SUMMARY
Cardiac hypertrophy and failure are characterized by transcriptional reprogramming of gene expression. Adult cardiomyocytes in mice express primarily α-myosin heavy chain (α-MHC), whereas embryonic cardiomyocytes express β-MHC. Cardiac stress triggers adult hearts to undergo hypertrophy and a shift from α-MHC to fetal β-MHC expression. Here we show that Brg1, a chromatin-remodeling protein, plays critical roles in regulating cardiac growth, differentiation and gene expression. In embryos, Brg1 promotes myocyte proliferation by maintaining BMP10 and suppressing p57kip2 expression. It preserves fetal cardiac differentiation by interacting with HDAC and PARP to repress α-MHC and activate β-MHC. In adults, Brg1 is turned off in cardiomyocytes. It is reactivated by cardiac stresses and complexes with its embryonic partners, HDAC and PARP, to induce a pathological α- to β-MHC shift. Preventing Brg1 re-expression decreases hypertrophy and reverses such MHC switch. Brg1 is activated in certain patients with hypertrophic cardiomyopathy, its level correlating with disease severity and MHC changes. Our studies show that Brg1 maintains cardiomyocytes in an embryonic state, and demonstrate an epigenetic mechanism by which three classes of chromatin-modifying factors, Brg1, HDAC and PARP, cooperate to control developmental and pathological gene expression.
Keywords: chromatin, heart development, myosin heavy chain, Brg1, BAF, BMP10, p57kip2, HDAC, PARP, MHC, cardiac hypertrophy, cardiomyopathy, heart failure, differentiation, proliferation
Myosin heavy chain (MHC) is the molecular motor in muscle cells, and two isoforms, α- and β-MHC, are expressed specifically in mammalian hearts. The α-MHC has higher ATPase activity than β-MHC, and their relative amount changes under different pathophysiological conditions1 (Supplementary Fig. 1a). The α- and β-MHC ratio correlates directly with the overall cardiac performance in animals2-4 as well as in patients with cardiomyopathy and heart failure5-8. MHC protein mutations also cause cardiac dysfunction in mice and humans9, 10. Pathologic hypertrophy of adult hearts is associated with α-MHC downregulation and β-MHC induction11, returning to a fetal state of MHC expression. However, hearts expressing α-MHC have better outcome under stress conditions than those expressing mainly β-MHC2-4. Thus, strategies to control MHC expression represent attractive approaches for heart failure therapy1, 12.
Chromatin remodeling offers one such control to modulate gene expression. The Brg1/Brm-associated-factor (BAF) complex, consisting of 12 protein subunits, is a major type of ATP-dependent chromatin-remodeling complexes in vertebrates13. Our studies show that Brg1, the essential ATPase subunit of the BAF complex14, interacts with two other classes of chromatin-modifying enzymes, histone deacetylase (HDAC)15 and poly (ADP-ribose) polymerase (PARP)16, to regulate gene expression during cardiac growth, differentiation and hypertrophy in mice (Supplementary Fig. 1b, c). Both HDACs and PARP1 are therapeutic targets for cardiac hypertrophy since pharmacologic inhibition of their activities or genetic mutations of class I HDACs and PARP1 in mice reduce hypertrophy17-22. Brg1's interaction with HDACs and PARP1, and its activation in certain patients with hypertrophic cardiomyopathy suggest that Brg1/BAF may be a target for treating cardiac hypertrophy and failure.
Growth of Brg1-null myocardium
We used the Sm22αCre transgene23 to remove floxed alleles of Brg1 (Brg1F)24 in the mouse myocardium (cardiomyocytes) by embryonic day 9.5 (E9.5)(Supplementary Fig. 2a-c). Sm22αCre;Brg1F/F embryos were grossly normal at E11.5, but died thereafter (Supplementary Fig. 2d-f). At E10.5 the heart had thin compact myocardium and failed to form interventricular septum (Fig. 1a, b; Supplementary Fig. 3a-c) while the trabeculation was normal23. At E11.5 trabeculation, cardiac jelly, and vasculature remained normal despite the thin myocardium and absent septum (Supplementary Fig. 3d-f; 4a-f; Supplementary text). Loss of compact myocardium could reduce cardiac output, causing embryonic lethality.
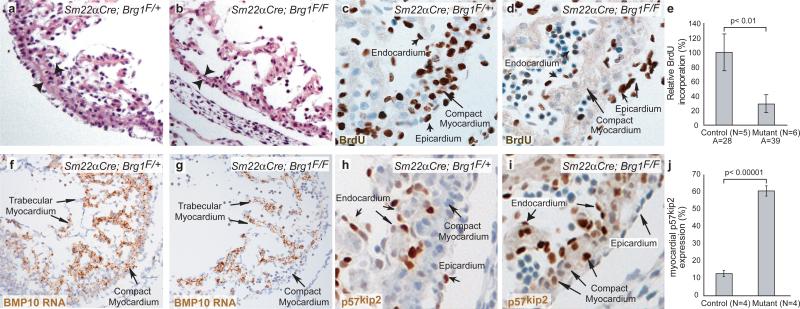
Figure 1. Brg1 promotes myocardial proliferation.
a, b, H&E sections of E10.5 compact myocardium, whose thickness is denoted by arrowheads.
c, d, BrdU immunostaining of E10.5 compact myocardium.
e, BrdU incorporation quantitation. A: number of areas examined. p-value: Student's t-test. Error bar: standard deviation.
f, g, BMP10 in situ hybridization of E10.5 hearts.
h, i, p57kip2 immunostaining of E10.5 hearts.
j, p57kip2 quantitation. p-value: Student's t-test. Error bar: standard deviation.
These embryos had almost no myocardial apoptosis (Supplementary Fig. 3g-l). The compact and septal primordial myocardium had dramatic decrease of cell proliferation at E10.5, while other heart layers were normal (Fig. 1c-e; Supplementary Fig. 3m-o). Therefore, Brg1 is required for cell proliferation to form the compact and septal myocardium.
To identify genes responsible for these defects, we used RNA in situ hybridization to survey crucial myocardial transcripts in E10-E11 Sm22αCre;Brg1F/F hearts, including Nkx2.5, Gata4, MEF2C, Tbx3, Tbx5, CX43, Irx1, Irx2, NPPA, and BMP10. We found no changes in these transcripts (Supplementary Fig. 5) except for BMP10, a key factor required for myocardial proliferation25. BMP10 expression was nearly abolished in the compact myocardium of E10.5 Sm22αCre;Brg1F/F (Fig. 1f, g). We next examined p57kip2, a cyclin-dependent kinase inhibitor, whose expression is normally suppressed by BMP1025. We found p57kip2 level correlated inversely with normal cardiac cell proliferation (Supplementary Fig. 6a). Also, p57kip2 appeared ectopically in E10.5 Sm22αCre;Brg1F/F myocardium (Fig. 1h-j; Supplementary Fig. 6b), correlating with BMP10 reduction and termination of myocardial cell proliferation. The BMP10/p57kip2-mediated proliferation was confirmed when we rescued myocardial proliferation in Sm22αCre;Brg1F/F with recombinant BMP10 by whole embryo cultures26 (Supplementary Fig. 6c-e).
