Abstract
Arsenic is a ubiquitous environmental poison that inhibits root elongation and seed germination to a variable extent depending on the plant species. To understand the molecular mechanisms of arsenic resistance, a genetic screen was developed to isolate arsenate overly sensitive (aos) mutants from an activation-tagged Arabidopsis (Arabidopsis thaliana) population. Three aos mutants were isolated, and the phenotype of each was demonstrated to be due to an identical disruption of plastidial LIPOAMIDE DEHYDROGENASE1 (ptLPD1), a gene that encodes one of the two E3 isoforms found in the plastidial pyruvate dehydrogenase complex. In the presence of arsenate, ptlpd1-1 plants exhibited reduced root and shoot growth and enhanced anthocyanin accumulation compared with wild-type plants. The ptlpd1-1 plants accumulated the same amount of arsenic as wild-type plants, indicating that the aos phenotype was not due to increased arsenate in the tissues but to an increase in the innate sensitivity to the poison. Interestingly, a ptlpd1-4 knockdown allele produced a partial aos phenotype. Two loss-of-function alleles of ptLPD2 in Arabidopsis also caused elevated arsenate sensitivity, but the sensitivity was less pronounced than for the ptlpd1 mutants. Moreover, both the ptlpd1 and ptlpd2 mutants were more sensitive to arsenite than wild-type plants, and the LPD activity in isolated chloroplasts from wild-type plants was sensitive to arsenite but not arsenate. These findings show that the ptLPD isoforms are critical in vivo determinants of arsenite-mediated arsenic sensitivity in Arabidopsis and possible strategic targets for increasing arsenic tolerance.
Arsenic (As) is a naturally occurring metalloid found in soil, water, and air, but anthropogenic activities, including smelting and fossil fuel combustion, have led to increased environmental exposure (Mandal and Suzuki, 2002). In the environment, As exists in both organic and inorganic forms. Arsenate [As(V)] is the principal inorganic form of As in aerobic soils, while arsenite [As(III)] is the main form found under anaerobic conditions (Marin et al., 1993; Onken and Hossner, 1995, 1996; Mandal and Suzuki, 2002; Masscheleyn et al., 2002).
Both As(V) and As(III) are toxic to plants, inducing symptoms ranging from poor seed germination and inhibited root growth to death (Meharg and Hartley-Whitaker, 2002; Lee et al., 2003; Ahsan et al., 2008; Smith et al., 2010). The modes of action of As(V) and As(III) differ, owing to their distinct chemical properties. As(V), with its structural similarity to phosphate, can compete with phosphate in oxidative phosphorylation, leading to the production of ADP-As(V) (Gresser, 1981). However, half-maximal stimulation of ADP-As(V) formation requires physiologically unlikely concentrations of approximately 0.8 mm As(V) (Moore et al., 1983). As(V) has been recently shown to enhance membrane fluidity, and thus membrane permeability, by binding and replacing phosphate or choline head groups (Tuan et al., 2008). The resulting damage to the membrane would disrupt the transport of mineral nutrients and water (Smith et al., 2010). As(V) can be promptly reduced in plants, including Arabidopsis (Arabidopsis thaliana), to As(III) by endogenous As(V) reductases, so that often more than 90% of As in plant cells is in the form of As(III) (Zhao et al., 2009). As(III) readily forms covalent bonds with sulfhydryl groups, especially vicinal dithiols. Binding to the free thiols of proteins is believed to be the basis of As(III) toxicity, either by inhibiting activity directly or by disrupting protein structure. Many enzymes have been proposed to be targets leading to As(III) toxicity, and the As(III) sensitivity of some of these enzymes has been investigated in nonplant systems (Adamson and Stevenson, 1981; Cavigelli et al., 1996; Lynn et al., 1997; Hu et al., 1998; Kitchin and Wallace, 2008). Of the many potential protein targets, only the pyruvate dehydrogenase complex (PDC) has been shown to be inactivated by physiologically relevant micromolar concentrations of As(III) (Hu et al., 1998), suggesting that PDC may be the primary target for As(III)-mediated cytotoxicity. However, little is known about the mechanism of As toxicity in vivo, especially in plants.
Although As is phytotoxic, some plants species are resistant to high levels of As through avoidance mechanisms, while species of the Pteridaceae family of ferns hyperaccumulate As without toxic effects (Verbruggen et al., 2009; Zhao et al., 2009). As an analog of phosphate, As(V) is readily taken up by plants through high-affinity phosphate transporters encoded by the PHOSPHATE TRANSPORTER1 (PHT1) gene family (Shin et al., 2004; González et al., 2005; Catarecha et al., 2007). Except for the hyperaccumulating ferns, avoidance of As toxicity by resistant species is often accomplished by a decrease in phosphate uptake activity (Meharg and Hartley-Whitaker, 2002). Unlike As(V), the transport of As(III) is facilitated by aquaporin nodulin 26-like intrinsic proteins (Bienert et al., 2008; Isayenkov and Maathuis, 2008; Ma et al., 2008; Kamiya et al., 2009). In roots and fronds of hyperaccumulating ferns, As(III) is sequestered in the vacuole (Lombi et al., 2002; Pickering et al., 2006). Much of the As(III) taken up by nonaccumulating resistant species may be released back to the rhizosphere through an undefined efflux pathway (Zhao et al., 2009). As(III) that remains in tissues reacts with thiol-containing molecules, such as glutathione or phytochelatins, both of which are usually produced in greater abundance in response to As (Grill et al., 1987; Sneller et al., 1999; Schmöger et al., 2000; Schulz et al., 2008). As(III)-glutathione adducts can be sequestered in the vacuole (Dhankher et al., 2002; Bleeker et al., 2006). However, increased synthesis of glutathione or phytochelatins alone is unlikely to confer a very high level of tolerance (Zhao et al., 2009).
To identify genes essential for As resistance in plants, we used a genetic screen to identify mutants of Arabidopsis that were hypersensitive to As(V). The screen was analogous to that used to isolate the salt overly sensitive (sos) mutants of Arabidopsis (Wu et al., 1996) that led to the identification of the SOS pathway for salt tolerance (Zhu, 2000, 2003). Our hypothesis was that arsenate overly sensitive (aos) mutants would reveal a different set of genes from those identified in mutants showing increased resistance to As(V).
RESULTS
Isolation of aos Mutants of Arabidopsis
Plants exposed to As(V) suffer a reduction in root and shoot growth. Arabidopsis is sensitive to moderate to high concentrations of As(V) (Quaghebeur and Rengel, 2004), and inhibition of root elongation is one of the most conspicuous developmental changes that occur during short-term exposure. We adapted the root-bending assay developed for isolating Arabidopsis mutants that are sensitive to toxic cations (Howden and Cobbett, 1992; Wu et al., 1996) to identify aos mutants. The optimal concentration of As(V) for isolating aos mutants was determined by exposing 5-d-old wild-type Arabidopsis seedlings of uniform size to various As(V) concentrations in solid growth medium. Plates were placed so that seedlings were vertically orientated with the root tip pointing upward. During root elongation, gravitropism caused the roots to bend downward, allowing the increase in root length since exposure to As(V) to be readily visualized and measured. Exposure to higher As(V) concentrations generally caused a stronger inhibition of root elongation (Supplemental Fig. S1A). The only exception was a stimulation of root elongation observed at 100 μm As(V), the lowest concentration tested. As(V) at 200 μm was the highest concentration tested that did not strongly inhibit wild-type root growth and was chosen for isolating aos mutants. It was expected that mutants could be recovered from the screen, because wild-type seedlings could be rescued after exposure to 1 mm As(V) for 5 d (data not shown).
Approximately 80,000 Arabidopsis seedlings representing a random selection of 40,000 activation-tagged M3 lines in the Columbia 2 (Col-2) background were screened for an aos phenotype. About 350 putative mutants were identified (Supplemental Fig. S1B), rescued, and grown to seed. During rescreening of progeny from each putative mutant, only three lines, 106, 107, and 116, were confirmed to have an aos phenotype. The aos phenotype of these three mutants was transmitted into both M5 and M6 generations, indicating that the phenotype was genetically stable.
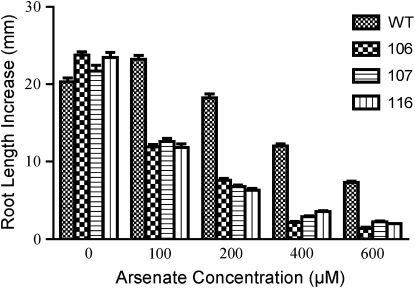
The phenotype of each aos mutant was characterized more fully by growing mutant and wild-type (Col-2) seedlings side by side on solid medium containing a range of As(V) concentrations. In the absence of As(V), the root growth of each mutant was similar to that of wild-type seedlings (Fig. 1; Supplemental Fig. S1C). Exposure to As(V) for 4 d caused a concentration-dependent inhibition of root growth in wild-type seedlings. At each As(V) concentration tested, root elongation for each mutant was inhibited similarly, but much more severely, than for wild-type seedlings (Fig. 1; Supplemental Fig. S1C). The As(V) concentration that inhibited root elongation by 50% (I50) compared with growth in the absence of As(V) was estimated roughly by examining the data in Figure 1 to be about 500 μm for wild-type seedlings and about 100 μm for each mutant.
