Abstract
Pollen grains of land plants have evolved remarkably strong outer walls referred to as exine that protect pollen and interact with female stigma cells. Exine is composed of sporopollenin, and while the composition and synthesis of this biopolymer are not well understood, both fatty acids and phenolics are likely components. Here, we describe mutations in the Arabidopsis (Arabidopsis thaliana) LESS ADHESIVE POLLEN (LAP5) and LAP6 that affect exine development. Mutation of either gene results in abnormal exine patterning, whereas pollen of double mutants lacked exine deposition and subsequently collapsed, causing male sterility. LAP5 and LAP6 encode anther-specific proteins with homology to chalcone synthase, a key flavonoid biosynthesis enzyme. lap5 and lap6 mutations reduced the accumulation of flavonoid precursors and flavonoids in developing anthers, suggesting a role in the synthesis of phenolic constituents of sporopollenin. Our in vitro functional analysis of LAP5 and LAP6 using 4-coumaroyl-coenzyme A yielded bis-noryangonin (a commonly reported derailment product of chalcone synthase), while similar in vitro analyses using fatty acyl-coenzyme A as the substrate yielded medium-chain alkyl pyrones. Thus, in vitro assays indicate that LAP5 and LAP6 are multifunctional enzymes and may play a role in both the synthesis of pollen fatty acids and phenolics found in exine. Finally, the genetic interaction between LAP5 and an anther gene involved in fatty acid hydroxylation (CYP703A2) demonstrated that they act synergistically in exine production.
Pollen grains of land plants are surrounded by complex cell walls that are divided into three layers: (1) an outer exine, itself a multilayered structure, primarily made of sporopollenin; (2) an inner intine, made primarily of cellulose; and (3) a lipid- and protein-rich pollen coat in the crevices of exine. The exine is morphologically diverse, provides protection against environmental stresses and bacterial and fungal attacks, and plays a role in species-specific adhesion (Zinkl et al., 1999; Edlund et al., 2004).
Several studies indicate that sporopollenin is a complex polymer composed of fatty acids and phenolic compounds (Guilford et al., 1988; Ahlers et al., 1999; Wiermann et al., 2001). Sporopollenin is remarkably strong and chemically resistant, making it difficult to determine its precise composition by direct chemical analysis. Ozonolysis has yielded simple straight- and branched-chain monocarboxylic acids, typical of fatty acid breakdown (Shaw, 1971), as well as phenolic acids, such as p-hydroxybenzoic, m-hydroxybenzoic, and protocatechuic acids. Additional evidence for phenolic compounds came from degradation experiments or studies showing the incorporation of radiolabeled Phe and p-coumaric acid into sporopollenin (Shulze Osthoff and Wiermann, 1987; Rittscher and Wiermann, 1988; Gubatz et al., 1993), while immunolocalization studies with anti-p-coumaric acid antibodies demonstrated the occurrence of phenols in exines of different plant species (Niester-Nyveld et al., 1997).
While a growing number of genes have been identified that are important for exine development, still relatively little is known about the genetic network that governs the formation of this structure, and the pathways that lead to its biosynthesis are far from being understood. In recent years, the importance of fatty acid-derived components in sporopollenin composition has been revealed through the identification of several Arabidopsis (Arabidopsis thaliana) genes, such as MALE STERILITY2 (MS2; Aarts et al., 1997), cytochrome P450 CYP703A2 (Morant et al., 2007), cytochrome P450 CYP704B1 (Dobritsa et al., 2009), and ACYL-CoA SYNTHETASE5 (ACOS5; de Azevedo Souza et al., 2009), which are important for exine production and involved in fatty acid metabolism. Less is known concerning the role of phenolics in sporopollenin biosynthesis, and the key synthetic and regulatory genes specifically associated with this aspect of sporopollenin biosynthesis are absent from the literature.
Phenolic compounds are a large class of secondary metabolites that play a variety of biological roles (Hahlbrock and Scheel, 1989). Most plant phenolics are products of phenylpropanoid metabolism, including lignins, coumarins, stilbenes, and flavonoids. A well-characterized biosynthetic pathway leads to the biosynthesis of flavonoids (Supplemental Fig. S1). Chalcone synthase (CHS) catalyzes the first committed step in this pathway using 4-coumaroyl-CoA provided by 4-coumaroyl:CoA ligase as a substrate. Flavonoids are important for pollen germination and plant fertility in several plant species (Coe et al., 1981; Taylor and Jorgensen, 1992; van der Meer et al., 1992; Fischer et al., 1997; Napoli et al., 1999), while a null mutation in the Arabidopsis CHS gene, TRANSPARENT TESTA4 (TT4), results in plants with normal fertility and an absence of flavonoids in the mature stamens (Burbulis et al., 1996; Ylstra et al., 1996). This suggests that flavonoids are either not required for Arabidopsis male fertility or that TT4-independent flavonoid synthesis occurs in anthers, perhaps transiently and at an earlier developmental stage, through a mechanism that has not been detected in previous experiments.
Recently, an anther-specific gene, ACOS5, was described that is essential for exine production and sporopollenin biosynthesis (de Azevedo Souza et al., 2009). ACOS5 is related to a phenylpropanoid enzyme, 4-coumaroyl:CoA ligase, but encodes a novel medium- to long-chain fatty acyl-CoA synthetase. In this study, we describe the identification and characterization of two highly conserved anther-specific genes that are involved in pollen exine development, likely participate in sporopollenin biosynthesis, and, similar to ACOS5, are related to, yet distinct from, an enzyme of the phenylpropanoid pathway. Our results provide further insight into the mechanism that leads to the formation of sporopollenin.
RESULTS
lap5 and lap6 Mutants Have Defective Exine Patterning
We recovered less adhesive pollen mutants lap5 and lap6 in a screen for genes that play a role in Arabidopsis pollen-stigma adhesion (Nishikawa et al., 2005). Both mutants had morphological defects in exine structure that were apparent under light microscopy or scanning electron microscopy (SEM; Fig. 1). The mutant anthers and pollen appeared glossy with a dissection stereomicroscope (Fig. 1, A–C). The pollen adhered tightly to the anthers and was not easily released. Under SEM, lap5 and lap6 pollen grains exhibited abnormal exine patterning (Fig. 1, D–I): lap5 pollen lacked the characteristic reticulate structure (Fig. 1H), and lap6 pollen grains had a more extensively covered surface with broader muri and smaller lacunae (Fig. 1I). Pollen grains of both mutants collapsed more easily under vacuum than the wild type, although the thickness of the exine layer was unaltered (data not shown).
Figure 1.
lap5 and lap6 mutants have defects in anther and pollen exine morphology. A to C, Compared with the wild-type (WT) anthers (A), anthers of lap5-1 (B) and lap6-1 (C) mutants appear glossy and do not easily shed pollen (arrowhead in A). D to I, SEM of pollen grains and exine. Wild-type grains (D and G) have a regular reticulate exine pattern, while lap5-1 (E and H) and lap6-1 (F and I) mutations cause pollen to collapse more easily and disrupt exine, changing the pattern or resulting in a more extensively covered surface. J to L, Similar to the wild type (J), lap5-1 (K) and lap6-1 (L) pollen does not demonstrate sensitivity to acetolysis. The lap6-1 grains, however, exhibit decreased reactivity to acetolysis-dependent staining. Bars = 100 μm (A–C), 10 μm (D–F and J–L), and 5 μm (G–I).
Defects in some Arabidopsis exine mutants, such as ms2 (Aarts et al., 1997), flp1 (Ariizumi et al., 2003), and cyp704b1 (Dobritsa et al., 2009), render pollen sensitive to acetolysis, an oxidative process often used by palynologists to prepare exine samples (Erdtman, 1960). We incubated lap5 and lap6 pollen in a mixture of acetic anhydrate and sulfuric acid but did not observe sensitivity. Interestingly, lap6 but not lap5 exine consistently showed reduced staining, suggesting that the two mutants are different in the quality or amounts of the exine constituents that react with the acetolysis mixture (Fig. 1, J–L).
Mapping LAP5 and LAP6
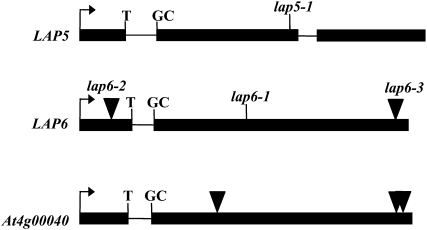
Both lap5 and lap6 mutations segregated as single-locus, homozygous recessive mutations with phenotypic ratios (wild-type:mutant) of 1,008:326 (lap5) and 740:219 (lap6), consistent with 3:1 segregation (χ2, P > 0.5 and 0.5 > P > 0.1, respectively). Using bulked segregant analysis (Michelmore et al., 1991), we mapped LAP5 to chromosome 4 and LAP6 to chromosome 1 (see “Materials and Methods”). Additional PCR-based markers were used to refine the locations to within 59 kb for LAP5 and 140 kb for LAP6. One gene in each interval, At4g34850 and At1g02050, respectively, contained mutations within open reading frames (ORFs) that cosegregated with the lap5 and lap6 mutant phenotypes. Both candidate genes encoded predicted CHS family proteins. The mutations in the genes resulted in missense codons, causing a G227E conversion in lap5 and an I132T conversion in lap6. In addition to the lap6-1 allele that we isolated, Arabidopsis stock centers contained two lines with T-DNA or transposon insertions in At1g02050 (SALK_134643 [lap6-2] and N172991 [lap6-3]; Fig. 2). We confirmed the positions of these insertions in the first exon (lap6-2) and in the second exon close to the end of the ORF (lap6-3). Both insertion alleles had pollen and anther phenotypes identical to those of lap6-1 (data not shown).
Figure 2.
The lap5 and lap6 defects map to At4g34850 and At1g02050, respectively. LAP5, LAP6, and At4g00040 have gene structures similar to CHS/STS family members, including the conserved position of an intron separating the first and second nucleotides in a Cys codon (TGC). Exons are shown as black rectangles. Positions of the lap5-1 and lap6-1 point mutations and the lap6-2, lap6-3, and At1g00040 insertions are indicated with black triangles.
To confirm the identity of the LAP5 and LAP6 genes, complementation constructs were created containing the entire At4g34850 or At1g02050 genes, including the native promoters, and transformed, respectively, into lap5-1 and lap6-1. Wild-type anther and pollen phenotypes were restored in 38 of 40 BASTA-resistant T1 plants for lap5. Of the remaining two plants, one had a lap5 phenotype and the other lacked pollen and was sterile. For lap6, 35 of 40 BASTA-resistant T1 plants had restored the wild-type phenotype. The remaining five plants had the lap6 phenotype. We confirmed the presence of the lap5-1 and lap6-1 mutations in the rescued lines using cleaved-amplified polymorphic sequence (CAPS) markers. These results indicate that wild-type copies of At4g34850 and At1g02050 can complement the lap5-1 and lap6-1 mutants, respectively, and that the constructs used contained sufficient cis-regulatory elements to drive expression in a spatially and temporally relevant manner.
