Abstract
A multi-angle lensfree holographic imaging platform that can accurately characterize both the axial and lateral positions of cells located within multi-layered micro-channels is introduced. In this platform, lensfree digital holograms of the micro-objects on the chip are recorded at different illumination angles using partially coherent illumination. These digital holograms start to shift laterally on the sensor plane as the illumination angle of the source is tilted. Since the exact amount of this lateral shift of each object hologram can be calculated with an accuracy that beats the diffraction limit of light, the height of each cell from the substrate can be determined over a large field of view without the use of any lenses. We demonstrate the proof of concept of this multi-angle lensless imaging platform by using light emitting diodes to characterize various sized microparticles located on a chip with sub-micron axial and lateral localization over ~60 mm2 field of view. Furthermore, we successfully apply this lensless imaging approach to simultaneously characterize blood samples located at multi-layered micro-channels in terms of the counts, individual thicknesses and the volumes of the cells at each layer. Because this platform does not require any lenses, lasers or other bulky optical/mechanical components, it provides a compact and high-throughput alternative to conventional approaches for cytometry and diagnostics applications involving lab on a chip systems.
1. Introduction
The rapid progress in micro and nano-technologies significantly improved our capabilities to handle, process and characterize cells in a high-throughput and cost-effective manner that was not possible a decade ago [1–7]. This recent progress made lab on a chip systems extremely powerful with various applications in bioengineering and medicine. On the other hand, an essential component of most lab on a chip systems involves optical detection, frequently in the form of microscopy, which unfortunately does not entirely match with the miniaturized and cost-effective format of lab on a chip devices. Therefore, there is still a significant need to further improve the simplicity, compactness and cost-effectiveness of optical imaging to make the most out of existing lab on a chip based cell assays.
To meet this demanding need and bring simplification and compactness to on-chip cell microscopy, one important route to consider is digital holographic imaging [8,9]. Quite importantly, over the last few years we have also experienced numerous new holographic microscope designs to advance various aspects of conventional microscopy making digital holography an important tool especially for biomedical research [10–26].
In this manuscript, along the same direction, we introduce a multi-angle lensfree digital holography platform that can measure, with sub-micron accuracy, both the axial and the lateral position of any given cell/particle within an imaging field of view (FOV) of 20–60 mm2. The key to this high-throughput and accurate performance in a lensfree configuration is the use of multiple angles of illumination combined with a novel digital processing scheme.
Unlike conventional lensless in-line holography approaches, here we utilize a spatially incoherent light source emanating from a large aperture (e.g., D ~50–100µm) with a unit fringe magnification, which enables an imaging field of view that is equivalent to the sensor chip active area [24–26]. In our hologram recording geometry, due to the use of an incoherent source and a large aperture size, the spatial coherence diameter at the sample plane is much smaller than the imaging field of view, but on the other hand is sufficiently large to record holograms of each cell/particle individually. Under this condition, the vertical illumination creates lensless in-line holograms of the cells on the sensor chip. To get more insight on holographic image reconstruction using an incoherent source emanating from a large aperture the reader can refer to references 25 and 26 which report various micro-objects’ reconstructed images under vertical illumination condition.
These digitally sampled cell holograms start to shift laterally on the sensor plane as the illumination angle of the source is tilted – for instance the cells at higher heights will shift laterally more than the cells located at lower heights (see e.g., Fig. 1(a)). In the presented approach, the exact amount of this lateral shift of each lensfree cell hologram is calculated with an accuracy that beats the diffraction limit of light and therefore, by quantifying the amount of this lateral shift on the sensor array as a function of the illumination angle, we can determine the height of each cell from the substrate over a large field of view without the use of any lenses. Such an accurate depth resolving capability when combined with the wide field of view of the presented approach may especially be significant for monitoring multi-layered microfluidic devices to improve the imaging throughput or for conducting micro-array imaging experiments to quantify e.g., on-chip DNA hybridization over a large field of view [27].
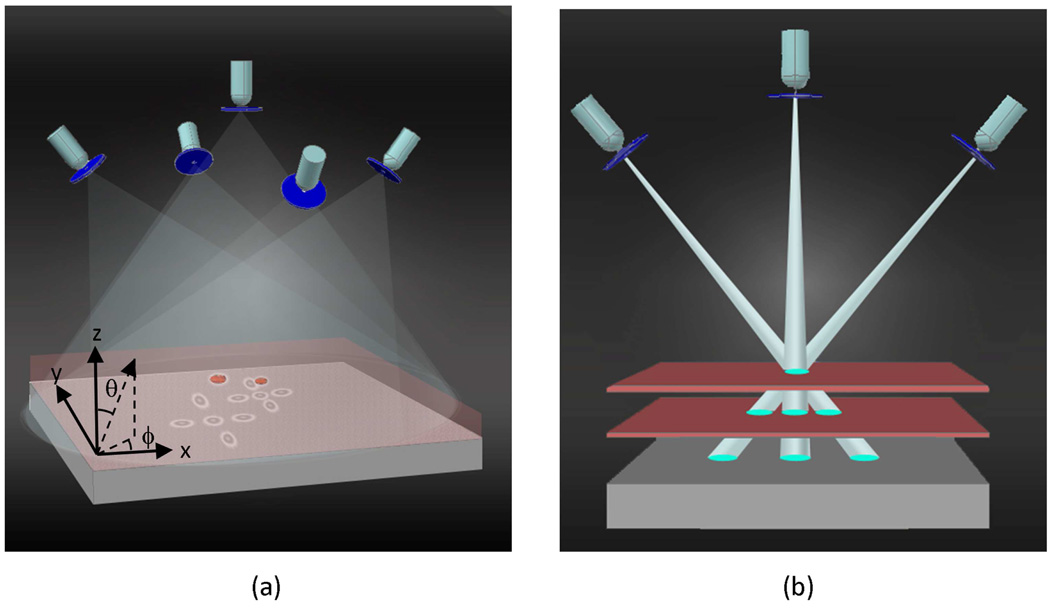
Fig. 1.
The schematic diagram illustrating the principles of multi-angle lensfree holographic imaging. For each illumination angle, a spatially incoherent source such as a light emitting diode is filtered by a large aperture (~0.05mm - 0.1mm diameter), which is placed ~6 cm away from the object plane. Note that unlike conventional in-line holography approaches, the sample plane is much closer to the detector plane with a vertical distance of ~1mm, such that the entire active area of the sensor becomes the imaging FOV. (a) The shadow of each cell shifts laterally on the sensor plane as a function of the illumination angle of the incoherent source, encoding its axial position. (b) Matching of the cells’ shadows acquired at different illumination angles can be achieved by forming imaginary rays between each cell shadow and the corresponding source.
When compared to lens-based or coherent holographic imaging approaches [28–30] that also have a high localization accuracy, the overall hardware complexity of this multi-angle lensfree imaging platform is considerably simplified. First, there is no use of lenses or any other wavefront shaping elements involved in the presented approach. In addition, the source requirement is also greatly simplified permitting the use of spatially incoherent light sources that are emitting through rather large apertures. By using a fringe magnification that is close to unity [24–26], over a few cm’s of free space propagation (e.g., over a distance of L), a spatially filtered incoherent source (regardless of its propagation angle) picks up partial spatial coherence that is sufficient to create individual holograms of each cell on the active area of the sensor. For the experiments to be reported in this manuscript the spatial coherence diameter (Dcoh ∝ Lλ/D) at the sample plane was controlled to be >400λ which was significantly larger than the object size. In our recording geometry, as another important advantage, the speckle noise and the multiple reflection interference noise are much weaker [13,25,26]. Further, because of the limited spatial coherence at the object plane, the coherent cross-talk among different cells of the same sample solution is significantly reduced, which is especially important for imaging of a highly dense cell solution such as whole blood samples. In other words, the scattered waves from each cell’s body cannot effectively interfere with the scattered fields of other cells located roughly outside of the coherence diameter which is advantageous since such cross-interference terms are considered to be noise as far as holography is concerned. Another advantage of using incoherent illumination through large apertures is that mechanical alignment requirements are significantly relaxed which makes it simpler and cost-effective to manufacture, align and operate without the need for light coupling optics and a micro-mechanical alignment interface between the source and the large aperture.
Overall, we believe that this incoherent multi-angle cell imaging platform should provide a simple and compact platform to conduct high-throughput cell analysis over a large FOV within multi-layered micro-channels, making it especially suitable for lab on a chip systems in cell biology and medicine.
