Abstract
Protein kinase C (PKC) isoforms comprise a family of lipid-activated enzymes that have been implicated in a wide range of cellular functions. PKCs are modular enzymes comprised of a regulatory domain (that contains the membrane-targeting motifs that respond to lipid cofactors, and in the case of some PKCs calcium) and a relatively conserved catalytic domain that binds ATP and substrates. These enzymes are coexpressed and respond to similar stimulatory agonists in many cell types. However, there is growing evidence that individual PKC isoforms subserve unique (and in some cases opposing) functions in cells, at least in part as a result of isoform-specific subcellular compartmentalization patterns, protein-protein interactions, and posttranslational modifications that influence catalytic function. This review focuses on the structural basis for differences in lipid cofactor responsiveness for individual PKC isoforms, the regulatory phosphorylations that control the normal maturation, activation, signaling function, and downregulation of these enzymes, and the intra-/intermolecular interactions that control PKC isoform activation and subcellular targeting in cells. A detailed understanding of the unique molecular features that underlie isoform-specific posttranslational modification patterns, protein-protein interactions, and subcellular targeting (i.e., that impart functional specificity) should provide the basis for the design of novel PKC isoform-specific activator or inhibitor compounds that can achieve therapeutically useful changes in PKC signaling in cells.
I. INTRODUCTION
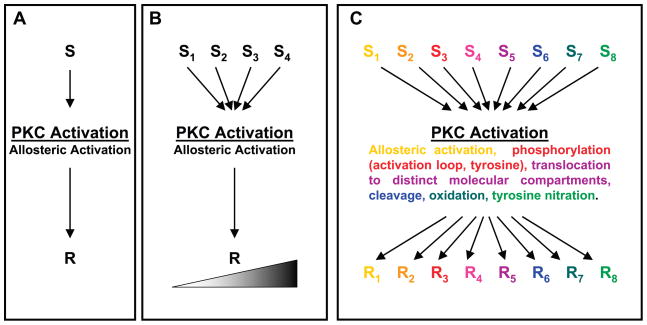
Protein kinase C (PKC) comprises a multigene family of related serine/threonine kinases that sit at the crossroads of many signal transduction pathways and are implicated in a wide range of G protein-coupled receptor and other growth factor-dependent cellular responses (41, 176). PKCs have traditionally been viewed as lipid-sensitive enzymes that are activated by growth factor receptors that stimulate phospholipase C (PLC), the enzyme that hydrolyzes phosphatidylinositol 4,5-bisphosphate (PIP2) to generate membrane-bound diacylglycerol (DAG) which activates PKC and inositol trisphosphate (IP3), which mobilizes intracellular calcium. Many PKCs are also pharmacologically activated by tumor-promoting phorbol esters such as phorbol 12-myristate 13-acetate (PMA) that anchor PKCs in their active conformations to membranes. According to the classical model of PKC activation, cellular PKC responses result from the ensemble actions of individual PKC isoforms (which traditionally are viewed as having only relatively limited in vitro substrate specificity) that are coexpressed in a particular cell type and localized to their distinctive subcellular compartments (in close proximity to their specific membrane substrate). However, the notion that PKCs act as generic kinases and achieve specificity only through translocation events has been challenged by recent studies showing that 1) PKCs can also be controlled through phosphorylations on both serine/threonine and tyrosine residues that influence the stability, protease/phosphatase resistance, protein-protein interactions, subcellular targeting, and activity (including substrate specificity) of the enzyme; 2) PKCs can be cleaved by caspases, generating a catalytically active kinase domain (in some cases, with altered enzymology) and a freed regulatory domain fragment that can act both as an inhibitor of the full-length enzyme and as an activator of certain signaling responses (i.e., under certain conditions, some PKC family members exert kinase-independent functions); and 3) PKCs can be activated by less traditional lipid cofactors (such as ceramide or arachidonic acid) or through lipid-independent mechanisms (such as oxidative modifications or tyrosine nitration) that allow for PKC signaling throughout the cell, not just at DAG-containing membranes. Since these concepts have only recently emerged in the mainstream literature, this review will focus on recent progress in understanding the structural determinants that dictate PKC isoform-specific differences in enzymology, subcellular targeting, protein-protein interactions, and downregulation. General reviews that examine the physiological consequences of PKC isoform activation in individual tissues have appeared elsewhere (21, 71, 119, 137, 139, 201, 230); these topics are beyond the scope of this review and will not be covered.
A. PKC Isoform Structure
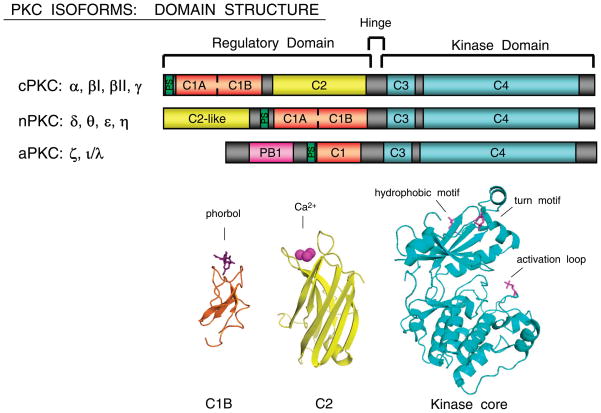
PKC isoforms are members of the AGC (PKA, PKG, PKC) family of protein kinases that share certain basic structural features (Fig. 1). These kinases contain a highly conserved catalytic domain (consisting of motifs required for ATP/substrate-binding and catalysis) and a regulatory domain that maintains the enzyme in an inactive conformation. PKC regulatory domains reside in the NH2 terminus of the protein and contain an autoinhibitory pseudosubstrate domain (a sequence that contains an alanine in place of the serine/threonine phosphoacceptor site, but otherwise resembles a PKC substrate) and two discrete membrane targeting modules, termed C1 and C2. PKC isoforms are broadly subdivided into three subfamilies based on differences in their NH2-terminal regulatory domain structure. The regulatory domains of conventional PKC isoforms (cPKCs; α, βI, and the alternatively spliced βII which contains an additional 43 residues at the NH2 terminus, and γ) contain a C1 domain (consisting of tandem ~50 residue long sequences, termed C1A and C1B, each with 6 cysteines and 2 histidines that coordinate two Zn2+) that functions as a DAG-/PMA-binding motif. cPKC regulatory domains also contain a C2 domain that binds anionic phospholipids in a calcium-dependent manner (Figs. 1–3). Novel PKCs (nPKCs, which can be further subdivided based on structural features into the related δ/θ and ε/η isoforms) also have twin C1 domains and a C2 domain (although the ordering of nPKC isoform C1 and C2 domains, along the linear sequence of the protein, is switched relative to the order in cPKCs; Fig. 1). Importantly, nPKC C2 domains lack the critical calcium-coordinating acidic residues (i.e., the determinants for calcium binding). This difference in C2 domain structure in large part underlies the distinct pharmacology of cPKC and nPKC isoforms. nPKCs are maximally activated by agonists that promote DAG accumulation or by PMA, without a calcium requirement. Atypical PKCs (aPKCs; ζ and ι/λ) lack a calcium-sensitive C2 domain; they contain an atypical C1 domain (with only one cysteine-rich membrane-targeting structure) that binds PIP3 or ceramide (not DAG or PMA) and a protein-protein interaction PB1 (Phox and Bem 1) domain that mediates interactions with other PB1-containing scaffolding proteins [including p62, partitioning defective-6 (PAR-6), and MEK5; Refs. 138, 139]. aPKC activity is regulated primarily by protein-protein interactions and phosphorylation by phosphoinositide-dependent kinase-1 (PDK-1, another AGC kinase family member that contains a PH domain that localizes the enzyme to PIP3-enriched membranes). While some PKC isoforms are expressed in a tissue-specific manner (i.e., PKCθ is expressed primarily by skeletal muscle, lymphoid organs, and hematopoietic cell lines and PKCγ is detected largely in neuronal tissues), most PKC isoforms are ubiquitous and many cells coexpress multiple PKC family members.
FIG. 1.
Domain structure of protein kinase C (PKC) isoforms. Top: PKCs have a conserved kinase domain (depicted in teal) and more variable regulatory domains. All PKC regulatory domains have a pseudosubstrate motif (shown in green) NH2 terminal to the C1 domain (shown in pink). Tandem C1 domains are the molecular sensors of phorbol 12-myristate 13-acetate (PMA)/diacylglycerol (DAG) in cPKC and nPKC isoforms, whereas the single aPKC C1 domain does not bind DAG/PMA. The C2 domains (in yellow) function as calcium-dependent phospholipid binding modules in cPKCs. nPKC C2 domains do not bind calcium; the PKCδ-C2-like domain is a phosphotyrosine interaction module. PKC isoform variable regions are shown in gray. Bottom: ribbon diagrams of PKC C1B domain, C2 domain, and kinase domain structures. Kinase domain phosphorylation sites are discussed in the text. (Figure courtesy of Dr. Alexandra Newton.)
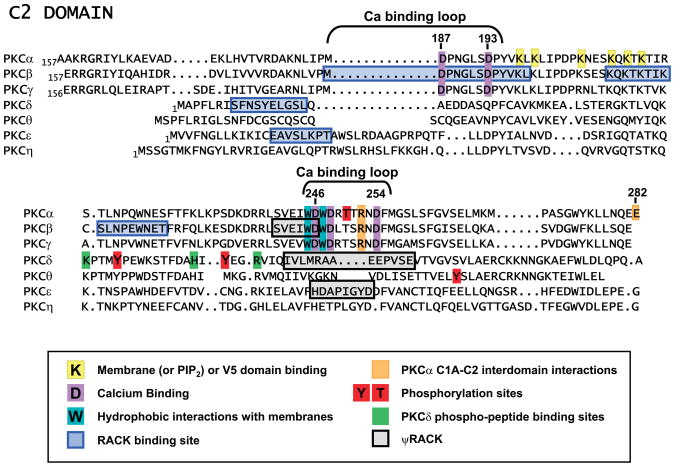
FIG. 3.
Alignment of PKC C2 domains.
B. PKC Activation and Localization Mechanisms in Cells
The traditional model of PKC activation derives from early studies of PKCα which localizes to the cytosol of resting cells; PKCα interacts only weakly/transiently with membranes in the absence of calcium or DAG. Agonists that promote phosphoinositide hydrolysis and IP3 generation lead to the mobilization of intracellular calcium, a soluble ligand that binds to the C2 domain and increases its affinity for membranes. This initial electrostatic interaction of the PKCα-C2 domain with membranes is relatively low affinity. However, once anchored to membranes, PKCα diffuses within the plane of the lipid bilayer and participates in a secondary C1A domain interaction with DAG (the membrane-restricted product of phosphoinositide hydrolysis). Membrane phosphatidylserine (PS) plays a critical role in this secondary membrane interaction, since PS disrupts a electrostatic C1A/C2 interdomain interaction, freeing the C1A domain so that it can penetrate the lipid bilayer and bind DAG (198). C1A binding to membranes also is relatively low affinity. However, the combined energy from the coordinate C1/C2 domain engagement with membranes leads to high-affinity cPKC binding to membranes and a conformational change that expels the autoinhibitory pseudosubstrate domain from the substrate-binding pocket and facilitates PKC activation.
PKC translocation to the plasma membrane generally has been considered the hallmark of activation (and frequently has been used as a surrogate measure of PKC isoform activation in cells). However, this simple model of PKC activation is not sufficient to explain the complex spatiotemporal controls of PKC localization in cells. For example, cPKCs (PKCα, PKCβI, and PKCβII) rapidly/transiently translocate to the plasma membrane via a mechanism that involves PLC-derived DAG accumulation. However, in cells that display a biphasic DAG response, PKCα and PKCβII (but not PKCβI) are released from the plasma membrane via a regulated process that requires PKC catalytic activity (and variably has been attributed to PKC autophosphorylation; Refs. 55, 56). PKCα/PKCβII then accumulate at a perinuclear site dubbed the “pericentron” (a subset of recycling endosomes containing the small GTPase Rab11; Refs. 10, 80) as a result of sustained DAG formation through a PLC-independent mechanism involving phospholipase D (PLD); PLD is a membrane-bound enzyme that generates phosphatidic acid (PA; through the hydrolysis of phosphatidylcholine), which is subsequently converted to DAG by PA phosphohydrolase (77). A different lipid, namely ceramide, acts as an inhibitory regulator of PKCα/PKCβII signaling at perinuclear membranes by inhibiting pericentron formation and stimulating a ceramide-activated protein phosphatase (tentatively identified as PP1) that reverses PKCα/PKCβII activation loop phosphorylation (a modification described in greater detail in sect. IVA; Refs. 11, 97). PKCα/PKCβII localized to the “pericentron” play an important role to “set” hormone responses by controlling the trafficking of continuously recycling membrane signaling proteins such as caveolin-1 and certain cell surface receptors. This chronic PKC signaling response has been implicated in the pathogenesis of clinical disorders such as oncogenic transformation by Ras, hyperglycemia, and Gq-triggered cardiac hypertrophy (220, 223).
PKC isoforms also translocate to specialized membrane compartments such as lipid rafts or caveolae (154). Lipid rafts are sphingolipid-/cholesterol-enriched plasma membrane microdomains that contribute to signal transduction by coalescing into large platforms that concentrate signaling complexes. Caveolae are the sphingolipid-/cholesterol-enriched detergent-resistant membranes that form flasklike invaginations of the plasma membrane in cells expressing caveolin. While the precise relationship between lipid rafts and caveolae remains the focus of considerable controversy (and is beyond the scope of this review), ceramide has recently emerged as an important PKC-regulated lipid in raft/caveolae. Ceramide (formed in lipid rafts) provides the driving force for raft fusion into platforms. PKC regulates ceramide formation. Acid sphingomyelinase (ASM), an enzyme that catalyzes the hydrolysis of sphingomyelin to form ceramide at the plasma membrane, is a PKCδ-activated enzyme (235). PKCδ binds ASM and promotes ASM-S508 phosphorylation, leading to ASM recruitment to the plasma membrane and ASM activation. This effect of PKCδ to promote local ceramide accumulation at the plasma membrane drives raft fusion (providing a nonspecific mechanism to localize signaling proteins such as PKCs to membrane rafts). Local ceramide formation also leads to the recruitment and activation (i.e., activation loop phosphorylation) of PKCζ in this compartment (140). 14-3-3 proteins are a family of scaffolding proteins that control cell growth/apoptosis pathways and are general inhibitors of PKCs that have been implicated in ceramide-dependent PKCζ activation; ceramide activates PKCζ at least in part by dissociating PKCζ-14-3-3 complexes. The 14-3-3 proteins interact with their binding partners through specific RSXpSXP or RXXXpSXP recognition domains (where pS represents phosphorylated Ser and X represents any amino acid; Ref. 143). While the molecular determinants for PKCζ-14-3-3 interactions have not been identified, two putative 14-3-3-binding motifs have been mapped to the NH2- and COOH-terminal sequences (101HKFRLHSYSSPT112 and 141RCVRSVPSLCG151, Fig. 2) that form the base of the PKCγ C1B domain, opposite from the lipid-binding surface. However, a serine phosphorylation that would control this 14-3-3 protein interaction has never been identified. Peptides based on these sequences compete with PKCγ for 14-3-3ε binding in vitro and promote PKCγ translocation to membranes in vivo (presumably by releasing PKCγ from the inhibitory constraints imposed by 14-3-3ε binding). These peptides can be used to activate PKCγ in lens epithelial cells, leading to increased PKCγ-dependent phosphorylation of the gap junction protein connexin 43 (Cx43 at S368) and disassembly of gap junction, suggesting that 14-3-3 proteins that control PKC isoform activation might be targeted for therapeutic advantage.
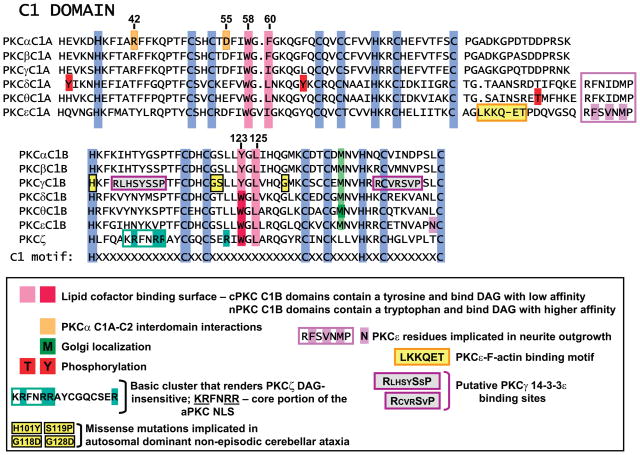
FIG. 2.
Alignment of PKC C1 domains. Numbering is based on PKCα. Conserved cysteine and histidine residues are in blue. Other structural determinants of C1 domain function are as indicated on the figure.
cPKCs and nPKCs are recovered at rest or following agonist activation in the caveolae fraction of several cell types (134, 175). Some caveolin isoforms (caveolin-1 and the muscle-specific caveolin-3, but not caveolin-2) physically interact with certain PKC isoforms (PKCα, PKCγ, and PKCζ, but not PKCε; Refs. 116, 148). Caveolin-1 binding serves to decrease PKCα and PKCζ catalytic activity. While some have speculated that this is attributable to a direct interaction between the caveolin scaffolding domain (a short membrane proximal region of the caveolin NH2 terminus that serves as a protein docking motif) and a caveolin interacting motif (ψXψXXXXψ or ψXXXXψXXψ, where ψ is a hydrophobic residue) found at similar locations in the kinase domains of PKCα and PKCζ, and that caveolin-bound PKC represents a pool of membrane-associated enzyme poised for activation, this model has never been directly verified (148) (and other mechanisms also are likely to anchor/regulate PKCs at this destination). We previously demonstrated that caveolae isolated from resting neonatal rat cardiomyocytes contain little to no phorbol ester-sensitive PKC isoforms and that PKCα, PKCδ, and PKCε accumulate in the caveolae fraction in cardiomyocytes treated with PMA (175). Our recent studies provide surprising evidence that α1-adrenergic receptor (α1-AR) activation does not lead to PKCδ/PKCε translocation to the caveolae fraction [(171) although α1-ARs induce a robust increase in PLC activity leading to DAG accumulation and PKCδ/PKCε translocation to a light sarcolemmal membrane fraction in this preparation; unpublished data]. These results suggest that 1) α1-ARs do not promote DAG accumulation in caveolae membranes, either because the density of α1-ARs or their downstream signaling partners is limiting in cardiomyocytes caveolae; or 2) α1-ARs increase DAG, but a locally active diacyglycerol kinase (DGK) converts DAG to PA, locally depleting DAG. In fact, norepinephrine has recently been reported to activate DGK-θ in the caveolae/raft fraction of rat mesenteric small arteries (32), although the relevance of this finding to PKC regulation in cardiomyocytes, where a different DGK isoform, namely, DGK-ζ, negatively regulates hypertrophic signaling responses, remains uncertain (5, 210). Irrespective of mechanism, these results emphasize that phorbol esters may be imperfect reagents to explore PKC localization to lipid raft membranes or the physiological controls of PKCs by lipid cofactors that are generated endogenously in growth factor-stimulated cells.
Certain PKCs also accumulate in the Golgi or in the nucleus. These other PKC localization mechanisms are discussed in greater detail in the sections that follow.
C. PKC Interactions With RACK Proteins
Models of PKC activation have generally focused on the intramolecular interaction between the pseudosubstrate domain and the catalytic pocket; PKC mutations that disrupt this intramolecular interaction (such as a pseudosubstrate domain deletion or an alanine→glutamate phosphomimetic substitution in the pseudosubstrate domain sequence) generate constitutively active forms of PKC that partition to membranes. However, research from the Mochly-Rosen laboratory has focused on a second intramolecular interaction that is based upon PKC interactions with receptors for activated C kinase (RACKs), a family of membrane-associated PKC anchoring proteins that act as molecular scaffolds to localize individual PKCs to distinct membrane microdomains in close proximity with their allosteric activators and unique intracellular substrates. These investigators have proposed that cells express a unique RACK (with a distinct subcellular localization) for each PKC isoform and that PKC-RACK interactions are essential for isoform-specific cellular responses. To date, proteins with characteristics of RACKs (i.e., proteins that selectively/saturably bind only the active conformation of the cognate PKC isoform and recruit the enzyme in an active conformation to a specific membrane compartment) have been identified for PKCβ (RACK1) and PKCε (RACK2 or β-COP; Table 1; Refs. 37, 128, 181). These RACK proteins share a seven-WD40-motif repeat structure, similar to the protein-protein binding motifs found in heterotrimeric G protein β-subunits. In the case of RACK1, the PKCβ binding site (SIKIWD) maps to the end of the sixth WD40 motif and is homologous to sequences in three other WD40 repeats in this protein (and distinct from the sequences found at similar regions of heterotrimeric G protein β-subunit structures; Ref. 168). The identity of the PKCδ RACK protein is less straightforward. Robles-Flores et al. (166) have identified p32/gC1qBP as a PKCδ binding partner that resides in the Triton-insoluble cell fraction and exhibits properties of a RACK. p32/gC1qBP associates only with allosterically activated PKCδ (and co-localizes with PKCδ to the nucleus) in PMA-treated cells (166). However, p32/gC1qBP constitutively interacts with PKCθ (i.e., it is a PKCθ binding partner, not a PKCθ-RACK). p32/gC1qBP and PKCθ translocate as a complex from the cytosol to the perinucelar region and the cell nucleus in response to PMA. Kheifets et al. (96) have recently identified an additional PKCδ localization mechanism involving annexin V, a cytoskeletal protein with a short stretch of sequence homology to PKCδ. These investigators used FRET technology to show that PMA promotes an interaction between PKCδ and annexin V in the cytosol and that this interaction precedes (and is required for) subsequent PKCδ translocation to membranes (96). These results have been taken as tentative evidence that annexin V serves as a shuttle protein on microtubules to transport PKCδ to its site of action (and its bone fide RACK) in the particulate fraction.
TABLE 1.
Structural basis for PKC isoform-RACK protein interactions
| PKC | Rack Protein | ψRack Sequence | Rack Binding Site |
|||
|---|---|---|---|---|---|---|
| C2 domain | V5 domain | |||||
| PKCβ | RACK1 SIKIWD |
241SVEIWD246 |
186MDPNGLSDPYVKL198 209KQKTKTIK216 218SLNPEWNET226 |
βI |
621ACGRNAE627 645QEVIRN650 660SFVNSEFLKPEVKS673 |
|
| PKCδ | Annexin V 157VVLIQANRDPDAG164 |
71IVLMRRAEDPMSE83 | 8SFNSYELGSL17 | βII | 645KLFIMN650 | |
| PKCε | p32/gC1qBP RACK2 or β′COP 285NNVALGYD292 |
85HDAPIGYD92 | 14EAVSLKPT21 | |||
Sequences in RACK1, annexin V, p32/gC1qBP, and RACK2 that bind their respective PKC isoforms, PKC-ψ RACK sequences (that are homologous to the sequences on RACK proteins that bind PKC), and RACK binding sites on PKCβ, PKCδ, and PKCε are provided. Charged residues that underlie RACK-ψ RACK interactions are in bold.
Mochly-Rosen and colleagues (180) have built on their model of PKC localization to membranes via isoform-specific interactions with RACK proteins to suggest that each PKC isoform contains both a RACK-binding sequence and a sequence that mimics the PKC binding site on the respective RACK protein (termed a ψ RACK sequence, Table 1) and that these sequences participate in an intramolecular interaction that maintains the enzyme in an inactive conformation at rest; this interaction must be disrupted for PKC activation. ψRACK sequences with close (albeit imperfect) homology to actual PKC binding sites on the respective RACK protein have been identified in PKCβ, PKCδ, and PKCε. In each case, a single charge change lowers the affinity of the intramolecular interaction, presumably allowing displacement of the ψRACK sequence and favor PKC binding to its bona fide membrane-anchored RACK protein upon PKC activation. Mutagenesis studies support this model, showing that a single 85H(D→N)APIGYD92 mutation in the PKCε ψRACK sequence (which generates a ψRACK sequence more closely mimicking the bona fide PKCε binding sequence in β′COP) renders PKCε resistant to proteolysis and agonist-dependent translocation to membranes (i.e., it increases the intramolecular interaction that maintains PKCε in a closed, inactive conformation), whereas a PKCε-85H(D→A)APIGYD92 mutation generates an enzyme that translocates to membrane more rapidly than WT-PKCε, suggesting that the D86A mutation weakens the intramolecular contact (180).
Peptides based on RACK binding sites have been developed as PKC isoform-selective translocation inhibitors that competitively inhibit PKC docking to its specific membrane anchoring RACK protein. Similarly, peptides based on ψRACK sequences have been used as allosteric PKC agonists; the notion being that the ψRACK sequence binds to the RACK-binding sites, interferes with the autoinhibitory intramolecular interaction between the RACK-binding site and the ψRACK sequence, and destabilizes the inactive “closed” conformation of the enzyme. The use of peptide modulators of PKC translocation offers an advantage over traditional overexpression strategies in that it alters PKC signaling without disturbing the natural stoichiometry of any given PKC isoform to its upstream activators or downstream substrates. However, it is important to note that RACK proteins may be key for some aspects of PKC localization and activation in cells, but there is ample evidence that PKCs localize in cells via RACK-independent interactions with cytoskeletal proteins (such as actin and tubulin) and true scaffolding proteins (such as caveolin and A-kinase anchoring proteins). Moreover, RACK proteins fulfill many cellular functions that are completely unrelated to PKC (i.e., as scaffolds that organize signaling complexes involving Src family kinases, G protein β γ subunits, dynamin, integrins, STAT1, the receptor protein tyrosine phosphatase PTPμ, and phosphodiesterase 4D5; Ref. 130). A complete understanding of the full biological consequences of treatments with peptides designed to modulate PKC translocation will require further studies.
Attempts to generate crystals of full-length PKC suitable for high-resolution three-dimensional structural analyses have not been successful. Nevertheless, high-resolution crystal structures of individual PKC regulatory motifs (including the C1 domain of PKCα, PKCγ, and PKCδ and the C2 domains of PKCα, PKCβ, PKCδ, and PKCε) as well as the catalytic domains of PKCβII, PKCθ, PKCι, and related AGC kinases (notably PKA and AKT) have been published (75, 100, 101, 133, 145, 151, 208, 215, 225, 226, 231, 237). Newer concepts regarding the molecular determinants of PKC isoform-specific functions based on these X-ray crystal structures as well as more traditional biochemical and mutagenesis approaches are reviewed in the sections that follow.
II. THE C1 DOMAIN
C1 domains are membrane-targeting modules that interact with tumor-promoting phorbol esters and lipid metabolites generated in response to growth factor stimulation (such as DAG, ceramide, and arachidonic acid). This section considers C1 domain-dependent mechanisms that localize full-length PKC to specific membrane compartments.
