 in PF-68742-sensitive strains and that certain residues were altered in PF-68742-resistant strains (i.e., A612 and L619 [highlighted in Fig. 7]). This observation suggested to us a possible role of the central ectodomain region of gp41 in conferring PF-68742 resistance. Residues near to the DSL region of gp41 have been previously implicated in the resistance of HIV-1 to a fusion inhibitor, RPR103611 (3, 38). Moreover, the DSL is reportedly functionally linked to the furin cleavage site of Env (5, 31, 57), the latter of which is proximal to position 514 in the FP of gp41. We had available to us in the lab for another purpose several mutants of a PF-68742-sensitive molecular clone, LAI-JRFL, that contain random substitutions in the DSL region and for which we had previously determined to be fusion competent. Thus, we used these DSL mutants to explore a hypothesis that the DSL may be proximal to a PF-68742 binding site on gp41 that is also close to, or includes, G514 (see Table 1 for DSL mutant sequences analyzed).
FIG. 7.
Sequence alignment of the gp41 fusion peptide (FP) and disulfide loop (DSL) region of HIV-1 strains tested in viral entry assays with PF-68742. Sequence alignment of Envs from different HIV-1 strains showing FP and DSL sequences. The number in parentheses following the laboratory or clinical isolate name is the Env numbering specific for that sequence. Boxed residues highlight G514, A612, and A619 residues discussed in the text. The antiviral EC50s were determined in by Monogram BioSciences using pseudotyped virus in a single cycle HIV-1 assay. The tropism of each pseudotyped virus is also shown.
TABLE 1.
Sequence of gp41 DSL region mutants in a HIV- 1LAI-JR-FL molecular clone background and mutant sensitivity to PF-68742
| Virus | Sequence (aa 592 to 610)a | Relative infectivity (%) | PF-68742 EC50 (nM) |
|---|---|---|---|
| LAI-JR-FL wild type | LLGIWGCSGKLICTTAVPW | 100 | 425 |
| PID3-19 | LLGIWGCSGKHVCPTSVEW | 164 | >30,000 |
| PID2-6 | LLGIWGCRGNLVCNTAVPW | 118 | >30,000 |
| PID2-18b | LLGIWGCTGNTICPTAVPW | 26 | 19,800 |
| PID2-26b | LLGIWGCSASFICTTAVPW | 81 | 2,220 |
| PID2-27b | LLGIWGCSAKHICHTAVPW | 99 | >30,000 |
| PID2-28b | LLGIWGCRGSLVCYTAVPW | 191 | >30,000 |
HxB2 numbering. aa, amino acids.
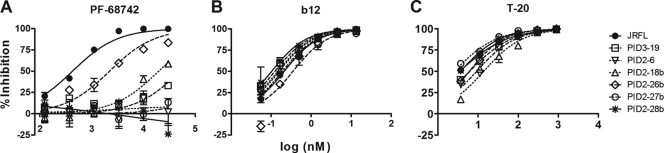
Thus, using a panel of HIV-1 (LAI-JR-FL) variants carrying DSL-proximal substitutions (in the context of G514), we identified several that were completely resistant to PF-68742 (EC50 >30 μM compared to an EC50 of ∼400 nM with the wild type; Fig. 8). Substitutions occurring C-terminal to the DSL produced the greatest resistance to PF-68742 (compare mutations PID3-19, PID2-6, PID2-27b, and PID2-28b to PID2-26b). We note that PF-68742 was slightly less potent against wild-type LAI-JR-FL molecular clone than against pseudotyped HIV-1JR-FL; however, we also observed this difference in sensitivity with other inhibitors and antibodies (data not shown). Importantly, none of the LAI-JR-FL DSL mutants showed any resistance to inhibition by the MAb b12. Moreover, T20 (enfuvirtide) inhibition was minimally affected, with at most a fourfold sensitivity loss (PID2-18b), and several mutants that were most resistant to PF-68742 showed no resistance at all to T20. The clear and specific impact of the DSL mutations on PF-68742 resistance, in addition to that observed with G514R, is consistent with a functional link between the DSL region, G514, and FP sequences proximal to the furin cleavage site.
FIG. 8.