2. Depth resolved imaging using multi-angle lensless holography
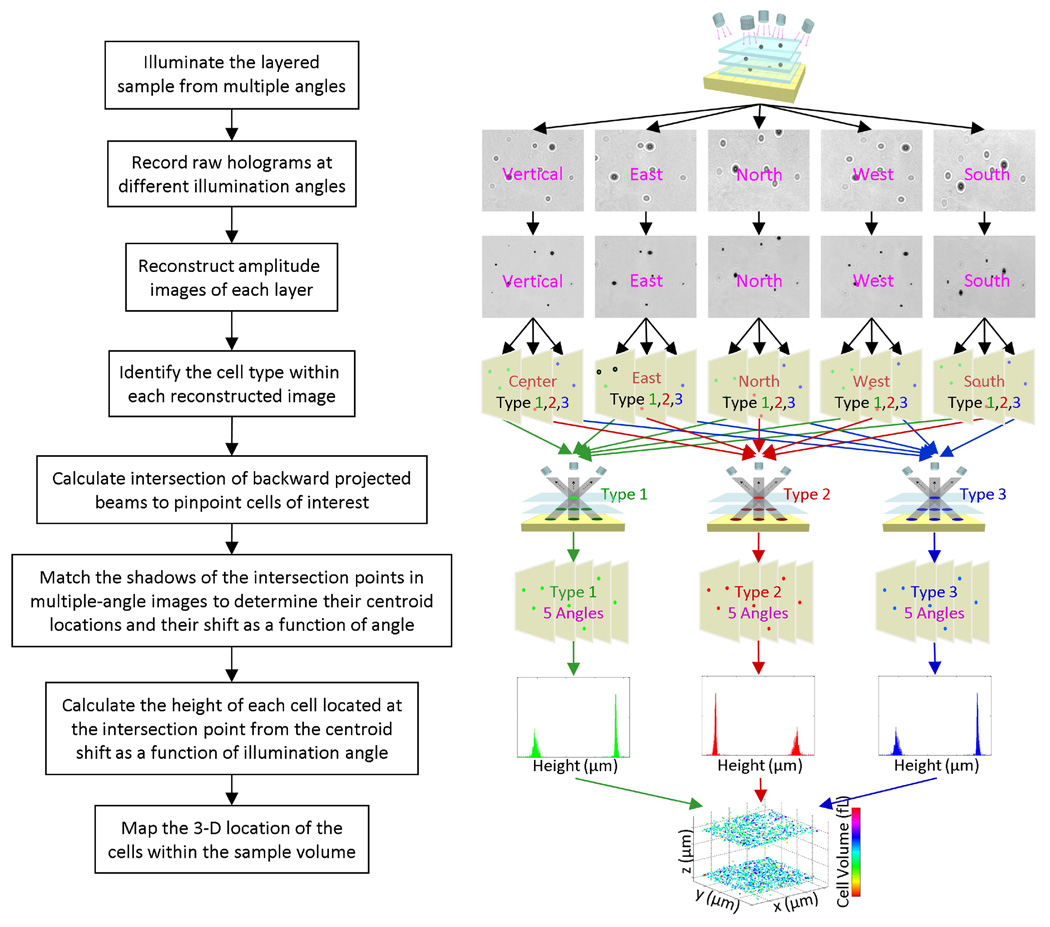
The details of the depth resolved imaging process using multi-angle lensfree holography is summarized in Fig. 2. After the capture of the multi-angle lensfree holographic images of the sample volume as illustrated in Fig. 1(a), each one of these raw holograms is then processed using an iterative twin image elimination and phase recovery technique [25,26,31,32], which can reconstruct amplitude and phase images of different cross sections of the sample volume. As a result of this numerical reconstruction process, we can distinguish overlapping lensfree holograms of the cells from each other, and therefore increase the density of the cells that we can work with.
Fig. 2.
The details of the depth resolved imaging process using multi-angle lensfree holography is summarized.
Before calculating the 3D location of each micro-object within the sample volume of interest, the coarse locations and the types of the objects need to be identified in each lensfree image. For this purpose, automated pattern matching algorithms [24] are used to identify the target objects within each lensfree image yielding the relative x–y coordinates of each object of interest. Repeating this automated identification step using statistical image libraries of different target cells, this platform digitally sorts out a heterogeneous cell mixture according to their cell types [24]. This initial screening step ensures that the depth calculations can be limited to only the cells of interest and the rest of the undesired micro-objects can simply be ignored. For this purpose the raw holograms and/or the reconstructed amplitude images of the cells can be used as long as the individual patterns are not severely overlapping.
Following this pre-screening process, imaginary rays (with a finite cross-section for each ray) are digitally formed by connecting the light source position to the rough x–y coordinate of the shadow or the reconstructed image of each target cell within the imaging field of view (see Fig. 1(b)). These solid rays, which are calculated for all the illumination angles (including the vertical one), are then combined to find their intersection points in 3D, where the total count of the intersecting rays at any given point is also recorded. To provide a rough estimate for the position of each target cell type within the 3D sample volume, a threshold is applied to this ray count – for instance under 5 different illumination angles a ray threshold of 3 implies that at least 3 rays can define a positive count towards a target cell type. More discussion on the effect of this threshold factor on characterization accuracy is provided in the Discussion Section. The localization accuracy of this initial interception algorithm is determined by the cross-sectional width of the rays that are used for back-projection of each shadow (Fig. 1(b)), and is practically on the order of ~5 µm. To achieve a much better depth accuracy (<1 µm) for each target micro-object within the sample volume, an additional calculation step is required, which will be discussed next.
At the end of the above discussed calculation step, for each one of the intersection points (that have a sufficient ray count above the threshold), all the shadows of the same cell at different illumination angles become digitally connected to each other. In some cases, if there is severe overlap between different shadows, some of these lensfree multi-angle images cannot be used. This, however, does not pose a limitation for our depth localization algorithm since 2 independent angles in principle would be sufficient to localize the depth of the micro-object with submicron accuracy, i.e., there is redundancy in the system to better handle dense cell solutions. This also implies that for a less dense solution of interest, fewer angles (for instance 2–3) would also be sufficient. More discussion on the effect of the cell density on characterization accuracy is provided in the Discussion Sections.
Following these initial steps, the fine axial position of each target object needs to be calculated all across the sample volume. Considering the fact that there is no fringe magnification or a lens involved in the presented approach and that the pixel size at the sensor chip is relatively large (i.e., a few microns), this task seems rather challenging. The key to achieve sub-micron depth localization accuracy over a large field of view is to accurately calculate the lateral shift of each one of the multi-angle shadows/holograms corresponding to the same cell. Since the earlier numerical steps already connected the multi-angle holograms of the same cell to each other, all one needs to do next is to accurately calculate the lateral shift amount of the cell hologram as a function of the illumination angle. This step involves calculation of the centroid location of each cell with an accuracy that is much better than the diffraction limit of light. The difference between the centroid locations of at least 2 shadows acquired under different illumination angles is sufficient to accurately estimate the relative heights of all the cells within the sample volume. Next, we will discuss the details of these centroid calculations for each acquired cell shadow.
Centroid calculations for lensfree depth localization of cells on a chip
Accuracy of depth localization is achieved by calculating the centroid location of each cell’s shadow/hologram, and determining the relative shift of the centroid position of the same cell as a function of the illumination angle. For this approach to work effectively, the cell holograms should exhibit minimum amount of overlap such that their centroid calculations remain accurate. While dealing with high cell densities (such as >10,000 cells/µL) there are two factors that help us maintain a good depth localization accuracy: (1) The amplitude reconstruction process enables resolving highly overlapping cell shadows from each other. This permits digital removal of the undesired effects of the other cell shadows on the centroid calculation of each target cell type. And (2) there is redundancy in the measurements such that if one illumination angle produces an overlap for certain cell shadows (which cannot be fully resolved by the amplitude reconstruction process), then the other illumination angles can still remain free from overlaps. All one needs is 2 independent illumination angles where the centroid of the same cell shadow can be calculated accurately for achieving depth localization.
Assuming that Iij represents the cell hologram or its reconstructed amplitude image, and (i, j) denotes the pixel numbers, initially we subtract a linearly fitted background image (IBij) from Iij such that pij ≡ Iij − IBij is calculated. This background profile (IBij) is automatically calculated for each illumination angle through linear regression analysis of Iij. Specifically, for each illumination angle, the background profile (IBij) is evaluated by fitting the edge pixels of the region of interest (ROI) with a linear 2D function. The pixel values of the background profile within the whole ROI are then replaced with the values generated by the fitted 2D function. This process serves to minimize the undesired effects of (1) non-uniform illumination of the sample volume and (2) the surface curvature and/or tilt of the substrates on the depth localization accuracy of the cells. This is crucial especially for achieving accurate depth localization over a large FOV as illustrated in Fig. 3. Following this background subtraction step, the centroid coordinates (xc, yc) of each cell are calculated as:
| (1) |
where is the square of the subtracted intensity.
Fig. 3.
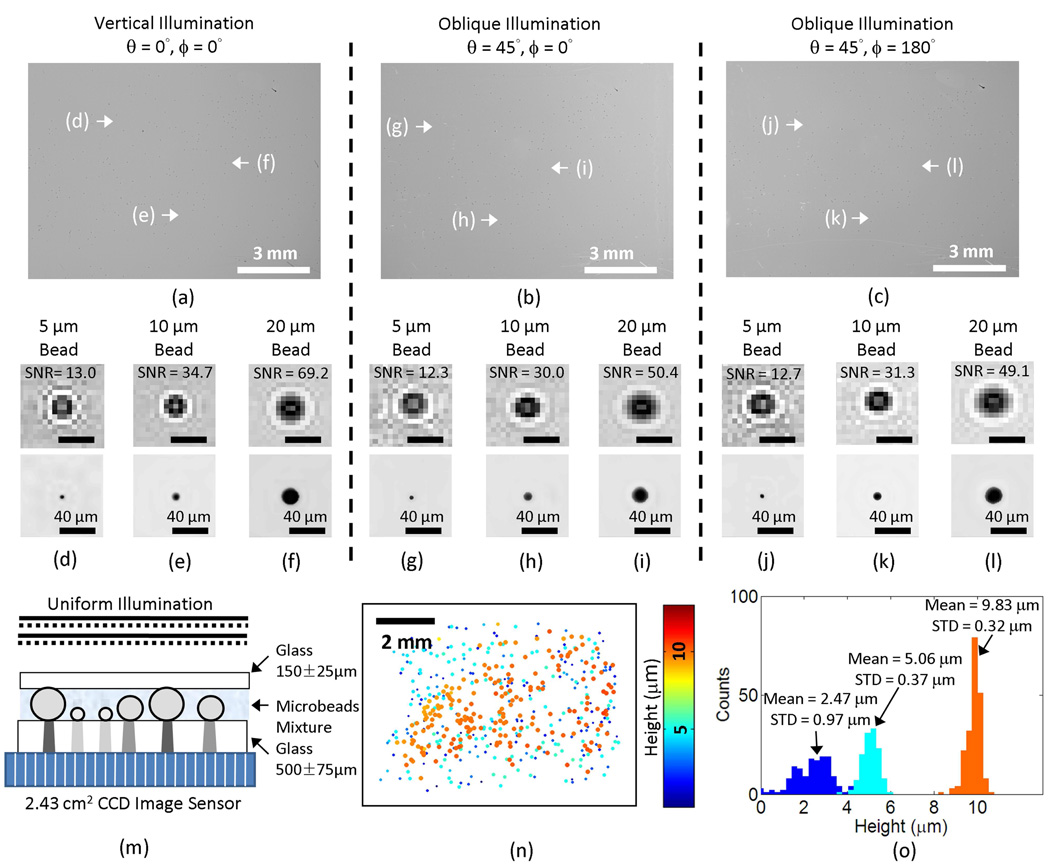
The validation of sub-micron localization performance over a large field of view of ~60 mm2. (a–b–c) Lensfree holograms captured with three different illumination angles are illustrated. (d–e–f) Raw hologram signatures (digitally cropped from the vertical illumination hologram shown in (a)) and their corresponding reconstructed amplitude images for (d) 5 µm, (e) 10 µm, and (f) 20 µm microbeads are illustrated. (g–h–i) Raw hologram signatures (digitally cropped from the oblique illumination hologram shown in (b)) and their corresponding reconstructed amplitude images for (g) 5 µm, (h) 10 µm, and (i) 20 µm microbeads are illustrated. (j–k–l) Raw hologram signatures (digitally cropped from the oblique illumination hologram shown in (c)) and their corresponding reconstructed amplitude images for (j) 5 µm, (k) 10 µm, and (l) 20 µm microbeads are also illustrated. The scale bars in (d–l) are 40 µm long. (m) The cross-sectional structure of the imaged sample is shown. (n) The 2-D distribution of the characterized microbeads is illustrated, with their physical size and the relative height coded by the spot size and the colormap, respectively. (o) The height histogram is calculated from (n) showing three distinct peaks for the 5 µm, 10 µm, 20 µm beads in the mixture. In (n) and (o), the relative height of the substrate surface is arbitrarily assumed to be 0 µm.