Figure 1.
Isolation and phenotypic characterization of aos mutants. Arabidopsis wild-type (WT) and aos mutant 106, 107, and 116 seeds were germinated on plates containing solid As-free medium and placed in a vertical orientation. Five-day-old seedlings were transferred in rows to growth medium containing As(V). Plates of seedlings were placed vertically, with the seedling roots pointing upward. Growth was allowed to continue for 4 d before root elongation was measured from the top of the “hook” to the root tip. Means ± se (n = 20–30 seedlings) are shown.
Genetic Characterization of the aos Mutants
Each aos mutant was crossed with a wild-type plant to determine the pattern of aos inheritance. None of the F1 progeny had the aos phenotype (Supplemental Table S1), demonstrating that the phenotype was recessive in each mutant. The recessive nature of the mutation indicated that the aos phenotype arose from a loss-of-function mutation caused by the T-DNA insertion per se, rather than a gain-of-function mutation caused by activation of a nearby gene by the enhancer elements present in the T-DNA. The mutant phenotype segregated in a 1:3 (aos:wild type) ratio in the progeny from selfed F1 plants from all three crosses, indicating the involvement of a single locus inherited according to Mendelian principles. Allelism among the three mutants was tested by crossing a homozygous mutant 106 plant with plants homozygous for the other two aos mutations. No phenotypic complementation was seen in the F1 progeny (Supplemental Table S1), indicating that the three aos mutations were allelic. One of the As(V)-sensitive F2 progeny from the mutant 107 × wild type cross was named plastidial lipoamide dehydrogenase1-1 (ptlpd1-1) based on its molecular characteristics (see below). This line was used for subsequent experimentation.
ptlpd1 Conferred Enhanced As(V) Sensitivity at the Whole Plant Level and at Germination
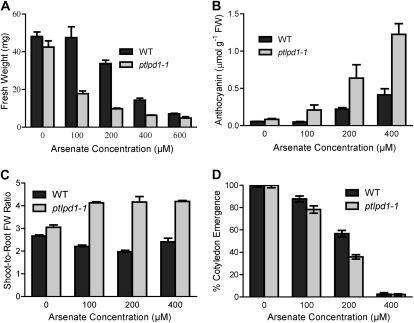
Exposure of ptlpd1-1 seedlings to As(V) for 12 d resulted in plants that were much smaller than wild-type plants (data not shown). When exposed to 100 μm As(V) in the growth medium, the fresh weight of ptlpd1-1 seedlings was 63% lower than that of wild-type seedlings, whose growth was unaffected (Fig. 2A). In the presence of 200 μm As(V), the fresh weight of wild-type seedlings was 30% lower than seedlings grown in the absence of As(V), while that of ptlpd1-1 seedlings was 77% lower. The ptlpd1-1 seedlings also accumulated more anthocyanin in the shoot than wild-type seedlings (Fig. 2B). Wild-type plants showed no change in anthocyanin concentration when exposed to 100 μm As(V), while the concentration in ptlpd1-1 seedlings increased 3-fold. The ptlpd1-1 seedlings had a higher shoot-to-root fresh weight ratio when exposed to As(V) than wild-type seedlings (Fig. 2C), indicating a shift in resource allocation toward shoot growth.
Figure 2.
Increased sensitivity of the ptlpd1-1 mutant to As(V). A to C, Five-day-old wild-type (WT) and ptlpd1-1 seedlings germinated on solid medium (see legend to Fig. 1) were transferred to medium containing the indicated As(V) concentrations and allowed to grow for 12 d in a vertical orientation. Mean values ± se (n = 4 plates; two to four seedlings from each plate were pooled for each replicate) are presented for fresh weight (A), shoot anthocyanin concentration (B), and shoot-to-root fresh weight (FW) ratio (C). D, ptlpd1-1 and wild-type seeds were germinated on solid medium containing the indicated As(V) concentrations and scored for the emergence of green cotyledons after 5 d. Values are means ± se (n = 4 plates of 50–100 seeds).
Seeds from ptlpd1-1 plants showed a significantly lower germination rate, scored as percentage cotyledon emergence (Lee et al., 2003), than seeds from wild-type plants when the medium contained either 100 or 200 μm As(V) (Fig. 2D). At both As(V) concentrations, the ptlpd1-1 germinants had shorter roots and smaller shoots than wild-type germinants (data not shown). Germination of both ptlpd1-1 and wild-type seeds was nearly completely inhibited by 400 μm As(V) in the medium.
Molecular Identification of the Genetic Lesion in the aos Mutants
The T-DNA insertion site in each mutant was localized by amplifying the genomic DNA sequences adjacent to the left border of the T-DNA insertion by thermal asymmetric interlaced (TAIL)-PCR (Liu et al., 1995). Two PCR products were obtained from mutants 107 and 116, while three products were detected for mutant 106 (data not shown). Among these TAIL-PCR products was a 900-bp amplicon common to all three mutants. Moreover, the three products obtained from mutant 106 were a combination of those obtained from mutants 107 and 116. DNA sequencing showed that the various TAIL-PCR products represented three unique T-DNA integration sites (Supplemental Fig. S2). The TAIL-PCR product common to all three lines corresponded to a T-DNA insertion precisely at the same location within the At3g17250/At3g17260 intergenic region (Supplemental Fig. S2). The presence of these T-DNA insertions was confirmed in each mutant by PCR using gene-specific primers in combination with either gene-specific primers expected to anneal on the far side of the T-DNA or a primer specific for the left border of the T-DNA (Supplemental Fig. S2). The absence of wild-type amplicons from the mutant genomic DNA templates indicated further that the T-DNA insertions were in the homozygous state in the three mutants.
The common insertion in the At3g17250/At3g17260 intergenic region in the three aos mutants indicated that this lesion was responsible for the aos phenotype. To confirm this conclusion, plants carrying T-DNA insertions only at this locus were isolated from the F2 progeny of crosses between wild-type Col-2 plants and mutants 107 and 116. Only seedlings homozygous for the T-DNA insertion found in the At3g17250/At3g17260 intergenic region had the aos phenotype, establishing genetic linkage. Linkage was verified by cosegregation analysis, where all 60 aos F2 progeny obtained from the wild type × mutant 107 cross were found by PCR to be homozygous for the T-DNA insertion in the At3g17250/At3g17260 intergenic region (data not shown).
The T-DNA insertion responsible for the aos phenotype was located 1.4 kb upstream of the annotated start codon of At3g17250 (Supplemental Fig. S2), suggesting that the insertion may interrupt transcription of this gene. At3g17250 encodes a PP2C-type protein phosphatase, a class of enzyme implicated in plant stress signaling (Schweighofer et al., 2004). However, no differences in At3g17250 transcript abundance were observed by semiquantitative reverse transcription (RT)-PCR between wild-type and mutant plants (data not shown). Additionally, plants carrying At3g17250 T-DNA insertional alleles had similar sensitivity to As(V) as wild-type plants (data not shown), further suggesting that the aos phenotype of ptlpd1-1 was not due to disruption of At3g17250 expression.
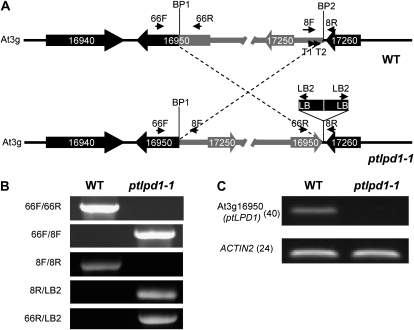
The possibility that the T-DNA insertion caused a chromosomal rearrangement (Nacry et al., 1998; Laufs et al., 1999; Tax and Vernon, 2001) was tested by TAIL-PCR. The primers used, T1 and T2, were specific for sequences in the At3g17250/At3g17260 intergenic region immediately adjacent to the putative right border of the T-DNA and pointing toward the insertion site (Fig. 3A). DNA sequencing of the single 1.0-kb TAIL-PCR product showed that a DNA rearrangement had occurred that linked the upstream region of At3g17250 directly to sequences internal to At3g16950 (Fig. 3A). PCR using primers specific for At3g16950 in combination with T-DNA-specific primers revealed the presence of at least two T-DNA insertions in a head-to-head orientation (Fig. 3, A and B) and that a 107-kb inversion of genomic DNA had occurred during the T-DNA integration event. RT-PCR analysis showed that ptldp1-1 lacked At3g16950 transcripts (Fig. 3C). Taken together, these results suggest that the interruption of At3g16950 was responsible for the aos phenotype. At3g16950 is the ptLPD1 gene, encoding an E3 subunit of the plastidial PDC (ptPDC).