Gp41 (DSL) mutants are resistant to PF-68742 but not to T20 or MAb b12. Viral entry assays in TZM-bl target cells were performed in the presence of PF-68742 (A), b12 (B), and T20 (C) against the LAI-JRFL molecular clone and a variety of corresponding mutants in the DSL region of gp41. Experiments were performed in triplicate. LAI-JRFL contains the Env ectodomain of HIV-1JR-FL in a HIV-1LAI backbone. Sequences of the gp41 (DSL) mutants are shown in Table 1.
DISCUSSION
The benzene sulfonamide PF-68742 was initially selected as an inhibitor of cell fusion that was equipotent whether fusion was mediated by NL4-3 Env (X4) or JR-FL Env (R5). HIV-1 pseudotyped with Env from SIV or VSV-G was not sensitive to PF-68742, whereas HIV-1 pseudotyped with JR-FL Env was potently inhibited. Inhibition of HIV-1 by PF-68742 was evident in both cell-cell fusion and virus-cell fusion assays, and Env was found to be the sole viral target of inhibition. PF-68742 acts as an entry inhibitor in the cell fusion assay, as evidenced by a gradual loss of efficacy as PF-68742 is added after mixing of Env cells and target cells. PF-68742 showed potent antiviral activity against HIV-1 BaL but did not block binding of soluble recombinant BaL gp120 to sCD4 and did not block the binding of sCD4:gp120 complexes to coreceptor-bearing cells. These data point to Env and the HIV-1 fusion reaction as likely targets in the mechanism of action of PF-68742. A recent study had suggested that a portion of the Env cytoplasmic tail is exposed and plays a role in fusion during the early postattachment stage (16). That this region could be removed with no impact on susceptibility ruled out the cytoplasmic tail of Env as a target.
Pools of PF-68742-resistant HIV-1 were selected and env genes were sequenced to reveal that a G514R mutation in the FP of gp41 was characteristic of all resistant viruses. When subcloned into a wild-type NL4-3 molecular clone, G514R alone conferred resistance to PF-68742. In fact, profound negative inhibition of the NL4-3G514R mutant virus was evident in the presence of modest concentrations of the compound. The negative inhibition seen with NL4-3G514R mutant virus offers important insight into a plausible model for the mechanism of action of PF-68742. Increasing the concentration of PF-68742 leads initially to a restoration of infectivity of the mutant virus, HIV-1NL4-3(G514R), strongly suggesting that G514R does not prevent PF-68742 binding to the cognate Env but rather modulates the functional outcome through its interaction with Env. Evidence for retention of PF-68742 binding is shown in the almost identical EC50s for inhibition and for “negative inhibition” of NL4-3 and NL4-3G514R viruses, respectively. Proximity of G514R to the gp120-gp41 interface would be expected to have a functional impact on Env fusogenicity following CD4/coreceptor engagement. Indeed, an ∼90% penalty to infectivity was observed with the NL4-3G514R mutant virus compared to the wild type, which may in turn explain why the G514R polymorphism is not found in clinical isolates of HIV-1. Such a model would be consistent with the mutation G514R impacting on the gp120-gp41 interface in such a way as to counteract the inhibitory effect of bound PF-68742. This route to resistance to PF-68742 is highly unusual among antiviral agents binding viral targets. Resistance to compounds that bind viral targets is nearly always generated via viral mutations that abrogate compound binding resulting in rightward shifts of EC50 values. This classical mechanism of resistance is exemplified by the DSL SDM mutants and is clearly in contrast to that observed for the NL4-3G514R virus discussed above. One notable precedent for viral escape involving modulation of the inhibitor-bound state is with the recently described T20-escape mutants of HIV-1 that depend on modest levels of T20 for optimal infectivity (2). We note also that our results with PF-68742 are consistent with a close functional relationship between the DSL region of gp41 and the FP, the latter of which abuts the furin cleavage site. Altogether, these elements may constitute a functional interface for gp120/gp41, as proposed by other groups (5, 25, 27, 31, 36, 46, 51, 57).
Breakthrough (resistant) virus in the initially selected pools did not show negative inhibition in the presence of PF-68742, in contrast to the NL4-3G514R mutant. Such a result may be due to the acquisition of compensatory mutations that evolved to increase viral fitness when G514R emerged in the resistant population. Because drug is always present and since drug binding is a reversible process, compensatory mutations that enhance fitness in both the drug-bound and unbound states will be selected preferentially. The NL4-3G514R mutant virus has much reduced fitness and an altered sensitivity (10× more sensitive) to T-20 compared to wild-type NL4-3 (Fig. 5B). We interpret this observation to mean that the G514R mutation disrupts Env conformation prior to fusion resulting in an overexposed T20 binding site. If compensatory mutations in the PF-68742 escape virus increase fitness, one might expect that Env disruption would be reduced and that the attendant hypersensitivity of the escape virus to T20 would also be diminished. We show that is indeed the case (Fig. 5B).
