Abstract
Background
Climate in alpine habitats has undergone extreme variation during Pliocene and Pleistocene epochs, resulting in repeated expansion and contraction of alpine glaciers. Many cold-adapted alpine species have responded to these climatic changes with long-distance range shifts. These species typically exhibit shallow genetic differentiation over a large geographical area. In contrast, poorly dispersing organisms often form species complexes within mountain ranges, such as the California endemic ice-crawlers (Grylloblattodea: Grylloblattidae: Grylloblatta). The diversification pattern of poorly dispersing species might provide more information on the localized effects of historical climate change, the importance of particular climatic events, as well as the history of dispersal. Here we use multi-locus genetic data to examine the phylogenetic relationships and geographic pattern of diversification in California Grylloblatta.
Results
Our analysis reveals a pattern of deep genetic subdivision among geographically isolated populations of Grylloblatta in California. Alpine populations diverged from low elevation populations and subsequently diversified. Using a Bayesian relaxed clock model and both uncalibrated and calibrated measurements of time to most recent common ancestor, we reconstruct the temporal diversification of alpine Grylloblatta populations. Based on calibrated relaxed clock estimates, evolutionary diversification of Grylloblatta occurred during the Pliocene-Pleistocene epochs, with an initial dispersal into California during the Pliocene and species diversification in alpine clades during the middle Pleistocene epoch.
Conclusions
Grylloblatta species exhibit a high degree of genetic subdivision in California with well defined geographic structure. Distinct glacial refugia can be inferred within the Sierra Nevada, corresponding to major, glaciated drainage basins. Low elevation populations are sister to alpine populations, suggesting alpine populations may track expanding glacial ice sheets and diversify as a result of multiple glacial advances. Based on relaxed-clock molecular dating, the temporal diversification of Grylloblatta provides evidence for the role of a climate-driven species pump in alpine species during the Pleistocene epoch.
Background
In the present climate, many low-latitude mountain ranges are environmentally isolated from one another by intervening low elevation habitat. Geological and paleoecological data indicate that cooling global climate led to the historical growth of alpine glaciers [1,2] and to down-slope range shifts of montane vegetation [3,4]. It has been argued that ice age climates provided environmental corridors between mountain ranges that enabled arctic and alpine populations to migrate into and among sky-islands [5,6]. This hypothesis suggests that the expansion of continental and alpine glaciers during recent climatic cycles of the Pliocene and Pleistocene epochs facilitated spatial expansion of populations in alpine environments and, subsequently, contraction of these glaciers led to the diversification of geographically isolated lineages.
Molecular genetic studies support this hypothesis in several ways. First, several wide-ranging arctic species also inhabit alpine habitats [7] and these isolated populations exhibit recent genetic divergence [8-11]. This is most easily explained by colonization events or gene flow during the last glacial cycle. Second, many alpine species comprise closely related populations separated on geographically distant mountain ranges. These patterns are evident in mountainous regions in North America [12,13], Japan [14,15], Europe [e.g. [16]], and New Zealand [17]. Finally, several molecular studies examining divergence patterns of populations on separate mountains have detected low levels of gene flow during the divergence process, implying connectivity during glacial periods [18,19]. Fossil remains also suggest that winged insects [20-22] and plants commonly exhibit widespread range shifts during glacial cycles [3,23].
Most of the alpine organisms exhibiting these spatial patterns are highly vagile species. In contrast, many alpine species that have low vagility are considered to be restricted endemics and often form species complexes within mountain ranges [24]. The phylogeographic history of low vagility organisms might therefore exhibit different patterns [25,26] and can offer several important contributions to the study of alpine biogeographic history. First, population diversification of poorly dispersing species might provide insight on the effects of alpine glaciations within mountain ranges, in particular by delineating the spatial location of climatic refugia. Second, the temporal pattern of diversification within and between these species might provide information on the relative impact of different glacial cycles because poorly dispersing populations are more likely to diverge genetically due to the stochastic effects of genetic drift [27]. Third, low vagility species provide an opportunity to understand the dispersal history of populations in alpine environments. For example, these species clarify whether shifts in elevation are necessary to the survival of alpine populations, or if populations managed to survive in high elevation refugia [i.e. ice-free nunataks; [28]]. Similarly, under a "progression-rule" biogeographic model [29], ancestral-descendent relationships could be used to infer the direction of colonization into mountain ranges by a dispersing population.
In this study, the evolutionary history of a low vagility species complex is examined within California. Grylloblatta species (Grylloblattodea: Grylloblattidae), or ice-crawlers, are members of an ancient lineage that evolved during the radiation of neopterous insects [30]. All Grylloblatta species are cryophilic, with an obligate link to stable near-freezing temperatures. Acute physiological temperature tolerance ranges from -8.5 to 10°C in an undescribed species on Mt. Ranier [31], and low temperature tolerance is a conserved trait in the genus [32]. Very few arthropods survive prolonged exposure to temperatures experienced on snow fields, but Grylloblatta have a foraging strategy utilizing snow-fields, where wind-blown insects and organic detritus are deposited (into the "Aeolian ecosystem") and form an energy rich resource for scavenging insects [33]. While five extant genera of Grylloblattidae are known, less than thirty species have been described. These species occur in northeastern Asia (Korea, Japan, China and Russia) and western North America (Canada and the western United States). A single genus, Grylloblatta, occurs in North America with eleven described species, five of which occur in California. In California, Grylloblatta species are restricted to montane areas, caves, and deep canyons, and within these habitats remain in close proximity to permanent ice and deep retreats in rock outcrops. Previous to this study, most of the California species were known from type localities and are extremely rare in entomological collections.
Kamp [34,35], working on populations of grylloblattids in northeastern California and the Canadian Rocky Mountains, proposed that contemporary populations were relicts of populations broadly distributed in periglacial habitat during glacial periods. He hypothesized that 1) populations expanded their geographic range during glacial cycles and that 2) alpine populations colonized high elevation habitat after the retreat of glacial ice. We set out to test Kamp's [34] ideas by studying the genetic relationships of California Grylloblatta populations in the context of alpine glacial cycles in California. We attempt to explicitly test whether the diversification of Grylloblatta in California is intimately tied to glacial-interglacial fluctuations during the Pleistocene. Previous studies have shown that low vagility organisms often exhibit deep genetic breaks due to stochastic processes [25,26]. While these genetic breaks can occur at random in continuously distributed populations [36], they also occur due to evolutionary and environmental events that impede gene flow [37]. Multi-locus phylogenies offer a way to distinguish between random and non-random genetic breaks [38]. A previous multi-locus phylogeny of grylloblattid populations in both the Pacific Northwest and northeastern Asia [39] showed a recurrent pattern of deep genetic subdivision over a small geographical area, suggesting that California grylloblattids might provide similar levels of phylogenetic substructure. We therefore utilize genetic sequence data from multiple independent loci to infer the population genetic history, phylogenetic relationships and diversification time of California Grylloblatta species. Our results support Kamp's hypothesis that Grylloblatta populations expanded spatially at the edges of glacial habitat. Furthermore, we provide evidence that Pleistocene climatic shifts have had a direct role in Grylloblatta species formation.
Results
Population Genetic Patterns of Grylloblatta
A total of 132 individuals of Grylloblatta from 30 sites (Figure 1; Additional file 1 Table 1S) were screened for genetic variation at four genes. There were 52 unique COII haplotypes, with one individual (Lillburn Cave) having two highly divergent copies. These copies were amplified with two different primer sets, never co-amplified, and yet translate into appropriate amino acid residues. Although it is unclear whether or not this individual has acquired a nuclear mitochondrial DNA gene (numt), or is heteroplasmic, analyses using alternate copies did not significantly alter phylogenetic results. The copy amplified from the least degenerate mitochondrial primers was chosen for subsequent divergence time analyses. There were 12 unique sequences of the nuclear histone (h3) gene, 16 unique sequences of the nuclear 18 S gene, and 18 unique sequences of the nuclear 28 S gene. The combined sequences included a total of 5,542 sites (with gaps), with considerable rate variation among genes (Table 1). All sequences have been deposited in NCBI's GenBank (Accessions: FJ918575-FJ918687, GU013769-GU013770; Additional file 2 Table 2S).
