SUMMARY
Understanding liver development should lead to greater insights into liver diseases and improve therapeutic strategies. In a forward genetic screen for genes regulating liver development in zebrafish, we identified a mutant – oliver – that exhibits liver-specific defects. In oliver mutants, the liver is specified, bile ducts form and hepatocytes differentiate. However, the hepatocytes die shortly after their differentiation, and thus the resulting mutant liver consists mainly of biliary tissue. We identified a mutation in the gene encoding translocase of the outer mitochondrial membrane 22 (Tomm22) as responsible for this phenotype. Mutations in tomm genes have been associated with mitochondrial dysfunction, but most studies on the effect of defective mitochondrial protein translocation have been carried out in cultured cells or unicellular organisms. Therefore, the tomm22 mutant represents an important vertebrate genetic model to study mitochondrial biology and hepatic mitochondrial diseases. We further found that the temporary knockdown of Tomm22 levels by morpholino antisense oligonucleotides causes a specific hepatocyte degeneration phenotype that is reversible: new hepatocytes repopulate the liver as Tomm22 recovers to wild-type levels. The specificity and reversibility of hepatocyte ablation after temporary knockdown of Tomm22 provides an additional model to study liver regeneration, under conditions where most hepatocytes have died. We used this regeneration model to analyze the signaling commonalities between hepatocyte development and regeneration.
INTRODUCTION
The liver is a crucial organ with a central role in metabolic homeostasis: it is responsible for the metabolism, storage and redistribution of nutrients and it also removes waste and xenobiotics by metabolic conversion and biliary excretion (for a review, see Saxena et al., 2003). Reduced liver function carries severe consequences and, if not reversed, can be a primary cause of death. A thorough understanding of liver development should provide a valuable basis for the comprehension of disease onset and progression, and help design successful therapeutic approaches for liver pathologies. Several studies in the mouse, chick, zebrafish (Danio rerio), frog (Xenopus laevis) and medaka (Oryzia latipes) have contributed to our understanding of this process and have made use of reverse and forward genetic approaches, as well as embryological and in vitro studies (for reviews, see Zhao and Duncan, 2005; Zaret and Grompe, 2008; Nakamura and Nishina, 2009). However, much is yet to be uncovered regarding the cellular and molecular mechanisms regulating liver development.
Analyzing organ regeneration can provide important insights into therapeutics for pathological conditions that can benefit from tissue repair. For example, a better understanding of liver regeneration should enable the development of therapeutic strategies to promote endogenous liver regeneration in patients with liver diseases and, in addition, help to increase the number of liver transplantations using partial livers of living donors. As the main detoxifying organ of the body, the liver is likely to be injured by ingested toxins. This organ has, however, a remarkable ability to regenerate in response to injury caused by toxic exposure, partial hepatectomy (PHx) or virus infection (for reviews, see Fausto et al., 1995; Michalopoulos and DeFrances, 1997). The mechanisms responsible for liver mass recovery seem to be highly dependent on the form of injury. After most forms of injury, including PHx, the regenerative process relies on the replication of the remaining hepatocytes, whereas after acute liver failure, a common clinical presentation of liver disease in humans, the liver repopulation process appears to involve the differentiation of intrahepatic progenitor cells (for a review, see Fausto et al., 2006). Despite intensive research in the last few decades, and although the process of liver regeneration has been described extensively, the molecular mechanisms underlying this phenomenon are still not clearly understood, and the exact origin of the newly formed cells is still debated.
A number of assays have been developed to damage the liver and subsequently study its regeneration. A 70% hepatectomy procedure (PHx resection) is commonly used in rodents to study this process (for a review, see Martins et al., 2008), and has also been applied successfully in zebrafish (Sadler et al., 2007; Goessling et al., 2008). This approach is, however, quite laborious and time consuming, and thus is mostly applicable to a limited number of animals and mainly used to uncover the role of genes that have already been identified. As an alternative approach, we have developed a genetic/pharmacological protocol to ablate hepatocytes specifically and in an inducible manner (Curado et al., 2007; Curado et al., 2008). This assay relies on the liver-specific expression of the bacterial nitroreductase (NTR) enzyme, which converts the non-toxic metronidazole (Mtz) into a noxious substance, ablating exclusively the cells that express NTR. The design of the NTR-Mtz system makes it suitable to damage a large number of larvae and it is therefore compatible with forward genetic and pharmacological screens. However, the NTR-Mtz system does not seem to be as efficient in ablating hepatocytes compared with other cell types, such as cardiomyocytes or pancreatic β cells (Curado et al., 2007; Pisharath et al., 2007), possibly because Mtz is being metabolized by the hepatocytes themselves, thereby lowering its effective concentration.
Here, we report a surprisingly specific role for the mitochondrial import gene tomm22 in hepatocyte survival. In addition, we use this finding to generate an alternative model of liver regeneration: the transient knockdown of Tomm22 results in hepatocyte ablation followed by recovery. Using this model, we identify a role for Wnt2b and fibroblast growth factor (Fgf) signaling in hepatocyte regeneration.
RESULTS
oliver mutants show a liver-specific phenotype
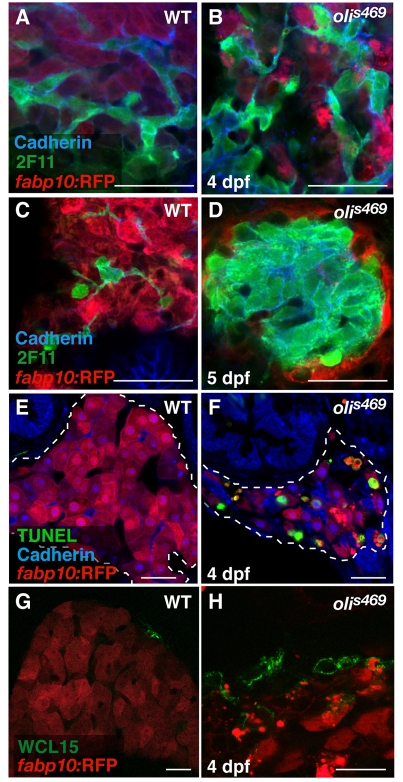
olivers469 (olis469) was identified in a forward genetic screen for genes regulating the development of endodermal organs, such as the liver, pancreas and gut (Ober et al., 2006). Screening the progeny of N-ethyl-N-nitrosourea (ENU)-mutagenized Tg(XlEef1a1:GFP)s854 zebrafish, which express green fluorescent protein (GFP) throughout the developing endoderm (Field et al., 2003), led to the identification of the recessive mutant olis469, which exhibits a smaller liver compared with wild-type (WT) siblings. We further examined the liver phenotype of olis469 mutants using the Tg(–2.8fabp10:EGFP)as3 line (Her et al., 2003), which expresses enhanced GFP (EGFP) specifically in differentiated hepatocytes, combining bright-field and fluorescence imaging. At 6 days post-fertilization (dpf), the mutant larvae (Fig. 1A) exhibit an o-shaped liver (o-liver) of drastically reduced size compared with their WT siblings (Fig. 1A). At the morphological level, only the liver appears to be affected in olis469 mutants; olis469 mutant larvae survive until 9–11 dpf without showing any other obvious phenotype. At this stage, most mutant larvae have inflated their swim bladder although they are thinner than their WT siblings, probably owing to the lack of a functional liver. To gain further insight into the liver defect, we stained 5-dpf Tg(–2.8fabp10:EGFP)as3; olis469 larvae with phalloidin and the nuclear marker TO-PRO. Whereas, at this stage, WT livers consist of multiple hepatocytes that are well organized in a sheet-like structure (Fig. 1B), olis469 mutant livers show a very distinct hepatic structure, where only a few hepatocytes can be detected (Fig. 1C). Confocal imaging of 5-dpf mutant larvae expressing red fluorescent protein (RFP) in hepatocytes [Tg(fabp10:RFP)gz12] (Farooq et al., 2008), stained with an antibody against cadherin and with TO-PRO, revealed that the cells that were surrounded by only a few hepatocytes (Fig. 1E) expressed high cadherin levels, which, at this stage in WT livers, are only present in the biliary duct cells (Fig. 1D). Moreover, analysis of the hepatic structure in olis469 mutants clearly shows that, not only is the overall size and organization of the liver affected in these mutants, but the ratio between bile duct cells and hepatocytes is also altered, with a much higher number of bile duct cells compared with hepatocytes.
