 Extracellular glycoprotein interactions are not detected by most high throughput assays creating “blind-spots” in protein interaction maps. This review examines this problem and discusses recent advances that have begun to address it.
Extracellular glycoprotein interactions are not detected by most high throughput assays creating “blind-spots” in protein interaction maps. This review examines this problem and discusses recent advances that have begun to address it.
Abstract
Individual cells within biological systems frequently coordinate their functions through signals initiated by specific extracellular protein interactions involving receptors that bridge the cellular membrane. Due to their biochemical nature, these membrane-embedded receptor proteins are difficult to manipulate and their interactions are characterised by very weak binding strengths that cannot be detected using popular high throughput assays. This review will provide a general outline of the biochemical attributes of receptor proteins focussing in particular on the biophysical properties of their transient interactions. Methods that are able to detect these weak extracellular binding events and especially those that can be used for identifying novel interactions will be compared. Finally, I discuss the feasibility of constructing a complete and accurate extracellular protein interaction map, and the methods that are likely to be useful in achieving this goal.
Introduction
The individual cells within metazoan organisms must communicate with one another to ensure that they function collectively as a coordinated biological system. Frequently, this intercellular communication is initiated by specific extracellular protein–protein interactions involving membrane-tethered receptors that subsequently trigger cytoplasmic signalling pathways to effect an appropriate cellular response. This communication is important both in the development of the organism—so that each cell behaves appropriately as a function of its position—and in the maintenance of the organism in response to changing physiological conditions. Extracellular recognition events are also important in infectious diseases since many pathogens use host cell surface proteins to initiate cellular invasion processes. Given their fundamental role in biology and infection, a comprehensive and accurate map of extracellular protein interactions would be an important resource for biomedical science.
Recent technical advances have made mapping complete and accurate protein interaction networks a realistic possibility. In particular, the yeast-two-hybrid (Y2H), and biochemical affinity purifications followed by mass spectrometry have emerged as the two main techniques that can be scaled for genome-wide studies. Both techniques, however, are generally regarded as unsuitable to detect transient interactions between extracellular proteins: structurally-important posttranslational modifications such as disulfide bonds and glycans are not added to proteins within the yeast nucleus and the stringent washing steps of biochemical purifications do not allow the detection of transient interactions. The ever-increasing scale with which these two techniques are being applied is therefore likely to create interaction maps which are underrepresented for extracellular proteins, making them both biased and incomplete. This is of particular concern since extracellular proteins and their interactions are easily accessible to systemically delivered drugs and are therefore considered therapeutically tractable.
This review will address some of the questions related to the identification of novel low affinity extracellular interactions. What biochemical properties make them refractory to detection using popular high throughput techniques? What makes a protein interaction transient and yet specific? How many of these recognition events are missing from current protein interaction maps? Which existing methods could be scaled to detect them in a high throughput setting? By providing answers to these questions, we can try to assess the feasibility of constructing a complete and accurate extracellular protein interaction map.
Properties of extracellular proteins and their interactions
The biochemical nature of extracellular proteins
Proteins that are located in the extracellular space are structurally diverse. They include both secreted ligands and membrane-embedded proteins such as receptors and transporters. Unsurprisingly, then, the interactions made by extracellular proteins are equally diverse, making any single interaction assay unlikely to be applicable to all protein classes. Paradigms within particular structural protein classes are important, however, to facilitate the development of assays that can then be applied more broadly to whole families of similar proteins. What then, have we learnt about the biochemical properties of extracellular proteins and their interactions?
Perhaps the main biochemical characteristics of proteins that occupy the extracellular region are that they contain structurally-important posttranslational modifications. The oxidising environment of the extracellular compartment causes the rapid oxidation of the sulfydryl groups on cysteine residues of polypeptide chains to form covalent disulfide bonds that are critically important for correct protein folding. These bonds are also used to covalently link two or more polypeptide chains together in proteins containing multiple subunits. In addition, extracellular proteins are often modified by the covalent addition of large hydrophilic sugar chains creating glycoproteins. These surface-exposed sugars can make up a large percentage of the molecular mass (a typical N-linked glycan has a mass of ∼3 kDa1) and one could envisage many extracellular proteins as a cloud of hydrophilic sugars with relatively small exposed bald patches of protein which are used as interaction surfaces. It is the necessary addition of these posttranslational modifications that make many convenient and scalable heterologous expression methods such as prokaryotic or cell-free systems unsuitable for producing extracellular proteins in an active conformation. Recently, improvements in cell-free translation systems such as the addition of protein disulfide isomerase and the lowering of reducing agent concentrations have offered hope for conveniently producing large numbers of active extracellular protein fragments.2
Hydrophobic residues in membrane-spanning regions of cell surface proteins create amphipathic molecules that are difficult to solubilise in aqueous solutions. A great deal of progress has been made in optimising protocols to manipulate insoluble membrane proteins for identification by mass spectrometry.3,4 Frequently, however, this requires the use of organic solvents or strongly ionic detergents that are incompatible with maintaining the protein in an active, native conformation—an absolute requirement for identifying physiologically-relevant protein interactions. Because solubilising a protein in an aqueous buffer is often a key initial step in detecting its interacting partners, the amphipathic nature of receptor proteins has made the identification of their binding partners technically very challenging.