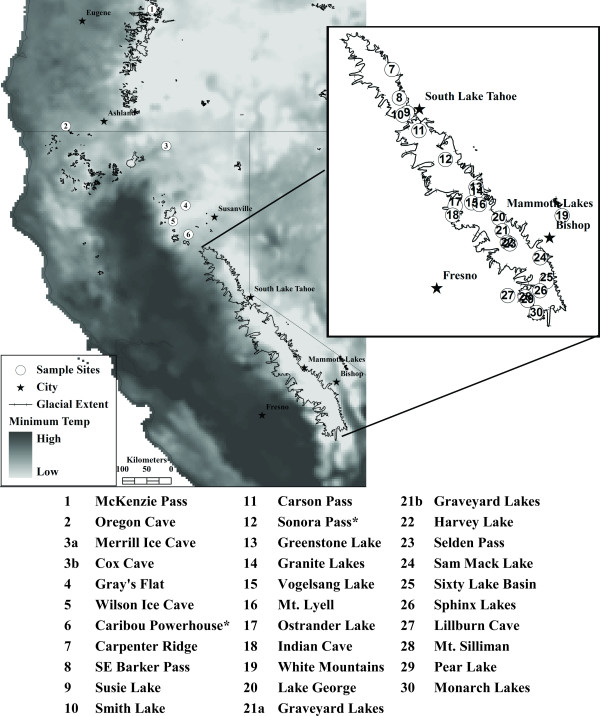
Figure 1.
Sampling locations of Grylloblatta. Location of sampled Grylloblatta populations in California and Oregon, overlaid on an mean minimum annual temperature surface for 1971-2000 [81] and an outline of glacial ice extent during the last glacial maximum [82]. Type localities of species indicated with an asterisk.
Table 1.
Properties of gene partitions used in phylogenetic analysis.
| Number of Basepairs | Variable Site % | Gap % | Relative Mean Rate of Substitution* | |
|---|---|---|---|---|
| COII | 742 | 19.27 | 0 | 1.033E-4 |
| Histone | 327 | 5.49 | 0 | 1.381E-5 |
| 18S | 2,052 | 3.21 | 0.11 | 3.216E-6 |
| 28S | 2,420 | 5.37 | 1.89 | 3.333E-6 |
*Relative substitution rates are based on a fixed mean rate of evolution across lineages (1/time × 10-4).
In five well-sampled alpine populations, within-population genetic variation is quite low (Table 2). Mitochondrial DNA variation approaches zero in the Sonora Pass, White Mountain and Graveyard Lakes populations (range θS per bp = 0.000-0.001, range θπ per bp = 0.000-0.001). In contrast, the Barker Pass and Susie Lake populations exhibit slightly higher mtDNA variation (range θS per bp = 0.004-0.007, range θπ per bp = 0.006). In all populations, variation in the nuclear gene histone is low and approaches zero. Tests for deviation from neutral equilibrium are non-significant in most populations, although some show a trend towards negative D and FS values. The Susie Lake population exhibits significant negative D and FS values in the histone locus, possibly indicating recent population expansion. Mismatch distribution tests for most populations across loci were consistent with a model of sudden population expansion, the exception being the Barker Pass population for COII. Under a spatial population expansion model, mismatch distributions were qualitatively similar (results not shown).
Table 2.
Population genetic summary statistics in five alpine populations of Grylloblatta.
| Barker Pass (n = 17) |
Susie Lake (n = 22) |
Sonora Pass (n = 12) |
White Mountains (n = 11) |
Graveyard Lakes (n = 12) |
|
|---|---|---|---|---|---|
| Cytochrome Oxidase subunit II | |||||
| θS(per bp) | 0.004 | 0.007 | 0.001 | 0.000 | 0.001 |
| θπ(per bp) | 0.006 | 0.006 | 0.001 | 0.000 | 0.001 |
| Tajima's D | 1.73 | -0.62 | 0.02 | -1.13 | -0.38 |
| Fu's FS | 4.31 | -2.21 | -0.62 | -0.41 | -0.36 |
| SSD | 0.21* | 0.03 | 0.02 | 0.02 | 0.02 |
| Raggedness | 0.33 | 0.07 | 0.17 | 0.44 | 0.20 |
| Histone | |||||
| θS(per bp) | 0.001 | 0.001 | 0.000 | 0.001 | 0.002 |
| θπ(per bp) | 0.001 | 0.000 | 0.000 | 0.001 | 0.004 |
| Tajima's D | -0.19 | -1.48* | 0 | -0.64 | 1.65 |
| Fu's FS | 0.28 | -2.86* | 0 | -0.18 | -1.13 |
| SSD | 0.32 | 0 | 0 | 0.02 | 0 |
| Raggedness | 0.37 | 0.68 | 0 | 0.46 | 0.08 |
*Statistical tests significant at α = 0.05 are labeled with an asterisk.
Estimates of Phylogenetic Relationships
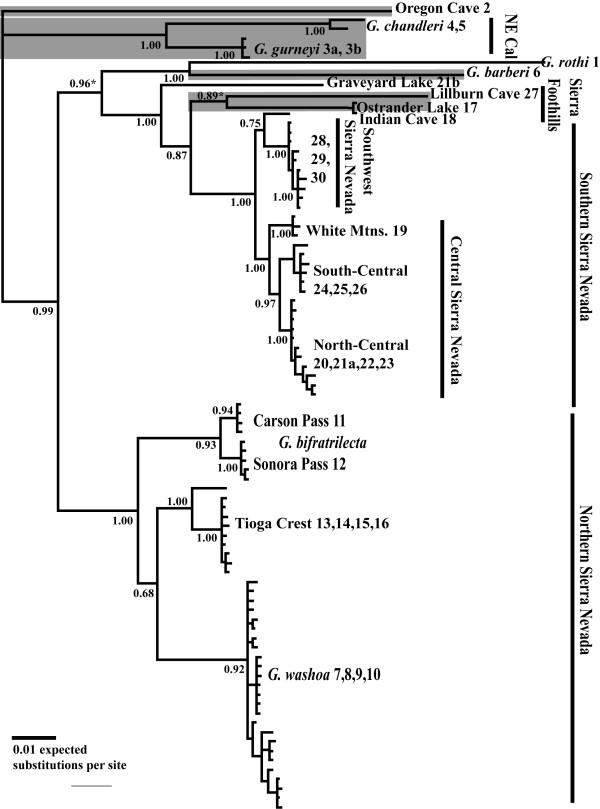
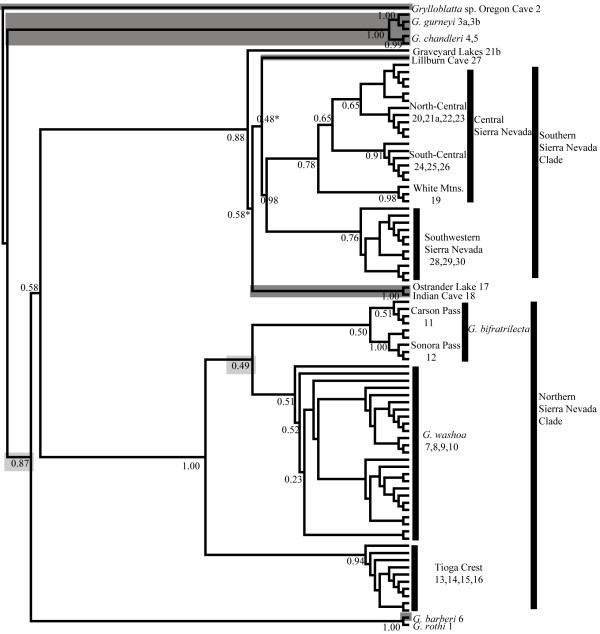
The concatenated phylogenies estimated from the unrooted Bayesian analysis (Figure 2) and the maximum likelihood bootstrap analysis (Figure 3) are very similar. Notable differences in the relationships of a few lineages (see gray highlighting) under these two methods largely concern the order of early branching events in the phylogeny. Separate Bayesian analyses of the mitochondrial data (Additional file 3 Figure 1S) and nuclear data (Additional file 4 Figure 2S) provide similar topologies to the Bayesian concatenated phylogeny, although fine scale geographic structure is only evident in the mitochondrial data. The concatenated phylogenetic analysis identifies three to four major lineages, with substantial phylogeographic structure within these lineages (Figure 4). The first lineage represents the Oregon Cave sample of southern Oregon. The second lineage includes two unique taxa, G. gurneyi and G. chandleri, sampled from northeastern California. The third lineage includes all individuals from the Sierra Nevada. In the maximum likelihood analysis, a fourth lineage is identified, which is nested within the Sierra Nevada lineage in the Bayesian analysis. This lineage includes G. rothi (Three Sisters, Oregon) and G. barberi (Feather River, California), which form a sister group in both analyses. Within the Sierra Nevada lineage, there is further subdivision into two major clades, including a northern Sierra Nevada clade and a southern Sierra Nevada clade (9.8% uncorrected mtDNA divergence). Within each of the Sierra Nevada clades there are strongly supported monophyletic groups that are geographically subdivided. The northern Sierra Nevada includes