Fig. 1.
oliver mutants show a liver-specific phenotype. (A) Bright-field images of 6-dpf Tg(–2.8fabp10:EGFP)as3WT (top) and olis469 mutant (bottom) larvae combined with fluorescence to reveal their liver (arrows). By 6 dpf, and compared with their WT siblings, olis469 mutants (bottom) exhibit an o-shaped liver of reduced size (arrow) in an otherwise phenotypically WT body. (B,C) Confocal images of 5-dpf Tg(–2.8fabp10:EGFP)as3WT (B) and olis469 mutant (C) livers stained with phalloidin (red) and TO-PRO (blue). EGFP expression (green) in hepatocytes reveals a well-organized structure in the WT liver, contrasting with the disarray observed in olis469 mutants. (D,E) Confocal images of 5-dpf Tg(fabp10:RFP)gz12WT (D) and olis469 mutant (E) livers stained for cadherin (green) and TO-PRO (blue). The cells surrounded by the few hepatocytes (red) that are detected in olis469 mutant livers express cadherin (green) at levels that, at this stage in WT livers, are characteristic of cells of the intrahepatic biliary tree (D, arrows). Bars, 20 μm.
To further understand the origin of the olis469 mutant liver phenotype, we examined the hepatic structure at earlier stages. At 4 dpf, both WT and olis469 mutant livers exhibit a mesh-like intrahepatic biliary duct structure, highlighted by cadherin and 2F11 staining (Fig. 2A,B). Twenty-four hours later, in olis469 mutant livers, this biliary duct structure forms a core of cadherin/2F11-positive cells surrounded by a few remaining hepatocytes (Fig. 2D), as opposed to the WT liver structure, where the hepatic biliary ducts and hepatocytes intermix (Fig. 2C).
Fig. 2.
The oliver mutant phenotype is caused by hepatocyte apoptosis. (A–D) Confocal images of 4-dpf (A,B) and 5-dpf (C,D) Tg(fabp10:RFP)gz12 WT (A,C) and olis469 mutant (B,D) livers stained for cadherin (blue) and 2F11 (green). At 4 dpf, both WT and olis469 mutant livers exhibit a distinct intrahepatic biliary tree (highlighted by the expression of cadherin and 2F11), intercalated by hepatocytes (red). However, 24 hours later (5 dpf), the biliary tree in olis469 mutants has turned into a dense cellular mass. (E,F) Confocal images of transverse sections of 4-dpf Tg(fabp10:RFP)gz12WT (E) and olis469 mutant (F) livers stained for cadherin (blue) and TUNEL (green); the livers are outlined by dashed lines. At 4 dpf, a large number of hepatocytes (red) undergo apoptosis in olis469 mutant livers (F), whereas no TUNEL staining can be detected in the livers of their WT siblings (E). (G,H) Confocal images of 4-dpf Tg(fabp10:RFP)gz12WT (G) and olis469 mutant (H) livers stained with the macrophage antibody WCL15 (green). As hepatocytes (red) undergo apoptosis in olis469 mutant livers, multiple macrophages (green) can be detected engulfing them (H), whereas in their WT siblings, macrophages can only very occasionally be detected in the periphery of the liver (G). Bars, 20 μm.
The oliver mutant phenotype is caused by hepatocyte apoptosis
Both bile duct cells and hepatocytes derive from hepatoblasts, which in turn originate from the primary liver bud endoderm (Zhao and Duncan, 2005). There are at least two possible explanations for the development of the abnormal bile duct cell:hepatocyte ratio that is observed in olis469 mutants: hepatocyte-specific apoptosis or a bias of hepatoblast differentiation towards the bile duct lineage. To test these hypotheses, we examined the extent of apoptosis using TUNEL (TdT-mediated dUTP nick-end labeling). Whereas no TUNEL staining was detected in WT livers (Fig. 2E), we observed many TUNEL-positive hepatocytes [marked by Tg(fabp10:RFP) expression] in olis469 mutants (Fig. 2F). Confocal imaging of 4-dpf TUNEL-stained sections clearly shows that, in tissues other than the liver (such as the gut), the low number of apoptotic cells detected in olis469 mutants is similar to that observed in WT siblings (supplementary material Fig. S1A,B). In addition, using the monoclonal antibody WCL15 (Romano et al., 1998; van der Sar et al., 2004), we detected several macrophages engulfing dying hepatocytes in olis469 mutants (Fig. 2H), whereas in WT livers macrophages can only very occasionally be observed in the periphery of the liver (Fig. 2G). In addition, to assess hepatocyte differentiation, we examined the distribution of the bile salt export pump Abcb11 (Gerloff et al., 1998; Sakaguchi et al., 2008) at 4 dpf, and observed clear labeling of mutant hepatocytes with this marker (data not shown). Altogether, these results indicate that the mutant hepatocytes are specified, differentiate and then undergo apoptosis. Although, at 4 dpf, olis469mutant livers exhibit a WT-like cellular organization with sheets of hepatocytes separated by biliary ducts, owing to hepatocyte apoptosis, their size is already much reduced compared with WT siblings.
tomm22 is essential for hepatocyte survival
To isolate the gene disrupted by the olis469 mutation we used a standard set of simple sequence length polymorphism (SSLP) markers and placed the olis469 locus on linkage group 12. Further mapping positioned this mutation within a 140-kb genomic interval containing eight genes (supplementary material Fig. S2A). Comparative cDNA sequence analysis revealed a nucleotide change from thymine in WT fish, to cytosine in olis469 mutants, in the translocase of the outer mitochondrial membrane 22 (tomm22) gene (supplementary material Fig. S2B). This nucleotide change results in the substitution of a stop codon with an arginine residue, leading to a predicted mutant protein that is 47 amino acids longer than the WT protein (supplementary material Fig. S2C).
Tomm22 is part of the multiprotein translocation complex of the outer mitochondrial membrane (OMM) that regulates protein traffic from the cytoplasm into mitochondria. Mitochondrial import is crucial, as the majority of mitochondrial proteins are encoded by nuclear DNA and thus need to be imported after cytosolic translation. Tomm22 acts as a central receptor and a trans binding site for this translocation, as well as a docking point for other peripheral Tomm components of the mitochondrial import pore (for a review, see Koehler, 2004). To date, only one zebrafish tomm22 gene has been identified.
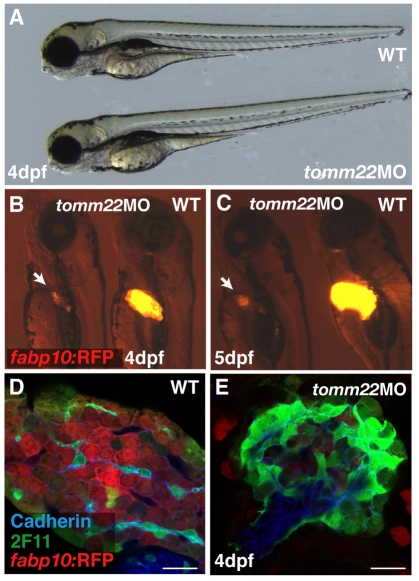
Interference with tomm22 translation (or knockdown of the Tomm22 protein), using 4 ng of an ATG-targeting morpholino antisense oligonucleotide (MO), accurately reproduced the olis469 phenotype (Fig. 3A–C). Confocal analysis of these morphants showed that their livers consisted of a mass of cadherin/2F11-positive cells surrounded by a few hepatocytes (Fig. 3E), as detected in olis469 mutants (Fig. 3D). These results confirm that the liver phenotype observed in olis469 mutants is caused by the disruption of the tomm22 gene.