To gain further insight into the antiviral activity of PF-68742, we tested the compound against a wide range of different HIV-1 Env variants. This analysis revealed seven PF-68742-sensitive and an equal number of PF-68742-resistant Envs (Fig. 7). X4, R5, and dualtropic Env sequences are represented in both groups, confirming the tropism-independent mechanism of PF-68742. The sequence alignment also showed total conservation of G514. Certain polymorphisms (A612 and L619) proximal to the DSL in the gp41 central ectodomain region were also more commonly observed in susceptible viruses. We note that other groups have previously reported that low-molecular-weight antiviral compounds targeting gp41 can bind to a pocket proximal to the DSL, identified by resistant mutations I595S and L602H in X4-tropic HIV-1 (38, 39, 49). The presence of wild-type G514 in PF-68742 resistant HIV-1 strains indicated that other subelements within Env can contribute to PF-68742 sensitivity. To study this further, we utilized available HIV-1 mutants with substitutions in the DSL (and adjacent sequences) in the context of a JR-FL backbone that contained wild-type G514. To our surprise, whereas certain centrally mutated DSL sequences produced only a modest (∼5-fold) increase in EC50 (e.g., PID2-26b), other substitutions to the DSL and to residues immediately C-terminal to it conferred complete resistance to PF-68742. Note that the dose-response curves with DSL mutant viruses resistant to PF-68742 displayed classical rightward shifts in EC50s characteristic of reduced PF-68742 affinity. Considering the previous identification of a binding pocket in this region, it is quite possible that sequences proximal to the DSL contribute to at least part of the PF-68742 binding pocket. A physical binding model could be tested through use of tracer-labeled PF-68742 analogues, which we are currently attempting to synthesize.
Our model of G514R resistance characterized by altered properties of inhibitor-bound Env may explain why we did not see the emergence of DSL mutations in breakthrough viruses. Thus, mutations in the DSL region would be expected to prevent compound binding and preclude emergence of NL4-3G514R viruses. Reciprocally, the emergence of G514R would provide a specific replication advantage in the presence of PF-68742 and preclude the outgrowth of mutations in the DSL region of gp41. Determination of the possible functional consequences of combining escape mutations in the FP and DSL of gp41 may help clarify whether specific interactions exist between these two regions.
The DSL region is important in the Env-mediated fusion process, as highlighted by three observations. First, the DSL has been shown to directly contribute to membrane binding by gp41 (36, 52). Second, hydrophobic amino acids on either side of the DSL form a hydrophobic patch that is required for interaction with gp120 and therefore play a role during the CD4/coreceptor induced conformational changes necessary for fusion (5, 46). Third, mutations in these hydrophobic patches impact on the formation of fusion pores, as measured by the exchange of cytoplasmic fluorescent dyes during cell-cell fusion (3). The DSL allows the reversal of peptide chain orientation necessary for the packing of the HR1 and HR2 motifs in the formation of the 6HB. Indeed, chain reversal and 6HB formation would likely follow the extrusion of the FP from other elements of the receptor-activated Env complex. Our finding that resistance to PF-68742 can involve either G514 in the FP and/or the DSL region reinforces the role of the latter region during viral entry. The gp120 subunit is in close spatial proximity with the DSL in the gp41 subunit (5, 27, 36), but it remains to be established whether gp120 sequences contribute to PF-68742 binding.
We have substantially narrowed the possibilities of both the site and mechanism of action of PF-68742, but the exact molecular details remain to be defined. It is still unclear, for example, whether PF-68742 prevents Env from adopting a particular fusogenic conformation; impacts gp120 disengagement from the receptor-bound Env complex; or uses yet another unappreciated mechanism involving gp41 to inhibit HIV-1 fusion. It has been well documented that T20 potency is altered as a result of either increased or decreased efficiency of Env-mediated fusion (54). Since the DSL-proximal mutations that confer resistance to PF-68742 have only limited effects on enfuvirtide (T20) EC50s, we can rule out that they cause a general change in fusion kinetics. In contrast, the G514R mutant shows increased sensitivity to T20, suggesting that G514R increases availability of the pre-hairpin intermediate to this inhibitor.