Reversible adhesion: the importance of low affinity interactions
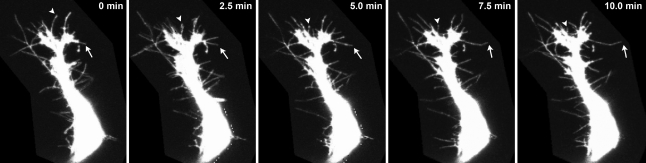
The biophysical properties of extracellular protein–protein interactions range from very high affinity interactions (equilibrium dissociation constants (K D) in the nM to pM range; for example, those made by soluble ligands) to extremely low affinity interactions (K D in μM to mM range, typically made between membrane receptor proteins).5 Soluble ligands bind their receptors with high affinities (∼pM) because they are usually present at very low concentrations in the interstitial fluid and have therefore evolved high binding affinities to ensure a good level of receptor occupancy to initiate a signal. This contrasts with the low affinities observed between membrane-embedded receptor proteins that often have half-lives of fractions of a second when measured in the monomeric state.5 One explanation for this dichotomy is that receptor proteins are confined within the plane of the plasma membrane, locally concentrating them. Any intercellular recognition event between two apposing membranes is therefore likely to involve large multivalent arrays comprising hundreds, possibly thousands, of receptors that will increase the overall avidity of the contact to a level sufficient to trigger a signalling event. Importantly, these adhesive events must be readily reversible. The use of fluorescent reporter proteins and improvements in time-resolved imaging techniques have revealed that the membranes of living cells in vivo are not static but highly dynamic, showing cells frenetically extending and retracting filopodia as they contact and communicate with their neighbours (Fig. 1). If cells are to retain this highly motile behaviour and also move freely, these adhesive bonds must be easily broken. Velcro™ provides a good analogy: thousands of arrayed hooks provide individually weak interaction forces that additively ensure strong adhesion between two surfaces which can still be easily separated. One consequence of the very transient nature of these interactions, however, is that they are very difficult to detect experimentally.
Fig. 1. Cellular membranes are highly dynamic in vivo. Time-resolved images of a migrating zebrafish intersomitic endothelial cell during vascular embryonic development. Note the rapid extension (arrows) and retraction (arrowheads) of filopodial processes. Images kindly provided by Dr Jon Leslie, Cancer Research UK, London.
How weak are functionally relevant extracellular interactions?
The affinity of interactions between cell surface receptor proteins can be very low, but is there a single affinity threshold below which any given interaction could not be functionally relevant? Answering such a general question is difficult because there are many other factors such as local protein abundance which could vary between individual receptor–ligand pairs in different biological contexts. There are, however, examples of functionally well-characterised interactions whose affinity have been measured and can therefore be used as a guide to address this question. The interactions between the T-cell co-receptors, CD8 and CD4, and their respective ligands, MHC class I and II, are important for both thymic development and activation of T lymphocytes in the periphery. The affinities of these important interactions have been shown to be remarkably low, K Ds in the range of 100–200 μM (human CD8αα–HLA-A2 = 200 μM6 and mouse CD4–MHCII ≥ 100 μM7). Other very weak interactions include the murine CD2–CD48 interaction8 (75 μM) and the homophilic interaction of the human SLAM molecule9 (∼200 μM). While the co-receptors CD4 and CD8 interact in the context of a multi-protein complex which could tolerate weaker interaction strengths, the functional significance of other very weak (>75 μM) interactions has been debated in relation to their two-dimensional affinity: a biologically more relevant quantitative parameter which accounts for the fact that the mobility of receptor proteins are restricted to the plane of a membrane.10 These calculations have shown that solution interaction strengths weaker than ∼50 μM are unlikely to be high enough to support spontaneous interactions at physiological surface densities.11
What makes a protein interaction transient, yet specific?
The physical interactions between extracellular receptor proteins are weak and yet highly specific. These seemingly contradictory qualities are governed by the underlying structural and thermodynamic properties of the interactions and have been the subject of many detailed investigations. Understanding any one interaction interface requires several pieces of independent data: firstly, a kinetic analysis of the interaction to identify the on (k on) and off-rate (k off) constants establish the rates at which the two proteins associate and dissociate. Secondly, a thermodynamic analysis of the interaction reveals the relative contributions of enthalpy (the binding energy released by the formation of chemical bonds) and entropy (the relative change in the disorder of the system upon binding) to the overall binding energy released when the proteins associate. Finally, the structures of both the bound and unbound proteins reveal conformational changes upon binding and the identity of amino acids involved at the binding interface. Given the difficulty in obtaining these data, only recently has a general view of the biophysical nature of these interactions begun to emerge.
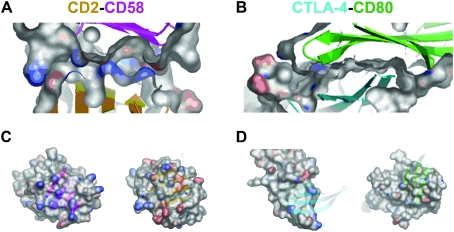
Protein interaction binding kinetics as measured by surface plasmon resonance has revealed that low affinity extracellular interactions are always characterised by very fast off-rate kinetics when measured in their monomeric form. Typically, off-rate constants (k off) are remarkably fast, in the region of 1 to 4 s–1 which corresponds to a half-life of a second or even less; on-rate constants (k on) are unremarkable at ∼105 M–1 s–1. Structurally, however, the interaction interfaces of low affinity extracellular interactions are heterogeneous as illustrated by the CD2–CD58, and CTLA-4–CD80 and CD86 interactions (Fig. 2). The interaction interface between human CD2 and CD58 (K D≈ 20 μm12) has been shown to be small (∼1200 Å2) and flat with a poor surface-shape complementarity13 (Fig. 2A). The majority of the contacts within the interface are salt bridges formed between oppositely-charged residues within the binding sites of the two proteins (Fig. 2C). Although these salt bridges have been shown to be energetically neutral (because energy is required to desolvate the hydration shell surrounding these residues in the unbound proteins), comprehensive mutational studies have revealed that the role of the charged residues is important to impart a high degree of specificity to the interaction. This suggested a general method by which interactions could be highly specific and yet weak.14,15 In almost complete contrast, however, the interaction interfaces between CTLA-4 and its two ligands CD80 (K D = 0.4 μm16) and CD86 (K D = 2.6 μm17), whilst also small (1200 to 1300 Å2), have a remarkably high degree of surface-shape complementarity (Fig. 2B) and are not charged but mainly composed of hydrophobic contacts18,19 (Fig. 2D). The biophysical nature of transient interactions is also divergent. In the three cases mentioned above, binding is primarily enthalpically-driven and restricted by an entropic “penalty” likely to be due to either a reduction in the conformational flexibility of the proteins upon binding or the trapping of water molecules within the binding interface. Where measured, however, other low affinity interactions including CD48–CD220 and FcγRIIa and b–IgG1Fc21 have been shown to have both favourable enthalpic and entropic components, suggesting differences in the overall structural mechanisms of binding. The answer, therefore, seems to be that despite having relatively small interaction surfaces and fast off-rate kinetics in common, cell surface proteins have evolved different structural and thermodynamic solutions to the problem of forming weak and yet specific interactions.