G. bifratrilecta, G. washoa, and a Tioga Crest subclade (5.6-7.1% uncorrected mtDNA divergence). The southern Sierra Nevada clade includes a southwest Sierra Nevada and a central Sierra Nevada subclade (3.1% uncorrected mtDNA divergence), as well as a deeply divergent subclade including Ostrander Lake-Indian Cave, Lillburn Cave, and a unique individual at Graveyard Lakes. The unusual Graveyard Lakes specimen is a juvenile which was sympatric and actively foraging with grylloblattids from the Central Sierra Nevada lineage, yet exhibits 6.6% uncorrected mtDNA divergence as well as unique nuclear alleles. The subclade including Ostrander Lake-Indian Cave, Lillburn Cave and the Graveyard Lake specimen are sister to the alpine populations in the southern Sierra Nevada. There is further geographic subdivision within the alpine subclades, notably north ("Carson Pass") to south ("Sonora Pass") in G. bifratrilecta and northeast ("White Mountains"), north ("North-Central"), and south ("South-Central") in the Central Sierra Nevada subclade.
Figure 2.
Bayesian phylogeny of California Grylloblatta. Concatenated phylogeny with Bayesian posterior probability support values shown at the nodes. Nodes conflicting with the maximum likelihood topology indicated by asterisks and low elevation populations indicated by shading in gray. Sample site numbers follow names at tips.
Figure 3.
Maximum likelihood phylogeny of California Grylloblatta. Maximum likelihood topology based on 1000 bootstrap replicates. Nodes conflicting with the Bayesian topology indicated by asterisks and low elevation populations indicated by shading in gray. Sample site numbers follow names at tips.
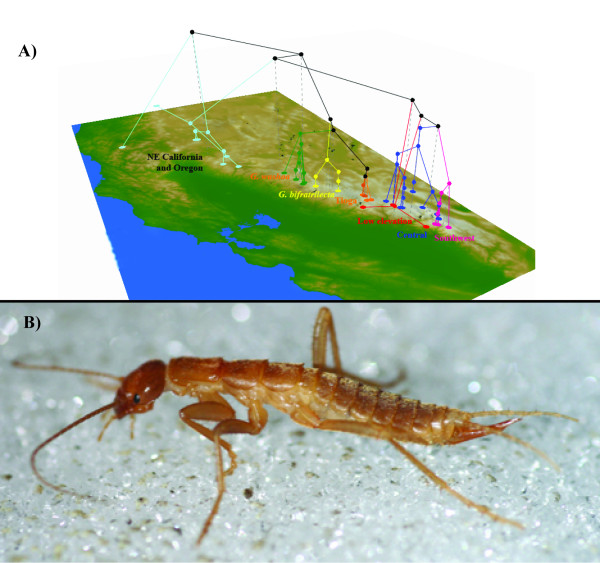
Figure 4.
Geographic distribution of alpine Grylloblatta lineages. A) Geographic distribution of genetically distinct alpine lineages in the Sierra Nevada overlaid on a topographic base map of California [82]. B) Adult female Grylloblatta sp. from the White Mountains.
Temporal Pattern of Diversification
Time to Most Recent Common Ancestor (TMRCA) was estimated using a Bayesian relaxed clock model with two different methods: an unscaled, fixed evolutionary rate and a calibrated molecular clock (Table 3). Unscaled estimates of TMRCA provide a representation of the tempo of diversification, unbiased by a prior on the molecular substitution rate. In the alpine populations, pulses of diversification are evident at several different time intervals, first in the diversification within the northern and southern Sierra Nevada clades, then in the diversification of subclades (G. bifratrilecta, G. washoa, Tioga Crest, Central Sierra Nevada, and Southwest Sierra Nevada), and finally in the diversification within populations of these subclades (White Mountains, Carson Pass and Sonora Pass). The time of divergence roughly doubles across these different levels and this is clear in the unscaled relaxed clock estimates, as well as the clock analysis. However, median estimates of TMRCA in both methods result in large confidence intervals and the lineage through time plot (Additional file 5 Figure 3S) does not indicate synchronous pulses of diversification. Instead, divergence rates are shown to be high in the past relative to a constant-rate model, with a subsequent decline and a more recent recovery.
Table 3.
Estimated Time to Most Recent Common Ancestor under a relaxed clock model.
| Unscaled TMRCA1 | COII Calibrated TMRCA2 | |||||
|---|---|---|---|---|---|---|
| Median | 95% Low | 95% High | Median | 95% Low | 95% High | |
| California | 696.17 | 372.43 | 1177.92 | 4.53MY | 2.59MY | 7.45MY |
| Northeastern California | 324.12 | 146.08 | 571.70 | 1.28MY | 498,100 | 2.47MY |
| G. gurneyi | 40.69 | 3.35 | 135.96 | 139,600 | 9,430 | 446,100 |
| G. chandleri | 110.81 | 27.18 | 228.72 | 239,600 | 38,680 | 595,500 |
| Division of Sierra Nevada Clades | 613.79 | 349.42 | 1142.14 | 3.94MY | 2.08MY | 6.62MY |
| Northern Sierra | 391.26 | 198.67 | 816.92 | 1.94MY | 994,800 | 3.36MY |
| G. bifratrilecta | 148.10 | 54.53 | 299.44 | 798,500 | 298,900 | 1.58MY |
| Carson Pass | 45.84 | 10.21 | 126.57 | 245,600 | 45,040 | 625,400 |
| Sonora Pass | 62.47 | 7.45 | 162.28 | 195,000 | 38,240 | 478,500 |
| G. washoa | 189.92 | 90.52 | 381.93 | 727,200 | 302,400 | 1.50MY |
| Tioga Crest | 219.01 | 63.87 | 459.42 | 967,300 | 364,200 | 1.85MY |
| Southern Sierra | 419.23 | 220.56 | 707.23 | 2.52MY | 1.32MY | 4.28MY |
| Low Elevation Sierra Populations | 417.75 | 241.96 | 730.51 | 2.54MY | 1.35MY | 4.31MY |
| Southwest Sierra Nevada | 189.90 | 74.48 | 350.74 | 806,100 | 330,400 | 1.52MY |
| Central Sierra Nevada | 213.16 | 108.11 | 372.05 | 814,600 | 380,000 | 1.45MY |