As previously noted, the betulinic acid derivative RPR130611 has been identified as binding to sequences proximal to the DSL region (38). This compound was initially characterized as a fusion inhibitor acting at a post-CD4 binding step (49). Further studies showed that gp41 is the target for RPR103611 but that the compound cannot inhibit R5-tropic viruses (39). Lai et al. (40) have recently shown that IC9564 (a stereoisomer of RPR103611) has a wider spectrum of activity but binds to the V3 loop of gp120. Most recently, it has been proposed that the antiviral mechanism of action for IC9564 is due to immobilization of gp120 conformational changes due to compound binding (30). There are clearly a number of differences between the benzene sulfonamide PF-68742 and the previously reported betulinic acid derived inhibitors, RPR103611 and IC9564. First, PF-68742 acts independently of tropism, unlike RPR103611 (39). Second, resistance to IC9564 required changes to V3 unlike PF-68742 (29, 30). A novel class of HIV-1 fusion inhibitors, also based on betulinic acid, was being developed at Panacos, but available resistance data to these undisclosed inhibitors suggest that they do not act in the same manner as PF-68742 (23).
It is remarkable that resistance mutations in FP also exist in breakthrough HIV-1 selected to escape CCR5 antagonists, which also bear substitutions proximal to the furin cleavage site in the context of specific Env sequences (1). These escape viruses also exhibit “negative inhibition” due to the existence of distinct forms of CCR5—each with various affinities for vicriviroc—and certain types of which may be used in the drug-bound form by resistant HIV-1 variants. The mechanism of action of PF-68742 is clearly different from CCR5 antagonists due to coreceptor independence and targeting a viral target (Env) rather than host target, CCR5. The observation from the present study and others that both PF-68742 and vicriviroc, with such different mechanisms of action, can generate escape viruses with FP mutations could imply a universal HIV escape strategy driven by the adaptability of Env to find alternate conformations to circumvent barriers imposed by different classes of entry inhibitors.
With resistance to existing HIV-1 drugs becoming an increasing problem in the clinic, novel targets for therapeutic intervention are needed. We have shown that PF-68742 works through a novel mechanism and could serve as a starting point for chemistry efforts to identify compounds that meet the current therapeutic standard of care. For example, additional contacts to conserved elements of the DSL may result in compounds with improved spectrum. The most interesting aspect of this mechanism is the selection of a resistant form of gp41 that maintains affinity but becomes functionally dependent on inhibitor binding. The concomitant increase in sensitivity to T20 provides further hope that specific drug combinations may combine to limit viral escape. Finally, PF-68742 serves as a useful chemical tool to learn more about viral fusion, still arguably one of the least understood aspects of HIV replication, possibly enabling entirely new therapeutic approaches to block this complex process.
Acknowledgments
We thank the NIH for support (T32AI007606 [D.P.L.], AI077381 and AI06993 [M.B.Z.]).
We thank Monogram Biosciences for the viral entry assays. We thank Andrew Bell (Pfizer) for the initial synthesis of PF-68742. We also thank Dennis Burton for IgG b12, Hermann Katinger for IgG 4E10, Michael Root for 5-helix, and the NIH ARRRP and Roche for enfuvirtide/T20.
Footnotes
Published ahead of print on 28 April 2010.