Fig. 3.
tomm22 MO injections reproduce the olis469 liver-specific phenotype. (A) Bright-field image of a 4-dpf uninjected control larva (top) and a 4-dpf larva injected with 4 ng of tomm22 MO (bottom). (B,C) Fluorescence images of 4-dpf (B) and 5-dpf (C) Tg(fabp10:RFP)gz12 larvae injected with 4 ng of tomm22 MO (left) and uninjected controls (right). The 4-dpf and 5-dpf tomm22 morphants show a liver-specific phenotype: the size of the liver is much reduced (arrows in B,C) in an otherwise WT-like body (A, bottom) compared with the uninjected controls. (D,E) Confocal images of 4-dpf Tg(fabp10:RFP)gz12 livers from uninjected (D) and tomm22 MO-injected (E) animals stained for cadherin (blue) and 2F11 (green). The hepatic structure of the tomm22 morphants, similar to that observed in olis469 mutants, consists of a mass of cadherin- and 2F11-positive cells surrounded by residual hepatocytes (in red) (E), in contrast to the cellular organization that is observed in uninjected larvae (D). Bars, 20 μm.
The tight genetic linkage, presence of a significant molecular lesion in the tomm22s469 coding sequence, and the tomm22 MO phenocopy indicate that a disruption in the tomm22 gene is the cause of the olis469 mutant phenotype and, therefore, that tomm22 plays a crucial role in hepatocyte survival.
Like the liver, the heart has a high demand for mitochondrial function. However, tomm22s469 mutant hearts show no obvious morphological defects or function impairment (data not shown).
Tomm22s469 mutant protein is correctly inserted into the yeast OMM
Like the majority of the mitochondrial proteins, Tomm22 is encoded by a nuclear gene. Tomm22 is initially translated in the cytosol, then imported into the mitochondria, and integrated in the OMM (Becker et al., 2009). To investigate whether the phenotype observed in tomm22s469 mutants was the result of inappropriate targeting of the Tomm22s469 mutant protein, we used a yeast in vitro import system (Hwang et al., 2007). Radiolabeled WT or Tomm22s469 mutant protein was imported into WT yeast mitochondria and the import reaction was split and carbonate-extracted at various pHs (supplementary material Fig. S3A). Carbonate removes proteins from the mitochondrial membranes (pellet fraction) with increasing pH, whereas soluble proteins and unimported proteins remain in the supernatant fraction. We found that, similar to the WT protein, the imported Tomm22s469 mutant protein remained associated with the membrane fraction even at the high pH of 12.5. In addition, a control of carbonate extraction in the absence of mitochondria showed that this pellet association was not simply the result of protein aggregation (data not shown). To further test that Tomm22s469 was indeed in the OMM, we performed import assays and then added trypsin to degrade the exposed proteins (supplementary material Fig. S3B). Tomm22s469, like WT Tomm22 and the yeast ortholog Tom22, was degraded in these experiments, indicating that it localizes to the OMM. Taken together, these results indicate that Tomm22s469 is correctly imported and localized to the yeast OMM, like the WT form. Thus, the lack of Tomm22s469 function in olis469 mutants is likely to be the result of an effect other than inappropriate targeting.
tomm22 is expressed highly in the endoderm
We next investigated tomm22 expression in WT larvae using a tomm22-specific probe, as well as a control sense probe. Throughout 3–5 dpf, tomm22 appears to be highly expressed in the endoderm-derived organs – the liver, gut and, although not as strongly, the pancreas – as compared with other tissues (supplementary material Fig. S4A–D). This staining pattern was clearly absent when using the sense control probe (supplementary material Fig. S4E–G’). tomm22 expression was also detected in the head, although weaker nonspecific staining in the head could also be detected in the sense control. This enriched expression in endodermal organs during embryogenesis is in contrast to the low level of ubiquitous expression that is seen in mature mammals (Unigene; www.ncbi.nlm.nih.gov/unigene).
Tomm22 transient knockdown provides an additional model to study liver regeneration
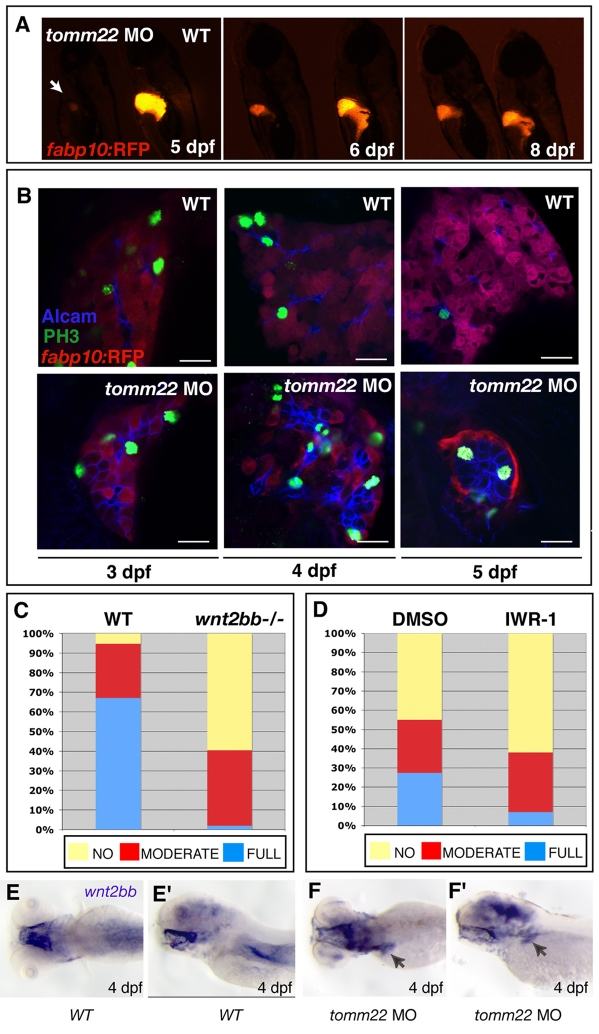
Injection of the tomm22 MO results in the specific apoptosis of hepatocytes, and can therefore be used as a tool to ablate hepatocytes. Contrary to the permanent absence of Tomm22 in tomm22s469mutant larvae, the knockdown of Tomm22 levels using a tomm22 MO is transient, as this reagent is gradually depleted. Thus, whereas the liver phenotype in tomm22s469 mutants is irreversible, once Tomm22 levels are re-established in tomm22 morphants, the degenerated liver recovers its size and structure. tomm22 morphants, which show a smaller liver with a reduced number of hepatocytes at 5 dpf, show a recovering hepatic mass at 6 dpf and, by 8 dpf, start resembling uninjected controls (Fig. 4A). The ability to ablate hepatocytes specifically using a tomm22 MO, together with the reversibility of its effect, make the tomm22 MO-based assay a useful model to study liver regeneration when hepatocyte replication is absent or impaired.