LAP5 and LAP6 Encode CHS Superfamily Proteins
The predicted LAP5 and LAP6 proteins are similar to enzymes in the CHS superfamily (Fig. 3). Members of this superfamily are plant-specific type III polyketide synthases (PKS) and catalyze condensing reactions that generate the backbone required for the synthesis of a large number of phenylpropanoid compounds, such as flavonoids and stilbenes (Schröder, 1997; Austin and Noel, 2003). BLAST results revealed that, in addition to these two genes and a well-characterized CHS TT4 (Shirley et al., 1995), Arabidopsis contains a third CHS-related gene, At4g00040.
Figure 3.
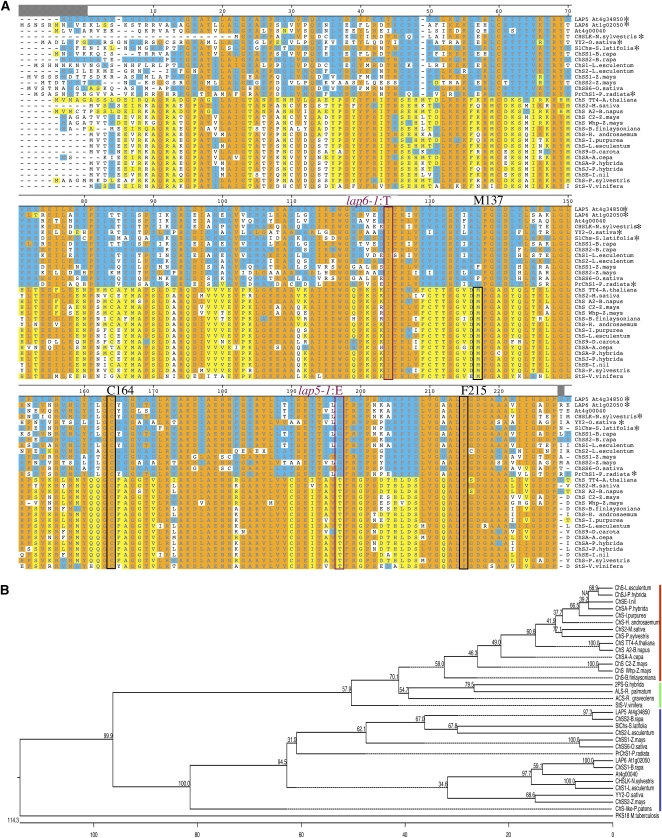
LAP5, LAP6, and At1g00040 belong to a male-specific clade of the CHS/STS family. A, Amino acid alignment of protein sequences from LAP5, LAP6, At1g00040, CHS, STS, and CHS/STS-like proteins from several plant species (two-thirds of the protein length is shown; the remainder shows a similar trend). Residues identical to LAP5 are shaded blue; those identical to the Arabidopsis CHS TT4 are shaded yellow; and those that match the majority consensus from 17 sequences are shaded orange. Amino acid numbers correspond to CHS2 from M. sativa. Proteins with demonstrated male-specific expression patterns are marked with asterisks; the lap5-1 and lap6-1 lesions are indicated, and two of the four amino acids critical for active center formation (C164 and F215) are boxed. M137, important for dimer formation, is conserved in the CHS-like enzymes (boxed) but replaced with L/F in enzymes more similar to LAP5. B, A phylogenetic tree demonstrates a clear segregation of the LAP5/LAP6-like proteins in a clade separate from the other members of the CHS superfamily. Red bar, CHS; blue bar, sequences similar to LAP5; green bar, four CHS-like proteins with demonstrated non-CHS activity (2PS, aloesone synthase [ALS], acridone synthase [ACS], and STS). Numerical values indicate bootstrap support for each node. PKS18 from M. tuberculosis was used as an outgroup.
CHSs (EC 2.3.1.74) from different plants often share 80% to 90% sequence identity (Niesbach-Klösgen et al., 1987). In contrast, LAP5, LAP6, and At4g00040 share only 40% sequence identity with the Arabidopsis CHS TT4 or with CHSs from other organisms. However, they share 64% (LAP5-LAP6), 76% (At4g00040-LAP5), and 62% (At4g00040-LAP6) identity with each other and are members of a male-organ-specific CHS-like protein clade, which includes proteins from dicots, monocots, and gymnosperms (BA42 from Brassica napus [Shen and Hsu, 1992], YY2 from rice [Oryza sativa; Hihara et al., 1996], SlCHS from Silene latifolia [Ageez et al., 2005], and CHSLK from Nicotiana sylvestris [Atanassov et al., 1998]; Fig. 3). The evolutionarily distant gymnosperm Pinus radiata also has a CHS-like protein, PrChS1, expressed in male cones (Walden et al., 1999), with 68%, 68%, and 64% sequence identity with LAP5, LAP6, and At4g00040, respectively. An ortholog of these male-organ-specific proteins exists in the genome of the moss Physcomitrella patens (National Center for Biotechnology Information XP_001781520), in addition to orthologs of a canonical CHS.
LAP5, LAP6, and At4g00040 have a predicted gene structure similar to that of CHS, stilbene synthase (STS), and related type III PKS genes, including a conserved Cys codon (Cys-60 in the numbering scheme of the well-characterized enzyme CHS2 from alfalfa [Medicago sativa]) that is split by a short intron between the first and second nucleotides (Figs. 2 and 3). In addition to this intron, LAP5, unlike the majority of this family's members, has a second intron. Four absolutely conserved residues (Cys-164, Phe-215, His-303, and Asn-336) that define the catalytic machinery in the active centers of CHS-related enzymes (Ferrer et al., 1999; Lanz et al., 1991; Jez et al., 2000b; Austin et al., 2004) are present in LAP5, LAP6, and At4g00040 as well as in male-specific CHS-like proteins from other species (Fig. 3). However, known male-specific CHS-like proteins, as well as LAP5, LAP6, and At4g00040, have a large number of amino acid changes specific to this clade (Fig. 3A).
The lap5-1 and lap6-1 point mutations affect highly conserved amino acids: Ile-132 in LAP6, replaced with Thr in lap6-1, is present in all 42 CHS, STS, and CHS-like proteins examined (Fig. 3A; data not shown). Gly-227 in LAP5, replaced with Glu in lap5-1, is conserved in all LAP5/LAP6-like proteins (Fig. 3A).
LAP5 and LAP6 Have Anther-Specific Expression Patterns
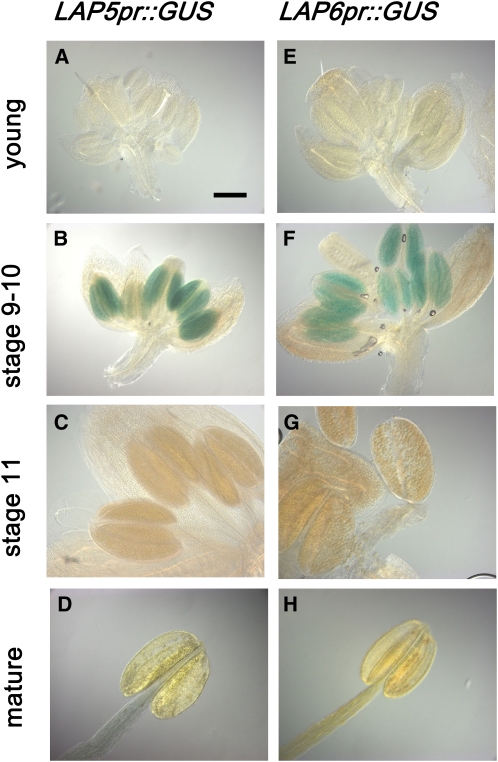
We searched publicly available expression databases, including Genevestigator (Zimmermann et al., 2004; https://www.genevestigator.ethz.ch/) and Massively Parallel Signature Sequencing (Meyers et al., 2004; http://mpss.udel.edu/at/), to identify LAP5 and LAP6 tissue expression patterns. According to microarray and Massively Parallel Signature Sequencing data, LAP5 and LAP6 were specifically expressed in young anthers and in flower buds (stages 9 and 10), and transcripts were absent in flower buds from stamen-lacking ap3 and ag mutants. In addition, a search of ATTED-II and BAR Expression Angler databases, which analyze microarray expression data and predicted cis-regulatory elements to build networks of genes that are likely to be coexpressed (Toufighi et al., 2005; Obayashi et al., 2007; http://www.atted.bio.titech.ac.jp/ and http://bbc.botany.utoronto.ca/ntools/cgi-bin/ntools_expression_angler.cgi), suggested that LAP5 and LAP6 were expressed together and highly coexpressed with several other genes involved in exine production, including ACOS5 (de Azevedo Souza et al., 2009), MS2 (Aarts et al., 1997), CYP703A2 (Morant et al., 2007), CYP704B1 (Dobritsa et al., 2009), and DIHYDROFLAVONOL 4-REDUCTASE-LIKE1 (DRL1; Tang et al., 2008). To confirm LAP5 and LAP6 expression patterns, we transformed wild-type plants with GUS under the control of the LAP5 and LAP6 promoters. In the resulting T1 plants, GUS staining was observed only in anthers of stage 9 and 10 buds (Fig. 4). Taken together, these data indicate that LAP5 and LAP6 are male-organ-specific members of the CHS family and are expressed in anthers coincident with the timing of exine formation.
Figure 4.