A. C1 Domain Structure
C1 domains were first identified as highly conserved DAG/PMA binding sites in PKCs with a characteristic HX12CX2CXnCX2CX4HX2CX7C motif, where H is histidine, C is cysteine, X is any other amino acid, and n is 13 or 14 (Fig. 2). While this motif duplicated in tandem is a characteristic feature of cPKCs and nPKCs, high-affinity PMA-binding C1 domains also are found in proteins that lack kinase domains such as the chimaerins (a family of Rac GTPase activating proteins), RasGRPs (Ras/Rap1 exchange factors), and Munc13 isoforms (scaffolding proteins involved in exocytosis). C1 domains that lack structural determinants for PMA binding also are found in aPKCs, Raf-1, DAG kinases, and Vav (93).
High-resolution crystal structures of PKCα, PKCγ, and PKCδ C1B domains (complexed with phorbol esters) have been published; these C1 domains adopt similar tertiary structures and function as hydrophobic switches to anchor PKCs to membranes (75, 225, 237). The upper third of the C1 domain forms a largely hydrophobic surface. Positively charged residues that interact with anionic phospholipids are exposed on the middle third of the C1 domain structure. The bottom third of the C1 domain contains the two Zn2+-coordinating sites (each formed by three cysteines and one histidine) that are required for proper C1 domain folding. C1 domain positively charged residues initially interact with electrostatically anionic membrane phospholipids. This initial interaction positions the C1 domain to penetrate the membrane bilayer and bind DAG, which is located more deeply within the membrane structure. Lipid cofactors such as DAG or PMA then bind to a narrow polar groove in the otherwise highly conserved hydrophobic surface at the top of the C1 domain (formed by the C1A-Trp58/Phe60 pair and the C1B-Tyr123/Leu125 pairs in PKCα-C1a and C1b domains, and cognate residues in other PKC isoforms, Fig. 2). By capping this hydrophilic ligand-binding pocket (i.e., generating a continuous hydrophobic surface), lipid cofactors act as “molecular glue” to increase C1 domain affinity for membranes (31, 78). Some C1 domains (such as the C1 domain of PKCα) appear to be buried in the resting state and exposed (becoming accessible to DAG/PMA) only following the conformational change that accompanies calcium-dependent membrane binding.
Early studies provided unambiguous evidence that a single C1 domain contains the structural determinants to support full-length PKC binding to DAG-/PMA-containing membranes (i.e., aPKCs are DAG/PMA-unresponsive due to an intrinsic difference in the properties of their C1 domains, and not because they have only one C1 domain). The structural features that distinguish DAG-/PMA-sensitive C1 domains in cPKCs and nPKCs from PMA-insensitive aPKC C1 domains have recently been identified. The NH2-terminal half of aPKC C1 domains contain a cluster of basic residues (HLFQAKRFNRRAYCGQCSERI) that are not found in PMA-sensitive C1 domains (Fig. 2). Modeling studies (based on the PKCδ-C1 domain structure) suggest that the adjacent Arg-Arg residues in the middle of this sequence are exposed on the protein surface. This portion of the PKCλC1 domain sequence functions as a nuclear localization sequence (NLS) when fused to GFP (155). Recent studies also indicate that this portion of the C1 domain also influences lipid responsiveness, since PKCζ can be converted to a PMA-sensitive enzyme simply by substituting the four arginine residues in this sequence to the corresponding (uncharged) residues from the PKCδ-C1b domain (160). Conversely, the PKCδ-C1b domain is rendered PMA-insensitive by arginine substitutions at these positions. The relative importance of the C1 domain basic cluster as a NLS (that drives full-length PKCλ to the nucleus) versus as a regulator of aPKC-lipid (both DAG and PIP3) interactions requires further study.
B. C1 Domain Lipid Binding Affinities
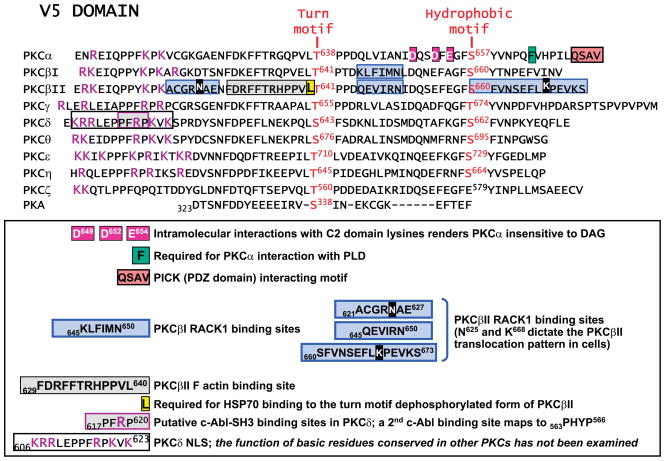
C1A and C1B domains of PKCγ and PKCε bind both DAG and phorbol esters with high affinity; both domains contribute to full-length PKCγ and PKCε binding to membranes (195). In contrast, other PKC-C1 domains display a wide range of affinities for lipid cofactors (ranging from 1 nM to >3 μM). In fact, the C1A and C1B domains of certain PKC isoforms have opposite intrinsic affinities for DAG and phorbol esters. For example, the C1A domains of PKCα and PKCδ have high affinity for DAG; mutagenesis studies implicate the C1A domain in DAG-dependent activation of full-length PKCα or PKCδ. In contrast, phorbol esters activate PKCα and PKCδ through a high-affinity interaction at C1B. While PKCδ and PKCε C1B domains are DAG-responsive, DAG binds with higher affinity to the PKCε C1A domain than to the PKCε C1B domain. PKCα and PKCβII C1B domain interactions with DAG are too weak to contribute to membrane binding under physiological conditions (47, 195, 196). However, while DAG activates full-length PKCα, PKCβII, PKCδ, and PKCε through a C1A domain-dependent mechanism, DAG interacts with high affinity to the PKCθ C1B (not C1A) domain (132). The structural basis for isoform-specific differences in C1B domain affinity for DAG was recently mapped to an invariant tryptophan residue that is conserved in the lipid-binding surface of nPKCs (that bind DAG-containing membranes with high affinity) and is replaced by a tyrosine in cPKC C1B domains (Y123 in PKCα). Dries et al. (47) recently used a cell imaging approach with YFP-tagged C1B constructs to show that a single Tyr-Trp substitution at this site converts the PKCβ-C1B domain into a DAG binding module. However, other studies identify mechanisms outside the C1 domain that regulate DAG binding. An intramolecular interaction between acidic V5 domain residues (D649, D652, and E654; Fig. 6) and C2 domain lysine-rich cluster stabilizes the closed DAG-insensitive conformation of PKCα. The DAG sensitivity of PKCα is enhanced by V5 domain truncation, overexpression of a V5 domain peptide (which is presumed to disrupt V5-dependent intramolecular interactions), V5 domain autophosphorylation site alanine substitutions, or mutations that reverse charge at the V5 domain acidic residue cluster (a maneuver that would disrupt this intramolecular interaction; Ref. 205).
FIG. 6.
Alignment of kinase domain V5 domain.
PKCs also interact with lipids such as ceramide and arachidonic acid. Ceramide is reported to drive PKCα from the cytoplasm to the membrane (79) and PKCδ/PKCε from the cytosol to the Golgi (91; although an effect of ceramide to release PKCδ and PKCε from membranes also has been reported, Ref. 178). Arachidonic acid mimics the effect of ceramide to translocate PKCε from the cytosol to the Golgi, whereas arachidonic acid does not alter the subcellular localization of PKCδ (and it inhibits PKCγ translocation to surface membranes, Ref. 144). Mutagenesis and domain swapping experiments indicate that ceramide activates PKCε and PKCδ via an interaction with the C1B domain, arachidonic acid activates PKCε via an interaction with the C1B domain (91), whereas arachidonic acid inhibits PKCγ via an interaction with the C1A domain (145).
Electrostatic interactions of individual PKC C1 domains with anionic phospholipids (i.e., the PS requirement for membrane binding and enzyme activity) also differ. The C1 domains of PKCα, PKCβ, and PKCδ show strong preference for PS over other lipid species, whereas the C1 domains of PKCγ and PKCε do not (64, 187, 195, 196, 198). Studies of PKCα indicate that PS plays an important role to disrupt intramolecular tethers (involving electrostatic Asp55-Arg252 and Arg42-Glu282 interactions) that maintain PKCα in a closed conformation in the resting state (Figs. 2 and 3). Conventional wisdom holds that PS releases these intramolecular tethers, allowing the C1A domain to partially penetrate the membrane, bind DAG, and trigger the conformational changes that lead to enzyme activation. In support of this model, single residue substitutions at any one of the four residues that form C1A-C2 tethering ion pairs (i.e., at Asp-55, Arg-252, Arg-42, or Glu-282) increase PKCα’s membrane affinity, increase its basal activity, and abrogate its PS requirement. Moreover, double charge reversals (Asp55Lys/Arg252Glu or Arg42Glu/Glu282Arg substitutions) largely restore WT membrane binding affinity (198). However, an inaccessible C1A domain is not a defining feature of all cPKC, since PKCγ C1A and C1B domains are reported to be conformationally flexible; they penetrate the membrane and bind DAG, without a PS requirement (3).
C. C1 Domain-Mediated Subcellular Targeting
The conventional model of PKC activation focuses on effects of DAG or PMA to anchor PKCs to the plasma membrane. However, it has become increasingly evident that PKCs have many destinations in cells. As noted in the previous section, the NH2-terminal half of aPKC C1 domains contain a basic cluster that acts as a NLS when fused to GFP (independent of any zinc finger structure). For PKCs that lack a C1 domain NLS, translocation patterns (to the plasma membrane, lipid rafts, nuclear membranes, Golgi, endoplasmic reticulum, and mitochondria) can vary substantially depending on the cell type and particular stimulus. Several laboratories have examined whether C1 domains (that inherently differ in their affinities for various lipid cofactors) play a specific role to drive full-length PKC to specific intracellular membranes with distinct lipid compositions. These studies implicate the tandem C1 domains in cPKC/nPKC isoforms in mechanisms that provide for a high level of spatiotemporal control of PKC localization. In general, the tandom C1 domains are not functionally redundant.
The Blumberg laboratory has demonstrated that PKCδ localizes to distinct subcellular compartments through C1 domain-mediated interactions that are influenced by the hydrophobicity (i.e., fatty acid side chain length) of the phorbol ester used in the experiment. They used phorbol ester derivatives with different fatty acid side chain lengths (that bind C1 domains with comparable affinities) to show that very hydrophobic phorbol ester derivatives drive PKCδ to the plasma membrane, derivatives with intermediate hydrophobicity drive PKCδ to the plasma membrane followed by redistribution of PKCδ to the nucleus, and more hydrophilic phorbol esters translocate PKCδ primarily to the nuclear membrane (hydrophilic phorbol esters do not drive PKCδ to the plasma membrane; Ref. 218). However, Cho and colleagues (196) performed similar studies and obtained a somewhat different result, showing that relatively hydrophilic short-chain DAGs (that distribute to both surface and internal cell membranes) selectively drive PKCδ to the PS-enriched plasma membrane (i.e., PS cooperates with DAG to localize PKCδ to surface membranes). The discrepant findings between these two laboratories are not readily reconciled and will require additional studies, including with newer technologies.
PKC targeting as a result of C1 domain interactions with proteins also has been reported. As noted in section IB, PKCγ localization to membranes is inhibited as a result of a C1B domain interaction with 14-3-3ε (143). Other studies show that the PKCα C1B domain interacts with fascin, an actin-bundling protein that plays a role in cell adhesion and spreading on fibronectin (4) and that the PKCβII C1A domain binds the centrosomal protein pericentrin (to control microtubule organization, spindle assembly, and chromosome segregation during cell division) (28). nPKCs (including in PKCδ, PKCε, PKCθ, and PKCη) also localize through their C1B domain to the Golgi complex where they play a role in specific cellular functions (i.e., PKCδ and the related PKCθ induce apoptosis, whereas PKCε modulates secretion from the Golgi complex; Refs. 37, 90, 183). nPKC C1A domains lack a Golgi localization signal and do not drive full-length PKCδ, PKCε, PKCθ, or PKCη to the Golgi complex (129, 182). While some studies have focused on ceramide as a lipid cofactor that localizes PKCδ and PKCε (through their C1B domains) to the Golgi (91; see sect. IB), Larsson and colleagues (183) have used sequence alignment, structural modeling, and mutagenesis studies to implicate a conserved C1B domain methionine (Met267 in the C1B domain of PKCθ) as a Golgi localization signal, showing that 1) this residue lies outside the C1 domain lipid binding surface; 2) a methionine or similar residue at this position is a highly conserved feature of C1 domains that localize to Golgi (including the C1B domains of PKCs, the C1A domain of PKD, and the C1 domain of β-chimaerin), but not C1 domains that accumulate at other cellular membranes; and 3) a single M267G substitution is sufficient to fully abrogate PKCθ localization to Golgi (and the resultant apoptosis) without interfering with PMA-dependent PKCθ translocation to the nucleus.
The C1B domain also has been implicated as a structural determinant of PKCγ trafficking and activity during oxidative stress in lens epithelial cells. Here, low H2O2 concentrations (100 μM) lead to oxidative modification of PKCγ, increased PKCγ activity, and PKCγ translocation to caveolae membranes, where PKCγ interacts with caveolin-1 and the junctional gap protein Cx43, increases Cx43-S368 phosphorylation, and inhibits gap junction activity (115). This mechanism results in decreased trans-cellular transmission of proapoptotic signals and is cytoprotective in lens epithelia. The importance of this PKCγ-dependent mechanism has been established in PKCγ knockout mice where oxidative stress leads to exaggerated lens opacification (113). Of note, missense mutations localized to key regions of the PKCγ C1B domain (H101Y, G118D, S119P, and G128D) disrupt C1B domain structure and are implicated in the neurological disorder autosomal dominant nonepisodic cerebellar ataxia (Fig. 2; Refs. 29, 214). Studies in a more reductionist cell culture model indicate that PKCγ harboring C1B domain mutations displays low basal activity and is not activated by H2O2; in fact, these mutant forms of PKCγ act in a dominant inhibitory manner to prevent WT-PKCγ activation by H2O2, leading to a loss of PKCγ-dependent gap junction inhibition and increased apoptosis in lens epithelial cells (114). Cx43 also has been implicated as a target (and binding partner) for PKC in cardiomyocytes (46). Here, Cx43 phosphorylation has been attributed to PKCε mapped to Cx43-S262. Cx43-S262 phosphorylation is implicated as a mechanisms that decreases gap junction permeability and intercellular communication (45).
D. C1A-C1B Interdomain Regions and PKC Targeting
Thuille et al. (212) recently identified NSRET219MF (which lies between the twin C1 domains) as a major autophosphorylation site in PKCθ. In contrast to the constitutive priming phosphorylations at the activation loop and hydrophobic motif that are retained in resting Jurkat and T cells (see sect. IVA), PKCθ is not phosphorylated at T219 in resting T cells. PKCθ-T219 phosphorylation is induced by phorbol 12,13-dibutyrate (PDBu), vanadate, and antigen receptor cross-linking. T219 phosphorylation is essential for PKCθ-dependent antigen receptor signaling responses; a PKCθ-T219A mutant exhibits WT in vitro catalytic activity and lipid binding activity, but PKCθ-T219A localizes aberrantly to the detergent-insoluble fraction of resting T cells, is not recruited to the T cell receptor (TCR) in lipid rafts, and fails to support NFκB activation. These results suggest that T219 phosphorylation is critical for proper PKCθ targeting to the TCR in lipid raft membranes. Of note, the T219 phosphorylation motif (NSRET219MF) in the C1A-C1B interdomain region of PKCθ is conserved in PKCδ (NSRDT218IF), but not in any other PKC isoform (although a potential regulatory role for PKCδ-T218 phosphorylation has never been considered).
PKCε does not share sequence homology with PKCδ/PKCθ at this interdomain position. Nevertheless, residues within the PKCε C1 interdomain region (LKKQET) bind F-actin and localize PKCε in an activated form to the cytoskeletal fraction (157, 158). Other studies have focused on a different RFX(V/I)XMP sequence that is immediately NH2-terminal to the C1B domain and is highly conserved in PKCδ, PKCε, and PKCθ (PKC isoforms that have the capacity to induce neurite outgrowth in neuroblastoma cells). This sequence is not conserved in cPKCs that do not induce neurite outgrowth (118, 158). Mutagenesis studies identify a critical role for the F237, V239, and M241 (numbering based upon PKCε); PKCε mutants harboring substitutions at these hydrophobic sites localize to the plasma membrane, but they do not induce neurite outgrowth. These results have fueled speculation that the region between the C1A and C1B domains represents an exposed surface on the active enzyme and functions as a protein-protein interaction “hot spot” that is critical for proper PKC targeting in cells. Indeed, recent studies also identify an Asn residue at the COOH terminus of the C1B domain that is unique to the PKCε and PKCη (i.e., is not found in other PKC isoforms) that is critical for neurite-inducing capacity; this Asn residue models to a position at the base of the C1 domain opposite from the lipid binding surface (117).
E. PKCδ-C1 Domain Tyrosine Phosphorylation
Two tyrosine residues in the PKCδ C1A domain have been identified as sites for functionally important phosphorylations. PKCδ-Y155 (which is flanked by the pseudosubstrate motif and the C1A domain and is unique to PKCδ) has been implicated in PKCδ-dependent growth responses. In certain heterologous overexpression systems, WT-PKCδ overexpression slows proliferation, whereas a single Y155F-substitution is sufficient to link PKCδ to an enhanced growth response (1, 104, 219). The mechanism(s) that control PKCδ-Y155 phosphorylation or the downstream signals linking the PKCδ-Y155F mutant to altered growth regulation have not been examined. PKCδ phosphorylation at Y187 (a site conserved in other nPKC isoforms) has been reported in some cells treated with PMA and platelet-derived growth factor (PDGF) (104, 111). PKCδ-Y187 phosphorylation does not influence PKCδ kinase activity (in vitro, using a pseudosubtrate domain peptide), and heterologously overexpressed PKCδ-Y187F mimics the effect of WT-PKCδ to inhibit growth in NIH3T3 and C6 glial cells and to induce the monocyte differentiation program in 32D myeloid progenitor cells (111, 219). However, PKCδ-Y187F does not mimic the effect of WT-PKCδ to promote differentiation in C6 glial cells (104). Again, the molecular mechanisms that account for altered signaling by the PKCδ-Y187F mutant have not been identified.
III. THE C2 DOMAIN
A. C2 Domain Structure and Lipid Binding
C2 domains were first described as ~130 residue sequences that function as calcium-dependent membrane-binding modules in the regulatory domain of cPKCs. C2 domains were subsequently identified in many proteins that participate in membrane trafficking and signal transduction. C2 domains share a common tertiary structure comprised of eight anti-parallel β-strands connected by variable loops. In general, C2 domains share more structural homology in the core β-sandwich portion of the domain (which play a more structural scaffolding role) than in the loop sequences, which are more divergent and dictate functional specificity. Detailed structural studies of PKCα suggest that two or three calcium ions bind in a highly cooperative manner to several highly conserved Asp residues in the calcium-binding loops (D187, D193, D246, D248, and D254 in PKCα) that connect the β-stands at the top of the domain structure (131). nPKC C2 domains lack these calcium-coordinating residues in their loop sequences and therefore bind membranes in a calcium-independent manner. Further structural studies of PKCα show that Asn189 plays an important role in the mechanisms that render PKCα selective for PS (over other anionic phospholipids), Arg249/Arg252 contribute to PKCα’s electrostatic interactions with the anionic membrane surface, Trp245/Trp247 are involved in hydrophobic interactions with membrane, and the highly basic lysine-rich β3- and β4-sheets localize active PKCα to PIP2-enriched membranes (34, 51, 54, 131, 197). While early studies from the Parker laboratory identified a Thr250 (a threonine residue that is strategically located in the calcium binding loop of PKCα and is highly conserved in other cPKCs) as an autophosphorylation site (142, 159), the importance of C2 domain threonine autophosphorylation as a mechanism to regulate PKCα interaction with membranes has not been revisited.
cPKC C2 domains play important roles as calcium-dependent membrane-targeting modules. However, the isolated PKCδ-C2 domain does not bind lipid (and the C2-domain deleted forms of full-length PKCδ shows no membrane targeting defect; Refs. 64, 196). Newton et al. (64) have speculated that the very high intrinsic affinity of the PKCδ-C1 domain for membranes (2 orders of magnitude higher than the C1 domain of cPKCs) may have evolved to compensate for the lipid-binding defect of the PKCδ C2 domain. Studies of PKCε C2 domain function are less consistent. The Corbalan-Garcia laboratory has presented evidence that the PKCε C2 domain acts as a lipid-binding domain to anchor PKCε to membranes containing DAG and phosphatidic acid (88, 145). In contrast, the Cho laboratory has reported that the PKCε C2 domain binds lipids only weakly (with an affinity that is more than 2 orders of magnitude lower than the affinity of the full-length protein) and that the C2 domain deleted forms of full-length PKCε show no membrane targeting defect (195).
B. PKC-C2 Domains as Protein-Protein Interaction Motifs
The Mochly-Rosen laboratory has used sequence homology analysis with synaptotagmin (a known RACK1 binding protein) to identify 186MDPNGLSDPYVKL198, 209KQKTKTIK216, and 218SLNPEWNET226 within the PKCβ-C2 domain (along with additional sites in the V5 domain, discussed further in sect. VIA) as RACK1-binding sites (167). These RACK1 binding sites lie on β-strands in the domain, with the 186MDPNGLSDPYVKL198 sequence on strand 3, adjacent and anti-parallel to a RACK1-mimetic sequence (dubbed ψβRACK, 241SVEIWD246) on strand 6. This spatial orientation would permit an intermolecular strand-strand interaction that stabilizes the β-sandwich structure (8). Peptides based on this PKCβ-C2 domain RACK1 binding sequences inhibit agonist-dependent PKCβII translocation to membranes when introduced into cardiomyocytes or Xenopus oocytes (167).
The C2 domain of PKCε (which is not required for membrane translocation) also contains sequence (14EAVSLKPT21) that binds to the εRACK β′COP, as well as sequence (85HDAPIGYD92) that is 75% homologous with sequence in β-COP (285NNVALGYD292), the εRACK that anchors PKCε at its intracellular site of action (19). These sequences are evolutionarily conserved in PKCε and are distinct from the cognate sequences in other PKC isoforms; as noted in section IC, these sequences are believed to participate in an intramolecular interaction that stabilizes PKCε in an inactive closed conformation that must be interrupted for PKCε activation (87, 180).
While the identity of the C2 domain sequence that acts as a ψRACKs (or RACK binding) site for PKCδ is less certain, the PKCδ C2 domain (which adopts a somewhat different conformation relative to other C2 domains) interacts with a number of PKCδ binding partners, including annexin V, actin, and GAP-43 (a PKC substrate involved in neurite outgrowth) (40, 123, 151). Very recent studies also implicate the PKCδ C2 domain as a novel phosphotyrosine binding motif that mediates the interaction between PKCδ and CDCP1 (a transmembrane protein that is tyrosine phosphorylated by Src and overexpressed in colon cancer) (12). Using a degenerated phosphotyrosine peptide library screen, Benes et al. (12) defined an optimal PKCδ C2 domain consensus binding sequence as (Y/F)-(S/A)-(V/I)-pY-(Q/R)-X-(Y/F) and showed that the PKCδ C2 domain binds to this sequence with an affinity that compares favorably with the affinities reported for SH2 and PTB domain binding (250–500 nM). However, unlike SH2 and PTB domains (which recognize phosphotyrosine in the context of either COOH- or NH2-terminal sequence), PKCδ’s C2 domain recognizes phosphotyrosines with aromatic residues at both the +3 and −3 positions (i.e., sequence specificity is defined by residues both NH2 and COOH terminal to the phosphotyrosine). These investigators have mapped the phosphopeptide binding sites in the PKCδ C2 domain to Lys48, His62, and Arg67 (which interact with the phosphate) as well as residues at the COOH and NH2 termini of the C2 domain (that interact with the peptide backbone). It is interesting to note that all of the residues that contribute to phosphopeptide binding are highly conserved in PKCθ (but not other PKC isoforms); a role for PKCθ C2 domain as a protein-protein interacting motif is likely, but has not yet been considered. Furthermore, tyrosine residues within or adjacent to the PKCδ C2 domain phosphopeptide binding surface (namely, Tyr52 and Tyr64) have been identified as targets for functionally important regulatory phosphorylations (89, 151, 191). Tyr52 (which is an in vitro substrate for Lyn, is phosphorylated via a Lyn-dependent mechanism in rat basophilic leukemia cells, and is conserved in PKCθ; Ref. 209) interacts with the +3 position Tyr in the phosphopeptide binding partner; PKCδ-Y52 phosphorylation is predicted to influence phosphopeptide binding. Similarly, Tyr64 (which is unique to PKCδ, and not found in PKCθ) is optimally positioned to interact with Arg67; a phosphotyrosine at this position in the C2 domain might compete with (and prevent) phosphopeptide binding to the C2 domain. This suggests a mechanism that might underlie previous observations that C2 domain tyrosine phosphorylation plays a critical role to influence PKCδ’s cellular actions and is likely to be a fruitful area for future research.
Finally, there is evidence that the C2 domain of PKCθ (which is structurally related to PKCδ) is tyrosine phosphorylated in antigen-stimulated T cells (122). Here, phosphorylation has been mapped to Y90, a tyrosine that is unique to the PKCθ calcium-insensitive C2 domain (and is not found in the related PKCδ C2 domain). Other C1 or C2 domain tyrosine residues (such as Y53 and Y188, which correspond to Y52 and Y187 in PKCδ) are not phosphorylated in antigen-stimulated T cells. PKCθ-Y90 phosphorylation has been attributed to Lck, which phosphorylates PKCθ in vitro and constitutively associates with the PKCθ regulatory domain in vivo; PKCθ is not tyrosine phosphorylated by other non-receptor tyrosine kinases such as Fyn, Zap-70, or Syk. A functional role for PKCθ-Y90 phosphorylation has begun to emerge in the literature. While Y90 phosphorylation is not required for PKCθ-Lck interactions and an initial mutagenesis study failed to link Y90 phosphorylation to changes in PKCθ targeting to membranes, a recent study implicated Y90 phosphorylation as a mechanism that enhances PKCθ affinity for lipid membranes (132), and there is evidence that a single Y90F mutation introduced into the constitutively active PKCθ-A148E mutant is sufficient to fully abrogate PKCθ biological functions (such as the induction of interleukin-2 promoter NFAT/AP-1 activity and cell proliferation, Ref. 122).
