Abstract
The survival and proliferation of the obligate intracellular malaria parasite Plasmodium falciparum require salvage of essential purines from the host. Genetic studies have previously shown that the parasite plasma membrane purine permease, PfNT1, plays an essential function in the transport of all naturally occurring purine nucleosides and nucleobases across the parasite plasma membrane. Here, we describe an intracellular permease, PfNT2. PfNT2 is, like PfNT1, a member of the equilibrative nucleoside transporter family. Confocal and immunoelectron microscopic analyses of transgenic parasites harboring green fluorescent protein- or hemagglutinin-tagged PfNT2 demonstrated endoplasmic reticulum localization. This localization was confirmed by colocalization with the endoplasmic reticulum marker PfBiP. Using yeast as a surrogate system, we show that targeting PfNT2 to the plasma membrane of fui1Δ cells lacking the plasma membrane nucleoside transporter Fui1 confers sensitivity to the toxic nucleoside analog 5-fluorouridine. This study provides the first evidence of an intracellular purine permease in apicomplexan parasites and suggests a novel biological function for the parasite endoplasmic reticulum during malaria infection.
Keywords: Endoplasmic Reticulum, Metabolism, Plasma Membrane, Purine, Yeast, Malaria, Plasmodium, Transporter
Introduction
Plasmodium parasites, the causative agents of malaria, are incapable of synthesizing the purine ring de novo. Instead, they rely on the salvage of purine-containing compounds from their environment. Within the mammalian host, Plasmodium parasites initially undergo replication in the liver before invading red blood cells, where they undergo further cycles of asexual reproduction. It is this intraerythrocytic cycle that causes the symptoms of disease, and it is also during this stage that most current drugs act. The pathways used by the parasite to salvage purines from the host erythrocyte and the host plasma are therefore of interest as potential drug targets.
Purines enter the host cell via a combination of endogenous transporters and “new permeation pathways” induced in the erythrocyte membrane as the parasite matures (1–5). From here, purines are presumed to enter the parasitophorous vacuole via nonspecific pores on the parasitophorous vacuole membrane (6). Once inside the parasitophorous vacuole, purines are taken up across the parasite plasma membrane. Studies on parasites “isolated” from their host cell have shown that both purine and pyrimidine nucleosides and nucleobases are transported across this membrane via a rapid, low affinity, equilibrative process (7, 8).
Searches of Plasmodium genome sequence data with the sequences of known protozoan and mammalian nucleoside transporters led to the identification of a Plasmodium protein, PfNT1 (9, 10), which is a member of the equilibrative nucleoside transporter (ENT)3 family and is localized to the parasite plasma membrane (11). Cloning and expression of the gene in Xenopus laevis oocytes revealed PfNT1 to be a low affinity, broad specificity transporter, capable of transporting a range of nucleosides and nucleobases (9, 10). The transport characteristics of PfNT1 expressed in Xenopus oocytes matched closely the characteristics of nucleoside/nucleobase transport across the parasite plasma membrane (7, 8). Functional analysis by genetic disruption in Plasmodium falciparum indicated that PfNT1 is essential for parasite survival under physiological purine concentrations (12). In a subsequent study it was reported that pfnt1Δ knock-out parasites were unable to transport most physiologically relevant purine nucleosides/nucleobases, although residual adenine transport was observed (13). Another recent study (14), involving transport analyses in both wild type and pfnt1Δ parasites, obtained somewhat different results; however, the interpretation of some of the uptake data has been questioned (15, 16).
The completion of the Plasmodium genome sequencing project (17) revealed three further ENT family members, PfNT2 (MAL8P1.32), PfNT3 (PF14_0662), and PfNT4 (PFA0160c). All have a predicted secondary structure typical of the ENT family (18), despite low sequence homology. The roles these proteins play in parasite physiology have not yet been elucidated.
Of the four P. falciparum members of the ENT family, PfNT2 is the largest and displays the highest sequence homology to PfNT1. Here, we show, using both immunofluorescence and immunoelectron microscopy, that this protein localizes not to the parasite plasma membrane, but to its endoplasmic reticulum (ER). Expression of a codon-optimized PfNT2 in yeast also revealed an intracellular localization. N-terminal modifications, leading to yeast plasma membrane expression, conferred 5-fluorouridine (5-FUrd) toxicity on otherwise resistant yeast strains, suggesting that PfNT2 is capable of mediating nucleoside/nucleobase transport.
EXPERIMENTAL PROCEDURES
Parasite Cell Culture
The 3D7 clone of P. falciparum was propagated in human erythrocytes at 2% hematocrit and 10% parasitemia by the method of Trager and Jensen (19) with the exception that the serum component in the culture medium was replaced with 0.5% Albumax (Invitrogen).
Plasmodium Plasmid Constructions
Transfection vectors encoding PfNT2 tagged with three tandem hemagglutinin (HA) epitopes at either the N or C terminus, or PfNT2 fused to GFP at the C terminus, were constructed using the Invitrogen Multisite Gateway system previously established for Plasmodium transfection (20, 21). In each case, the PfNT2 coding region was amplified from 3D7 cDNA. Promoters used were that of PfCRT (for HA-tagged PfNT2) or HSP86 (PfNT2-GFP). All expression vectors contained the hDHFR selectable marker, which confers resistance to the drug WR99210.