Fig. 4.
tomm22 MO provides a model to study liver regeneration and uncovers an involvement of the Wnt2b signaling pathway. (A) Fluorescence images of 5-, 6- and 8-dpf (left, middle and right images, respectively) Tg(fabp10:RFP)gz12 larvae injected with 4 ng of tomm22 MO (left side of each image) and uninjected control larvae (right side of each image). At 5 dpf, tomm22 morphants show a smaller liver (arrow); as the tomm22 MO is diluted and Tomm22 levels recover, the tomm22 morphant livers start recovering their size and structure, which is already obvious at 6 dpf, and are comparable to uninjected controls at 8 dpf. (B) Confocal images of the livers of Tg(fabp10:RFP)gz12WT (top) and tomm22 MO-injected (bottom) larvae at 3, 4 and 5 dpf (left, middle and right panels, respectively) stained for PH3 (green) and Alcam (blue); hepatocytes are stained red. Cellular proliferation occurs at a higher rate in post-ablated livers compared with WT livers. Bars, 20 μm. (C) Comparison of hepatic recovery post-tomm22 MO-mediated ablation in the WT (left bar, n=41) versus wnt2bb−/− (right bar, n=42) background. tomm22 morphants, which at 4 dpf showed severe hepatic ablation, were allowed to recover and at 6 dpf their liver recovery was classified as (1) ‘no’ change in liver size, (2) ‘moderate’ recovery of hepatic volume, or (3) ‘full’ recovery, i.e. comparable to that observed at 6 dpf in most WT larvae injected with 4 ng of tomm22 MO. In the absence of wnt2bb function, ‘full’ recovery (blue) decreased from 67.1% to 1.9%. Conversely, ‘no’ recovery (yellow) increased from 5.2% in WT larvae to 59.5% in wnt2bb−/− larvae. ‘Moderate’ recovery changed from 27.7% in WT larvae to 38.6% in wnt2bb−/− larvae. (D) Comparison of hepatic recovery post-tomm22 MO-mediated ablation in larvae that were allowed to recover in carrier solution (DMSO; left bar, n=73) compared with in the presence of 5 μM of the Wnt–β-catenin pathway inhibitor IWR-1 (right bar, n=82). Recovery rates were classified as in C. In the presence of the Wnt–β-catenin pathway inhibitor IWR-1, ‘full’ recovery decreased compared with DMSO (from 27.5% to 7%), whereas the ‘no’ recovery rate increased from 45% (DMSO) to 62% (5 μM IWR-1). No clear difference was observed in the ‘moderate’ recovery category (27.5% to 31%). (E–F’) Expression of wnt2bb in 4-dpf uninjected (E,E’) and tomm22 MO-injected (F,F’) larvae. E and F are dorsal views; E’ and F’ are lateral views. wnt2bb hepatic expression appeared to be upregulated in many 4-dpf tomm22 morphants (n=9/17) (F, F’) compared with uninjected larvae (n=0/17) (E, E’) (similar numbers were observed at 5 dpf).
Although it is known that mature hepatocytes are the main source of regenerating cells following liver damage such as PHx (for a review, see Fausto et al., 2006), in conditions where there is low or no mature hepatocyte replication, the source of new hepatic cells is still highly debated. Following tomm22 MO-induced ablation, we followed cellular proliferation at 3, 4 and 5 dpf and observed a higher number of proliferating [phospho-histone H3 (PH3)-positive] cells in recovering tomm22 morphants compared with WT livers, and found that the proliferating cells were also Alcam positive (Fig. 4B). This result suggests that, in conditions where mature hepatocyte replication is absent or impaired, cells expressing Alcam, which at this stage in WT livers corresponds to biliary epithelial cells (Sakaguchi et al., 2008), may be responsible for replenishing the liver with new hepatocytes.
The Wnt2b signaling pathway positively regulates hepatocyte regeneration
The Wnt–β-catenin signaling pathway has been implicated in the regeneration process of a number of tissues, including the liver (Monga et al., 2001; Sodhi et al., 2005; Tan et al., 2006; Goessling et al., 2008). However, these studies have been carried out after PHx. To investigate whether the Wnt–β-catenin signaling pathway is also involved in the regeneration process after severe hepatocyte ablation, we used the tomm22 MO-based regeneration assay in the wnt2bbs403 mutant background (Ober et al., 2006).
tomm22 MO-injected larvae (WT, wnt2bb+/– and wnt2bb−/−), exhibiting a small liver at 4 dpf, were analyzed at 6 dpf for liver repopulation. At this stage, under control conditions, the liver mass of tomm22 morphants has recovered significantly (Fig. 4A). Hepatic recovery was classified into three categories: ‘full’ recovery (comparable to recovery in the WT background), ‘moderate’ recovery (new hepatocytes formed but to a lesser extent than in ‘full’ recovery) and ‘no’ recovery (the liver size remained the same as at 4 dpf). To investigate the correlation between liver recovery and wnt2bb function, larvae were sorted at 6 dpf into one of the three categories and then genotyped for the wnt2bbs403 mutation. We observed that, in the absence of wnt2bb function, ‘full’ recovery was rarely observed (67.1% in non-mutants, 1.9% in wnt2bb−/− mutants), and conversely, ‘no’ recovery increased (5.2% in nonmutants, 59.5% in wnt2bb−/− mutants) (Fig. 4C).
wnt2bb mutant larvae exhibit early liver development defects, however, liver size becomes comparable to WT by 4 dpf (the time point when tomm22 MO-injected larvae were selected, based on their liver size, to follow hepatic regeneration). Nevertheless, to further test the involvement of the Wnt–β-catenin signaling pathway in this hepatocyte recovery model, we exposed tomm22 morphants to the Wnt–β-catenin inhibitor IWR-1 (Chen et al., 2009), starting at 4 dpf. Inhibition of the Wnt–β-catenin signaling pathway using IWR-1 also resulted in the inhibition of hepatic regeneration. In the presence of 5 μM of IWR-1, the ‘full’ recovery rate decreased (27.5% in controls, 7% in 5 μM IWR-1-treated larvae), whereas the ‘no’ recovery rate increased (45% in controls, 62% in 5 μM IWR-1-treated larvae) (Fig. 4D). These results indicate a role for the Wnt–β-catenin signaling pathway in hepatic regeneration post-tomm2 MO-induced hepatocyte ablation and are consistent with the recovery results observed in the absence of wnt2bb function. To further investigate the role of Wnt–β-catenin signaling during liver regeneration, we examined tomm22 morphants at 4 and 5 dpf, and observed an upregulation of wnt2bb expression in their hepatic region compared with controls (Fig. 4E–F’). Thus, wnt2bb expression appears to be induced following hepatocyte apoptosis in tomm22 morphants. Similar to the results obtained using the Wnt–β-catenin inhibitor IWR-1, we observed a consistent inhibitory effect on hepatic regeneration when recovering larvae were exposed to the Fgf receptor (FgfR) inhibitor SU5402 (supplementary material Fig. S5). However, when using the bone morphogenetic protein (Bmp) signaling pathway inhibitor dorsomorphin (Yu et al., 2008), we did not observe an obvious effect on hepatic regeneration (data not shown).
DISCUSSION
Our study revealed an unexpectedly specific requirement for Tomm22 in hepatocyte survival. In tomm22 mutants, the liver is initially specified, bile ducts form and hepatocytes differentiate, but hepatocytes die shortly thereafter, leading to a small liver consisting mainly of biliary tissue.
Mitochondria provide eukaryotic cells with energy and play other crucial roles in cell signaling, the metabolism of amino acids and lipids, the biosynthesis of heme and iron-sulfur clusters, and apoptosis. In humans, only 13 mitochondrial proteins are encoded by the mitochondrial genome, the remaining roughly 99% are encoded by nuclear genes and, thus after their synthesis on cytosolic ribosomes, need to be imported into the four different mitochondrial compartments (outer membrane, inner membrane, intermembrane space and matrix) (Bolender et al., 2008). Most mitochondrial proteins are translocated across the OMM using the TOM (translocase of the outer membrane) complex, which is the entry gate of the mitochondria (Endo et al., 2003; Koehler, 2004; Rehling et al., 2004; Neupert and Herrmann, 2007). Multiple studies on mitochondrial import, carried out mainly in yeast or in vitro, have shown that the TOM complex includes the general import pore (GIP) and is formed by a complex of proteins: Tomm22, Tomm20, Tomm70, Tomm40, Tomm5, Tomm6 and Tomm7. Tomm22 is a well-conserved translocase that, in yeast, has been shown to function as an essential receptor for the proteins that are recognized initially by Tomm20 and Tomm70. Tomm22 then transfers these proteins to the import pore and, as they pass through the Tomm40 channel, they interact with the Tomm22 intermembrane space domain. In addition, Tomm22 is essential for stabilizing the integrity of the import pore (for a review, see Pfanner and Wiedemann, 2002). Limited loss-of-function studies of translocases carried out in multicellular organisms have revealed defects in multiple tissues. In zebrafish, timm50 morphants exhibit multiple developmental defects including dysmorphic hearts, neurodegeneration, reduced motility and, eventually, lethality at 4 dpf (Guo et al., 2004); knockdown of three different translocases of the inner mitochondrial membrane (Timm) in Caenorhabditis elegans was shown to result in several developmental defects, affecting mostly cell size, and partial lethality (Curran et al., 2004); and Timm23 knockout mice show a neurological phenotype and reduced life span (Ahting et al., 2009). Our finding that the absence of Tomm22, a protein thought to be a major player in the general import process common to all mitochondria, results in the specific apoptosis of hepatocytes is unexpected and intriguing.