Fig. 2. Contrasting structural properties of low affinity extracellular protein interactions. The interfaces of the human CD2–CD58 and CTLA-4–CD80 co-crystal structures are shown side by side to highlight the differences in their structural properties. (A) The CD2–CD58 interface has a poor surface-shape complementarity—notice the large gaps between the two interacting proteins—whereas that between CTLA-4–CD80, shown in (B) is comparatively good. In (C) and (D), the two proteins in the complex have been separated and rotated to reveal the charge distribution at the interacting surfaces. In each case, the backbone of the binding partner is shadowed as a partly-transparent structure in ribbon format. The CD2–CD58 interaction interface (C) is highly charged whereas the CTLA-4–CD80 interface is composed almost entirely of hydrophobic residues (D). The coordinates for CD2–CD58 and CTLA-4–CD80 co-crystal structures (PDB accession numbers 1qa9 and 1i8l, respectively) were rendered using the program OpenAstexViewer™ 3.0. Key: ribbon backbones: CD2, orange; CD58, magenta; CTLA-4, cyan; CD80, green; charge polarity: blue, positive; red, negative.
Detecting low affinity extracellular protein interactions
Why don’t some popular methods detect extracellular protein interactions?
The biochemical properties of secreted and membrane receptors make their interactions difficult to detect by most protein interaction techniques, which often do not add the structurally-important posttranslational modifications, or require extensive washing steps that preclude detection of transient interactions. For example, the widespread yeast-two-hybrid (Y2H) system necessitates that both binding partners interact within the reducing environment of the yeast nucleus, precluding the addition of glycosylation and disulfide bonds. Also, unless removed, the N-terminal signal peptide found on many membrane receptors will direct the proteins through the export pathway and exclude them from the nucleus. Finally, the transmembrane region(s) will render the protein insoluble and therefore unable to fold into its native structure if retained within the yeast nucleus. Extracellular interactions are therefore underrepresented in Y2H interactome maps.22,23
Affinity purification followed by the identification of interacting partners by mass spectrometry (AP-MS) is another very popular and successful technique for identifying protein interactions. This approach co-purifies tagged proteins and their binding partners expressed in their endogenous environment and should therefore not suffer the same context-dependent problems inherent to heterologous expression. This approach requires that receptor proteins are solubilised in a detergent without disrupting the interaction, which is difficult to achieve due to the amphipathic nature of membrane proteins. Also, many protocols of affinity purification require very stringent washing steps—often for several hours—which are therefore unsuitable to detect transient interactions.
Despite these technical difficulties, the importance of extracellular interactions in cellular recognition and signal initiation has led researchers to develop several alternative techniques. A brief overview of some of the most popular and successful is presented, focusing, in particular, on those that can be used to screen orphan receptors for novel interactions.
Detecting extracellular interactions: cell-based methods
The first assays used to detect low affinity interactions mirror the in vivo situation by displaying the two interacting proteins on the surface of different cell populations. This was first used when human T-lymphocytes, expressing the CD2 cell surface marker, were shown to form “rosettes” with erythrocytes expressing CD58, the binding partner for CD2.24,25 Similar erythrocyte rosetting assays have been used to demonstrate the binding of Plasmodium falciparum merozoite surface proteins to human erythrocyte receptors.26 Clearly, this rosetting technique is limited to receptors that have ligands expressed on erythrocytes; however, an approach based on the same principles can be employed using cellular aggregation. Here, cells that are normally non-adherent are separately transfected with the two proteins under study and differentially labelled. A positive interaction is inferred if the labelled cells are capable of forming aggregates with each other but not with untransfected cells; both heterophilic27 and homophilic28 adhesion pairs have been identified with this technique. By using insect or mammalian cell lines, the receptors contain appropriate posttranslational modifications and are displayed as multimeric arrays within the context of a lipid bilayer. The results of this approach need to be interpreted with caution, however, since one is not directly monitoring the physical interaction of the two proteins. Biological effects such as the up or down regulation of endogenous pro- or anti-adhesive receptors following transfection, or signalling derived from the exogenously expressed genes, are all possible sources of confounding factors. Additional technical difficulties such as controlling the varying expression levels of transfected protein and quantifying the extent of cellular aggregation make this a challenging technique to implement.