IV. THE KINASE DOMAIN
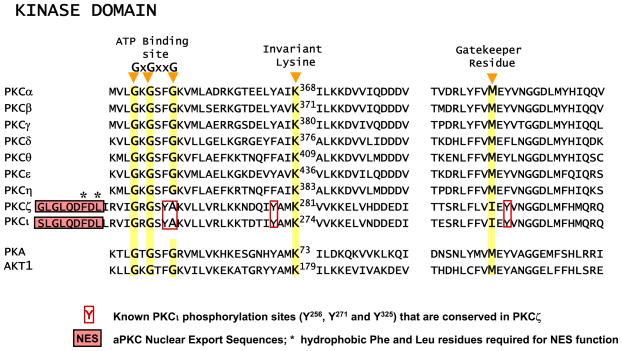
Early attempts to understand the structural determinants of PKC isoform function relied on X-ray crystallographic data derived from studies of closely related AGC family members such as PKA or AKT (100, 133, 226, 231), since crystal structures for PKC kinase domains were not available prior to 2004. With the publication of X-ray crystal structures for three bacterial-expressed, phosphorylated PKC catalytic domains (PKCβ II complexed with 2-methyl-1H-indol-3yl-BIM-1, PKCθ bound to staurosporine, and PKCι complexed with BIM-1, i.e, a PKC isoform representative of each subfamily; Refs. 72, 133, 226), it has been possible to validate many of the assumptions made in these early studies. PKC catalytic regions share high overall structural alignment with PKA, with most of the residues that are invariant across AGC kinase family members clustering at sites of nucleotide binding or catalysis. Like other AGC kinases, PKC catalytic domains contain a smaller NH2-terminal lobe that is comprised mainly of β-sheets and contains the characteristic glycine-rich ATP-binding loop with the consensus GXGXXG sequence (a structural hallmark of protein kinases and nucleotide binding proteins) and an invariant Lys which structures the enzyme for phosphoryl-transfer (and is generally mutated to generate kinase-inactive mutants, Fig. 4). The COOH-terminal lobe of the kinase domain is predominantly α-helical and contains the activation loop segment that positions magnesium and peptide substrates for catalysis followed by the V5 domain at the NH2 terminus (Figs. 5 and 6). A “gatekeeper” residue located in sequence connecting the two lobes of the kinase domain controls access to a preexisting cavity in the ATP binding pocket (Fig. 4). This residue is conserved as a large hydrophobic amino acid in the human kinome; a glycine substitution at this site generates an enzyme that uniquely binds unnatural ATP analogs with bulky substitutions (that do not bind any closely related endogenous enzymes). Gatekeeper residue mutations are the basis of the chemical genetic approaches that have been used to engineer kinases that are uniquely sensitive to certain unnatural inhibitor or activator ATP analogs and provide a powerful and elegant strategy to resolve the physiological substrates of individual kinases in cells (42). Moreover, there is growing evidence that the gatekeeper residue also may play a structural role to constrain the flexibility and autocatalytic activation of certain enzymes (53, 179). Finally, the activation loop and V5 domain contain highly conserved priming/regulatory phosphorylation sites that play a critical role to structure the catalytic pocket; these phosphorylation events are considered in this section, whereas the more structurally divergent V5 domain features (that impart certain aspects of PKC isoform specificity) are considered in greater detail in section VI.
FIG. 4.
Alignment of the ATP binding site, invariant lysine, and gatekeeper residues in PKC kinase domains. NES sequences NH2 terminal to aPKC ATP binding sites are depicted.
FIG. 5.
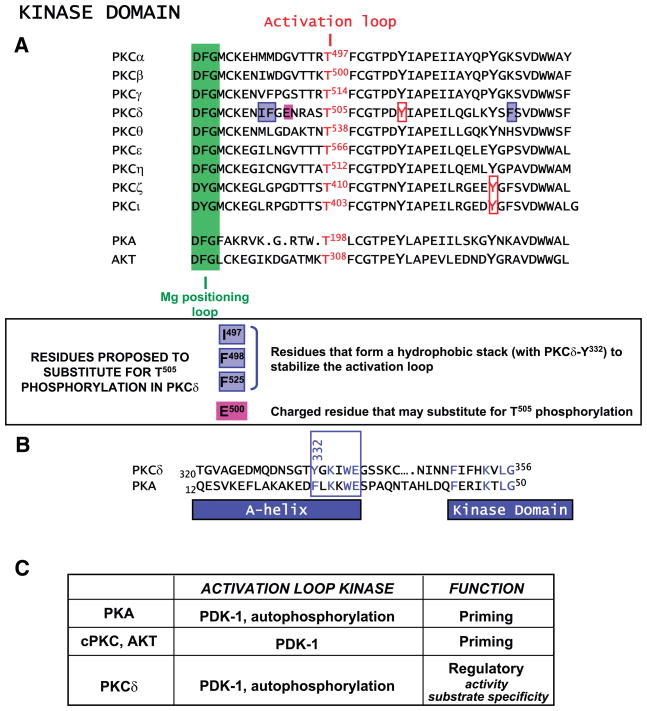
Alignment of kinase domain activation loops. A: Mg positioning loop (in green), activation loop phosphorylation site, invariant tyrosines that are phosphorylated in certain PKCs, and the unique structural features of PKCδ that recently have been considered a mechanism to render this isoform catalytically competent without activation loop phosphorylation are depicted. B: sequence alignment for the A-helix in PKCδ and protein kinase A (PKA) (see text). C: activation loop phosphorylation mechanisms and functions.
A. cPKC Priming Phosphorylations
Traditional models of PKC activation have focused on allosteric activation by calcium and DAG. The “priming” Ser/Thr phosphorylations that lock the enzyme in a closed, stabilized, catalytically-competent, phosphatase/protease-resistant conformation have only recently become the focus of research interest (141, 153). PKCs are first phosphorylated at a threonine residue in the “activation loop,” a highly conserved 20–30 residue sequence in the cleft of the kinase domain that is flexible (assumes a different orientation in the active and inactive enzyme) and forms part of the peptide substrate binding surface (Fig. 5). PKC activation loop phosphorylation introduces a negative charge that aligns residues in the catalytic pocket and stabilizes the active conformation of the enzyme. PKC activation loop phosphorylation is generally attributed to PDK-1. However, it is worth noting that the PKA activation loop site can be phosphorylated either by a heterologous kinase with properties resembling PDK-1 or through an autocatalytic mechanism (depending on the in vivo environment). In fact, the observation that PKA activation loop phosphorylation and enzyme activity are similar in PDK1+/+ and PDK1−/− ES cells suggests that PDK1 activity is not necessarily rate limiting for PKA activation loop phosphorylation (23, 136, 222). These results are pertinent to our recent studies of PKCδ, which undergoes activation loop autophosphorylation under certain circumstances (170; see sect. IVB2, Fig. 5C).
Newly synthesized PKCs are believed to adopt an open conformation that allows PDK-1 access to the exposed unphosphorylated hydrophobic motif sequence in the COOH-terminal V5 domain (48, 62, 192). The prevailing model (based primarily on detailed studies of related AGC kinases) holds that PDK-1 must dock to the hydrophobic motif to be activated and positioned for activation loop phosphorylation. However, it is worth noting that PKCε truncation mutants that lack the hydrophobic motif can interact with PDK-1, and they display activation loop phosphorylation (238); these results are surprising and suggest caution when extrapolating conclusions derived from studies of other AGC kinases to PKCs.
Once PDK-1 phosphorylates the activation loop, PDK-1 must be released from the COOH terminus for full maturation of the enzyme (since persistent PDK-1 binding to the COOH terminus would prevent hydrophobic motif autophosphorylation). The cPKCs and nPKCs then undergo two additional phosphorylations at conserved motifs in the COOH-terminal V-5 domain: at a (generally proline-flanked) conserved “turn motif” and at a FXXFS/TF/Y motif (a phosphorylation site bracketed by hydrophobic residues) 19 residues COOH terminal to the “turn motif” (Fig. 6). For cPKCs, these are autophosphorylation reactions that are stable modifications (i.e., completed during the maturation of the enzyme and typically retained during normal culture conditions); the mechanism for nPKCs is less straightforward (and is discussed in sect. IVB3). While hydrophobic motif phosphorylations are a feature of cPKCs and nPKCs, the Ser/Thr phosphorylation site is replaced by a phosphomimetic Glu in aPKCs, and PKA terminates at Phe (the residue immediately preceding the hydrophobic motif S/T phosphorylation site in cPKC/nPKC structures, Fig. 6). X-ray crystal structures of PKA, AKT, and PKCθ identify an interaction between this phenylalanine (preceding the hydrophobic motif phosphorylation site) and a hydrophobic pocket on the back side of the active site; this interaction stabilizes the catalytic core structure in a position that allows for high-affinity interactions with ATP and substrate (i.e., is favorable for catalysis), suggesting a mechanism whereby COOH-terminal phosphorylations regulate catalytic activity. However, COOH-terminal phosphorylations also appear to control enzyme activity by influencing the thermal stability, detergent solubility, and protease/phosphatase susceptibility of the enzyme. Additional long range effects of the hydrophobic motif to influence regulatory C2 domain interactions with calcium also have been reported (50, 226). Collectively, these three functions for the hydrophobic motif (to dock PDK-1 and thereby support activation loop phosphorylation, to structure the catalytic pocket, and to participate in long-range interactions with the C2 domain) are critical to generate a fully phosphorylated catalytically competent enzyme that is maintained in an autoinhibited conformation in the cytosol, poised to be activated by second messengers. Of note, once fully phosphorylated, cPKCs no longer require a negative charge at the activation loop for catalysis. PKCβII retains catalytic activity when selectively dephosphorylated at the activation loop. However, a turn motif phosphorylation is required for catalytic competence; PKCβII is rendered catalytically inactive when it is expressed as the turn motif, T641A, mutant (or when it is dephosphorylated selectively at the turn motif or generally at all three priming phosphorylation sites; Refs. 16, 49). Isolated reports showing that PKCα tolerates a “turn motif” mutation without disrupting catalytic activity (i.e., PKCα-T638A is catalytically active) do not necessarily undermine this conclusion; apparently compensatory phosphorylations in the vicinity of the T→A mutations can functionally substitute for the turn motif phosphorylation (emphasizing that some single residue mutations lead to uninformative phenotypes; Ref. 49).
Apart from its role to maintain the enzyme in a conformation that is favorable for catalysis, COOH-terminal phosphorylations have been implicated as mechanisms that control PKC binding to proteins or membranes. PKC isoforms (and other AGC kinases such as AKT and PKA) interact via their COOH terminus with both PDK-1 and the heat shock protein HSP70 (60). PDK-1 binds unphosphorylated PKC and leads to maturation of the nascent enzyme, whereas HSP70 binding to the dephosphorylated turn motif protects PKCβII from degradation. In keeping with the general notion that HSP70 preferentially binds hydrophobic residues, recent studies implicate an invariant Leu immediately preceding the turn motif phosphorylation site in HSP70 binding (61). Maneuvers that disrupt PKCβII-HSP70 interactions facilitate PKCβII dephosphorylation and ubiquitination and lead to the accumulation of PKCβII in the detergent-insoluble cell fraction (i.e., prevent PKCβII downregulation). PKC-HSP70 interactions may be particularly important in cancer, where high levels of HSP70 expression would prevent activation-dependent PKC dephosphorylation/downregulation.
Hydrophobic motif autophosphorylations also have been viewed as a mechanism that regulates cPKC binding to membranes. As noted in section IB, PKCα and PKCβ translocation to the plasma membrane can be transient, and followed by subsequent translocation to a perinuclear compartment; hydrophobic motif autophosphorylation has emerged as a mechanism that regulates the release of PKCα and PKCβII from the plasma membranes (16, 17, 55, 56, 94). This mechanism appears to be particularly important for PKCα, which anchors to DAG-containing membranes only when it harbors an alanine substitution at its autophosphorylation sites or it is catalytically inactive; DAG does not drive WT-PKCα to membranes (205). These results have been taken as evidence that stoichiometric PKCα autophosphorylation at the hydrophobic motif can tonically inhibit PKCα actions at membranes under some conditions.
Finally, while most studies have focused on the kinases that phosphorylate PKCs, a ceramide-activated protein phosphatase (tentatively identified as PP1) is reported to dephosphorylate PKCα/PKCβII at their activation loop sites (97). Other phosphatase mechanisms undoubtedly regulate PKC phosphorylations under many physiologically relevant conditions, but have largely been ignored in the literature.
B. nPKC Serine/Threonine Phosphorylations
Although nPKC priming phosphorylations were initially considered to follow the mechanisms described for cPKC isoforms, closer inspection exposed three notable differences: 1) PKCδ is unique among PKC family members in its ability to function as a kinase even without activation loop (Thr505) phosphorylation; the closely related nPKCθ isoform and other cPKC and aPKC isoforms are catalytically inactive without activation loop phosphorylation (121). 2) Some nPKC phosphorylations are dynamically regulated (and not constitutive) phosphorylations (173). 3) nPKC turn and hydrophobic motif phosphorylations have variably been characterized as autophosphorylation and transphosphorylation mechanisms (depending on the isoform and cell context). The text that follows elaborates further on these three aspects of nPKC regulation and function.
1. The molecular determinants of PKCδ activity: activation loop phosphorylation versus other mechanisms
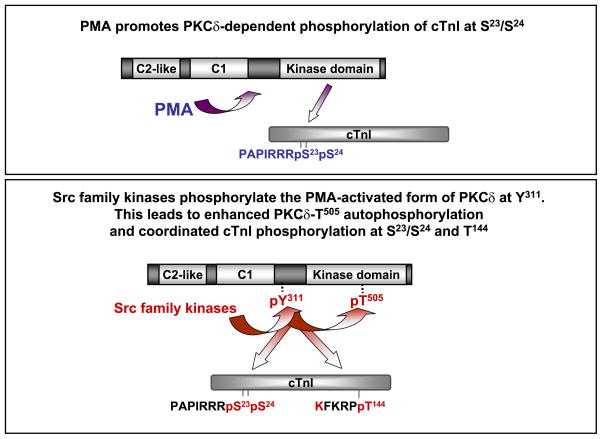
Early studies from Stempka et al. (204) focused on E500, showing that this acidic residue in PKCδ’s activation loop assumes the role of the activation loop phospho-Thr in cPKCs (Fig. 5). However, Liu et al. (120) recently identified another molecular feature of PKCδ that allows for activity without activation loop phosphorylation. In attempting to explain the fact that PKCδ is catalytically competent without activation loop phosphorylation, but the closely related PKCθ is not, these investigators noted that residues 320–345 of PKCδ (NH2 terminal to the catalytic domain and outside of the conserved catalytic core) models to an A-helix in PKA that spans the surface of both lobes of the catalytic core structure and is conserved in a number of other protein kinases (66, 74, 216; Fig. 5B). Trp30 and Phe26 in PKA (corresponding to Trp336 and Tyr332 in PKCδ) fill a deep hydrophobic pocket between the two lobes of the kinase domain; these residues participate in intramolecular interactions that stabilize the PKA structure. The PKCδ A-helix models as a structure that binds directly to the activation loop through hydrophobic interactions involving Tyr332 and three aromatic residues in or near the PKCδ activation loop, namely, Ile497, Phe498, and Phe525. These residues are not found in PKA or PKCθ; although these residues are found individually in certain other PKC isoforms (i.e., Phe498 is conserved in PKCγ, Phe525 is conserved in PKCζ, and Ile497 is conserved in PKCβ and other nPKC isoforms), PKCδ is the only enzyme that contains all four structural determinants. This hydrophobic stack functionally substitutes for activation loop Thr505 phosphorylation as a mechanism to generate a catalytically competent enzyme. These results suggest that PKCδ is equipped with a unique mechanism for activation loop stabilization that is not available to other PKC isoforms. However, PKCδ’s substrate specificity differs somewhat (particularly toward substrates with basic residues in positions P-4 and P-5) depending on whether it is stabilized through an activation loop phosphorylation or this hydrophobic stack. As a result, the PKCδ-T505A mutant mimics only some WT-PKCδ responses in cells; it triggers apoptosis, but it does not mimic the effect of WT-PKCδ to activate NFκB or AP-1 reporter constructs in Jurkat T cells (120). Of note, Cheng et al. (30) recently identified a role for PKCδ-T505 phosphorylation in the formyl-methionyl-leucyl-phenylalanine pathway leading to NADPH activation in phagocytes, showing that a single T505A substitution abrogates PKCδ phosphorylation of p47phox (but not other substrates such as histone). These results are consistent with our recent studies showing that WT-PKCδ and PKCδ-T505A differ with respect to the phosphorylation of individual sites on the myofibrillar protein cardiac troponin I (206), which are described in greater detail in section VC. The notion that activation loop phosphorylation regulates PKCδ substrate specificity (rather than absolute PKCδ activity) could underlie previous discrepancies in the literature regarding the role of Thr505 phosphorylation in the control of PKCδ catalytic function (106, 173, 203). Studies to date have relied heavily on in vitro kinase assays with specific “model” substrates and would be profoundly influenced by the specific assay conditions, including the choice of substrate (see sect. VIIIC). Finally, these results underscore the importance of coordinate translocation events (that regulate PKCδ’s intracellular localization) and phosphorylation events that “fine tune” PKCδ’s enzymology in cells.
2. The controls of nPKC activation loop phosphorylation
The original studies from the Parker laboratory combined in vitro and overexpression approaches to characterize PDK-1 as a PKCδ binding partner and PKCδ activation loop kinase (106). Subsequent studies from the Newton laboratory identified a similar effect of PDK-1 to act as a PKCε activation loop kinase, showing that PDK-1 stimulates PKCε activation loop (T566) phosphorylation and triggers subsequent PKCε autophosphorylation at the COOH-terminal turn and hydrophobic motif sites (24). This model is analogous to the model set forth for cPKC isoforms. In contrast, we recently identified a different mechanism that contributes to the dynamic control of PKCδ activation loop (T505) phosphorylation (170). We showed that PKCδ retains little-to-no T505 phosphorylation in resting cardiomyocyte cultures; PKCδ-T505 phosphorylation is induced by G protein-coupled receptor agonists that promote DAG accumulation (i.e., the α1-AR agonist norepinephrine, endothelin, and PGF2α) or by PMA through a mechanism that requires nPKC activity. These results implicate either a PKCδ autocatalytic mechanism or PKCδ phosphorylation in trans by another nPKC isoform (such as PKCε) as the mechanisms for the dynamic agonist-dependent increase in PKCδ-T505 phosphorylation; PDK-1 does not contribute to this process. With regard to a potential PKCδ autophosphorylation mechanism, it is interesting to note that PKCδ and PKCθ activation loop phosphorylation sites reside in PKC consensus phosphorylation motifs (i.e., RAST505F and KTNT538F in PKCδ and PKCθ, respectively; Fig. 5A). Other PKC isoforms lack a positively charged residue at the −3 position. Moreover, while early in vitro studies from Parker’s laboratory did not detect PKCδ-T505 autophosphorylation, these studies used a bacterially expressed PKCδ preparation that retained only limited catalytic activity and would not detect a PKCδ activation loop autophosphorylation (106).
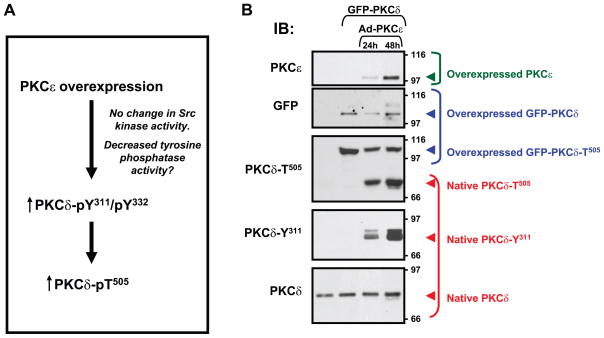
Studies using adenoviral-mediated PKCε overexpression have recently exposed a novel form of PKCε-PKCδ cross-talk, showing that PKCε overexpression leads to a robust increase in PKCδ-T505 phosphorylation (without any changes in PDK-1 protein/activity) in cardiomyocytes and several other cell types (170). While these results could be construed as evidence that PKCε acts as a direct PKCδ-T505 kinase, a more detailed analysis revealed that 1) PKCε overexpression does not lead to an increase in PKCδ-T505 phosphorylation in cells treated with PP1 (a Src kinase inhibitor); 2) the PKCε-dependent increase in PKCδ-T505 phosphorylation is not detected in SYF cells, that lack the major Src family kinases (Src, Yes, and Fyn) and display a defect in PKCδ-Tyr311/Tyr332 phosphorylation; and 3) the PKCε-dependent increase in PKCδ-T505 phosphorylation is restored by Src reexpression (which restores PKCδ-Tyr311/Tyr332 phosphorylation) in the Src+ cell line (170). These results indicate that PKCε promotes PKCδ-T505 phosphorylation via an indirect mechanism that requires Src activity (and PKCδ tyrosine phosphorylation, Fig. 7A). In vitro kinase assays exposed the underlying mechanism, showing that PKCδ undergoes a T505 autophosphorylation reaction that is facilitated when assays are performed in the presence of active Src (under conditions leading to PKCδ-Tyr311/Tyr332 phosphorylation). Collectively, these results add a new dimension to models of PKCδ signaling, showing that Src is a physiologically relevant PKCδ-Tyr311/Tyr332 kinase in cells and that Src-dependent PKCδ tyrosine phosphorylation controls PKCδ-T505 autophosphorylation. According to this revised model of PKCδ signaling, PKCδ is uniquely positioned to sense signaling inputs from both Src and PKCε pathways. However, the mechanism(s) linking PKCε signaling to increased PKCδ tyrosine (Tyr311/Tyr332) phosphorylation remains uncertain. Studies to date show that PKCε overexpression does not lead to gross changes in the abundance or activity of Src family kinases; studies to determine whether PKCε regulates PKCδ tyrosine phosphorylation by inhibiting a PKCδ-targeted phosphatase are ongoing.
FIG. 7.
PKCε overexpression leads to PKCδ-Tyr311/Tyr332 phosphorylation and increased PKCδ-Thr505 autophosphorylation; the mechanisms that control activation loop phosphorylation on native PKCδ do not influence the heterologously overexpressed enzyme. A: schematic of PKCε-PKCδ cross-talk in cells (see text). B: green fluorescent protein (GFP)-tagged PKCδ was heterologously overexpressed in COS7 cells (lanes 2–4). PKCδ overexpression was without (lane 2) or with PKCε overexpression for 24 h (lane 3) or 48 h (lane 4). Samples were subjected to SDS-PAGE and immunblotting for PKCδ protein and PKCδ-Thr505 phosphorylation. The heterologously overexpressed protein (labeled in blue) is constitutively Thr505-phosphorylated; GFP-PKCδ-Thr505 phosphorylation is not increased by PKCε overexpression. Note, while this experiment shows a minor decrease in PKCδ protein and Thr505 phosphorylation in the context of PKCε overexpression for 24 h, this was not a consistent finding in replicates of this experiment. In contrast, native PKCδ (labeled in red, which migrates more rapidly than the GFP-tagged PKCδ transgene) is not phosphorylated at Thr505 in resting cultures that do not overexpress PKCε; PKCε overexpression induces Thr505 (and Tyr311) phosphorylation on native PKCδ.
3. The controls and consequences of nPKC COOH-terminal phosphorylations
nPKCs also undergo COOH-terminal phosphorylations at conserved turn and hydrophobic motifs. nPKC COOH-terminal phosphorylations have been attributed to either autophosphorylation or transphosphorylation mechanisms (24, 152). Early studies from the Parker laboratory attributed PKCδ hydrophobic motif phosphorylation to the actions of a heterologous kinase complex comprised of mTOR and an atypical PKC isoform (152, 239). However, subsequent studies have identified a hydrophobic motif phosphorylation defect for KD-PKCδ and KD-PKCθ; this result is more consistent with a hydrophobic motif autophosphorylation (121). In contrast, KD-PKCδ and KD-PKCθ are recovered with detectable (albeit reduced) turn motif phosphorylation, suggesting that the turn motif is the preferred target for phosphorylation in trans (121).
No single model is sufficient to describe the effect of nPKC COOH-terminal phosphorylations on enzyme function. PKCδ is reported to require turn motif phosphorylation for full catalytic function, whereas PKCθ appears to require hydrophobic motif phosphorylation (rather than turn motif phosphorylation) for catalytic activity (24, 112, 121). The general consensus (based on structural models of PKA and PDK-1) is that the hydrophobic motif participates in an intramolecular interaction that stabilizes the enzyme in an optimal conformation for catalysis (121). However, the functional role of individual PKC priming phosphorylations has been difficult to resolve, in large part due to the fact that PKC mutants lacking phosphorylation at either the activation loop or the COOH terminus become targets of phosphatases. While the fully phosphorylated enzyme is relatively resistant to dephosphorylation by phosphatases (since the hydrophobic motif negative charge protects the activation loop from dephosphorylation, and the activation loop phosphorylation exerts a reciprocal effect to protect the hydrophobic motif from dephosphorylation) (16, 17), the PKCε-S729A hydrophobic motif mutant displays an activation loop phosphorylation defect, and the PKCθ-T538A activation motif mutant displays a hydrophobic loop phosphorylation defect. Both of these secondary phosphorylation defects are reversed by treatment with a Ser/Thr phosphatase inhibitor (24, 121). These reciprocal regulatory controls render the analysis of PKC phosphorylation mechanisms quite challenging.
C. Kinase Domain Tyrosine Residues
There is a limited but generally consistent literature indicating that certain catalytic domain tyrosine residues (that are highly conserved across AGC kinase family members) influence enzyme activity. The initial studies implicated Tyr204 in PKA (corresponding to PKCδ-Tyr512) in intramolecular interactions that structure the catalytic pocket for phosphoryl transfer (232). While the cognate residue in PKCδ is phosphorylated in the context of oxidative or genotoxic stress, PKA tyrosine phosphorylation has never been considered. Rather, most studies have focused on PKCδ, which is tyrosine phosphorylated (including at Tyr512 and Tyr523) and activated in cells treated with H2O2. The Nishizuka laboratory has reported that single residue substitutions at Tyr512 or Tyr523 attenuate (and the combined Tyr512/Tyr523 mutation abrogates) H2O2-dependent PKCδ activation (albeit without completely blocking H2O2-dependent PKCδ tyrosine phosphorylation) (102, 103). The Kufe laboratory has also implicated PKCδ-Tyr512 phosphorylation in mechanisms that contribute to oxidative stress responses involving c-Abl, showing that H2O2 promotes PKCδ translocation and a local increase in PKCδ-Tyr512 phosphorylation in the mitochondrial compartment. In this case, PKCδ-Tyr512 phosphorylation is attributed to c-Abl (which also localizes and is activated via a PKCδ-dependent mechanism in mitochondria of H2O2-treated cells). This local PKCδ-Abl amplification loop leads to an increase in PKCδ activity which contributes to the mitochondrial death response (105, 207). While studies from the Kufe laboratory failed to detect c-Abl-dependent PKCδ-Tyr523 phosphorylation, aPKC phosphorylation at the equivalent tyrosine residue (PKCζ-Tyr428, PKCι-Tyr421, sites that model to a position close to the active site) is reported to increase enzyme activity (162). The notion that PKC phosphorylation at highly conserved tyrosine residues in the activation loop might constitute an alternate (threonine phosphorylation-independent) activation mechanism for certain PKC isoforms (in particular PKCδ or aPKCs) requires further study. Finally, the Blumberg laboratory identified Tyr565, along with Tyr155 and Tyr52, as in vitro Lyn-dependent phosphorylation sites in PKCδ, although the functional importance of PKCδ-Tyr565 phosphorylation as a mechanism to alter PKCδ catalytic activity was not examined (and the functional significance of this event is uncertain, since other studies link Lyn activation in activated RBL-2H3 cells to PKCδ phosphorylation at Tyr52 in the C2 domain, and not Tyr565) (209).