We further used the Mef2cCre line27 to delete Brg1 in the right ventricular myocardium, leaving Brg1 intact in the endocardium and left ventricle (Supplementary Fig. 7a, b). Mef2cCre;Brg1F/F embryos had hypoplastic outflow tract and right ventricle (Supplementary Fig. 7c-f; 7i, j). The Brg1-null right ventricle phenocopied the defects in Sm22αCre;Brg1F/F ventricles, namely BMP10 downregulation, ectopic p57kip2 expression, and proliferation reduction (Supplementary Fig. 7c-h) while the Brg1-positive left ventricle was normal, demonstrating a primary and myocardially-specific regulation of BMP10-p57kip2 by Brg1.
Differentiation of Brg1-null myocardium
We examined if the early proliferation termination in Sm22αCre;Brg1F/F myocardium was coupled with premature differentiation. We analyzed the myofibril formation of E10.5 cardiomyocytes by α-actinin immunostaining and electron microscopy (EM). While control compact myocardial cells showed diffuse distribution of α-actinin, a component of z-lines that demarcate sarcomeres, those of Sm22αCre;Brg1F/F began to show striated patterns (Supplementary Fig. 9a, b). EM verified that Sm22αCre;Brg1F/F displayed consecutive sarcomeres while controls only had short myofibrils (Fig. 2a, b).
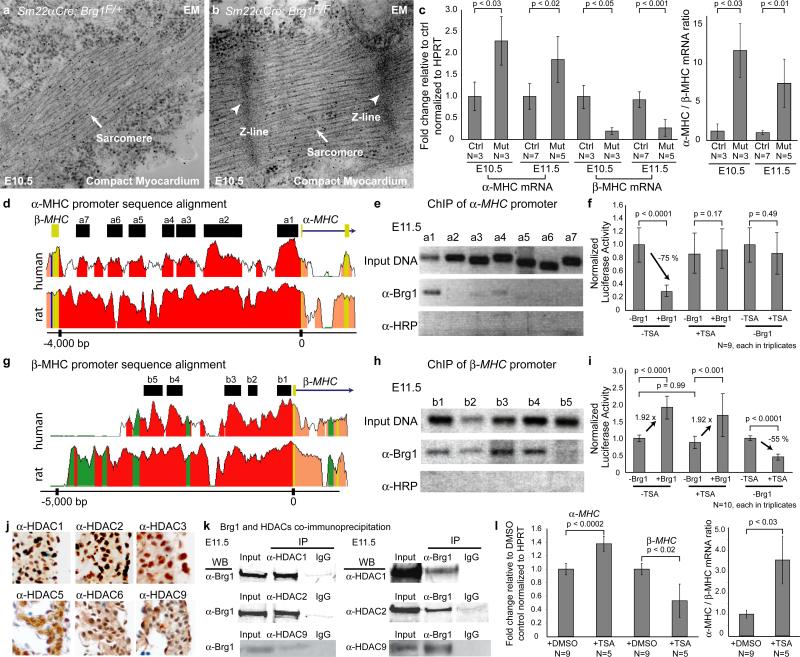
Figure 2. Brg1 suppresses myocardial differentiation.
a, b, EM of the compact myocardium of E10.5 embryos.
c, Quantitative RT-PCR of ventricular α- and β-MHC at E10.5 and E11.5. Ctrl: control. Mut: Sm22αCre;Brg1F/F. p-value: Student's t-test. Error bar: standard deviation.
d, Sequence alignment of the α-MHC locus from mouse, human, and rat. Peak heights indicate degree of sequence homology. Black boxes (a1-a7) are regions of high sequence homology and further analyzed by ChIP. Red: promoter elements. Salmon: introns. Yellow: untranslated regions.
e, PCR of Brg1-immunoprecipitated chromatin from E11.5 hearts. α-HRP: anti-horse radish peroxidase antibody.
f, Luciferase reporter assay of the proximal α-MHC promoter (-462 to +192) in SW13 cells. p-value: Student's t-test. Error bar: standard deviation.
g, Sequence alignment of the β-MHC locus from mouse, human, and rat. Black boxes (b1-b5) are regions of high sequence homology and further analyzed by ChIP. Green: transposons/simple repeats.
h, PCR analysis of Brg1-immunoprecipitated chromatin from E11.5 hearts.
i, Luciferase reporter assays of the β-MHC proximal promoter (-835 to +222) in SW13 cells. p-value: Student's t-test. Error bar: standard deviation.
j, Immunostaining of HDAC1, 2, 3, 5, 6 and 9 (brown) in E11.5 hearts.
k, Co-immunoprecipitation of Brg1 with HDAC1, 2 and 9 in E11.5 hearts.
l, Quantitative RT-PCR of α- and β-MHC of cultured embryos treated with DMSO or TSA. p-value: Student's t-test. Error bar: standard deviation.
We then quantified mRNA expression of the two MHC isoforms, α-MHC, which is mainly expressed by adult hearts, and β-MHC, which is expressed primarily by embryonic hearts. E10.5 and E11.5 Sm22αCre;Brg1F/F ventricles highly expressed α-MHC, and down-regulated β-MHC, thereby increasing α-MHC/β-MHC ratio by 7-12 folds (Fig. 2c). Together with myofibril analysis, these data indicate that Brg1-null myocardial cells are highly differentiated and thus support a role of Brg1 in maintaining myocardial cells in an embryonic state of differentiation.
To test if Brg1 directly regulates MHC expression, we examined Brg1 binding to MHC promoters. With sequence alignment (www.dcode.org), we identified 7 evolutionarily conserved regions (a1-a7) in the mouse intergenic ~4 kb α-MHC promoter28 among mouse, rat, and human (Fig. 2d). Chromatin immunoprecipitation (ChIP) assay using E11.5 hearts with J1 anti-Brg1 antibody23 showed that of the 7 regions, Brg1 was strongly associated with the proximal promoter (a1) of α-MHC (Fig. 2e)(Supplementary text). In contrast to α-MHC, none of the 4 conserved regions in the 5kb upstream promoter of BMP10 was associated with Brg1 (Supplementary Fig. 8a, b). To test Brg1/BAF transcriptional activity, we cloned different regions of the α-MHC promoter into chromatinized episomal reporter pREP429 and then transfected the constructs into SW13 cells23, which lack Brg1 and Brm30 (Supplementary Fig. 8d). We found that restoring Brg1 expression caused approximately 65-75% reduction in the α-MHC reporter activity, and the proximal promoter (a1) was critical for α-MHC repression (Fig. 2f; Supplementary Fig. 8d). These observations support a direct repression of α-MHC by Brg1.