LAP5 and LAP6 are expressed specifically in developing anthers. LAP5pr::GUS (A–D) and LAP6pr::GUS (E–H) promoter fusion constructs are expressed in anthers of stage 9 and 10 buds (B and F) but not at earlier stages (A and E) or later stages (stage 11 [C and G] and mature anthers [D and H]). All images are to the same magnification. Bar = 100 μm.
lap5 lap6 Double Mutants Have Enhanced Exine Defects
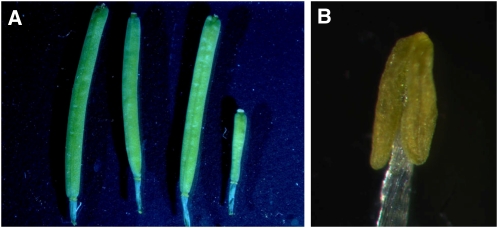
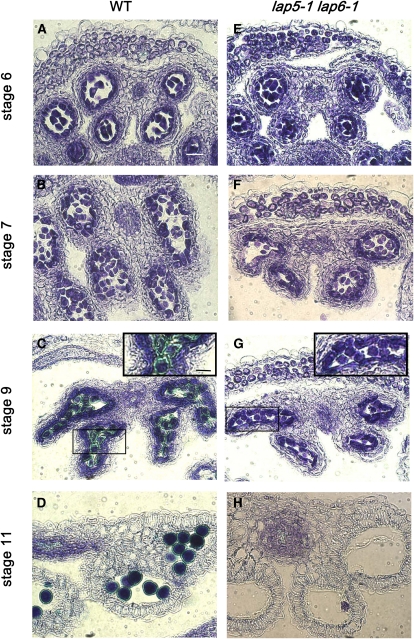
A lap5-1 lap6-1 double mutant was created to explore the relationships between the LAP5 and LAP6 genes in pollen development. While both single mutations resulted in fertile pollen with abnormally patterned exine, the double mutant was sterile, produced brown shriveled anthers with almost no pollen, and failed to produce elongated seed pods or siliques (Fig. 5). Sections of developing lap5-1 lap6-1 anthers showed normal pollen development until stage 9 (Fig. 6) but lacked the exine typically found in the wild type at stage 9 (green staining around the pollen grains, Fig. 6C; Sanders et al., 1999). Pollen in the double mutant subsequently collapses, with almost no pollen remaining by the stage 11 (Fig. 6H).
Figure 5.
A lap5-1 lap6-1 double mutant is sterile and lacks pollen. A, Left to right: siliques from the wild type, lap5-1, lap6-1, and a lap5-1 lap6-1 double mutant. B, Mature anther from lap5-1 lap6-1 demonstrates complete absence of pollen grains (compare with Fig. 1A for a wild-type anther).
Figure 6.
Pollen produced by a lap5-1 lap6-1 double mutant lacks exine and degenerates. Anther development is shown in the wild-type (WT) Arabidopsis (A–D) and a lap5-1 lap6-1 double mutant (E–H). Anther locules at stages 6 (A and E), 7 (B and F), 9 (C and G), and 11 (D and H) are shown. Exine is deposited around developing pollen grains at stage 9 in the wild type (green halo around the pollen grains; C) but not in lap5 lap6 (G). Insets in C and G are higher magnification views of boxed areas. Little pollen is visible in lap5 lap6 by stage 11 (H). Bars = 20 μm in A to H and 10 μm in the insets.
Neither At4g00040 nor TT4 Can Substitute for LAP5 or LAP6
Given the high similarity of the predicted At4g00040 protein to LAP5, LAP6, and male-specific CHS-like enzymes from several organisms, we tested whether this gene also plays a role in pollen development. Three lines with DNA insertions in At4g00040, including one with an insertion in the middle of the predicted ORF (Fig. 2), were available from Arabidopsis stock centers. We confirmed the insertion positions in each line and found that each contained anthers and pollen indistinguishable from the wild type.
Because gene expression databases indicated that At4g00040 was not male specific but instead was expressed in various organs and stages of development, we next tested whether driving expression of this gene in stage 9 to 10 anthers would rescue lap5 or lap6 defects. We placed a wild-type copy of At4g00040 under the control of a LAP5 or LAP6 promoter and introduced these constructs into lap5-1 and lap6-1 mutants. Ten BASTA-resistant T1 plants were analyzed for each of the four resulting combinations, but none had reversed the mutant phenotype. Thus, despite its high similarity to male-specific CHS-like proteins, At4g00040 apparently is unable to substitute for LAP5 or LAP6 in pollen development.
In addition, we tested if expression of Arabidopsis CHS TT4 in stage 9 to 10 anthers would rescue lap5 lap6 defects by expressing it under the control of a LAP5 promoter and introducing it into the lap5-1/+ lap6-1/lap6-1 plants. None of the 36 BASTA-resistant T1 plants analyzed (11 lap5-1/lap5-1 lap6-1/lap6-1 and 18 lap5-1/+ lap6-1/lap6-1; 7 lap6-1/lap6-1) had mutant phenotypes reversed. Thus, canonical CHS is also unable to substitute for LAP5 and LAP6 in pollen development.
lap5-1 and lap6-1 Affect the Production of Anther Chalcone, Naringenin, Other Flavonoids, and Additional Metabolites
Because LAP5 and LAP6 resemble CHS proteins, we hypothesized that they might catalyze the production of phenolic components needed to form sporopollenin. We collected metabolic profiles from stage 9/10 anther extracts from the wild type, lap5-1, lap6-1, and lap5-1 lap6-1 (see “Materials and Methods”). Using ultra-high-performance liquid chromatography and gas chromatography coupled to mass spectrometry (UPLC-MS and GC-MS), a broad spectrum of primary and secondary metabolites were separated, detected, and characterized. A total of 83 metabolites were characterized with UPLC coupled to a quadrupole time-of-flight mass spectrometer (QTOF-MS; Supplemental Tables S1–S4), and 45 of these were annotated using available authentic standards or comparison with previously published data. The annotated metabolites predominantly belonged to three chemical groups: phenolics (mainly flavonoids), glucosinolates, and long-chain fatty acids.
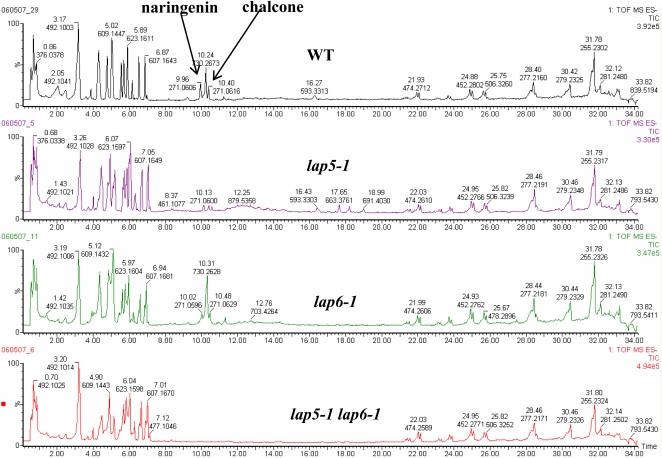
The levels of the phenolics, chalcone and naringenin, formed in the first committed step of the flavonoid pathway, were significantly reduced in the lap mutants (11.8-fold for lap5 and 5.6-fold for lap6) and were undetectable in the double mutant (Fig. 7; Supplemental Table S1). In addition, several other flavonoids were reduced in both mutants, including dihydrokaempferol and isorhamnetin 3-sophoroside. Kaempferol-3-O-gentiobioside-7-O-rhamnoside was not detected in lap5, and kaempferol 3-glucoside-7-p-coumarylglucoside was not detected in lap6. Levels of kaempferol-3-galactoside-7-rhamnoside, luteolin 7-O-rhamnoside, and 6-methoxytaxifolin were significantly reduced in lap6, and 6-methoxytaxifolin was not detected in the double mutant. Robinin was reduced in lap5 and almost absent in the double mutant samples. Surprisingly, increases (almost 6-fold) of quercetin 3-glucoside-7-rhamnoside were observed in the lap6 mutant, and another flavone, apigenin-6-C-glucoside, was detected only in lap6.
Figure 7.
Levels of chalcone and naringenin are affected in the lap5 and lap6 mutants. Examples of UPLC-QTOF-MS results obtained from lap5-1, lap6-1, the lap5-1 lap6-1 double mutant, and wild-type (WT) anther extracts are shown.
In addition to changes in the levels of phenolics, we also detected changes in the levels of other compounds in mutant anther extracts. For instance, several glucosinolate compounds were observed only in the mutant plants, as were some fatty acids, such as α-eleosteric acid and 10,16-dihydroxypalmitic acid (Supplemental Tables S2 and S3). Given the possible role of 16-hydroxypalmitic acid in the biosynthesis of sporopollenin (Dobritsa et al., 2009), accumulation of this fatty acid in the lap6 and lap5 lap6 mutants could be a consequence of the overall changes in sporopollenin composition in the mutant anthers, which could result in changes in fatty acid polymerization. Significant changes in several unidentified compounds were also found in the mutant extracts (Supplemental Table S4).
The relative abundances of 96 molecules of known chemical structures, including amino acids, Krebs cycle intermediates, carbohydrates, fatty acid derivatives, and several other primary metabolites, were determined by GC-MS (Supplemental Table S5). Several primary metabolites were altered in the mutant lines, the most noticeable being a general increase in the levels of amino acids and their catabolic products. Levels of l-Asn, l-Val, and 4-aminobutyric acid were increased in all mutant lines, with the most pronounced changes observed in the double mutant samples. Additionally, levels of pipecolic acid, a degradation product of l-Lys, were increased 13.6-fold in lap5 and 6.1-fold in the double mutant. Several changes in the levels of carbohydrates were observed. The most evident change was a dramatic reduction of maltose levels in all three mutant samples. Because maltose is a starch hydrolysis product, this suggests a reduction in starch pools.
Enzymatic Activity of LAP5 and LAP6
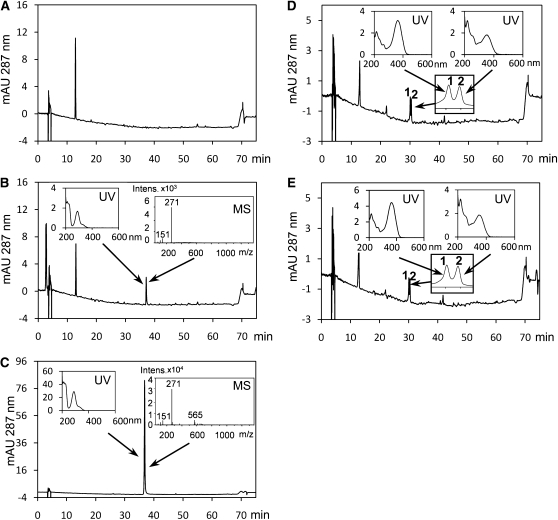
The metabolic profiling data of the lap5 and lap6 mutants revealed decreased flavonoid levels, suggesting that LAP5 and LAP6 may play a role in anther flavonoid biosynthesis. Heterologous gene expression and in vitro enzymatic assays were performed to investigate the functions of LAP5 and LAP6 proteins. Enzymatic reactions were performed in the presence of 4-coumaroyl-CoA and malonyl-CoA, CHS substrates. The reaction products were analyzed by HPLC-UV/visible-ion trap mass spectrometry (ITMS; Fig. 8). The empty-vector extract and the Arabidopsis CHS TT4 were used as the negative and positive controls, respectively (Fig. 8, A and B). The product of TT4/CHS (Fig. 8B) was identified as naringenin by comparing its retention time and UV and mass spectra with those obtained with the authentic naringenin standard (Fig. 8C). Naringenin, the expected CHS product, was not present in the LAP5 or LAP6 assay. However, two other products, peaks 1 and 2, were found (Fig. 8, D and E), which were absent in both the positive and negative controls. The mass spectra of the two peaks observed in the LAP5 and LAP6 product profiles were almost identical and dominated by three major ions, mass-to-charge ratio (m/z) 229, 185, and 143, with slightly different abundance ratios (Supplemental Fig. S2). These two peaks were unambiguously identified as trans- and cis-bisnoryangonin (i.e. 4-coumaroylacetic acid lactone), based upon the observed ions at m/z 229 [M-H]–185 [M-H-CO2]–and 143 [retro Diels-Alder reaction: M-H-C3H2O3]–as described previously (Jindaprasert et al., 2008), and characteristic UV absorption maxima at 365 nm (Shiokawa et al., 2000). The MS assignments were further confirmed by MS/MS of the molecular ion m/z 229, which gave two fragment ions, m/z 185 and 143 (Akiyama et al., 1999). The in vitro activities of LAP5 and LAP6 were also investigated at different pH levels (pH 5.0, 8.0, and 9.0). Bisnoryangonin was found to be the only product in both LAP5 and LAP6 assays at all pH levels. We also tested whether combining both LAP5 and LAP6 would result in a different activity, because CHS and STS are known to function as homodimers (Ferrer et al., 1999; Shomura et al., 2005) and are also capable of forming in vitro heterodimers with partial catalytic activities (Tropf et al., 1995). However, bisnoryangonin was again the only detected product.