For each target cell within the imaging field of view, the choice of the region of interest (ROI) to define Iij is made through a fast iterative algorithm such that the centroid coordinates (xc, yc) eventually match with the geometric center of the ROI for each cell signature. At each step of this iterative algorithm, the center of the ROI was shifted to the position of the calculated centroid and the same centroid calculation was repeated until their positions matched with each other.
After the calculation of all the centroid coordinates of a target cell type under different illumination angles, the lateral shift of each cell’s signature was calculated by taking the difference of the centroid coordinates. This lateral shift was then transferred into a projected height with a known oblique illumination angle, where the angle was already calibrated by a glass substrate with a measured thickness. Due to the non-uniformity of the illumination light and the surface curvature and/or tilt of the substrates, these projected height values needed to be further corrected by a quadratic surface fitting. The same process was performed separately with all the oblique illumination angles, and then these corrected height values from different angles were averaged to accurately determine the axial position (i.e., the z coordinate) of each cell within the sample volume. As for the lateral location (i.e., the x–y coordinates), the centroid coordinates calculated on the lensfree image with the vertical illumination was used without further modification. The reader can refer to the Appendix for further details.
3. Experimental results
To validate the depth resolving performance of our multi-angle holographic imaging platform over a large field of view of ~60 mm2, we conducted an experiment with a mixture of 5, 10 and 20 µm diameter polystyrene beads (Monosized microsphere size standards, Thermo Scientific) suspended in DI water. The micro-particle suspension liquid was dispensed on a 0.5 mm thick glass substrate and covered by a No.1 glass cover slip (~150 µm thick) as shown in Fig. 3(m). To quantify the accuracy of our axial localization results over the entire imaging area of the sensor, before being imaged, the samples were kept still for >10 minutes, allowing the suspended micro-particles to fully settle on the substrates. This ensured that the recovered height (i.e., the axial position) of the particles can be related to the well-controlled radii of the micro-particles, enabling cross validation of our results. During this settlement time period, holograms of the samples were periodically recorded using the vertical illumination to track the trajectories of individual particles. These trajectories were then analyzed to ensure that the displacement had decreased to a stable level (below the Brownian motion limit).
To record the digital holograms of the micro particles distributed over ~60 mm2 field-of-view and their lateral shifts as a function of the illumination angle, three optical fibers with a core diameter of D = 50 µm each were utilized to illuminate the sample placed on the bare surface of a CCD image sensor chip with a pixel size of 5.4 µm (KAF-8300, Kodak). The protective glass of the sensor chip has been removed to minimize the distance between the sample and the sensor surface and maximize signal-to-noise ratio of the lensfree holograms. The fibers are individually butt-coupled to three cyan light-emitting diodes (LEDs, LXHL-LE3C, Luxeon) and their tips are placed approximately 6 cm away from the sample to provide one vertical and two other tilted illuminations with (ϕ= 0°; θ = 45°) and (ϕ = 180°; θ = 45°) as illustrated in Fig. 1. The center wavelength (λ) of the LEDs is 505 nm and the FWHM spectral width is ~30 nm.
Note that this fiber with a core diameter of D = 50 µm introduces partial spatial coherence before its exit aperture. However, this is not a requirement for the presented approach, since the distance between fiber-end and the object plane (L~6 cm) is sufficiently large to create a coherence diameter (Dcoh ∝ Lλ/D) that is significantly wider than the micro-object diameter (<50λ for all objects reported in this manuscript) for recording of their holograms individually. We further validated this by using an LED that is directly butt-coupled to a pinhole (D = 0.1 mm) to effectively record similar holograms. Therefore, the fiber length and the degree of spatial coherence that the light picks up within the fiber is not of crucial importance in this study as the spatial coherence at the sample plane is sufficiently large even for a completely spatially incoherent source that is filtered by a similar aperture size of 0.05–0.1 mm.
Lensfree height characterization results of these particles are summarized in Fig. 3. Figures 3(a–c) illustrate the raw lensfree holograms that are captured under each illumination angle over an imaging FOV of >1 cm2. Smaller Figs. 3(d–l) focus on the individual holographic signatures, digitally taken from Figs. 3(a–c); and the reconstructed amplitude images (created by iterative holographic reconstruction [25,26]) of these representative particle holograms are also shown under different illumination conditions. As expected, in these figures the raw holograms of the tilted illumination conditions show an elongated texture, parallel to the tilt direction. Based on digital processing of these multi-angle lensfree holograms as described in the previous section, we recovered the height distribution of the micro-particles from the substrate surface as illustrated in Fig. 3(n), where for convenience the relative height of the substrate surface is assumed to be 0 µm (the physical size and the height of the particles are coded by each spot size and the colormap, respectively). Figure 3(o) also reports the height histogram calculated from Fig. 3(n), which clearly resolves 3 different particle types from each other based on their relative heights (i.e., radii). For 20 µm and 10 µm particles our results estimate the mean height of these particles from the substrate surface as 9.83 µm and 5.06 µm, with a standard deviation of 0.32 µm and 0.37 µm, respectively, whereas for the smaller particle (5 µm diameter), the mean height from the surface was estimated to be 2.47 µm with a standard deviation of 0.97 µm. These results are in close agreement with the height values that one would expect from the radii of these particles, i.e., 10 µm, 5 µm and 2.5 µm, respectively. One could attribute the differences between our characterization results (9.83, 5.06 and 2.47 µm) and the known radii of the particles (10, 5 and 2.5 µm) to the unavoidable surface curvature of the substrate over the large imaging field of view (~60 mm2) and to the standard deviation of the particle radii, which is reported by the manufacturer (Thermo Scientific) to be ± 1% for each particle type. The relatively worse performance (with a standard deviation of 0.97 µm) of the smaller sized particle (5 µm) is related to a reduced hologram signal-to-noise ratio (SNR) (refer to the individual hologram signatures and the SNR values that are provided in Fig. 3(d–l)). This is also a topic that we will also address in the Discussion Section.
We further validated our multi-angle lensless holography approach by imaging a two-layered micro-channel containing red blood cells (RBCs) (see Fig. 4(f)). Whole blood samples were mixed with the anticoagulant EDTA at a ratio of 2 mg of EDTA per ml of blood (EDTA tubes, BD). The blood was kept still for ~20 minutes until the RBCs settled. After sedimentation, RBCs were extracted from the bottom of the sediment and diluted with cell culture medium (RPMI 1640, Invitrogen) to a concentration of ~15,000 cells/µL. A small number of polystyrene microbeads with a diameter of 20 µm were then added to the suspension (~40 beads/µL), serving as mechanical spacers in the multi-layer structure shown in Fig. 4(f).
Fig. 4.