Transfection of P. falciparum
Expression plasmids were transfected into ring-stage parasites using electroporation as previously described (12). Transfected parasites were cultured for 48 h without drug pressure, and thereafter the medium was supplemented with 5 nm WR99210. Resistant parasites were first observed within 3 weeks of WR99210 addition.
Yeast Plasmid Construction
Codon-optimized PfNT2 (PfNT2CO) was synthesized using 72 overlapping 50-nucleotide oligonucleotides (for details, see supplemental Table 1). PfNT2CO was first assembled and amplified as three small fragments that were subsequently used as templates to amplify the whole gene. The full-length PfNT2CO was cloned into the commercially available vector pCR2.1 (Invitrogen). From here, PfNT2CO was inserted into the yeast expression vector pYES2.1-/V5-His-TOPO (Invitrogen), so that it was under the transcriptional control of the GAL1 promoter generating pYES2.1-PfNT2CO-V5. The sequence of PfNT2CO was verified by DNA sequencing.
To generate PfNT2fcy2, encoding PfNT2 fused to the N-terminal domain of the yeast plasma membrane protein Fcy2, the 279-bp fragment encoding the first 93 amino acids of Fcy2 was cloned from yeast DNA and fused with a PCR product of PfNT2CO that lacked the first 210 nucleotides (encoding the first 70 amino acids, i.e. those N-terminal residues before the start of the predicted first transmembrane domain) and inserted into the pYES2.1-/V5-His-TOPO vector.
PfNT2CO-fcy2 was subcloned into the pBEVY-U vector (22) using the restriction enzyme sites BamHI and SalI such that it was under the control of the glyceraldehyde-3-phosphate dehydrogenase (GPD) promoter, which leads to constitutive levels of transcription at lower levels than that of the GAL1 promoter.
Yeast Strains and Growth Conditions
Saccharomyces cerevisiae strains (BY4741: Mata his3Δ1 leu2Δ0 met15Δ0 ura3Δ0; fui1Δ: Mata his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 fui1Δ::Kanr and end3Δ: Mata his3Δ1 leu2Δ0 met15Δ0 ura3Δ0, end3Δ::Kanr) were grown in minimal medium (1.7% yeast nitrogen base, 0.5% ammonium sulfate, containing either 2% dextrose, 2% galactose, or 2% galactose plus 1% raffinose). Supplements were added as required to maintain cell growth. For localization studies and growth assays, PfNT2 constructs were under the transcriptional control of the glyceraldehyde-3-phosphate dehydrogenase (GPD) promoter (in pBEVY-U) (22).
Yeast Growth Assays
Yeast cultures were grown overnight in 5 ml of medium. Cells were harvested by centrifugation, washed once in dideoxy H2O, and diluted to a working concentration of 104 cells/ml. The cells were inoculated in medium with or without 1 mg/ml 5-FUrd. Cells were grown at 30 °C with agitation (210 rpm) and growth was monitored using OD660.
Microscopy
Infected red blood cells were fixed and labeled using a previously described formaldehyde glutaraldehyde protocol (23). Rat anti-HA (Roche) and rabbit anti-PfBiP (MR4), antisera were used at concentrations of 1:200 and 1:1000, respectively, whereas rabbit anti-GFP antibody (AbCAM) was used at 1:250. Secondary antibodies (Alexa Fluor 488 and Alexa Fluor 594; Invitrogen) were used at a concentration of 1:1000. Parasite nuclei were labeled with Hoescht 33258 (Invitrogen). Images were collected on a Leica TCS SP2 confocal microscope or a Nikon Eclipse TE2000-E fluorescent microscope. Immunoelectron microscopy on P. falciparum parasites was carried out as previously described by Rager et al. (11). For immunofluorescence in yeast, wild type and end3Δ cells harboring expression vectors containing full-length PfNT2 or PfNT2fcy2 were grown to OD660 0.5–1.0 in minimal medium containing glucose or galactose. The cells were fixed by addition of formaldehyde to a final concentration of 3.7% (v/v) and incubated at room temperature for 2 h. After centrifugation at 700 × g for 2 min, the cells were resuspended in 10 ml of 0.1 m potassium phosphate, pH 6.8, and 0.5 mm MgCl2. Cells were then centrifuged and washed with 10 ml of solution containing 0.1 m potassium phosphate, pH 6.8, 0.5 mm MgCl2, and 1.2 m sorbitol. After centrifugation, the cells were resuspended in a 0.5 ml of solution containing 0.1 m potassium phosphate, pH 6.8, 0.5 mm MgCl2, and 1.2 m sorbitol and incubated overnight at 4 °C. To digest the yeast cell wall, cells were incubated with 2.5 μl of β-mercaptoethanol and 10 μl of lyticase (12 mg/ml) at 37 °C for ∼30 min. The resulting spheroplasts were centrifuged at 4200 × g for 7 min and resuspended in 5 ml of 0.1 m potassium phosphate, pH 6.8, 0.5 mm MgCl2, and 1.2 m sorbitol. Cells were centrifuged at 700 × g for 7 min and resuspended in 1 ml of a solution containing 0.1 m potassium phosphate, pH 6.8, 0.5 mm MgCl2, and 1.2 m sorbitol. Cell permeabilization was performed following addition of 15 μl of 0.2% Triton X-100 in PBS for 10 min. Cells were washed three times with PBS before blocking for 10 min at room temperature in blocking buffer (0.5% w/v bovine serum albumin in PBS) then for 2 h (at room temperature) in blocking buffer containing mouse anti-V5 antibody (1:500; Invitrogen), rabbit anti-Kar2 (1:1000; Santa Cruz Biotechnology), and rabbit anti-Pma1 antibody (1:1000; Santa Cruz Biotechnology). After washing four times in PBS, a blocking buffer containing Alexa Fluor 488 anti-mouse (1:1000) and Alexa Fluor 594 anti-rabbit (1:2000) was added. The slides were then washed with PBS, air dried, and visualized by a Nikon Eclipse TE2000-E fluorescent microscope.