Detecting extracellular interactions: recombinant proteins
The experimental tractability of extracellular receptor binding was vastly improved when it was shown that entire ectodomain fragments of receptors, when expressed as soluble recombinant proteins in appropriate expression systems, generally retained their extracellular binding function. This approach simultaneously removed the difficulties associated with the insoluble hydrophobic transmembrane region and enabled the addition of protein tags, making the extracellular binding regions easier to biochemically manipulate, quantitate and purify. Perhaps most importantly, this provided a way of multimerising the ectodomain fragments so as to increase its avidity in binding assays—an essential step to overcome the transient nature of many extracellular interactions. In general, there are two methods for multimerising proteins for interaction detection: one is to fuse the protein of interest to a tag which forms multimers; the other is to produce arrays of proteins by clustering them around a scaffold such as a bead. Selecting an appropriate multimerising tag for interaction screening experiments is difficult because there is no simple relationship between the affinity (the strength of a single, monovalent interaction) and the avidity (the overall interaction strength of a multivalent binding reagent). In addition, the interaction stoichiometry and the steric manner in which a protein interacts with its binding partner—which are usually unknown—will also have an overall effect on the interaction avidity. To illustrate these points, I compare the use of several protein tags that have been used to multimerise the ectodomain fragments of receptor protein for ligand screening.
Increasing avidity: multimerising protein tags
By far the most convenient method of multimerising soluble receptor ectodomains is to recombinantly add a protein tag that is able to spontaneously form multimers in solution (Fig. 3). Perhaps the most commonly used tag for this purpose is the Fc region of human IgG to form an “immunoadhesin”. Typically, this involves the production of a soluble recombinant cell adhesion molecule that forms functional dimers through the inter-chain disulfide bridges present in the hinge region of IgG.29 The recombinant protein can then be purified using well-established antibody purification protocols (such as with immobilised protein G) and is compatible with a large range of commercially available labelled secondary antibody reagents. The resulting immunoadhesin can also be used in functional studies as well as biochemical binding studies.29 In the cases where interaction strengths have been measured, however, the increase in avidity using the dimeric Fc tag is significant but sometimes not sufficient to enable ligand detection using standard protocols that involve wash steps. For example, the interaction between rat CD2 and CD48 (monomeric interaction half-life = 0.2 s) could not be detected using a CD48 dimeric IgG (interaction half-life increased to 23 minutes) on cell lines expressing high levels of CD2 and using different washing stringencies.30 Further increases in avidity to produce decameric multimers using the IgM constant regions (increased half-life to ∼1 hour) were again not sufficient to detect the interaction.30 Large but undefined aggregates can be made by using anti-Fc antibodies to pre-cluster immunoadhesins prior to binding tests, sometimes increasing the avidity sufficiently to enable the detection of an interaction.31 One major disadvantage of using the constant regions of immunoglobulins when working on immune cells is that they themselves can bind and activate Fc receptors causing unwanted positive binding signals or biological effects.
Fig. 3. Schematic representation of fusion proteins containing recombinant multimerising protein tags. The spatial arrangement (N-terminus, dark to C-terminus, light shading) of ectodomains from a typical type I two immunoglobulin superfamily domain-containing cell surface receptor multimerised using different tags are drawn approximately to scale.
Other dimerising tags such as glutathione-S-transferase32 and placental secreted alkaline phosphatase33 have been used successfully. The alkaline phosphatase tag has been particularly useful because the protein can be detected directly using a wide range of readily available phosphatase substrates, thus reducing the need for an intermediate wash step. This tag was first used to identify ligands for the Mek4 and Sek receptor tyrosine kinases34 and subsequently allowed the discovery of additional receptor–ligand pairs such as the Neuropilin–Sema3 interaction35 and the leptin receptor.36 In these examples, the molecular identity of the receptor molecule was isolated through an expression cloning approach following the detection of a positive binding reaction.
Comparatively short stretches of coiled-coil α-helices arranged in a parallel fashion can also be used to form higher-order oligomers of recombinant proteins and used to engineer spontaneously-forming trimers or pentamers (Fig. 3). Trimers can be formed by the addition of a triplex-forming collagen-like peptide,37 isoleucine-zippers38 or cartilage matrix protein.39,40 Similarly, pentamers can be produced by using a 46 amino acid sequence from the rat cartilage oligomeric matrix protein (COMP).41 This tag seems to be particularly effective in increasing interaction avidities, possibly due to the parallel bundling of the peptide chains so that the tagged proteins are presented at the same end of the molecule, making them all available for binding (Fig. 3). Where measured, the increases in avidity can be quite striking—the mouse CD200–CD200R interaction, which has a monomeric half-life of ∼2 s,42 is increased to several hours by pentamerisation with the COMP tag.43
Increasing avidity: clustering supports
A vast increase in the amount of clustered proteins can also be achieved by presenting proteins as multivalent arrays around a solid support, most commonly a microsphere. Using this method, site densities of up to 40 000 molecules per μm2 can be achieved, which exceeds the typical physiological site densities of 100 to 1000 molecules per μm2.30,44 By immobilising proteins as multivalent arrays around fluorescent microspheres, highly avid binding reagents can be produced, which can then be used to demonstrate interactions between candidate receptors45,46 or probe for novel receptor interactions by presenting them to different cell types. The molecular identity of the receptor can then be determined by expression cloning or the isolation of antibodies that can block the interaction.47,48 Soluble supports such as streptavidin to form tetramers49 and dextran to form larger complexes have also been successfully employed.50,51 The increases in avidity due to multimerisation around the streptavidin scaffold have been quantified using the binding of MHC II-Ek–moth cytochrome C peptide to its receptor, the T-cell receptor clone 2B4. The calculated half-lives were 9.5 s (monomer), 3.1 minutes (dimer) and 32 minutes (tetramer) showing an approximately 200-fold decrease in dissociation rate.52 The large avidity gain by using these methods enables the robust detection of interactions as weak as 60 μM.30,53
Finally, there are several quantitative techniques that have the sensitivity to detect low affinity interactions including optical (surface plasmon resonance) and acoustic (resonant acoustic profiling) biosensors,54,55 isothermal calorimetry56 and ultracentrifugation.57 These techniques, however, generally require that both binding partners are known and often require large amounts of purified protein for accurate measurements, making them broadly unsuitable for novel interaction discovery.