| White Mountains | 52.10 | 6.35 | 145.69 | 179,800 | 17,320 | 505,900 |
1. Based on a fixed mean rate of evolution across lineages (1/time × 10-4).
2. Based on a mean rate calibration for COII (1.5%/million years).
The calibrated molecular clock estimates have large confidence intervals, but the median estimates of TMRCA suggest Grylloblatta species inhabited California before the onset of the Pleistocene (~4.53 million years B.P., Table 3). TMRCA of major lineages in the Sierra Nevada are estimated to be in the Early Pleistocene (~1.94 MY for the Northern Sierra Nevada clade, ~2.52 for the Southern Sierra Nevada clade) and during the Early to Middle Pleistocene for geographically isolated subclades in the Sierra Nevada.
Discussion
Phylogenetic Relationships among California Grylloblatta
Previous phylogenetic analysis [39] demonstrated that North American Grylloblatta are monophyletic and derived from a northeast Asian grylloblattid ancestor, and that Grylloblatta was split into two deeply divergent lineages occupying the northern Cascades and southern Oregon-California. By increasing the taxonomic and spatial sampling of Grylloblatta in California, our analysis reveals a deep split in the Sierra Nevada with a high degree of local geographic subdivision. Both Bayesian and maximum likelihood methods identify a basal polytomy of three major lineages. These lineages include an Oregon Cave lineage, a northeastern California lineage, and a Sierra Nevada lineage. A fourth lineage is identified in the maximum likelihood analysis, comprised of southern Cascades G. rothi and northern Californian G. barberi, but is alternatively nested in the Sierra Nevada lineage in the Bayesian analysis. This conflict might result from differences in methodological assumptions and/or long-branch effects. All currently recognized species in California form discrete monophyletic groups and correspond to subclades within the major lineages. The northeastern California lineage includes G. gurneyi and G. chandleri. The northern Sierra Nevada clade includes G. bifratrilecta and G. washoa. The monophyly of these groups indicates that morphological criteria have been useful in delineating distinct evolutionary lineages. However, other monophyletic groups of similar levels of genetic divergence are present in the Sierra Nevada, including populations along the Tioga Crest, the southwest Sierra, and the central Sierra. Several other specimens from unique low elevation sites (Ostrander Lake and Lillburn Cave) are highly divergent from and sister to alpine subclades. Finally, one specimen collected at Graveyard Lakes is genetically distinct, with unique alleles at all loci (unshared by 12 other individuals at that site). This specimen may represent a sympatric population that has come into contact but is reproductively isolated from the Central Sierra Nevada lineage. Most of the Sierra Nevada populations sampled in our study represent new localities and several monophyletic clades may represent undescribed species.
Biogeographic Patterns of Lineage Diversification
The diversification of grylloblattids in North America has taken place over a large geographic region that has experienced a complex geological and climatic history [1]. Despite dramatic environmental events, including periodic volcanic eruptions, Grylloblatta have managed to persist and recolonize alpine habitats [40]. They are currently found in small, relictual populations where local microenvironments provide cold temperature retreats. The major division of North American Grylloblatta occurs between a northern Cascades-Rocky Mountain clade and a California-Southern Oregon clade [39]. This provides little information about the geographic origin of Californian taxa, but indicates that a common ancestor diversified across western North America in the distant past. Furthermore, a basal polytomy in our phylogenetic analysis leaves the relationship of major California lineages unresolved. The Oregon Cave lineage is isolated in the lower slopes of the Siskiyou Mountains, which stretch south into the California Trinity Alps and Mount Shasta region. This population is at the southern end of the Cascades Range and represents the most likely source of colonization into California. However, the unusual relationship and placement of G. rothi and G. barberi suggests that there may have been multiple dispersal events between Oregon and California, perhaps utilizing other biogeographic corridors.
Relationships of the remaining California taxa are particularly interesting, because these species are a mix of low elevation relict populations and alpine populations. Kamp [34] proposed that taxa in northeastern California were relicts of populations broadly distributed in periglacial habitat during glacial periods. The geographical location of widely separated populations of G. chandleri supports this hypothesis, as these populations lie within the periglacial zone of Mt. Lassen. G. chandleri and G. gurneyi form a highly divergent monophyletic group in northeastern California, suggesting that these low elevation populations have been isolated for a long period of time. Time to Most Recent Common Ancestor, based on fossil-derived substitution rates, suggests diversification within these species (~239,600 years B.P. for G. chandleri and ~139,600 for G. gurneyi) was initiated during the early stages of glacial cooling phases [41]. Other low elevation populations in our study, including G. barberi, Ostrander Lake, and Lillburn Cave, are distinct from and sister to alpine lineages. These taxa do not appear to be recent remnant populations of alpine populations that moved down-slope during glacial periods, but instead seem to share ancestry with populations that gave rise to alpine lineages. All populations in the Sierra Nevada share a single common ancestor, suggesting one dispersal event gave rise to a diverse group of alpine lineages.
Within the alpine lineages, there is a high degree of substructure reflecting the dispersal limitation of populations and, perhaps, the existence of geographical barriers. Some subclades (Central Sierra Nevada, Southwest Sierra Nevada, G. barberi, Tioga Crest) have unique mitochondrial haplotypes at all geographical localities sampled, but other well-sampled populations (i.e. G. washoa) have shared haplotypes across sites. Current sampling does not clearly elucidate whether nunataks (ice-free high elevation habitat) were important in the persistence of alpine populations and the preservation of genetic variation. It is clear that alpine populations are subdivided north to south, with unique lineages occupying major drainage basins. These drainages were the principal course of alpine glacial growth and it is probable that Grylloblatta populations tracked suitable habitat along the periglacial edge environment of these growing glaciers. The White Mountains, lying east of the Sierra Nevada at the edge of the Great Basin, has a population of Grylloblatta closely related to Central Sierra Nevada populations. Currently, a distance of roughly 30 miles separates the two ranges in the Long Valley region (elevation >2,125 m). Historically, a high plateau connected these mountain ranges, but subsidence along the Owens Valley fault (beginning 3 million years ago and reaching its present state around 300,000 years ago) separated high elevation habitat of both mountain ranges [42,43]. Our dating analysis suggests that Grylloblatta populations from the White Mountains (TMRCA median estimate ~179,800, 95% CI 17,320-505,900) may have been isolated during this period of subsidence. These are the first data to suggest multiple refugia existed in the Sierra Nevada, and on both sides of the crest, during glacial phases.