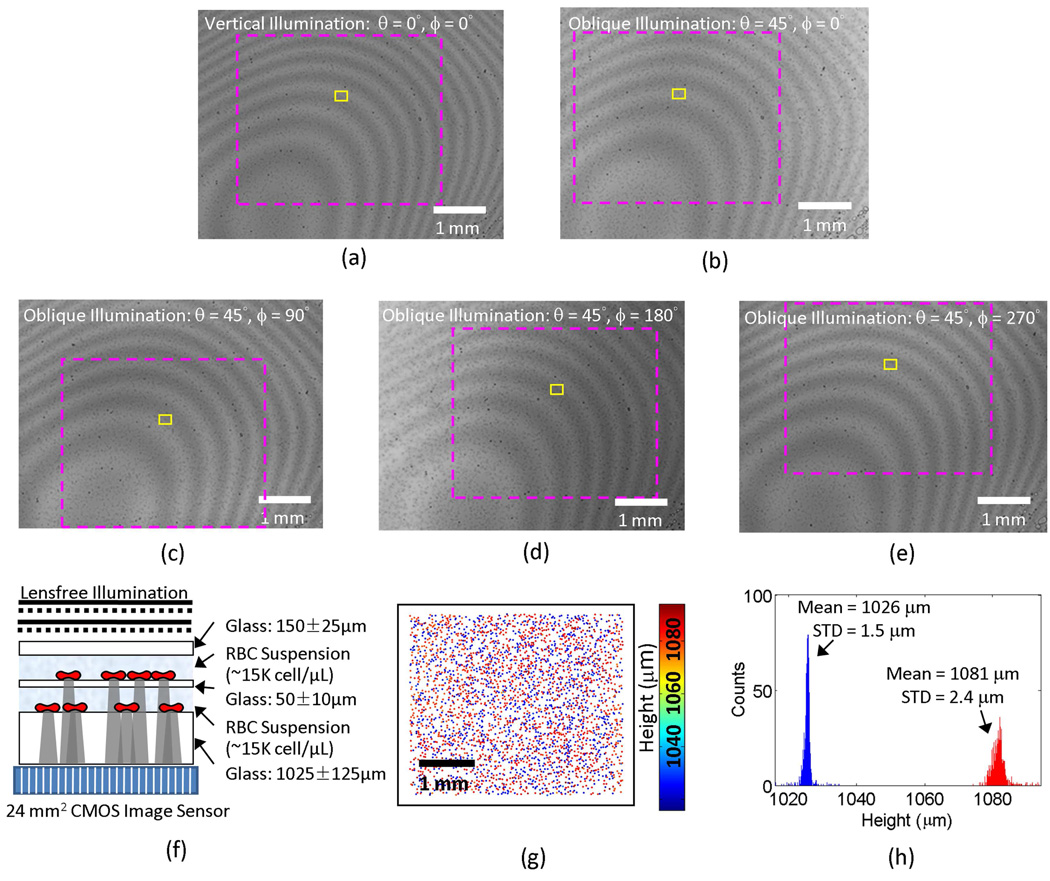
Lensless multi-angle characterization of RBCs located within two-layered micro-channels. (a)–(e) Lensfree holograms are captured with five different illumination angles. The magenta dashed rectangles in (a)–(e) are the regions corresponding to the field-of-view shown in (g); and the yellow rectangles define the regions corresponding to the field of view of the images in Fig. 6. (f) The cross-sectional structure of the 2-layered sample is shown. (g) The 2D distribution of the RBCs located in both vertical channels is calculated with their height coded by the colormap. (h) shows the histogram of the cell heights over the entire field of view, which exhibits a double peaked behavior, as expected, resolving the 2 vertical micro-channels. In (g) and (h), the relative height is arbitrarily assumed to be 0 µm at the surface of the sensor.
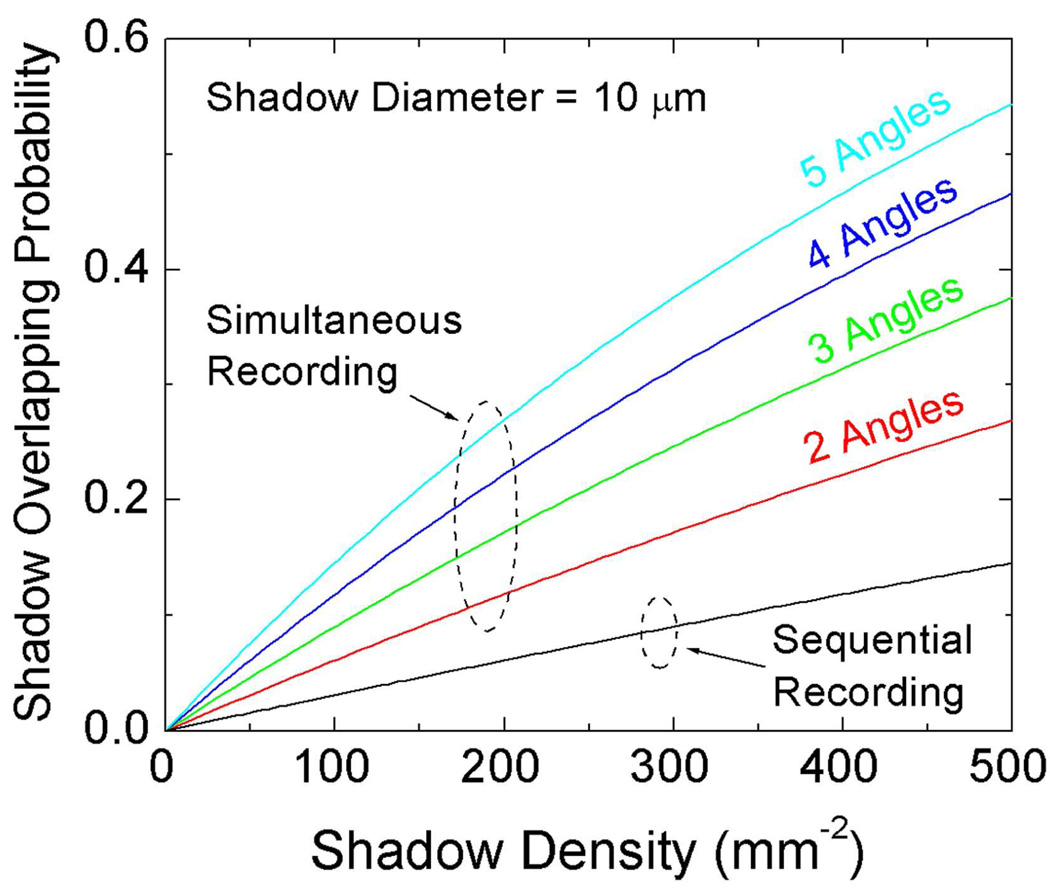
The holograms of the cells were recorded by placing the samples directly on the image sensor chip as shown in Fig. 4(f). A CMOS image sensor with a pixel size of 2.2 µm and an active area of 24.4 mm2 (MT9P031, Aptina) was used for imaging the RBC suspension sample. After settlement for >10 minutes, the samples were illuminated from different angles sequentially and the lensfree holograms with different illumination angles were recorded separately as illustrated in Fig. 1. Alternatively, this image acquisition process could also be done in parallel by turning all the multi-angle sources on at the same time rather than sequentially. However the overall density of cells that can be imaged with parallel illumination is lower than sequential imaging, which is further quantified in the Discussion Section (Figs. 7–8).
Fig. 7.
The shadow overlap probability plotted as a function of both the shadow density (i.e., the throughput) and the multi-angle hologram recording method (parallel vs. sequential). The diameter of the holographic shadows for all the angles and all the vertical layers is assumed to be ~10 µm.
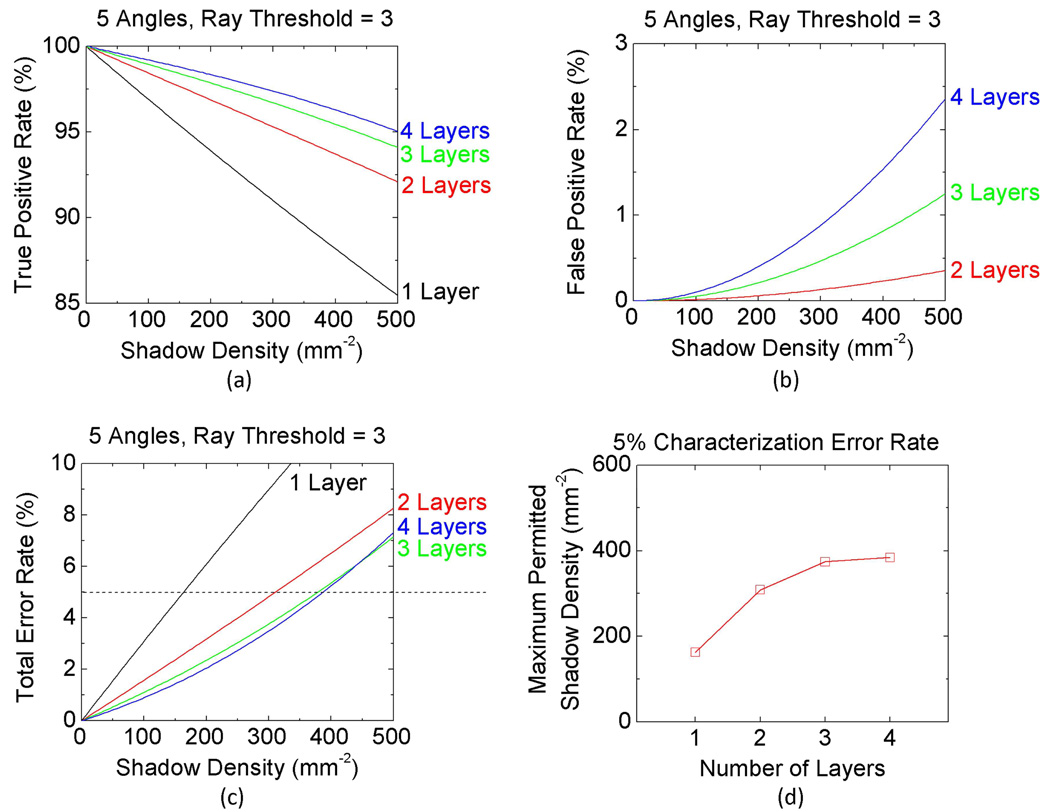
Fig. 8.
Quantified performance comparison of the multi-angle lensfree holographic cell characterization platform as a function of the shadow density and the number of vertical layers on the sensor chip. (a) The true positive rate, (b) the false positive rate, and (c) the total error rate for different cell densities distributed to 1, 2, 3, or 4 vertical layers/channels, where the total error rate includes both the missed cells and the false positives. (d) The maximum permitted shadow density (i.e., the maximum permitted throughput) at the sensor plane is plotted as a function of the number of vertical layers when the total error rate is maintained at a level of 5%. The cells are assumed to be illuminated from 5 different angles as shown in Fig. 1(a), and the ray threshold value is set to 3 for detecting each cell’s 3-D location (refer to Section 2). The shadow width at the sensor plane is assumed to be 10 µm for these numerical simulations.