Since HDACs are chromatin modifiers that mediate transcriptional repression15, we tested if Brg1 requires HDACs to repress α-MHC. Indeed, Brg1 failed to repress α-MHC reporter in SW13 cells treated with trichostatin A (TSA), an HDAC inhibitor (Fig. 2f). Neither could HDAC repress the α-MHC reporter without Brg1 (Fig. 2f). HDAC proteins, including Class I HDAC1, 2, 3 and Class II HDAC5, 6, 9 were present in myocardial nuclei (Fig. 2j), and Brg1 co-immunoprecipitated with HDAC1, 2, 3 and 9 in E11.5 ventricles (Fig. 2k; data not shown). These findings indicate Brg1 and HDACs co-repress α-MHC in the embryonic myocardium.
We also analyzed the 5.5 Kb β-MHC promoter28, where we identified 5 highly conserved regions (b1 to b5 in Fig. 2g). Brg1 was widely associated with four of the five regions by ChIP experiments (Fig. 2h). Restoring Brg1 expression in SW13 cells activated β-MHC reporters (Fig. 2i). Deletional analysis of β-MHC promoter showed the proximal promoter (b1) was necessary for the Brg1-mediated β-MHC activation (Fig. 2i; Supplementary Fig. 8e). This activation of β-MHC did not require HDAC activity (Fig. 2i). However, HDAC is necessary for the basal activity of β-MHC since HDAC inhibition resulted in significant reduction of β-MHC promoter activity (Fig. 2i).
We then asked if HDAC inhibition in embryos causes premature α/β-MHC switches as observed in Brg1-null myocardium. Indeed, TSA-treated cultured embryos significantly up-regulated α-MHC while down-regulated β-MHC (Fig. 2l; Supplementary Fig. 9c, d). Overall, the biochemical studies, reporter assays, and embryo culture experiments support Brg1 and HDACs co-repress α-MHC, but independently activate β-MHC.
Although TSA causes MHC switches in embryos, HDAC inhibition did not produce reduction in myocardial proliferation (Supplementary Fig. 9e-g). Conversely, BMP10 rescued myocardial proliferation of Sm22αCre;Brg1F/F, but did not influence α/β-MHC expression (Supplementary Fig. 9h, i). Therefore, Brg1 governs two parallel pathways to independently control myocardial growth and differentiation in embryos.
Cardiac hypertrophy and MHC changes in adult
We asked if Brg1 is also critical for cardiac growth and differentiation in stressed adult hearts. To bypass embryonic lethality, we used the doxycycline-inducible Tnnt2-rtTA;Tre-Cre mouse line31 to effect adult myocardial gene deletion. A 5 day-doxycycline treatment was sufficient to activate a β-galactosidase reporter (Supplementary Fig. 10a). We surgically constricted the transverse aorta (TAC) to pressure-overload the heart and induce cardiac hypertrophy in control and Tnnt2-rtTA;Tre-Cre;Brg1F/F littermates. The transgene and doxycycline alone did not cause hypertrophy (Fig. 3a; Supplementary Fig. 10f). Four weeks after surgery, the control and Tnnt2-rtta;Tre-Cre;Brg1F/F mice fed with normal diet developed severe cardiac hypertrophy with increased cardiomyocyte size (Fig. 3a; Supplementary Fig. 10b, c, f), ventricular/body weight ratio (Supplementary Fig. 10f), and cardiac fibrosis (Supplementary Fig. 10g, i). In contrast, doxycycline-treated Tnnt2-rtTA;Tre-Cre;Brg1F/F mice exhibited only mild cardiac hypertrophy with slight increase in cardiomyocytes size (Fig. 3a; Supplementary Fig. 10d, e) and ventricular/body weight ratio (Supplementary Fig. 10f), and without fibrosis (Supplementary Fig. 10h, j). Overall, Brg1-null myocardium had a 63-73% reduction of cardiac hypertrophy. Thus, Brg1 is essential for the pressure-induced cardiac hypertrophy.
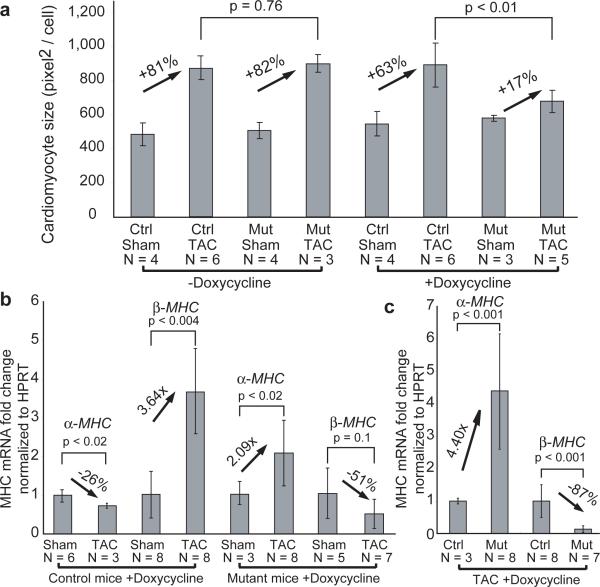
Figure 3. Brg1 is required for cardiac hypertrophy.
a, Cardiomyocyte size quantitation. Ctrl: control. Mut: Tnnt2-rtTA;Tre-Cre;Brg1F/F. p-value: Student's t-test. Error bar: standard deviation.
b, c, Quantitative RT-PCR of α- and β-MHC in cardiac ventricles of doxycycline-treated control and Tnnt2-rtTA;Tre-Cre;Brg1F/F mice 4 weeks after sham/TAC operation. p-value: Student's t-test. Error bar: standard deviation.
We next investigated whether Brg1 regulates MHC expression in hypertrophic hearts. Control hypertrophic hearts underwent canonical MHC changes, namelyα-MHC downregulation and β-MHC upregulation (Fig. 3b). In contrast, doxycycline-treated Tnnt2-rtTA;Tre-Cre;Brg1F/F mice showed a 2.1-fold increase of α-MHC and a 51% reduction of β-MHC (Fig. 3b). Consequently, the pressure-stressed Brg1-null myocardium expressed 4.4-fold α-MHC and 0.13 fold β-MHC as the control myocardium (Fig. 3c). This reversal of MHC was not caused by reduced hypertrophy, which could only lessen, but not reverse, canonical MHC changes. Therefore, Brg1 is critical for α/β-MHC switch in hypertrophic hearts.