REFERENCES
- 1.Anastassopoulou, C. G., T. J. Ketas, P. J. Klasse, and J. P. Moore. 2009. Resistance to CCR5 inhibitors caused by sequence changes in the fusion peptide of HIV-1 gp41. Proc. Natl. Acad. Sci. U. S. A. 106:5318-5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldwin, C. E., R. W. Sanders, Y. Deng, S. Jurriaans, J. M. Lange, M. Lu, and B. Berkhout. 2004. Emergence of a drug-dependent human immunodeficiency virus type 1 variant during therapy with the T20 fusion inhibitor. J. Virol. 78:12428-12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bar, S., and M. Alizon. 2004. Role of the ectodomain of the gp41 transmembrane envelope protein of human immunodeficiency virus type 1 in late steps of the membrane fusion process. J. Virol. 78:811-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bianchi, E., M. Finotto, P. Ingallinella, R. Hrin, A. V. Carella, X. S. Hou, W. A. Schleif, M. D. Miller, R. Geleziunas, and A. Pessi. 2005. Covalent stabilization of coiled coils of the HIV gp41 N region yields extremely potent and broad inhibitors of viral infection. Proc. Natl. Acad. Sci. U. S. A. 102:12903-12908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Binley, J. M., R. W. Sanders, B. Clas, N. Schuelke, A. Master, Y. Guo, F. Kajumo, D. J. Anselma, P. J. Maddon, W. C. Olson, and J. P. Moore. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74:627-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bjorndal, A., H. Deng, M. Jansson, J. R. Fiore, C. Colognesi, A. Karlsson, J. Albert, G. Scarlatti, D. R. Littman, and E. M. Fenyo. 1997. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J. Virol. 71:7478-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradley, J., J. Gill, F. Bertelli, S. Letafat, R. Corbau, P. Hayter, P. Harrison, A. Tee, W. Keighley, M. Perros, G. Ciaramella, A. Sewing, and C. Williams. 2004. Development and automation of a 384-well cell fusion assay to identify inhibitors of CCR5/CD4-mediated HIV virus entry. J. Biomol. Screen. 9:516-524. [DOI] [PubMed] [Google Scholar]
- 8.Burton, D. R., J. Pyati, R. Koduri, S. J. Sharp, G. B. Thornton, P. W. Parren, L. S. Sawyer, R. M. Hendry, N. Dunlop, P. L. Nara, et al. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024-1027. [DOI] [PubMed] [Google Scholar]
- 9.Cai, L., and M. Gochin. 2007. A novel fluorescence intensity screening assay identifies new low-molecular-weight inhibitors of the gp41 coiled-coil domain of human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 51:2388-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cavacini, L., M. Duval, L. Song, R. Sangster, S. H. Xiang, J. Sodroski, and M. Posner. 2003. Conformational changes in env oligomer induced by an antibody dependent on the V3 loop base. AIDS 17:685-689. [DOI] [PubMed] [Google Scholar]
- 11.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 12.Debnath, A. K. 6 June 2006. Antiviral composites comprising heterocyclic substituted phenyl furans and related compounds. Patent application WO/2006/138118 A2.
- 13.De Clercq, E. 2003. The bicyclam AMD3100 story. Nat. Rev. Drug Discov. 2:581-587. [DOI] [PubMed] [Google Scholar]
- 14.Dimitrov, A. 2007. Ibalizumab, a CD4-specific MAb to inhibit HIV-1 infection. Curr. Opin. Invest. Drugs 8:653-661. [PubMed] [Google Scholar]
- 15.Dimitrov, A. S., A. Jacobs, C. M. Finnegan, G. Stiegler, H. Katinger, and R. Blumenthal. 2007. Exposure of the membrane-proximal external region of HIV-1 gp41 in the course of HIV-1 envelope glycoprotein-mediated fusion. Biochemistry 46:1398-1401. [DOI] [PubMed] [Google Scholar]
- 16.Dimmock, N. J. 2005. The complex antigenicity of a small external region of the C-terminal tail of the HIV-1 gp41 envelope protein: a lesson in epitope analysis. Rev. Med. Virol. 15:365-381. [DOI] [PubMed] [Google Scholar]
- 17.Doranz, B. J., S. S. Baik, and R. W. Doms. 1999. Use of a gp120 binding assay to dissect the requirements and kinetics of human immunodeficiency virus fusion events. J. Virol. 73:10346-10358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dorr, P., M. Westby, S. Dobbs, P. Griffin, B. Irvine, M. Macartney, J. Mori, G. Rickett, C. Smith-Burchnell, C. Napier, R. Webster, D. Armour, D. Price, B. Stammen, A. Wood, and M. Perros. 2005. Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob. Agents Chemother. 49:4721-4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eckert, D. M., and P. S. Kim. 2001. Design of potent inhibitors of HIV-1 entry from the gp41 N-peptide region. Proc. Natl. Acad. Sci. U. S. A. 98:11187-11192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 21.Eckert, D. M., V. N. Malashkevich, L. H. Hong, P. A. Carr, and P. S. Kim. 1999. Inhibiting HIV-1 entry: discovery of D-peptide inhibitors that target the gp41 coiled-coil pocket. Cell 99:103-115. [DOI] [PubMed] [Google Scholar]
- 22.Este, J. A., and A. Telenti. 2007. HIV entry inhibitors. Lancet 370:81-88. [DOI] [PubMed] [Google Scholar]
- 23.Finnegan, C. M., I. Burinski, T. Jackson, A. Castillo, N. Kilgore, M. Reddick, A. Yunus, M. Zuiderhoff, T. Nitz, C. Wild, G. Allaway, and K. Salzwedel. 2006. Validation of a novel approach for HIV-1 fusion inhibitor drug discovery. XVI International AIDS Conference, Toronto, Ontario, Canada.