Figure 8.
HPLC-UV-MS results of the enzymatic assays with 4-coumaroyl-CoA and malonyl-CoA as substrates. A, Empty-vector control. B, TT4/CHS. Insets are UV and MS spectra of the enzymatic reaction product (naringenin). C, Naringenin standard. The insets are the UV and MS spectra. D, LAP5. The insets are closeup views of peaks 1 and 2 and their UV spectra. E, LAP6. The insets are closeup views of peaks 1 and 2 and their UV spectra.
In vitro formation of trans- and cis-bisnoryangonin has been observed with other recombinant CHSs (Akiyama et al., 1999; Shiokawa et al., 2000). It is formed through the condensation of one molecule of 4-coumaroyl-CoA and two molecules of malonyl-CoA (Supplemental Fig. S3). Like coumaroyltriacetic acid lactone (a condensation product of one molecule of 4-coumaroyl-CoA and three molecules of malonyl-CoA), bisnoryangonin has been reported as an enzymatic “derailment” product (i.e. an incomplete biosynthetic product that detaches from the enzyme prior to completion of all the enzymatic steps) of CHS or CHS-like proteins (Kreuzaler and Hahlbrock, 1975; Shiokawa et al., 2000). The formation of the derailment products probably reflects the differences between the in vitro and the in vivo environments and was attributed to the lack of subsequent reactions in the in vitro environments (Eckermann et al., 2003). Targeted analyses of the UPLC-QTOF-MS data did not show the presence of bisnoryangonin or its glycosides in the wild-type plants, indicating that bisnoryangonin is unlikely the in vivo physiological product of LAP5 and LAP6.
LAP5 and LAP6 belong to the type III PKS superfamily, which produces a wide variety of secondary metabolites using different starter CoA esters, different elongation cycles, and different cyclization mechanisms (Austin and Noel, 2003; Flores-Sanchez and Verpoorte, 2009). Acetyl-CoA is one of the most common starters in polyketide synthesis and leads to the formation of both aromatic and pyrone polyketides. To determine whether LAP5 and LAP6 proteins have PKS activity in the presence of acetyl-CoA, 4-coumaroyl-CoA was replaced with acetyl-CoA in a separate experiment. However, no product was observed, indicating that acetyl-CoA is unlikely a physiological substrate.
While these experiments were in progress, another group reported on the biochemical characterization of the LAP5 and LAP6 proteins (Mizuuchi et al., 2008). The proteins were shown in vitro to catalyze condensation reactions using medium- and long-chain fatty acyl-CoA and malonyl-CoA as starter molecules, resulting in the formation of triketide and tetraketide α-pyrone lipids. We repeated these experiments using a variety of acyl-CoAs (chain length C4–C18) as substrates. The empty-vector extract was used as a negative control. Similar to Mizuuchi et al. (2008), we found that LAP5 and LAP6 are also capable of accepting fatty acyl-CoA as starters and performing condensation reactions with malonyl-CoA to form triketide (major product) and tetraketide (minor product) α-pyrone lipids (Fig. 9; Supplemental Figs. S4–S6; data for one protein are shown; the other demonstrated identical activity). However, unlike Mizuuchi et al. (2008), who observed conversion of C4 to C20 fatty acyl-CoA, only C6 to C12 were converted into the corresponding triketide or tetraketide pyrones under our experimental conditions. In addition to LAP5 and LAP6, TT4/CHS was also assayed with fatty acyl-CoA as a substrate. We found that it also possessed an in vitro activity comparable to that of LAP5 and LAP6 toward fatty acyl-CoAs, accepting C6 to C10 substrates and catalyzing condensation reactions to form triketide and tetraketide pyrones (Fig. 9; Supplemental Figs. S5 and S6). In reaction with hexanoyl-CoA catalyzed by TT4/CHS, a minor product was detected that was identified as tetraketide resorcylic acid (Supplemental Fig. S5), based on its mass spectrum and its retention time relative to triketide pyrone (Zhou et al., 2008). In contrast, tetraketides found in octanoyl-CoA (Fig. 9) and decanoyl-CoA (Supplemental Fig. S6) experiments were tetraketide pyrones rather than tetraketide resorcylic acid, suggesting that tetraketides can undergo different cyclization in solutions (Supplemental Fig. S4).
Figure 9.
HPLC-ITMS UV analyses of the products of the enzymatic reactions with octanoyl-CoA and malonyl-CoA as substrates. A to C, UV chromatograms (287 nm) of the products of enzymatic reactions in the presence of empty-vector control (A), LAP5 (B), or TT4/CHS (C). D, Mass spectrum of the major enzymatic product, 4-hydroxy-6-heptyl-2-pyrone, of the reaction catalyzed by LAP5. E, Mass spectrum of the major enzymatic product, 4-hydroxy-6-heptyl-2-pyrone, of the reaction catalyzed by TT4.
To determine if LAP5/LAP6 serve as multifunctional proteins that could generate cross-linked phenolics and fatty acids in exine, we have performed a multisubstrate assay for LAP5 and TT4/CHS using hexanoyl-CoA, 4-coumaroyl-CoA, and malonyl-CoA at a 1:1:3 molar ratio as substrates. 4-Hydroxy-6-pentyl-2-pyrone and bisnoryangonin were obtained with LAP5 (at a molar ratio of 32:1), and 4-hydroxy-6-pentyl-2-pyrone and naringenin were obtained with TT4/CHS (at a molar ratio 11:1; Supplemental Fig. S7). No novel products different from the compounds obtained in separate experiments were detected, with the exception of bisnoryangonin found in the reaction catalyzed by TT4/CHS.
Mutations Affecting LAP5 and a Gene from the Fatty Acid Hydroxylation Pathway Have Synergistic Effects
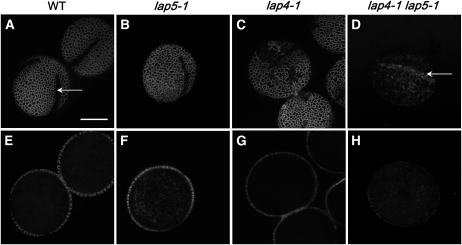
A recently described gene (CYP703A2) is necessary for normal exine development and encodes a protein that hydroxylates lauric acid, potentially providing fatty acid building blocks required for sporopollenin biosynthesis (Morant et al., 2007). We created a double mutant between lap5-1 and a hypomorphic lap4-1 allele of CYP703A2 (Dobritsa et al., 2009). The lap4-1 mutant causes plants to produce functional pollen that, like lap5 and lap6, appears glossy and strongly adheres to anthers. When this pollen was stained with the exine-specific fluorescent dye auramine O and examined by laser scanning confocal microscopy, we observed a strong reduction in the thickness of exine, which nonetheless exhibited a reticulate pattern (Fig. 10, C and G). Patches of exine were often missing on the surface of the lap4-1 pollen (Fig. 10C). The lap4-1 lap5-1 double mutant was fertile but produced glossy pollen that did not easily separate from the anthers. When we examined the lap4-1 lap5-1 pollen grains by laser scanning confocal microscopy, we found that they exhibited a more severe defect in their surface structure than either lap5-1 or lap4-1 single mutant. The pollen surface showed no reticulate pattern and almost no auramine O fluorescence (Fig. 10, D and H). When optical sections through the middle of the grains were obtained to determine exine thickness, we found that in the lap4-1 lap5-1 double mutant exine was virtually missing (Fig. 10H). Interestingly, in many of those pollen grains, areas of somewhat increased fluorescence were located longitudinally (Fig. 10D).
Figure 10.
Exine from the lap5-1 and cyp703a2 (lap4-1) double mutant has defects stronger than either single mutant. Confocal images of the exine surface from wild-type (A and E), lap5-1 (B and F), lap4-1 (C and G), and lap4-1 lap5-1 double mutant (D and H) pollen grains stained with auramine O. The arrow in A indicates an aperture; the arrow in D indicates a longitudinal area of increased fluorescence. E and F show optical sections through the middle of pollen grains, allowing visualization of exine thickness. All images are to the same magnification. Bar = 10 μm.
DISCUSSION
Defects in LAP5 and LAP6 Proteins Have Broad Metabolic Consequences
This study underscores that a disruption of a single gene can have wide-ranging consequences on the metabolic composition, revealing the intricacy of interactions among metabolic pathways. Collected metabolomics data indicate that alterations in the expression of LAP5 and LAP6 genes result in the differential accumulation of several flavonoids and of two lipids (10,16-dihydropalmitic acid and α-eleosteric acid) and profoundly affect the biosynthesis of a variety of metabolites outside the fatty acid or phenylpropanoid pathway. It has been previously suggested (Rohde et al., 2004) that primary and secondary metabolism are interlocked through the Phe pool. Thus, it is possible that a reduced carbon flux into the phenylpropanoid pathway affects the homeostasis of amino acid and carbohydrate metabolism. Possibly, part of the surplus carbon resulting from the reduced phenolic flux in the lap5 and lap6 mutants is redirected toward increased amino acids. Under normal conditions, 20% of all fixed carbon is directed into the shikimate pathway that generates aromatic amino acids (Herrmann, 1995). One of the major sinks of this pathway is the conversion of Phe to cinnamic acid via Phe lyase, which is the first enzyme in the phenylpropanoid pathway. Interestingly, the reduction in LAP5 and LAP6 activity has dramatic consequences for the accumulation of several amino acids, not only the aromatic ones. Similarly, increases in the abundance of multiple amino acids were previously observed in mutants with defective Phe lyase activity (Rohde et al., 2004). Also, depletion of Tyr and Trp pools via overexpression of Trp and Tyr decarboxylases alters levels of both aromatic and nonaromatic amino acids (Guillet et al., 2000). These results led to a proposed mechanism that controls amino acid homeostasis across individual biosynthetic pathways (Guillet et al., 2000; Rohde et al., 2004).