Figure 3.
A paracentric inversion underlies the ptlpd1-1 phenotype. A, Schematic representation of the paracentric chromosomal inversion in ptlpd1-1. The top diagram represents the gene organization in the region of wild-type (WT) genomic DNA involved in the inversion event. Genes are represented by thick arrows indicating the direction of transcription. BP1 and BP2 are the break points of the chromosomal inversion. For clarity, the fragment involved in the inversion is shown in gray. The bottom diagram represents the gene arrangement in the ptlpd1-1 mutant. LB represents the left border of the T-DNA in pSKI015. The T-DNA insert is not drawn to scale. Small arrows indicate the annealing sites of primers used to confirm the genome organization. B, PCR-based confirmation of the genomic inversion in ptlpd1-1. Fragments were amplified using the primer pairs indicated on the left and wild-type or ptlpd1-1 mutant genomic DNA as the template. The annealing position of each primer used is shown in A. C, Abundance of At3g16950 (ptLPD1) transcripts in ptlpd1-1 mutant and wild-type seedlings. At3g18780 (ACTIN2) was used as an amplification control. Numbers in parentheses are the numbers of cycles used in the PCR.
Confirmation That the aos Phenotype Is Due to Mutation of ptLPD1
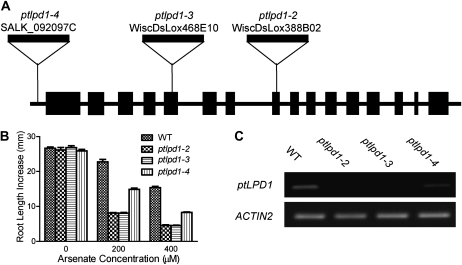
To confirm that the disruption of ptLPD1 caused the increased As(V) sensitivity of the ptlpd1-1 mutant, we determined the As(V) sensitivity of three T-DNA insertional mutants obtained from the publicly available stocks (Arabidopsis Biological Resource Center). These lines were WiscDsLox388B02 (designated ptlpd1-2), WiscDsLox468E10 (designated ptlpd1-3), and SALK_092097C (designated ptlpd1-4). The ptlpd1-4 mutant contained an insertion in the 5′ untranslated region of ptLPD1, while the other two lines contained insertions within the exons of ptLPD1 (Fig. 4A). Homozygous mutants were isolated from a segregating population of each insertion line. In the absence of As(V), the phenotype of seedlings carrying any one of these alleles was indistinguishable from that of the corresponding wild-type (Col-0) seedlings. When grown in the presence of 200 μm As(V), the mutants showed the aos phenotype of compromised root elongation (Fig. 4B; Supplemental Fig. S3A). Root elongation in ptlpd1-2 and ptlpd1-3 seedlings was as sensitive to As(V) as that of ptlpd1-1 seedlings, while that of ptlpd1-4 seedlings was intermediate between the wild type and the other mutants. To understand the intermediate nature of the ptlpd1-4 phenotype, the abundance of ptLPD1 transcripts was determined in ptlpd1-2, ptlpd1-3, ptlpd1-4, and wild-type seedlings. The ptLPD1 transcripts in ptlpd1-2 or ptlpd1-3 seedlings were greatly reduced compared with wild-type seedlings (Fig. 4C). These results, combined with the location of the T-DNA insert in the middle of the ptLPD1 open reading frame in both mutants and their phenotypic similarities to ptlpd1-1, indicated that ptlpd1-2 and ptlpd1-3 are loss-of-function alleles. In ptlpd1-4 seedlings, the ptLPD1 transcript abundance was intermediate between that of the wild type and the other mutant seedlings, indicating that the T-DNA insert in the 5′ untranslated region of ptLPD1 is only partially inhibitory.
Figure 4.
As(V) sensitivity of root elongation in seedlings with ptlpd1 alleles. A, Schematic diagram showing the T-DNA insertion sites in the ptlpd1-2, ptlpd1-3, and ptlpd1-4 alleles of At3g16950. Black boxes and lines represent exons and introns/intergenic regions, respectively, with the direction of transcription being left to right. B, Five-day-old seedlings (see legend to Fig. 1) of the wild type (WT), ptlpd1-2, ptlpd1-3, and ptlpd1-4 were transferred to solid medium containing As(V). Increases in root length were measured after growth in the presence of As(V) for 4 d in a vertical orientation. Values are means ± se (n = 15–20 seedlings). C, RT-PCR determination of ptLPD1 transcript abundance in plants with the wild-type, ptlpd1-2, ptlpd1-3, or ptlpd1-4 allele. Total RNA was extracted from 5-d-old seedlings and reverse transcribed. ACTIN2 (At3g18780) was used as an amplification control.
Like ptlpd1-1 seedlings, ptlpd1-2, ptlpd1-3, and ptlpd1-4 seedlings exposed to As(V) were smaller (Supplemental Fig. S3B), had lower fresh weight (Supplemental Fig. S3C), increased shoot anthocyanin concentration (Supplemental Fig. S3D), and increased shoot-to-root fresh weight ratio (Supplemental Fig. S3E) compared with wild-type seedlings. In addition, ptlpd1-2, ptlpd1-3, and ptlpd1-4 seeds showed decreased germination rates relative to the wild type at all but the highest As(V) concentration tested (Supplemental Fig. S3F). In all these experiments, the ptlpd1-4 mutant phenotype was intermediate between the wild type and the knockout mutants, consistent with the view that ptlpd1-4 is a knockdown allele. Taken together, these results suggest that ptLPD expression quantitatively controls Arabidopsis As(V) sensitivity.
To verify that the enhanced As(V) sensitivity is due to the disruption of the ptLPD1 gene, we crossed homozygous ptlpd1-1 with ptlpd1-2 and ptlpd1-3. The F1 progeny of these crosses showed decreased root growth similar to each homozygous mutant (data not shown). This result confirms that the mutations in all three lines are allelic. Moreover, the aos phenotype of ptlpd1-1 was functionally complemented by transforming the mutant with a 5,464-bp genomic DNA fragment that included the wild-type ptLPD1 gene (Supplemental Fig. S4; Supplemental Materials and Methods S1). These data provided further evidence that the enhanced As(V) sensitivity of ptlpd1-1 was due to disruption of ptLPD1.
The ptlpd1-1 Allele Does Not Enhance As Accumulation
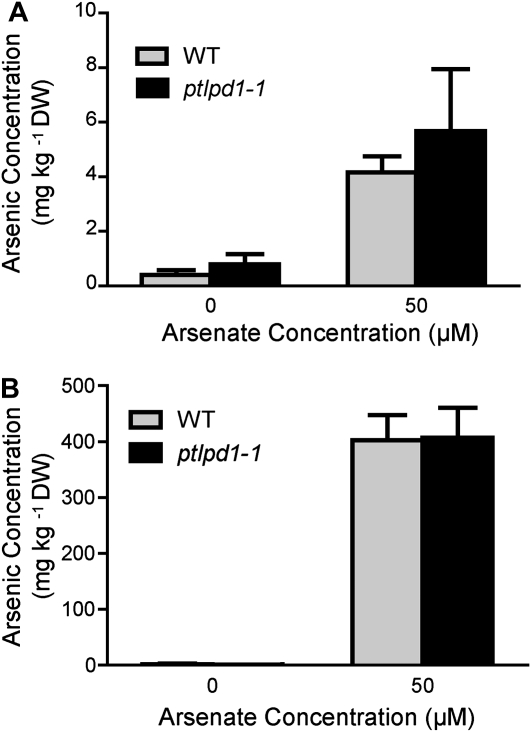
One possible mechanism for the enhanced As(V) sensitivity of ptlpd1 seedlings is an increased accumulation of As in the tissues. The As concentrations in both shoots and roots of wild-type and ptlpd1-1 plants exposed to 50 μm As(V) for 3 d did not differ significantly (Fig. 5), indicating that the aos phenotype conferred by ptlpd1-1 was not due to enhanced As accumulation. Another mechanism for increased As(V) sensitivity could be alteration of the phosphate status of the mutant (Lee et al., 2003). As(V), as an analog of phosphate, can compete with phosphate for uptake (Asher and Reay, 1979; Meharg and Macnair, 1992; Clark et al., 2000) and can replace it in some biochemical reactions (Hughes, 2002; Tseng, 2004). The total phosphate concentrations in shoot and root tissues of the ptlpd1-1 mutant in both the presence and absence of As(V) were similar to those of wild-type plants (data not shown), indicating that the increased As(V) sensitivity in the ptlpd1-1 mutants was not due to a change in tissue phosphate concentration.