There can be several explanations for the tissue-specific phenotype in tomm22s469 mutants. We observed that this gene is expressed highly in the developing endodermal organs – the liver, gut and, although at a lower level, the pancreas – and thus there may be a higher requirement for Tomm22 in these organs. Although no obvious phenotype was detected in the gut of tomm22 mutants or morphants, some of these embryos showed a slightly smaller pancreas and/or lower Tg(XlEef1a1:GFP)s854 pancreatic expression. Previous work has reported that, across 14 different adult mouse tissues, the heart has the highest mitochondrial density, whereas the liver showed a third of that density and less than many other organs including the large and small intestine, kidney, skeletal muscle and neural tissues (Pagliarini et al., 2008). These findings point to a qualitative, rather than quantitative, difference in mitochondrial tissue requirement as the cause of the tissue-specific phenotype in tomm22s469 mutants.
Indeed, mitochondria perform a variety of different functions whose relevance depends on the specific tissue, or even on their specific location within the cell (Kuznetsov et al., 2004). Pagliarini et al. (Pagliarini et al., 2008) also assessed the relative abundance of each mitochondrial protein across 14 different murine tissues and revealed that approximately only a third of the 1098 mitochondrial proteins are present across all the sampled tissues, whereas most of the mitochondrial proteins show some tissue specificity. The finding that liver mitochondria contain mitochondrial proteins that are different from the ones present in other tissues can help explain the specificity of the phenotype observed in tomm22 mutants. Thus, we hypothesize that Tomm22 is essential for the import of a specific protein or set of proteins required for a particular mitochondrial function that is necessary for hepatocyte survival. Of course, we cannot exclude the possibility of functional redundancy in non-hepatic tissues by (1) an alternative bypass mechanism that is independent of the TOM complex [such as the one shown for cytochrome P450 import in yeast (Anandatheerthavarada et al., 2009)], or (2) the existence of a second tomm22 gene in the zebrafish genome, or of another gene that functions like tomm22.
The specific hepatocyte apoptosis observed in tomm22s469 mutants and tomm22 MO-injected larvae; the regeneration of hepatocytes once the Tomm22 levels are re-established; and the high expression levels of tomm22 detected in the endodermal tissues, such as the liver, point to a specific role for tomm22. Most studies on mitochondrial protein import in multicellular organisms have been carried out in vitro or ex vivo, and only a few have addressed the effect of the loss of function of a mitochondrial import protein in vivo. To our knowledge, the finding that the absence of vertebrate Tomm22 function appears to affect mainly the maturing hepatocytes has not been described before.
Tomm22 liver regeneration model introduces another application for morpholino-based assays
In this study, we showed that the temporary knockdown of Tomm22 levels through the use of tomm22 MO resulted in the specific ablation of hepatocytes followed by hepatic tissue recovery. MOs have been mostly used in developmental studies, as a tool for reverse genetics to uncover the role of a specific target protein in different animal models, but also in therapeutics, targeting bacteria or viruses (Nasevicius and Ekker, 2000; Heasman, 2002; Geller, 2005; Deas et al., 2007). Our tomm22 MO-based assay provides a model to study liver regeneration, highlighting an additional application for the MO technology. The use of MO-based assays, such as the one described here, can be quite powerful when applied to larger-scale regeneration studies, where larger numbers of samples can be tested. This approach may allow the screening of a large number of pharmacological substances for their potential to enhance or impair regeneration or, in forward genetic screens, for the identification of genes that modify this process. Moreover, MO-based regeneration assays may prove to be especially useful for later developmental stages when using the emerging caged MO technology. In the latter approach, the MO is photoactivatable, which allows spatiotemporal protein downregulation (Shestopalov et al., 2007). Alternatively, a conditional transgenic rescue of the tomm22 mutation may also allow late-onset hepatocyte apoptosis followed by regeneration.
Wnt2b signaling is involved in liver regeneration
Several studies have suggested that Wnt–β-catenin signaling is involved in the regeneration of different tissues, such as the newt lens (Hayashi et al., 2008), bone (Zhong et al., 2006), muscle (Polesskaya et al., 2003) and zebrafish tail fin (Stoick-Cooper et al., 2007). The Wnt–β-catenin pathway has also been shown to be activated during liver regeneration after PHx in rodents (Monga et al., 2001) and zebrafish (Goessling et al., 2008), and downregulation of β-catenin has been shown to partially inhibit this process in the rat and mouse (Sodhi et al., 2005; Tan et al., 2006). In contrast to PHx, the hepatic recovery assay that we developed and used in this study is likely to be progenitor cell dependent as, after hepatocyte ablation, only very few hepatocytes remain. Our proliferation assays suggest that, in the tomm22 MO hepatocyte ablation model, the newly formed hepatocytes do not derive from other hepatocytes but rather from biliary duct cells or other Alcam-positive cells. Lineage experiments with appropriate Cre-expressing lines will be required to test this hypothesis rigorously. In previous work, Wnt2b signaling has been shown to positively regulate liver development (Ober et al., 2006; McLin et al., 2007; Tan et al., 2008). Liver specification is initially impaired in zebrafish wnt2bb mutants, although the liver eventually forms in most of these animals (Ober et al., 2006). Using our assay, we observed that Wnt2b signaling plays a role in the regeneration of the ablated hepatic tissue, suggesting that it may also play a role in a progenitor-dependent liver regeneration process. The decrease in liver regeneration rate in the presence of a Wnt–β-catenin pathway inhibitor (IWR-1), as well as the upregulation of wnt2bb in the liver region in post-ablation recovering larvae, provides additional support for the involvement of this pathway in the hepatic regeneration process following tomm22 MO-mediated hepatocyte ablation.
Development and regeneration
Several aspects of tissue development are thought to be recapitulated during the process of regeneration, as both processes involve common mechanisms such as proliferation, differentiation, patterning and growth control. However, the extent of the similarity between the two processes remains unclear. Whereas some studies suggest parallels between the two processes, as shown by the similarity of the antigenic and biochemical profiles of pluripotent progenitors in the liver during development and regeneration (Zhang et al., 2008), a recent study reported that progenitor cells involved in muscle development (embryonic muscle progenitors) and during regeneration (adult satellite cells) show significant differences in their genetic requirements (Lepper et al., 2009).
The role of Wnt2b and Fgf signaling in the tomm22 MO regeneration model suggests a certain level of similarity between the pathways regulating liver development and regeneration. These findings are in agreement with a promoting role for Wnt2b and Fgf signaling in liver specification (Jung et al., 1999; Rossi et al., 2001; Ober et al., 2006; Shin et al., 2007), as well as in hepatocyte proliferation-based recovery post-PHx (Monga et al., 2001; Steiling et al., 2003; Sturm et al., 2004; Goessling et al., 2008; Kan et al., 2009). Although the Bmp pathway has also been shown to be involved in liver development (Rossi et al., 2001), as well as in regeneration post-PHx (Steiling et al., 2003; Sturm et al., 2004; Sugimoto et al., 2007; Kan et al., 2009) or following tetrachloride-induced injury (Nakatsuka et al., 2007), we observed no clear role for Bmp signaling based on dorsomorphin treatments. In the future, it will be important to dissect the specific roles of these signaling pathways in various models of liver regeneration. The tomm22 MO assay will be a powerful tool in such studies, as it is easily scalable.