Immunoblotting
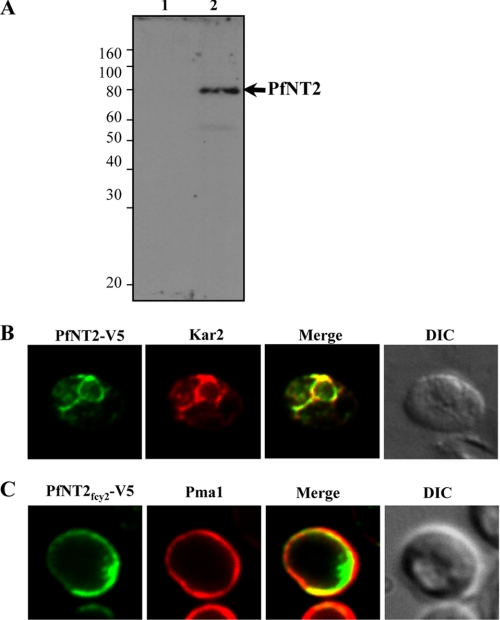
S. cerevisiae transformants harboring pYES2.1 or pYES2.1-PfNT2CO-V5 were grown to mid-log phase on minimal medium containing galactose. Y-PER (Pierce) reagent was used to extract the soluble protein fractions from the S. cerevisiae strains. Proteins were separated by SDS-PAGE, transferred to a nitrocellulose membrane, and analyzed by immunoblotting using anti-V5 primary antibody (1:5000; Invitrogen).
RESULTS
PfNT2 Localizes to the ER of P. falciparum
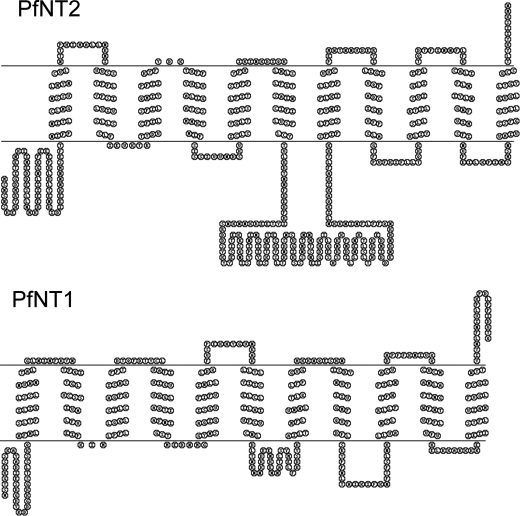
The P. falciparum protein PfNT2 is a polypeptide of 585 amino acids that shares 15% identity (and 31% similarity) with the parasite's primary plasma membrane purine transporter PfNT1 (supplemental Fig. 1). The predicted topology of PfNT2 is typical of ENT family members, with 11 transmembrane domains, a cytoplasmic N terminus, and the large intracellular loop between helices 6 and 7 that is characteristic of members of this family (Fig. 1). The N-terminal domain contains a dileucine motif similar to that of the intracellular human purine transporter hENT3 (supplemental Fig. 1), which has been localized to the lysosomal compartment (24). To establish the subcellular localization of PfNT2 within the P. falciparum-infected erythrocyte, wild type parasites were transfected with plasmids encoding either: (i) PfNT2 with a C-terminal GFP tag under the regulatory control of the HSP86 promoter; (ii) PfNT2 tagged at the C terminus with three tandem HA epitopes; or (iii) PfNT2 tagged at the N terminus with three tandem HA epitopes. Both HA constructs were under the control of the PfCRT promoter, which drives low levels of expression (25). The different constructs and alternate promoters were employed to ensure that the observed localization was not due to overexpression of the protein or to epitope-induced masking of localization sequences within the protein. Transgenic parasites 3D7-HA-PfNT2, 3D7-PfNT2-HA, and 3D7-PfNT2-GFP harboring these expression vectors were selected, and the expression of the tagged proteins was confirmed by immunoblotting using anti-GFP and anti-HA antibodies (data not shown).
FIGURE 1.
Schematic representation of the predicted topology of PfNT1 and PfNT2. The predicted topology is based on a consensus of three programs, and the illustrations were generated using the TOPO2 program.