Evolutionary History of Alpine Endemics
Grylloblatta lineages, clades and subclades in California are characterized by broadly overlapping periods of diversification. Using unscaled relaxed clock methods, median estimates of TMRCA reveal pulses at multiple nested levels in the phylogeny. However there is inherent error in these estimates and a lineage through time plot does not show synchronous pulses of diversification. Instead, the lineage through time analysis shows a high early diversification rate, a pronounced decline, and a very recent recovery in Grylloblatta diversification. The calibrated molecular clock rate places diversification times within Sierra Nevada Grylloblatta lineages during the Pleistocene glacial stages. Although some median estimates are in accord with specific events in the glacial chronology in the Sierra Nevada, confidence intervals are broad and overlap glacial-interglacial cycles. For example, median age estimates of within-population variation in alpine populations is ~150,000 years B.P., during the glacial maximum of the Illinoian advance, which was followed by an interglacial warm phase [23], but confidence intervals include the most recent glacial maximum (~25,000 years B.P.). Hence, the inherent uncertainty in the timing of diversification does not provide clear resolution to whether grylloblattid populations diversify during glacial advances or, alternatively, diversify during the retreat of alpine glaciers. Population genetic statistical tests provide equivocal support for recent population expansion in several well-sampled alpine Grylloblatta populations, although this may be an artifact of limited sample sizes. Furthermore, glacial phases in the Sierra Nevada are known to involve multiple glacial advances and retreats [44], which caused dynamic shifts in vegetation over short periods of time [23,45]. The pace and scale of such changes may not have favored periods of stable growth in Grylloblatta populations.
An older period of lineage diversification, corresponding to the TMRCA of recognized morphological species, dates to ~700,000-1,000,000 years B.P., well within the Middle Pleistocene epoch. Morphological criteria for distinguishing these species involve differences in male and female reproductive morphology, as well as differences in sensory structures and body size attributes. Changes in reproductive morphology are known to impede reproductive compatibility in insects [46], which suggests that multiple alpine Grylloblatta lineages have undergone recent speciation. The TMRCA of all California and all Sierra Nevada populations (~3.9-4.5 MYA) predates the Pleistocene. This suggests that Grylloblatta species may have colonized California during glacial advances of Pliocene epoch. It is likely that climatic oscillations led to the formation of discrete genetic populations, morphologically distinguishable taxa [47,48], and the first dispersal event into the Sierra Nevada during the evolution of California Grylloblatta.
Conclusions
Recent speciation and the formation of multiple endemic lineages may be a common feature of alpine landscapes. This has been shown repeatedly in alpine taxa, as demonstrated in examples of North American grasshoppers [49], New Zealand plants and insects [50], and European plants [51]. These studies provide evidence that Pleistocene climate change has acted as a species pump [52] in alpine environments. California Grylloblatta provide an example of lineage diversification during glacial expansion events on a very small geographical scale. This highlights the importance of multiple refugia in mountains as sources of contemporary genetic variation in alpine species.
Methods
Genetic sampling
Sampling was done between 2005 and 2008 throughout California, with a particular focus on alpine populations in the Sierra Nevada. Despite systematic efforts to sample Grylloblatta [[34,53]; Schoville, S.D. unpublished data, [54]], populations are highly fragmented and local population densities appear to be low. Genetic samples comprise thirty sites and a total of 134 individuals (Figure 1; Additional file 1 Table 1S; Additional file 2 Table 2S). The majority of populations sampled are new locality records, including most Sierra Nevada collecting sites. Relevant genetic samples were taken from Jarvis and Whiting [39], including four California sites and two sites from Oregon. An individual of Grylloblatta from Oregon Cave, Oregon is used as an out-group based on the phylogenetic results of Jarvis and Whiting [39]. When possible, individuals were assigned to species based on previous phylogenetic hypotheses [34,39] and using morphological criteria [47,48,55], but populations from new sites within the Sierra Nevada have not been assigned to previously described species. All specimens have been deposited in the Essig Museum of Entomology, University of California, Berkeley.
Molecular techniques and data preparation
Genomic DNA was extracted from 1-2 legs of each individual with a Qiagen DNEasy Blood and Tissue Kit (Qiagen) with reduced elution volumes. To maintain consistency with previously collected datasets [39], genetic data were collected from one mitochondrial and three nuclear genes. A region of approximately 790 bp representing the entire mitochondrial cytochrome oxidase subunit II (COII) gene was amplified and sequenced in both directions with the primers COII-F-Leucine and COII-R-Lysine [56]. Three nuclear genes were also amplified, including histone (H3) and the 18 S and 28 S ribosomal genes. A 380 bp region of the histone gene was amplified and sequenced in both directions using the primers HexA-F and HexA-R [56]. 2,100 bp of 18 S and 2,450 bp of 28 S were amplified using several overlapping primer combinations [57]. PCR products were sequenced in both directions on an ABI 3730 capillary sequencer using Big Dye v3.1 chemistry (Applied Biosystems).
DNA sequences were manually edited using the program SEQUENCHER v4.8 (Gene Codes Corporation). Some individuals were heterozygous at histone (H3) and haplotypes phase was estimated using the program PHASE v2 [58]. Visual alignments were made for each gene in the program MACLADE v4.08 [59]. Gaps were evident in both 18 S and 28 S and these were retained in subsequent phylogenetic analysis.
Population Genetic Analysis of Alpine Populations
Changes in demographic history are known to affect the frequency of alleles, the distribution of mutations, and the coalescent times of gene copies. Population summary statistics examine genetic variability within populations and can be used to test for departures from demographic equilibrium. Statistical tests using Tajima's D, Fu's FS, and the mismatch distribution SSD and Raggedness Index were calculated in ARLEQUIN v3.11 [60]. The D and FS neutrality tests are sensitive to changes in demography or selection [61,62]. Significant negative values indicate population growth or purifying expansion, while significant positive values indicate population bottlenecks or balancing selection. Mismatch distributions model the frequency distribution of pairwise differences in a sudden population expansion [63], as well as under a spatial population expansion model [64,65]. Significant SSD or Raggedness Index values indicate that an expansion model is rejected by the data. Although most of the Grylloblatta sample sites have too few individuals to calculate population genetic summary statistics, five alpine localities contain sample sizes of greater than ten individuals (Barker Pass, Susie Lake, Sonora Pass, White Mountains and Graveyard Lakes). Each of the summary statistics was calculated for COII and histone in these five localities. Significance values for all four statistical tests were calculated using 1,000 bootstrap replicates.
Phylogenetic Analysis
Phylogenetic relationships of California Grylloblatta were estimated using an unrooted Bayesian method and a maximum likelihood bootstrapping method. The four genes were concatenated and partitioned in each analysis. COII was further partitioned by codon position. MRMODELTEST2 v2.3 [66] was used to estimate models of molecular evolution for each partition. Based on Akaike Information Criteria [AIC, [67]], the best models for each partition were: COIIpos1 GTR+G+I, COIIpos2 GTR+I, COIIpos3 GTR+G+I, histone HKY+I, 18 S GTR+I, and 28 S GTR+I. Histone was not partitioned by codon due to the lack of variable sites at first and second codon positions.
The program MRBAYES v3.1.2 [68] was used to estimate a Bayesian phylogeny of the concatenated and partitioned dataset (n = 87). Polymorphic sites at nuclear genes were treated as base ambiguities. Two independent analyses were run under the following conditions: 15 million steps, four chains, and genealogies sampled every 1000 steps. Performance of the Monte Carlo Markov Chain simulations was assessed for convergence using AWTY [69]. The first 4,000 samples were discarded as burn-in samples and a 50% majority rule consensus tree of the two independent runs was produced with posterior probability values equal to bipartition frequencies. A maximum-likelihood tree was also estimated using a rapid bootstrap heuristic in RAxML v7.0.4 [70]. A single run was conducted with 1000 bootstrap replicates followed by a full maximum likelihood search, using the partitioned data with the GTR+G+I model selected for each partition and the Oregon Cave sample selected as an out-group. Additionally, an mtDNA gene tree and a concatenated nuclear gene tree were estimated using MRBAYES with the following conditions: data partitioned by codon, two separate runs of 20 million steps each, four chains, and genealogies sampled every 1000 steps. The first 5,000 samples were discarded as burn-in samples and a 50% majority rule consensus tree of the two runs was produced with posterior probability values equal to bipartition frequencies.