To generate these lensfree cell holograms with different illumination angles, five optical fibers with a core diameter of 50 µm each were mounted with their tips approximately 6 cm away from the samples. Except the vertical illumination case, the illumination angles were 0°, 90°, 180° and 270° azimuthally and the polar angles were all 45° from the normal direction of the imaging plane, as shown in Fig. 1. The fibers are connected to a Xenon lamp (6258, Newport Corp.) filtered by a monochromator (Cornerstone T260, Newport Corp.), where the central wavelength of the monochromator was set to ~500 nm and the FWHM spectral width was ~10 nm.
By processing all these raw holograms acquired at different illumination angles as discussed in the previous section, we recovered the height distribution of the RBCs located at both of the vertical channels as illustrated in Fig. 4(g). Figure 4(h) also shows the histogram of the cell heights over the entire field of view, which exhibits a double peaked behavior, as expected, resolving the 2 vertical micro-channels. Because the cells were permitted to sediment on the surface of each micro-layer, we obtained a very narrow height distribution at each channel as a result of our fine depth resolving power. For the upper micro-channel, the standard deviation of the cell height (2.4 µm) is larger than the lower channel one (1.5 µm), which is due to the surface curvature of the spacer glass. In other words, because the spacer glass between the vertical channels is much thinner than the substrate of the bottom layer, it exhibits a significantly larger surface curvature over the imaging field of view which increased the height variations as observed in the upper channel cell height histogram (Fig. 4(h)). Meanwhile, for the lower channel, the substrate was chosen to be >0.5 mm thick and therefore the cell height histogram showed a much better accuracy with a standard deviation of 1.5 µm in relative height of the cells.
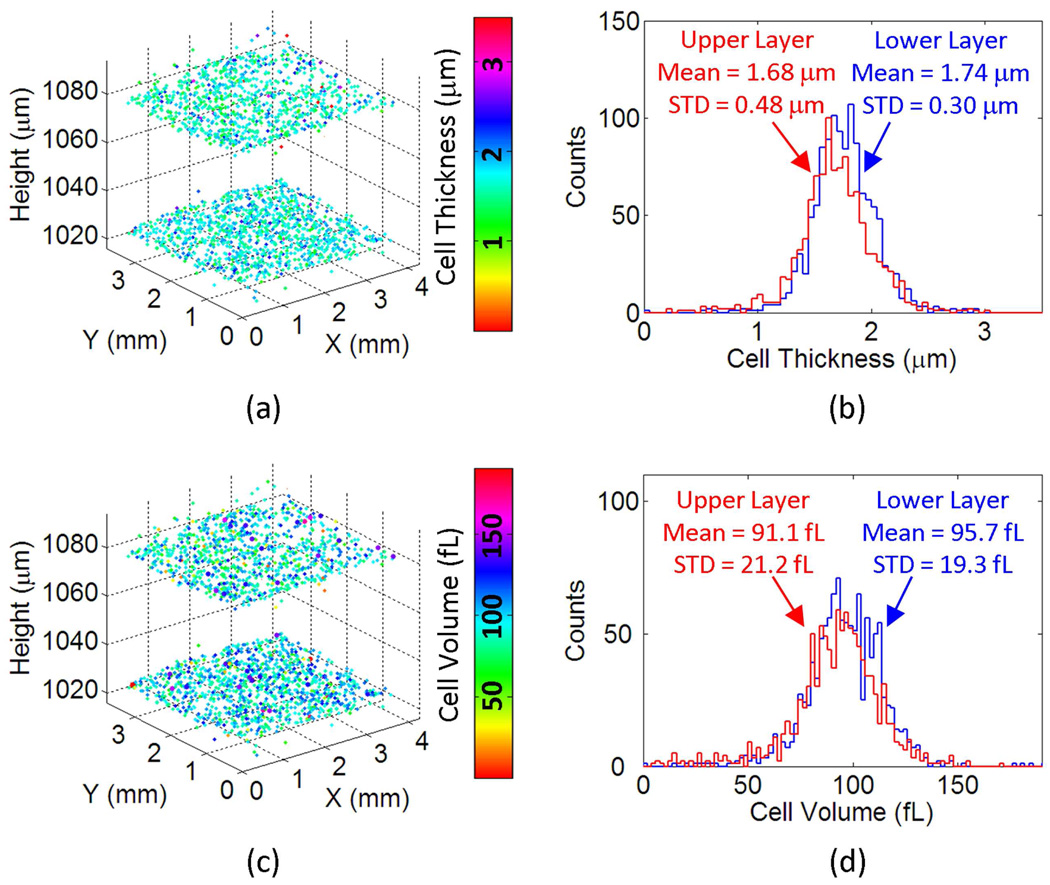
Once the axial and lateral locations of the cells are accurately determined within this multi-layered structure (Fig. 4(f)), we can also characterize other properties of the cells in 3D such as the thickness or the volume of each cell. Figures 5(a) and 5(c) report the thickness and the volume maps, respectively, of each one of the red blood cells that are characterized in Fig. 4. In these figures the colormaps code the measured thickness (µm) and volume (fL) of each cell. Figure 5(b) and 5(d) also plot the thickness and volume histograms of the red blood cells at each vertical channel, which predict a mean RBC thickness of 1.74 µm and 1.68 µm for the bottom and top channels, respectively; and a mean RBC volume of 95.7 fL and 91.1 fL for the bottom and top channels, respectively. These results are in good agreement with standard values of healthy red blood cells, further validating our results [33]. The computation time required to generate the presented results in Figs. 4 and 5 is ~30 minutes on a 2.2 GHz Opteron CPU. However, this computation time can be significantly reduced by further optimizing the code and performing the tasks on a graphic processing unit (GPU) since most computations in this technique can be highly parallelized [26].
Fig. 5.
Thickness and volume of each cell within the two-layered micro-channels are calculated over a field of view of >15 mm2. (a) The 3D distribution of the RBCs in both of the vertical channels is illustrated with their cell thickness value coded by the colormap. (b) The thickness histograms of the RBCs in both the upper and lower micro-channels are shown. (c) The 3D distribution of the RBCs in both vertical channels is illustrated with their cell volume coded by the colormap. (d) The volume histograms of the RBCs in both the upper and lower micro-channels are shown.
The key to estimate each cell’s thickness and volume properties individually over the entire imaging FOV is the iterative twin image elimination algorithm that permits digital reconstruction of the phase and amplitude images of each cell from its lensfree hologram [25,26,31,32]. To relate the recovered optical phase of each cell to a physical thickness, we assumed that red blood cells are phase only objects with an average refractive index of 1.40 in a solution with refractive index 1.33 [34]. Under these assumptions the thickness of the RBC is directly proportional to its phase recovered from the iterative twin-image elimination algorithm. The areas of the cells were estimated by a simple global thresholding of the recovered phase images, and the volume of each cell was estimated by the product of its thickness and area. These imaging results are quite important as they enable lensfree on-chip characterization of a 3D distribution of cells over a much larger volume than a regular microscope could enable (note that the lateral scale bars and grid size in Figs. 4(g), 5(a) and 5(c) are all 1 mm).
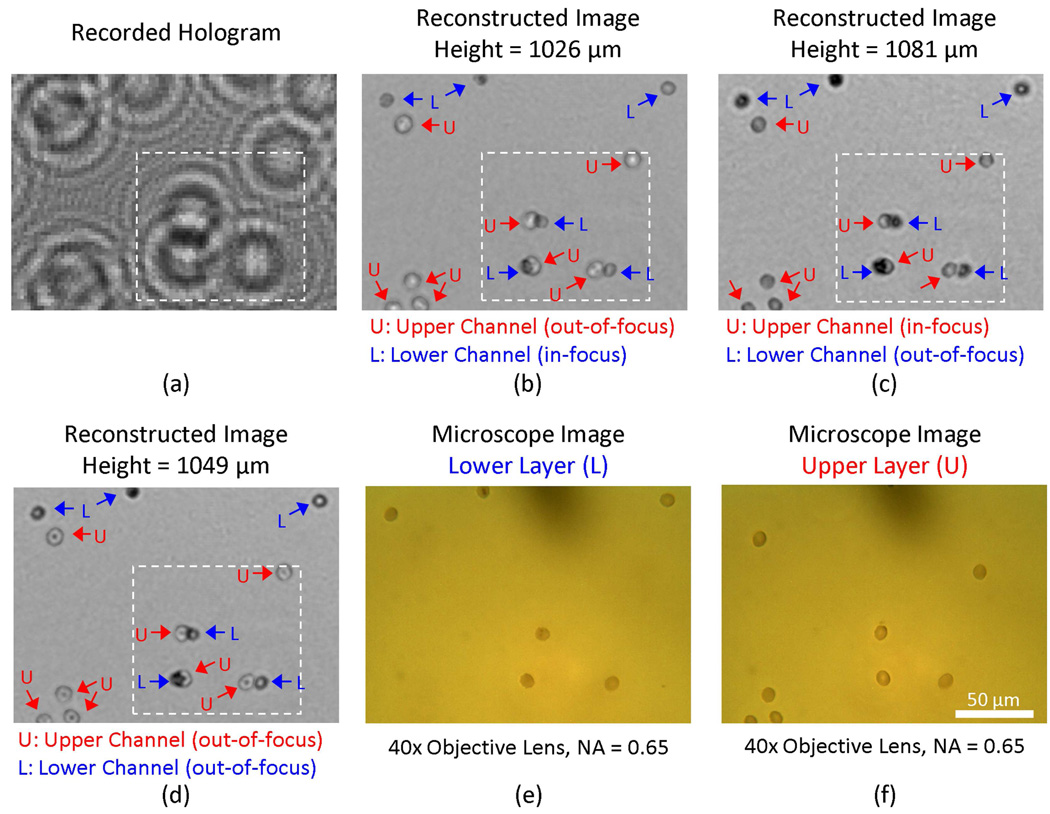
For the experiments reported in Figs. 4–5, the cell density at each layer was ~15,000 cells/µL. To achieve the reported depth accuracy in 3D for such a high concentration of cells, we made use of two key factors: (1) we used 5 illumination angles (see Fig. 4) which reduced the likelihood of the events where all the shadows corresponding to a single cell were overlapping with other cells for all the illumination angles. And (2) the image reconstruction process enabled resolving densely packed cell shadows from each other. A good example of the success of this digital reconstruction process is illustrated in Fig. 6, where 3 red blood cells from the top micro-channel overlap at the sensor plane with the holograms of 3 different red blood cells located at the bottom micro-channel (refer to the holograms within the white dashed rectangle of Fig. 6(a) which correspond to these 6 RBCs at both layers). Figure 6(b) and 6(c) illustrate the reconstructed amplitude images at the bottom and top channel surfaces, respectively. Figure 6(d) also illustrates the digital reconstruction results at an intermediate plane between the bottom and the top micro-channels. To independently confirm our reconstruction results, two microscope images of the bottom and top micro-channels (corresponding to the same FOV as in Fig. 6(a)) are also provided in Figs. 6(e) and (f), respectively. Here we would like to also emphasize that the lensfree holographic image and its reconstructions that are reported in Figs. 6(a–d) are digitally taken from a much larger field of view shown in Fig. 4(a), which illustrates ~2 orders of magnitude increased FOV of our approach when compared to conventional optical microscope images (Figs. 6(e–f)).