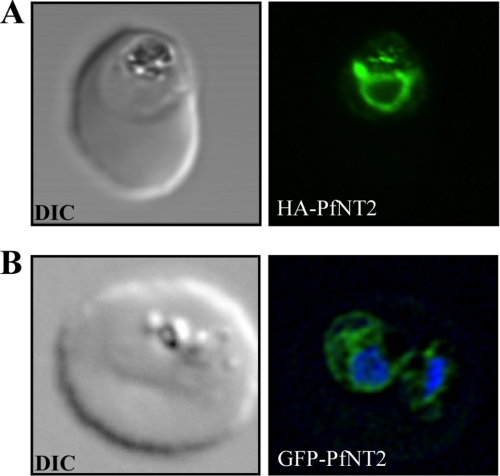
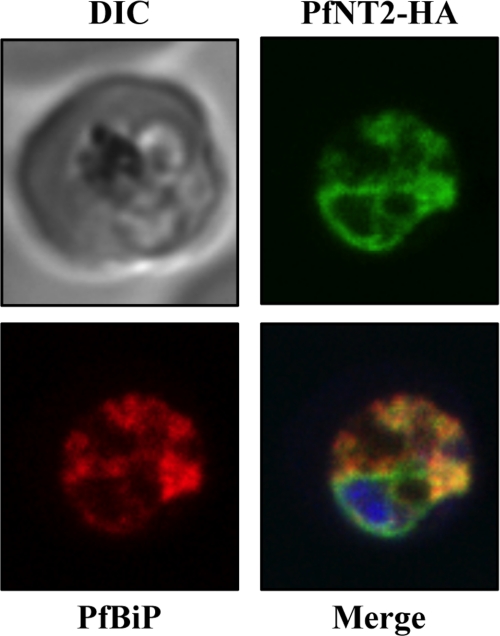
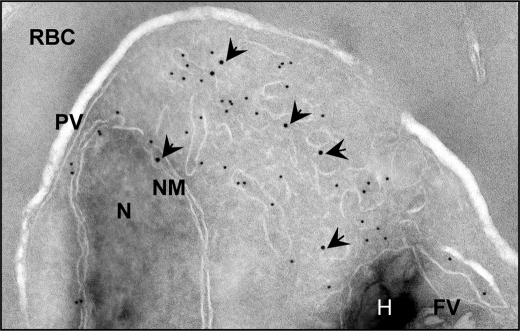
To determine the cellular localization of PfNT2 in these parasites, immunofluorescence assays were performed on fixed, parasitized red blood cells using anti-HA and anti-GFP antibodies. These studies showed that, unlike PfNT1, PfNT2 is not a plasma membrane protein (Fig. 2). The perinuclear localization observed in these parasites suggested an ER location of PfNT2 (21). To investigate this possibility, colocalization assays were carried out using an antibody directed at PfBiP, an endoplasmic reticulum marker for P. falciparum (26), in conjunction with anti-HA or anti-GFP antibodies. As shown in Fig. 3, PfNT2-HA exhibited a high degree of colocalization with PfBiP. Similar colocalization was seen for both other constructs (data not shown). This colocalization was confirmed by immunoelectron microscopic analyses on transgenic parasites expressing PfNT2-GFP, using antibodies against GFP and PfBiP. Gold particles marking PfNT2 were found near those marking PfBiP, on membranous structures extending from the nuclear membrane (typical of the ER), thus confirming an ER localization for PfNT2 (Fig. 4). Deletion of the N-terminal domain of PfNT2 (first 30 amino acids preceding the first transmembrane domain) in the PfNT2-GFP construct abolished the ER localization of PfNT2 and resulted in diffuse cytoplasmic staining within the parasite (not shown).
FIGURE 2.
Immunolocalization of HA-PfNT2 and GFP-PfNT2. Anti-HA (A) or anti-GFP (B) antibody (green) localizes to a compartment within transgenic parasites expressing either HA-PfNT2 (A) or GFP-PfNT2 (B). The differential contrast image (DIC) images show the location of the parasites within the erythrocyte. The nucleic acids of the GFP-PfNT2 parasites were visualized using Hoescht 33258 (blue).
FIGURE 3.
Immunolocalization of C-terminal HA-tagged PfNT2 (PfNT2-HA) to the ER. Anti-BiP (red) colocalizes with anti-HA (green). Yellow fluorescence in merged images indicates areas of red (BiP) and green (PfNT2-HA) colocalization. The nucleic acids of the parasite were visualized using Hoescht 33258 (blue). DIC, differential contrast image.
FIGURE 4.
Immunoelectron microscopy indicates that PfNT2-GFP colocalizes with the ER marker PfBiP. Transmission electron micrograph shows ultrathin cryosections of the intraerythrocytic trophozoite stage of P. falciparum PfNT2-GFP transgenic parasites using anti-GFP and anti-PfBiP antibodies. The image shows immunogold labeling of anti-GFP (18-nm gold particles; indicated with arrowheads) and anti-PfBiP (12-nm gold particles) antibodies bound to intraerythrocytic P. falciparum. N, nucleus; NM, nuclear membrane; PV, parasitophorous vacuole; RBC, red blood cell cytoplasm; FV, food vacuole; H, hemozoin.