- 24.Frey, G., S. Rits-Volloch, X. Q. Zhang, R. T. Schooley, B. Chen, and S. C. Harrison. 2006. Small molecules that bind the inner core of gp41 and inhibit HIV envelope-mediated fusion. Proc. Natl. Acad. Sci. U. S. A. 103:13938-13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guilhaudis, L., A. Jacobs, and M. Caffrey. 2002. Solution structure of the HIV gp120 C5 domain. Eur. J. Biochem. 269:4860-4867. [DOI] [PubMed] [Google Scholar]
- 26.Harrison, S. C. 2008. Viral membrane fusion. Nat. Struct. Mol. Biol. 15:690-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helseth, E., U. Olshevsky, C. Furman, and J. Sodroski. 1991. Human immunodeficiency virus type 1 gp120 envelope glycoprotein regions important for association with the gp41 transmembrane glycoprotein. J. Virol. 65:2119-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho, H. T., L. Fan, B. Nowicka-Sans, B. McAuliffe, C. B. Li, G. Yamanaka, N. Zhou, H. Fang, I. Dicker, R. Dalterio, Y. F. Gong, T. Wang, Z. Yin, Y. Ueda, J. Matiskella, J. Kadow, P. Clapham, J. Robinson, R. Colonno, and P. F. Lin. 2006. Envelope conformational changes induced by human immunodeficiency virus type 1 attachment inhibitors prevent CD4 binding and downstream entry events. J. Virol. 80:4017-4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holz-Smith, S. L., I. C. Sun, L. Jin, T. J. Matthews, K. H. Lee, and C. H. Chen. 2001. Role of human immunodeficiency virus (HIV) type 1 envelope in the anti-HIV activity of the betulinic acid derivative IC9564. Antimicrob. Agents Chemother. 45:60-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang, L., W. Lai, P. Ho, and C. H. Chen. 2007. Induction of a nonproductive conformational change in gp120 by a small molecule HIV type 1 entry inhibitor. AIDS Res. Hum. Retrovir. 23:28-32. [DOI] [PubMed] [Google Scholar]
- 31.Jacobs, A., J. Sen, L. Rong, and M. Caffrey. 2005. Alanine scanning mutants of the HIV gp41 loop. J. Biol. Chem. 280:27284-27288. [DOI] [PubMed] [Google Scholar]
- 32.Jiang, S., K. Lin, N. Strick, and A. R. Neurath. 1993. HIV-1 inhibition by a peptide. Nature 365:113. [DOI] [PubMed] [Google Scholar]
- 33.Jiang, S., H. Lu, S. Liu, Q. Zhao, Y. He, and A. K. Debnath. 2004. N-substituted pyrrole derivatives as novel human immunodeficiency virus type 1 entry inhibitors that interfere with the gp41 six-helix bundle formation and block virus fusion. Antimicrob. Agents Chemother. 48:4349-4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kadkhodayan, S., L. O. Elliott, G. Mausisa, H. A. Wallweber, K. Deshayes, B. Feng, and W. J. Fairbrother. 2007. Evaluation of assay technologies for the identification of protein-peptide interaction antagonists. Assay Drug Dev. Technol. 5:501-513. [DOI] [PubMed] [Google Scholar]
- 35.Kilgore, N. R., K. Salzwedel, M. Reddick, G. P. Allaway, and C. T. Wild. 2003. Direct evidence that C-peptide inhibitors of human immunodeficiency virus type 1 entry bind to the gp41 N-helical domain in receptor-activated viral envelope. J. Virol. 77:7669-7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim, S., H. B. Pang, and M. S. Kay. 2008. Peptide mimic of the HIV envelope gp120-gp41 interface. J. Mol. Biol. 376:786-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koshiba, T., and D. C. Chan. 2003. The prefusogenic intermediate of HIV-1 gp41 contains exposed C-peptide regions. J. Biol. Chem. 278:7573-7579. [DOI] [PubMed] [Google Scholar]