The accumulation of amino acids in lap5 and lap6 mutants may have additional effects. β-Ala and the l-Val biosynthetic intermediate 2-oxoisovalerate are used in the formation of pantothenic acid (White et al., 2001), which is subsequently converted to CoA (Kupke et al., 2003). We interpret the accumulation of branched-chain amino acids (such as Val and Leu), putrescine, γ-aminobutyrate, and β-Ala in the lap5 and lap6 anthers as a result of altered CoA biosynthesis. A similar metabolic model was previously suggested by Broeckling et al. (2005) to describe experimentally observed changes of several metabolites. CoA serves as a carrier for organic acids, including acetic acid (utilized in fatty acid biosynthesis, glycolysis, Krebs cycle, and amino acid biosynthesis), 4-coumaric acid, and malonic acid, that are used in flavonoid biosynthesis. Thus, levels of CoA might, in turn, provide feedback control for the production of flavonoids and fatty acids.
Amino acids also provide a link between phenylpropanoids and glucosinolates, another class of secondary metabolites that was increased in lap5 and lap6 mutants. Glucosinolates are derived from various amino acids (Halkier and Gershenzon, 2006), and their close metabolic connection (Knill et al., 2008) might account for the observed increases in glucosinolate levels in our mutants.
LAP5 and LAP6 Are Paralogs Required for Exine Biosynthesis
Here, we have shown that LAP5 and LAP6 are essential for the production of pollen exine. Both genes are specifically expressed during the period of exine synthesis in developing anthers and belong to a male-specific clade of the CHS superfamily. Point mutations lap5-1 and lap6-1 result in the formation of fertile pollen with aberrant exine patterning; available insertions (likely null alleles) in lap6 show an identical phenotype. Double mutants (lap5-1 lap6-1) in both genes abolish exine deposition, followed by pollen collapse and male sterility. Given the sequence similarity of these genes and the results of in vitro enzymatic assays, a possible interpretation of the double mutant phenotype is that the genes are functionally redundant, with the single mutants reducing the levels of the same compound(s). On the other hand, since the lap5 and lap6 mutants have different exine morphologies, responses to acetolysis, and metabolite profiles, it is conceivable that their functions are somewhat distinct.
In addition to LAP5 and LAP6, Arabidopsis has a third paralog, At4g00040, which, despite sequence similarity with known male-specific CHS family proteins, does not exhibit anther-specific expression or appear to participate in pollen development. Several other plant species also have multiple orthologs of LAP5 and LAP6. Poplar (Populus trichocarpa) has three orthologous proteins (http://genome.jgi-psf.org/Poptr1/Poptr1.home.html), and at least two orthologs are present in the Brassica rapa, tomato (Solanum lycopersicum), maize (Zea mays), and rice genomes. Classical CHS and STS are known to function as homodimers (Ferrer et al., 1999; Shomura et al., 2005). However, an in vitro study with mutated proteins that were inactive as homodimers demonstrated that CHS and STS are capable of forming heterodimers with partial catalytic activities (Tropf et al., 1995). It would be interesting to determine if LAP5 and LAP6 can function as heterodimers, thus potentially expanding the repertoire of reaction substrates and products. Although our experiments in combining LAP5 and LAP6 in the CHS reaction have not resulted in the change of activity in a heterodimer, it is probable that the substrates used were not physiological.
LAP5 and LAP6: Novel Type III PKSs
Canonical CHSs, such as Arabidopsis TT4 or alfalfa CHS2, are the most ubiquitous and best characterized members of the plant type III PKS family, which is often referred to as the CHS family. They condense a CoA ester from the phenylpropanoid pathway (usually 4-coumaroyl-CoA) with three molecules of malonyl-CoA to form a tetraketide intermediate and a new aromatic ring of chalcone. STSs, which are found in a relatively small number of plants, share approximately 70% sequence identity with CHS and use the same substrates (4-coumaroyl-CoA and malonyl-CoA) to perform three condensation reactions. The final steps of STS tetraketide ring closure are mechanistically different from those of CHS, enabling the formation of stilbenes such as resveratrol. Other members of the type III PKS superfamily, such as acridone synthase, benzophenone synthase, and valerophenone synthase, also perform three condensation reactions but use different substrates, while still other type III PKS enzymes utilize a different number of condensation reactions (Austin and Noel, 2003; Karppinen et al., 2008).
Sequence comparisons among different members of the CHS superfamily provide few clues to their enzymatic functions, and amino acid residues that cause differences between catalytic activities remain unknown. For instance, replacement of only eight amino acids is sufficient to convert alfalfa CHS to STS (Austin et al., 2004). Yet, there is no apparent STS consensus sequence, as eight STS-derived mutations in Pinus sylvestris that can convert CHS to STS are not conserved among STS enzymes from different species. Of the two CHS-like proteins from Pinus strobus that share 87.6% identity, only one has CHS activity, whereas the other one is completely inactive with any starter CoA esters from the phenylpropanoid pathway or with linear CoA starters. Instead, it performs a single condensation reaction with a diketide derivative and methylmalonyl-CoA (Schröder et al., 1998). Out of the five CHS-like genes in hop (Humulus lupulus), only one encodes CHS activity, whereas the others prefer substrates unrelated to flavonoid metabolism (Novak et al., 2006). Additionally, a 2-pyrone synthase (2PS) from Gerbera hybrida shares 74% sequence identity with two CHSs from the same species yet exhibits a very different activity: it uses acetyl-CoA as a substrate, performs only two condensation reactions, and folds the triketide intermediate into a pyrone ring structure (Eckermann et al., 1998). Replacement of only three amino acids in CHS is sufficient to convert it to 2PS (Jez et al., 2000a). To complicate matters further, members of the CHS family are promiscuous in vitro. CHS is capable of accepting nonphysiological substrates such as benzoyl-CoA or aliphatic acyl-CoA (Schüz et al., 1983); use of isovaleryl-CoA as a substrate for CHS from Pinus sylvestris was sufficient to convert it to pyrone synthase that produces a pyrone derivative (Zuurbier et al., 1998), and G. hybrida 2PS is able to produce pyrone using isovaleryl-CoA or benzoyl-CoA as substrate (Eckermann et al., 1998).
LAP5, LAP6, and other members of their clade in the CHS superfamily share approximately 40% sequence identity with CHS. At the amino acid level, they are much more dissimilar to the canonical CHS than 2PS and other members of this superfamily with demonstrated non-CHS activity. While there are limitations to primary structure analysis for members of this family, several notable sequence differences could be important for LAP5 and LAP6 enzymatic activity. (1) The conserved residue, Thr-197, is important for determining the volume of the coumaroyl-binding pocket in the active center of CHS and possibly for the preference for 4-coumaroyl-CoA as a substrate (Jez et al., 2000a). In addition, the hydroxyl of Thr-197 interacts with the carbonyl of the chalcone product (Ferrer et al., 1999). LAP5, LAP6, and other members of the male-specific CHS-like clade contain an invariant Gly at the corresponding position, potentially affecting a substrate preference. The lap5-1 mutation highlights the importance of this residue, replacing it with the larger and negatively charged amino acid, Glu. (2) Lys-62, an amino acid that sits at the entrance to the CoA-binding tunnel of CHS2 and forms a hydrogen bond with a phosphate of CoA (Ferrer et al., 1999), is replaced with Thr or Ser in LAP5, LAP6, and related proteins. (3) Homodimers of CHS have two independent active sites, each of which is composed almost entirely of residues from a single monomer. The only residue that comes from a different monomer is Met-137, which protrudes into a hole on the surface of the adjoining monomer to form a part of the cyclization pocket (Ferrer et al., 1999). This Met residue is conserved in all CHS, STS, and most other members of the superfamily with identified biochemical activity. Met-137 is replaced by Leu or Phe in the LAP5/LAP6-related proteins.
The physiological role of LAP5 and LAP6, as that of many other members of the type III PKS superfamily, remains unclear. Given that very few amino acid changes are sufficient to convert CHS into a protein with a different activity and the level of LAP5/LAP6 divergence from CHS, we did not readily expect LAP5 and LAP6 to catalyze CHS-type reactions. This assumption is consistent with the results of the in vitro protein assays and with the apparent inability of the TT4/CHS to rescue the lap5 and lap6 pollen defects. Interestingly, despite the fact that LAP5 and LAP6 do not exhibit in vitro CHS activity, the metabolic profiles of the lap5 and lap6 mutants revealed clear reduction in anther chalcones and flavonoids. It is unknown at present whether this reduction is directly caused by the absence of LAP5 and/or LAP6 activities or whether this is an indirect consequence of the general changes stemming from altered sporopollenin synthesis and exine development. Thus, a strong possibility remains that LAP5 and LAP6 function in the production of phenylpropanoid precursors of sporopollenin.
The biochemistry results presented here and by Mizuuchi et al. (2008) demonstrated that, in vitro, LAP5 and LAP6 are also able to accept a variety of fatty acyl-CoAs and convert them to corresponding pyrone lipids in the condensation reactions with malonyl-CoA. Homology modeling of LAP5 and LAP6 predicted that, while they share the three-dimensional overall fold with other plant type III PKS proteins, they have an unusually long substrate-binding cavity (Mizuuchi et al., 2008), consistent with their ability to accept long substrates. This feature resembles the long substrate-binding tunnel of the bacterial PKS-18 from Mycobacterium tuberculosis, whose crystal structure was resolved (Sankaranarayanan et al., 2004) and which also produces long-chain pyrones from fatty acyl-CoA starters (Saxena et al., 2003). Interestingly, however, we also observed similar activity toward fatty acid derivatives with the bona fide CHS, TT4, which is known to accept other aliphatic acyl-CoA substrates (Schüz et al., 1983). This further highlights the in vitro multifunctionality of the type III PKS superfamily enzymes and demonstrates that conclusions on the physiological roles of these proteins from in vitro studies should be approached with caution.
If long-chain pyrones were produced in the developing anthers by LAP5 and LAP6, one would expect to see a differential accumulation of these compounds in the wild type and the lap5 and lap6 mutants. However, the metabolite analyses did not identify long-chain pyrones in the developing anthers. One possibility is that these compounds are among the metabolites that were not positively identified (Supplemental Table S4), some of which were significantly reduced in mutants. Another possibility is that they undergo rapid polymerization and incorporation into sporopollenin and, therefore, escape detection during a metabolomics analysis. It remains to be demonstrated that fatty acyl-CoA molecules serve as in vivo substrates for LAP5 and LAP6.