D. Kinase Domain Features That Distinguish aPKCs From cPKC/nPKC Isoforms
aPKC isoforms are generally distinguished from phorbol ester-sensitive cPKC and nPKC isoforms on the basis of their truncated C1 domains. However, three rather striking differences between aPKC and phorbol ester-sensitive cPKC/nPKC kinase domains have been identified. These differences are likely to have important implications for the development of PKC isoform-specific therapeutics.
cPKC and nPKC must be phosphorylated at a highly conserved COOH-terminal hydrophobic motif to achieve catalytic competence; as noted above, aPKCs contain a phosphomimetic Glu in place of the phosphorylatable hydrophobic motif Ser/Thr residue and do not require this processing mechanism.
aPKCs have a rather distinctive nucleotide binding loop (P loop) structure. The vast majority of protein kinases contain a consensus GXGXXG motif that plays a critical role to anchor and position the nontransferable phosphates of ATP (Fig. 4). Single residue substitutions at any one of the conserved glycines generally results in an enzyme defect; in some cases, this contributes to the etiology of human disease. For example, a Gly-Val substitution at the third Gly of the insulin receptor leads to insulin resistance (147), and a Gly-Val substitution at the second Gly is the basis for the Ras-V12 oncogene (211). aPKCs contain an alanine in place of the third conserved Gly (GRGSYA), which is presumed to underlie their somewhat different ATP binding properties and ~100-fold lower affinity for GF109203X (a competitive ATP inhibitor that acts by displacing ATP from this ATP binding pocket), relative to cPKC and nPKC isoforms (72). A tyrosine (GRGSYA) in the aPKC ATP binding loop represents a second structural feature that could impart signaling specificity; cPKC and nPKC isoforms contain a phenylalanine at this position (GRGSFG). Nucleotide binding loop tyrosine phosphorylations control the catalytic activity of certain cyclin-dependent kinases (which are inactive when tyrosine phosphorylated and activated by cellular phosphatases prior to the initiation of mitosis, Ref. 146) and Bcr-Abl (169, 186). Similarly, nerve growth factor (NGF) activates PKCι via a mechanism that involves the activation of Src, enhanced PKCι-Src complex formation (via an interaction mapped to the Src-SH3 domain and a proline-rich 98VFPSIPEQPGMPCPGE114 sequence just NH2 terminal to the PKCι regulatory domain pseudosubstrate domain), and Src-dependent PKCι-Y256 phosphorylation. The functional importance of this phosphorylation is suggested by the further observation that Y256 phosphorylation increases PKCι binding to importin-β (presumably as a result of a phosphorylation-dependent conformational change that exposes an otherwise hidden arginine-rich functional NLS in the aPKC C1 domain) and leads to enhanced PKCι entry into the nucleus (155, 221). NGF also increases Src-dependent PKCι phosphorylation at two other tyrosine residues, namely, Tyr271 (which is at the −3 position relative to the invariant Lysine) and Tyr325 (which is +2 to the kinase domain gatekeeper residue) (224). A single Y→F mutation at Tyr325 (but not Tyr256 or Tyr271) reduces Src-dependent PKCι activation and NGF-dependent survival in PC12 cells (224). While a tyrosine at the +2 position relative to the gatekeeper residue is conserved in many kinases, a general role for regulatory phosphorylations at this site in other kinases has not been considered.
The invariant Lys residue that generally is targeted to generate a catalytically inactive (dominant-negative) kinase appears to play a distinctive role in aPKCs, relative to other AGC kinases. Studies of PKA indicate that the invariant Lys72 residue interacts with the α- and β-phosphates of ATP (to orient ATP for catalysis); it also participates in intramolecular interactions that stabilize the catalytically active conformation of the kinase. Single residue substitutions at Lys72 of PKA (including mutations to Arg or His that conserved charge, mutations to Met that conserve approximate size, or mutations to Ala) result in nonproductive ATP binding modes and a catalytically inactive enzyme (84, 85). However, PKCι tolerates an Lys→Arg replacement at this site; the PKCι-K274R mutant binds ATP, is catalytically active, and displays an affinity for GF109203X that is identical to the WT enzyme (194). PKCι activity is disrupted by a K274W substitution. These results provide tentative evidence that the ATP binding pockets of aPKCs are distinctive (relative to other protein kinases) and that these unique structural features might provide the basis for the development of highly specific aPKC isoform inhibitors that do not inhibit other PKCs or unrelated protein kinases.
V. THE HINGE REGION
The hinge regions of certain PKCs have been identified as targets for caspase-dependent cleavage, protein-protein interactions, and tyrosine phosphorylations. These mechanisms are reviewed in this section.
A. Caspase Cleavage at the Hinge Region
PKCδ, PKCθ, PKCε, and PKCζ undergo caspase-dependent cleavage in response to a range of apoptogenic stimuli; the atypical PKCλ and -ι isoforms have no sequence homology with the PKCζ hinge region, lack caspase cleavage sites, and do not appear to be regulated in this manner (58). Hinge region cleavage by caspase results in the release of a catalytic domain fragment that is freed from autoinhibitory regulatory domain constraints. While this has been viewed as a model to activate PKC, this model is valid only if kinase domains are intrinsically catalytically competent (i.e., endowed with all of the structural determinants required for catalytic activity); most studies that examine cleavage-dependent PKC activation mechanisms have ignored a potential requirement for activation loop transphosphorylation. In fact, as noted in section IVB1, this assumption is justified for PKCδ, which is active without activation loop phosphorylation, although differences in the enzymology of full-length PKCδ (which is inhibited by sphingosine) and the freed PKCδ catalytic domain (which acts as a sphingosine-dependent kinase to phosphorylate 14-3-3 proteins) have been noted (73). The freed PKCε catalytic domain is recovered as an active kinase, suggesting that activation loop phosphorylation (which is required for PKCε activity) is a constitutive modification that is retained following caspase-dependent cleavage. In contrast, two laboratories have reported that the free catalytic domain of PKCζ is catalytically inactive (58, 188). This result was not predicted based on a sequence comparison with PKCδ. As noted in section IVB1, an acidic Glu500 immediately preceding the PDK-1 phosphorylation motif in PKCδ has been viewed as a structural feature that might functionally substitute for activation loop phosphorylation (leading to autonomous PKCδ activity without Thr505 phosphorylation). Atypical PKCs (such as PKCζ, as well as PKCθ, but not PKCε and other PKC isoforms) also contain acidic aspartyl residues at this position. PKCζ also has a phosphomimetic glutamate at the conserved hydrophobic motif autophosphorylation site, in theory making it the PKC isoform that is most favorably configured to be constitutively active without phosphorylation. However, the recent evidence that phosphatase treatment, T410A substitution, or mutations at the PDK-1 docking motif generate a catalytically inactive enzyme argues strongly that both full-length PKCζ and the freed PKCζ kinase domain require activation loop Thr410 phosphorylation for catalytic activity (188).
Proteolytic PKCδ activation by caspases (that contribute to proapoptotic responses) is a prominent mechanism in many, but not all, cell types. Recent studies expose alternative splicing mechanisms for both human and rodent PKCδ that increase PKCδ V3 domain structural diversity by introducing inserts that disrupt the caspase-3 cleavage site [at DIL↓DNNGTY332 in mouse PKCδ (177) and DMQ↓DNSGTY334 in human PKCδ (86)], prevent proteolytic PKCδ activation, and protect cells from pro-apoptotic stimuli. Studies exploring the functional importance of V3 domain alternative splicing mechanisms are confined to neurons, where the differentiation factor retinoic acid regulates PKCδ alternative splicing. This leads to a shift in the relative abundance of full-length PKCδ versus the PKCδ COOH-terminal catalytic fragment (which exert diametrically opposite effects to prevent or induce cellular apoptosis, respectively) leading to important changes in neurogenesis.
B. PKCα-D294G Mutation
The PKCα-Asp294Gly mutation is found in a sub-population of particularly aggressive pituitary tumors, although the precise role of this mutation in tumor progression remains uncertain (161, 213). Studies to date have focused on a trafficking defect. WT-PKCα does not translocate to the surface membrane in isolated cells pituitary GH3B6 cells (i.e., cells that lack cell-cell contacts); G protein-coupled receptor agonists and PMA direct WT-PKCα to cell-cell contacts, but not to the remainder of the surface membrane. However, the PKCα-D294G mutant (which exhibits grossly normal catalytic activity) translocates to the entire surface membrane of both single and apposed cells; the PKCα-D294G mutant is not confined to cell-cell contacts. This phenotype suggests that the PKCα-V3 region is important for binding to specialized membranes (161).
C. PKCδ-Tyr311 and -Tyr332 Phosphorylation
The functional consequences of PKCδ-Tyr311 and/or -Tyr332 phosphorylation are just beginning to emerge, largely because previous literature had focused on the role of PKCδ tyrosine phosphorylation in general (and not necessarily Tyr311 or Tyr332 phosphorylation) and the results were inconclusive; tyrosine phosphorylation was variably implicated as a mechanism that increases, decreases, or does not change PKCδ catalytic activity. The absence of any consensus in the literature is presumed to reflect the presence of many tyrosine residues that are targets for independently regulated phosphorylation events throughout PKCδ’s structure. The precise configuration of tyrosines phosphorylated on PKCδ varies according to the inciting stimulus and likely dictates the functional consequences with respect to enzyme activity. In general, the highly conserved tyrosines in or near the activation loop (Tyr512 and perhaps Tyr523) have been implicated in the control of catalytic function (see sect. IVC), whereas tyrosine phosphorylation in the regulatory region (Tyr52, Tyr64, Tyr155, Tyr187) appears to be more important for PKCδ-dependent changes in gene expression and/or growth/apoptosis responses; regulatory domain phosphorylations generally have not been linked to changes in PKCδ kinase activity.
Our recent studies have focused on PKCδ-Tyr311 and -Tyr332 phosphorylation in cardiomyocytes; two distinct mechanisms have been identified. We showed that H2O2 activates Src family kinases (SFKs, Src and the related Fyn, Lyn, and Yes kinases) and induces a global increase in PKCδ phosphorylation at Tyr311 and Tyr332 in both soluble and particulate cellular compartments; H2O2-dependent PKCδ-Tyr311/Tyr332 phosphorylation is via a PP1-sensitive pathway, suggesting a role for Src or a related SFK (172). In contrast, PMA promotes PKCδ tyrosine phosphorylation by delivering PKCδ to caveolae (a SFK-enriched membrane fraction); PMA-dependent PKCδ-Tyr311 phosphorylation is confined to the caveolae fraction and disrupted by cyclodextrin (a cholesterol-binding agent that undermines caveolae structural integrity). Of note, PMA-dependent PKCδ-Tyr311 phosphorylation also is via a PP1-sensitive mechanism, but PMA does not increase Src family kinase activity (171, 172). In vitro kinase assays suggest a possible mechanism for the PMA-dependent increase in PKCδ tyrosine phosphorylation, showing that Src preferentially phosphorylates the active conformation of PKCδ (i.e., PMA induces a conformational change that renders PKCδ a better substrate for Src kinases, that constitutively localize to caveolae membranes; Ref. 172). However, the identity of the PMA-activated (PP1-sensitive) kinase that phosphorylates PKCδ at Tyr311, but not Tyr332, remains uncertain. While studies in SYF cells (a continuous fibroblast cell line generated from the embryos of mice lacking the three major SFKs, Src, Yes, and Fyn) and Src+ cells (a SYF cell derivative engineered to overexpress Src) provide unambiguous evidence that Src is required for PKCδ-Tyr311 and -Tyr332 phosphorylation under certain stimulatory conditions (171) (and Tyr311 and Tyr332 appear to be the major in vivo PKCδ phosphorylation sites in many H2O2-treated cell types), a role for Src (and related SFKs) in the PMA-dependent pathway that leads to PKCδ phosphorylation at Tyr311 (but not Tyr332) in cardiomyocytes caveolae is not obvious. In vitro kinase assays followed by immunoblot analysis with phosphosite specific antibodies or peptide sequencing approaches identify similar effects of other SFKs, such as Fyn, Lyn, and Yes to phosphorylate PKCδ at both Tyr311 and Tyr332 (206). Other tyrosine kinases that might signal downstream from Src (i.e., act in a PP1-sensitive manner) and might be localized to caveolae (including PDGFRs, FAK, and JAK2) exhibit little to no PKCδ-Tyr311/Tyr332 kinase activity (171). Rather, our recent studies implicate c-Abl as a selective PKCδ-Tyr311 kinase; c-Abl does not phosphorylate PKCδ at Tyr332 (171). However, c-Abl is not detected in cardiomyocyte caveolae, and pharmacological studies with Gleevec (a very potent c-Abl inhibitor) establish that PMA-dependent PKCδ-Tyr311 phosphorylation does not require c-Abl activity (although c-Abl does appear to contribute to H2O2-dependent PKCδ-Tyr311 phosphorylation, Ref. 171). The molecular components of the PMA-dependent PP1-sensitive PKCδ-Tyr311 phosphorylation pathway in cardiomyocyte caveolae remain the focus of ongoing studies. Finally, it is important to note that G protein-coupled receptor agonists (such as norepinephrine and endothelin) do not increase PKCδ-Tyr311 phosphorylation; these agonists also fail to promote PKCδ translocation to caveolae (171). This cannot be attributed to differences in PKCδ activation by PMA and DAG (the endogenous lipid cofactor), since DAG analogs support Src- (or c-Abl-) dependent PKCδ tyrosine phosphorylation in vitro and DAG mimics the effect of PMA to recruit PKCδ to caveolae and increase PKCδ-Tyr311 (and Thr505) phosphorylation in cardiomyocytes. The observation that DAG analogs effectively substitute for PMA both in vivo and in vitro argues that α1-ARs do not increase PKCδ tyrosine phosphorylation because they do not promote DAG accumulation in caveolae membranes, either because the density of α1-ARs or their downstream signaling partners is limiting in cardiomyocyte caveolae (171).
Some studies implicate PKCδ phosphorylation at Tyr332 (and perhaps also Tyr311, sites that flank a caspase cleavage site) as a mechanism that facilitates PKCδ activation via caspase-dependent cleavage in cells undergoing apoptosis (125). However, insofar as cleaved forms of PKCδ are not convincingly identified in H2O2-treated cardiomyocyte cultures (unpublished data) and certain other cellular contexts, studies of hinge region tyrosine phosphorylation have focused on other cellular processes. Early studies established that Tyr311/Tyr332-phosphorylated PKCδ is recovered as a lipid-independent enzyme in both the soluble and particulate fraction of H2O2-treated cells (including cardiomyocytes); this lipid-independent form of PKCδ is predicted to mediate PKCδ responses throughout the cell, not just on lipid membranes (102, 103, 172). However, recent studies expose a considerably more elaborate role for Src (and hinge region tyrosine phosphorylation) in the control of PKCδ activity. Building on the recent evidence that Src (and PKCδ-Tyr311/Tyr332 phosphorylation) increases PKCδ-Thr505 autophosphorylation (170; as noted in sect. IVB2), we showed that Src also regulates PKCδ-dependent phosphorylation of a physiologically important cardiac substrate, namely, cardiac troponin I (cTnI). cTnI is the “inhibitory” subunit of the troponin complex that contributes to Ca2+-dependent regulation of myofilament function in the heart (Fig. 8). Our recent studies establish that PKCδ phosphorylates cTnI at Ser23/Ser24 (a site traditionally viewed as a PKA target) when it is allosterically activated by PMA; functional studies on skinned muscle fibers show that lipid cofactor-activated PKCδ sensitizes the myofibril to sub-maximal Ca2+, but has no significant effect on maximum Ca2+-activated tension (the predicted effect of cTnI-Ser23/Ser24 phosphorylation). However, PKCδ acquires the ability to also phosphorylate cTnI at Thr144 (in addition to Ser23/Ser24) when it is tyrosine phosphorylated by Src; functionally, this abrogates calcium sensitization (the effect of selective cTnI phosphorylation at S23/S24) and leads to a decrease in maximum Ca2+-activated tension, the predicted effect of cTnI-Thr144 phosphorylation (based on previous literature, Ref. 22). A mutagenesis approach was used to map the structural determinants for Src-dependent changes in PKCδ substrate specificity. These studies showed that the Src-dependent change in PKCδ substrate specificity (enabling PKCδ to act as a cTnI-T144 kinase) is abrogated by either a Y→F substitution at Tyr311 (but not Tyr332) or a T→A substitution at Thr505; Y311F and T505A substitutions had no effect on cTnI-S23/S24 phosphorylation by PKCδ in the presence of PMA. These results identify a role for Src-dependent PKCδ-Tyr311 phosphorylation and PKCδ-Thr505 autophosphorylation as novel regulators of PKCδ’s substrate specificity, leading to enhanced PKCδ activity toward only selected sites on cTnI and a functionally important change in contractile function. Of note, these results conform to recent observations by Lui et al. (120) showing that PKCδ acquires activity toward substrates with basic residues at positions P-4 and/or P-5 when phosphorylated at Thr505. Wild-type PKCδ (which is Thr505-phosphorylated in assays with Src) phosphorylates cTnI at KFKRPT141 (a site that contains a basic residue at position P-5 relative to the phosphorylation sites), whereas the PKCδ-T505A mutant does not; its actions are confined to PAPIRRRS23S24.
FIG. 8.
Src-dependent PKCδ tyrosine phosphorylation leads to changes in PKCδ phosphorylation of cardiac troponin I; PKCδ-Tyr311 and Thr505 critically regulate PKCδ substrate specificity.
Tyr332 phosphorylation has also emerged as a mechanism that influences PKCδ’s interactions with other signaling proteins. Tyr332 is one of two tyrosines in PKCδ with a +3 position Ile that conforms to a consensus binding sequence for the SH2 domain of Shc, an adapter protein that nucleates signaling to the MAPK cascade. PKCδ-Tyr332 phosphorylation in RBL-2H3 cells leads to the formation of PKCδ-Shc complexes and a physiologically important indirect interaction between PKCδ and SHIP [an SH2-domain-containing inositol 5′-phosphatase that also binds Shc, dephosphorylates PI(3,4,5)P3 to PI(3,4)P2, and provides a mechanism to negatively regulate AKT phosphorylation] (110, 191). Our recent studies indicate that PKCδ-Shc complexes accumulate in a detergent-insoluble cytoskeletal fraction (that is enriched in intermediate filament proteins such as vimentin, vinculin, and desmin) in H2O2-treated cardiomyocytes. WT-PKCδ overexpression leads to an increase in H2O2-dependent Shc localization to this fraction, whereas KD-PKCδ (which constitutively localizes to the cytoskeletal fraction) leads to increased Shc localization to the cytoskeleton, even without an oxidative stress stimulus. These results expose a kinase-independent role for PKCδ to regulate the subcellular localization of Shc (and potentially other binding partners). While PKCδ accumulates in the cytoskeleton of H2O2-treated cardiomyocytes as a Tyr332-phosphorylated protein (and KD-PKCδ is constitutively Tyr332-phosphorylated in this fraction), the role of PKCδ-Tyr332 phosphorylation in this process in cardiomyocytes is not convincing. Pharmacological inhibitors that prevent PKCδ tyrosine phosphorylation do not disrupt PKCδ-Shc interactions or prevent PKCδ-dependent Shc localization to the cytoskeletal fraction in H2O2-treated cardiomyocytes. The molecular determinants of PKCδ-Shc interactions are the focus of ongoing studies.
VI. THE V5 DOMAIN
V5 domains are 50- to 70-amino acid sequences COOH-terminal to the catalytic core of the enzyme (C3 and C4 domains) that contain the highly conserved turn and hydrophobic phosphorylation motifs as well as an additional 7–21 residues at the extreme COOH terminus (beyond the hydrophobic motif) that are highly variable both in their length and sequence. The extreme COOH-terminal regions of the V5 domain that share little to no sequence homology have been exploited as epitopes to raise PKC isoform-specific antibodies for Western blotting and immunolocalization studies. These regions were otherwise generally ignored in early studies exploring the structural determinants of PKC isoform function. However, V5 domains have recently emerged as structures that impart important determinants of PKC isoform-specific targeting and function, suggesting that V5 domains might represent novel targets for pharmaceuticals designed to regulate PKC isoform-specific signaling in cells.
A. PKCβV5 Domain Splice Variants With Distinct Subcellular Targeting and Signaling Functions
PKCβ is expressed as differentially spliced versions of the same gene (termed PKCβI and PKCβII) that differ only at their COOH-terminal V5 regions. PKCβI and PKCβII are expressed in a tissue-specific and developmentally regulated manner (26). Endogenous PKCβ isoforms target to distinct subcellular compartments (both at rest and following activation) and have been linked to distinct cellular functions (in some cases, even in a single cell type). For example, individual PKCβ isoforms exert functionally opposing actions in A10 vascular smooth muscle cells. The effects of PKCβI to stimulate and PKCβII to inhibit A10 vascular smooth muscle growth constitute perhaps the most convincing evidence that PKC isoforms can decode second messenger signals in a highly specialized manner as a result of subtle differences in V5 domain structures (228).
Both native and heterologously overexpressed PKCβI and PKCβII exhibit distinct subcellular localization patterns. In cardiomyocytes, endogenous PKCβI localizes to the cytosol and perinuclear region under basal conditions and translocates to the nucleus following treatment with PMA, whereas native PKCβII decorates fibrillar cytoskeletal structures at rest and translocates to the cell periphery and the perinuclear region (where it colocalizes with the PKC anchoring protein RACK1) following treatment with PMA (44, 167). In PMA-treated NIH3T3 fibroblasts, both PKCβI and PKCβII are detected at membrane ruffles, but only PKCβII (which contains a V5 domain actin binding site somewhat homologous with the actin binding site of troponin I) accumulates in the cytoskeleton at actin-rich microfilaments (15, 68; Fig. 6). PKCβII tethered to cytoskeletal F-actin displays increased autophosphorylation (including in the absence of lipid cofactors), enhanced trans-phosphorylation of selected physiological substrates such as vimentin (but not necessarily all substrates, suggesting a change in substrate specificity), and resistance to PMA-dependent downregulation (15). As noted is previous sections, PMA transiently/reversibly translocates PKCβI and PKCβII to the plasma membrane, but only PKCβII translocates in a more sustained (irreversible) manner to a juxtanuclear site containing a subset of recycling endosomes during chronic agonist exposure (11).
PKCβI and PKCβII display distinct oscillation patterns (between the cytosol and membranes) in response to metabotropic glutamate receptor-1a activation (i.e., a single stimulus) in HEK293 cells (6, 68). Here, two nonconserved residues in PKCβII’s V5 domain (Asn625 and Lys668, that lie within two of the three V5 domain sequences that have been implicated as RACK1-binding motifs; Refs. 6, 68, 200) were mapped as structural determinants of the distinct translocation patterns. Single N625G or K668G substitutions at either site convert PKCβII to an enzyme that translocates in a PKCβI-like manner (6).
RACK1 anchors PKCβII (but not PKCβI) to the perinuclear region; a PKCβI selective RACK protein has not yet been identified. While RACK1 binding sites were initially mapped to the PKCβ C2 domain, C2 domain RACK1 binding sites (which are common to PKCβI and PKCβII) do not explain the in vivo specificity of RACK1 for PKCβII. Rather, RACK1-binding specificity has been attributed to protein-protein interaction motifs in the V5 domain that are unique to PKCβII (621ACGRNAE627, 645QEVIRN650, and 660SFVNSEFLKPEVKS673) and not found in PKCβI. A peptide based on the unique RACK1 binding sequence in PKCβII (645QEVIRN650) acts as PKCβII-selective translocation inhibitor when introduced into permeabilized neonatal cardiomyocyte cultures; similarly, a peptide based on the putative PKCβI-RACK binding site (the unique 645KLFIMN650 sequence in PKCβI) selectively inhibits PKCβI translocation. The observation that PMA-dependent cardiomyocyte hypertrophy is inhibited by either the PKCβI or the PKCβII translocation inhibitor peptides suggests that both PKCβ isoforms contribute to cardiac growth responses (200).
Other studies have exploited a chimeric approach to show that the COOH-terminal 13 amino acids of PKCβII are sufficient to confer a “PKCβII-specific” function (namely localization to the nucleus and lamin B phosphorylation) to PKCα (65). While these results are consistent with the notion that the PKCβII V5 region contains the molecular determinants for nuclear localization, studies using chimeric constructs must be interpreted with caution given the growing recognition that the phosphorylated hydrophobic motif forms stabilizing intramolecular contacts with other regions of the enzyme (including the C2 domain and the catalytic core) and that cooperative intramolecular interactions fulfill a crucial function to regulate affinity for substrate and cofactors as well as calcium-dependent binding to acidic membranes (95). Foreign V5 domains that cannot participate in cooperative intramolecular interactions might not necessarily produce informative phenotypes.
B. The V5 Domain and PKC Catalytic Function
Yeong et al. (233) recently published surprising evidence that the very distal portion of the V5 domain (COOH terminal to the hydrophobic motif phosphorylation site, that is absent in PKA) is critical for PKCα catalytic activity and function in cells (233). These investigators used a deletion approach to show that a single residue truncation at the extreme COOH terminus leads to a 60% reduction in enzyme activity (without any activation loop phosphorylation defect); truncation of the last 10 amino acids results in a PKCα mutant that lacks activation loop phosphorylation and is essentially catalytically inactive. A similar seven-residue COOH-terminal truncation abrogates the catalytic activity of PKCε (in this case, without fully blocking activation loop phosphorylation, Ref. 238). In each case, modeling studies identified an intramolecular interaction between the V5 domain and the N-lobe of the kinase that structures the ATP-binding pocket and influences enzyme activity. Further studies showed that an antibody directed against the PKCα extreme COOH terminus immunoprecipitates PKCα protein, but relatively little PKCα activity; in contrast, an antibody directed against an NH2-terminal determinant (in this case a tag) immunoprecipitates catalytically active PKCα. These results have been interpreted as evidence that protein-protein interactions involving the PKC extreme COOH terminus might prevent V5 domain interactions with the catalytic pocket (i.e., mimic the effect of a truncation) and inhibit catalytic activity (233). The implications of these results to the interpretation of the PKC literature (which has relied heavily on immune kinase assays performed with antibodies directed against PKC-V5 domain epitopes) and the full functional significance of PKC V5 domain epitopes as determinant of enzyme activity (that could be targeted to generate isoform-specific PKC antagonists) require further study. Finally, it is worth noting that the PKCα extreme COOH terminus is the only PKC isoform that ends in a COOH-terminal PDZ (PSD-95, disheveled and ZO-1) domain consensus binding motif (QSAV). Staudinger et al. (199) have demonstrated that PKCα interacts (through this COOH-terminal motif) with the PDZ domain-containing protein PICK1. PICK1 self-associates and forms oligomers with other PDZ domain containing proteins at membranes, providing a rationale to consider PICK1 as a scaffold that localizes PKCα with other signaling partners at specialized membranes.