Figure 5.
As accumulation in wild-type (WT) and ptlpd1-1 plants. Arabidopsis plants (38 d old) grown in the absence of As (see “Materials and Methods”) were transferred to medium containing 0 or 50 μm As(V) and grown for a further 3 d. The As concentrations in the shoots (A) and roots (B) were determined. Means ± se (n = 4 plants) are shown. DW, Dry weight.
ptlpd1 Mutants Were Also Sensitive to As(III)
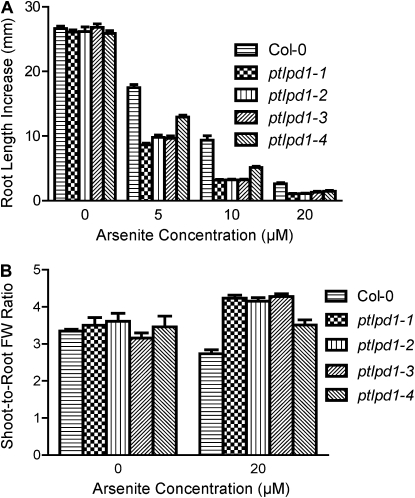
To investigate whether the aos phenotype is As(V) specific, the sensitivity to As(III) of seedlings carrying various ptlpd1 alleles was determined using the root-bending assay. Col-0 was used as the wild-type control, as there was no difference between Col-0 and Col-2 in response to As(III) (data not shown). Root elongation in wild-type Arabidopsis was inhibited by As(III), with the I50 roughly estimated from the data in Figure 6A to be 5 to 10 μm. The ptlpd1 alleles caused a dramatic increase in the sensitivity of root elongation to As(III) (Fig. 6A), with an I50 for As(III) of less than 5 μm for the knockout alleles. As(III) caused a slight decrease in shoot-to-root fresh weight ratio in wild-type seedlings (Fig. 6B). In contrast, seedlings carrying ptlpd1 alleles had an increased shoot-to-root fresh weight ratio when exposed to As(III) (Fig. 6B). As was the case for As(V), ptlpd1-4 seedlings showed a response to As(III) that was intermediate between the wild type and the ptlpd1 knockout alleles (Fig. 6).
Figure 6.
ptlpd1 alleles confer increased sensitivity to As(III). Five-day-old seedlings (see legend to Fig. 1) of wild-type Col-0, ptlpd1-1, ptlpd1-2, ptlpd1-3, or ptlpd1-4 were transferred to solid medium containing As(III) and grown in a vertical orientation. A, Root length increase was measured after 4 d of growth in the presence of As(III). Means ± se (n = 10–15 seedlings) are shown. B, Shoot-to-root fresh weight (FW) ratios were determined after 10 d of growth on As(III)-containing medium. Means ± se (n = 4 plates; two to four seedlings from a plate were pooled for each replicate) are shown.
The specificity of the aos phenotype to As anions was examined by exposing wild-type and ptlpd1-1 mutant seedlings to various concentrations of Cd2+, Zn2+, Ni2+, and Cu2+. After 4 d of exposure, the root elongation of wild-type and ptlpd1-1 mutant seedlings was similar (data not shown), indicating that the aos phenotype was specific for As anions and that mutation of ptLPD1 did not affect the sensitivity of Arabidopsis to divalent heavy metal cations in the growth medium.
ptlpd2 Mutants Also Demonstrate an aos Phenotype
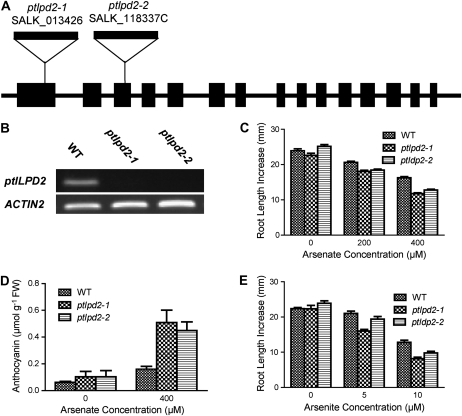
There are two ptLPD genes in the Arabidopsis genome. Having found that loss of ptLPD1 function caused increased sensitivity to As(V) and As(III), the relationship between lipoamide dehydrogenase (LPD) and As was further explored in two Arabidopsis lines where the second gene, ptLPD2 (At4g16155), was disrupted by T-DNA insertion. The T-DNA insertions in the alleles designated ptlpd2-1 (SALK_013426) and ptlpd2-2 (SALK_118337C) were located in the first and third exons of ptLPD2, respectively (Fig. 7A). There were no detectable ptLPD2 transcripts in seedlings homozygous for either ptLPD2 allele (Fig. 7B), indicating that both ptlpd2-1 and ptlpd2-2 were knockout alleles.
Figure 7.
Response of ptlpd2 mutants to As(V) and As(III). A, Schematic diagram showing the T-DNA insertions in the ptlpd2-1 and ptlpd2-2 alleles of At4g16155. Black boxes and lines represent exons and introns/intergenic regions, respectively, with the direction of transcription being left to right. The size of T-DNA is not drawn to scale. B, Semiquantitative RT-PCR determination of ptLPD2 transcript abundance in plants with the wild-type (WT), ptlpd2-1, or ptlpd2-2 allele. Total RNA was extracted from 5-d-old seedlings and reverse transcribed. ACTIN2 (At3g18780) was used as an amplification control. C and D, Five-day-old seedlings (see legend to Fig. 1) homozygous for wild-type, ptlpd2-1, and ptlpd2-2 alleles were transferred to medium supplemented with As(V). Plants were grown for 4 d with plates in a vertical position, then for 10 d with plates in a horizontal position. C, Root growth was measured 4 d after transfer. Means ± se (n = 15–20 seedlings) are shown. D, Anthocyanin accumulation in the shoots of the wild type and ptlpd2 mutants was determined 14 d after transfer. Means ± se (n = 4 plates; two to four seedlings from a plate were pooled for each replicate) are shown. FW, Fresh weight. E, Root elongation response of ptlpd2-1, ptlpd2-2, and the wild type to As(III). Five-day-old seedlings were transferred to solid medium containing As(III). After 4 d of exposure, the increase in root length was measured. Means ± se (n = 15–20 seedlings) are shown.
Similar to the ptlpd1 knockout mutants, the ptlpd2-1 and ptlpd2-2 mutants did not have an apparent phenotype in the absence of As(V), while root elongation was more severely inhibited by As(V) in both mutants than in wild-type seedlings (Fig. 7C). The presence of As(V) in the growth medium also enhanced anthocyanin accumulation in the shoots of both ptlpd2-1 and ptlpd2-2 seedlings compared with that in wild-type seedlings (Fig. 7D). Interestingly, root elongation in the ptlpd2 mutants was less sensitive to As(III) and As(V) compared with that in the ptlpd1 mutants, including the ptlpd1-4 knockdown mutant.
ptLPD Is Sensitive to As(III) But Not As(V)
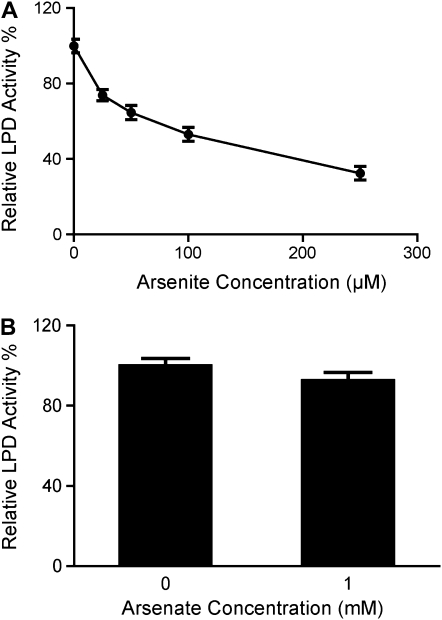
As(III) is a well-known thiol group reagent that has been widely used to identify the reactive disulfide group in LPDs (Massey and Veeger, 1960; Marcinkeviciene and Blanchard, 1997). All known LPD protein sequences, including those of ptLPD1 and ptLPD2 from Arabidopsis, contain two absolutely conserved Cys residues (data not shown) that participate in catalysis through disulfide bridge formation. To further understand the enhanced As(V) sensitivity of the ptlpd knockout mutants, the effect of As on ptLPD activity was determined. As(III) inhibited ptLPD activity in chloroplasts isolated from leaves of wild-type Arabidopsis (Fig. 8A). The I50 for As(III) on the ptLPD activity was estimated from the data in Figure 8A to be about 100 μm. In contrast, As(V), even at a concentration of 1 mm, did not significantly inhibit ptLPD activity in these extracts (Fig. 8B).