Functional Characterization of PfNT2 in Yeast
Despite previous success at expressing Plasmodium transport proteins in Xenopus oocytes (9, 10, 27), initial attempts at the functional expression of PfNT2 in this system were unsuccessful (data not shown). We therefore used yeast as a heterologous expression system to investigate the function of PfNT2. To facilitate its successful expression in yeast, the full codon sequence of the PfNT2 cDNA was redesigned to reduce its A+T content from 76.8% to 61.2%. A C-terminal V5 epitope tag was then added to allow localization in yeast. An immunoblot using anti-V5 antibody showed that the PfNT2 protein was expressed in yeast cells transformed with pYES2.1-PfNT2CO-V5 (Fig. 5A). Immunofluorescence analyses using anti-V5 monoclonal antibody and antibodies against the yeast ER marker Kar2 showed that full-length PfNT2 colocalizes with Kar2 (Fig. 5B).
FIGURE 5.
Immunofluorescence analysis of PfNT2 in yeast cells. A, imunoblotting using extracts from yeast cells transformed with either pYES2.1 (lane 1) or pYES2.1-PfNT2CO-V5 (lane 2). Cell lysates were subjected to SDS-PAGE, transferred to nitrocellulose, and probed with an anti-V5 antibody. B and C, immunofluorescence assays in yeast cells transformed with the plasmid encoding V5-tagged PfNT2CO (B) or V5-tagged PfNT2fcy2 (C). Colocalization studies were performed using anti-Kar2 (B) and anti-Pma1 (C) antibodies. Images represent localization of full-length PfNT2 and chimeric PfNT2fcy2 in end3Δ yeast cells. Similar results were obtained in wild type cells (data not shown). DIC, differential contrast image.
To evaluate PfNT2 as a nucleoside/nucleobase transporter, its coding region was modified with the aim of redirecting it to the yeast plasma membrane. Previous studies on the human (h)ENT3 protein revealed that the N-terminal peptide sequence was responsible for its intracellular localization (24). Accordingly, we substituted the PfNT2 N-terminal domain with that of the yeast plasma membrane protein Fcy2 (creating PfNT2fcy2) and expressed the chimeric protein in yeast. Immunofluorescence analysis showed that PfNT2fcy2 localizes to the yeast plasma membrane. This localization pattern is similar to that of Pma1, a yeast plasma membrane marker (28) (Fig. 5C). The images shown in Fig. 5 were obtained with end3Δ yeast cells which lack the END3 gene involved in endocytosis (29). Very similar results were obtained using wild type cells (not shown).
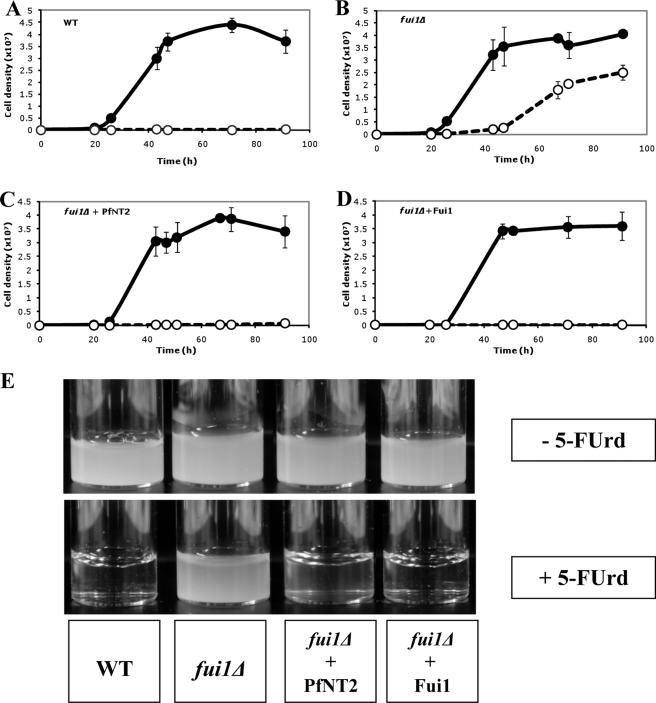
Wild type yeast are sensitive to the toxic nucleoside analog 5-FUrd. The yeast strain fui1Δ lacks the high affinity plasma membrane uridine permease Fui1; it is, as a result, deficient in uridine transport and, consequently, resistant to 5-FUrd (30). We used the PfNT2fcy2 fusion protein to determine whether expression of PfNT2 at the surface of the yeast cell can, by complementation, reverse the 5-FUrd insensitivity seen in fui1Δ yeast cells. Wild type, fui1Δ, fui1Δ+PfNT2fcy2, and fui1Δ+Fui1 yeast strains inoculated at an approximate density of 104 cells/ml and grown in the absence of 5-FUrd exhibited similar growth patterns, achieving cell densities of greater than 9 × 107 cells/ml at 43 h of growth after which the growth of these strains remained unchanged as the cells entered the stationary phase (Fig. 6, A–D). In the presence of 5-FUrd, the growth of wild type, fui1Δ-PfNT2fcy2, and fui1Δ-Fui1 strains was severely inhibited, with cells reaching a density of ∼1.5 × 105 cells/ml, ∼5.5 × 105 cells/ml, and ∼1.2 × 105 cells/ml, respectively, at 43 h and ∼1.5 × 105 cells/ml, ∼5.5 × 105 cells/ml and ∼1.2 × 105 cells/ml, respectively, after 91 h of incubation (Fig. 6, A, C, and D). By contrast, the fui1Δ strain reached a cell density of ∼1.9 × 106 cells/ml after 43 h and ∼2.5 × 107 cells/ml after 91 h (Fig. 6B). This finding suggests that expression of PfNT2fcy2 on the plasma membrane of fui1Δ cells resulted in the increased uptake of 5-FUrd by these (i.e. the fui1Δ-PfNT2fcy2) cells.