While Brg1 is highly expressed in embryonic hearts23, it is turned off in adult myocardium with some expression in endothelial or interstitial cells (Fig. 4a). Brg1 became detectable in cardiomyocytes within 7 days after TAC by immunostaining (Fig. 4b), confirmed by western blot (Fig. 4d); while it remained absent in Tnnt2-rtTA;Tre-Cre;Brg1F/F myocardium (Fig. 4c). Also, Brg1 mRNA increased by 1.8 fold within 2 weeks after TAC (Fig. 4e), indicating Brg1 reactivation by stress signals is essential for the hypertrophic process.
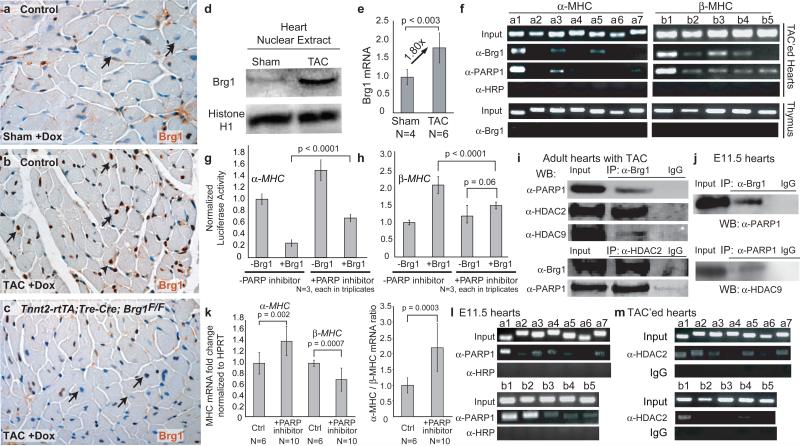
Figure 4. MHC regulation by Brg1, PARP and HDAC.
a, b, c, Brg1 immunostaining in ventricular myocardium of doxycycline-treated control and Tnnt2-rtTA;Tre-Cre;Brg1F/F mice 1 week after sham/TAC operation. Arrows: cardiomyocyte nuclei.
d, Brg1 immunoblot of cardiac nuclear extracts from wildtype mice 2 weeks after TAC.
e, Quantitative RT-PCR of Brg1 mRNA in wildtype mice 2 weeks after TAC. p-value: Student's t-test. Error bar: standard deviation.
f, PCR of Brg1- and PARP1- immunoprecipitated chromatin from thymus and adult hearts 2 weeks after TAC.
g, h, Luciferase reporter assays of α-MHC (g) and β-MHC (h) promoter in SW13 cells with PARP inhibition. p-value: Student's t-test. Error bar: standard deviation.
i, j, Co-immunoprecipitation of Brg1, PARP1, HDAC2 and 9 in TAC-treated adult hearts (i) and in E11.5 hearts (j).
k, Quantitative RT-PCR of α- and β-MHC of PJ34-treated cultured embryos. p-value: Student's t-test. Error bar: standard deviation.
l, PCR of PARP1- immunoprecipitated chromatin from E11.5 hearts.
m, PCR of HDAC2- immunoprecipitated chromatin from adult hearts 2 weeks after TAC.
We then determined if reactivated Brg1 controls MHC expression through direct binding of MHC promoters. ChIP analysis of TAC-treated hearts showed Brg1 was highly enriched in the proximal promoters of both α-MHC and β-MHC, but not BMP10 (Fig. 4f; Supplementary Fig. 8a, c). Furthermore, Brg1 binding to MHC promoters was detectable only in TAC-treated, but not sham-operated, hearts (Supplementary Fig. 10k), consistent with Brg1 reactivation by pressure overload. The binding pattern was similar to that in embryonic hearts (Fig. 2e, 2h), indicating a common mechanism underlying the Brg1-mediated MHC control in embryonic and hypertrophic hearts.
MHC regulation by Brg1, HDAC and PARP
Besides HDACs18-20, PARP1 is the only other chromatin-modifying enzyme16 known to regulate cardiac hypertrophy17, 22. However, it is unknown if PARP1 binds to MHC promoters. ChIP analysis of TAC-treated hearts showed PARP1 bound to the proximal promoters of α-MHC and β-MHC, but not BMP10 in a pattern similar to that of Brg1 (Fig. 4f; Supplementary Fig. 11a). Like Brg1, PARP1 binding occurred only in TAC-treated, but not sham-operated, hearts (data not shown). Furthermore, inhibiting PARP1 activity by PJ-3432 reduced both Brg1-mediated α-MHC repression and β-MHC activation in reporter assays in SW13 cells, indicating Brg1 and PARP1 cooperate to regulate MHC (Fig. 4g, h). Indeed, PARP1 and Brg1 co-immunoprecipitated in both TAC-treated hearts and E11.5 hearts (Fig. 4i, j). Embryos cultured with PJ-34 had normal myocardial proliferation, but exhibited α/β-MHC switches characteristic of Brg1-null myocardium (Supplementary Fig. 11b; Fig. 4k). Immunostaining and ChIP analyses of E11.5 hearts showed PARP1 was present in myocardial nuclei and bound to the proximal promoters of α- and β-MHC, but not BMP10, in a pattern similar to that of Brg1 (Fig. 4l; Supplementary Fig. 11c, d). These findings indicate Brg1 complexes with PARP1 to regulate MHC in embryonic and stressed adult hearts.
Brg1 and PARP1 co-immunoprecipitated with HDAC1, 2 or 9 in E11.5 and stressed adult hearts (Fig. 2k; 4i, j; Supplementary Fig. 11e). We asked if HDACs are present on MHC promoters. Using ChIP with two cross-linking steps33 in TAC-treated hearts, we found HDAC2 and HDAC9 were enriched in the α-MHC promoter, but bound minimally to the β-MHC promoter (Fig. 4m; Supplementary Fig. 11f), suggesting direct α-MHC and indirect β-MHC regulation by HDACs. Together with reporter assays (Fig. 2f, i; 4g, h), these biochemical studies suggest that Brg1, PARP, and HDAC physically form a chromatin-remodeling complex on the α-MHC promoter to repress α-MHC, while Brg1 complexes with PARP on the β-MHC promoter to activate β-MHC.