Figure 8.
Inhibition of ptLPD by As(III) (A) and As(V) (B). Chloroplasts were isolated from 4-week-old rosette leaves of wild-type Arabidopsis grown in soil. Twelve micrograms of heat-treated chloroplast protein was used to determine the effects of As(III) and As(V) on LPD activity. Means ± se (n ≥ 3) are shown.
DISCUSSION
Three aos mutants were isolated from about 40,000 activation-tagged lines of Arabidopsis. The root growth of aos seedlings was about five times more sensitive to As(V) than that of the wild-type seedlings. Genetic and molecular analyses showed that the aos phenotype in all three mutants was due to a single mutational event that produced a recessive allele that we have designated ptlpd1-1. While T-DNA activation-tagging mutagenesis was designed to produce dominant mutations (Weigel et al., 2000), the isolation of a recessive mutation was not unexpected. T-DNA activation tagging not only has the ability to activate genes near the insertion site but also to disrupt a gene at the insertion site (Weigel et al., 2000).
The ptlpd1-1 mutation was mapped to the ptLPD1 gene. The T-DNA insertion event responsible for this mutation caused the inversion of a 107-kb fragment of chromosome 3, producing a break point in exon 6 of ptLPD1. Genomic DNA rearrangements associated with T-DNA insertions are common. For example, Castle et al. (1993) reported that seven out of 36 T-DNA-tagged embryonic mutants had indications of chromosomal translocations. Several lines of evidence demonstrated that the aos phenotype was caused by disruption of ptLPD1. First, the T-DNA causing the inversion that disrupted ptLPD1 cosegregated with the aos phenotype. Second, transcripts from ptLPD1 were absent from ptlpd1-1 mutant seedlings. Third, two independent ptLPD1 loss-of-function alleles produced phenotypes identical to that of the ptlpd1-1 allele when plants were exposed to As(V). The aos phenotype in the presence of As(V) included reduced root elongation, increased anthocyanin accumulation, and decreased fresh weight compared with wild-type plants. Consistent with this, the ptlpd1-4 knockdown allele, where the T-DNA insertion is located upstream of the coding region, caused a phenotype intermediate between the wild type and the loss-of-function alleles. Fourth, F1 seedlings derived from crosses between ptlpd1-1 and ptlpd1-2 or ptlpd1-3 mutants had a mutant phenotype, demonstrating allelism for these mutations. Finally, the As(V) sensitivity of ptlpd1-1 mutant seedlings was restored to wild-type levels through the introduction of a genomic DNA fragment containing the ptLPD1 gene.
The ptLPD1 gene encodes an isoform of ptLPD, a key component of ptPDC. In Arabidopsis, two genes encode ptLPD, ptLPD1 (At3g16950) and ptLPD2 (At4g16155; Lutziger and Oliver, 2000; Drea et al., 2001). Another two genes, mitochondrial LIPOAMIDE DEHYDROGENASE1 (mtLPD1; At1g48030) and mtLPD2 (At3g17240), encode the mitochondrial forms of LPD (Lutziger and Oliver, 2001) that are found in the mitochondrial pyruvate, α-ketoglutarate, and branched-chain α-ketoacid dehydrogenase complexes and in the Gly decarboxylase complex. In nonphotosynthetic eukaryotes, LPD is exclusively located in mitochondria.
The underlying cause of the increased As(V) sensitivity of the ptlpd1 mutants was not the overaccumulation of As, because the roots and shoots of the ptlpd1 mutant and wild-type plants exhibited similar concentrations of As. It was also not the result of changes in the phosphate status of the plants. Instead, we hypothesize that the higher sensitivity was due to As directly inhibiting residual ptLPD activity, thus inhibiting ptPDC activity. Consistent with this idea, we showed that As(III), but not As(V), inhibited LPD activity, in agreement with studies on LPD activity from bacteria and animals (Massey and Veeger, 1961; Searls et al., 1961; Matthews and Reed, 1963; Ide et al., 1967; Marcinkeviciene and Blanchard, 1997). The mechanism of LPD inhibition is through the binding of As(III) to a catalytic dithiol group formed by two absolutely conserved and nearly adjacent Cys residues within the LPD protein. Plants can rapidly produce As(III) through the reduction of acquired As(V) by endogenous As(V) reductase activity (Zhao et al., 2009). Therefore, the reduced amounts of ptLPD in the plastids of ptlpd1 mutants would provide fewer targets for As(III) binding, leading to a stronger inhibition by As(III) and, consequently, reduced ptPDC activity.
It is unclear how As enters the plastids to exert its toxic effect on ptLPD. Both As(V) and As(III) may be able to cross the plastid envelope. As(V) might enter plastids via phosphate transporters, in a manner analogous to its crossing the plasma membrane. Biochemical studies (Fliege et al., 1978; Borchert et al., 1993) showed that As(V) suppresses phosphate uptake into plastids, suggesting that As(V) and phosphate compete for the same transporters. PHT2;1, a plastid-localized phosphate transporter in Arabidopsis (Versaw and Harrison, 2002), may facilitate the import of As(V) to plastids. Once As(V) is in plastids, it might be reduced to As(III) through an unknown mechanism. It is also possible that As(III) is reduced outside the plastid and then imported by an unknown mechanism.
The available evidence suggests that the aos phenotype of the ptlpd mutants is caused by lowering of ptPDC activity below a critical threshold. ptPDC provides two essential substrates, acetyl-CoA and NADH, for de novo fatty acid biosynthesis (Mooney et al., 2002). Since the plastid is the predominant site of fatty acid biosynthesis in plants, inhibition of ptPDC would have a profound effect on fatty acid production. Fatty acids and fatty acid-derived complex lipids are essential for plants, as for other organisms. They are the components of cellular membranes, the precursors of cellular signaling molecules, such as jasmonic acid, and major energy reserves in storage tissues, such as seeds. Fatty acids and their derivatives are so important that an Arabidopsis knockout mutant of the E2 subunit of ptPDC is early-embryo lethal in the homozygous state (Lin et al., 2003).
The two Arabidopsis genes encoding ptLPDs are likely to be paralogs resulting from a relatively recent gene duplication (Mooney et al., 2002). ptLPD1 and ptLPD2 have similar tissue-dependent transcript profiles, although the relative abundance of transcripts from the two genes differs somewhat among some tissues (Lutziger and Oliver, 2000). The lack of an apparent phenotype for ptLPD1 and ptLPD2 loss-of-function mutants grown in soil or on plates in the absence of As(V) indicates that, under these growth conditions, each ptLPD gene can compensate for the loss of the other. However, the As(V) hypersensitive phenotype of the ptlpd1 knockout mutants indicates that ptLPD2 cannot fully compensate for the loss of ptLPD1 in the presence of As(V). A similar situation exists in the ptlpd2 knockout mutant, although the hypersensitivity is much less severe. Microarray data (Genevestigator; Zimmermann et al., 2004) indicate that ptLPD1 transcripts are more abundant in the root tip than are ptLPD2 transcripts. Thus, the ptlpd1 mutants may be more sensitive to As(V) than the ptlpd2 mutants simply because ptLPD1 is more highly expressed than ptLPD2 in root tips.
The presence of both ptLPD genes is vital for maximizing As resistance, despite their appearance of being largely redundant under normal growth conditions. The absence of ptLPD transcriptional induction in an analysis of Arabidopsis responses to As(V) using microarrays (Abercrombie et al., 2008) suggests that the compensatory effect of one ptLPD gene in the absence of the other is due to the additive nature of their expression, rather than an ability to substitute for one another. The ptlpd1-4 knockdown mutant, which produces ptLPD transcripts, but in lower abundance than wild-type seedlings, displayed an intermediate sensitivity to As(V), further suggesting that loss of ptLPD1 expression cannot be compensated for by an endogenous change in ptLPD2 expression. Therefore, knockout of ptLPD1, with its higher contribution to the ptLPD transcript pool in the root, resulted in greater sensitivity to As(V) than the loss of ptLPD2, which contributes proportionally less to the ptLPD transcript pool. This study has shown that the amount of ptLPD probably plays an essential role in determining the As resistance of Arabidopsis. Thus, the possibility that an increase in the amount of ptLPD through transgenic overexpression would result in greater As tolerance warrants further investigation.
In conclusion, the ptlpd1 mutant is hypersensitive to both As(V) and As(III), due to a loss-of-function mutation in ptLPD1. ptLPD1 and ptLPD2 play largely redundant but additive roles, as loss of function of either ptLPD gene only results in a phenotype when exposed to As(V) or As(III). It is likely that a lower amount of ptLPD in the ptlpd1 mutants resulted in their greater sensitivity to As and that As sensitivity is caused by As(III) binding directly to the catalytic dithiol group of ptLPD. To our knowledge, this is the first study identifying a molecular target for As toxicity in plants. Future overexpression of ptLPD should shed light on its possible ability to confer As tolerance.