FIGURE 6.
Growth of different yeast strains in the absence or presence of 5-FUrd. Cultures were inoculated with 104 cells/ml of yeast cells in Gal/Raf medium alone (filled symbols) or Gal/Raf medium supplemented with 3.8 mm 5-FUrd (open symbols), and yeast multiplication was monitored by measuring optical density at 660 nm. Measurements were taken in triplicate and are shown as the average ± S.D. (error bars). The yeast strains used were wild type (WT) (BY4741) harboring an empty vector (A), fui1Δ cells harboring an empty vector (B), fui1Δ-PfNT2fcy2 strain (C), or fui1Δ cells expressing Fui1 (D). Growth experiments were performed several times (at least five times), and all studies demonstrated an increase in sensitivity of the fui1Δ-PfNT2fcy2 strain to 5-FUrd. E, photographs of liquid cultures taken at the 91-h time point.
DISCUSSION
The localization of PfNT2 to the ER, although unexpected, was not unprecedented, as several nucleoside transporters are known to function as intracellular transporters. Yeast FUN26 and mammalian ENT3 proteins are found on lysosomal membranes where they are thought to be responsible for the release of nucleosides from intracellular compartments following nucleic acid breakdown (30, 24). The human ENT4 protein localizes to intracellular vesicles as well as the plasma membrane, and this localization appears to be dynamic (31).
Mutations in the lysosomal hENT3 protein are associated with H-syndrome, an autosomal recessive disorder characterized by various symptoms including, but not limited to, hyperpigmentation, hypertrichosis, heart anomalies, and hearing loss (32), and with the related pigmented hypertrichotic dermatosis with insulin-dependent diabetes (PHID) syndrome (33). The fact that the only mammalian nucleoside transporter yet to be linked to any disorders resides on intracellular membranes suggests that intracellular nucleoside transport could be an important physiological phenomenon.
Recently, a splice variant of human concentrative nucleoside transporter 3 was localized exclusively to the endoplasmic reticulum where it was shown to mediate the accumulation of nucleosides and nucleobases (34). Our finding that PfNT2 localizes to the ER is a first for a member of the ENT family and suggests a novel role for this organelle in parasite biology.
Despite several attempts, we were unable to generate antibodies to localize the native PfNT2 protein. We therefore used an epitope tag-based approach, which has been used with much success in the localization of other Plasmodium proteins (20, 27, 35, 36). We used multiple tagged constructs (GFP, N- and C-terminal HA) to address the possibility that the addition of a tag to the protein resulted in mislocalization. The observation that all three tagged PfNT2 constructs localized to the ER when expressed in transgenic parasites suggests that the tags did not cause the protein to be mistargeted. Deletion of the N-terminal domain of PfNT2 abolished ER localization in P. falciparum and resulted in diffuse staining in the parasite cytoplasm (not shown). The N-terminal domain may therefore play an important role in ER targeting or retention. Further investigations of this are warranted.
The intracellular localization of a full-length, epitope-tagged PfNT2 protein expressed in yeast cells (Fig. 5B) is consistent with our intracellular localization of epitope-tagged PfNT2 in Plasmodium parasites. Substitution of the N-terminal domain of PfNT2 with that of the yeast Fcy2 resulted in successful localization of the chimeric protein to the yeast plasma membrane. The observation of similar localization patterns in wild type cells and end3Δ yeast cells lacking the END3 gene (29) suggests that PfNT2fcy2 membrane localization is not controlled by an End3-dependent endocytic or degradation machinery.
PfNT2fcy2 plasma membrane localization resulted in 5-FUrd sensitivity on otherwise resistant fui1Δ yeast cells, consistent with PfNT2 mediating the entry of this compound into the yeast cells. Attempts to measure the transport of labeled uridine as well as other purine nucleosides and nucleobases across the yeast plasma membrane of fui1Δ and fui1Δ-PfNT2fcy2 cells were unsuccessful, with no uptake detected despite incubation periods of up to 1 h. Whether the absence of detectable uridine transport in fui1Δ-PfNT2fcy2 cells was due to low expression levels or due to uridine being a poor substrate for this transporter is unclear.