Implication in human cardiomyopathy
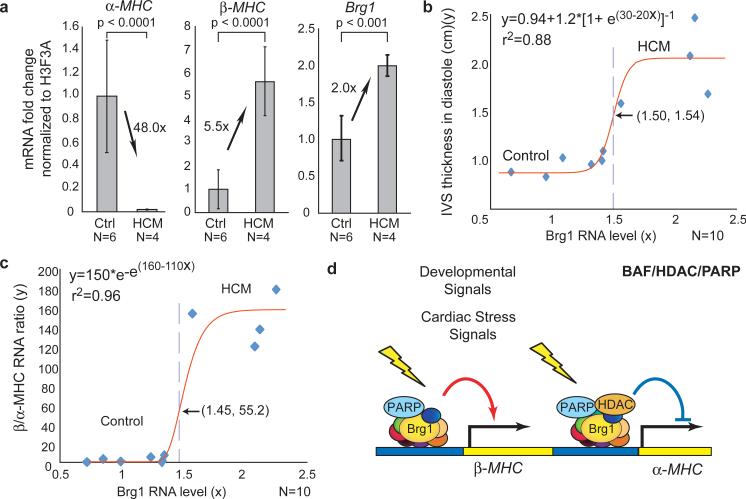
To investigate if Brg1 is activated in human hypertrophic hearts, we studied patients with hypertrophic cardiomyopathy (HCM) of unknown etiology, who required surgical myectomy34 to relieve cardiac obstruction caused by prominent ventricular or septal hypertrophy (Supplementary Fig. 12a). HCM severity was measured by the maximal thickness of interventricular septum during diastole (IVSd). IVSd in HCM patients was 2.02 folds of the control (Supplementary Fig. 12b, c). Quantitative RT-PCR analyses showed HCM hearts had 48-fold reduction of α-MHC, 5.5-fold increase of β-MHC and 2-fold increase of Brg1 expression (Fig. 5a). The loss of α-MHC, gain of β-MHC, and activation of Brg1 resembled the changes observed in mice with hypertrophy, suggesting a similar role of Brg1 in human disease. Consistent with this notion, the IVSd and β/α-MHC ratio correlated well with Brg1 level between control and HCM subjects, with sigmoidal regression curves inflecting at 1.50-fold and 1.45-fold of Brg1, respectively (Fig. 5b, c; Supplementary Fig. 12d, e). At the inflection point of 1.50-fold Brg1, IVSd equals 1.54 cm, coinciding with a clinical criterion (IVSd >1.50 cm) in HCM diagnosis35. We therefore speculate a 50% increase of Brg1 may be a threshold for disease development in certain patients.
Figure 5. Brg1 activation in human cardiomyopathy.
a, Quantitative RT-PCR of α-MHC, β-MHC, and Brg1 expression in normal and HCM subjects. p-value: Student's t-test. Error bar: standard deviation.
b, IVSd (y) plotted against the Brg1 RNA level (x). Red: regression curve. e: the base of natural logarithm (~2.718). Arrow and dashed line: the inflection point.
c, The β /α-MHC RNA ratio (y) plotted against the Brg1 RNA level (x).
d, Model of developmentally-activated and stress-induced assembly of BAF/HDAC/PARP complexes on the α-MHC, and BAF/PARP complex on the β-MHC promoter.
Discussion
Brg1/BAF may have regenerative and therapeutic implications given its newly identified roles in both embryonic and adult cardiomyocytes. Similar mechanisms are directed by Brg1 to control cardiac growth, differentiation and gene expression under developmental and pathological conditions (Supplementary Fig. 1b, c). The stress-dependent assembly of a developmental complex to modify chromatins in adult hearts provides a molecular explanation for fetal gene activation in the diseased adult myocardium. HDAC, PARP and now Brg1/BAF are the only classes of chromatin-modifying factors known to regulate cardiac hypertrophy. Cardiac stresses activate Brg1, which then assembles a BAF/HDAC/PARP chromatin complex on MHC promoters (Fig. 5d), where they may interact with transcription factors such as TR, TEF1, MEF2, SRF, GATA4 and NFAT36, 37 to control MHC expression. The induction of Brg1 by hypertrophic stimuli suggests that chromatin may ultimately be where all the stress-response signals converge for the regulation of MHC genes, a critical step in the myopathic process.
Brg1/BAF, besides thyroid hormone receptors36, provides the only known direct mechanism that antithetically regulates α-MHC and β-MHC. This opposite MHC regulation may underlie the on-off, rather than graded, switching of MHC in individual cardiomyocytes of hypertrophic hearts28. Exactly how HDACs and PARPs contribute to this BAF-mediated process awaits further investigations. HDACs and PARPs may covalently modify histones and thereby help anchor BAF to certain sites of MHC promoters. BAF proteins may also be modified through acetylation/deacetylation or poly-ADP-ribosylation by HDACs and PARPs. Such modifications may decide the composition of BAF complex and how BAF interacts with chromatin as well as other MHC regulators such as miR-2081. Elucidating these issues will help determine how the chromatin and target specificities of BAF may be established by its possible combinatorial assembly with the large families of 17 PARP16 and 18 HDAC15 proteins.
METHODS
Sm22αCre, Brg1F/F, Mef2cCre, R26R and Tnnt2-rtTA;Tre-Cre mice have been described23, 24, 27, 31, 38. Immunostaining, RNA in situ hybridization, quantitative RT-PCR, and whole embryo culture were performed as described23, 26. TAC was modified from previous descriptions20. The pressure load caused by TAC was verified by the pressure gradient across the aortic constriction measured by echocardiography. Only mice with a pressure gradient > 30 mmHg were analyzed for cardiac hypertrophy and gene expression. Curve modeling was performed with the Levenburg-Marquardt non-linear regression method and XLfit software. Detailed methods can be found in the Supplementary Information.
Supplementary Material
Acknowledgements
We thank B. Black for providing Mef2cCre mice; G.R. Crabtree, D. Bernstein, M. Rabinovitch, V. Christoffels, W. Shou, J. Wysocka, and K. Zhao for materials or discussions; M. Zhao, B. Wu, A. Sun, J. Lehrer-Graiwer, M. Zeini, W. Li, CY Lin, CJ Lin and K. Stankunas for technical advice. C.P.C. was supported by funds from NIH, American Heart Association (AHA), Children's Heart Foundation, March of Dimes Foundation, Office of the University of California, California Institute of Regenerative Medicine, Kaiser Foundation, Baxter Foundation, Oak Foundation, and Stanford Cardiovascular Institute. C.T.H was supported by predoctoral training fellowships from NIH and AHA; H.L.C. by a visiting scholar fellowship; C.S. by Kirschstein-NRSA Postdoctoral Fellowship; B.Z. by NIH and AHA grants.
Methods
Mice
Sm22αCre, Brg1F/F, Mef2cCre, R26R and Tnnt2-rtTA;Tre-Cre mice have been described previously23, 24, 27, 31, 38. The date of observing of a vaginal plug was set as embryonic day E0.5 by convention. Embryonic development was confirmed by ultrasonography39 prior to sacrificing pregnant mice and by examination of embryo morphology and somite number. The use of mice for studies is in compliance with the regulations of Stanford University and National Institute of Health.
Histology, RNA in situ hybridization and Immunostaining
Histological analysis, immunostaining and RNA in situ hybridization were performed as described 23, 26. The probes used for the RNA in situ hybridization were described in the text. The following primary antibodies were used for immunostaining: J1 antibody23 (Supplementary Figure 2b, c), G7 Brg1 (Santa Cruz Biotechnology, CA)(Figure 4a-c), p57kip2 (Lab Vision, Fremont, CA), α-actinin (Sigma-Aldrich, St. Louis, MO), troponin T (Hybridoma Bank, University of Iowa), HDAC1 (Abcam, ab7028), HDAC2 (Abcam, ab7029), HDAC3 (Cell Signaling Technology 2632), HDAC5 (Abcam, ab1439), HDAC6 (Upstate 07-732), HDAC9 (Abcam ab59718, Abcam ab18970) and activated caspase 3 (Sigma C8487).