The concatenated phylogeny was plotted on a digital elevation base map of North American using GENGIS v.1.05 [71].
Timing of Lineage Diversification
Coalescent theory can be used to estimate the age of a monophyletic lineage by calculating the time to most recent common ancestor (TMRCA). This approach was used to calculate the age of the Grylloblatta lineages, as well as the time to a most recent common ancestor of sister clades. The estimation of absolute ages requires translation of molecular substitution rates into average rates per year. These rates cannot currently be estimated using fossil calibration because no there are no known fossils within the extant Grylloblattidae or of recent ancestral stem groups [72]. Mitochondrial substitution rates are available from fossil-based studies of other insect lineages [73,74]. These studies have shown a convergence in cytochrome oxidase subunit I mutation rates at 1.5% per million years. Molecular substitution rates are similar between cytochrome oxidase subunit I and cytochrome oxidase subunit II in insects, and the same rate is often applied to both genes [e.g [75,76]].
In order to explicitly address the uncertainty in using imported substitution rates, a relaxed clock model was selected for a Bayesian phylogenetic analysis in BEAST v1.5.3 [77,78]. Under this model, substitution rates are allowed to vary within a lineage and across lineages within the dataset under the constraints of a prior distribution. Two alternative priors were chosen to explore the divergence time of California Grylloblatta. First, the mean rate of evolution across the whole tree fixed to a mean of 1 in order to provide unscaled TMRCA estimates [78]. Second, a mean substitution rate estimate of 1.5% per million years was applied to COII. Uncertainty was added to the substitution rate by setting the standard deviation to 0.5% (mean = 0.015, 95% interval range: 0.00678 to 0.0232 per million years). The multi-locus dataset was partitioned by gene using the following substitution models: COII GTR+G+I, histone HKY+I, 18 S GTR+I, and 28 S GTR+I. The relaxed clock model with an uncorrelated log-normal distribution was chosen as a prior for rate variation among lineages and the Yule speciation process was chosen as a tree prior. Independent analyses were conducted under the following conditions: 100 million steps, genealogies sampled every 1000 steps, and a burn-in period of 10 million steps. Median estimates of time to most recent common ancestor (TMRCA) and 95% confidence intervals were calculated within the phylogeny using TRACER v1.5.
Lineage through time plots were constructed using the unscaled ultrametric phylogeny from the relaxed clock analysis. The library APE [79] in the software R v2.10.1 [80] was used to plot log-transformed lineage diversification versus time, relative to null model with a constant rate of speciation.
Authors' contributions
SDS conducted the field work, laboratory work, genetic analysis and wrote the manuscript. GKR provided critical comments on the analysis and drafting of the manuscript. Both authors read and approved the final manuscript.
Authors' Information
Sean Schoville's research focuses on population history, climate change and conservation of alpine insects. This work was completed as part of his PhD research on alpine biogeography. George Roderick is a Professor in the Department of Environmental Science, Policy and Management. His research interests include historical population dynamics, invasion biology, and biodiversity, mostly of insects.
Supplementary Material
Table 1S. Collecting localities of Grylloblatta. Latitude and longitude reported in the WGS84 datum. Type localities indicated with an asterisk.
Table 2S. GenBank Accession numbers of California Grylloblatta collections.
Figure 1S. Mitochondrial gene tree of California Grylloblatta species. Bayesian posterior probability support values are shown at the base of the nodes.
Figure 2S. Concatenated nuclear gene tree of California Grylloblatta species. Bayesian posterior probability support values are shown at the base of the nodes.
Figure 3S. Lineage through time plot of California Grylloblatta species. Plot of log-lineage diversification (solid line) over unscaled time, compared to a null model (dashed line) of constant diversification.
Contributor Information
Sean D Schoville, Email: schoville@berkeley.edu.
George K Roderick, Email: roderick@berkeley.edu.
Acknowledgements
S.D. Schoville would like to dedicate this manuscript to the memory of Stephanie Yeun Kim and thank her for her support and companionship during this research. We are also grateful to the following people for help with field work or for providing specimens: Jean Krejca (Zara Environmental LLC), Tina Cheng, Danny Cluck, Sean Rovito, Matt Medeiros and Tate Tunstall. Thanks to Will Richardson for providing us with the Barker Pass locality and to Karl Jarvis for his assistance with Grylloblatta natural history. This research was supported by grants to SD Schoville from the Yosemite Fund, White Mountain Research Station, Magy Fellowship, and Walker Fund. All specimens were collected under permits granted by Sequoia and Kings National Parks (Study# SEKI-0091), Yosemite National Park (Study# YOSE-00093), and the California Fish and Game Department (#SC-006997). Our thanks to three anonymous reviewers and the editor for comments on our manuscript.
References
- Kaufman DS, Porter SC, Gillespie AR. In: The Quaternary Period in the United States, 1. Gillespie AR, Porter SC, Atwater BF, editor. Vol. 1. Amsterdam, The Netherlands: Elsevier; 2004. Quaternary alpine glaciation in Alaska, the Pacific Northwest, Sierra Nevada, and Hawaii; pp. 77–103. [Rose J (Series Editor): Developments in Quaternary Science] [Google Scholar]
- Ehlers J, Gibbard PL. Quaternary Glaciations. Extent and Chronology: Part I Europe. Den Haag: Elsevier; 2004. [Google Scholar]
- Thompson RS, Anderson KH. Biomes of western North America at 18,000, 6000 and 0 C-14 yr BP reconstructed from pollen and packrat midden data. Journal of Biogeography. 2000;27:555–584. doi: 10.1046/j.1365-2699.2000.00427.x. [DOI] [Google Scholar]
- Frenzel B. History of flora and vegetation during the Quaternary North America. Progress in Botany. 2005;66:409–440. full_text. [Google Scholar]
- Chabot BF, Billings WD. Origins and ecology of the Sierran alpine flora and vegetation. Ecological Monographs. 1972;42:163–199. doi: 10.2307/1942262. [DOI] [Google Scholar]
- DeChaine EG, Martin AP. Historic cycles of fragmentation and expansion in Parnassius smintheus (Papilionidae) inferred using mitochondrial DNA. Evolution. 2004;58:113–127. doi: 10.1111/j.0014-3820.2004.tb01578.x. [DOI] [PubMed] [Google Scholar]
- Billings WD. Arctic and alpine vegetations: similarities, differences, and susceptibility to disturbance. Bioscience. 1973;23:697–704. doi: 10.2307/1296827. [DOI] [Google Scholar]
- Holderegger R, Abbott RJ. Phylogeography of the Arctic-Alpine Saxifraga oppositifolia (Saxifragaceae) and some related taxa based on cpDNA and ITS sequence variation. American Journal of Botany. 2003;90:931–936. doi: 10.3732/ajb.90.6.931. [DOI] [PubMed] [Google Scholar]
- Brochmann C, Gabrielsen TM, Nordal I, Jon YL, Elven R. Glacial Survival or tabula rasa? The History of North Atlantic Biota Revisited. Taxon. 2003;52:417–450. doi: 10.2307/3647444. [DOI] [Google Scholar]