Our finding that PfNT2 is an ER nucleoside transporter raises the possibility that this organelle may act as an intracellular purine store. The concept of the ER accumulating nucleosides and nucleobases is supported by the recent localization of a hCNT3 splice variant to this organelle (34). In some lower eukaryotes, specialized organelles and specific transport proteins mediate detoxification processes for compounds such as transition metals and purine analogs (37–39). As such, the possibility that the ER, together with PfNT2, may play a role in purine or purine product detoxification in P. falciparum warrants further investigation. Future genetic analyses will aim to delete PfNT2 to evaluate its importance in the metabolism, storage, or detoxification of purines and their products, as well as to assess its function during parasite development, multiplication and survival.
PfNT2 homologs are found in other apicomplexa; the protein shows 55–60% identify and 70–80% similarity with orthologs from Plasmodium species P. knowlesi, P. vivax, P. berghei, and P. chabaudi; and 18% identity and 38% similarity with a protein in Toxoplasma gondii (GenBank no. EEE 22737.1). The function of these proteins might therefore be conserved among apicomplexan parasites.
Supplementary Material
Acknowledgments
We thank Dr. Seong-Kyoun Kim for assistance with plasmid construction and Plasmodium transfection. We are grateful to the late Dr. Kylie Mullin and to Prof. Geoff McFadden for help with parasite immunofluorescence assays and to the Australian Red Cross Blood Transfusion Service for the provision of blood to the Australian National University authors.
This work was supported, in whole or in part, by National Institutes of Health Grant AI51507 through the NIAID (to C. B. M.), Department of Defense Grant PR033005 (to C. B. M.), by the Burroughs Wellcome Fund (to C. B. M), and by grants from the Australian Research Council and National Health and Medical Research Council (to K. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1 and Table 1.
- ENT
- equilibrative nucleoside transporter
- ER
- endoplasmic reticulum
- 5-FUrd
- 5-fluorouridine
- HA
- hemagglutinin
- GFP
- green fluorescent protein
- PBS
- phosphate-buffered saline.
REFERENCES
- 1.Quashie N. B., Ranford-Cartwright L. C., de Koning H. P. (2010) Malar. J. 9, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gero A. M., Bugledich E. M., Paterson A. R., Jamieson G. P. (1988) Mol. Biochem. Parasitol. 27, 159–170 [DOI] [PubMed] [Google Scholar]
- 3.Gati W. P., Lin A. N., Wang T. I., Young J. D., Paterson A. R. (1990) Biochem. J. 272, 277–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirk K., Horner H. A., Elford B. C., Ellory J. C., Newbold C. I. (1994) J. Biol. Chem. 269, 3339–3347 [PubMed] [Google Scholar]
- 5.Upston J. M., Gero A. M. (1995) Biochim. Biophys. Acta 1236, 249–258 [DOI] [PubMed] [Google Scholar]
- 6.Desai S. A., Krogstad D. J., McCleskey E. W. (1993) Nature 362, 643–646 [DOI] [PubMed] [Google Scholar]
- 7.Downie M. J., Saliba K. J., Howitt S. M., Bröer S., Kirk K. (2006) Mol. Microbiol. 60, 738–748 [DOI] [PubMed] [Google Scholar]
- 8.Downie M. J., Saliba K. J., Bröer S., Howitt S. M., Kirk K. (2008) Int. J. Parasitol. 38, 203–209 [DOI] [PubMed] [Google Scholar]
- 9.Carter N. S., Ben Mamoun C., Liu W., Silva E. O., Landfear S. M., Goldberg D. E., Ullman B. (2000) J. Biol. Chem. 275, 10683–10691 [DOI] [PubMed] [Google Scholar]
- 10.Parker M. D., Hyde R. J., Yao S. Y., McRobert L., Cass C. E., Young J. D., McConkey G. A., Baldwin S. A. (2000) Biochem. J. 349, 67–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rager N., Mamoun C. B., Carter N. S., Goldberg D. E., Ullman B. (2001) J. Biol. Chem. 276, 41095–41099 [DOI] [PubMed] [Google Scholar]
- 12.El Bissati K., Zufferey R., Witola W. H., Carter N. S., Ullman B., Ben Mamoun C. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 9286–9291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El Bissati K., Downie M. J., Kim S. K., Horowitz M., Carter N., Ullman B., Ben Mamoun C. (2008) Mol Biochem. Parasitol. 161, 130–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quashie N. B., Dorin-Semblat D., Bray P. G., Biagini G. A., Doerig C., Ranford-Cartwright L. C., De Koning H. P. (2008) Biochem. J. 411, 287–295 [DOI] [PubMed] [Google Scholar]
- 15.Kirk K., Howitt S. M., Broer S., Saliba K. J., Downie M. J. (2009) Trends Parasitol. 25, 246–249 [DOI] [PubMed] [Google Scholar]