Despite the overlapping in vitro biochemical activity, TT4/CHS does not rescue the lap5 and lap6 pollen phenotypes when expressed under the control of the LAP5 promoter. It is possible that the intracellular environment may affect the in vivo activity of this enzyme or that TT4/CHS does not get access to the same substrates as LAP5 and LAP6 because it is sequestered in a metabolon dedicated to flavonoid metabolism (Winkel, 2004).
Phenylpropanoid Pathway and Sporopollenin Biosynthesis Pathway: Possible Parallels
While the precise structure of sporopollenin remains undetermined, chemical studies have suggested that it is composed of fatty acid and phenolic compounds (Guilford et al., 1988; Wiermann et al., 2001). Several genes and corresponding mutants have been described that confirm the important role of fatty acid metabolism in sporopollenin production. CYP703A2 and CYP704B1 are cytochromes P450 that have fatty acid hydroxylase activities and use lauric acid and long-chain fatty acids, respectively, as preferred substrates (Morant et al., 2007; Dobritsa et al., 2009), whereas MS2 is an anther-specific gene encoding a fatty acyl reductase (Aarts et al., 1997; Doan et al., 2009). Mutations in each of these genes severely impair pollen wall production. A recently described gene, ACOS5, encodes a fatty acyl-CoA synthetase that has an in vitro preference for medium- to long-chain fatty acids (de Azevedo Souza et al., 2009). ACOS5 protein is related to 4-coumaroyl-CoA ligase, an enzyme of the general phenylpropanoid pathway that acts directly upstream of and provides the substrate (4-coumaroyl-CoA) for CHS. Thus, it is very tempting to speculate that LAP5 and LAP6, which are related to CHS and are able to utilize medium-chain fatty acyl-CoA in vitro, function immediately downstream of ACOS5. This is in agreement with mutant analysis of these three genes. An insertion in ACOS5 results in the formation of brown shriveled anthers that lack pollen, causing male sterility (de Azevedo Souza et al., 2009; A.A. Dobritsa, unpublished data). Sections of developing anthers from this mutant revealed that pollen develops normally until stage 9, when an exine deficiency was observed, followed by subsequent collapse of the pollen grains (de Azevedo Souza et al., 2009; Supplemental Fig. S8). This phenotype is identical to that of the lap5 lap6 double mutant and is very distinct from the phenotype of the cyp703a2 cyp704b1 ms2 triple mutant, which has viable pollen despite its lack of the normal exine layer and a characteristic zebra exine phenotype (Dobritsa et al., 2009). The more severe phenotypes point to a more central role that the ACOS5/LAP5/LAP6 pathway plays in the sporopollenin biosynthesis compared with the fatty acid-modifying CYP703A2, CYP704B1, and MS2.
Similar to CYP703A2, CYP704B1, and MS2 (Morant et al., 2007; Dobritsa et al., 2009), ACOS5 (de Azevedo Souza et al., 2009) and LAP5/LAP6 are highly conserved in plant lineages, with homologs present in the genomes of angiosperms, gymnosperms, and bryophytes but absent in green algae. The ability to synthesize sporopollenin is believed to be a critical evolutionary innovation at a time when plants colonized land, which allowed plants to protect spores from harmful effects of UV irradiation and desiccation (Bowman et al., 2007; Morant et al., 2007; de Azevedo Souza et al., 2009). The existence of ACOS5 and LAP5/LAP6 orthologs in the genome of the moss P. patens indicates that this possible variation of the phenylpropanoid pathway was an ancient adaptation.
DRL1, another anther-specific protein in Arabidopsis related to a protein from the phenylpropanoid pathway (dihydroflavonol 4-reductase; Supplemental Fig. S1), whose function is unknown at present, has also been recently found to be important for exine development (Tang et al., 2008). DRL1 is similarly conserved in land plants. Expression analysis of microarray data from the wild type and several transcription factor mutants with abnormal anther and pollen development (Wijeratne et al., 2007; Yang et al., 2007; Xing and Zachgo, 2008; Xu et al., 2010) demonstrates that DRL1 is coexpressed with ACOS5, LAP5, LAP6, CYP703A2, CYP704B1, and MS2. This makes DRL1 another strong candidate for an enzyme that is involved in the biochemical pathway leading to sporopollenin biosynthesis.
MATERIALS AND METHODS
Plant Material
Arabidopsis (Arabidopsis thaliana) lap5-1, lap6-1, and the lap4-1 allele of CYP703A2 were isolated as described (Nishikawa et al., 2005) from CS6242, a cer6-2 Landsberg erecta (Ler) variety, mutagenized with 1.25 mm ethyl nitrosourea. lap6-2 (SALK_134643) and lap6-3 (N172991) were from the Arabidopsis Biological Resource Center and the European Arabidopsis Stock Center (NASC), respectively. T-DNA insertions in At4g00040 were from NASC (N180467 and N162784; both in the 3′ end of the ORF) and from the Arabidopsis Biological Resource Center (SAIL_829_D04; in the middle of the ORF). A transposon insertion in At1g62940 (N123936) was from NASC. Plants were grown in a greenhouse at 22°C with a 16-h-light/8-h-dark photoperiod.
Genetic Mapping
Positional cloning was used to map lap5-1 and lap6-1 (backcrossed three times to CER6 Ler) in populations generated by crossing to wild-type Columbia. Bulked segregant analysis demonstrated linkage between lap5-1 and CIW7 and NGA1107 on the bottom of chromosome 4 and between lap6-1 and ZFPG on the top of chromosome 1 (markers are available at The Arabidopsis Information Resource; http://www.arabidopsis.org/). Fine mapping of lap5-1 in 657 F2 plants localized it to a 59-kb region between markers T4L20E2 and F11I11P1; similarly, mapping analysis of 378 F2 plants localized lap6-1 to 140 kb between T1N6A and T7I23G1. Candidate genes were sequenced, and polymorphisms relative to wild-type Ler were found in At4g34850 for lap5-1 and At1g02050 for lap6-1. The lap5-1 mutation removes a BamHI site. A corresponding CAPS marker was designed (lap5-850-CF, 5′-GAGCCTGCATCAAGAACTGG-3′; lap5-850-CR, 5′-CGGTTCAAGATGGCTGGTC-3′) to score the lap5-1 alleles in the segregating populations. To score the lap6-1 alleles, a dCAPS marker was designed (lap6-2050-CF, 5′-GCAGCTCCAACTAGGTCG-3′; lap6-2050-BR1, 5′-GGTTGCATCAAGGAATGGGGAAGGCCAGTGGAAGATA-3′; EcoRV cleaves the wild-type allele).
Complementation Constructs
LAP5 (from 631 nucleotides upstream of the start codon to 264 nucleotides downstream of the stop codon) was amplified with Platinum Pfx DNA polymerase (Invitrogen) using lap5-850-EF2 (5′-GTGTACCGGTCATCAGTGGGAAATGTCTCTCAG-3′) and lap5-850-ER (5′-CCATGGATGAGAAGATCCAACTCCTGGAC-3′). LAP6 (from 479 nucleotides upstream of the start codon to 148 nucleotides downstream of the stop codon) was amplified using lap6-2050-ER2 (5′-GTGTACCGGTTGAAGGATAGACCGTGCATCTGG-3′) and lap6-2050-EF (5′-CCATGGGCACTTGCTTTAACGACACGTGGTGC-3′). PCR products were digested with AgeI/NcoI and incorporated into the modified binary vector pGreenII02229 (Hellens et al., 2000; von Besser et al., 2006). lap5-1 and lap6-1 plants were transformed by the floral dip method (Clough and Bent, 1998), and transgenic plants were selected with BASTA.
GUS, At4g00040, and TT4 Expression Constructs
LAP5 and LAP6 promoters were amplified (lap5-850-EF3, 5′-GGATCCCATCAGTGGGAAATGTCTCTCAG-3′; lap5-850-FR, 5′-ACCGGTTTGTATCTGTTTGTTGAAGACTC-3′; lap6-2050-DF2, 5′-ACCGGTTTCTTTTCAAGCGCAGAAGG-3′; lap6-2050-ER2), digested with BamHI/AgeI, and incorporated into the modified vector pGreenII02229, creating LAP5pr-pGR111 or LAP6pr-pGR111. The GUS gene was amplified with GUS-AF (5′-CACCATGGATGTTACGTCCTGTAGAAACCC-3′) and GUS-CR (5′-CCACTAGTTCATTGTTTGCCTCCCTGCTGC-3′), digested with NcoI/SpeI, and cloned into NcoI/SpeI-cut LAP5pr-pGR111 or LAP6pr-pGR111. TT4 was amplified with TT4-AF (5′-TCTCCCATGGATGGTGATGGCTGGTGCTTC-3′) and TT4-CR (5′-AGGGACTAGTTTAGAGAGGAACGCTGTGC-3′) and similarly cloned into LAP5pr-pGR111. At4g00040 was amplified with 040-AF (5′-ACCGGTATGTTGGTGTCCGCAAGGGTAGAG-3′) and 040-AR2 (5′-CCATGGGTCAATAGCATTGAGTTGTAAATAAGTCTC-3′), digested with AgeI/NcoI, and cloned into AgeI/NcoI-digested LAP5pr-pGR111 or LAP6pr-pGR111.
Phylogenetic Analysis
CHS family proteins analyzed and their accession numbers from the Entrez Protein database are as follows: Brassica napus CHS A2 (AAC31912), Zea mays CHS C2 (P24825) and CHS Whp (P24824), Bromheadia finlaysonia CHS (O23729), Hypericum androsaemum CHS (Q9FUB7), Ipomoea purpurea CHS-D (AAK39111), Solanum lycopersicum CHS (P23419), Pinus sylvestris CHS (P30079), Medicago sativa CHS2 (P30074), Daucus carota CHS9 (Q9SB26), Allium cepa CHS (AAO63020), Petunia × hybrida CHSA (P08894) and CHSJ (P22928), Ipomaea nil CHS (O22046), Gerbera hybrida 2-PS (P48391), Ruta graveolens ACS (Q9FSC0), Rheum palmatum ALS (AAS87170), Pinus radiata PrCHS1(AAB80804), Silene latifolia SlCHS (BAE80096), Vitis vinifera STS (CAA54221), Nicotiana sylvestris CHSLK (CAA74847), Oryza sativa YY2 (BAA23618) and CHSS6 (BAC21541), and Physcomitrella patens CHS-like (XP_001781520). Additional sequences were obtained from The Institute for Genomic Research (http://www.tigr.org): Z. mays CHSS1 and CHSS2, Brassica rapa CHSS1 and CHSS2, and S. lycopersicum CHS1 and CHS2. Predicted proteins were aligned using ClustalW (MegAlign, DNAStar, Lasergene). A phylogenetic tree was created using the neighbor-joining method; bootstrap values were obtained (trials = 1,000, seeds = 111). PKS18 from Mycobacterium tuberculosis (A70958) was used as an outgroup.