METHODS
Zebrafish strains
Zebrafish embryos, larvae and adult fish were raised under standard laboratory conditions at 28°C (Westerfield, 2000). We used the following mutant and transgenic lines: wnt2bbs403 (Ober et al., 2006), Tg(fabp10:RFP, ela3l:EGFP)gz12 (Farooq et al., 2008) [for simplicity, this line is referred to as Tg(fabp10:RFP)gz12 in this manuscript], Tg(–2.8fabp10:EGFP)as3 (Her et al., 2003) and Tg(XlEef1a1:GFP)s854 (Field et al., 2003).
Sectioning, immunohistochemistry and microscopy
Larvae were fixed overnight at 4°C with 3% formaldehyde and then immunostained in whole mounts or following sectioning. For sectioning, larvae were embedded in low-melting agarose and cut into 200-μm sections with a vibratome. Antibody staining was carried out in PBST (0.1% Triton X-100, 4% BSA, 0.02% NaN3 in PBS, pH 7.3). We used the following antibodies: polyclonal antibody against cadherin (rabbit, 1:1000; Sigma); 2F11 monoclonal antibody (mouse, 1:400; a generous gift from Julian Lewis, Cancer Research, UK), 2F11 is used as a marker of the intrahepatic biliary duct in the zebrafish liver and for secretory cells in the zebrafish gut (Crosnier et al., 2005); WCL15 monoclonal antibody (mouse, 1:100; a generous gift from Jan Rombout, University of Tuscia, Italy); Zn8 monoclonal antibody against Alcam (mouse, 1:10; Developmental Studies Hybridoma Bank, University of Iowa); anti-phospho-histone H3 (PH3) antibody (rabbit, 1:100; Upstate); and fluorescently conjugated secondary antibodies from Molecular Probes (1:200). For F-actin or nuclear staining, whole larvae and sections were stained with rhodamine phalloidin (1:100, Molecular Probes) or TO-PRO 3 (1:5000, Molecular Probes), respectively. Images were acquired with a Zeiss 510 confocal or Zeiss Lumar microscope and prepared for publication using Adobe Photoshop and Illustrator.
TUNEL cell death assay
The TUNEL cell death assay was performed using an In Situ Cell Death Detection Kit, Fluorescein (Roche; cat. no. 11 684 795 001). After fixation of zebrafish larvae in 3% formaldehyde and sectioning, transverse sections of the liver were pre-incubated in PBST and then labeled with the TUNEL kit for 1 hour at 37°C. Sections were washed with PBT (0.1% Triton X-100 in PBS, pH 7.3) and visualized by confocal imaging. For co-immunostaining with cadherin, sections were first incubated with primary antibodies, then with TUNEL solutions, and finally with secondary antibodies.
Mapping and genotyping of the tomm22s469 allele
The mutation in olis469 was mapped to linkage group 12 using a set of SSLP markers. For fine mapping, 1363 mutant larvae were tested for SSLPs in the critical region, followed by isolation of cDNA from olis469 larvae and siblings, and sequence analysis of candidate genes within the critical interval, including tomm22.
Detection of the tomm22s469 allele was performed by PCR amplification of genomic DNA using the forward 5′-CAGAGGAGAGATAATCAAACAGGT-3′ and reverse 5′-TGGACGTCAGTCTATGCAGAA-3′ primers, followed by digestion with Fnu4HI, generating digestion products of 305, 73, 17 and 120 bp in the tomm22s469 allele, and products of 378, 17 and 120 bp in the WT.
Morpholino antisense oligonucleotide design and injection
The tomm22 morpholino oligonucleotide (tomm22 MO) was targeted against the translational site of tomm22 with the following sequence: 5′-GAGAAAGCTCCTGGATCGTAGCCAT-3′ (Open Biosystems). Embryos were injected with 4 ng of tomm22 MO at the one-cell stage.
Purification of yeast mitochondria and in vitro import assays
WT GA74 yeast mitochondria were isolated from cultures grown at 30°C, to an OD600 of 2, in rich lactate medium (1% yeast extract, 2% tryptone, 0.05% dextrose, 2% lactic acid, 3.4 mM CaCl2, 8.5 mM NaCl, 2.95 mM MgCl2, 7.35 mM KH2PO4, 18.7 mM NH4Cl). Mitochondria were isolated as described previously (Daum et al., 1982). WT or mutant Tomm22 in pCS2 (SP6), or Tom22 in pcGlobin2 (T7), were translated using a TnT kit (Promega) in the presence of 35S-methionine. Radiolabeled precursors were incubated at 30°C with isolated mitochondria in import buffer (1 mg/ml BSA, 0.6 M sorbitol, 150 mM KCl, 10 mM MgCl2, 2.5 mM EDTA, 2 mM ATP, 2 mM NADH, 20 mM Hepes-KOH, pH 7.4) for 30 minutes. Outer membrane proteins were digested with 1 μg/ml trypsin for 30 minutes on ice. Trypsin was inhibited with 10 μg/ml of soybean trypsin inhibitor. Mitochondria were pelleted at 14,000 × g for 5 minutes at 4°C, and resuspended in SDS-PAGE sample buffer. For carbonate extraction, a single import reaction was divided into four tubes. Mitochondria were collected by spinning at 8000 × g for 5 minutes at 4°C and then resuspended in 0.1 M sodium carbonate, at various pHs, or in Thorner buffer (10% glycerol, 8 M urea, 5% SDS, 40 mM Tris, pH 6.8, 4 mg/ml Bromophenol Blue, 5% β-mercaptoethanol). After 30 minutes on ice, the mitochondria were pelleted at 14,000 × g for 10 minutes at 4°C. The pellet containing the membrane fraction was resuspended in Thorner buffer, and the supernatants were precipitated with 20% trichloroacetic acid for 30 minutes on ice. The supernatants were then spun at 14,000 × g for 10 minutes at 4°C. These pellets were then resuspended in Thorner buffer, resolved by 17.5% SDS-PAGE, and visualized by autoradiography.
Whole-mount in situ hybridization
Whole-mount in situ hybridization was performed, as described previously (Alexander et al., 1998), using the wnt2bb (Ober et al., 2006) and tomm22 probes. To generate the tomm22 in situ probe, the last exon and 3′-UTR of tomm22 were cloned into the pGEMT Easy vector (Promega).
Chemical treatments
tomm22 MO-injected Tg(fabp10:RFP)gz12 larvae exhibiting a degenerating liver at 4 dpf were treated with 5 μM of the Wnt–β-catenin signaling pathway inhibitor IWR-1 (a gift from Lawrence Lum, University of Texas Southwestern Medical Center) (Chen et al., 2009) in 0.05% dimethyl sulfoxide (DMSO), or with 0.05% DMSO (carrier control) in embryo medium, at 28°C. A 1 mM stock of the Fgf inhibitor SU5402 (Calbiochem) was prepared in 100% DMSO and diluted to 1 and 3 μM with egg water; the 0.1% DMSO solution in egg water was used as a control.
Supplementary Material
Acknowledgments
We thank Nikolaus Pfanner, Duc Dong, Michel Bagnat, Cagla Eroglu and members of the Stainier lab for discussions; Markus Bussen, Isla Cheung and Stephanie Woo for comments on the manuscript; Bruce Draper, Natasha Zvenigorodsky, Donghun Shin, Linda Setiawan, Koraboshka Brand, Chong Hyun Shin, as well as Sandra Huling and the UCSF Liver Center, for experimental assistance; Ana Ayala and Milagritos Alva for expert help with the fish; Julian Lewis and Jan Rombout for antibodies; and Lawrence Lum and Jim Amatruda for the IWR-1 compound. P.C-H. thanks Jodi Nunnari for support. This work was supported in part by postdoctoral fellowships from the CVRI (S.C.), the American Heart Association (E.A.O.) and the Human Frontier Science Program (H.V.), and grants from the NIH ( DK60322) and the Packard Foundation to D.Y.R.S. Deposited in PMC for release after 12 months.