C. The V5 Domain and PLD Activation
The PKCα V5 domain has been implicated in the activation of PLD. PKCα activates PLD through a protein-protein interaction (i.e., a kinase-independent mechanism); while PLD is a target for PKCα-dependent phosphorylation, this is an inhibitory mechanism that reduces PLD activity (76). Recent literature suggests that PLD does not bind to a single region of PKCα; rather, the PKCα-PLD interaction involves many points of contact between these proteins (76, 185). Nevertheless, the PKCα-V5 domain appears to be particularly critical for in vivo PLD activation. A COOH-terminal 10-amino acid deletion or a single V5 domain substitution at Phe663 (10 residues from the COOH terminus) fully abrogates in vivo PKCα-dependent PLD activation (76). PKCβII (which shares a high level of V5 domain structural homology with PKCα) also activates PLD, whereas PKCβI (with a structurally more divergent V5 domain) does not.
D. V5 Domain and Nuclear Localization Signals
In some cells, PKCδ localizes to the nucleus early in apoptosis. The structural requirements for PKCδ entry into the nucleus have been mapped to six basic amino acid residues that reside in a functional NLS in the COOH terminus (611KRKVEPPFKPKVK623, Fig. 6) (43). While many of these basic residues are conserved in other cPKC and nPKC isoforms, their role in the nuclear localization of other PKCs has not been examined. The PKCδ NLS directs rapid nuclear import of full-length PKCδ. Studies with chimeric PKCs show that the PKCδ V5 domain contains all of the molecular determinants required for nuclear localization; a chimeric PKCε-V5δ construct accumulates in the nucleus and induces apoptosis, whereas PKCε itself does not accumulate in the nucleus under identical conditions (43).
E. V5 Domain Tyr Phosphorylation
A single study identifies a tyrosine phosphorylation of a COOH-terminal site on PKCα (Tyr658) and PKCβI (Tyr662) that is mediated by the non-receptor tyrosine kinase Syk (and not Lyn or BtK) and requires prior phosphorylation of adjacent hydrophobic motif serine residues (S657 in PKCα and S661 in PKCβI) (92). Once phosphorylated, this tyrosine residue docks the SH2 domain of Grb2, leading to Grb2/SOS binding and the activation of the Ras/ERK pathway. In theory, PKCγ and PKCε (which also contain tyrosine residues at the +1 position following the hydrophobic motif phosphorylation site) also might nucleate a Grb2/SOS/Ras activation complex through this mechanism, whereas PKCβII and PKCδ lack tyrosines at this position. The importance of this COOH-terminal structural feature as a mechanism that might underlie the distinct signaling properties of individual PKC isoforms has not been explored in any subsequent studies.
VII. NONTRADITIONAL PKC ACTIVATION MECHANISMS
A. Cellular Actions of Freed Kinase Domains
Several PKCs are cleaved by caspase during apoptosis. PKCε cleavage in cells subjected to serum starvation, chemotherapeutic agents, or tumor necrosis factor (TNF)- α has generally been attributed to caspase 3 and mapped to SSPD383↓G in the hinge region (although caspase 7 can substitute for caspase 3 in MCF-7 cells that lack caspase 3 activity, Refs. 9, 150). PKCε exerts a dichotomous role on cell function. Full-length PKCε prevents the proapoptotic effects of TNF-α, whereas the freed PKCε catalytic fragment (that is generated in cells treated with TNF-α) promotes apoptosis. This form of apoptosis is attenuated by WT-PKCε overexpression and completely abrogated by overexpression of the caspase-insensitive PKCε-D383A mutant (9, 150).
PKCδ also regulates apoptosis in a cell- and stimulus-specific manner. PKCδ contributes to the proapoptotic actions of etoposide, UV and γ-irradiation via a mechanism that typically involves the cleavage of PKCδ in the hinge region by caspase 3 and the generation of a 40-kDa catalytic domain fragment of PKCδ that is a constitutively active (phospholipid-independent) enzyme, since it is freed from the autoinhibitory constraints imposed by the NH2-terminal regulatory domain. The freed PKCδ catalytic domain serves as both a marker and effector of apoptosis. The PKCδ catalytic domain lacks regulatory region membrane targeting modules; it accumulates in the nucleus through its NLS. Nuclear targeted catalytic domain PKCδ is sufficient to induce apoptosis in certain cell types (52, 135). There is some evidence that PKCδ cleavage by caspase and proapoptotic signaling responses are facilitated by PKCδ phosphorylation at Tyr64/Tyr187 or Tyr332 (depending on the model and inciting stimulus, Refs. 14, 124). However, PKCδ is also reported to protect glioma cells from the proapoptotic effects of TRAIL through a mechanism that involves TRAIL-dependent PKCδ-Tyr155 phosphorylation leading to PKCδ translocation to the endoplasmic reticulum, caspase-dependent PKCδ cleavage, and the activation of the cytoprotective enzyme AKT (149). These opposing roles for PKCδ in the apoptosis pathways induced by etoposide and TRAIL emphasize the challenges associated with efforts to expose the specific cellular actions of this enzyme.
Differences in the enzymology of the freed PKCδ catalytic domain fragment and full-length PKCδ also have been noted; the freed PKCδ catalytic domain fragment acts as a sphingosine-dependent kinase to phosphorylate 14-3-3 proteins, whereas full-length PKCδ is inhibited by sphingosine (73). These results could suggest that full-length PKCδ and the PKCδ catalytic domain fragment play distinct roles to phosphorylate distinct cellular targets at different stages of apoptosis. In support of this concept, p73β (a structural/functional homolog of p53 that interacts with c-Abl and activates transcription from p53 promoters, Ref. 163) and nuclear DNA-dependent protein kinase (an enzyme essential for the repair of double-stranded DNA breaks, Ref. 13) have been identified as nuclear PKCδ substrates that are preferentially phosphorylated by the freed PKCδ catalytic fragment. Other substrates that might contribute to PKCδ’s nuclear actions include hRad9 (key component of the genotoxin-activated checkpoint signaling complex which also binds antiapoptosis Bcl-2 family proteins and mediates apoptotic responses to DNA damage when phosphorylated by PKCδ, Ref. 234), lamin B (a nuclear structural protein that is cleaved upon phosphorylation, leading to the disassembly of the nuclear lamina, Ref. 35), and c-Abl (a kinase activated by apoptotic stimuli that forms complexes with Lyn, DNA-PK and PKCδ in the nucleus).
PKCθ is cleaved at a DEVD354K site in the hinge region by caspase 3 in response to various agents that induce apoptosis (38). The catalytic fragment of PKCθ is an active enzyme; overexpression of the cleaved kinase-active (but not full-length PKCθ or the kinase-inactive PKCθ catalytic fragment) is sufficient to induce nuclear fragmentation and cell death.
B. Kinase-Independent Actions of Freed Regulatory Domains
PKC regulatory domains (RDs) are generally viewed as dominant-negative inhibitors of the full-length enzyme, since they are predicted to compete with the cognate full-length enzyme for docking proteins at sites of action. However, certain PKC-RDs mimic the actions of the full-length protein (i.e., certain cellular responses are mediated by PKC regulatory domain and are kinase independent). For example, PKCε colocalizes through its RD with F-actin at the cortical cytoskeleton where it functions to induce neurite outgrowth in neuroblastoma cells and plays an important role in neuronal differentiation (118, 236). PKCε and PKCθ localize via their RD-C1 motif to the Golgi where PKCε regulates glycosaminoglycan sulfation and PKCθ induces apoptosis (108, 182).
C. PKC Regulation by Reactive Oxygen Species
While the focus of most studies in the literature has been on the effects of oxidative stress to activate Src family kinases and alter PKC signaling mechanisms through PKC tyrosine phosphorylation, PKCs are also direct targets for redox modifications (69). Mild oxidative stress promotes the oxidation of C1 domain cysteine residues, leading to the release of Zn ions, reduced autoinhibitory function, and cofactor-independent PKC activation; PKCγ activation through this mechanism is discussed in section IIC. In contrast, intense oxidative stress modifies reactive cysteines in the kinase domain and abrogates PKC activity.
D. PKC Regulation by Reactive Nitrogen Species
The canonical nitric oxide (NO) signaling pathway involves the activation of guanylyl cyclase and the formation of cGMP. However, recent studies have exposed an additional guanylyl cyclase-independent effect of reactive nitrogen species to modify a range of biologically important molecular targets, including proteins, lipids, and DNA. NO and NO-derived reactive nitrogen species such peroxynitrate (or ONOO−, which is formed when NO reacts with superoxide) react with reduced cysteines or tyrosines, forming cysteine S-nitrosothiols or 3-nitrotyrosines, respectively. These posttranslational modifications have been linked to changes in the function of a variety of cellular proteins. Insofar as NO and PKCs share many signaling responses, including in the heart where both NO and certain PKCs have been implicated in cardioprotection, it was perhaps inevitable that there would be interest in a possible role of PKCs as NO targets (7). Studies to date have focused on PKC tyrosine nitration; PKC modification through S-nitrosylation has not yet been considered.
Knapp et al. (99) reported that high concentrations of ONOO− (100 μM) lead to the tyrosine nitration of PKCα and PKCβII. These investigators reported that low ONOO− concentrations (1 μM, which does not promote PKC tyrosine nitration) increase PKC activity via a redox-dependent mechanism, whereas high ONOO− concentrations inhibit cofactor-dependent PKCα, PKCβII, PKCε, and PKCζ activities (99). However, high ONOO− concentrations also have been linked to increased intracellular protease activity and proteolytic activation of PKCα (which is tyrosine nitrated under these conditions) in other studies (25).
PKC nitration has been reported to increase PKC signaling by enhancing PKC-protein complex formation. There is evidence that NO donors increase PKCε tyrosine nitration and PKCε binding to its anchoring protein RACK2 (or β′-COP, without changing the availability of RACK2 in membranes), leading to increased PKCε protein and activity in the particulate fraction of rabbit cardiomyocytes (7). Protein nitration also has been identified as a PKCδ activation mechanism in substantia nigra-derived SN4741 dopaminergic cells, where sodium nitroprusside (SNP) promotes PKCδ nitration, PKCδ activation, enhanced PKCδ-p53 complex formation, and p53 phosphorylation at Ser15 (a residue adjacent to the p53-MDM2 binding site); SNP does not promote p53 phosphorylation at other serine residues (107). An effect of p53-S15 phosphorylation to disrupt ubiquitin-dependent p53 degradation (by preventing MDM2 binding) would increase apoptosis, and might have relevance to the pathophysiology of Parkinson’s disease. However, it is important to note that the role of PKCs as targets for functionally important posttranslational modifications by NO-derived reactive nitrogen species remains tentative, since studies to date have not used mutagenesis strategies to map a PKC nitration site and unambiguously implicate a specific nitration event to a change in PKC function (activity, protein-protein interaction, targeting).
VIII. OBSTACLES THAT STYMIE EFFORTS TO RESOLVE PKC ISOFORM-SPECIFIC FUNCTIONS
PKC functions and substrates in cells have traditionally been identified using a pharmacological strategy (with activators or inhibitors) or a molecular strategy (involving targeted deletion or overexpression of individual PKC isoforms or translocation modifier peptides). The uncertainties associated with each of these standard investigative approaches are widely recognized. For example, some commonly used PKC inhibitors exert PKC-independent cellular actions. Most notably, chelerythrine inhibits Bcl-XL interactions with BH3-containing proteins such as Bax and it induces apoptosis through a PKC-independent mechanism (27, 33, 229). Rottlerin uncouples mitochondrial respiration from oxidative phosphorylation; the notion that rottlerin acts as a selective PKCδ inhibitor is seriously undermined by evidence that rottlerin exerts inhibitory actions in PKCδ −/− cells and that rottlerin is not a particularly effective in vitro PKCδ inhibitor (39, 109, 189, 190). Even Ro 318220 and GF109203X, which are relatively specific inhibitors of PKC isoforms, exert off target actions to inhibit RSK and p70 S6 kinase in certain cellular contexts (2, 164, 165). Molecular overexpression strategies also must be interpreted with caution given the growing recognition that the subcellular compartmentalization and phosphorylation patterns of overexpressed PKCs (particularly kinase-inactive PKCs) may not faithfully recapitulate the properties of the native enzyme. Kinase-inactive PKCs aberrantly localized to detergent-insoluble cell fractions can promiscuously interact with nonphysiological binding partners (and may not be properly positioned to act as competitive inhibitors of the endogenous enzyme, Ref. 173). Furthermore, early studies from the Parker laboratory exposed effects of catalytically inactive (activation loop Thr→Ala) PKCδ, PKCε, and PKCζ mutants to inhibit a common step in the processing/activation of all PKC isoforms, leading to a dominant-inhibitory effect on both the cognate enzymes and on PKCα (63). These results emphasize that isoform-specific inhibition may be inherently trickier than generally acknowledged in the literature. While RNA interference gene silencing strategies may avoid some of the limitations inherent in overexpression studies, PKC knockout mouse models may be more problematic. Our recent studies suggest that the phenotype described for the PKCε−/− mouse reflects the compensatory changes resulting from chronic deficiency of PKCε from an early developmental stage (and not necessarily the physiological role of PKCε in differentiated tissues, see sect. VIIIA3).
The other major strategy to interrogate PKC signaling function involves the overexpression of peptides designed to either promote or inhibit PKC translocation in cells. While these peptides certainly exert pronounced biological actions, the precise molecular mechanism(s) that underlie their actions may not necessarily be as straightforward as generally assumed for several reasons: 1) PKCδ is released from the membranes as a lipid-independent enzyme during oxidant stress. A peptide inhibitor that prevents RACK-driven PKCδ compartmentation to membranes would not prevent substrate phosphorylations in the soluble fraction of H2O2-treated cells. 2) Many PKCs localize to multiple subcellular compartments in a single cell (including surface membranes, lipid rafts, mitochondria, and nuclei). There is no a priori reason to assume that a single RACK anchors its cognate PKC isoform at all subcellular compartments (i.e., that a single peptide inhibitor should block PKC isoform translocation to distinct subcellular compartments). 3) There also is no a priori reason to assume that translocation modifier peptides selectively block PKC-RACK interactions, without blocking PKC interactions with non-RACK binding partners (for example, a PKCδ interaction with Src and PKCδ tyrosine phosphorylation). This entirely different interpretation for PKC inhibitor or activator peptide actions has never been adequately considered.
In addition to these well-established caveats that raise general concerns regarding standard investigative approaches routinely used to define PKC isoform specific functions, several caveats pertaining to studies of PKC isoforms that have not been sufficiently emphasized in the literature (and are underappreciated by the general research community) are discussed in the sections that follow.
A. PKC Isoform Cross-Talk in Cells
There is growing evidence that PKC isoforms do not always operate in isolation. PKC isoform cross-talk may be key for the functional integration of signaling networks in cells and may undermine the interpretation of knockout or overexpression studies.
1. PKC activity controls the spatiotemporal dynamics of PKC signaling in cells
There is a small (but consistent) literature that PKC activity itself influences PKC translocation and downregulation in cells (67). PKC activity is critical for the reversible cycling of cPKCs between the cytosol and plasma membrane in response to physiological stimuli such as G protein-coupled receptor agonists; PKC activity is less important for irreversible pharmacological responses to PMA (55). PKCs accumulate in cells treated with PKC inhibitors at least in part due to a defect in COOH-terminal autophosphorylation. However, chronic PKC inhibition also leads to a defect in PKCε-dependent phosphorylation of the intermediate filament protein vimentin (which is required for vesicular transport and protein trafficking) and the accumulation of PKCε in large β1-integrin-containing cytoplasmic vesicles, leading to abnormalities in integrin recycling to the plasma membrane and cell migration on a β1-integrin substrate (82, 83).
2. PKC activity regulates lipid metabolic enzymes that control DAG accumulation in cells
As noted in section VIC, certain PKC isoforms have been linked to the activation of PLD and the generation of PA and DAG (potentially amplifying PKC signaling responses). PKCs also regulate diacylglycerol kinases (DGKs), a family of nine mammalian enzymes that catalyze the phosphorylation of DAG to produce PA (a mechanism that terminates cPKC/nPKC signaling responses). DGKs have been subdivided into five groups based on regulatory domain motifs; individual DGK family members display distinct tissue expression patterns, catalytic properties, and modes of regulation, presumably allowing individual DGK enzymes to subserve distinct biological functions. Several DGK family members have been implicated as PKC binding partners or downstream targets. For example, PKCγ colocalizes with class I DGKγ at the plasma membrane in CHO-K1 cells; PKCγ-dependent DGKγ-S776/S779 phosphorylation results in increased DGKγ activity and feedback inhibition of PKC signaling responses (227). Class II DGKs (δ and η) contain an NH2-terminal PH domain that contributes to membrane translocation and is the target of a cPKC-dependent regulatory phosphorylation that interferes with membrane translocation (81). The recent evidence that DGKδ gene inactivation leads to elevated DAG levels, increased PKC activity, and enhanced phosphorylation of PKC targets suggests that DGKδ inactivation may be a particularly important mechanism to amplify PKC signaling responses (36). Finally, class IV DGKs (ζ, ι) contain a NLS motif that shares homology with the phosphorylation domain of the MARCKs protein (a prominent PKC substrate) and is phosphorylated by conventional PKC isoforms. DGKζ is recovered in complexes with PKCα and acts to negatively regulate PKCα signaling in cells. However, a reciprocal effect of PKCα to phosphorylate the DGKζ-MARCKs domain disrupts the PKCα-DGKζ interaction, interferes with DGKζ nuclear localization, and reduces DGKζ activity. In this manner, PKCα reverses the inhibitory effects of DGKζ and contributes to its own activation (126, 127). While there is still only limited information on the functional role of DGK-PKC interactions in physiologically relevant disease models, functionally important DGKζ-PKC cross-talk has been identified in the heart, where DGKζ overexpression prevents hypertrophic growth responses in both cardiomyocyte culture and transgenic mouse models (5, 210).
3. PKCε-PKCδ cross-talk in cell culture models and genetically engineered mice
Studies in PKCε−/− mice suggest that PKCε exerts an inhibitory control on PKCδ (that is lost in the PKCε−/− mouse). Klein et al. (98) reported that PKCδ protein expression and Thr505 phosphorylation are increased in PKCε−/− (but not normal) hearts subjected to pressure overload. While Klein et al. did not identify changes in baseline PKCδ protein or phosphorylation in the absence of a hypertrophic stimulus, Gray et al. (70) identified increased PKCδ expression and PKCδ localization to perinuclear structures (a sign of chronic PKCδ activation) in resting cardiomyocytes isolated from PKCε−/− mice.
Our recent studies using an adenoviral-mediated overexpression strategy also unambiguously place PKCδ downstream from a PKCε signaling pathway in cardiomyocytes. However, the nature of this regulatory control runs counter to the expectation based on results obtained in genetically engineered mouse models; as noted in section IVB2, adenoviral-mediated PKCε overexpression leads to a coordinate increase in PKCδ-Thr505/Tyr311/Tyr332 phosphorylation in cardiomyocyte cultures (170). This is a general mechanism that also is observed in other cell types, including cardiac fibroblasts and PKCε−/− MEFs (202). These studies emphasize our still very rudimentary understanding of mechanisms that control nPKC isoform cross-talk. The results also emphasize that results obtained in PKCε−/− mice (which describe the manner in which a particular mouse strain compensates for total body knockout of a signaling protein from embryonic life onward) should be interpreted with caution. Some aspects of a knockout phenotype may be pharmacological curiosities and not necessarily inform studies examining the physiological controls of nPKCs in highly differentiated tissues. These results also seriously undermine confidence in the literature that has exploited an adenoviral overexpression strategy to discriminate nPKC isoform functions in cells. In general, compensatory changes in protein abundance have been used as the sole criteria to exclude any PKC isoform cross-talk. Our results emphasize that this is not sufficient to exclude physiologically relevant forms of nPKC cross-talk, perhaps explaining some of the ambiguous results obtained in previous studies that have utilized this experimental strategy (156).
Our attempts to interrogate the molecular determinants of PKCε-PKCδ cross-talk in cells also exposed a technical limitation imposed by conventional PKC overexpression strategies that is not widely recognized and deserves comment. A typical experiment, depicted in Figure 7B, shows that PKCε overexpression leads to a time-dependent increase in native PKCδ-Thr505 and Tyr311 phosphorylation. However, the GFP-PKCδ fusion construct heterologously overexpressed in the same cells (that can be resolved from native PKCδ due to its slower migration in SDS-PAGE) is constitutively Thr505-phosphorylated and is not influenced by PKCε overexpression (i.e., PKCε-PKCδ cross-talk is not detected on the heterologously overexpressed enzyme). This feature of PKC overexpression studies is underappreciated and may limit its utility as a method to interrogate the physiological controls of PKC phosphorylation in highly differentiated tissues.
B. Some PKC-Directed Antibodies Are State Specific
Western blotting and immunoprecipitation experiments are predicated upon the assumption that anti-PKC antibodies recognize proteins of interest regardless of conformation or posttranslational modifications (i.e., that antibodies do not discriminate pools of enzyme with different phosphorylation patterns). However, we recently reported that a commercially available anti-PKCδ monoclonal antibody (from BD Transduction Laboratories, that has been used widely for almost a decade to detect and immunoprecipitate PKCδ) recognizes PKCδ in resting cardiomyocytes, but artifactually perceives a decline in PKC immunoreactivity under conditions of PMA activation (without any change in PKCδ protein expression, Ref. 174). We showed that PKCδ immunoreactivity is preserved when in vivo PMA treatment is performed in the presence of the PKC inhibitor GF109203X and that PKCδ immunoreactivity is restored by in vitro acid phosphatase treatment, suggesting the recognition epitope for the BD Transduction Laboratory anti-PKCδ antibody encompasses a site for PKC-dependent phosphorylation (or a site that is concealed as a result of a PKC-dependent phosphorylation elsewhere in the protein that alters conformation). Hence, the BD anti-PKCδ antibody is particularly ill-suited for studies that compare PKCδ expression levels in resting and PMA- (or growth factor-) activated samples or studies that examine the relationship between PKCδ phosphorylation and PKCδ downregulation (since it artifactually perceives PKCδ phosphorylation as a decline in total PKCδ protein). These findings suggest caution with regard to the interpretation of published literature using this antibody.
While conformationally sensitive anti-PKC antibodies represent obstacles that can confound research efforts, they also might offer a novel strategy that can be exploited to interrogate PKC functions. Based on the assumption that RACK binding sites are cryptic in the inactive conformation and exposed only following PKC activation, Souroujon et al. (193) used this approach to generate an antibody (termed 14E6, directed against residues 2–145 of PKCε, encompassing the RACK binding site) that selectively recognizes antigenic determinants that are exposed following PKCε activation by lipid cofactors; 14E6 does not recognize the inactive conformation of PKCε (193). This antibody competes with εRACK/β′-COP (425–905, which contains the PKCε binding site) for PKCε binding. By comparing 14E6 immunoreactivity to that of a commercial anti-PKCε antibody (against the terminal 15 residues in the PKCε-V5 domain), these investigators demonstrated that 1) PKCε is localized in an inactive conformation that is not recognized by 14E6 in the nucleus of resting cardiomyocytes, 2) PKCε localizes mainly to perinuclear and cross-striated structures upon activation (this activated form of PKCε is recognized by 14E6), and 3) the activation-dependent increase in PKCε staining with 14E6 is transient, presumably because the 14E6 epitope becomes masked when PKCε binds εRACK in cells.
C. Results of PKC Activity Assays Can Vary, Depending on Assay Conditions
The literature has generally viewed PKCs as generic kinases that phosphorylate substrates in a stereotypical manner (with PKC activity controlled exclusively by translocation events that control access to substrate, rather than any regulatory event that alters the enzyme). Many laboratories have used generic immunocomplex kinase assay kits (that are available from a number of commercial sources) to track PKC activity in cells. However, even a casual review of the literature reveals many inconsistencies (even for ostensibly identical experiments), suggesting that “the devil is in the details.” Variables that can markedly alter the results of in vitro kinase assays and deserve greater attention in future studies include the following: 1) uncertainties with regard to the specificity and/or efficacy of antibodies used in immunoprecipitation experiments. Most commercially available antibodies have been characterized only superficially. While antibodies are generally assumed to recognize all of the enzyme in the cell (regardless of posttranslational modifications), recent experience in our laboratory and by others suggests otherwise. Many anti-PKC antibodies (particularly for PKCδ) do not quantitatively recover all of the immunoreactive enzyme from a cell lysate (at least in part because antibodies that interact with PKCs in a conformationally sensitive manner may be considerably more common than heretofore appreciated) (174). Antibody bias that can lead to the selective recovery of pools of enzyme with distinct posttranslational modifications and catalytic properties; this would seriously undermine the results of an experiment and is only rarely considered. The recent report that antibody binding to the PKCα COOH terminus induces a conformational change that impacts on catalytic activity (i.e., antibody binding is sufficient to inhibit activity, Ref. 233) also could impact on the results of immune complex kinase assays and should be considered. 2) PKCs localize to detergent-insoluble subcellular compartments. The cytoskeleton is an important detergent-insoluble signaling compartment for several PKC isoforms. For example, PKCδ localizes as a tyrosine phosphorylated protein to the detergent-insoluble fraction of H2O2-treated cardiomyocytes. PKC isoforms that reside in detergent-insoluble compartments are not recovered in the extracts used for immunocomplex kinase assays; this pool of enzyme is not captured (and is not monitored) by traditional in vitro kinase assays. 3) Many PKCs coimmunoprecipitate with other serine-threonine kinases or regulatory proteins; the identity of the kinase activity in an immune complex kinase assay must be validated. The interaction between PKCδ and PKD (or Src kinases) constitutes just one example of a PKC-protein interaction that could impact on the results of immunocomplex kinase assays. Other serine/threonine kinases and phosphatases that also might coprecipitate and confound the interpretation of experiments must be considered. 4) The phosphorylation patterns of native and heterologously overexpressed PKCs frequently differ. Current concepts of the controls of PKC phosphorylation are based largely on studies of heterologously overexpressed enzymes. However, depicted in Figure 7B, native and heterologously overexpressed PKC phosphorylation patterns can differ dramatically even in the same cell. 5) Certain posttranslational modifications alter PKC enzymology (including cofactor requirements and substrate specificity). Most PKC kinase assays are predicated on the assumption that PKCs act as generic enzymes to phosphorylate target substrates in a stereotypical manner. Until quite recently, there was only limited anecdotal evidence that PKC activity is subject to more complex and elaborate controls (103). However, our recent studies identify a specific mechanism (involving Tyr311/Thr505 phosphorylation) that leads to a dramatic change in PKCδ-dependent phosphorylation of a physiologically relevant substrate, namely, cTnI (see sect. VC, Fig. 8). These studies emphasize that PKCδ kinase assays are influenced by the choice of the substrate. Since some stimuli alter PKCδ cofactor requirements, results may vary according to the composition of the assay buffer. Mechanisms that control the PKC enzymology in cells generally have not heretofore been considered; these newer concepts identify a real challenge for studies that track PKC activity in cells and emphasize the need for new technologies.