Fig. 6.
Demonstration of overlapping RBCs from two vertical micro-channels being digitally resolved by the holographic reconstruction process. (a) The raw lensfree hologram of the digitally zoomed region specified with the yellow rectangle in Fig. 2 (a) is illustrated. The amplitude images (b), (c), and (d) were reconstructed from (a) at a height of 1026 µm, 1081 µm, and 1049 µm respectively. In these reconstructed images, “L” and “U” refer to the RBCs located at the lower and upper micro-channels, respectively. The same field of view is also imaged using a 40X objective lens (0.65 NA) by focusing on both the lower (e) and the upper (f) micro-channels for comparison purposes. Note that the field of view that is imaged with Fig. 4 constitutes ~2 orders of magnitude improvement over the 40X microscope images shown in (e–f).
4. Discussion
The holographic shadow of a cell has a 2D texture that contains both the phase and amplitude of the scattered fields. This information, when sampled by a dense array of 2D detectors, permits digital recognition of the cell type, and it also enables digital reconstruction of microscopic images of the same cells through e.g., iterative phase recovery algorithms [24–26,31,32] as illustrated in the Results Section. In the presented multi-angle lensfree holography approach, since the fringe magnification is unity, the effective numerical aperture with existing sensor arrays is limited to ≤0.2 which results in a poor axial position accuracy. As a result of this, the holographic signatures of two cells that are vertically separated by <50–100 µm do not have sufficient information to reliably tell apart their axial positions. To solve this issue, we devised the use of multiple angles of illumination as depicted in Fig. 1(a). Under vertical lensfree illumination, each cell casts a 2D shadow on the sensor array. This holographic shadow, however, shifts laterally by a certain distance once the angle of illumination is tilted. The lateral shift amount on the sensor array is directly related to the relative height of the cell within the sample. And a key observation is that the centroid (x–y) location of each cell under any given illumination condition can be determined with an accuracy that beats the diffraction limit of light. The accuracy of this centroid calculation (which is detailed in the Appendix) is only limited by detection signal to noise ratio, and can easily achieve sub-micron localization accuracy.
Besides the accuracy of cell/particle localization, another important parameter that needs more discussion is the characterization error rate (i.e., the overall percentage of false positives and missed cells within the sample volume), which surely is dependent on the density of the objects to be imaged. To better investigate and quantify this dependence, we have performed numerical simulations, the results of which are summarized in Figs. 7–8 and Supplementary Fig. S1, Fig. S2, and Fig. S3. In these simulations, we report the density of cells in terms of the density of shadows that they create at the sensor plane under the vertical illumination. Therefore, this density of shadows at the sensor plane is equivalent to the number of independent micro-objects to be imaged per frame, which is a direct measure of the throughput of imaging.
According to our simulation results, as the density of shadows at the sensor chip increases, the overlap probability among cell shadows also increases as illustrated in Fig. 7. However, as one would expect, sequential imaging with different illumination angles copes much better with increasing shadow density when compared to parallel (i.e., simultaneous) imaging with all the angles (see Fig. 7). The cost of this improvement that comes with sequential lensfree imaging is a reduction in the speed of data capture since more frames need to be captured to characterize the same volume of interest.
Overlapping shadows at the sensor plane also cause missed (i.e., un-identified) objects as well as false characterized (i.e., false positive) objects. To better understand this error process, Fig. 8 quantifies the performance of our algorithm for such characterization errors as a function of the shadow density at the sensor chip. Accordingly, Figs. 8(a) and 8(b) quantify the True Positive Rate and the False Positive Rate of our algorithm as a function of the shadow density, respectively; and Fig. 8(c) summarizes the Total Error Rate (in %) which combines both the missed and the false characterized target objects in its results. These figures are calculated for 5 angles of illumination and a ray threshold of 3 was used to make a depth characterization decision. Figure 8(c) concludes that for the same total number of cells to be imaged per FOV (such that for achieving the same characterization throughput) utilizing 3 or 4 vertical micro-channels performs roughly the same in terms of their characterization error rate, whereas 2 vertical micro-channels (although performing much better than a single channel having all the cells) perform worse than either 3 or 4 vertical channels. The same conclusion is also summarized in Fig. 8(d) in a different format: for a total error rate of 5% (which includes both the missed cells and the false positives) a 3 or 4 layered microfluidic device will have roughly the same level of maximum permitted shadow density at the sensor plane, whereas 2 vertical channels can accept a smaller shadow density implying a reduced throughput of imaging.
Another way of restating this last conclusion is that for characterization of a certain number of cells per FOV, distributing these cells over 3 or 4 vertical channels would perform much better than a single or 2 layered micro-device in terms of the total error rate. More detailed investigation of the behavior of 2, 3 and 4 vertical channels for handling increasing shadow densities (i.e., increasing throughput) as a function of the number of illumination angles and ray threshold factor is provided in Supplementary Figures. S1, S2, and S3, respectively. These detailed Supplementary Figures also support the conclusions of Fig. 8, and further shed light on the choice of the optimum conditions for a given number of illumination angles to achieve a desired imaging throughput at an acceptable characterization error rate.
Based on the above discussion we can conclude that the presented multi-angle holographic imaging platform can achieve a much higher throughput using multi-layered micro-fluidic devices. The compactness and increased throughput of this platform could greatly benefit point-of-care cytometry and diagnostics applications, where within a simple multi-layered micro-fluidic device, sample fluids from multiple patients can be simultaneously processed and characterized in isolated vertical channels without the risk of cross-contamination.
5. Conclusions
In conclusion, we have introduced a multi-angle lensfree holographic imaging platform that can accurately characterize both the axial and lateral positions of cells and micro-particles located within multi-layered micro-channels. We have demonstrated a depth localization accuracy of ~300–400 nm, which is limited by detection noise, over ~60 mm2 field of view by utilizing three LEDs illuminating various sized microparticles located on a chip from one vertical and two oblique angles. Furthermore, we successfully applied this lensless multi-angle imaging approach to simultaneously characterize whole blood samples located at multi-layered micro-channels in terms of the counts, individual thicknesses and the volumes of the cells at each layer. Because this multi-angle on-chip holography platform does not require any lenses, lasers or other bulky optical/mechanical components, it provides a high-throughput alternative to conventional approaches for cytometry and diagnostics applications involving lab on a chip systems.
Supplementary Material
Acknowledgments
A. Ozcan gratefully acknowledges the support of the Office of Naval Research (through a Young Investigator Award 2009) and the NIH Director's New Innovator Award (DP2OD006427 from the Office of the Director, NIH). The authors also acknowledge support of the Okawa Foundation, Vodafone Americas Foundation, the Defense Advanced Research Project Agency's Defense Sciences Office (grant 56556-MS-DRP), the National Science Foundation BISH Program (awards 0754880 and 0930501), the National Institutes of Health (NIH, under grant 1R21EB009222-01) and AFOSR (under project 08NE255).
Appendix - Analysis of the Depth Localization Accuracy
The accuracy of the centroid-based localization method described in the previous sections has two fundamental limiting factors in a lensfree holographic configuration: the detection noise and the pixelation error. Brownian motion of particles is a key factor only when the micro-objects are suspended in the liquid, which can then be handled by reducing the integration time at the sensor chip or by using simultaneous multi-angle imaging where all the illumination angles are used at the same time. Once the micro-objects settle on the substrate surface, the friction between the objects and the surface provides enough anchoring force to significantly limit the Brownian motion.
In this section, we will derive the governing theory to quantify the effects of the detection noise and the pixelation error on the accuracy of our depth localization calculations. Next we start with the quantification of the detection noise.
The effect of the detection noise on the calculation of the centroid coordinates of the particles under the vertical illumination can be analyzed by quantifying the error propagation from the noise at individual pixels to the centroid calculations [35,36]. Following the same notation presented in Section 2, the centroid coordinates of a particle shadow created by the vertical illumination can be re-written as:
| (2) |
where ux ≡ ∑ij xij nij, uy ≡ ∑ij yij nij, and v ≡ ∑ij nij.