MATERIALS AND METHODS
Plant Materials
Seeds of wild-type Arabidopsis (Arabidopsis thaliana), Col-0 and Col-2 accessions, as well as T-DNA insertion lines were obtained from the Arabidopsis Biological Resource Center (http://www.arabidopsis.org/abrc/). The mutant population used in this study was activation-tagged (Weigel et al., 2000) M3 seeds in Col-2 background. Individual T-DNA insertion lines used in this study were WiscDsLox388B02 (ptlpd1-2) and WiscDsLox468E10 (ptlpd1-3) from the Wisconsin collection (Krysan et al., 1999; Woody et al., 2007) and SALK_092097C (ptlpd1-4), SALK_013426 (ptlpd2-1), and SALK_118337C (ptlpd2-2) from the Salk collection (Alonso et al., 2003).
Plant Growth Conditions and Mutant Isolation
Seeds were surface sterilized in 70% (v/v) ethanol for 2 min and in 5% (v/v) hypochlorite with 0.1% (v/v) Tween 20 for 10 min followed by rinsing five times with sterile, distilled water. All plants were grown at 22°C with a 16-h-light (70–100 μmol photons m–2 s–1)/8-h-dark cycle.
A modified root-bending assay (Howden and Cobbett, 1992; Wu et al., 1996) was used to screen for aos mutants. Surface-sterilized seeds were suspended in 0.1% (w/v) agarose and stratified at 4°C for 2 to 3 d. Stratified seeds were sown in rows on plates containing two-thirds-strength Gamborg B-5 basal salts medium (Phytotechnology Laboratories) supplemented with 1% (w/v) agar and 3% (w/v) Suc (Amresco). Plates were placed vertically to allow roots to grow along the surface of the agar medium to facilitate transfer to fresh plates. Four- to 5-d-old seedlings were transferred, one by one, to germination medium supplemented with As(V) (NaH2AsO4) or As(III) (NaAsO2) and placed vertically with root tips pointing upward. After 3 to 4 d of exposure to As(V), seedlings with apparently inhibited root growth were recovered by transferring to fresh germination medium lacking As for about 2 weeks before transfer to soil.
For hydroponic growth, 7-d-old seedlings grown vertically on plates of germination medium were transferred to fresh medium and grown for another 3 weeks with the plate in a horizontal position. Plants were transferred to hydroponics and grown in nutrient solution containing 1.25 mm KNO3, 1.5 mm Ca(NO3)2, 0.75 mm MgSO4, 1 mm KH2PO4, 50 μm KCl, 50 μm H3BO3, 10 μm MnSO4, 2.0 μm ZnSO4, 1.5 μm CuSO4, 0.075 μm (NH4)6Mo7O24, 0.1 mm Na2SiO3, and 25 μm FeNaEDTA. Solutions were changed every 3 d. After 10 d in hydroponics, plants were transferred for 3 d to fresh nutrient solution with or without supplementation with 50 μm As(V).
Growth Measurements
In the root-bending assay, the length of newly elongated root was measured with a ruler from the top of the curl to the root tip after exposure of plants to As-containing medium. For measuring shoot and root fresh weights, shoots were excised in the middle of the hypocotyls. Shoots and roots from a number of plants were pooled and treated as one biological replicate. The seed survival rate was determined according to Lee et al. (2003), except that the plates containing seeds on growth medium were placed vertically.
Choloplast Isolation
Chloroplasts were isolated according to Kunst (1998) with modifications. Four-week-old Arabidopsis leaves were soaked in ice-cold water for 20 min, blotted dry, and homogenized in 0.45 m sorbitol, 20 mm Tricine-KOH, pH 8.4, 10 mm EDTA, 10 mm NaHCO3, 0.1% (w/v) bovine serum albumin, 5 mm Cys, and 5 mm glutathione. The homogenate was filtered through four layers of Miracloth (Calbiochem), and chloroplasts were collected by centrifugation at 2,000g for 5 min. The chloroplast pellet was gently resuspended in ice-cold resuspension buffer (0.3 m sorbitol, 20 mm Tricine-KOH, pH 7.6, 5 mm MgCl2, and 2.5 mm EDTA), followed by layering of the suspension onto a preformed linear density gradient made by centrifugation of 50% (v/v) Percoll, 0.3 m sorbitol, 20 mm Tricine-KOH, pH 7.6, 5 mm MgCl2, 2.5 mm EDTA, and 0.6 mm glutathione at 43,000g for 30 min followed by deceleration without a brake. Chloroplasts were separated by centrifugation at 12,100g for 20 min, withdrawn in the lower band, collected by centrifugation 2,000g for 5 min, and washed twice with resuspension buffer.
LPD Activity Determination
Isolated chloroplasts were incubated for 1 h at 65°C in the presence of 1% (v/v) Triton X-100, and the lysate was clarified by centrifugation at 20,000g for 20 min to remove pigment and protein aggregates (Conner et al., 1996). The protein concentration in the supernatant was determined using a commercial kit (Coomassie Plus Reagent; Pierce Chemical) according to the supplier's instructions. LPD activity was determined as NADH consumption at 25°C by measuring the change in A340 (Cary 300 Biospectrophotometer; Varian). The reaction mixture contained 50 mm TES-KOH, pH 7.5, 2 mm MgCl2, 0.05 mm NADH, 1 mm dl-α-lipoamide (Sigma-Aldrich), and 12 μg mL−1 clarified chloroplast protein. The reaction was initiated by adding the clarified chloroplast protein.
Anthocyanin Extraction and Estimation
Total anthocyanins were extracted according to Lange et al. (1971). Shoots of seedlings were weighed fresh and placed in 1.5 mL of 1-propanol:HCl:water (18:1:81). Samples were submerged in boiling water for 3 min, followed by incubation in the dark at 22°C for 24 h. Insoluble material was removed by centrifugation at 14,000g for 15 min at ambient temperature, before the optical density of the supernatant was measured at 535 and 650 nm. The absorbance due to anthocyanins was calculated as A = A535 – 0.24 A650 (Lange et al., 1971). The quantity of anthocyanins was determined from the corrected absorbance using a molar extinction coefficient of 38,000 L mol−1 cm−1 and normalized to the fresh weight of each sample.
Measurement of As
Shoot and root tissues were harvested separately, rinsed three times with distilled water, and dried at 70°C for 3 d. Dried tissues were weighed and digested with nitric acid at 250°C for 5 min and at 500°C for 5 min in a microwave extraction system (Microwave Laboratory Station Mileston ETHOS 900; Milestone). The As content of the digested samples was analyzed by inductively coupled plasma optical emission spectrometry and normalized to the dry weight of each sample.
PCR and DNA Sequencing
DNA samples were amplified in a thermal cycler (Eppendorf) using a thermostable DNA polymerase (GoTaq Flexi DNA Polymerase; Promega) according to the enzyme manufacturer's instructions. Unless specified otherwise, the thermal cycling parameters used were as follows: one cycle of 95°C for 2 min; 23 to 40 cycles of 30 s at 95°C, 30 s at 55°C, and 1 to 3 min at 72°C; and one cycle of 10 min at 72°C. PCR products were separated by electrophoresis on agarose gels and purified from excised gel slices using a commercial kit (AxyPrep DNA Gel Extraction Kit; Axygen). The purified PCR products were used in dye-terminator sequencing reactions using a commercial kit (BigDye Terminator Version 3.1 Sequencing Kit; Applied Biosystems) according to the manufacturer's instructions followed by separation and analysis (Lottery West Biomedical Facility: Genomics).
TAIL-PCR
TAIL-PCR was done according to Liu et al. (1995). Genomic DNA was prepared using a commercial kit (PhytoPure Plant Genomic DNA Extraction Kit; Amersham Biosciences) according to the manufacturer's instructions. The primers (Supplemental Table S2) used to recover genomic DNA sequences flanking the T-DNA left border of pSKI015 were LB1, LB2, and LB3, in combination with DEG1. In other TAIL-PCR experiments, T1 and T2 were used in combination with DEG1. PCR conditions were those of Liu et al. (1995).
Verification of T-DNA Insertion Sites
The annotated positions of T-DNA insertions were confirmed by PCR using the T-DNA insert-specific primers LBa1 for the SALK lines, p745 for the Wisconsin (Wisc) lines, and LB3 for the activation-tagged mutants, in combination with the sequence-specific primers 66F and 66R for genotyping WiscDsLox388B02 and WiscDsLox468E10; 67F and 67R for genotyping SALK_118337C; 8F and 8R for genotyping ptlpd1-1; and 69F and 69R for genotyping SALK_118337C and SALK_013426 (Supplemental Table S2).