Microscopy
Dissection stereomicroscopy (50×–90× magnification) was used to assess anther/pollen phenotypes. Anther images were captured using Zeiss SteReo Lumar V12 (80× magnification) fitted with a Zeiss AxioCam MRc5 digital camera. SEM was performed as described (Nishikawa et al., 2005). For developmental analysis, inflorescences were fixed in 4% paraformaldehyde, 0.5% glutaraldehyde, and 0.05 m phosphate buffer, pH 7.2, for 2 h (room temperature), washed in phosphate buffer (four times, 15 min), dehydrated in ethanol (15%, 30%, 45%, 60%, 70%), ethanol:n-butanol (50%:35%, 40%:55%:, 25%:75%, v/v), and 100% n-butanol, infiltrated with paraffin, and cut into 5-μm sections. Slides were processed in two changes of xylene, gradually rehydrated in an ethanol series followed by water, and stained with 0.1% toluidine blue. Auramine O staining, GUS staining, and acetolysis were performed as described (Dobritsa et al., 2009).
Metabolomics
Anthers from stage 9 and 10 flower buds (staged according to Smyth et al. [1990]) were collected on dry ice; three replicates composed of 60 anthers each were analyzed per genotype. Samples were lyophilized, homogenized, and extracted in 50 μL of 80% methanol with the internal standard 2 μg of umbelliferone for 2 h. Half the sample was used for UPLC-QTOF-MS, using an ACQUITY UPLC system (Waters) equipped with a binary solvent delivery system and an autosampler. The mobile phases were 0.1% acetic acid in water (eluent A) and 100% acetonitrile (eluent B). UPLC separations used a Waters 2.1- × 100-mm, BEH C18 column with 1.7-μm particles, a linear gradient of 95%:5% to 30%:70% eluent A:B, a flow rate of 0.56 mL min−1, column temperature of 60°C, and sample temperature of 4°C (autosampler). Each wash cycle used 0.8 mL of 100% methanol and 2 mL of 0.1% acetic acid in water. A blank (80% methanol) was injected between every five samples. Mass spectrometry was performed using a Waters QTOF-MS Premier system operating in the negative electrospray ionization mode, with the nebulization gas at 850 L h−1 (350°C) and the cone gas at 50 L h−1 (120°C). QTOF-MS data were collected between 50 and 2,000 m/z and were acquired using an independent reference lock-mass ion via the LockSpray interface, with the LockSpray frequency set at 10 s in the centroid mode. Raffinose (m/z 503.1612) was used as the reference compound and delivered at a concentration of 50 fmol mL−1 and a flow rate of 0.2 mL h−1.
GC-MS was performed on the remaining portion of the anther extracts (three replicates, approximately 25 μL each). A total of 500 μL of 80% methanol containing an internal standard of 2.5 μg of ribitol was added and samples were incubated for 1 h (50°C), equilibrated to room temperature, and incubated for 1 h at 50°C following the addition of 500 μL of chloroform containing 1 μg of docosanol (an internal standard). Water (400 μL) was added, and phases were separated by centrifugation at 2,900g (30 min, 4°C). Equal amounts of each phase were collected, transferred to 2-mL vials, and dried at room temperature under nitrogen (organic phase) or in air (aqueous phase). Polar extracts were methoximated in pyridine with 20 μL of 1.5% methoxyamine-HCl, briefly sonicated, suspended at 50°C, and incubated with 20 μL of commercial derivatization solution containing N-methyl-N-trimethylsilyltrifluoroacetamide + 1% trimethylchlorosilane (1 h, 50°C). Nonpolar extracts were suspended in 0.5 mL of chloroform, hydrolyzed with 0.5 mL of 1.25 m HCl in methanol (4 h, 50°C), and then evaporated under nitrogen. Polar and nonpolar extracts were analyzed with an Agilent 6890 GC apparatus coupled to a 5973 MSD. Samples were injected at a 1:1 split ratio, and the inlet and transfer lines were maintained at 280°C. Separation was achieved with a temperature program of 80°C for 2 min, ramped at 5°C min−1 to 315°C, and held for 12 min on a 60-m DB-5MS column (J&W Scientific; 0.25 mm i.d., 0.25 μm film thickness) and a constant flow of 1 mL min−1 helium. Mass spectra were collected from 50 to 800 m/z. For each compound, a Student's t test was performed to assess statistical differences.
Protein Expression and Purification
LAP5, LAP6, and TT4 ORFs were PCR amplified from cDNA that was prepared from RNA isolated from stage 9 to 10 buds (Dobritsa et al., 2009). The following primers were used: lap5-start1 (5′-CACCATGGGTAGCATCGACGCAGCAGTGTTGGGT-3′) and lap5-stop1 (5′-TCAGACATCAAGGTTTCGAGC-3′), lap6-start1 (5′-CACCATGTCTAATTCTCGTATGAATGGTGTTGAG-3′) and lap6-stop1 (5′-TTAGGAAGAGGTGAGGCTGCGGATG-3′), TT4-start1 (5′-CACCATGGTGATGGCTGGTGCTTCTTC-3′) and TT4-stop1 (5′-TTAGAGAGGAACGCTGTGCAAGAC-3′). Start codons (ATG) are underlined in the start primers; stop codons are underlined in the stop primers. Boldface corresponds to the mutagenized nucleotides that introduce silent mutations for improved expression in Escherichia coli). The PCR products were cloned into pET100-D/Topo vector from Invitrogen, and resulting constructs were transformed into E. coli BL21 Star (DE3). Cultures (500 mL) of transformed E. coli were grown at 37°C in Luria-Bertani medium containing 50 μg mL−1 kanamycin until optical density at 600 nm = 0.8. After induction with 0.3 mm isopropyl 1-thio-β-d-galactopyranoside, the cultures were grown for 16 h at 16°C. Cells were pelleted, harvested, and resuspended in extraction buffer (50 mm NaH2PO4, pH 8.0, 500 mm NaCl, 10% glycerol, 10 mm β-mercaptoethanol, and 10 mm imidazole) supplemented with protease inhibitors. After brief sonication, cells were homogenized in an EmulsiFlex-C3 high-pressure homogenizer (Avestin). The homogenate was clarified at 30,000g and 50,000g for 20 min each, incubated with Talon Metal Affinity Resin (Clontech) for 1 h at 4°C, and then loaded onto a disposable column. After a 50-mL extraction buffer wash, proteins were eluted with Talon elution buffer (extraction buffer supplemented with 250 mm imidazole) and 1-mL fractions were collected. Protein purity was analyzed by 12.5% SDS-PAGE. Protein-rich elution fractions (1–6) were pooled (fractions 1–6 were similarly pooled for vector-only control; fractions 1–4 from vector-only control contained proteins detectable by Coomassie Brilliant Blue staining), dialyzed in 2 L of storage buffer (20 mm HEPES, 1 mm EDTA, 200 mm KCl, 0.01% NaN3, and 10% [v/v] glycerol, pH 7.4) overnight, concentrated using Centriprep filters (Millipore) to approximately 1 mg mL−1, and stored on ice. Similarly processed extracts from vector-only-transformed E. coli were used as negative controls.
Synthesis of 4-Coumaroyl-CoA
4-Coumaroyl-CoA was synthesized as described previously (Stoeckigt and Zenk, 1975). The N-hydroxysuccinimide ester of 4-coumaric acid was first synthesized from 4-coumaric acid (C-9008; Sigma-Aldrich) and N-hydroxysuccinimide (130672; Sigma-Aldrich) in the presence of N,N′-dicyclohexylcarbodiimide (36650; Sigma-Aldrich). After purification, the N-hydroxysuccinimide ester of 4-coumaric acid was then converted into 4-coumaroyl-CoA through thioester exchange between the N-hydroxysuccinimide ester and CoA. The resulting 4-coumaroyl-CoA was purified on a diethylaminoethyl-Sephacel (GE Healthcare) column (1.6 × 20 cm) and lyophilized.
Protein Activity Assays
The activities of LAP5 and LAP6 were determined by monitoring the enzymatic reaction products with HPLC-UV/visible-ITMS. The TT4 gene product, TT4/CHS, was used as a positive control, and the empty-vector extract was used as a negative control. The enzymatic reactions were performed in a 100-μL reaction buffer (100 mm potassium phosphate buffer, pH 7.0) containing 15 μm 4-coumaroyl-CoA, 45 μm of malonyl-CoA (M4263; Sigma-Aldrich), and 50 μg of the purified proteins. In the separate assays, 4-coumaroyl-CoA was replaced with acetyl-CoA (A2181; Sigma-Aldrich) with other conditions remaining unchanged. Reactions were allowed to continue for 4 h at 37°C, quenched with the addition of 20 μL of formic acid, and extracted with 200 μL of ethyl acetate twice. The mixtures were vortexed and briefly centrifuged to assist in phase partitioning. The ethyl acetate phases were collected, combined and evaporated to dryness under nitrogen stream, redissolved in methanol (35 μL), and subjected to HPLC-UV/visible-ITMS analyses as described previously (Farag et al., 2007). Briefly, 30-μL aliquots of the reaction products were injected onto an Agilent 1100 series II HPLC system (Hewlett-Packard) that was coupled to a Bruker Esquire ion-trap mass spectrometer. Separations were achieved using a reverse-phase C18 column (5 μm, 4.6 mm i.d. × 250 mm; J.T. Baker) and a linear gradient of 5% to 70% B (v/v) over 60 min at a flow rate of 0.8 mL min−1. The mobile phases consisted of solvent A (0.1% acetic acid [v/v] in water) and solvent B (acetonitrile). UV spectra were obtained by scanning from 200 to 600 nm, and the mass spectra were acquired in negative electrospray ionization mode with a mass scan range of 50 to 2,200 m/z using an electrospray ionization voltage of 4.0 kV. MS/MS was performed with manual selection of precursor ion and the fragmentation energy set at 1.5. To determine the activities of LAP5, LAP6, and TT4/CHS toward fatty acyl-CoA, 4-coumaroyl-CoA was replaced with eight n-alkyl fatty acyl-CoAs (butyryl-, hexanoyl-, octanoyl-, decanoyl-, lauroyl-, myristoyl-, palmitoyl-, and stearoyl-CoA; Sigma-Aldrich), with the reaction conditions, product processing, and analyses remaining unchanged.
The Arabidopsis Genome Initiative locus numbers for the genes analyzed in this article are as follows: LAP5 (At4g34850), LAP6 (At1g02050), At4g00040, CYP703A2 (At1g01280), and TT4 (At5g13930).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Biosynthesis of plant phenolic compounds.