By using the law of error propagation, the variance of xc and yc (, respectively) can then be evaluated as:
| (3) |
where is the variance of ux, is the variance of uy, is the variance of v, Suvx ≡ 〈ux v〉 − 〈ux〉 〈v〉 is the covariance of ux and v, Suvy ≡ 〈uy v〉 − 〈uy〉 〈v〉 is the covariance of uy and v; and 〈〉 denotes the expectation operator. With these definitions of ux, uy, and v, their variance and covariance can be expressed as:
| (4) |
where is the variance of nij and Sijkl ≡ 〈(nij − 〈nij〉) (nkl − 〈 nkl〉) 〉 is the covariance of nij at two pixels. Since the noise on individual pixels can be assumed to be uncorrelated, Sijkl can be dropped and the variance of the centroid coordinates, xc and yc, (under the vertical illumination) can be simplified as:
| (5) |
where .
Since the detection noise, without loss of generality, can be assumed to have a probability density function with normal distribution, the variance of can be evaluated as:
| (6) |
where is the variance of pij. As expected, Eq. (6) predicts that the noise level of is not only directly linked to the noise level of pij but also modulated by the 2D profile of pij. In summary: Eqs. (5) and (6) determine the effect of the detection noise on the accuracy of the lateral centroid calculations for a measured pattern of Iij under the vertical lensfree illumination.
The same analysis can also be applied to calculate the variation of the centroid coordinates for patterns imaged under oblique illumination angles, such that can also be quantified in a similar fashion. Given (xco, yco) as the centroid coordinates of an oblique shadow, and as the lateral shift along this oblique illumination angle, then the variance of the lateral shift and the variance of the projected height can be estimated by the law of error propagation as such:
| (7) |
and
| (8) |
where dx ≡ (xco − xc), dy ≡ (yco − yc), Sdxdy is the covariance of dx and dy with its value close to zero, θg is the oblique illumination’s refractive angle within the substrate, and are the variances of dx, dy, xc, yc, xco, yco, respectively.
After calculating the variance values of the projected height from all the oblique illumination angles with Eqs. (7) and (8), the standard deviation of the averaged particle height is estimated by:
| (9) |
where na is the number of the oblique illumination angles involved in calculating the particle height and is the variance of the projected height for an individual oblique illumination angle. Therefore Eqs. (5)–(9) quantify the contribution of the detection noise to the final depth resolution of the proposed multi-angle lensfree holography platform.
Next we will investigate the impact of the second major source of error in our depth localization calculations, such that the effect of the pixelation error at the sensor array will now be quantified. For the vertical lensfree illumination case, the effect of pixelation error on determining the lateral centroid coordinates of the particle can be estimated by analyzing its spatial pattern sampled at the sensor array [37]. Assuming that e(x, y) is the 2D continuous profile of the vertical projection pattern without noise (i.e., it represents the optical intensity profile of the particle’s holographic shadow on the image sensor before being sampled); and that fe(x, y) represents the convolution of e(x, y) with a square function whose width is the pixel size (Δ) of the image sensor, then the centroid coordinates xc and yc of the sampled pattern of the vertical illumination case can be calculated as:
| (10) |
where fex(x) ≡ ∫ fe(x, y) dy and fey(y) ≡ ∫ fe(x, y)dx. Assuming that ηx and ηy define the offset of the centroid position from the center of a pixel along the x and y directions, respectively, then the centroid coordinate estimation errors δx (ηx) and δy (ηy)can be expressed as [37]:
| (11) |
where Fex (u) and Fey (v) represent the Fourier transforms of fex(x) and fey(y), respectively; and Fex′ (u) and Fey′ (v) are the first derivatives of Fex (u) and Fey (v), respectively. Without loss of generality, we can confidently assume that ηx and ηy are both uniformly distributed between −Δ/2 and Δ/2. Accordingly, the standard deviation of the 2D localization errors arising from pixelation noise can be calculated as such:
| (12) |
Eq. (12) quantifies the impact of pixelation error on the accuracy of the lateral centroid coordinate calculations for a pattern measured under the vertical lensfree illumination. The same procedures that we discussed above for the detection noise analysis can also be used to calculate the variances of the centroid coordinates for all the oblique illumination angles due to pixelation error. After calculating the variances of all illumination angles, the height deviation contributed by pixelation error can then be evaluated the same way as the height deviation due to the detection noise was calculated above (refer to Eqs. (7)–(9)).
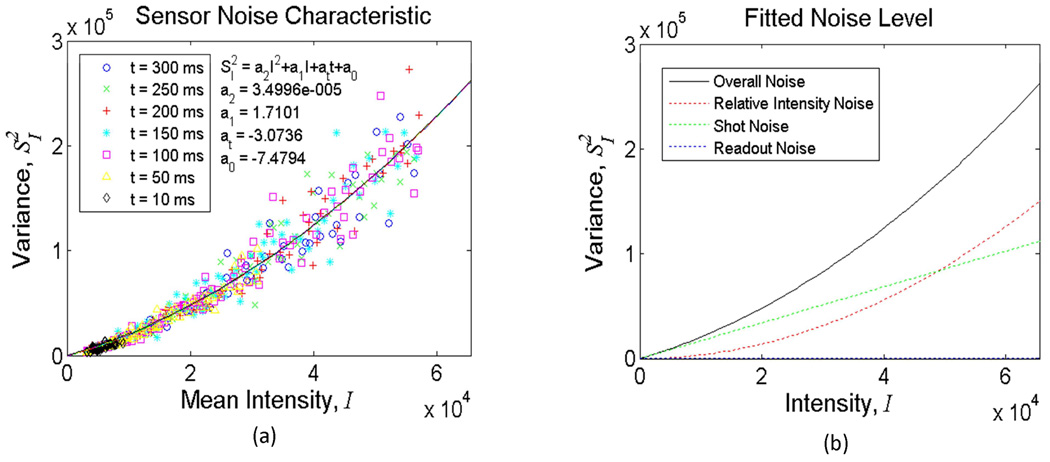
To better quantify the nature of the detection noise in our set-up, we experimentally characterized the noise statistics of one of our sensors (Kodak, CCD KAF-8300) under the same illumination conditions that we reported in the Experimental Results Section (Fig. 3). In these characterization experiments, our goal was to estimate the relative strengths of different noise terms in our experimental set-up to permit an accurate comparison of our results against the theoretical limits. Toward this end, Fig. A1 reports the variance values of the pixels of the sensor chip measured with different integration times as a function of the illumination intensity. To quantify the individual contributions of different noise processes, we used a noise model given by:
| (13) |
where I is the mean value of the measured pixels; is the variance of the pixel values; t is the integration time; a2 a1, at, and a0 a are the parameters for Relative Intensity Noise (RIN), Shot Noise (SN), Dark Leakage Noise (DLN), and Readout Noise (RN), respectively. Multi-variable fitting results showed that the detection noise at the sensor chip was mostly dominated by RIN and SN (see the Fig. A1 for the decomposition of the noise terms as a function of the illumination intensity). These results enabled us to assess the relative magnitudes of different detection noise terms and their total contribution to the localization error in our lensfree measurements.
Fig. A1.
Noise characteristics of the CC D image sensor used in Fig. 3 are quantified. (a) reports the variance values of the pixels of the sensor chip measured with different integration times as a function of the illumination intensity. (b) quantifies the decomposition of various noise terms as a function of the illumination intensity. The fitted strengths of individual noise terms in (b) were calculated based on the parameter values estimated in (a). The results indicate that dominant detection noise sources were RIN and SN in our experiments reported in Fig. 3.
After these characterization steps, using Eqs. (7)–(12) we estimated the individual contributions of both the detection noise and the pixelation error on the accuracy of our centroid calculations for the depth localization experiments reported in Fig. 3 (see Tables A1–A2). In our experiments, since a total distance of ~500–1000 µm has been used between the micro-objects and the sensor plane, the detected holograms are spread out over at least 6–8 pixels, which greatly suppressed the pixelation error. Therefore, the relative weight of the pixelation error on centroid calculation accuracy is much smaller than the detection noise contribution as also quantified in Tables A1 and A2. The overall lateral localization errors reported in Table A1 (Sxc,all and Syc,all) are purely based on the vertical lensfree illumination measurements, while the height localization errors reported in Table A2 are calculated from all the oblique illumination angles together with the vertical one (see Fig. 3(a–c)).
Table A1.
Theoretical breakdown of the lateral localizationa
| Estimated Lateral Localization Error (µm) |
||||||
|---|---|---|---|---|---|---|
| Beads Diameter (um) |
Det. Noise |
Pixelation |
Overall |
|||
| Sxc | Syc | Sxc,px | Syc,px | Sxc,all | Syc,all | |
| 5 | 0.30 | 0.31 | 0.13 | 0.11 | 0.32 | 0.33 |
| 10 | 0.09 | 0.10 | 0.04 | 0.02 | 0.10 | 0.10 |
| 20 | 0.04 | 0.04 | 0.00 | 0.00 | 0.04 | 0.04 |
Standard deviation value is evaluated for each micro-particle type using the lensfree holograms of Figs. 3 (a), (b), and (c)
Table A2.
Theoretical breakdown of the height localization errors and comparison to measurement resultsa
| Beads Diameter (um) |
Height Deviation (µm) |
|||||
|---|---|---|---|---|---|---|
| Detection Noise (SN + RIN) |
Pixelation Error |
Beads Size STD |
Theoretical STDb |
Measured STD |
Systematic STD |
|
| 5 | 0.66 |
0.05 |
0.03 |
0.67 | 0.97 | 0.71 |
| 10 | 0.20 |
0.01 |
0.05 |
0.21 | 0.37 | 0.31 |
| 20 | 0.08 | 0.00 | 0.14 | 0.16 | 0.32 | 0.28 |
Standard deviation values of both the theoretical and experimental results are evaluated with the lensfree holograms of Figs. 3 (a), (b), and (c).