IX. CHALLENGES AND OPPORTUNITIES FOR FUTURE RESEARCH: IDENTIFICATION OF CELLULAR TARGETS FOR INDIVIDUAL PKC ISOFORMS
Despite years of intense research, there is still relatively little information regarding the specific actions (and cellular substrates) of individual PKC isoforms in cells. Novel avenues for PKC research that take advantage of chemical genetic approaches and fluorescence resonance energy transfer (FRET)-based technologies are summarized briefly in this section.
A. Genetically Engineered Kinases
Through a mutagenesis approach directed at the gatekeeper residue, the ATP binding sites of various kinases have been engineered to accept structurally modified ATP analogs with bulky substitutions (that do not bind to endogenous enzymes in the cell; see sect. IV and Fig. 4) (184). This chemical genetic approach (which already has been applied to studies of PKA, Ref. 179) should provide a powerful and elegant strategy to resolve the cellular substrates of individual PKC isoforms.
B. FRET-Based Reporters That Track Intracellular PKC Activity
The Tsien laboratory (217) has developed a FRET-based reporter for PKC (CKAR) consisting of a PKC substrate (GGSGGRFRRFQTLKIKAKAGGSGG, which conforms to a PKC consensus phosphorylation motif and is a less favorable substrate for other kinases) tethered to a phosphothreonine-binding domain (the FHA2 domain from the yeast checkpoint protein rad53p) flanked by yellow- and cyan-shifted green fluorescent protein (YFP and CFP). PKC activation leads to the phosphorylation of the PKC substrate and an intramolecular interaction with the FHA2 phosphothreonine-binding domain, leading to a conformational change that increases FRET from CFP to YFP. Since the FHA2 binds the phosphorylated PKC substrate with relatively low affinity (10 μM), the intramolecular CKAR interaction is reversible, providing a continuous readout of PKC activity in cells (217). Forms of CKAR with targeting sequences that drive the reporter to specific subcellular localizations, including the plasma membrane, the Golgi, mitochondria, nucleus, and the cytosol have been generated and used to expose receptor-dependent PKC activation mechanisms that are dynamically regulated in time and space. Studies to date identify rapid/transient increases in calcium-sensitive PKC (presumably cPKC isoform activity) at the plasma membrane and in the cytosol that are followed by a slower and more sustained increase in PKC (tentatively identified as nPKCs) at the Golgi. These studies also identify a heretofore underestimated role for cellular phosphatases to control PKC activity, particularly in the nuclear compartment (59).
The Blumberg laboratory has taken a different approach to track PKCδ activity, generating a chimeric fusion consisting of PKCδ flanked by YFP and CFP at the NH2 and COOH termini. These studies build on the notion that conformational changes associated with PKC activation alter the distance between the PKCδ NH2 and COOH termini, leading to changes in FRET (as a measure of PKCδ activity) (20). This FRET-based PKCδ activity reporter (which shows native or near-native biological and biochemical activity, including in in vitro lipid cofactor binding assays and in translocation studies in cells) can be used to track PKCδ activity in cells and can be applied to high-throughput screens to evaluate prospective PKCδ ligands.
X. CONCLUSIONS
The term signal transduction was first coined in 1969 by Martin Rodbell in an attempt to describe how cells receive, process, and ultimately transmit information from external “signals,” such as hormones, drugs, or even light. Rodbell viewed signal transduction as a process in which discriminators (or receptors) on the cell surface receive information from outside the cell and transmit this information to an amplifier (or effector such as adenylyl cyclase) via some form of go-between. Borrowing from computer science terminology at the time, he coined the term transducer for the molecule that affects this type of information processing, setting the stage for the identification of G proteins (and his 1994 Nobel Prize in Physiology and Medicine). Rodbell originally viewed “signal transducers” (such as G proteins, or as pertains to this review, PKC isoforms) as “on-off switches” (Fig. 9A). However, this simple model was subsequently revised to incorporate new science showing that many signaling proteins act as “rheostats” to control the amplitude of signaling output (Fig. 9B). Recent studies outlined in this review suggest an even higher level of molecular control, with certain signaling proteins (such as PKCδ) acting as “mini-processors” to integrate input from multiple cellular stimuli, using functionally important conformational changes in response to cofactors, translocation events, and post-translational modifications (phosphorylation, tyrosine nitration, etc.) as a “language” to sense the intracellular microenvironment and adjust signaling output (Fig. 9C). This even more elaborate and nuance type of regulatory control is probably a general mechanism that pertains to many signaling proteins, suggesting that the field of signal transduction is still in a relatively early stage of the discovery process.
FIG. 9.
Evolving concepts of signaling pathway activation. A: the original concepts of linear signal transduction pathways held that stimuli evoke responses via the actions of transducers (such as G proteins or PKC isoforms). B: models of signal transduction were first revised to allow for the presence of multiple stimuli that can converge on a single signaling pathway and alter the amplitude of a signaling response. This could reflect the activation of different pools of a single enzyme. Alternatively, amplitude control might be due to the differential stimulatory properties of full or partial agonists. C: more current models of signaling transduction allow for a high level of signal integration by proteins (such as PKCδ) that are regulated through conformational changes, translocation events, and various posttranslational modifications (phosphorylation, oxidation, tyrosine nitration, etc.) that can underlie stimulus-specific signaling responses.
Acknowledgments
GRANTS
This work was supported by National Heart, Lung, and Blood Institute Grant HL-77860.
References
- 1.Aces P, Beheshti M, Szallasi Z, Li L, Yuspa SH, Blumberg PM. Effect of a tyrosine 155 to phenylalanine mutation of protein kinase C-δ on the proliferative and tumorigenic properties of NIH 3T3 fibroblasts. Carcinogenesis. 2000;21:887–891. doi: 10.1093/carcin/21.5.887. [DOI] [PubMed] [Google Scholar]
- 2.Alessi DR. The protein kinase C inhibitors Ro 318220 and GF 109203X are equally potent inhibitors of MAPKAP kinase-1β (Rsk-2) and p70 S6 kinase. FEBS Lett. 1997;402:121–123. doi: 10.1016/s0014-5793(96)01510-4. [DOI] [PubMed] [Google Scholar]
- 3.Ananthanarayanan B, Stahelin RV, Digman MA, Cho W. Activation mechanisms of conventional protein kinase C isoforms are determined by the ligand affinity and conformational flexibility of their C1 domains. J Biol Chem. 2003;278:46886–46894. doi: 10.1074/jbc.M307853200. [DOI] [PubMed] [Google Scholar]
- 4.Anilkumar N, Parsons M, Monk R, Ng T, Adams JC. Interaction of fascin and protein kinase Cα: a novel intersection in cell adhesion and motility. EMBO J. 2003;22:5390–5402. doi: 10.1093/emboj/cdg521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arimoto T, Takeishi Y, Takahashi H, Shishido T, Niizeki T, Koyama Y, Shiga R, Nozaki N, Nakajima O, Nishimaru K, Abe J, Endoh M, Walsh RA, Goto K, Kubota I. Cardiac-specific overexpression of diacylglycerol kinase-ζ prevents Gq protein-coupled receptor agonist-induced cardiac hypertrophy in transgenic mice. Circulation. 2006;113:60–66. doi: 10.1161/CIRCULATIONAHA.105.560771. [DOI] [PubMed] [Google Scholar]
- 6.Babwah AV, Dale LB, Ferguson SS. Protein kinase C isoform-specific differences in the spatial-temporal regulation and decoding of metabotropic glutamate receptor1a-stimulated second messenger responses. J Biol Chem. 2003;278:5419–5426. doi: 10.1074/jbc.M211053200. [DOI] [PubMed] [Google Scholar]
- 7.Balafanova Z, Bolli R, Zhang J, Zheng Y, Pass JM, Bhatnagar A, Tang XL, Wang O, Cardwell E, Ping P. Nitric oxide induces nitration of PKCε, facilitating PKCε translocation via enhanced PKCε-RACK2 interactions: a novel mechanism of NO-triggered activation of PKCε. J Biol Chem. 2002;277:15021–15027. doi: 10.1074/jbc.M112451200. [DOI] [PubMed] [Google Scholar]
- 8.Banci L, Cavallaro G, Kheifets V, Mochly-Rosen D. Molecular dynamics characterization of the C2 domain of protein kinase Cβ. J Biol Chem. 2002;277:12988–12997. doi: 10.1074/jbc.M106875200. [DOI] [PubMed] [Google Scholar]
- 9.Basu A, Lu D, Sun B, Moor AN, Akkaraju GR, Huang J. Proteolytic activation of protein kinase C-ε by caspase-mediated processing and transduction of antiapoptotic signals. J Biol Chem. 2002;277:41850–41856. doi: 10.1074/jbc.M205997200. [DOI] [PubMed] [Google Scholar]
- 10.Becker KP, Hannun YA. cPKC-dependent sequestration of membrane-recycling components in a subset of recycling endosomes. J Biol Chem. 2003;278:52747–52754. doi: 10.1074/jbc.M305228200. [DOI] [PubMed] [Google Scholar]
- 11.Becker KP, Hannun YA. Isoenzyme-specific translocation of PKC-βII and not PKCβI to a juxtanuclear subset of recycling endosomes: involvement of phospholipase D. J Biol Chem. 2004;279:28251–28256. doi: 10.1074/jbc.M400770200. [DOI] [PubMed] [Google Scholar]
- 12.Benes CH, Wu N, Elia AE, Dharia T, Cantley LC, Soltoff SP. The C2 domain of PKCδ is a phosphotyrosine binding domain. Cell. 2005;121:271–280. doi: 10.1016/j.cell.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 13.Bharti A, Kraeft SK, Gounder M, Pandey P, Jin S, Yuan ZM, Lees-Miller SP, Weichselbaum R, Weaver D, Chen LB, Kufe D, Kharbanda S. Inactivation of DNA-dependent protein kinase by protein kinase Cδ: implications for apoptosis. Mol Cell Biol. 1998;18:6719–6728. doi: 10.1128/mcb.18.11.6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blass M, Kronfeld I, Kazimirsky G, Blumberg PM, Brodie C. Tyrosine phosphorylation of protein kinase Cδ is essential for its apoptotic effect in response to etoposide. Mol Cell Biol. 2002;22:182–195. doi: 10.1128/MCB.22.1.182-195.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blobe GC, Stribling DS, Fabbro D, Stabel S, Hannun YA. Protein kinase C βII specifically binds to and is activated by F-actin. J Biol Chem. 1996;271:15823–15830. doi: 10.1074/jbc.271.26.15823. [DOI] [PubMed] [Google Scholar]
- 16.Bornancin F, Parker PJ. Phosphorylation of threonine 638 critically controls the dephosphorylation and inactivation of protein kinase Cα. Curr Biol. 1996;6:1114–1123. doi: 10.1016/s0960-9822(02)70678-7. [DOI] [PubMed] [Google Scholar]
- 17.Bornancin F, Parker PJ. Phosphorylation of protein kinase C-α on serine 657 controls the accumulation of active enzyme and contributes to its phosphatase-resistant state. J Biol Chem. 1997;272:3544–3549. doi: 10.1074/jbc.272.6.3544. [DOI] [PubMed] [Google Scholar]
- 18.Bourbon NA, Yun J, Kester M. Ceramide directly activates protein kinase C-ζ to regulate a stress-activated protein kinase signaling complex. J Biol Chem. 2000;275:35617–35623. doi: 10.1074/jbc.M007346200. [DOI] [PubMed] [Google Scholar]
- 19.Brandman R, Disatnik MH, Churchill E, Mochly-Rosen D. Peptides derived from the C2 domain of protein kinase Cε modulate εPKC activity and identify potential protein-protein interaction surfaces. J Biol Chem. 2007;282:4113–4123. doi: 10.1074/jbc.M608521200. [DOI] [PubMed] [Google Scholar]
- 20.Braun DC, Garfield SH, Blumberg PM. Analysis by fluorescence resonance energy transfer of the interaction between ligands and protein kinase Cδ in the intact cell. J Biol Chem. 2005;280:8164–8171. doi: 10.1074/jbc.M413896200. [DOI] [PubMed] [Google Scholar]
- 21.Bright R, Mochly-Rosen D. The role of protein kinase C in cerebral ischemic and reperfusion injury. Stroke. 2005;36:2781–2790. doi: 10.1161/01.STR.0000189996.71237.f7. [DOI] [PubMed] [Google Scholar]
- 22.Burkart EM, Sumandea MP, Kobayashi T, Nili M, Martin AF, Homsher E, Solaro RJ. Phosphorylation or glutamic acid substitution at protein kinase C sites on cardiac troponin I differentially depress myofilament tension and shortening velocity. J Biol Chem. 2003;278:11265–11272. doi: 10.1074/jbc.M210712200. [DOI] [PubMed] [Google Scholar]
- 23.Cauthron RD, Carter KB, Liauw S, Steinberg RA. Physiological phosphorylation of protein kinase A at Thr-197 is by a protein kinase A kinase. Mol Cell Biol. 1998;18:1416–1423. doi: 10.1128/mcb.18.3.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cenni V, Doppler H, Sonnenburg ED, Maraldi N, Newton AC, Toker A. Regulation of novel protein kinase Cε by phosphorylation. Biochem J. 2002;363:537–545. doi: 10.1042/0264-6021:3630537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chakraborti T, Das S, Chakraborti S. Proteolytic activation of protein kinase Cα by peroxynitrite in stimulating cytosolic phospholipase A2 in pulmonary endothelium: involvement of a pertussis toxin sensitive protein. Biochemistry. 2005;44:5246–5257. doi: 10.1021/bi0477889. [DOI] [PubMed] [Google Scholar]
- 26.Chalfant CE, Mischak H, Watson JE, Winkler BC, Goodnight J, Farese RV, Cooper DR. Regulation of alternative splicing of protein kinase C-β by insulin. J Biol Chem. 1995;270:13326–13332. doi: 10.1074/jbc.270.22.13326. [DOI] [PubMed] [Google Scholar]
- 27.Chan SL, Lee MC, Tan KO, Yang LK, Lee AS, Flotow H, Fu NY, Butler MS, Soejarto DD, Buss AD, Yu VC. Identification of chelerythrine as an inhibitor of BclXL function. J Biol Chem. 2003;278:20453–20456. doi: 10.1074/jbc.C300138200. [DOI] [PubMed] [Google Scholar]
- 28.Chen D, Purohit A, Halilovic E, Doxsey SJ, Newton AC. Centrosomal anchoring of protein kinase CβII by pericentrin controls microtubule organization, spindle function, cytokinesis. J Biol Chem. 2004;279:4829–4839. doi: 10.1074/jbc.M311196200. [DOI] [PubMed] [Google Scholar]
- 29.Chen DH, Brkanac Z, Verlinde CL, Tan XJ, Bylenok L, Nochlin D, Matsushita M, Lipe H, Wolff J, Fernandez M, Cimino PJ, Bird TD, Raskind WH. Missense mutations in the regulatory domain of PKCβ: a new mechanism for dominant nonepisodic cerebellar ataxia. Am J Hum Genet. 2003;72:839–849. doi: 10.1086/373883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng N, He R, Tian J, Dinauer MC, Ye RD. A critical role of protein kinase Cδ activation loop phosphorylation in formyl-methionyl-leucyl-phenylalanine-induced phosphorylation of p47phox and rapid activation of nicotinamide adenine dinucleotide phosphate oxidase. J Immunol. 2007;179:7720–7728. doi: 10.4049/jimmunol.179.11.7720. [DOI] [PubMed] [Google Scholar]
- 31.Cho W. Membrane targeting by C1 and C2 domains. J Biol Chem. 2001;276:32407–32410. doi: 10.1074/jbc.R100007200. [DOI] [PubMed] [Google Scholar]
- 32.Clarke CJ, Ohanian V, Ohanian J. Noradrenaline and endothelin activate diacylglycerol kinases in caveolae/rafts of rat mesenteric arteries. Am J Physiol Heart Circ Physiol. 2007;292:H2248–H2256. doi: 10.1152/ajpheart.01170.2006. [DOI] [PubMed] [Google Scholar]
- 33.Clerk A. Death by protein kinase C inhibitor. J Mol Cell Cardiol. 2001;33:1773–1776. doi: 10.1006/jmcc.2001.1458. [DOI] [PubMed] [Google Scholar]
- 34.Corbalán-García S, Garcia-García J, Rodríguez-Alfaro JA, Gómez-Fernández JC. A new phosphatidylinositol 4,5-bisphosphate-binding site located in the C2 domain of protein kinase Cα. J Biol Chem. 2003;278:4972–4980. doi: 10.1074/jbc.M209385200. [DOI] [PubMed] [Google Scholar]
- 35.Cross T, Griffiths G, Deacon E, Sallis R, Gough M, Watters D, Lord JM. PKC-δ is an apoptotic lamin kinase. Oncogene. 2000;19:2331–2337. doi: 10.1038/sj.onc.1203555. [DOI] [PubMed] [Google Scholar]
- 36.Crotty T, Cai J, Sakane F, Taketomi A, Prescott SM, Topham MK. Diacylglycerol kinase δ regulates protein kinase C and epidermal growth factor receptor signaling. Proc Natl Acad Sci USA. 2006;103:15485–15490. doi: 10.1073/pnas.0604104103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Csukai M, Chen CH, De Matteis MA, Mochly-Rosen D. The coatomer protein β′-COP, a selective binding protein (RACK) for protein kinase Cθ. J Biol Chem. 1997;272:29200–29206. doi: 10.1074/jbc.272.46.29200. [DOI] [PubMed] [Google Scholar]
- 38.Datta R, Kojima H, Yoshida K, Kufe D. Caspase-3-mediated cleavage of protein kinase Cθ in induction of apoptosis. J Biol Chem. 1997;272:20317–20320. doi: 10.1074/jbc.272.33.20317. [DOI] [PubMed] [Google Scholar]
- 39.Davies SP, Reddy H, Caivano M, Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dekker LV, Parker PJ. Regulated binding of the protein kinase C substrate GAP-43 to the V0/C2 region of protein kinase C-δ. J Biol Chem. 1997;272:12747–12753. doi: 10.1074/jbc.272.19.12747. [DOI] [PubMed] [Google Scholar]
- 41.Dempsey EC, Newton AC, Mochly-Rosen D, Fields AP, Reyland ME, Insel PA, Messing RO. Protein kinase C isozymes and the regulation of diverse cell responses. Am J Physiol Lung Cell Mol Physiol. 2000;279:L429–L438. doi: 10.1152/ajplung.2000.279.3.L429. [DOI] [PubMed] [Google Scholar]
- 42.Denzel A, Hare KJ, Zhang C, Shokat K, Jenkinson EJ, Anderson G, Hayday A. Cutting edge: a chemical genetic system for the analysis of kinases regulating T cell development. J Immunol. 2003;171:519–523. doi: 10.4049/jimmunol.171.2.519. [DOI] [PubMed] [Google Scholar]
- 43.DeVries TA, Neville MC, Reyland ME. Nuclear import of PKCδ is required for apoptosis: identification of a novel nuclear import sequence. EMBO J. 2002;21:6050–6060. doi: 10.1093/emboj/cdf606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Disatnik MH, Buraggi G, Mochly-Rosen D. Localization of protein kinase C isozymes in cardiac myocytes. Exp Cell Res. 1994;210:287–297. doi: 10.1006/excr.1994.1041. [DOI] [PubMed] [Google Scholar]
- 45.Doble BW, Dang X, Ping P, Fandrich RR, Nickel BE, Jin Y, Cattini PA, Kardami E. Phosphorylation of serine 262 in the gap junction protein connexin-43 regulates DNA synthesis in cell-cell contact forming cardiomyocytes. J Cell Sci. 2004;117:507–514. doi: 10.1242/jcs.00889. [DOI] [PubMed] [Google Scholar]
- 46.Doble BW, Ping P, Kardami E. The ε subtype of protein kinase C is required for cardiomyocyte connexin-43 phosphorylation. Circ Res. 2000;86:293–301. doi: 10.1161/01.res.86.3.293. [DOI] [PubMed] [Google Scholar]
- 47.Dries DR, Gallegos LL, Newton AC. A single residue in the C1 domain sensitizes novel protein kinase C isoforms to cellular diacylglycerol production. J Biol Chem. 2007;282:826–830. doi: 10.1074/jbc.C600268200. [DOI] [PubMed] [Google Scholar]
- 48.Dutil EM, Toker A, Newton AC. Regulation of conventional protein kinase C isozymes by phosphoinositide-dependent kinase 1. Curr Biol. 1998;8:1366–1375. doi: 10.1016/s0960-9822(98)00017-7. [DOI] [PubMed] [Google Scholar]
- 49.Edwards AS, Faux MC, Scott JD, Newton AC. Carboxyl-terminal phosphorylation regulates the function and subcellular localization of protein kinase C-βII. J Biol Chem. 1999;274:6461–6468. doi: 10.1074/jbc.274.10.6461. [DOI] [PubMed] [Google Scholar]
- 50.Edwards AS, Newton AC. Phosphorylation at conserved carboxyl-terminal hydrophobic motif regulates the catalytic and regulatory domains of protein kinase C. J Biol Chem. 1997;272:18382–18390. doi: 10.1074/jbc.272.29.18382. [DOI] [PubMed] [Google Scholar]
- 51.Edwards AS, Newton AC. Regulation of protein kinase C βII by its C2 domain. Biochemistry. 1997;36:15615–15623. doi: 10.1021/bi9718752. [DOI] [PubMed] [Google Scholar]
- 52.Emoto Y, Manome Y, Meinhardt G, Kisaki H, Kharbanda S, Robertson M, Ghayur T, Wong WW, Kamen R, Weichselbaum R, Kufe D. Proteolytic activation of protein kinase C δ by an ICE-like protease in apoptotic cells. EMBO J. 1995;14:6148–6156. doi: 10.1002/j.1460-2075.1995.tb00305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Emrick MA, Lee T, Starkey PJ, Mumby MC, Resing KA, Ahn NG. The gatekeeper residue controls autoactivation of ERK2 via a pathway of intramolecular connectivity. Proc Natl Acad Sci USA. 2006;103:18101–18106. doi: 10.1073/pnas.0608849103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Evans JH, Murray D, Leslie CC, Falke JJ. Specific translocation of protein kinase Cα to the plasma membrane requires both Ca2+ and PIP2 recognition by its C2 domain. Mol Biol Cell. 2006;17:56–66. doi: 10.1091/mbc.E05-06-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feng X, Becker KP, Stribling SD, Peters KG, Hannun YA. Regulation of receptor-mediated protein kinase C membrane trafficking by autophosphorylation. J Biol Chem. 2000;275:17024–17034. doi: 10.1074/jbc.275.22.17024. [DOI] [PubMed] [Google Scholar]
- 56.Feng X, Hannun YA. An essential role for autophosphorylation in the dissociation of activated protein kinase C from the plasma membrane. J Biol Chem. 1998;273:26870–26874. doi: 10.1074/jbc.273.41.26870. [DOI] [PubMed] [Google Scholar]
- 57.Fox TE, Houck KL, O’Neill SM, Nagarajan M, Stover TC, Pomianowski PT, Unal O, Yun JK, Naides SJ, Kester M. Ceramide recruits and activates protein kinase C-ζ within structured membrane microdomains. J Biol Chem. 2007;282:12450–12457. doi: 10.1074/jbc.M700082200. [DOI] [PubMed] [Google Scholar]
- 58.Frutos S, Moscat J, Diaz-Meco MT. Cleavage of ζPKC but not λιPKC by caspase-3 during UV-induced apoptos. J Biol Chem. 1999;274:10765–10770. doi: 10.1074/jbc.274.16.10765. [DOI] [PubMed] [Google Scholar]
- 59.Gallegos LL, Kunkel MT, Newton AC. Targeting protein kinase C activity reporter to discrete intracellular regions reveals spatiotemporal differences in agonist-dependent signaling. J Biol Chem. 2006;281:30947–30956. doi: 10.1074/jbc.M603741200. [DOI] [PubMed] [Google Scholar]
- 60.Gao T, Newton AC. The turn motif is a phosphorylation switch that regulates the binding of Hsp70 to protein kinase C. J Biol Chem. 2002;277:31585–31592. doi: 10.1074/jbc.M204335200. [DOI] [PubMed] [Google Scholar]
- 61.Gao T, Newton AC. Invariant leu preceding turn motif phosphorylation site controls the interaction of protein kinase C with hsp70. J Biol Chem. 2006;281:32461–32468. doi: 10.1074/jbc.M604076200. [DOI] [PubMed] [Google Scholar]
- 62.Gao T, Toker A, Newton AC. The carboxyl terminus of protein kinase C provides a switch to regulate its interaction with the phosphoinositide-dependent kinase, PDK-1. J Biol Chem. 2001;276:19588–19596. doi: 10.1074/jbc.M101357200. [DOI] [PubMed] [Google Scholar]
- 63.Garcia-Paramio P, Cabrerizo Y, Bornancin F, Parker PJ. The broad specificity of dominant inhibitory protein kinase C mutants infers a common step in phosphorylation. Biochem J. 1998;333:631–636. doi: 10.1042/bj3330631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Giorgione JR, Lin JH, McCammon JA, Newton AC. Increased membrane affinity of the C1 domain of protein kinase Cδ compensates for the lack of involvement of its C2 domain in membrane recruitment. J Biol Chem. 2006;281:1660–1669. doi: 10.1074/jbc.M510251200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gökmen-Polar Y, Fields AP. Mapping of a molecular determinant for protein kinase C-βII isozyme function. J Biol Chem. 1998;273:20261–20266. doi: 10.1074/jbc.273.32.20261. [DOI] [PubMed] [Google Scholar]