RT-PCR
Total RNA was extracted from 7-d-old seedlings using a commercial kit (RNeasy Plant Mini Prep Kit; Qiagen) according to the manufacturer's instructions and treated with DNA-free DNase I (Ambion) according to the manufacturer's instructions. First-strand cDNA was synthesized from 1 μg of total RNA with an oligo(dT) primer using a commercial kit (MonsterScript 1st-Strand cDNA Synthesis Kit; Epicentre Biotechnologies) according to the manufacturer's instructions and the primers ptLPDF and ptLPD1R for ptLPD1, ptLPDF and ptLPD2R for ptLPD2, and actin2F and actin2R for ACTIN2 (Supplemental Table S2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. As(V) inhibition of Arabidopsis root elongation.
Supplemental Figure S2. T-DNA integration sites in the ptlpd1-1 mutant lines.
Supplemental Figure S3. As(V) sensitivity of seedlings with ptlpd1 alleles.
Supplemental Figure S4. Transformation of ptlpd1-1 with a genomic DNA fragment encompassing the ptLPD1 gene restores wild-type As(V) sensitivity.
Supplemental Table S1. Genetic analysis of ptlpd1-1 mutant lines in Arabidopsis.
Supplemental Table S2. Primer sequences used in this study.
Supplemental Materials and Methods S1. Plasmid construction and transformation of Arabidopsis.
Supplementary Material
Acknowledgments
We thank Michael Smirk (University of Western Australia) for his assistance in determining the As concentrations and Fazilah Abd Manan (University of Western Australia) for her help in isolating chloroplasts. We are grateful to Oliver Berkowitz (Murdoch University) for his critical reading of the manuscript.
References
- Abercrombie JM, Halfhill MD, Ranjan P, Rao MR, Saxton AM, Yuan JS, Stewart CN. (2008) Transcriptional responses of Arabidopsis thaliana plants to As (V) stress. BMC Plant Biol 8: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamson SR, Stevenson KJ. (1981) Inhibition of pyruvate dehydrogenase multienzyme complex from Escherichia coli with a bifunctional arsenoxide: selective inactivation of lipoamide dehydrogenase. Biochemistry 20: 3418–3424 [DOI] [PubMed] [Google Scholar]
- Ahsan N, Lee DG, Alam I, Kim PJ, Lee JJ, Ahn YO, Kwak SS, Lee IJ, Bahk JD, Kang KY, et al. (2008) Comparative proteomic study of arsenic-induced differentially expressed proteins in rice roots reveals glutathione plays a central role during As stress. Proteomics 8: 3561–3576 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Asher CJ, Reay PF. (1979) Arsenic uptake by barley seedlings. Aust J Plant Physiol 6: 459–466 [Google Scholar]
- Bienert GP, Thorsen M, Schussler MD, Nilsson HR, Wagner A, Tamas MJ, Jahn TP. (2008) A subgroup of plant aquaporins facilitate the bi-directional diffusion of As(OH)3 and Sb(OH)3 across membranes. BMC Biol 6: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleeker PM, Hakvoort HWJ, Bliek M, Souer E, Schat H. (2006) Enhanced arsenate reduction by a CDC25-like tyrosine phosphatase explains increased phytochelatin accumulation in arsenate-tolerant Holcus lanatus. Plant J 45: 917–929 [DOI] [PubMed] [Google Scholar]
- Borchert S, Harborth J, Schunemann D, Hoferichter P, Heldt HW. (1993) Studies of the enzymic capacities and transport properties of pea root plastids. Plant Physiol 101: 303–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle LA, Errampalli D, Atherton TL, Franzmann LH, Yoon ES, Meinke DW. (1993) Genetic and molecular characterization of embryonic mutants identified following seed transformation in Arabidopsis. Mol Gen Genet 241: 504–514 [DOI] [PubMed] [Google Scholar]
- Catarecha P, Segura MD, Franco-Zorrilla JM, García-Ponce B, Lanza M, Solano R, Paz-Ares J, Leyva A. (2007) A mutant of the Arabidopsis phosphate transporter PHT1;1 displays enhanced arsenic accumulation. Plant Cell 19: 1123–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavigelli M, Li WW, Lin AN, Su B, Yoshioka K, Karin M. (1996) The tumor promoter arsenite stimulates AP-1 activity by inhibiting a JNK phosphatase. EMBO J 15: 6269–6279 [PMC free article] [PubMed] [Google Scholar]
- Clark GT, Dunlop J, Phung HT. (2000) Phosphate absorption by Arabidopsis thaliana: interactions between phosphorus status and inhibition by arsenate. Aust J Plant Physiol 27: 959–965 [Google Scholar]
- Conner M, Krell T, Lindsay JG. (1996) Identification and purification of a distinct dihydrolipoamide dehydrogenase from pea chloroplasts. Planta 200: 195–202 [DOI] [PubMed] [Google Scholar]
- Dhankher OP, Li Y, Rosen BP, Shi J, Salt D, Senecoff JF, Sashti NA, Meagher RB. (2002) Engineering tolerance and hyperaccumulation of arsenic in plants by combining arsenate reductase and γ-glutamylcysteine synthetase expression. Nat Biotechnol 20: 1140–1145 [DOI] [PubMed] [Google Scholar]
- Drea SC, Mould RM, Hibberd JM, Gray JC, Kavanagh TA. (2001) Tissue-specific and developmental-specific expression of an Arabidopsis thaliana gene encoding the lipoamide dehydrogenase component of the plastid pyruvate dehydrogenase complex. Plant Mol Biol 46: 705–715 [DOI] [PubMed] [Google Scholar]
- Fliege R, Flugge UI, Werdan K, Heldt HW. (1978) Specific transport of inorganic phosphate, 3-phosphoglycerate and triosephosphates across the inner membrane of the envelope in spinach chloroplasts. Biochim Biophys Acta 502: 232–247 [DOI] [PubMed] [Google Scholar]
- González E, Solano R, Rubio V, Leyva A, Paz-Ares J. (2005) PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 is a plant-specific SEC12-related protein that enables the endoplasmic reticulum exit of a high-affinity phosphate transporter in Arabidopsis. Plant Cell 17: 3500–3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresser MJ. (1981) ADP-arsenate: formation by submitochondrial particles under phosphorylating conditions. J Biol Chem 256: 5981–5983 [PubMed] [Google Scholar]
- Grill E, Winnacker EL, Zenk MH. (1987) Phytochelatins, a class of heavy-metal binding peptides from plants, are functionally analogous to metallothioneins. Proc Natl Acad Sci USA 84: 439–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden R, Cobbett CS. (1992) Cadmium-sensitive mutants of Arabidopsis thaliana. Plant Physiol 100: 100–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Su L, Snow ET. (1998) Arsenic toxicity is enzyme specific and its effects on ligation are not caused by the direct inhibition of DNA repair enzymes. Mutat Res 408: 203–218 [DOI] [PubMed] [Google Scholar]
- Hughes MF. (2002) Arsenic toxicity and potential mechanisms of action. Toxicol Lett 133: 1–16 [DOI] [PubMed] [Google Scholar]
- Ide S, Hayakawa T, Okabe K, Koike M. (1967) Lipoamide dehydrogenase from human liver. J Biol Chem 242: 54–60 [PubMed] [Google Scholar]
- Isayenkov SV, Maathuis FJM. (2008) The Arabidopsis thaliana aquaglyceroporin AtNIP7;1 is a pathway for arsenite uptake. FEBS Lett 582: 1625–1628 [DOI] [PubMed] [Google Scholar]
- Kamiya T, Tanaka M, Mitani N, Ma JF, Maeshima M, Fujiwara T. (2009) NIP1;1, an aquaporin homolog, determines the arsenite sensitivity of Arabidopsis thaliana. J Biol Chem 284: 2114–2120 [DOI] [PubMed] [Google Scholar]
- Kitchin KT, Wallace K. (2008) The role of protein binding of trivalent arsenicals in arsenic carcinogenesis and toxicity. J Inorg Biochem 102: 532–539 [DOI] [PubMed] [Google Scholar]
- Krysan PJ, Young JC, Sussman MR. (1999) T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11: 2283–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst L. (1998) Preparation of physiologically active chloroplasts from Arabidopsis. Martínez-Zapater JM, Salinas J, , Methods in Molecular Biology: Arabidopsis Protocols. Humana Press, Totowa, NJ, pp 43–48 [DOI] [PubMed] [Google Scholar]
- Lange H, Shropshire W, Jr, Mohr H. (1971) An analysis of phytochrome-mediated anthocyanin synthesis. Plant Physiol 47: 649–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufs P, Autran D, Traas J. (1999) A chromosomal paracentric inversion associated with T-DNA integration in Arabidopsis. Plant J 18: 131–139 [DOI] [PubMed] [Google Scholar]