High throughput identification of low affinity extracellular interactions
The recent improvements in laboratory automation, clone resources, genome annotation and mass spectrometry technology have seen an explosion in the number of novel interactions detected using the Y2H and AP-MS techniques. These techniques have been so successful that currently ∼85% of all interactions in the IntAct protein interaction database have been discovered using either of these two techniques or variations of them (Fig. 4A). Whilst these advances have made interaction screening a realistic possibility for a large proportion of soluble cytosolic proteins, there are classes of protein interaction including those involving extracellular proteins that cannot be detected using these techniques creating “blind-spots” within protein interaction maps. The number of genes encoding a protein that is secreted or in some way associated with the membrane in the human genome is estimated at ∼7000, corresponding to ∼30% of all protein-coding genes.58 The relative proportion of these proteins in different functional categories is estimated in Fig. 4B. Given the large number of potential interactions that are currently refractory to detection using Y2H and AP-MS, new techniques that are able to detect these interactions on a large scale need to be developed if a complete extracellular receptor interaction network is to be achieved.
Fig. 4. (A) The relative rates of novel protein interaction discovery according to detection method and protein localisation. All metazoan binary protein interactions were extracted from the IntAct database (a total of 106 415 interactions) and then grouped into three broader categories depending on the method by which they were identified: (1) Y2H methods, (2) biochemical purification strategies and (3) all other methods. The cumulative number of interactions for each method per year is expressed as a percentage of the final number of metazoan interactions in the database. (B) The relative proportions of membrane-associated and secreted proteins according to their functional class. Functional classes were defined according to their Pfam domain annotation. The number of protein-coding genes containing these Pfam domains in the human genome was extracted from the Ensembl database.
Despite the importance of this problem, there are only a handful of published large scale systematic screens for novel extracellular receptor–ligand pairs and, with the exception of two studies, the detailed screening results were not described but only individual positive interactions were reported. Lin et al. 59 used a proprietary expression library of undisclosed size to create a panel of COS7 cells each transfected with a different receptor protein displayed on the cell surface. These cells were then probed with a similar library of Fc fusion proteins and interactions detected using immunohistochemistry. Using this approach, a single interaction, that between netrin-G1 and NGL-1, was reported. Similarly, a library of >2000 soluble ectodomains were systematically screened against a BTLA-Fc fusion protein for novel interactions using surface plasmon resonance implemented in a BIAcore machine.60 Again, out of the >2000 interactions screened, only a single interaction was described, making it difficult to assess the suitability of these approaches as a general method for systematic extracellular interaction screening. There have been two recent studies, both based on an ELISA-style assay, where the results of a large scale complete systematic extracellular interaction screen were reported. The first method aimed to establish the binding specificity of the many splice variants of the Drosophila Dscam protein and involved screening 3442 interactions within 92 proteins separated into three groups.61 All the proteins were produced as soluble ectodomain fragments in S2 cells as both Fc- and alkaline phosphatase fusion proteins. The systematic nature of the screen clearly demonstrated that 95% of the splice variants demonstrated isoform-specific homophilic binding. The second method described a scalable method dedicated to identifying novel low affinity extracellular interactions called AVEXIS (for AVidity-based EXtracellular Interaction Screen). This method was used to test almost 10 000 individual interactions within a library of 110 ectodomains from zebrafish immunoglobulin superfamily receptors. Proteins were expressed as soluble fusion proteins in mammalian cells either as a monomeric biotinylated bait for arrayed capture on streptavidin-coated microtitre plates or as a multimeric β-lactamase-tagged prey using the COMP peptide. This approach was able to detect interactions with half-lives of at least a tenth of a second when measured in their monomeric form.62
Future perspectives: mapping the whole extracellular interactome
Performing systematic binary extracellular interaction screens that could be implemented on a genome-wide scale in vertebrates would require throughputs in the order of millions of pairwise interaction tests. From the methods outlined above, the ELISA-style assays which can be readily miniaturised in microtitre plates and microarray formats hold the most promise for efficiently managing this increase in scale. Large reagent collections of suitably tagged secreted and ectodomain protein fragments will be needed which, while they exist in the commercial sector,60,63 are not generally available. Advances in mammalian protein expression technologies have made the production of large libraries of recombinant proteins that contain the appropriate posttranslational modifications possible.64,65 An important consideration to bear in mind when taking a systematic, parallelised approach to protein interaction detection is that frequently, a single stringency threshold must be set that is applied to all interactions being screened. If this threshold is set high, there will be a greater proportion of false negatives, if set low, false positives are likely to be a problem. As I have discussed, extracellular protein interactions can vary significantly in their biophysical properties, making the setting of this stringency threshold a key factor in the design of any high throughput approach. The most appropriate way of setting this threshold is to have an internal set of well-characterised physiologically-relevant interactions as positive controls and, ideally, a set of known negatives as demonstrated in Bushell et al.62 The use of secreted recombinant proteins in interaction screening is an advantage because their expression levels can be normalised prior to screening, which significantly contributes to the setting of a consistent stringency threshold.