- 66.Gonfloni S, Williams JC, Hattula K, Weijland A, Wierenga RK, Superti-Furga G. The role of the linker between the SH2 domain and catalytic domain in the regulation and function of Src. EMBO J. 1997;16:7261–7271. doi: 10.1093/emboj/16.24.7261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goode NT, Hajibagheri N, Parker PJ. Protein kinase C (PKC)-induced PKC down-regulation: association with up-regulation of vesicle traffic. J Biol Chem. 1995;270:2669–2673. doi: 10.1074/jbc.270.6.2669. [DOI] [PubMed] [Google Scholar]
- 68.Goodnight JA, Mischak H, Kolch W, Mushinski JF. Immunocytochemical localization of eight protein kinase C isozymes over-expressed in NIH 3T3 fibroblasts. Isoform-specific association with microfilaments, Golgi, endoplasmic reticulum, nuclear and cell membranes. J Biol Chem. 1995;270:9991–10001. doi: 10.1074/jbc.270.17.9991. [DOI] [PubMed] [Google Scholar]
- 69.Gopalakrishna R, Jaken S. Protein kinase C signaling and oxidative stress. Free Radic Biol Med. 2000;28:1349–1361. doi: 10.1016/s0891-5849(00)00221-5. [DOI] [PubMed] [Google Scholar]
- 70.Gray MO, Zhou HZ, Schafhalter-Zoppoth I, Zhu P, Mochly-Rosen D, Messing RO. Preservation of base-line hemodynamic function and loss of inducible cardioprotection in adult mice lacking protein kinase C-ε. J Biol Chem. 2004;279:3596–3604. doi: 10.1074/jbc.M311459200. [DOI] [PubMed] [Google Scholar]
- 71.Griner EM, Kazanietz MG. Protein kinase C and other diacylglycerol effectors in cancer. Nat Rev Cancer. 2007;7:281–294. doi: 10.1038/nrc2110. [DOI] [PubMed] [Google Scholar]
- 72.Grodsky N, Li Y, Bouzida D, Love R, Jensen J, Nodes B, Nonomiya J, Grant S. Structure of the catalytic domain of human protein kinase C βII complexed with a bisindolylmaleimide inhibitor. Biochemistry. 2006;45:13970–13981. doi: 10.1021/bi061128h. [DOI] [PubMed] [Google Scholar]
- 73.Hamaguchi A, Suzuki E, Murayama K, Fujimura T, Hikita T, Iwabuchi K, Handa K, Withers DA, Masters SC, Fu H, Hakomori S. Sphingosine-dependent protein kinase-1 (SDK1), directed to 14-3-3, is identified as the kinase domain of PKCδ. J Biol Chem. 2003;278:41557–41565. doi: 10.1074/jbc.M305294200. [DOI] [PubMed] [Google Scholar]
- 74.Herberg FW, Zimmermann B, McGlone M, Taylor SS. Importance of the A-helix of the catalytic subunit of cAMP-dependent protein kinase for stability and for orienting subdomains at the cleft interface. Protein Sci. 1997;6:569–579. doi: 10.1002/pro.5560060306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hommel U, Zurini M, Luyten M. Solution structure of a cysteine rich domain of rat protein kinase C. Nat Struct Biol. 1994;1:383–387. doi: 10.1038/nsb0694-383. [DOI] [PubMed] [Google Scholar]
- 76.Hu T, Exton JH. Mechanisms of regulation of phospholipase D1 by protein kinase Cα. J Biol Chem. 2003;278:2348–2355. doi: 10.1074/jbc.M210093200. [DOI] [PubMed] [Google Scholar]
- 77.Hu T, Exton JH. Protein kinase Cα translocates to the perinuclear region to activate phospholipase D1. J Biol Chem. 2004;279:35702–35708. doi: 10.1074/jbc.M402372200. [DOI] [PubMed] [Google Scholar]
- 78.Hurley JH, Misra S. Signaling and subcellular targeting by membrane-binding domains. Annu Rev Biophys Biomol Struct. 2000;29:49–79. doi: 10.1146/annurev.biophys.29.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huwiler A, Fabbro D, Pfeilschifter J. Selective ceramide binding to protein kinase C-α and -δ isoenzymes in renal mesangial cells. Biochemistry. 1998;37:14556–14562. doi: 10.1021/bi981401i. [DOI] [PubMed] [Google Scholar]
- 80.Idkowiak-Baldys J, Becker KP, Kitatani K, Hannun YA. Dynamic sequestration of the recycling compartment by classical protein kinase C. J Biol Chem. 2006;281:22321–22331. doi: 10.1074/jbc.M512540200. [DOI] [PubMed] [Google Scholar]
- 81.Imai S, Kai M, Yamada K, Kanoh H, Sakane F. The plasma membrane translocation of diacylglycerol kinase δ1 is negatively regulated by conventional protein kinase C-dependent phosphorylation at Ser-22 and Ser-26 within the pleckstrin homology domain. Biochem J. 2004;382:957–966. doi: 10.1042/BJ20040681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ivaska J, Vuoriluoto K, Huovinen T, Izawa I, Inagaki M, Parker PJ. PKCε-mediated phosphorylation of vimentin controls integrin recycling and motility. EMBO J. 2005;24:3834–3845. doi: 10.1038/sj.emboj.7600847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ivaska J, Whelan RD, Watson R, Parker PJ. PKC-ε controls the traffic of β1-integrins in motile cells. EMBO J. 2002;21:3608–3619. doi: 10.1093/emboj/cdf371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Iyer GH, Garrod S, Woods VL, Jr, Taylor SS. Catalytic independent functions of a protein kinase as revealed by a kinase-dead mutant: study of the Lys72His mutant of cAMP-dependent kinase. J Mol Biol. 2005;351:1110–1122. doi: 10.1016/j.jmb.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 85.Iyer GH, Moore MJ, Taylor SS. Consequences of lysine 72 mutation on the phosphorylation and activation state of cAMP-dependent kinase. J Biol Chem. 2005;280:8800–8807. doi: 10.1074/jbc.M407586200. [DOI] [PubMed] [Google Scholar]
- 86.Jiang K, Apostolatos AH, Ghansah T, Watson JE, Vickers T, Cooper DR, Epling-Burnette PK, Patel NA. Identification of a novel antiapoptotic human protein kinase c delta isoform, PKC-deltaVIII in NT2 cells. Biochemistry. 2008;47:787–797. doi: 10.1021/bi7019782. [DOI] [PubMed] [Google Scholar]
- 87.Johnson JA, Gray MO, Chen CH, Mochly-Rosen D. A protein kinase C translocation inhibitor as an isozyme-selective antagonist of cardiac function. J Biol Chem. 1996;271:24962–24966. doi: 10.1074/jbc.271.40.24962. [DOI] [PubMed] [Google Scholar]
- 88.Jose Lopez-Andreo M, Gomez-Fernandez JC, Corbalan-Garcia S. The simultaneous production of phosphatidic acid and diacylglycerol is essential for the translocation of protein kinase Cε to the plasma membrane in RBL-2H3 cells. Mol Biol Cell. 2003;14:4885–4895. doi: 10.1091/mbc.E03-05-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Joseloff E, Cataisson C, Aamodt H, Ocheni H, Blumberg P, Kraker AJ, Yuspa SH. Src family kinases phosphorylate protein kinase C-δ on tyrosine residues and modify the neoplastic phenotype of skin keratinocytes. J Biol Chem. 2002;277:12318–12323. doi: 10.1074/jbc.M111618200. [DOI] [PubMed] [Google Scholar]
- 90.Kajimoto T, Shirai Y, Sakai N, Yamamoto T, Matsuzaki H, Kikkawa U, Saito N. Ceramide-induced apoptosis by translocation, phosphorylation, activation of protein kinase Cδ in the Golgi complex. J Biol Chem. 2004;279:12668–12676. doi: 10.1074/jbc.M312350200. [DOI] [PubMed] [Google Scholar]
- 91.Kashiwagi K, Shirai Y, Kuriyama M, Sakai N, Saito N. Importance of C1B domain for lipid messenger-induced targeting of protein kinase C. J Biol Chem. 2002;277:18037–18045. doi: 10.1074/jbc.M111761200. [DOI] [PubMed] [Google Scholar]
- 92.Kawakami Y, Kitaura J, Yao L, McHenry RW, Kawakami Y, Newton AC, Kang S, Kato RM, Leitges M, Rawlings DJ, Kawakami T. A Ras activation pathway dependent on Syk phosphorylation of protein kinase C. Proc Natl Acad Sci USA. 2003;100:9470–9475. doi: 10.1073/pnas.1633695100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kazanietz MG. Novel “nonkinase” phorbol ester receptors: the C1 domain connection. Mol Pharmacol. 2002;61:759–767. doi: 10.1124/mol.61.4.759. [DOI] [PubMed] [Google Scholar]
- 94.Keranen LM, Dutil EM, Newton AC. Protein kinase C is regulated in vivo by three functionally distinct phosphorylations. Curr Biol. 1995;5:1394–1403. doi: 10.1016/s0960-9822(95)00277-6. [DOI] [PubMed] [Google Scholar]
- 95.Keranen LM, Newton AC. Ca2+ differentially regulates conventional protein kinase Cs′ membrane interaction and activation. J Biol Chem. 1997;272:25959–25967. doi: 10.1074/jbc.272.41.25959. [DOI] [PubMed] [Google Scholar]
- 96.Kheifets V, Bright R, Inagaki K, Schechtman D, Mochly-Rosen D. Protein kinase Cδ-annexin V interaction: a required step in δPKC translocation and function. J Biol Chem. 2006;281:23218–23226. doi: 10.1074/jbc.M602075200. [DOI] [PubMed] [Google Scholar]
- 97.Kitatani K, Idkowiak-Baldys J, Hannun YA. Mechanism of inhibition of sequestration of protein kinase C-α/βII by ceramide: roles of ceramide-activated protein phosphatases and phosphorylation/dephosphorylation of protein kinase C-α/β II on threonine 638/641. J Biol Chem. 2007;282:20647–20656. doi: 10.1074/jbc.M609162200. [DOI] [PubMed] [Google Scholar]
- 98.Klein G, Schaefer A, Hilfiker-Kleiner D, Oppermann D, Shukla P, Quint A, Podewski E, Hilfiker A, Schroder F, Leitges M, Drexler H. Increased collagen deposition and diastolic dysfunction but preserved myocardial hypertrophy after pressure overload in mice lacking PKCε. Circ Res. 2005;96:748–755. doi: 10.1161/01.RES.0000161999.86198.1e. [DOI] [PubMed] [Google Scholar]
- 99.Knapp LT, Kanterewicz BI, Hayes EL, Klann E. Peroxynitrite-induced tyrosine nitration and inhibition of protein kinase C. Biochem Biophys Res Commun. 2001;286:764–770. doi: 10.1006/bbrc.2001.5448. [DOI] [PubMed] [Google Scholar]
- 100.Knighton DR, Zheng JH, Ten Eyck LF, Ashford VA, Xuong NH, Taylor SS, Sowadski JM. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253:407–414. doi: 10.1126/science.1862342. [DOI] [PubMed] [Google Scholar]
- 101.Knighton DR, Zheng JH, Ten Eyck LF, Xuong NH, Taylor SS, Sowadski JM. Structure of a peptide inhibitor bound to the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253:414–420. doi: 10.1126/science.1862343. [DOI] [PubMed] [Google Scholar]
- 102.Konishi H, Tanaka M, Takemura Y, Matsuzaki H, Ono Y, Kikkawa U, Nishizuka Y. Activation of protein kinase C by tyrosine phosphorylation in response to H2O2. Proc Natl Acad Sci USA. 1997;94:11233–11237. doi: 10.1073/pnas.94.21.11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Konishi H, Yamauchi E, Taniguchi H, Yamamoto T, Matsuzaki H, Takemura Y, Ohmae K, Kikkawa U, Nishizuka Y. Phosphorylation sites of protein kinase Cδ in H2O2-treated cells and its activation by tyrosine kinase in vitro. Proc Natl Acad Sci USA. 2001;98:6587–6592. doi: 10.1073/pnas.111158798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kronfeld I, Kazimirsky G, Lorenzo PS, Garfield SH, Blumberg PM, Brodie C. Phosphorylation of protein kinase Cδ on distinct tyrosine residues regulates specific cellular functions. J Biol Chem. 2000;275:35491–35498. doi: 10.1074/jbc.M005991200. [DOI] [PubMed] [Google Scholar]
- 105.Kumar S, Bharti A, Mishra NC, Raina D, Kharbanda S, Saxena S, Kufe D. Targeting of the c-Abl tyrosine kinase to mitochondria in the necrotic cell death response to oxidative stress. J Biol Chem. 2001;276:17281–17285. doi: 10.1074/jbc.M101414200. [DOI] [PubMed] [Google Scholar]
- 106.Le Good JA, Ziegler WH, Parekh DB, Alessi DR, Cohen P, Parker PJ. Protein kinase C isotypes controlled by phosphoinositide 3-kinase through the protein kinase PDK1. Science. 1998;281:2042–2045. doi: 10.1126/science.281.5385.2042. [DOI] [PubMed] [Google Scholar]
- 107.Lee SJ, Kim DC, Choi BH, Ha H, Kim KT. Regulation of p53 by activated protein kinase C-δ during nitric oxide-induced dopaminergic cell death. J Biol Chem. 2006;281:2215–2224. doi: 10.1074/jbc.M509509200. [DOI] [PubMed] [Google Scholar]
- 108.Lehel C, Olah Z, Jakab G, Anderson WB. Protein kinase C-ε is localized to the Golgi via its zinc-finger domain and modulates Golgi function. Proc Natl Acad Sci USA. 1995;92:1406–1410. doi: 10.1073/pnas.92.5.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Leitges M, Elis W, Gimborn K, Huber M. Rottlerin-independent attenuation of pervanadate-induced tyrosine phosphorylation events by protein kinase C-δ in hemopoietic cells. Lab Invest. 2001;81:1087–1095. doi: 10.1038/labinvest.3780321. [DOI] [PubMed] [Google Scholar]
- 110.Leitges M, Gimborn K, Elis W, Kalesnikoff J, Hughes MR, Krystal G, Huber M. Protein kinase C-δ is a negative regulator of antigen-induced mast cell degranulation. Mol Cell Biol. 2002;22:3970–3980. doi: 10.1128/MCB.22.12.3970-3980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li W, Chen XH, Kelley CA, Alimandi M, Zhang J, Chen Q, Bottaro DP, Pierce JH. Identification of tyrosine 187 as a protein kinase C-δ phosphorylation site. J Biol Chem. 1996;271:26404–26409. doi: 10.1074/jbc.271.42.26404. [DOI] [PubMed] [Google Scholar]
- 112.Li W, Zhang J, Bottaro DP, Pierce JH. Identification of serine 643 of protein kinase C-δ as an important autophosphorylation site for its enzymatic activity. J Biol Chem. 1997;272:24550–24555. doi: 10.1074/jbc.272.39.24550. [DOI] [PubMed] [Google Scholar]
- 113.Lin D, Barnett M, Lobell S, Madgwick D, Shanks D, Willard L, Zampighi GA, Takemoto DJ. PKCγ knockout mouse lenses are more susceptible to oxidative stress damage. J Exp Biol. 2006;209:4371–4378. doi: 10.1242/jeb.02524. [DOI] [PubMed] [Google Scholar]
- 114.Lin D, Shanks D, Prakash O, Takemoto DJ. Protein kinase C-γ mutations in the C1B domain cause caspase-3-linked apoptosis in lens epithelial cells through gap junctions. Exp Eye Res. 2007;85:113–122. doi: 10.1016/j.exer.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lin D, Takemoto DJ. Oxidative activation of protein kinase Cγ through the C1 domain. Effects on gap junctions. J Biol Chem. 2005;280:13682–13693. doi: 10.1074/jbc.M407762200. [DOI] [PubMed] [Google Scholar]
- 116.Lin D, Zhou J, Zelenka PS, Takemoto DJ. Protein kinase Cγ regulation of gap junction activity through caveolin-1-containing lipid rafts. Invest Ophthalmol Vis Sci. 2003;44:5259–5268. doi: 10.1167/iovs.03-0296. [DOI] [PubMed] [Google Scholar]
- 117.Ling M, Sunesson L, Larsson C. Comparison of the PKCα and the PKCε C1b domains: identification of residues critical for PKCε-mediated neurite induction. J Mol Biol. 2007;368:951–965. doi: 10.1016/j.jmb.2007.02.073. [DOI] [PubMed] [Google Scholar]
- 118.Ling M, Troller U, Zeidman R, Stensman H, Schultz A, Larsson C. Identification of conserved amino acids N-terminal of the PKC-ε C1b domain crucial for protein kinase C-ε-mediated induction of neurite outgrowth. J Biol Chem. 2005;280:17910–17919. doi: 10.1074/jbc.M412036200. [DOI] [PubMed] [Google Scholar]
- 119.Liu XJ, He AB, Chang YS, Fang FD. Atypical protein kinase C in glucose metabolism. Cell Signal. 2006;18:2071–2076. doi: 10.1016/j.cellsig.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 120.Liu Y, Belkina NV, Graham C, Shaw S. Independence of protein kinase C-δ activity from activation loop phosphorylation: structural basis and altered functions in cells. J Biol Chem. 2006;281:12102–12111. doi: 10.1074/jbc.M600508200. [DOI] [PubMed] [Google Scholar]
- 121.Liu Y, Graham C, Li A, Fisher RJ, Shaw S. Phosphorylation of the protein kinase C-θ activation loop and hydrophobic motif regulates its kinase activity, but only activation loop phosphorylation is critical to in vivo nuclear-factor-κB induction. Biochem J. 2002;361:255–265. doi: 10.1042/bj3610255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Liu Y, Witte S, Liu YC, Doyle M, Elly C, Altman A. Regulation of protein kinase Cθ function during T cell activation by Lck-mediated tyrosine phosphorylation. J Biol Chem. 2000;275:3603–3609. doi: 10.1074/jbc.275.5.3603. [DOI] [PubMed] [Google Scholar]
- 123.Lopez-Lluch G, Bird MM, Canas B, Godovac-Zimmerman J, Ridley A, Segal AW, Dekker LV. Protein kinase C-δ C2-like domain is a binding site for actin and enables actin redistribution in neutrophils. Biochem J. 2001;357:39–47. doi: 10.1042/0264-6021:3570039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lu W, Finnis S, Xiang C, Lee HK, Markowitz Y, Okhrimenko H, Brodie C. Tyrosine 311 is phosphorylated by c-Abl and promotes the apoptotic effect of PKCδ in glioma cells. Biochem Biophys Res Commun. 2007;352:431–436. doi: 10.1016/j.bbrc.2006.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lu W, Lee HK, Xiang C, Finniss S, Brodie C. The phosphorylation of tyrosine 332 is necessary for the caspase 3-dependent cleavage of PKCδ and the regulation of cell apoptosis. Cell Signal. 2007;19:2165–2173. doi: 10.1016/j.cellsig.2007.06.015. [DOI] [PubMed] [Google Scholar]
- 126.Luo B, Prescott SM, Topham MK. Association of diacylglycerol kinase-ζ with protein kinase C-α: spatial regulation of diacylglycerol signaling. J Cell Biol. 2003;160:929–937. doi: 10.1083/jcb.200208120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Luo B, Prescott SM, Topham MK. Protein kinase C-α phosphorylates and negatively regulates diacylglycerol kinase-ζ. J Biol Chem. 2003;278:39542–39547. doi: 10.1074/jbc.M307153200. [DOI] [PubMed] [Google Scholar]
- 128.Mackay K, Mochly-Rosen D. Localization, anchoring, functions of protein kinase C isozymes in the heart. J Mol Cell Cardiol. 2001;33:1301–1307. doi: 10.1006/jmcc.2001.1400. [DOI] [PubMed] [Google Scholar]
- 129.Maeda Y, Beznoussenko GV, Van Lint J, Mironov AA, Malhotra V. Recruitment of protein kinase D to the trans-Golgi network via the first cysteine-rich domain. EMBO J. 2001;20:5982–5990. doi: 10.1093/emboj/20.21.5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.McCahill A, Warwicker J, Bolger GB, Houslay MD, Yarwood SJ. The RACK1 scaffold protein: a dynamic cog in cell response mechanisms. Mol Pharmacol. 2002;62:1261–1273. doi: 10.1124/mol.62.6.1261. [DOI] [PubMed] [Google Scholar]
- 131.Medkova M, Cho W. Mutagenesis of the C2 domain of protein kinase C-α. Differential roles of Ca2+ ligands and membrane binding residues. J Biol Chem. 1998;273:17544–17552. doi: 10.1074/jbc.273.28.17544. [DOI] [PubMed] [Google Scholar]
- 132.Melowic HR, Stahelin RV, Blatner NR, Tian W, Hayashi K, Altman A, Cho W. Mechanism of diacylglycerol-induced membrane targeting and activation of protein kinase Cθ. J Biol Chem. 2007;282:21467–21476. doi: 10.1074/jbc.M700119200. [DOI] [PubMed] [Google Scholar]
- 133.Messerschmidt A, Macieira S, Velarde M, Badeker M, Benda C, Jestel A, Brandstetter H, Neuefeind T, Blaesse M. Crystal structure of the catalytic domain of human atypical protein kinase Cι reveals interaction mode of phosphorylation site in turn motif. J Mol Biol. 2005;352:918–931. doi: 10.1016/j.jmb.2005.07.060. [DOI] [PubMed] [Google Scholar]
- 134.Mineo C, Ying YS, Chapline C, Jaken S, Anderson RG. Targeting of protein kinase Cα to caveolae. J Cell Biol. 1998;141:601–610. doi: 10.1083/jcb.141.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mizuno K, Noda K, Araki T, Imaoka T, Kobayashi Y, Akita Y, Shimonaka M, Kishi S, Ohno S. The proteolytic cleavage of protein kinase C isotypes, which generates kinase and regulatory fragments, correlates with Fas-mediated and 12-O-tetradecanoyl-phorbol-13-acetate-induced apoptosis. Eur J Biochem. 1997;250:7–18. doi: 10.1111/j.1432-1033.1997.00007.x. [DOI] [PubMed] [Google Scholar]
- 136.Moore MJ, Kanter JR, Jones KC, Taylor SS. Phosphorylation of the catalytic subunit of protein kinase A. Autophosphorylation versus phosphorylation by phosphoinositide-dependent kinase-1. J Biol Chem. 2002;277:47878–47884. doi: 10.1074/jbc.M204970200. [DOI] [PubMed] [Google Scholar]
- 137.Morgan A, Burgoyne RD, Barclay JW, Craig TJ, Prescott GR, Ciufo LF, Evans GJ, Graham ME. Regulation of exocytosis by protein kinase C. Biochem Soc Trans. 2005;33:1341–1344. doi: 10.1042/BST0331341. [DOI] [PubMed] [Google Scholar]
- 138.Moscat J, Diaz-Meco MT, Albert A, Campuzano S. Cell signaling and function organized by PB1 domain interactions. Mol Cell. 2006;23:631–640. doi: 10.1016/j.molcel.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 139.Moscat J, Rennert P, Diaz-Meco MT. PKCζ at the crossroad of NF-κB and Jak1/Stat6 signaling pathways. Cell Death Differ. 2006;13:702–711. doi: 10.1038/sj.cdd.4401823. [DOI] [PubMed] [Google Scholar]
- 140.Muller G, Ayoub M, Storz P, Rennecke J, Fabbro D, Pfizenmaier K. PKC-ζ is a molecular switch in signal transduction of TNF-α, bifunctionally regulated by ceramide and arachidonic acid. EMBO J. 1995;14:1961–1969. doi: 10.1002/j.1460-2075.1995.tb07188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Newton AC, Johnson JE. Protein kinase C: a paradigm for regulation of protein function by two membrane-targeting modules. Biochim Biophys Acta. 1998;1376:155–172. doi: 10.1016/s0304-4157(98)00003-3. [DOI] [PubMed] [Google Scholar]
- 142.Ng T, Squire A, Hansra G, Bornancin F, Prevostel C, Hanby A, Harris W, Barnes D, Schmidt S, Mellor H, Bastiaens PI, Parker PJ. Imaging protein kinase Cα activation in cells. Science. 1999;283:2085–2089. doi: 10.1126/science.283.5410.2085. [DOI] [PubMed] [Google Scholar]
- 143.Nguyen TA, Takemoto LJ, Takemoto DJ. Inhibition of gap junction activity through the release of the C1B domain of protein kinase Cγ from 14-3-3: identification of PKCγ-binding sites. J Biol Chem. 2004;279:52714–52725. doi: 10.1074/jbc.M403040200. [DOI] [PubMed] [Google Scholar]
- 144.Oancea E, Teruel MN, Quest AF, Meyer T. Green fluorescent protein (GFP)-tagged cysteine-rich domains from protein kinase C as fluorescent indicators for diacylglycerol signaling in living cells. J Cell Biol. 1998;140:485–498. doi: 10.1083/jcb.140.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ochoa WF, Garcia-Garcia J, Fita I, Corbalan-Garcia S, Verdaguer N, Gomez-Fernandez JC. Structure of the C2 domain from novel protein kinase Cε A membrane binding model for Ca2+-independent C2 domains. J Mol Biol. 2001;311:837–849. doi: 10.1006/jmbi.2001.4910. [DOI] [PubMed] [Google Scholar]
- 146.O’Connell MJ, Raleigh JM, Verkade HM, Nurse P. Chk1 is a wee1 kinase in the G2 DNA damage checkpoint inhibiting cdc2 by Y15 phosphorylation. EMBO J. 1997;16:545–554. doi: 10.1093/emboj/16.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Odawara M, Kadowaki T, Yamamoto R, Shibasaki Y, Tobe K, Accili D, Bevins C, Mikami Y, Matsuura N, Akanuma Y. Human diabetes associated with a mutation in the tyrosine kinase domain of the insulin receptor. Science. 1989;245:66–68. doi: 10.1126/science.2544998. [DOI] [PubMed] [Google Scholar]
- 148.Oka N, Yamamoto M, Schwencke C, Kawabe JI, Ebina T, Ohno S, Couet J, Lisanti MP, Ishikawa Y. Caveolin interaction with protein kinase C: isoenzyme-dependent regulation of kinase activity by the caveolin scaffolding domain peptide. J Biol Chem. 1997;272:33416–33421. doi: 10.1074/jbc.272.52.33416. [DOI] [PubMed] [Google Scholar]
- 149.Okhrimenko H, Lu W, Jiang C, Ju D, Blumberg PM, Gomel R, Kazimirsky G, Brodie C. Roles of tyrosine phosphorylation and cleavage of PKCδ in its protective effect against trail-induced apoptosis. J Biol Chem. 2005;280:23643–23652. doi: 10.1074/jbc.M501374200. [DOI] [PubMed] [Google Scholar]
- 150.Okhrimenko H, Lu W, Xiang C, Hamburger N, Kazimirsky G, Brodie C. Protein kinase C-ε regulates the apoptosis and survival of glioma cells. Cancer Res. 2005;65:7301–7309. doi: 10.1158/0008-5472.CAN-05-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Pappa H, Murray-Rust J, Dekker LV, Parker PJ, McDonald NQ. Crystal structure of the C2 domain from protein kinase C-δ. Structure. 1998;6:885–894. doi: 10.1016/s0969-2126(98)00090-2. [DOI] [PubMed] [Google Scholar]
- 152.Parekh D, Ziegler W, Yonezawa K, Hara K, Parker PJ. Mammalian TOR controls one of two kinase pathways acting upon nPKCδ and nPKCε. J Biol Chem. 1999;274:34758–34764. doi: 10.1074/jbc.274.49.34758. [DOI] [PubMed] [Google Scholar]