The theoretical standard deviation (STD) in height of each micro-particle type is evaluated by summing the height deviation contributed by detection noise, pixelation error, and bead size deviation.
Table A2 indicates that the level of our experimental characterization accuracy is quite close to the theoretical limit that is calculated based on the measured noise characteristics of the set-up. This fairly close comparison between our characterization results and the theoretical values supports the validity of our error analysis as well as the depth localization algorithm. Furthermore, the statement that the decreased depth localization accuracy with smaller micro-objects (as observed in Fig. 3) is due to the decrease of the detection signal-to-noise ratio is also validated with this comparison reported in Table A1.
Footnotes
OCIS codes: (090.1995) Digital holography; (100.6890) Three-dimensional image processing; (170.1530) Cell analysis.
References and links
- 1.Whitesides GM. The origins and the future of microfluidics. Nature. 2006;442(7101):368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 2.Squires TM, Quake SR. Microfluidics: Fluid physics at the nanoliter scale. Rev. Mod. Phys. 2005;77(3):977–1026. [Google Scholar]
- 3.Meldrum DR, Holl MR. Tech.Sight. Microfluidics. Microscale bioanalytical systems. Science. 2002;297(5584):1197–1198. doi: 10.1126/science.297.5584.1197. [DOI] [PubMed] [Google Scholar]
- 4.El-Ali J, Sorger PK, Jensen KF. Cells on chips. Nature. 2006;442(7101):403–411. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- 5.Psaltis D, Quake SR, Yang C. Developing optofluidic technology through the fusion of microfluidics and optics. Nature. 2006;442(7101):381–386. doi: 10.1038/nature05060. [DOI] [PubMed] [Google Scholar]
- 6.Yager P, Edwards T, Fu E, Helton K, Nelson K, Tam MR, Weigl BH. Microfluidic diagnostic technologies for global public health. Nature. 2006;442(7101):412–418. doi: 10.1038/nature05064. [DOI] [PubMed] [Google Scholar]
- 7.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450(7173):1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goodman JW. Introduction to Fourier Optics. Roberts & Company Publishers; 2005. [Google Scholar]
- 9.Brady DJ. Optical Imaging and Spectroscopy. Wiley; 2009. [Google Scholar]
- 10.Haddad WS, Cullen D, Solem JC, Longworth JW, McPherson A, Boyer K, Rhodes CK. Fourier-transform holographic microscope. Appl. Opt. 1992;31(24):4973–4978. doi: 10.1364/AO.31.004973. [DOI] [PubMed] [Google Scholar]
- 11.Xu W, Jericho MH, Meinertzhagen IA, Kreuzer HJ. Digital in-line holography for biological applications. Proc. Natl. Acad. Sci. U.S.A. 2001;98(20):11301–11305. doi: 10.1073/pnas.191361398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pedrini G, Tiziani HJ. Short-coherence digital microscopy by use of a lensless holographic imaging system. Appl. Opt. 2002;41(22):4489–4496. doi: 10.1364/ao.41.004489. [DOI] [PubMed] [Google Scholar]
- 13.Repetto L, Piano E, Pontiggia C. Lensless digital holographic microscope with light-emitting diode illumination. Opt. Lett. 2004;29(10):1132–1134. doi: 10.1364/ol.29.001132. [DOI] [PubMed] [Google Scholar]
- 14.Mann C, Yu L, Lo CM, Kim M. High-resolution quantitative phase-contrast microscopy by digital holography. Opt. Express. 2005;13(22):8693–8698. doi: 10.1364/opex.13.008693. [DOI] [PubMed] [Google Scholar]
- 15.Javidi B, Moon I, Yeom SK, Carapezza E. Three-dimensional imaging and recognition of microorganism using single-exposure on-line (SEOL) digital holography. Opt. Express. 2005;13(12):4492–4506. doi: 10.1364/opex.13.004492. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Sucerquia J, Xu W, Jericho MH, Kreuzer HJ. Immersion digital in-line holographic microscopy. Opt. Lett. 2006;31(9):1211–1213. doi: 10.1364/ol.31.001211. [DOI] [PubMed] [Google Scholar]
- 17.Ferraro P, Alferi D, De Nicola S, De Petrocellis L, Finizio A, Pierattini G. Quantitative phase-contrast microscopy by a lateral shear approach to digital holographic image reconstruction. Opt. Lett. 2006;31(10):1405–1407. doi: 10.1364/ol.31.001405. [DOI] [PubMed] [Google Scholar]
- 18.Park Y, Popescu G, Badizadegan K, Dasari RR, Feld MS. Diffraction phase and fluorescence microscopy. Opt. Express. 2006;14(18):8263–8268. doi: 10.1364/oe.14.008263. [DOI] [PubMed] [Google Scholar]
- 19.DaneshPanah M, Javidi B. Tracking biological microorganisms in sequence of 3D holographic microscopy images. Opt. Express. 2007;15(17):10761–10766. doi: 10.1364/oe.15.010761. [DOI] [PubMed] [Google Scholar]
- 20.Choi W, Fang-Yen C, Badizadegan K, Oh S, Lue N, Dasari RR, Feld MS. Tomographic phase microscopy. Nat. Methods. 2007;4(9):717–719. doi: 10.1038/nmeth1078. [DOI] [PubMed] [Google Scholar]
- 21.Mir M, Wang Z, Tangella K, Popescu G. Diffraction Phase Cytometry: Blood on a CD-Rom. Opt. Express. 2009;17(4):2579–2585. doi: 10.1364/oe.17.002579. [DOI] [PubMed] [Google Scholar]
- 22.Popescu G. Quantitative phase imaging of nanoscale cell structure and dynamics. In: Jena B, editor. Methods in Cell Biology. Elsevier; 2008. [DOI] [PubMed] [Google Scholar]
- 23.Rosen J, Brooker G. Non-scanning motionless fluorescence three-dimensional holographic microscopy. Nat. Photonics. 2008;2(3):190–195. [Google Scholar]
- 24.Seo S, Su TW, Tseng DK, Erlinger A, Ozcan A. Lensfree holographic imaging for on-chip cytometry and diagnostics. Lab Chip. 2009;9(6):777–787. doi: 10.1039/b813943a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oh C, Isikman SO, Khademhosseinieh B, Ozcan A. On-chip differential interference contrast microscopy using lensless digital holography. Opt. Express. 2010;18(5 Issue 5):4717–4726. doi: 10.1364/OE.18.004717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Isikman SO, Sencan I, Mudanyali O, Bishara W, Oztoprak C, Ozcan A. Color and monochrome lensless on-chip imaging of Caenorhabditis elegans over a wide field-of-view. Lab Chip. 2010;10(9):1109–1112. doi: 10.1039/c001200a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clack NG, Salaita K, Groves JT. Electrostatic readout of DNA microarrays with charged microspheres. Nat. Biotechnol. 2008;26(7):825–830. doi: 10.1038/nbt1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheng J, Malkiel E, Katz J. Digital holographic microscope for measuring three-dimensional particle distributions and motions. Appl. Opt. 2006;45(16):3893–3901. doi: 10.1364/ao.45.003893. [DOI] [PubMed] [Google Scholar]
- 29.Lee S, Roichman Y, Yi G, Kim S, Yang S, van Blaaderen A, van Oostrum P, Grier DG. Characterizing and tracking single colloidal particles with video holographic microscopy. Opt. Express. 2007;15(26):18275–18282. doi: 10.1364/oe.15.018275. [DOI] [PubMed] [Google Scholar]
- 30.Soulez F, Denis L, Fournier C, Thiebaut E, Goepfert C. Inverse-problem approach for particle digital holography: accurate location based on local optimization. J. Opt. Soc. Am. A. 2007;24(4):1164–1171. doi: 10.1364/josaa.24.001164. [DOI] [PubMed] [Google Scholar]
- 31.Situ G, Sheridan JT. Holography: an interpretation from the phase-space point of view. Opt. Lett. 2007;32(24):3492–3494. doi: 10.1364/ol.32.003492. [DOI] [PubMed] [Google Scholar]
- 32.Fienup JR. Reconstruction of an object from the modulus of its Fourier transform. Opt. Lett. 1978;3(1):27–29. doi: 10.1364/ol.3.000027. [DOI] [PubMed] [Google Scholar]
- 33.Canham PB, Burton AC. Distribution of size and shape in populations of normal human red cells. Circ. Res. 1968;22(3):405–422. doi: 10.1161/01.res.22.3.405. [DOI] [PubMed] [Google Scholar]
- 34.Shvalov AN, Soini JT, Chernyshev AV, Tarasov PA, Soini E, Maltsev VP. Light-scattering properties of individual erythrocytes. Appl. Opt. 1999;38(1):230–235. doi: 10.1364/ao.38.000230. [DOI] [PubMed] [Google Scholar]
- 35.Ares J, Arines J. Influence of thresholding on centroid statistics: full analytical description. Appl. Opt. 2004;43(31):5796–5805. doi: 10.1364/ao.43.005796. [DOI] [PubMed] [Google Scholar]
- 36.Morgan JS, Slater DC, Timothy JG, Jenkins EB. Centroid position measurements and subpixel sensitivity variations with the MAMA detector. Appl. Opt. 1989;28(6):1178–1192. doi: 10.1364/AO.28.001178. [DOI] [PubMed] [Google Scholar]
- 37.Alexander BF, Ng KC. Elimination of systematic error in subpixel accuracy centroid estimation. Opt. Eng. 1991;30(9):1320–1331. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.