Conclusions
The recent success of implementing the Y2H and AP-MS methods on an ever-increasing scale to detect novel protein–protein interactions has meant that classes of interactions such as those made by extracellular proteins, which are not detected using these techniques, are becoming increasingly underrepresented within known interaction networks. Since extracellular protein interactions are considered therapeutically tractable, it is important that the pace of interaction discovery within this important class of proteins matches that achieved for others. Recent advances in mammalian expression technologies, the availability of clone collections and the adaptation of interaction assays in a scalable format have offered the possibility of systematically screening for extracellular interactions on a genome-wide scale. The development of other scalable interaction assays that are dedicated to detecting particular classes of protein interaction will be critical in defining a complete and accurate human protein interaction map.
Acknowledgments
I am grateful to Henning Hermjakob and Sam Kerrien for help with IntAct database queries, Robert Finn for expert technical assistance for Fig. 2 and Cécile Crosnier for critical reading of the manuscript.
Biography
Gavin Wright is an Investigator at the Wellcome Trust Sanger Institute in Cambridge, UK. His main research interests are taking large scale systematic approaches to identify novel extracellular receptor–ligand pairs that initiate signalling pathways in biological systems.
Footnotes
†This article is part of a Molecular BioSystems themed issue on Computational and Systems Biology.
References
- Kornfeld R., Kornfeld S. Annu. Rev. Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- Goshima N., Kawamura Y., Fukumoto A., Miura A., Honma R., Satoh R., Wakamatsu A., Yamamoto J., Kimura K., Nishikawa T., Andoh T., Iida Y., Ishikawa K., Ito E., Kagawa N., Kaminaga C., Kanehori K., Kawakami B., Kenmochi K., Kimura R., Kobayashi M., Kuroita T., Kuwayama H., Maruyama Y., Matsuo K., Minami K., Mitsubori M., Mori M., Morishita R., Murase A., Nishikawa A., Nishikawa S., Okamoto T., Sakagami N., Sakamoto Y., Sasaki Y., Seki T., Sono S., Sugiyama A., Sumiya T., Takayama T., Takayama Y., Takeda H., Togashi T., Yahata K., Yamada H., Yanagisawa Y., Endo Y., Imamoto F., Kisu Y., Tanaka S., Isogai T., Imai J., Watanabe S., Nomura N. Nat. Methods. 2008;5:1011–1017. doi: 10.1038/nmeth.1273. [DOI] [PubMed] [Google Scholar]
- Josic D., Clifton J. G. Proteomics. 2007;7:3010–3029. doi: 10.1002/pmic.200700139. [DOI] [PubMed] [Google Scholar]
- Macher B. A., Yen T. Y. Mol. BioSyst. 2007;3:705–713. doi: 10.1039/b708581h. [DOI] [PubMed] [Google Scholar]
- van der Merwe P. A., Barclay A. N. Trends Biochem. Sci. 1994;19:354–358. doi: 10.1016/0968-0004(94)90109-0. [DOI] [PubMed] [Google Scholar]
- Wyer J. R., Willcox B. E., Gao G. F., Gerth U. C., Davis S. J., Bell J. I., van der Merwe P. A., Jakobsen B. K. Immunity. 1999;10:219–225. doi: 10.1016/s1074-7613(00)80022-9. [DOI] [PubMed] [Google Scholar]
- Weber S., Karjalainen K. Int. Immunol. 1993;5:695–698. doi: 10.1093/intimm/5.6.695. [DOI] [PubMed] [Google Scholar]
- van der Merwe P. A., Brown M. H., Davis S. J., Barclay A. N. EMBO J. 1993;12:4945–4954. doi: 10.1002/j.1460-2075.1993.tb06188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavaddat N., Mason D. W., Atkinson P. D., Evans E. J., Gilbert R. J., Stuart D. I., Fennelly J. A., Barclay A. N., Davis S. J., Brown M. H. J. Biol. Chem. 2000;275:28100–28109. doi: 10.1074/jbc.M004117200. [DOI] [PubMed] [Google Scholar]
- Davis S. J., Ikemizu S., Wild M. K., van der Merwe P. A. Immunol. Rev. 1998;163:217–236. doi: 10.1111/j.1600-065x.1998.tb01199.x. [DOI] [PubMed] [Google Scholar]
- Dustin M. L., Golan D. E., Zhu D. M., Miller J. M., Meier W., Davies E. A., van der Merwe P. A. J. Biol. Chem. 1997;272:30889–30898. doi: 10.1074/jbc.272.49.30889. [DOI] [PubMed] [Google Scholar]
- van der Merwe P. A., Barclay A. N., Mason D. W., Davies E. A., Morgan B. P., Tone M., Krishnam A. K., Ianelli C., Davis S. J. Biochemistry. 1994;33:10149–10160. doi: 10.1021/bi00199a043. [DOI] [PubMed] [Google Scholar]
- Wang J. H., Smolyar A., Tan K., Liu J. H., Kim M., Sun Z. Y., Wagner G., Reinherz E. L. Cell. 1999;97:791–803. doi: 10.1016/s0092-8674(00)80790-4. [DOI] [PubMed] [Google Scholar]