- 153.Parekh DB, Ziegler W, Parker PJ. Multiple pathways control protein kinase C phosphorylation. EMBO J. 2000;19:496–503. doi: 10.1093/emboj/19.4.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Parton RG, Simons K. The multiple faces of caveolae. Nat Rev Mol Cell Biol. 2007;8:185–194. doi: 10.1038/nrm2122. [DOI] [PubMed] [Google Scholar]
- 155.Perander M, Bjorkoy G, Johansen T. Nuclear import and export signals enable rapid nucleocytoplasmic shuttling of the atypical protein kinase C-λ. J Biol Chem. 2001;276:13015–13024. doi: 10.1074/jbc.M010356200. [DOI] [PubMed] [Google Scholar]
- 156.Porter MJ, Heidkamp MC, Scully BT, Patel N, Martin JL, Samarel AM. Isoenzyme-selective regulation of SERCA2 gene expression by protein kinase C in neonatal rat ventricular myocytes. Am J Physiol Cell Physiol. 2003;285:C39–C47. doi: 10.1152/ajpcell.00461.2002. [DOI] [PubMed] [Google Scholar]
- 157.Prekeris R, Hernandez RM, Mayhew MW, White MK, Terrian DM. Molecular analysis of the interactions between protein kinase C-ε and filamentous actin. J Biol Chem. 1998;273:26790–26798. doi: 10.1074/jbc.273.41.26790. [DOI] [PubMed] [Google Scholar]
- 158.Prekeris R, Mayhew MW, Cooper JB, Terrian DM. Identification and localization of an actin-binding motif that is unique to the epsilon isoform of protein kinase C and participates in the regulation of synaptic function. J Cell Biol. 1996;132:77–90. doi: 10.1083/jcb.132.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Prevostel C, Alice V, Joubert D, Parker PJ. Protein kinase Cα actively downregulates through caveolae-dependent traffic to an endosomal compartment. J Cell Sci. 2000;113:2575–2584. doi: 10.1242/jcs.113.14.2575. [DOI] [PubMed] [Google Scholar]
- 160.Pu Y, Peach ML, Garfield SH, Wincovitch S, Marquez VE, Blumberg PM. Effects on ligand interaction and membrane translocation of the positively charged arginine residues situated along the C1 domain binding cleft in the atypical protein kinase C isoforms. J Biol Chem. 2006;281:33773–33788. doi: 10.1074/jbc.M606560200. [DOI] [PubMed] [Google Scholar]
- 161.Quittau-Prevostel C, Delaunay N, Collazos A, Vallentin A, Joubert D. Targeting of PKCα and ε in the pituitary: a highly regulated mechanism involving a GD(E)E motif of the V3 region. J Cell Sci. 2004;117:63–72. doi: 10.1242/jcs.00832. [DOI] [PubMed] [Google Scholar]
- 162.Ranganathan S, Wang Y, Kern FG, Qu Z, Li R. Activation loop phosphorylation-independent kinase activity of human protein kinase C-ζ. Proteins. 2007;67:709–719. doi: 10.1002/prot.21348. [DOI] [PubMed] [Google Scholar]
- 163.Ren J, Datta R, Shioya H, Li Y, Oki E, Biedermann V, Bharti A, Kufe D. p73β is regulated by protein kinase Cδ catalytic fragment generated in the apoptotic response to DNA damage. J Biol Chem. 2002;277:33758–33765. doi: 10.1074/jbc.M110667200. [DOI] [PubMed] [Google Scholar]
- 164.Roberts NA, Haworth RS, Avkiran M. Effects of bisindolylmaleimide PKC inhibitors on p90RSK activity in vitro and in adult ventricular myocytes. Br J Pharmacol. 2005;145:477–489. doi: 10.1038/sj.bjp.0706210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Roberts NA, Marber MS, Avkiran M. Specificity of action of bisindolylmaleimide protein kinase C inhibitors: do they inhibit the 70kDa ribosomal S6 kinase in cardiac myocytes? Biochem Pharmacol. 2004;68:1923–1928. doi: 10.1016/j.bcp.2004.07.040. [DOI] [PubMed] [Google Scholar]
- 166.Robles-Flores M, Rendon-Huerta E, Gonzalez-Aguilar H, Mendoza-Hernandez G, Islas S, Mendoza V, Ponce-Castaneda MV, Gonzalez-Mariscal L, Lopez-Casillas F. p32 (gC1qBP) is a general protein kinase C (PKC)-binding protein. J Biol Chem. 2002;277:5247–5255. doi: 10.1074/jbc.M109333200. [DOI] [PubMed] [Google Scholar]
- 167.Ron D, Luo J, Mochly-Rosen D. C2 region-derived peptides inhibit translocation and function of β-protein kinase C in vivo. J Biol Chem. 1995;270:24180–24187. doi: 10.1074/jbc.270.41.24180. [DOI] [PubMed] [Google Scholar]
- 168.Ron D, Mochly-Rosen D. An autoregulatory region in protein kinase C: The pseudoanchoring site. Proc Natl Acad Sci USA. 1995;92:492–496. doi: 10.1073/pnas.92.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Roumiantsev S, Shah NP, Gorre ME, Nicoll J, Brasher BB, Sawyers CL, Van Etten RA. Clinical resistance to the kinase inhibitor STI-571 in chronic myeloid leukemia by mutation of Tyr-253 in the Abl kinase domain P-loop. Proc Natl Acad Sci USA. 2002;99:10700–10705. doi: 10.1073/pnas.162140299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Rybin VO, Guo J, Gertsberg Z, Elouardighi H, Steinberg SF. Protein kinase Cε and Src control PKCδ activation loop phosphorylation in cardiomyocytes. J Biol Chem. 2007;282:23631–23638. doi: 10.1074/jbc.M701676200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Rybin VO, Guo J, Gertsberg Z, Feinmark SJ, Steinberg SF. PMA-dependent PKCδ-Y311 phosphorylation in cardiomyocyte caveolae. J Biol Chem. 2008;283:17777–17788. doi: 10.1074/jbc.M800333200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Rybin VO, Guo J, Sabri A, Elouardighi H, Schaefer E, Steinberg SF. Stimulus-specific differences in protein kinase C-δ localization and activation mechanisms in cardiomyocytes. J Biol Chem. 2004;279:19350–19361. doi: 10.1074/jbc.M311096200. [DOI] [PubMed] [Google Scholar]
- 173.Rybin VO, Sabri A, Short J, Braz JC, Molkentin JD, Steinberg SF. Cross regulation of nPKC isoform function in cardiomyocytes: role of PKCδ in activation loop phosphorylations and PKCε in hydrophobic motif phosphorylations. J Biol Chem. 2003;278:14555–14564. doi: 10.1074/jbc.M212644200. [DOI] [PubMed] [Google Scholar]
- 174.Rybin VO, Steinberg SF. Immunoblotting PKCδ: a cautionary note from the bench. Am J Physiol Cell Physiol. 2006;290:C750–C756. doi: 10.1152/ajpcell.00395.2005. [DOI] [PubMed] [Google Scholar]
- 175.Rybin VO, Xu X, Steinberg SF. Activated protein kinase C isoforms target to cardiomyocyte caveolae. Circ Res. 1999;84:980–988. doi: 10.1161/01.res.84.9.980. [DOI] [PubMed] [Google Scholar]
- 176.Sabri A, Steinberg SF. Protein kinase C isoform-selective signals that lead to cardiac hypertrophy and the progression of heart failure. Mol Cell Biochem. 2003;251:97–101. [PubMed] [Google Scholar]
- 177.Sakurai Y, Onishi Y, Animator Y, Kizaki H. Novel protein kinase C delta isoform insensitive to caspase-3. Biol Pharm Bull. 2001;24:973–977. doi: 10.1248/bpb.24.973. [DOI] [PubMed] [Google Scholar]
- 178.Sawai H, Okazaki T, Takeda Y, Tashima M, Sawada H, Okuma M, Kishi S, Umehara H, Domae N. Ceramide-induced translocation of protein kinase C-δ and -ε to the cytosol: implications in apoptosis. J Biol Chem. 1997;272:2452–2458. doi: 10.1074/jbc.272.4.2452. [DOI] [PubMed] [Google Scholar]
- 179.Schauble S, King CC, Darshi M, Koller A, Shah K, Taylor SS. Identification of ChChd3 as a novel substrate of the cAMP-dependent protein kinase (PKA) using an analog-sensitive catalytic subunit. J Biol Chem. 2007;282:14952–14959. doi: 10.1074/jbc.M609221200. [DOI] [PubMed] [Google Scholar]
- 180.Schechtman D, Craske ML, Kheifets V, Meyer T, Schechtman J, Mochly-Rosen D. A critical intramolecular interaction for protein kinase Cε translocation. J Biol Chem. 2004;279:17617–17624. doi: 10.1074/jbc.M310696200. [DOI] [PubMed] [Google Scholar]
- 181.Schechtman D, Mochly-Rosen D. Adaptor proteins in protein kinase C-mediated signal transduction. Oncogene. 2001;20:6339–6347. doi: 10.1038/sj.onc.1204778. [DOI] [PubMed] [Google Scholar]
- 182.Schultz A, Jonsson JI, Larsson C. The regulatory domain of protein kinase Cθ localises to the Golgi complex and induces apoptosis in neuroblastoma and Jurkat cells. Cell Death Differ. 2003;10:662–675. doi: 10.1038/sj.cdd.4401235. [DOI] [PubMed] [Google Scholar]
- 183.Schultz A, Ling M, Larsson C. Identification of an amino acid residue in the protein kinase C C1b domain crucial for its localization to the Golgi network. J Biol Chem. 2004;279:31750–31760. doi: 10.1074/jbc.M313017200. [DOI] [PubMed] [Google Scholar]
- 184.Shah K, Shokat KM. A chemical genetic approach for the identification of direct substrates of protein kinases. Methods Mol Biol. 2003;233:253–271. doi: 10.1385/1-59259-397-6:253. [DOI] [PubMed] [Google Scholar]
- 185.Singer WD, Brown HA, Jiang X, Sternweis PC. Regulation of phospholipase D by protein kinase C is synergistic with ADP-ribosylation factor and independent of protein kinase activity. J Biol Chem. 1996;271:4504–4510. doi: 10.1074/jbc.271.8.4504. [DOI] [PubMed] [Google Scholar]
- 186.Skaggs BJ, Gorre ME, Ryvkin A, Burgess MR, Xie Y, Han Y, Komisopoulou E, Brown LM, Loo JA, Landaw EM, Sawyers CL, Graeber TG. Phosphorylation of the ATP-binding loop directs oncogenicity of drug-resistant BCR-ABL mutants. Proc Natl Acad Sci USA. 2006;103:19466–19471. doi: 10.1073/pnas.0609239103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Slater SJ, Seiz JL, Cook AC, Buzas CJ, Malinowski SA, Kershner JL, Stagliano BA, Stubbs CD. Regulation of PKC-α activity by C1–C2 domain interactions. J Biol Chem. 2002;277:15277–15285. doi: 10.1074/jbc.M112207200. [DOI] [PubMed] [Google Scholar]
- 188.Smith L, Smith JB. Lack of constitutive activity of the free kinase domain of protein kinase C-ζ. Dependence on transphosphorylation of the activation loop. J Biol Chem. 2002;277:45866–45873. doi: 10.1074/jbc.M206420200. [DOI] [PubMed] [Google Scholar]
- 189.Soltoff SP. Rottlerin is a mitochondrial uncoupler that decreases cellular ATP levels and indirectly blocks PKCδ tyrosine phosphorylation. J Biol Chem. 2001;276:37986–37992. doi: 10.1074/jbc.M105073200. [DOI] [PubMed] [Google Scholar]
- 190.Soltoff SP. Rottlerin: an inappropriate and ineffective inhibitor of PKCδ. Trends Pharmacol Sci. 2007;28:453–458. doi: 10.1016/j.tips.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 191.Song JS, Swann PG, Szallasi Z, Blank U, Blumberg PM, Rivera J. Tyrosine phosphorylation-dependent and -independent associations of protein kinase C-δ with Src family kinases in the RBL-2H3 mast cell line. Oncogene. 1998;16:3357–3368. doi: 10.1038/sj.onc.1201886. [DOI] [PubMed] [Google Scholar]
- 192.Sonnenburg ED, Gao T, Newton AC. The phosphoinositide-dependent kinase, PDK-1, phosphorylates conventional protein kinase C isozymes by a mechanism that is independent of phosphoinositide 3-kinase. J Biol Chem. 2001;276:45289–45297. doi: 10.1074/jbc.M107416200. [DOI] [PubMed] [Google Scholar]
- 193.Souroujon MC, Yao L, Chen H, Endemann G, Khaner H, Geeraert V, Schechtman D, Gordon AS, Diamond I, Mochly-Rosen D. State-specific monoclonal antibodies identify an intermediate state in ε-protein kinase C activation. J Biol Chem. 2004;279:17617–17624. doi: 10.1074/jbc.M400962200. [DOI] [PubMed] [Google Scholar]
- 194.Spitaler M, Villunger A, Grunicke H, Uberall F. Unique structural and functional properties of the ATP-binding domain of atypical protein kinase C-ι. J Biol Chem. 2000;275:33289–33296. doi: 10.1074/jbc.M002742200. [DOI] [PubMed] [Google Scholar]
- 195.Stahelin RV, Digman MA, Medkova M, Ananthanarayanan B, Melowic HR, Rafter JD, Cho W. Diacylglycerol-induced membrane targeting and activation of protein kinase Cε: mechanistic differences between protein kinases Cδ and Cε. J Biol Chem. 2005;280:19784–19793. doi: 10.1074/jbc.M411285200. [DOI] [PubMed] [Google Scholar]
- 196.Stahelin RV, Digman MA, Medkova M, Ananthanarayanan B, Rafter JD, Melowic HR, Cho W. Mechanism of diacylglycerol-induced membrane targeting and activation of protein kinase Cδ. J Biol Chem. 2004;279:29501–29512. doi: 10.1074/jbc.M403191200. [DOI] [PubMed] [Google Scholar]
- 197.Stahelin RV, Rafter JD, Das S, Cho W. The molecular basis of differential subcellular localization of C2 domains of protein kinase C-α and group IVa cytosolic phospholipase A2. J Biol Chem. 2003;278:12452–12460. doi: 10.1074/jbc.M212864200. [DOI] [PubMed] [Google Scholar]
- 198.Stahelin RV, Wang J, Blatner NR, Rafter JD, Murray D, Cho W. The origin of C1A-C2 interdomain interactions in protein kinase Cα. J Biol Chem. 2005;280:36452–36463. doi: 10.1074/jbc.M506224200. [DOI] [PubMed] [Google Scholar]
- 199.Staudinger J, Zhou J, Burgess R, Elledge SJ, Olson EN. PICK1: a perinuclear binding protein and substrate for protein kinase C isolated by the yeast two-hybrid system. J Cell Biol. 1995;128:263–271. doi: 10.1083/jcb.128.3.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Stebbins EG, Mochly-Rosen D. Binding specificity for RACK1 resides in the V5 region of βII protein kinase C. J Biol Chem. 2001;276:29644–29650. doi: 10.1074/jbc.M101044200. [DOI] [PubMed] [Google Scholar]
- 201.Steinberg SF. Distinctive activation mechanisms and functions for protein kinase C-δ. Biochem J. 2004;384:449–459. doi: 10.1042/BJ20040704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Steinberg SF, Sussman MA. Cardiac hypertrophy served with protein kinase C-ε: δ isoform substitution available at additional cost. Circ Res. 2005;96:711–713. doi: 10.1161/01.RES.0000164185.96546.82. [DOI] [PubMed] [Google Scholar]
- 203.Stempka L, Girod A, Muller HJ, Rincke G, Marks F, Gschwendt M, Bossemeyer D. Phosphorylation of protein kinase Cδ at threonine 505 is not a prerequisite for enzymatic activity. J Biol Chem. 1997;272:6805–6811. doi: 10.1074/jbc.272.10.6805. [DOI] [PubMed] [Google Scholar]
- 204.Stempka L, Schnolzer M, Radke S, Rincke G, Marks F, Gschwendt M. Requirements of protein kinase C δ for catalytic function. Role of glutamic acid 500 and autophosphorylation on serine 643. J Biol Chem. 1999;274:8886–8892. doi: 10.1074/jbc.274.13.8886. [DOI] [PubMed] [Google Scholar]
- 205.Stensman H, Raghunath A, Larsson C. Autophosphorylation suppresses whereas kinase inhibition augments the translocation of protein kinase Cα in response to diacylglycerol. J Biol Chem. 2004;279:40576–40583. doi: 10.1074/jbc.M405560200. [DOI] [PubMed] [Google Scholar]
- 206.Sumandea M, Rybin VO, Wang C, Hinken A, Kobayashi T, Harleton E, Sievert G, Feinmark SJ, Balke CW, Solaro RJ, Steinberg SF. Tyrosine phosphorylation modifies PKCδ phosphorylation of cardiac troponin I. Mol Cell. doi: 10.1074/jbc.M802396200. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Sun X, Wu F, Datta R, Kharbanda S, Kufe D. Interaction between protein kinase Cδ and the c-Abl tyrosine kinase in the cellular response to oxidative stress. J Biol Chem. 2000;275:7470–7473. doi: 10.1074/jbc.275.11.7470. [DOI] [PubMed] [Google Scholar]
- 208.Sutton RB, Sprang SR. Structure of the protein kinase Cβ phospholipid-binding C2 domain complexed with Ca2+ Structure. 1998;6:1395–1405. doi: 10.1016/s0969-2126(98)00139-7. [DOI] [PubMed] [Google Scholar]
- 209.Szallasi Z, Denning MF, Chang EY, Rivera J, Yuspa SH, Lehel C, Olah Z, Anderson WB, Blumberg PM. Development of a rapid approach to identification of tyrosine phosphorylation sites. Biochem Biophys Res Commun. 1995;214:888–894. doi: 10.1006/bbrc.1995.2370. [DOI] [PubMed] [Google Scholar]
- 210.Takahashi H, Takeishi Y, Seidler T, Arimoto T, Akiyama H, Hozumi Y, Koyama Y, Shishido T, Tsunoda Y, Niizeki T, Nozaki N, Abe J, Hasenfuss G, Goto K, Kubota I. Adenovirus-mediated overexpression of diacylglycerol kinase-ζ inhibits endothelin-1-induced cardiomyocyte hypertrophy. Circulation. 2005;111:1510–1516. doi: 10.1161/01.CIR.0000159339.00703.22. [DOI] [PubMed] [Google Scholar]
- 211.Taparowsky E, Suard Y, Fasano O, Shimizu K, Goldfarb M, Wigler M. Activation of the T24 bladder carcinoma transforming gene is linked to a single amino acid change. Nature. 1982;300:762–765. doi: 10.1038/300762a0. [DOI] [PubMed] [Google Scholar]
- 212.Thuille N, Heit I, Fresser F, Krumbock N, Bauer B, Leuthaeusser S, Dammeier S, Graham C, Copeland TD, Shaw S, Baier G. Critical role of novel Thr-219 autophosphorylation for the cellular function of PKCθ in T lymphocytes. EMBO J. 2005;24:3869–3880. doi: 10.1038/sj.emboj.7600856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Vallentin A, Lo TC, Joubert D. A single point mutation in the V3 region affects protein kinase Cα targeting and accumulation at cell-cell contacts. Mol Cell Biol. 2001;21:3351–3363. doi: 10.1128/MCB.21.10.3351-3363.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Verbeek DS, Warrenburg BP, Hennekam FA, Dooijes D, Ippel PF, Verschuuren-Bemelmans CC, Kremer HP, Sinke RJ. Gly118Asp is a SCA14 founder mutation in the Dutch ataxia population. Hum Genet. 2005;117:88–91. doi: 10.1007/s00439-005-1278-z. [DOI] [PubMed] [Google Scholar]
- 215.Verdaguer N, Corbalan-Garcia S, Ochoa WF, Fita I, Gomez-Fernandez JC. Ca2+ bridges the C2 membrane-binding domain of protein kinase Cα directly to phosphatidylserine. EMBO J. 1999;18:6329–6338. doi: 10.1093/emboj/18.22.6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Veron M, Radzio-Andzelm E, Tsigelny I, Ten Eyck LF, Taylor SS. A conserved helix motif complements the protein kinase core. Proc Natl Acad Sci USA. 1993;90:10618–10622. doi: 10.1073/pnas.90.22.10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Violin JD, Zhang J, Tsien RY, Newton AC. A genetically encoded fluorescent reporter reveals oscillatory phosphorylation by protein kinase C. J Cell Biol. 2003;161:899–909. doi: 10.1083/jcb.200302125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Wang QJ, Fang TW, Fenick D, Garfield S, Bienfait B, Marquez VE, Blumberg PM. The lipophilicity of phorbol esters as a critical factor in determining the pattern of translocation of protein kinase C delta fused to green fluorescent protein. J Biol Chem. 2000;275:12136–12146. doi: 10.1074/jbc.275.16.12136. [DOI] [PubMed] [Google Scholar]
- 219.Watanabe T, Ono Y, Taniyama Y, Hazama K, Igarashi K, Ogita K, Kikkawa U, Nishizuka Y. Cell division arrest induced by phorbol ester in CHO cells overexpressing protein kinase C-δ subspecies. Proc Natl Acad Sci USA. 1992;89:10159–10163. doi: 10.1073/pnas.89.21.10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Way KJ, Katai N, King GL. Protein kinase C and the development of diabetic vascular complications. Diabetes Med. 2001;18:945–959. doi: 10.1046/j.0742-3071.2001.00638.x. [DOI] [PubMed] [Google Scholar]
- 221.White WO, Seibenhener ML, Wooten MW. Phosphorylation of tyrosine 256 facilitates nuclear import of atypical protein kinase C. J Cell Biochem. 2002;85:42–53. [PubMed] [Google Scholar]
- 222.Williams MR, Arthur JS, Balendran A, van der KJ, Poli V, Cohen P, Alessi DR. The role of 3-phosphoinositide-dependent protein kinase 1 in activating AGC kinases defined in embryonic stem cells. Curr Biol. 2000;10:439–448. doi: 10.1016/s0960-9822(00)00441-3. [DOI] [PubMed] [Google Scholar]
- 223.Wolfman A, Macara IG. Elevated levels of diacylglycerol and decreased phorbol ester sensitivity in ras-transformed fibroblasts. Nature. 1987;325:359–361. doi: 10.1038/325359a0. [DOI] [PubMed] [Google Scholar]
- 224.Wooten MW, Vandenplas ML, Seibenhener ML, Geetha T, Diaz-Meco MT. Nerve growth factor stimulates multisite tyrosine phosphorylation and activation of the atypical protein kinase C’s via a src kinase pathway. Mol Cell Biol. 2001;21:8414–8427. doi: 10.1128/MCB.21.24.8414-8427.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Xu RX, Pawelczyk T, Xia TH, Brown SC. NMR structure of a protein kinase C-γ phorbol-binding domain and study of protein-lipid micelle interactions. Biochemistry. 1997;36:10709–10717. doi: 10.1021/bi970833a. [DOI] [PubMed] [Google Scholar]
- 226.Xu ZB, Chaudhary D, Olland S, Wolfrom S, Czerwinski R, Malakian K, Lin L, Stahl ML, Joseph-McCarthy D, Benander C, Fitz L, Greco R, Somers WS, Mosyak L. Catalytic domain crystal structure of protein kinase Cθ. J Biol Chem. 2004;279:50401–50409. doi: 10.1074/jbc.M409216200. [DOI] [PubMed] [Google Scholar]
- 227.Yamaguchi Y, Shirai Y, Matsubara T, Sanse K, Kuriyama M, Oshiro N, Yoshino K, Yonezawa K, Ono Y, Saito N. Phosphorylation and up-regulation of diacylglycerol kinase-γ via its interaction with protein kinase C-γ. J Biol Chem. 2006;281:31627–31637. doi: 10.1074/jbc.M606992200. [DOI] [PubMed] [Google Scholar]
- 228.Yamamoto M, Acevedo-Duncan M, Chalfant CE, Patel NA, Watson JE, Cooper DR. The roles of protein kinase C-βI and -βII in vascular smooth muscle cell proliferation. Exp Cell Res. 1998;240:349–358. doi: 10.1006/excr.1998.3999. [DOI] [PubMed] [Google Scholar]
- 229.Yamamoto S, Seta K, Morisco C, Vatner SF, Sadoshima J. Chelerythrine rapidly induces apoptosis through generation of reactive oxygen species in cardiac myocytes. J Mol Cell Cardiol. 2001;33:1829–1848. doi: 10.1006/jmcc.2001.1446. [DOI] [PubMed] [Google Scholar]
- 230.Yan SF, Harja E, Andrassy M, Fujita T, Schmidt AM. Protein kinase C-β/early growth response-1 pathway: a key player in ischemia, atherosclerosis, restenosis. J Am Coll Cardiol. 2006;48:A47–A55. doi: 10.1016/j.jacc.2006.05.063. [DOI] [PubMed] [Google Scholar]
- 231.Yang J, Cron P, Thompson V, Good VM, Hess D, Hemmings BA, Barford D. Molecular mechanism for the regulation of protein kinase B/Akt by hydrophobic motif phosphorylation. Mol Cell. 2002;9:1227–1240. doi: 10.1016/s1097-2765(02)00550-6. [DOI] [PubMed] [Google Scholar]
- 232.Yang J, Garrod SM, Deal MS, Anand GS, Woods VL, Jr, Taylor S. Allosteric network of cAMP-dependent protein kinase revealed by mutation of Tyr204 in the P+1 loop. J Mol Biol. 2005;346:191–201. doi: 10.1016/j.jmb.2004.11.030. [DOI] [PubMed] [Google Scholar]
- 233.Yeong SS, Zhu Y, Smith D, Verma C, Lim WG, Tan BJ, Li QT, Cheung NS, Cai M, Zhu YZ, Zhou SF, Tan SL, Duan W. The last 10 amino acid residues beyond the hydrophobic motif are critical for the catalytic competence and function of protein kinase Cα. J Biol Chem. 2006;281:30768–30781. doi: 10.1074/jbc.M511278200. [DOI] [PubMed] [Google Scholar]
- 234.Yoshida K, Wang HG, Miki Y, Kufe D. Protein kinase Cδ is responsible for constitutive and DNA damage-induced phosphorylation of Rad9. EMBO J. 2003;22:1431–1441. doi: 10.1093/emboj/cdg134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Zeidan YH, Hannun YA. Activation of acid sphingomyelinase by protein kinase Cδ-mediated phosphorylation. J Biol Chem. 2007;282:11549–11561. doi: 10.1074/jbc.M609424200. [DOI] [PubMed] [Google Scholar]
- 236.Zeidman R, Lofgren B, Pahlman S, Larsson C. PKCε, via its regulatory domain and independently of its catalytic domain, induces neurite-like processes in neuroblastoma cells. J Cell Biol. 1999;145:713–726. doi: 10.1083/jcb.145.4.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Zhang G, Kazanietz MG, Blumberg PM, Hurley JH. Crystal structure of the cys2 activator-binding domain of protein kinase C-δ in complex with phorbol ester. Cell. 1995;81:917–924. doi: 10.1016/0092-8674(95)90011-x. [DOI] [PubMed] [Google Scholar]
- 238.Zhu Y, Smith D, Verma C, Lim WG, Tan BJ, Armstrong JS, Zhou S, Chan E, Tan SL, Zhu YZ, Cheung NS, Duan W. The very C-terminus of protein kinase Cε is critical for the full catalytic competence but its hydrophobic motif is dispensable for the interaction with 3-phosphoinositide-dependent kinase-1. Cell Signal. 2006;18:807–818. doi: 10.1016/j.cellsig.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 239.Ziegler WH, Parekh DB, Le Good JA, Whelan RD, Kelly JJ, Frech M, Hemmings BA, Parker PJ. Rapamycin-sensitive phosphorylation of PKC on a carboxy-terminal site by an atypical PKC complex. Curr Biol. 1999;9:522–529. doi: 10.1016/s0960-9822(99)80236-x. [DOI] [PubMed] [Google Scholar]