Footnotes
COMPETING INTERESTS
The authors declare no competing financial interests.
AUTHOR CONTRIBUTIONS
S.C. designed and performed experiments, analyzed the data and wrote the manuscript. E.A.O. analyzed data and contributed reagents and to the writing of the manuscript. S.W. designed and performed experiments, analyzed data and contributed to the writing of the manuscript. P.C.-H. contributed with data analysis. H.V. contributed with reagents. C.M.K. designed experiments and analyzed data. D.Y.R.S. designed experiments, analyzed the data, wrote the manuscript and was the principal supervisor. All authors read and commented on the manuscript.
SUPPLEMENTARY MATERIAL
Supplementary material for this article is available at http://dmm.biologists.org/lookup/suppl/doi:10.1242/dmm.004390/-/DC1
REFERENCES
- Ahting U, Floss T, Uez N, Schneider-Lohmar I, Becker L, Kling E, Iuso A, Bender A, de Angelis MH, Gailus-Durner V, et al. (2009). Neurological phenotype and reduced lifespan in heterozygous Tim23 knockout mice, the first mouse model of defective mitochondrial import. Biochim Biophys Acta 1787, 371–376 [DOI] [PubMed] [Google Scholar]
- Alexander J, Stainier DY, Yelon D. (1998). Screening mosaic F1 females for mutations affecting zebrafish heart induction and patterning. Dev Genet. 22, 288–299 [DOI] [PubMed] [Google Scholar]
- Anandatheerthavarada HK, Sepuri NB, Avadhani NG. (2009). Mitochondrial targeting of cytochrome P450 proteins containing NH2-terminal chimeric signals involves an unusual TOM20/TOM22 bypass mechanism. J Biol Chem. 284, 17352–17363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Gebert M, Pfanner N, van der Laan M. (2009). Biogenesis of mitochondrial membrane proteins. Curr Opin Cell Biol. 21, 484–493 [DOI] [PubMed] [Google Scholar]
- Bolender N, Sickmann A, Wagner R, Meisinger C, Pfanner N. (2008). Multiple pathways for sorting mitochondrial precursor proteins. EMBO Rep. 9, 42–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Dodge ME, Tang W, Lu J, Ma Z, Fan CW, Wei S, Hao W, Kilgore J, Williams NS, et al. (2009). Small molecule-mediated disruption of Wntdependent signaling in tissue regeneration and cancer. Nat Chem Biol. 5, 100–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosnier C, Vargesson N, Gschmeissner S, Ariza-McNaughton L, Morrison A, Lewis J. (2005). Delta-Notch signalling controls commitment to a secretory fate in the zebrafish intestine. Development 132, 1093–1104 [DOI] [PubMed] [Google Scholar]
- Curado S, Anderson RM, Jungblut B, Mumm J, Schroeter E, Stainier DY. (2007). Conditional targeted cell ablation in zebrafish: a new tool for regeneration studies. Dev Dyn. 236, 1025–1035 [DOI] [PubMed] [Google Scholar]
- Curado S, Stainier DY, Anderson RM. (2008). Nitroreductase-mediated cell/tissue ablation in zebrafish: a spatially and temporally controlled ablation method with applications in developmental and regeneration studies. Nat Protoc. 3, 948–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran SP, Leverich EP, Koehler CM, Larsen PL. (2004). Defective mitochondrial protein translocation precludes normal Caenorhabditis elegans development. J Biol Chem. 279, 54655–54662 [DOI] [PubMed] [Google Scholar]
- Daum G, Gasser SM, Schatz G. (1982). Import of proteins into mitochondria. Energy-dependent, two-step processing of the intermembrane space enzyme cytochrome b2 by isolated yeast mitochondria. J Biol Chem. 257, 13075–13080 [PubMed] [Google Scholar]
- Deas TS, Bennett CJ, Jones SA, Tilgner M, Ren P, Behr MJ, Stein DA, Iversen PL, Kramer LD, Bernard KA, et al. (2007). In vitro resistance selection and in vivo efficacy of morpholino oligomers against West Nile virus. Antimicrob Agents Chemother. 51, 2470–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T, Yamamoto H, Esaki M. (2003). Functional cooperation and separation of translocators in protein import into mitochondria, the double-membrane bounded organelles. J Cell Sci. 116, 3259–3267 [DOI] [PubMed] [Google Scholar]
- Farooq M, Sulochana KN, Pan X, To J, Sheng D, Gong Z, Ge R. (2008). Histone deacetylase 3 (hdac3) is specifically required for liver development in zebrafish. Dev Biol. 317, 336–353 [DOI] [PubMed] [Google Scholar]
- Fausto N, Laird AD, Webber EM. (1995). Liver regeneration. 2. Role of growth factors and cytokines in hepatic regeneration. FASEB J. 9, 1527–1536 [DOI] [PubMed] [Google Scholar]
- Fausto N, Campbell JS, Riehle KJ. (2006). Liver regeneration. Hepatology 43, S45–S53 [DOI] [PubMed] [Google Scholar]
- Field HA, Ober EA, Roeser T, Stainier DYR. (2003). Formation of the digestive system in zebrafish. I. Liver morphogenesis. Dev Biol. 253, 279–290 [DOI] [PubMed] [Google Scholar]
- Geller BL. (2005). Antibacterial antisense. Curr Opin Mol Ther. 7, 109–113 [PubMed] [Google Scholar]
- Gerloff T, Stieger B, Hagenbuch B, Madon J, Landmann L, Roth J, Hofmann AF, Meier PJ. (1998). The sister of P-glycoprotein represents the canalicular bile salt export pump of mammalian liver. J Biol Chem. 273, 10046–10050 [DOI] [PubMed] [Google Scholar]
- Goessling W, North TE, Lord AM, Ceol C, Lee S, Weidinger G, Bourque C, Strijbosch R, Haramis AP, Puder M, et al. (2008). APC mutant zebrafish uncover a changing temporal requirement for wnt signaling in liver development. Dev Biol. 320, 161–174 [DOI] [PubMed] [Google Scholar]
- Guo Y, Cheong N, Zhang Z, De Rose R, Deng Y, Farber SA, Fernandes-Alnemri T, Alnemri ES. (2004). Tim50, a component of the mitochondrial translocator, regulates mitochondrial integrity and cell death. J Biol Chem. 279, 24813–24825 [DOI] [PubMed] [Google Scholar]
- Hayashi T, Mizuno N, Kondoh H. (2008). Determinative roles of FGF and Wnt signals in iris-derived lens regeneration in newt eye. Dev Growth Differ. 50, 279–287 [DOI] [PubMed] [Google Scholar]
- Heasman J. (2002). Morpholino oligos: making sense of antisense? Dev Biol. 243, 209–214 [DOI] [PubMed] [Google Scholar]
- Her GM, Chiang CC, Chen WY, Wu JL. (2003). In vivo studies of liver-type fatty acid binding protein (L-FABP) gene expression in liver of transgenic zebrafish (Danio rerio). FEBS Lett. 538, 125–133 [DOI] [PubMed] [Google Scholar]
- Hwang DK, Claypool SM, Leuenberger D, Tienson HL, Koehler CM. (2007). Tim54p connects inner membrane assembly and proteolytic pathways in the mitochondrion. J Cell Biol. 178, 1161–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J, Zheng M, Goldfarb M, Zaret KS. (1999). Initiation of mammalian liver development from endoderm by fibroblast growth factors. Science 284, 1998–2003 [DOI] [PubMed] [Google Scholar]
- Kan NG, Junghans D, Izpisua Belmonte JC. (2009). Compensatory growth mechanisms regulated by BMP and FGF signaling mediate liver regeneration in zebrafish after partial hepatectomy. FASEB J. 23, 3516–3525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler CM. (2004). New developments in mitochondrial assembly. Annu Rev Cell Dev Biol. 20, 309–335 [DOI] [PubMed] [Google Scholar]
- Kuznetsov AV, Usson Y, Leverve X, Margreiter R. (2004. Subcellular heterogeneity of mitochondrial function and dysfunction: evidence obtained by confocal imaging. Mol. Cell Biochem. 256–257, 359–365 [DOI] [PubMed] [Google Scholar]