- Davis S. J., Davies E. A., Tucknott M. G., Jones E. Y., van der Merwe P. A. Proc. Natl. Acad. Sci. U. S. A. 1998;95:5490–5494. doi: 10.1073/pnas.95.10.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney A., Avramovic A., Castro M. A., Carmo A. M., Davis S. J., van der Merwe P. A. J. Biol. Chem. 2007;282:13160–13166. doi: 10.1074/jbc.M700829200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Merwe P. A., Bodian D. L., Daenke S., Linsley P., Davis S. J. J. Exp. Med. 1997;185:393–403. doi: 10.1084/jem.185.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins A. V., Brodie D. W., Gilbert R. J., Iaboni A., Manso-Sancho R., Walse B., Stuart D. I., van der Merwe P. A., Davis S. J. Immunity. 2002;17:201–210. doi: 10.1016/s1074-7613(02)00362-x. [DOI] [PubMed] [Google Scholar]
- Schwartz J. C., Zhang X., Fedorov A. A., Nathenson S. G., Almo S. C. Nature. 2001;410:604–608. doi: 10.1038/35069112. [DOI] [PubMed] [Google Scholar]
- Stamper C. C., Zhang Y., Tobin J. F., Erbe D. V., Ikemizu S., Davis S. J., Stahl M. L., Seehra J., Somers W. S., Mosyak L. Nature. 2001;410:608–611. doi: 10.1038/35069118. [DOI] [PubMed] [Google Scholar]
- Evans E. J., Castro M. A., O'Brien R., Kearney A., Walsh H., Sparks L. M., Tucknott M. G., Davies E. A., Carmo A. M., van der Merwe P. A., Stuart D. I., Jones E. Y., Ladbury J. E., Ikemizu S., Davis S. J. J. Biol. Chem. 2006;281:29309–29320. doi: 10.1074/jbc.M601314200. [DOI] [PubMed] [Google Scholar]
- Maenaka K., van der Merwe P. A., Stuart D. I., Jones E. Y., Sondermann P. J. Biol. Chem. 2001;276:44898–44904. doi: 10.1074/jbc.M106819200. [DOI] [PubMed] [Google Scholar]
- Futschik M. E., Chaurasia G., Herzel H. Bioinformatics. 2007;23:605–611. doi: 10.1093/bioinformatics/btl683. [DOI] [PubMed] [Google Scholar]
- Xia Y., Lu L. J., Gerstein M. J. Mol. Biol. 2006;357:339–349. doi: 10.1016/j.jmb.2005.12.067. [DOI] [PubMed] [Google Scholar]
- Moingeon P., Chang H. C., Sayre P. H., Clayton L. K., Alcover A., Gardner P., Reinherz E. L. Immunol. Rev. 1989;111:111–144. doi: 10.1111/j.1600-065x.1989.tb00544.x. [DOI] [PubMed] [Google Scholar]
- Plunkett M. L., Sanders M. E., Selvaraj P., Dustin M. L., Springer T. A. J. Exp. Med. 1987;165:664–676. doi: 10.1084/jem.165.3.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappe S. H., Noe A. R., Fraser T. S., Blair P. L., Adams J. H. Proc. Natl. Acad. Sci. U. S. A. 1998;95:1230–1235. doi: 10.1073/pnas.95.3.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehon R. G., Kooh P. J., Rebay I., Regan C. L., Xu T., Muskavitch M. A., Artavanis-Tsakonas S. Cell. 1990;61:523–534. doi: 10.1016/0092-8674(90)90534-l. [DOI] [PubMed] [Google Scholar]
- Guttinger M., Sutti F., Panigada M., Porcellini S., Merati B., Mariani M., Teesalu T., Consalez G. G., Grassi F. J. Cell Biol. 1998;141:1061–1071. doi: 10.1083/jcb.141.4.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamow S. M., Ashkenazi A. Trends Biotechnol. 1996;14:52–60. doi: 10.1016/0167-7799(96)80921-8. [DOI] [PubMed] [Google Scholar]
- Brown M. H., Preston S., Barclay A. N. Eur. J. Immunol. 1995;25:3222–3228. doi: 10.1002/eji.1830251204. [DOI] [PubMed] [Google Scholar]
- Venkatesh K., Chivatakarn O., Lee H., Joshi P. S., Kantor D. B., Newman B. A., Mage R., Rader C., Giger R. J. J. Neurosci. 2005;25:808–822. doi: 10.1523/JNEUROSCI.4464-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan W., Husler P., Klump H., Erhardt J., Sluis-Cremer N., Dirr H. Protein Sci. 1997;6:399–406. doi: 10.1002/pro.5560060216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan J. G., Leder P. Cell. 1990;63:185–194. doi: 10.1016/0092-8674(90)90299-t. [DOI] [PubMed] [Google Scholar]
- Cheng H. J., Flanagan J. G. Cell. 1994;79:157–168. doi: 10.1016/0092-8674(94)90408-1. [DOI] [PubMed] [Google Scholar]
- He Z., Tessier-Lavigne M. Cell. 1997;90:739–751. doi: 10.1016/s0092-8674(00)80534-6. [DOI] [PubMed] [Google Scholar]
- Tartaglia L. A., Dembski M., Weng X., Deng N., Culpepper J., Devos R., Richards G. J., Campfield L. A., Clark F. T., Deeds J., Muir C., Sanker S., Moriarty A., Moore K. J., Smutko J. S., Mays G. G., Wool E. A., Monroe C. A., Tepper R. I. Cell. 1995;83:1263–1271. doi: 10.1016/0092-8674(95)90151-5. [DOI] [PubMed] [Google Scholar]
- Fan C. Y., Huang C. C., Chiu W. C., Lai C. C., Liou G. G., Li H. C., Chou M. Y. FASEB J. 2008;22:3795–3804. doi: 10.1096/fj.08-111484. [DOI] [PubMed] [Google Scholar]
- Stark S., Flaig R. M., Sandusky M., Watzl C. J. Immunol. Methods. 2005;296:149–158. doi: 10.1016/j.jim.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Coussen F., Choquet D., Sheetz M. P., Erickson H. P. J. Cell Sci. 2002;115:2581–2590. doi: 10.1242/jcs.115.12.2581. [DOI] [PubMed] [Google Scholar]