- 16.Riegelhaupt P. M., Cassera M. B., Frohlich R. F., Hazleton K. Z., Hefter J. J., Schramm V. L., Akabas M. H. (2010) Mol. Biochem. Parasitol. 169, 40–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gardner M. J., Hall N., Fung E., White O., Berriman M., Hyman R. W., Carlton J. M., Pain A., Nelson K. E., Bowman S., Paulsen I. T., James K., Eisen J. A., Rutherford K., Salzberg S. L., Craig A., Kyes S., Chan M. S., Nene V., Shallom S. J., Suh B., Peterson J., Angiuoli S., Pertea M., Allen J., Selengut J., Haft D., Mather M. W., Vaidya A. B., Martin D. M., Fairlamb A. H., Fraunholz M. J., Roos D. S., Ralph S. A., McFadden G. I., Cummings L. M., Subramanian G. M., Mungall C., Venter J. C., Carucci D. J., Hoffman S. L., Newbold C., Davis R. W., Fraser C. M., Barrell B. (2002) Nature 419, 498–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin R. E., Henry R. I., Abbey J. L., Clements J. D., Kirk K. (2005) Genome Biol. 6, R26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trager W., Jensen J. B. (1976) Science 193, 673–675 [DOI] [PubMed] [Google Scholar]
- 20.Mullin K. A., Lim L., Ralph S. A., Spurck T. P., Handman E., McFadden G. I. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 9572–9577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tonkin C. J., Pearce J. A., McFadden G. I., Cowman A. F. (2006) Curr. Opin. Microbiol. 9, 381–387 [DOI] [PubMed] [Google Scholar]
- 22.Miller C. A., 3rd, Martinat M. A., Hyman L. E. (1998) Nucleic Acids Res. 26, 3577–3583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tonkin C. J., van Dooren G. G., Spurck T. P., Struck N. S., Good R. T., Handman E., Cowman A. F., McFadden G. I. (2004) Mol. Biochem. Parasitol. 137, 13–21 [DOI] [PubMed] [Google Scholar]
- 24.Baldwin S. A., Yao S. Y., Hyde R. J., Ng A. M., Foppolo S., Barnes K., Ritzel M. W., Cass C. E., Young J. D. (2005) J. Biol. Chem. 280, 15880–15887 [DOI] [PubMed] [Google Scholar]
- 25.van Dooren G. G., Marti M., Tonkin C. J., Stimmler L. M., Cowman A. F., McFadden G. I. (2005) Mol. Microbiol. 57, 405–419 [DOI] [PubMed] [Google Scholar]
- 26.Kumar N., Koski G., Harada M., Aikawa M., Zheng H. (1991) Mol. Biochem. Parasitol. 48, 47–58 [DOI] [PubMed] [Google Scholar]
- 27.Saliba K. J., Martin R. E., Bröer A., Henry R. I., McCarthy C. S., Downie M. J., Allen R. J., Mullin K. A., McFadden G. I., Bröer S., Kirk K. (2006) Nature 443, 582–585 [DOI] [PubMed] [Google Scholar]
- 28.Serrano R., Kielland-Brandt M. C., Fink G. R. (1986) Nature 319, 689–693 [DOI] [PubMed] [Google Scholar]
- 29.Benedetti H., Raths S., Crausaz F., Riezman H. (1994) Mol. Biol. Cell 5, 1023–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vickers M. F., Yao S. Y., Baldwin S. A., Young J. D., Cass C. E. (2000) J. Biol. Chem. 275, 25931–25938 [DOI] [PubMed] [Google Scholar]
- 31.Barnes K., Dobrzynski H., Foppolo S., Beal P. R., Ismat F., Scullion E. R., Sun L., Tellez J., Ritzel M. W., Claycomb W. C., Cass C. E., Young J. D., Billeter-Clark R., Boyett M. R., Baldwin S. A. (2006) Circ. Res. 99, 510–519 [DOI] [PubMed] [Google Scholar]
- 32.Molho-Pessach V., Lerer I., Abeliovich D., Agha Z., Abu Libdeh A., Broshtilova V., Elpeleg O., Zlotogorski A. (2008) Am. J. Hum. Genet. 83, 529–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cliffe S. T., Kramer J. M., Hussain K., Robben J. H., de Jong E. K., de Brouwer A. P., Nibbeling E., Kamsteeg E. J., Wong M., Prendiville J., James C., Padidela R., Becknell C., van Bokhoven H., Deen P. M., Hennekam R. C., Lindeman R., Schenck A., Roscioli T., Buckley M. F. (2009) Hum. Mol. Genet. 18, 2257–2265 [DOI] [PubMed] [Google Scholar]
- 34.Errasti-Murugarren E., Molina-Arcas M., Casado F. J., Pastor- Anglada M. (2009) FASEB J. 23, 172–182 [DOI] [PubMed] [Google Scholar]
- 35.Henry R. I., Martin R. E., Howitt S. M., Kirk K. (2007) Biochem. Biophys. Res. Commun. 363, 288–291 [DOI] [PubMed] [Google Scholar]
- 36.Martin R. E., Marchetti R. V., Cowan A. I., Howitt S. M., Broer S., Kirk K. (2009) Science 325, 1680–1682 [DOI] [PubMed] [Google Scholar]
- 37.Ortiz D. F., Kreppel L., Speiser D. M., Scheel G., McDonald G., Ow D. W. (1992) EMBO J. 11, 3491–3499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ko N., Nishihama R., Pringle J. R. (2008) Yeast 25, 155–160 [DOI] [PubMed] [Google Scholar]
- 39.Lauer C. M., Jr., Bonatto D., Mielniczki-Pereira A. A., Schuch A. Z., Dias J. F., Yoneama M. L., Pegas Henriques J. A. (2008) FEMS Microbiol. Lett. 285, 79–88 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.