Quantitative analysis of myocardial development
H&E- stained sections were used for the morphometric analysis. The following parameters were measured: compact myocardial thickness, number of trabeculi directly connected to compact myocardium, and trabecular area normalized to ventricular size23, 25. p-values were calculated by the Student-t test. Error bars indicate one standard deviation.
Proliferation and apoptosis analysis
Pregnant mice were injected with BrdU (Sigma, 100 μg/g, intraperitoneal injection) for 6 hours prior to embryo isolation at E10.5 or E11.5. Incorporated BrdU was stained in the tissue-section of the heart according to manufacturer's protocol (Zymed Laboratories, South San Francisco, CA). BrdU incorporation was quantitated by the percentage of BrdU-positive cells in the myocardium. p-values were calculated by the Student-t test. Error bars indicate one standard deviation. Apoptosis of embryos were analyzed by TUNEL staining using a kit (Roche, Applied Science, Indianapolis, IN) and by activated caspase 3 immunostaining (Sigma C8487).
Whole embryo culture
Experiments were performed as described23, 26. Embryos were grown from E8.75 or E9.5 to E10.5 for harvest. Following were used: BMP10 (10 nM, RD Systems, Minneapolis, MN), BSA (125 ng/ml), TSA (100-500 nM, Sigma-Aldrich, St. Louis, MO), BrdU (30 μg/mL, Simga-Aldrich), PJ-34 (20 uM, Axxora, San Diego, CA).
Quantitative RT-PCR analysis
Quantitative RT-PCR analyses were performed as described previously23. The following primer sequences were used: Murine α-MHC, 5’ primer ACGGTGACCATAAAGGAGGA, 3’ primer TGTCCTCGATCTTGTCGAAC. Murine β-MHC, 5’ primer GCCCTTTGACCTCAAGAAAG, 3’ primer CTTCACAGTCACCGTCTTGC. Murine HPRT, 5’ primer GCTGGTGAAAAGGACCTCT, 3’ primer CACAGGACTAGAACACCTGC. Murine Brg1, 5’ primer CACCTAACCTCACCAAGAAGATGA, 3’ primer CTTCTTGAAGTCCACAGGCTTTC. Human H3F3A (histone 3), 5’ primer AAAACAGATCTGCGCTTCCA, 3’ primer TTGTTACACGTTTGGCATGG. Human Brg1, 5’ primer AGTGCTGCTGTTCTGCCAAAT, 3’ primer GGCTCGTTGAAGGTTTTCAG. Human α-MHC and β-MHC were by Taqman probes (Applied Biosciences, Foster City, CA). RT-PCR reactions were performed using SYBR green master mix (BioRad, Hercules, CA) or Taqman reagents (Applied Biosciences) with an Eppendorf realplex, and the primer sets were tested to be quantitative. Threshold cycles and melting curve measurements were performed with software. p-values were calculated by the Student-t test. Error bars indicate one standard deviation.
Chromatin Immunoprecipitation (ChIP)
Hearts from approximately eight litters of E10.5-E11.5 Swiss-Webster mice (80-100 embryonic hearts per ChIP experiment) were dissected in chilled PBS, and subsequently fixed with 1% PFA and washed with 0.125M glycine. Adult hearts were minced before fixing with 1% PFA (for Brg1 or PARP1 ChIP) or with 1% PFA and 2 mM disuccinimidyl glutarate (Sigma) for HDAC ChIP. Cells were lysed by cell lysis buffer (10 mM Hepes pH 7.5, 85 mM KCl, 0.5% NP-40, protease inhibitor (#78410, Pierce, Rockford, IL). Nuclei were isolated by disruption using a B dounce, and washed with SDS lysis buffer (1% SDS, 10mM EDTA and 50mM Tris, pH 8.1). Chromatin was sonicated to generate average fragment sizes of 100-300 bp, and immunoprecipitated using anti-Brg1 J1 antibody23, anti-PARP1 antibody (N-20, sc-1561, Santa Cruz Biotechnology, Santa Cruz, CA), anti-HDAC2 (H54; Santa Cruz Biotechnology, Santa Cruz, CA), HDAC9 (ab59718; Abcam, Cambridge, MA), or anti-HRP (AffiniPure rabbit anti-horse radish peroxidase, 323-005-021, Jackson Immunoresearch, West Grove, PA). Isolation of immunoprecipitated chromatin was done according to the manufacturer's protocol (Upstate). The precipitates were PCR amplified for 35 cycles in embryonic heart ChIP, and 32-35 cycles in adult heart ChIP analyses. PCR primers (a1-a7, b1-b5, p1-p4) were designed to amplify the following regions in the α-MHC (a1-a7), β-MHC (b1-b5) and BMP10 (p1-p4) promoters: a1 (-357 to -463); a2 (-1092 to -1237); a3 (-1775 to -1908); a4 (-2141 to -2290); a5 (-2997 to -3121); a6 (-3378 to -3486); a7 (-3569 to -3714); b1 (-64 to -205); b2 (-912 to -1061); b3 (-1374 to -1518); b4 (-2284 to -2409); b5 (-2690 to -2827); p1(-264 to -411); p2 (-1430 to -1579); p3 (-1724;-1868); p4 (-4420 to -4529). The DNA positions are denoted relative to the transcriptional start site (+1).
Cloning and Luciferase Reporter Assay
The reporter constructs were transfected into SW13 cells with lipofectamine 2000 (Invitrogen, CA) along with pREP7-RL as a transfection efficiency control and Brg1 expression vector with the appropriate empty vector control. Luciferase activity was measured and normalized to that of Renilla luciferase construct using the Dual-Luciferase Reporter System (Promega, WI). Specific truncations for the reporter constructs are as follows: for α-MHC promoter, full length promoter spans from 4243 bp upstream and 192 bp downstream of the transcription start site (-4243, +192), serially truncated versions span (-2537, +192), (-1802, +192), (-462, +192), and (-4243, -464). Likewise, β-MHC promoters span (-3561, +222), (-1770, +222), (-835, +222), and (-835, -206). p-values were calculated by the Student-t test. Error bars indicate one standard deviation.