- 38.Labrosse, B., O. Pleskoff, N. Sol, C. Jones, Y. Henin, and M. Alizon. 1997. Resistance to a drug blocking human immunodeficiency virus type 1 entry (RPR103611) is conferred by mutations in gp41. J. Virol. 71:8230-8236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Labrosse, B., C. Treboute, and M. Alizon. 2000. Sensitivity to a nonpeptidic compound (RPR103611) blocking human immunodeficiency virus type 1 Env-mediated fusion depends on sequence and accessibility of the gp41 loop region. J. Virol. 74:2142-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lai, W., L. Huang, P. Ho, Z. Li, D. Montefiori, and C. H. Chen. 2008. Betulinic acid derivatives that target gp120 and inhibit multiple genetic subtypes of human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 52:128-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lalezari, J. P., K. Henry, M. O'Hearn, J. S. Montaner, P. J. Piliero, B. Trottier, S. Walmsley, C. Cohen, D. R. Kuritzkes, J. J. Eron, Jr., J. Chung, R. DeMasi, L. Donatacci, C. Drobnes, J. Delehanty, and M. Salgo. 2003. Enfuvirtide, an HIV-1 fusion inhibitor, for drug-resistant HIV infection in North and South America. N. Engl. J. Med. 348:2175-2185. [DOI] [PubMed] [Google Scholar]
- 42.Leaman, D. P., H. Kinkead, and M. B. Zwick. 2010. In-solution virus capture assay helps deconstruct heterogeneous antibody recognition of human immunodeficiency virus type 1. J. Virol. 84:3382-3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee-Huang, S., P. L. Huang, D. Zhang, J. W. Lee, J. Bao, Y. Sun, Y. T. Chang, and J. Zhang. 2007. Discovery of small-molecule HIV-1 fusion and integrase inhibitors oleuropein and hydroxytyrosol. II. Integrase inhibition. Biochem. Biophys. Res. Commun. 354:879-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu, S., S. Wu, and S. Jiang. 2007. HIV entry inhibitors targeting gp41: from polypeptides to small-molecule compounds. Curr. Pharm. Des. 13:143-162. [DOI] [PubMed] [Google Scholar]
- 45.Louis, J. M., I. Nesheiwat, L. Chang, G. M. Clore, and C. A. Bewley. 2003. Covalent trimers of the internal N-terminal trimeric coiled-coil of gp41 and antibodies directed against them are potent inhibitors of HIV envelope-mediated cell fusion. J. Biol. Chem. 278:20278-20285. [DOI] [PubMed] [Google Scholar]
- 46.Maerz, A. L., H. E. Drummer, K. A. Wilson, and P. Poumbourios. 2001. Functional analysis of the disulfide-bonded loop/chain reversal region of human immunodeficiency virus type 1 gp41 reveals a critical role in gp120-gp41 association. J. Virol. 75:6635-6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mariani, R., G. Rutter, M. E. Harris, T. J. Hope, H. G. Krausslich, and N. R. Landau. 2000. A block to human immunodeficiency virus type 1 assembly in murine cells. J. Virol. 74:3859-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez-Cajas, J. L., and M. A. Wainberg. 2008. Antiretroviral therapy: optimal sequencing of therapy to avoid resistance. Drugs 68:43-72. [DOI] [PubMed] [Google Scholar]
- 49.Mayaux, J. F., A. Bousseau, R. Pauwels, T. Huet, Y. Henin, N. Dereu, M. Evers, F. Soler, C. Poujade, E. De Clercq, et al. 1994. Triterpene derivatives that block entry of human immunodeficiency virus type 1 into cells. Proc. Natl. Acad. Sci. U. S. A. 91:3564-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 51.Pancera, M., S. Majeed, Y. E. Ban, L. Chen, C. C. Huang, L. Kong, Y. D. Kwon, J. Stuckey, T. Zhou, J. E. Robinson, W. R. Schief, J. Sodroski, R. Wyatt, and P. D. Kwong. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc. Natl. Acad. Sci. U. S. A. 107:1166-1171. [DOI] [PMC free article] [PubMed]