- Lee DA, Chen A, Schroeder JI. (2003) ars1, an Arabidopsis mutant exhibiting increased tolerance to arsenate and increased phosphate uptake. Plant J 35: 637–646 [DOI] [PubMed] [Google Scholar]
- Lin M, Behal R, Oliver DJ. (2003) Disruption of plE2, the gene for the E2 subunit of the plastid pyruvate dehydrogenase complex, in Arabidopsis causes an early embryo lethal phenotype. Plant Mol Biol 52: 865–872 [DOI] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF. (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8: 457–463 [DOI] [PubMed] [Google Scholar]
- Lombi E, Zhao FJ, Fuhrmann M, Ma LQ, McGrath SP. (2002) Arsenic distribution and speciation in the fronds of the hyperaccumulator Pteris vittata. New Phytol 156: 195–203 [DOI] [PubMed] [Google Scholar]
- Lutziger I, Oliver DJ. (2000) Molecular evidence of a unique lipoamide dehydrogenase in plastids: analysis of plastidic lipoamide dehydrogenase from Arabidopsis thaliana. FEBS Lett 484: 12–16 [DOI] [PubMed] [Google Scholar]
- Lutziger I, Oliver DJ. (2001) Characterization of two cDNAs encoding mitochondrial lipoamide dehydrogenase from Arabidopsis. Plant Physiol 127: 615–623 [PMC free article] [PubMed] [Google Scholar]
- Lynn S, Lai HT, Gurr JR, Jan KY. (1997) Arsenite retards DNA break rejoining by inhibiting DNA ligation. Mutagenesis 12: 353–358 [DOI] [PubMed] [Google Scholar]
- Ma JF, Yamaji N, Mitani N, Xu XY, Su YH, McGrath SP, Zhao FJ. (2008) Transporters of arsenite in rice and their role in arsenic accumulation in rice grain. Proc Natl Acad Sci USA 105: 9931–9935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal BK, Suzuki KT. (2002) Arsenic round the world: a review. Talanta 58: 201–235 [PubMed] [Google Scholar]
- Marcinkeviciene J, Blanchard JS. (1997) Catalytic properties of lipoamide dehydrogenase from Mycobacterium smegmatis. Arch Biochem Biophys 340: 168–176 [DOI] [PubMed] [Google Scholar]
- Marin AR, Masscheleyn PH, Patrick WH., Jr (1993) Soil redox-pH stability of arsenic species and its influence on arsenic uptake by rice. Plant Soil 152: 245–253 [Google Scholar]
- Masscheleyn PH, Delaune RD, Patrick WH., Jr (2002) Effect of redox potential and pH on arsenic speciation and solubility in a contaminated soil. Environ Sci Technol 25: 1414–1419 [Google Scholar]
- Massey V, Veeger C. (1960) On the reaction mechanism of lipoyl dehydrogenase. Biochim Biophys Acta 40: 184–185 [DOI] [PubMed] [Google Scholar]
- Massey V, Veeger C. (1961) Studies on reaction mechanism of lipoyl dehydrogenase. Biochim Biophys Acta 48: 33–47 [DOI] [PubMed] [Google Scholar]
- Matthews J, Reed LJ. (1963) Purification and properties of a dihydrolipoic dehydrogenase from Spinacia oleracea. J Biol Chem 238: 1869–1876 [PubMed] [Google Scholar]
- Meharg AA, Hartley-Whitaker J. (2002) Arsenic uptake and metabolism in arsenic resistant and nonresistant plant species. New Phytol 154: 29–43 [Google Scholar]
- Meharg AA, Macnair MR. (1992) Suppression of the high affinity phosphate uptake system: a mechanism of arsenate tolerance in Holcus lanatus L. J Exp Bot 43: 519–524 [Google Scholar]
- Mooney BP, Miernyk JA, Randall DD. (2002) The complex fate of α-ketoacids. Annu Rev Plant Biol 53: 357–375 [DOI] [PubMed] [Google Scholar]
- Moore SA, Moennich DM, Gresser MJ. (1983) Synthesis and hydrolysis of ADP-arsenate by beef heart submitochondrial particles. J Biol Chem 258: 6266–6271 [PubMed] [Google Scholar]
- Nacry P, Camilleri C, Courtial B, Caboche M, Bouchez D. (1998) Major chromosomal rearrangements induced by T-DNA transformation in Arabidopsis. Genetics 149: 641–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onken BM, Hossner LR. (1995) Plant uptake and determination of arsenic species in soil solution under flooded conditions. J Environ Qual 24: 373–381 [Google Scholar]
- Onken BM, Hossner LR. (1996) Determination of arsenic species in soil solution under flooded conditions. Soil Sci Soc Am J 60: 1385–1392 [Google Scholar]
- Pickering IJ, Gumaelius L, Harris HH, Prince RC, Hirsch G, Banks JA, Salt DE, George GN. (2006) Localizing the biochemical transformations of arsenate in a hyperaccumulating fern. Environ Sci Technol 40: 5010–5014 [DOI] [PubMed] [Google Scholar]
- Quaghebeur M, Rengel Z. (2004) Arsenic uptake, translocation and speciation in pho1 and pho2 mutants of Arabidopsis thaliana. Physiol Plant 120: 280–286 [DOI] [PubMed] [Google Scholar]
- Schmöger MEV, Oven M, Grill E. (2000) Detoxification of arsenic by phytochelatins in plants. Plant Physiol 122: 793–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz H, Härtling S, Tanneberg H. (2008) The identification and quantification of arsenic-induced phytochelatins: comparison between plants with varying As sensitivities. Plant Soil 303: 275–287 [Google Scholar]
- Schweighofer A, Hirt H, Meskiene I. (2004) Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci 9: 236–243 [DOI] [PubMed] [Google Scholar]
- Searls RL, Peters JM, Sanadi DR. (1961) Alpha-ketoglutaric dehydrogenase. X. On the mechanism of dihydrolipoyl dehydrogenase reaction. J Biol Chem 236: 2317–2322 [PubMed] [Google Scholar]
- Shin H, Shin HS, Dewbre GR, Harrison MJ. (2004) Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J 39: 629–642 [DOI] [PubMed] [Google Scholar]
- Smith SE, Christophersen HM, Pope S, Smith FA. (2010) Arsenic uptake and toxicity in plants: integrating mycorrhizal influences. Plant Soil 327: 1–21 [Google Scholar]
- Sneller FEC, Heerwaarden LMV, Kraaijeveld-Smit FJL, Bookum WMT, Koevoets PLMHS, Verkleij JAC. (1999) Toxicity of arsenate in Silene vulgaris, accumulation and degradation of arsenate-induced phytochelatins. New Phytol 144: 223–232 [Google Scholar]
- Tax FE, Vernon DM. (2001) T-DNA-associated duplication/translocations in Arabidopsis: implications for mutant analysis and functional genomics. Plant Physiol 126: 1527–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng CH. (2004) The potential biological mechanisms of arsenic-induced diabetes mellitus. Toxicol Appl Pharmacol 197: 67–83 [DOI] [PubMed] [Google Scholar]
- Tuan LQ, Huong TTT, Hong PTA, Kawakami T, Shimanouchi T, Umakoshi H, Kuboi R. (2008) Arsenic (V) induces a fluidization of algal cell and liposome membranes. Toxicol In Vitro 22: 1632–1638 [DOI] [PubMed] [Google Scholar]
- Verbruggen N, Hermans C, Schat H. (2009) Mechanisms to cope with arsenic or cadmium excess in plants. Curr Opin Plant Biol 12: 364–372 [DOI] [PubMed] [Google Scholar]
- Versaw WK, Harrison MJ. (2002) A chloroplast phosphate transporter, PHT2;1, influences allocation of phosphate within the plant and phosphate-starvation responses. Plant Cell 14: 1751–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blazquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrandiz C, Kardailsky I, Malancharuvil EJ, Neff MM, et al. (2000) Activation tagging in Arabidopsis. Plant Physiol 122: 1003–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woody S, Austin-Phillips S, Amasino R, Krysan P. (2007) The WiscDsLox T-DNA collection: an Arabidopsis community resource generated by using an improved high-throughput T-DNA sequencing pipeline. J Plant Res 120: 157–165 [DOI] [PubMed] [Google Scholar]
- Wu SJ, Ding L, Zhu JK. (1996) SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell 8: 617–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao FJ, Ma JF, Meharg AA, McGrath SP. (2009) Arsenic uptake and metabolism in plants. New Phytol 181: 777–794 [DOI] [PubMed] [Google Scholar]
- Zhu JK. (2000) Genetic analysis of plant salt tolerance using Arabidopsis. Plant Physiol 124: 941–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. (2003) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.