Co-Immuunoprecipitation
Embryonic hearts (E10.5) were homogenized in cold NP-40 lysis buffer (25 mM K-HEPES pH 7.5, 250 mM KCl, 12.5 mM MgCl2, 0.5% NP-40, 8% glycerol, 1 mM DTT, protease and phosphatase inhibitors cocktail). The lysates was pre-cleared with protein A/G beads (Pierce, Thermo Scientific, Rockford, IL). After removal of beads, the lysates were incubated with 1 μl primary antibody Brg1 (H88, Santa Cruz Biotechnology, Santa Cruz, CA); HDAC1 (ab7028, Abcam Cambridge, MA); HDAC2 (H54, Santa Cruz Biotechnology); HDAC3 (2632, Cell Signaling, Danvers, MA); HDAC5 (ab1439, Abcam); HDAC9 (ab59718, Abcam); PARP1 (N-20, sc-1561, Santa Cruz Biotechnology) with rotation at 4°C, overnight. Lysates were incubated with Protein A/G beads to precipitate complexes, for 2 hours at 4°C with rotation. The immunocomplexes were recovered by centrifugation and washed twice with lysis buffer. SDS buffer was added to the precipitate and boiled for 10 min. Proteins were size separated in SDS-PAGE. The gels were blotted onto an Immobilon-P membrane (Millipore, Bedford, MA), blocked with 5% non-fat dry milk and incubated with the previously described antibodies. HRP-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA) were used for detection using ECL method (GE Healthcare Bio-Sciences Corp Piscataway, NJ).
Western Immunoblot Analysis
Whole hearts were collected and washed once with ice-cold phosphate-buffered saline (PBS) and homogenized in buffer A (25 mM Hepes, pH 7.0, 25 mM KCl, 5 mM MgCl2, 0.05mM EDTA, 10% glycerol, 0.1%NP-40). The nuclear pellets were resuspended in buffer B (50mM Tris-Hcl, pH 6.8, 2%SDS, 100mM DTT, 10%glycerol). After boiling and centrifugation, the supernatants were collected. The blots were reacted with antibodies for Brg1 (Santa Cruz, sc-17796) and Histone H1 (Santa Cruz, sc-34464), followed by horseradish peroxidase (HRP)-conjugated anti-mouse IgG or HRP-conjugated anti-goat IgG (Jackson). Chemiluminescence was detected with ECL Western blot detection kits (GE) according to the supplier's recommendations.
Transaortic Constriction (TAC)
Mice were fed with doxycycline food five days prior to TAC operation to induce deletion of Brg1. Surgeries were adapted from 20 and were performed on adult mice of 11–12 weeks of age and between 25 and 30 grams of weight. Mice were fed with doxycycline food pellets (6 gm doxycycline/kg of food, Bioserv, Frenchtown, NJ) five days prior to the TAC operation. Mice were anesthetized with ketamin (40mg /kg, ip), xylazine (10mg/kg, ip) and isoflurane (2-3%, inhalation). Mice were then intubated with a 20-gauge intravenous catheter and ventilated with a mouse ventilator (model Minivent, Harvard Apparatus, Inc). Anesthesia was maintained with inhaled isoflurane (1-2%). A longitudinal 5-mm incision of the skin was made with scissors at midline of sternum. The chest cavity was opened by a small incision at the level of the second intercostal space 2–3 mm from the left sternal border. While opening the chest wall, the chest retractor was gently inserted to spread the wound 4–5 mm in width. The transverse portion of the aorta was bluntly dissected with curved forceps. Then, 6–0 silk was brought underneath the transverse aorta between the left common carotid artery and the brachiocephalic trunk. One 26-gauge needle was placed directly above and parallel to the aorta. The loop was then tied around the aorta and needle, and secured with a second knot. The needle was immediately removed to create a lumen with a fixed stenotic diameter. The chest cavity was closed by 6–0 silk suture. Sham-operated mice underwent similar surgical procedures, including isolation of the aorta, looping of aorta, but without tying of the suture. The pressure load caused by TAC was verified by the pressure gradient across the aortic constriction measured by echocardiography. Only mice with a pressure gradient > 30 mmHg were analyzed for cardiac hypertrophy and gene expression.
Morphometric Analysis of Cardiomyocytes
Paraffin sections of the heart were immunostained with a fluoresecin isothiocyanate -conjugated Wheat Germ Agglutinin (WGA) antibody (F49, Biomeda, Foster City, CA) that highlighted the cell membrane of cardiomyocytes. Cellular areas outlined by WGA were determined by the number of pixels enclosed using the NIS element software (Nikon). Approximately 250 cardiomyocytes of the papillary muscle at the mid left ventricular cavity were measured to determine the size distribution. p-values were calculated by the Student-t test. Error bars indicate one standard deviation.
Human heart tissue collection and analysis
Human heart tissue from patients with HCM was acquired at the time of cardiac transplantation or surgical myotomy and myectomy34, which was performed for relief of the left ventricular outflow tract obstruction in HCM. An aortotomy approach was used, and 3-8g of tissue was removed from the proximal septum after retraction of the mitral valve. Normal tissue was acquired from heart transplant donors where the heart was not used for transplantation or by biopsy immediately following implantation. Heart tissue was flash frozen in liquid nitrogen immediately and stored at -80°C. For analysis, the tissues were thawed, RNA extracted and subject to quantitative RT-PCR analysis. The use of human tissues is in compliance with Stanford University regulation.
Construction of regression curves and derivation of equations
The regression curve, equation and r-square (r2) for Figure 5b and 5c are derived using the Levenburg-Marquardt non-linear regression method and a statistics/curve modelling software (XLfit, 2005). Different curve models ranging from linear, polynomial, exponential/log, power series, hyperbolic and sigmoidal curves were tested. Curves that fitted best with the data points based on r2 and F-test are shown in Figure 5. The mathematical derivations of the inflection points are described in Supplementary Figure 12d and 12e.