- Reisch C. Glacial history of Saxifraga paniculata (Saxifragaceae): molecular biogeography of a disjunct arctic-alpine species from Europe and North America. Biological Journal of the Linnean Society. 2008;93:385–398. [Google Scholar]
- Schonswetter P, Popp M, Brochmann C. Rare arctic-alpine plants of the European Alps have different immigration histories: the snow bed species Minuartia biflora and Ranunculus pygmaeus. Molecular Ecology. 2006;15:709–720. doi: 10.1111/j.1365-294X.2006.02821.x. [DOI] [PubMed] [Google Scholar]
- Nice CC, Shapiro AM. Patterns of morphological, biochemical, and molecular evolution in the Oeneis chryxus complex (Lepidoptera: Satyridae): A test of historical biogeographical hypotheses. Molecular Phylogenetics and Evolution. 2001;20:111–123. doi: 10.1006/mpev.2001.0951. [DOI] [PubMed] [Google Scholar]
- Hafner DJ, Sullivan RM. Historical and ecological biogeography of Nearctic pikas (Lagomorpha: Ochotonidae) Journal of Mammology. 1995;76:302–321. doi: 10.2307/1382343. [DOI] [Google Scholar]
- Ikeda H, Senni K, Fujii N, Setoguchi H. Post-glacial range fragmentation is responsible for the current distribution of Potentilla matsumurae Th. Wolf (Rosaceae) in the Japanese archipelago. Journal of Biogeography. 2008;35:791–800. doi: 10.1111/j.1365-2699.2007.01828.x. [DOI] [Google Scholar]
- Fujii N. Phylogeographic studies in Japanese alpine plants based on intraspecific chloroplast DNA variations. Bulletin of the Biogeographical Society of Japan. 1997;52:59–69. [Google Scholar]
- Tribsch A, Stuessy TF. Evolution and phylogeography of arctic and alpine plants in Europe: Introduction. Taxon. 2003;52:415–416. doi: 10.2307/3647443. [DOI] [Google Scholar]
- Buckley TR, Simon C, Shimodaira H, Chambers GK. Evaluating hypotheses on the origin and evolution of the New Zealand alpine cicadas (Maoricicada) using multiple-comparison tests of tree topology. Molecular Biology and Evolution. 2001;18:223–234. doi: 10.1093/oxfordjournals.molbev.a003796. [DOI] [PubMed] [Google Scholar]
- Eckert AJ, Tearse BR, Hall BD. A phylogeographical analysis of the range disjunction for foxtail pine (Pinus balfouriana, Pinaceae): the role of Pleistocene glaciation. Molecular Ecology. 2008;17:1983–1997. doi: 10.1111/j.1365-294X.2008.03722.x. [DOI] [PubMed] [Google Scholar]
- Schoville SD, Roderick GK. Alpine biogeography of Parnassian butterflies during Quaternary climate cycles in North America. Molecular Ecology. 2009;18:3471–3485. doi: 10.1111/j.1365-294X.2009.04287.x. [DOI] [PubMed] [Google Scholar]
- Ashworth AC. The response of arctic Carabidae (Coleoptera) to climate change based on the fossil record of the Quaternary period. Annales Zoologici Fennici. 1996;33:125–131. [Google Scholar]
- Elias SA. Insects and climate change. Bioscience. 1991;41:552–559. doi: 10.2307/1311608. [DOI] [Google Scholar]
- Potito AP, Porinchu DF, MacDonald GM, Moser KA. A late Quaternary chironomid-inferred temperature record from the Sierra Nevada, Califomia, with connections to northeast Pacific sea surface temperatures. Quaternary Research. 2006;66:356–363. doi: 10.1016/j.yqres.2006.05.005. [DOI] [Google Scholar]
- Woolfenden WB. A 180,000-year pollen record from Owens Lake, CA: terrestrial vegetation change on orbital scales. Quaternary Research. 2003;59:430–444. doi: 10.1016/S0033-5894(03)00033-4. [DOI] [Google Scholar]
- Mani MS. Ecology and Biogeography of High Altitude Insects. The Hague: Dr. W. Junk N.V. Publishers; 1968. [Google Scholar]
- Hugall A, Moritz C, Moussalli A, Stanisic J. Reconciling paleodistribution models and comparative phylogeography in the Wet Tropics rainforest land snail Gnarosophia bellendenkerensis (Brazier 1875) Proceedings of the National Academy of Sciences of the United States of America. 2002;99:6112–6117. doi: 10.1073/pnas.092538699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jockusch EL, Wake DB. Falling apart and merging: diversification of slender salamanders (Plethodontidae: Batrachoseps) in the American West. Biological Journal of the Linnean Society. 2002;76:361–391. doi: 10.1111/j.1095-8312.2002.tb01703.x. [DOI] [Google Scholar]
- Wright S. Isolation by distance. Genetics. 1943;28:114–138. doi: 10.1093/genetics/28.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehlik I. Nunataks and peripheral refugia for alpine plants during quaternary glaciation in the middle part of the Alps. Botanica Helvetica. 2000;110:25–30. [Google Scholar]
- Hennig W. Phylogenetic Systematics. Urbana: University of Illinois Press (translated by D. D. Davis and R. Zangerl); 1966. [Google Scholar]
- Terry MD, Whiting MF. Mantophasmatodea and phylogeny of the lower neopterous insects. Cladistics. 2005;21:240–257. doi: 10.1111/j.1096-0031.2005.00062.x. [DOI] [Google Scholar]
- Edwards JS. In: Biology of the Notoptera. Ando H Nagano, editor. Japan: Kashiyo-Insatsu Co. Ltd; 1982. Habitat, behavior and neurobiology of an American Grylloblattid; pp. 19–28. [Google Scholar]
- Henson WR. Temperature preference of Grylloblatta campodeiformis (Walker) Nature. 1957;637:179. [Google Scholar]
- Edwards JS. Arthropods of alpine aeolian ecosystems. Annual Review of Entomology. 1987;32:163–179. doi: 10.1146/annurev.en.32.010187.001115. [DOI] [Google Scholar]
- Kamp JW. Descriptions of two new species of Grylloblattidae and of the adult of Grylloblatta barberi, with an interpretation of their geographical disbtribution. Annals of the Entomological Society of America. 1963;56:53–68. [Google Scholar]
- Kamp JW. Taxonomy, distribution, and zoogeographic evolution of Grylloblatta in Canada (Insecta: Notoptera) Canadian Entomologist. 1979;111:27–38. doi: 10.4039/Ent11127-1. [DOI] [Google Scholar]
- Irwin DE. Phylogeographic breaks without geographic barriers to gene flow. Evolution. 2002;56:2383–2394. doi: 10.1111/j.0014-3820.2002.tb00164.x. [DOI] [PubMed] [Google Scholar]
- Avise JC. Phylogeography: The History and Formation of Species. Cambridge: Harvard University Press; 2000. [Google Scholar]
- Kuo CH, Avise J. Phylogeographic breaks in low-dispersal species: the emergence of concordance across gene trees. Genetica. 2005;124:179–186. doi: 10.1007/s10709-005-2095-y. [DOI] [PubMed] [Google Scholar]
- Jarvis KJ, Whiting MF. Phylogeny and biogeography of ice crawlers (Insecta: Grylloblattodea) based on six molecular loci: Designating conservation status for Grylloblattodea species. Molecular Phylogenetics and Evolution. 2006;41:222–237. doi: 10.1016/j.ympev.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Sugg PM, Edwards JS. Pioneer aeolian community development on pyroclastic flows after the eruption of Mount St. Helens, Washington, U.S.A. Arctic and Alpine Research. 1998;30:400–407. doi: 10.2307/1552013. [DOI] [Google Scholar]
- Lisiecki LE, Raymo ME. A Pliocene-Pleistocene stack of 57 globally distributed benthic δ18O records. Paleoceanography. 2005;20 [Google Scholar]
- Stockli DF, Dumitru TA, McWilliams MO, Farley KA. Cenozoic tectonic evolution of the White Mountains, California and Nevada. Geological Society of America Bulletin. 2003;115:788–816. doi: 10.1130/0016-7606(2003)115<0788:CTEOTW>2.0.CO;2. [DOI] [Google Scholar]
- Bachman SB. Pliocene-Pleistocene break-up of the Sierra Nevada-White-Inyo Mountains block and formation of Owens Valley. Geology. 1978;6:461–463. doi: 10.1130/0091-7613(1978)6<461:PBOTSN>2.0.CO;2. [DOI] [Google Scholar]