- Lepper C, Conway SJ, Fan CM. (2009). Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature 460, 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins PN, Theruvath TP, Neuhaus P. (2008). Rodent models of partial hepatectomies. Liver Int. 28, 3–11 [DOI] [PubMed] [Google Scholar]
- McLin VA, Rankin SA, Zorn AM. (2007). Repression of Wnt/beta-catenin signaling in the anterior endoderm is essential for liver and pancreas development. Development 134, 2207–2217 [DOI] [PubMed] [Google Scholar]
- Michalopoulos GK, DeFrances MC. (1997). Liver regeneration. Science 276, 60–66 [DOI] [PubMed] [Google Scholar]
- Monga SP, Pediaditakis P, Mule K, Stolz DB, Michalopoulos GK. (2001). Changes in WNT/beta-catenin pathway during regulated growth in rat liver regeneration. Hepatology 33, 1098–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Nishina H. (2009). Liver development: lessons from knockout mice and mutant fish. Hepatol Res. 39, 633–644 [DOI] [PubMed] [Google Scholar]
- Nakatsuka R, Taniguchi M, Hirata M, Shiota G, Sato K. (2007). Transient expression of bone morphogenic protein-2 in acute liver injury by carbon tetrachloride. J Biochem. 141, 113–119 [DOI] [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC. (2000). Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet. 26, 216–220 [DOI] [PubMed] [Google Scholar]
- Neupert W, Herrmann JM. (2007). Translocation of proteins into mitochondria. Annu Rev Biochem. 76, 723–749 [DOI] [PubMed] [Google Scholar]
- Ober EA, Verkade H, Field HA, Stainier DY. (2006). Mesodermal Wnt2b signalling positively regulates liver specification. Nature 442, 688–691 [DOI] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, et al. (2008). A mitochondrial protein compendium elucidates complex I disease biology. Cell 134, 112–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanner N, Wiedemann N. (2002). Mitochondrial protein import: two membranes, three translocases. Curr Opin Cell Biol. 14, 400–411 [DOI] [PubMed] [Google Scholar]
- Pisharath H, Rhee JM, Swanson MA, Leach SD, Parsons MJ. (2007). Targeted ablation of beta cells in the embryonic zebrafish pancreas using E. coli nitroreductase. Mech Dev. 124, 218–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polesskaya A, Seale P, Rudnicki MA. (2003). Wnt signaling induces the myogenic specification of resident CD45+ adult stem cells during muscle regeneration. Cell 113, 841–852 [DOI] [PubMed] [Google Scholar]
- Rehling P, Brandner K, Pfanner N. (2004). Mitochondrial import and the twinpore translocase. Nat Rev Mol Cell Biol. 5, 519–530 [DOI] [PubMed] [Google Scholar]
- Romano N, Picchietti S, Taverne-Thiele JJ, Taverne N, Abelli L, Mastrolia L, Verburg-van Kemenade BM, Rombout JH. (1998). Distribution of macrophages during fish development: an immunohistochemical study in carp (Cyprinus carpio, L.). Anat Embryol. 198, 31–41 [DOI] [PubMed] [Google Scholar]
- Rossi JM, Dunn NR, Hogan BL, Zaret KS. (2001). Distinct mesodermal signals, including BMPs from the septum transversum mesenchyme, are required in combination for hepatogenesis from the endoderm. Genes Dev. 15, 1998–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler KC, Krahn KN, Gaur NA, Ukomadu C. (2007). Liver growth in the embryo and during liver regeneration in zebrafish requires the cell cycle regulator, uhrf1. Proc Natl Acad Sci USA 104, 1570–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi TF, Sadler KC, Crosnier C, Stainier DY. (2008). Endothelial signals modulate hepatocyte apicobasal polarization in zebrafish. Curr Biol. 18, 1565–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena R, Zucker SD, Crawford JM. (2003. Hepatology: A Textbook of Liver Disease. Philadelphia: Saunders [Google Scholar]
- Shestopalov IA, Sinha S, Chen JK. (2007). Light-controlled gene silencing in zebrafish embryos. Nat Chem Biol. 3, 650–651 [DOI] [PubMed] [Google Scholar]
- Shin D, Shin CH, Tucker J, Ober EA, Rentzsch F, Poss KD, Hammerschmidt M, Mullins MC, Stainier DY. (2007). Bmp and Fgf signaling are essential for liver specification in zebrafish. Development 134, 2041–2050 [DOI] [PubMed] [Google Scholar]
- Sodhi D, Micsenyi A, Bowen WC, Monga DK, Talavera JC, Monga SP. (2005). Morpholino oligonucleotide-triggered beta-catenin knockdown compromises normal liver regeneration. J Hepatol. 43, 132–141 [DOI] [PubMed] [Google Scholar]
- Steiling H, Wustefeld T, Bugnon P, Brauchle M, Fassler R, Teupser D, Thiery J, Gordon JI, Trautwein C, Werner S. (2003). Fibroblast growth factor receptor signalling is crucial for liver homeostasis and regeneration. Oncogene 22, 4380–4388 [DOI] [PubMed] [Google Scholar]
- Stoick-Cooper CL, Weidinger G, Riehle KJ, Hubbert C, Major MB, Fausto N, Moon RT. (2007). Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development 134, 479–489 [DOI] [PubMed] [Google Scholar]
- Sturm J, Keese M, Zhang H, Bonninghoff R, Magdeburg R, Vajkoczy P, Dono R, Zeller R, Gretz N. (2004). Liver regeneration in FGF-2-deficient mice: VEGF acts as potential functional substitute for FGF-2. Liver Int. 24, 161–168 [DOI] [PubMed] [Google Scholar]
- Sugimoto H, Yang C, LeBleu VS, Soubasakos MA, Giraldo M, Zeisberg M, Kalluri R. (2007). BMP-7 functions as a novel hormone to facilitate liver regeneration. FASEB J. 21, 256–264 [DOI] [PubMed] [Google Scholar]
- Tan X, Behari J, Cieply B, Michalopoulos GK, Monga SP. (2006). Conditional deletion of beta-catenin reveals its role in liver growth and regeneration. Gastroenterology 131, 1561–1572 [DOI] [PubMed] [Google Scholar]
- Tan X, Yuan Y, Zeng G, Apte U, Thompson MD, Cieply B, Stolz DB, Michalopoulos GK, Kaestner KH, Monga SP. (2008). Beta-catenin deletion in hepatoblasts disrupts hepatic morphogenesis and survival during mouse development. Hepatology 47, 1667–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Sar AM, Appelmelk BJ, Vandenbroucke-Grauls CM, Bitter W. (2004). A star with stripes: zebrafish as an infection model. Trends Microbiol. 12, 451–457 [DOI] [PubMed] [Google Scholar]
- Westerfield M. (2000. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio), 4th edn Eugene, OR: University of Oregon Press [Google Scholar]
- Yu PB, Hong CC, Sachidanandan C, Babitt JL, Deng DY, Hoyng SA, Lin HY, Bloch KD, Peterson RT. (2008). Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat Chem Biol. 4, 33–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Grompe M. (2008). Generation and regeneration of cells of the liver and pancreas. Science 322, 1490–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Theise N, Chua M, Reid LM. (2008). The stem cell niche of human livers: symmetry between development and regeneration. Hepatology 48, 1598–1607 [DOI] [PubMed] [Google Scholar]
- Zhao R, Duncan SA. (2005). Embryonic development of the liver. Hepatology 41, 956–967 [DOI] [PubMed] [Google Scholar]
- Zhong N, Gersch RP, Hadjiargyrou M. (2006). Wnt signaling activation during bone regeneration and the role of Dishevelled in chondrocyte proliferation and differentiation. Bone 39, 5–16 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.