Footnotes
Author Information
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
REFERENCES
- 1.van Rooij E, et al. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–9. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 2.Herron TJ, McDonald KS. Small amounts of alpha-myosin heavy chain isoform expression significantly increase power output of rat cardiac myocyte fragments. Circ Res. 2002;90:1150–2. doi: 10.1161/01.res.0000022879.57270.11. [DOI] [PubMed] [Google Scholar]
- 3.Krenz M, Robbins J. Impact of beta-myosin heavy chain expression on cardiac function during stress. J Am Coll Cardiol. 2004;44:2390–7. doi: 10.1016/j.jacc.2004.09.044. [DOI] [PubMed] [Google Scholar]
- 4.James J, et al. Forced expression of alpha-myosin heavy chain in the rabbit ventricle results in cardioprotection under cardiomyopathic conditions. Circulation. 2005;111:2339–46. doi: 10.1161/01.CIR.0000164233.09448.B1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyata S, Minobe W, Bristow MR, Leinwand LA. Myosin heavy chain isoform expression in the failing and nonfailing human heart. Circ Res. 2000;86:386–90. doi: 10.1161/01.res.86.4.386. [DOI] [PubMed] [Google Scholar]
- 6.Abraham WT, et al. Coordinate changes in Myosin heavy chain isoform gene expression are selectively associated with alterations in dilated cardiomyopathy phenotype. Mol Med. 2002;8:750–60. [PMC free article] [PubMed] [Google Scholar]
- 7.Lowes BD, et al. Myocardial gene expression in dilated cardiomyopathy treated with beta-blocking agents. N Engl J Med. 2002;346:1357–65. doi: 10.1056/NEJMoa012630. [DOI] [PubMed] [Google Scholar]
- 8.Blaxall BC, Tschannen-Moran BM, Milano CA, Koch WJ. Differential gene expression and genomic patient stratification following left ventricular assist device support. J Am Coll Cardiol. 2003;41:1096–106. doi: 10.1016/s0735-1097(03)00043-3. [DOI] [PubMed] [Google Scholar]
- 9.Geisterfer-Lowrance AA, et al. A mouse model of familial hypertrophic cardiomyopathy. Science. 1996;272:731–4. doi: 10.1126/science.272.5262.731. [DOI] [PubMed] [Google Scholar]
- 10.Schmitt JP, et al. Cardiac myosin missense mutations cause dilated cardiomyopathy in mouse models and depress molecular motor function. Proc Natl Acad Sci U S A. 2006;103:14525–30. doi: 10.1073/pnas.0606383103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lowes BD, et al. Changes in gene expression in the intact human heart. Downregulation of alpha-myosin heavy chain in hypertrophied, failing ventricular myocardium. J Clin Invest. 1997;100:2315–24. doi: 10.1172/JCI119770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKinsey TA, Olson EN. Toward transcriptional therapies for the failing heart: chemical screens to modulate genes. J Clin Invest. 2005;115:538–46. doi: 10.1172/JCI24144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–84. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bultman S, et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6:1287–95. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- 15.Backs J, Olson EN. Control of cardiac growth by histone acetylation/deacetylation. Circ Res. 2006;98:15–24. doi: 10.1161/01.RES.0000197782.21444.8f. [DOI] [PubMed] [Google Scholar]
- 16.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–28. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 17.Bartha E, et al. PARP inhibition delays transition of hypertensive cardiopathy to heart failure in spontaneously hypertensive rats. Cardiovasc Res. 2009;83:501–10. doi: 10.1093/cvr/cvp144. [DOI] [PubMed] [Google Scholar]
- 18.Kong Y, et al. Suppression of class I and II histone deacetylases blunts pressure-overload cardiac hypertrophy. Circulation. 2006;113:2579–88. doi: 10.1161/CIRCULATIONAHA.106.625467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Antos CL, et al. Dose-dependent blockade to cardiomyocyte hypertrophy by histone deacetylase inhibitors. J Biol Chem. 2003;278:28930–7. doi: 10.1074/jbc.M303113200. [DOI] [PubMed] [Google Scholar]
- 20.Trivedi CM, et al. Hdac2 regulates the cardiac hypertrophic response by modulating Gsk3 beta activity. Nat Med. 2007;13:324–31. doi: 10.1038/nm1552. [DOI] [PubMed] [Google Scholar]
- 21.Kee HJ, et al. Inhibition of histone deacetylation blocks cardiac hypertrophy induced by angiotensin II infusion and aortic banding. Circulation. 2006;113:51–9. doi: 10.1161/CIRCULATIONAHA.105.559724. [DOI] [PubMed] [Google Scholar]
- 22.Pillai JB, et al. Poly(ADP-ribose) polymerase-1-deficient mice are protected from angiotensin II-induced cardiac hypertrophy. Am J Physiol Heart Circ Physiol. 2006;291:H1545–53. doi: 10.1152/ajpheart.01124.2005. [DOI] [PubMed] [Google Scholar]
- 23.Stankunas K, et al. Endocardial Brg1 represses ADAMTS1 to maintain the microenvironment for myocardial morphogenesis. Dev Cell. 2008;14:298–311. doi: 10.1016/j.devcel.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sumi-Ichinose C, Ichinose H, Metzger D, Chambon P. SNF2beta-BRG1 is essential for the viability of F9 murine embryonal carcinoma cells. Mol Cell Biol. 1997;17:5976–86. doi: 10.1128/mcb.17.10.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen H, et al. BMP10 is essential for maintaining cardiac growth during murine cardiogenesis. Development. 2004;131:2219–31. doi: 10.1242/dev.01094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang CP, et al. A field of myocardial-endocardial NFAT signaling underlies heart valve morphogenesis. Cell. 2004;118:649–63. doi: 10.1016/j.cell.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 27.Verzi MP, McCulley DJ, De Val S, Dodou E, Black BL. The right ventricle, outflow tract, and ventricular septum comprise a restricted expression domain within the secondary/anterior heart field. Dev Biol. 2005;287:134–45. doi: 10.1016/j.ydbio.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 28.Pandya K, et al. Discordant on/off switching of gene expression in myocytes during cardiac hypertrophy in vivo. Proc Natl Acad Sci U S A. 2008;105:13063–8. doi: 10.1073/pnas.0805120105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu R, et al. Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell. 2001;106:309–18. doi: 10.1016/s0092-8674(01)00446-9. [DOI] [PubMed] [Google Scholar]
- 30.Muchardt C, Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. Embo J. 1993;12:4279–90. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu B, et al. Inducible cardiomyocyte-specific gene disruption directed by the rat Tnnt2 promoter in the mouse. Genesis. 2009;48:63–72. doi: 10.1002/dvg.20573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szabo G, et al. Poly(ADP-Ribose) polymerase inhibition reduces reperfusion injury after heart transplantation. Circ Res. 2002;90:100–6. doi: 10.1161/hh0102.102657. [DOI] [PubMed] [Google Scholar]
- 33.Wang Z, et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–31. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morrow AG, Brockenbrough EC. Surgical treatment of idiopathic hypertrophic subaortic stenosis: technic and hemodynamic results of subaortic ventriculomyotomy. Ann Surg. 1961;154:181–9. doi: 10.1097/00000658-196108000-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Braunwald E. HEART DISEASE: A Texbook of Cardiovascular Medicine. W.B. Saunders Company; 1997. [Google Scholar]
- 36.Kinugawa K, et al. Regulation of thyroid hormone receptor isoforms in physiological and pathological cardiac hypertrophy. Circ Res. 2001;89:591–8. doi: 10.1161/hh1901.096706. [DOI] [PubMed] [Google Scholar]
- 37.Molkentin JD, et al. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93:215–28. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boucher P, Gotthardt M, Li WP, Anderson RG, Herz J. LRP: role in vascular wall integrity and protection from atherosclerosis. Science. 2003;300:329–32. doi: 10.1126/science.1082095. [DOI] [PubMed] [Google Scholar]
- 39.Chang CP, Chen L, Crabtree GR. Sonographic staging of the developmental status of mouse embryos in utero. Genesis. 2003;36:7–11. doi: 10.1002/gene.10186. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.