- Bischoff JL, Cummins K. Wisconsin glaciation of the Sierra Nevada (79,000-15,000 yr B.P.) as recorded by rock flour in sediments of Owens Lake, California. Quaternary Research. 2001;55:14–24. doi: 10.1006/qres.2000.2183. [DOI] [Google Scholar]
- Jennings SA, Elliott-Fisk DL. Pack-rat midden evidence of Late Quaternary vegetation change in the White Mountains, California-Nevada. Quaternary Research. 1993;39:214–221. doi: 10.1006/qres.1993.1024. [DOI] [Google Scholar]
- Eberhard WG. Sexual Selection and Animal Genitalia. Cambridge, Mass.: Harvard University Press; 1985. [Google Scholar]
- Gurney AB. Recent advances in the taxonomy and distribution of the Grylloblattidae. Journal of the Washington Academy of Sciences. 1953;43:325–332. [Google Scholar]
- Gurney AB. Further advances in the taxonomy and distribution of Grylloblattidae. Proceedings of the Biological Society of Washington. 1961;74:67–76. [Google Scholar]
- Knowles LL. Did the Pleistocene glaciations promote divergence? Tests of explicit refugial models in montane grasshopprers. Molecular Ecology. 2001;10:691–701. doi: 10.1046/j.1365-294x.2001.01206.x. [DOI] [PubMed] [Google Scholar]
- Buckley TR, Simon C. Evolutionary radiation of the cicada genus Maoricicada Dugdale (Hemiptera: Cicadoidea) and the origins of the New Zealand alpine biota. Biological Journal of the Linnean Society. 2007;91:419–435. doi: 10.1111/j.1095-8312.2007.00807.x. [DOI] [Google Scholar]
- Comes HP, Kadereit JW. Spatial and temporal patterns in the evolution of the flora of the European alpine system. Taxon. 2003;52:451–462. doi: 10.2307/3647445. [DOI] [Google Scholar]
- Weir JT, Schluter D. Ice sheets promote speciation in boreal birds. Proceedings of the Royal Society B: Biological Sciences. 2004;271:1881–1887. doi: 10.1098/rspb.2004.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann DH, Edwards JS, Gara RI. Diel activity patterns in snowfield foraging invertebrates on Mount Ranier, Washington. Arctic and Alpine Research. 1980;12:359–368. doi: 10.2307/1550722. [DOI] [Google Scholar]
- Huggard DJ, Klenner W. Grylloblattids in managed forests of south-central British Columbia. Northwest Science. 2003;77:12–18. [Google Scholar]
- Gurney AB. The taxonomy and distribution of the Grylloblattidae. Proceedings of the Entomological Society of Washington. 1948;50:86–102. [Google Scholar]
- Svenson GJ, Whiting MF. Phylogeny of Mantodea based on molecular data: evolution of a charismatic predator. Syst Entomol. 2004;29:359–370. doi: 10.1111/j.0307-6970.2004.00240.x. [DOI] [Google Scholar]
- Whiting MF. Mecoptera is paraphyletic: multiple genes and phylogeny of Mecoptera and Siphonaptera. Zoologica Scripta. 2002;31:93–104. doi: 10.1046/j.0300-3256.2001.00095.x. [DOI] [Google Scholar]
- Stephens M, Smith N, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison DR, Maddison WP. Book MacClade 4: Analysis of Phylogeny and Character Evolution, Version 4.08. (Editor ed.^eds.) 4.08. City: Sinauer Associates, Inc; 2008. MacClade 4: Analysis of Phylogeny and Character Evolution, Version 4.08. [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical-method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers A, Harpending H. Population growth makes waves in the distribution of pairwise genetic differences. Molecular Biology and Evolution. 1992;9:552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- Ray N, Currat M, Excoffier L. Intra-deme molecular diversity in spatially expanding populations. Molecular Biology and Evolution. 2003;20:76–86. doi: 10.1093/molbev/msg009. [DOI] [PubMed] [Google Scholar]
- Excoffier L. Patterns of DNA sequence diversity and genetic structure after a range expansion: lessons from the infinite-island model. Molecular Ecology. 2004;13:853–864. doi: 10.1046/j.1365-294X.2003.02004.x. [DOI] [PubMed] [Google Scholar]
- Nylander JAA. Book MrModeltest v2 (Editor ed.^eds.) City: Program distributed by the author; 2004. MrModeltest v2. [Google Scholar]
- Akaike H. Information theory and an extension of the maximum likelihood principle. Book Information theory and an extension of the maximum likelihood principle (Editor ed.^eds.). City. 1973.
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Nylander JAA, Wilgenbusch JC, Warren DL, Swofford DL. AWTY (are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics. 2008;24:581–583. doi: 10.1093/bioinformatics/btm388. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web-servers. Systematic Biology. 2008;75:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Parks DH, Porter M, Churcher S, Wang S, Blouin C, Whalley J, Brooks S, Beiko RG. GenGIS: A geospatial information system for genomic data. Genome Research. 2009;19:1896–1904. doi: 10.1101/gr.095612.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi D, Engel MS. Evolution of the Insects. Cambridge: Cambridge University Press; 2005. [Google Scholar]
- Gaunt MW, Miles MA. An insect molecular clock dates the origin fo the insects and accords with paleontological and biogeographic landmarks. Molecular Biology and Evolution. 2002;19:748–761. doi: 10.1093/oxfordjournals.molbev.a004133. [DOI] [PubMed] [Google Scholar]
- Quek S-P, Davies SJ, Itino T, Pierce NE. Codiversification in an ant-plant mutualism: stem texture and the evolution of host use in Crematogaster (Formicidae: Myrmicinae) Inhabitants of Macaranga (Euphorbiaceae) Evolution. 2004;58:554–570. [PubMed] [Google Scholar]
- Caterino MS, Sperling FAH. Papilio phylogeny based on mitochondrial cytochrome oxidase I and II genes. Molecular Phylogenetics and Evolution. 1999;11:122–137. doi: 10.1006/mpev.1998.0549. [DOI] [PubMed] [Google Scholar]
- Pellmyr O, Leebens-Mack J. Forty million years of mutualism: Evidence for Eocene origin of the yucca-yucca moth association. Proceedings of the National Academy of Sciences. 1999;96:9178–9183. doi: 10.1073/pnas.96.16.9178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biology. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. Book R version 2.10.1 (Editor ed.^eds.) City: The R Foundation for Statistical Computing; 2010. R version 2.10.1. [Google Scholar]
- PRISM Group. Oregon State University, http://www.prism.oregonstate.edu/ Book Oregon State University, (Editor ed.^eds.). City. 2007.
- Gillespie AR, Zehfuss PH. In: Quaternary Glaciations-Extent and Chronology, Part II. Ehlers J, Gibbard PL, editor. Amsterdam: Elsevier; 2004. Glaciations of the Sierra Nevada, California, USA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table 1S. Collecting localities of Grylloblatta. Latitude and longitude reported in the WGS84 datum. Type localities indicated with an asterisk.
Table 2S. GenBank Accession numbers of California Grylloblatta collections.
Figure 1S. Mitochondrial gene tree of California Grylloblatta species. Bayesian posterior probability support values are shown at the base of the nodes.
Figure 2S. Concatenated nuclear gene tree of California Grylloblatta species. Bayesian posterior probability support values are shown at the base of the nodes.
Figure 3S. Lineage through time plot of California Grylloblatta species. Plot of log-lineage diversification (solid line) over unscaled time, compared to a null model (dashed line) of constant diversification.