- Holler N., Kataoka T., Bodmer J. L., Romero P., Romero J., Deperthes D., Engel J., Tschopp J., Schneider P. J. Immunol. Methods. 2000;237:159–173. doi: 10.1016/s0022-1759(99)00239-2. [DOI] [PubMed] [Google Scholar]
- Malashkevich V. N., Kammerer R. A., Efimov V. P., Schulthess T., Engel J. Science. 1996;274:761–765. doi: 10.1126/science.274.5288.761. [DOI] [PubMed] [Google Scholar]
- Hatherley D., Cherwinski H. M., Moshref M., Barclay A. N. J. Immunol. 2005;175:2469–2474. doi: 10.4049/jimmunol.175.4.2469. [DOI] [PubMed] [Google Scholar]
- Voulgaraki D., Mitnacht-Kraus R., Letarte M., Foster-Cuevas M., Brown M. H., Barclay A. N. Immunology. 2005;115:337–346. doi: 10.1111/j.1365-2567.2005.02161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganpule G., Knorr R., Miller J. M., Carron C. P., Dustin M. L. J. Immunol. 1997;159:2685–2692. [PubMed] [Google Scholar]
- Wojtowicz W. M., Flanagan J. J., Millard S. S., Zipursky S. L., Clemens J. C. Cell. 2004;118:619–633. doi: 10.1016/j.cell.2004.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata M., Weiner J. A., Sanes J. R. Cell. 2002;110:649–660. doi: 10.1016/s0092-8674(02)00910-8. [DOI] [PubMed] [Google Scholar]
- Vernon-Wilson E. F., Kee W. J., Willis A. C., Barclay A. N., Simmons D. L., Brown M. H. Eur. J. Immunol. 2000;30:2130–2137. doi: 10.1002/1521-4141(2000)30:8<2130::AID-IMMU2130>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Wright G. J., Puklavec M. J., Willis A. C., Hoek R. M., Sedgwick J. D., Brown M. H., Barclay A. N. Immunity. 2000;13:233–242. doi: 10.1016/s1074-7613(00)00023-6. [DOI] [PubMed] [Google Scholar]
- McMichael A. J., O'Callaghan C. A. J. Exp. Med. 1998;187:1367–1371. doi: 10.1084/jem.187.9.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causey L. D., Dwyer D. S. Nat. Biotechnol. 1996;14:348–351. doi: 10.1038/nbt0396-348. [DOI] [PubMed] [Google Scholar]
- Parish C. R., Recny M. A., Knoppers M. H., Waldron J. C., Warren H. S. J. Immunol. 1993;150:4833–4843. [PubMed] [Google Scholar]
- Boniface J. J., Rabinowitz J. D., Wulfing C., Hampl J., Reich Z., Altman J. D., Kantor R. M., Beeson C., McConnell H. M., Davis M. M. Immunity. 1998;9:459–466. doi: 10.1016/s1074-7613(00)80629-9. [DOI] [PubMed] [Google Scholar]
- Sandrin M. S., Mouhtouris E., Vaughan H. A., Warren H. S., Parish C. R. J. Immunol. 1993;151:4606–4613. [PubMed] [Google Scholar]
- Cooper M. A. J. Mol. Recognit. 2004;17:286–315. doi: 10.1002/jmr.675. [DOI] [PubMed] [Google Scholar]
- van der Merwe P. A., Barclay A. N. Curr. Opin. Immunol. 1996;8:257–261. doi: 10.1016/s0952-7915(96)80065-3. [DOI] [PubMed] [Google Scholar]
- Leavitt S., Freire E. Curr. Opin. Struct. Biol. 2001;11:560–566. doi: 10.1016/s0959-440x(00)00248-7. [DOI] [PubMed] [Google Scholar]
- Lebowitz J., Lewis M. S., Schuck P. Protein Sci. 2002;11:2067–2079. doi: 10.1110/ps.0207702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehn M., Bhattacharya R., Botstein D., Brown P. O. PLoS Genet. 2006;2:e11. doi: 10.1371/journal.pgen.0020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. C., Ho W. H., Gurney A., Rosenthal A. Nat. Neurosci. 2003;6:1270–1276. doi: 10.1038/nn1148. [DOI] [PubMed] [Google Scholar]
- Gonzalez L. C., Loyet K. M., Calemine-Fenaux J., Chauhan V., Wranik B., Ouyang W., Eaton D. L. Proc. Natl. Acad. Sci. U. S. A. 2005;102:1116–1121. doi: 10.1073/pnas.0409071102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtowicz W. M., Wu W., Andre I., Qian B., Baker D., Zipursky S. L. Cell. 2007;130:1134–1145. doi: 10.1016/j.cell.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushell K. M., Sollner C., Schuster-Boeckler B., Bateman A., Wright G. J. Genome Res. 2008;18:622–630. doi: 10.1101/gr.7187808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Lee E., Hestir K., Leo C., Huang M., Bosch E., Halenbeck R., Wu G., Zhou A., Behrens D., Hollenbaugh D., Linnemann T., Qin M., Wong J., Chu K., Doberstein S. K., Williams L. T. Science. 2008;320:807–811. doi: 10.1126/science.1154370. [DOI] [PubMed] [Google Scholar]
- Backliwal G., Hildinger M., Chenuet S., Wulhfard S., De Jesus M., Wurm F. M. Nucleic Acids Res. 2008;36:e96. doi: 10.1093/nar/gkn423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durocher Y., Perret S., Kamen A. Nucleic Acids Res. 2002;30:E9. doi: 10.1093/nar/30.2.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]