- 52.Pascual, R., M. Contreras, A. Fedorov, M. Prieto, and J. Villalain. 2005. Interaction of a peptide derived from the N-heptad repeat region of gp41 Env ectodomain with model membranes: modulation of phospholipid phase behavior. Biochemistry 44:14275-14288. [DOI] [PubMed] [Google Scholar]
- 53.Reed, S. E., E. M. Staley, J. P. Mayginnes, D. J. Pintel, and G. E. Tullis. 2006. Transfection of mammalian cells using linear polyethylenimine is a simple and effective means of producing recombinant adeno-associated virus vectors. J. Virol. Methods 138:85-98. [DOI] [PubMed] [Google Scholar]
- 54.Reeves, J. D., S. A. Gallo, N. Ahmad, J. L. Miamidian, P. E. Harvey, M. Sharron, S. Pohlmann, J. N. Sfakianos, C. A. Derdeyn, R. Blumenthal, E. Hunter, and R. W. Doms. 2002. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc. Natl. Acad. Sci. U. S. A. 99:16249-16254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Root, M. J., M. S. Kay, and P. S. Kim. 2001. Protein design of an HIV-1 entry inhibitor. Science 291:884-888. [DOI] [PubMed] [Google Scholar]
- 56.Rusconi, S., A. Scozzafava, A. Mastrolorenzo, and C. T. Supuran. 2007. An update in the development of HIV entry inhibitors. Curr. Top. Med. Chem. 7:1273-1289. [DOI] [PubMed] [Google Scholar]
- 57.Sen, J., A. Jacobs, H. Jiang, L. Rong, and M. Caffrey. 2007. The disulfide loop of gp41 is critical to the furin recognition site of HIV gp160. Protein Sci. 16:1236-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Si, Z., N. Madani, J. M. Cox, J. J. Chruma, J. C. Klein, A. Schon, N. Phan, L. Wang, A. C. Biorn, S. Cocklin, I. Chaiken, E. Freire, A. B. Smith III, and J. G. Sodroski. 2004. Small-molecule inhibitors of HIV-1 entry block receptor-induced conformational changes in the viral envelope glycoproteins. Proc. Natl. Acad. Sci. U. S. A. 101:5036-5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Summa, V., A. Petrocchi, F. Bonelli, B. Crescenzi, M. Donghi, M. Ferrara, F. Fiore, C. Gardelli, O. Gonzalez Paz, D. J. Hazuda, P. Jones, O. Kinzel, R. Laufer, E. Monteagudo, E. Muraglia, E. Nizi, F. Orvieto, P. Pace, G. Pescatore, R. Scarpelli, K. Stillmock, M. V. Witmer, and M. Rowley. 2008. Discovery of raltegravir, a potent, selective orally bioavailable HIV-integrase inhibitor for the treatment of HIV-AIDS infection. J. Med. Chem. 51:5843-5855. [DOI] [PubMed] [Google Scholar]
- 60.Trkola, A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ullman, E. F., H. Kirakossian, S. Singh, Z. P. Wu, B. R. Irvin, J. S. Pease, A. C. Switchenko, J. D. Irvine, A. Dafforn, C. N. Skold, et al. 1994. Luminescent oxygen channeling immunoassay: measurement of particle binding kinetics by chemiluminescence. Proc. Natl. Acad. Sci. U. S. A. 91:5426-5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.UNAIDS. 2008, posting date. Report on the global AIDS epidemic: executive summary. UNAIDS, New York, NY. [Online.] http://data.unaids.org/pub/GlobalReport/2008/jc1511_gr08_executivesummary_en.pdf.
- 63.Van Acker, K. L. A., et al. 22 March 2007. Entry inhibitors of the HIV virus. U.S. patent 20070066623 A1.
- 64.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zwick, M. B., A. F. Labrijn, M. Wang, C. Spenlehauer, E. O. Saphire, J. M. Binley, J. P. Moore, G. Stiegler, H. Katinger, D. R. Burton, and P. W. Parren. 2001. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 75:10892-10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zwick, M. B., E. O. Saphire, and D. R. Burton. 2004. gp41: HIV's shy protein. Nat. Med. 10:133-134. [DOI] [PMC free article] [PubMed] [Google Scholar]