Supplemental Figure S2. Mass spectra of the two products from the LAP6 enzymatic assay.
Supplemental Figure S3. In vitro formation of trans- and cis-bisnoryangonin.
Supplemental Figure S4. Condensation of fatty acyl-CoA with malonyl-CoA leads to the formation of triketide pyrones, tetraketide pyrones, and tetraketide resorcylic acid.
Supplemental Figure S5. HPLC-ITMS UV analyses of the products of the enzymatic reactions with hexanoyl-CoA and malonyl-CoA as substrates.
Supplemental Figure S6. HPLC-ITMS UV analyses of the products of the enzymatic reactions with decanoyl-CoA and malonyl-CoA as substrates.
Supplemental Figure S7. HPLC chromatograms of the enzymatic assays with multiple substrates (coumaroyl-CoA, hexanoyl-CoA, and malonyl-CoA).
Supplemental Figure S8. Defects of pollen and anther development in the acos5 mutant strongly resemble those in lap5-1 lap6-1.
Supplemental Table S1. UPLC-MS analysis of levels of phenylpropanoid metabolites in wild-type, lap5, lap6, and lap5 lap6 anthers.
Supplemental Table S2. UPLC-MS analysis of levels of glucosinolates in wild-type, lap5, lap6, and lap5 lap6 anthers.
Supplemental Table S3. UPLC-MS analysis of levels of fatty acid metabolites in wild-type, lap5, lap6, and lap5 lap6 anthers.
Supplemental Table S4. UPLC-MS analysis of levels of unknown metabolites in wild-type, lap5, lap6, and lap5 lap6 anthers.
Supplemental Table S5. GC-MS analysis of metabolite levels in wild-type, lap5, lap6, and lap5 lap6 anthers.
Supplementary Material
Acknowledgments
We are grateful to J.J. Lado and A. Shicoff for technical help, to B. Scott for advice and help with protein purification, and to E. Bray, M. Jones-Rhoades, R. Swanson, and A. Fiebig for critical reading of the manuscript.
References
- Aarts MG, Hodge R, Kalantidis K, Florack D, Wilson ZA, Mulligan BJ, Stiekema WJ, Scott R, Pereira A. (1997) The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. Plant J 12: 615–623 [DOI] [PubMed] [Google Scholar]
- Ageez A, Kazama Y, Sugiyama R, Kawano S. (2005) Male-fertility genes expressed in male flower buds of Silene latifolia include homologs of anther-specific genes. Genes Genet Syst 80: 403–413 [DOI] [PubMed] [Google Scholar]
- Ahlers F, Thom I, Lambert J, Kuckuk R, Wiermann R. (1999) 1H NMR analysis of sporopollenin from Typha angustifolia. Phytochemistry 5: 1095–1098 [Google Scholar]
- Akiyama T, Shibuya M, Liu HM, Ebizuka Y. (1999) p-Coumaroyltriacetic acid synthase, a new homologue of chalcone synthase, from Hydrangea macrophylla var. thunbergii. Eur J Biochem 263: 834–839 [DOI] [PubMed] [Google Scholar]
- Ariizumi T, Hatakeyama K, Hinata K, Sato S, Kato T, Tabata S, Toriyama K. (2003) A novel male-sterile mutant of Arabidopsis thaliana, faceless pollen-1, produces pollen with a smooth surface and an acetolysis-sensitive exine. Plant Mol Biol 53: 107–116 [DOI] [PubMed] [Google Scholar]
- Atanassov I, Russinova E, Antonov L, Atanassov A. (1998) Expression of an anther-specific chalcone synthase-like gene is correlated with uninucleate microspore development in Nicotiana sylvestris. Plant Mol Biol 38: 1169–1178 [DOI] [PubMed] [Google Scholar]
- Austin MB, Bowman ME, Ferrer JL, Schröder J, Noel JP. (2004) An aldol switch discovered in stilbene synthases mediates cyclization specificity of type III polyketide synthases. Chem Biol 11: 1179–1194 [DOI] [PubMed] [Google Scholar]
- Austin MB, Noel JP. (2003) The chalcone synthase superfamily of type III polyketide synthases. Nat Prod Rep 20: 79–110 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Floyd SK, Sakakibara K. (2007) Green genes: comparative genomics of the green branch of life. Cell 129: 229–234 [DOI] [PubMed] [Google Scholar]
- Broeckling CD, Huhman DV, Farag MA, Smith JT, May GD, Mendes P, Dixon RA, Sumner LW. (2005) Metabolic profiling of Medicago truncatula cell cultures reveals the effects of biotic and abiotic elicitors on metabolism. J Exp Bot 56: 323–336 [DOI] [PubMed] [Google Scholar]
- Burbulis IE, Iacobucci M, Shirley BW. (1996) A null mutation in the first enzyme of flavonoid biosynthesis does not affect male fertility in Arabidopsis. Plant Cell 8: 1013–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coe EH, McCormick SM, Modena SA. (1981) White pollen in maize. J Hered 72: 318–320 [Google Scholar]
- de Azevedo Souza C, Kim SS, Koch S, Kienow L, Schneider K, McKim SM, Haughn GW, Kombrink E, Douglas CJ. (2009) A novel fatty acyl-CoA synthetase is required for pollen development and sporopollenin biosynthesis in Arabidopsis. Plant Cell 21: 507–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan TTP, Carlsson AS, Hamberg M, Bülow L, Stymne S, Olsson P. (2009) Functional expression of five Arabidopsis fatty acyl-CoA reductase genes in Escherichia coli. J Plant Physiol 166: 787–796 [DOI] [PubMed] [Google Scholar]
- Dobritsa AA, Shrestha J, Morant M, Pinot F, Matsuno M, Swanson R, Møller BL, Preuss D. (2009) CYP704B1 is a long-chain fatty acid ω-hydroxylase essential for sporopollenin synthesis in pollen of Arabidopsis. Plant Physiol 151: 574–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckermann C, Schroder G, Eckermann S, Strack D, Schmidt J, Schneider B, Schroder J. (2003) Stilbenecarboxylate biosynthesis: a new function in the family of chalcone synthase-related proteins. Phytochemistry 62: 271–286 [DOI] [PubMed] [Google Scholar]
- Eckermann S, Schröder G, Schmidt J, Strack D, Edrada RA, Helaruitta Y, Elomaa P, Kotilainen M, Kilpeläinen I, Proksch P, et al. (1998) New pathway to polyketides in plants. Nature 396: 387–390 [Google Scholar]
- Edlund AF, Swanson R, Preuss D. (2004) Pollen and stigma structure and function: the role of diversity in pollination. Plant Cell (Suppl) 16: S84–S97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdtman G. (1960) The acetolysis method. Sven Bot Tidskr 54: 561–564 [Google Scholar]
- Farag MA, Huhman DV, Lei Z, Sumner LW. (2007) Metabolic profiling and systematic identification of flavonoids and isoflavonoids in roots and cell suspension cultures of Medicago truncatula using HPLC-UV-ESI-MS and GC-MS. Phytochemistry 68: 342–354 [DOI] [PubMed] [Google Scholar]
- Ferrer JL, Jez JM, Bowman ME, Dixon RA, Noel JP. (1999) Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nat Struct Biol 6: 775–784 [DOI] [PubMed] [Google Scholar]
- Fischer R, Budde I, Hain R. (1997) Stilbene synthase gene expression causes changes in flower color and male sterility in tobacco. Plant J 11: 489–498 [Google Scholar]
- Flores-Sanchez IJ, Verpoorte R. (2009) Plant polyketide synthases: a fascinating group of enzymes. Plant Physiol Biochem 47: 167–174 [DOI] [PubMed] [Google Scholar]
- Gubatz S, Rittscher M, Meuter A, Nagler A, Wiermann R. (1993) Tracer experiments on sporopollenin biosynthesis: an overview. Grana (Suppl) 1: 12–17 [Google Scholar]
- Guilford WJ, Schneider DM, Labovitz J, Opella SJ. (1988) High resolution solid state 13C NMR spectroscopy of sporopollenins from different plant taxa. Plant Physiol 86: 134–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillet G, Poupart J, Basurco J, De Luca V. (2000) Expression of tryptophan decarboxylase and tyrosine decarboxylase genes in tobacco results in altered biochemical and physiological phenotypes. Plant Physiol 122: 933–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahlbrock K, Scheel D. (1989) Physiology and molecular biology of phenylpropanoid metabolism. Annu Rev Plant Physiol Plant Mol Biol 40: 347–369 [Google Scholar]
- Halkier BA, Gershenzon J. (2006) Biology and biochemistry of glucosinolates. Annu Rev Plant Biol 57: 303–333 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM. (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Herrmann KM. (1995) The shikimate pathway: early steps in the biosynthesis of aromatic compounds. Plant Cell 7: 907–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hihara Y, Hara C, Uchimiya H. (1996) Isolation and characterization of two cDNA clones for mRNAs that are abundantly expressed in immature anthers of rice (Oryza sativa L.). Plant Mol Biol 30: 1181–1193 [DOI] [PubMed] [Google Scholar]
- Jez JM, Austin MB, Ferrer JL, Bowman ME, Schröder J, Noel JP. (2000a) Structural control of polyketide formation in plant-specific polyketide synthases. Chem Biol 7: 919–930 [DOI] [PubMed] [Google Scholar]
- Jez JM, Ferrer JL, Bowman ME, Dixon RA, Noel JP. (2000b) Dissection of malonyl-CoA decarboxylation from polyketide formation in the reaction mechanism of a plant polyketide synthase. Biochemistry 39: 890–902 [DOI] [PubMed] [Google Scholar]
- Jindaprasert A, Springob K, Schmidt J, De-Eknamkul W, Kutchan TM. (2008) Pyrone polyketides synthesized by a type III polyketide synthase from Drosophyllum lusitanicum. Phytochemistry 69: 3043–3053 [DOI] [PubMed] [Google Scholar]
- Karppinen K, Hokkanen J, Mattila S, Neubauer P, Hohtola A. (2008) Oktaketide-producing type III polyketide synthase from Hypericum perforatum is expressed in dark glands accumulating hypericins. FEBS J 275: 4329–4342 [DOI] [PubMed] [Google Scholar]
- Knill T, Schuster J, Reichelt M, Gershenzon J, Binder S. (2008) Arabidopsis branched-chain aminotransferase 3 functions in both amino acid and glucosinolate biosynthesis. Plant Physiol 146: 1028–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzaler F, Hahlbrock K. (1975) Enzymic synthesis of an aromatic ring from acetate units. Eur J Biochem 56: 205–213 [DOI] [PubMed] [Google Scholar]
- Kupke T, Hernandez-Acosta P, Culianez-Macia FA. (2003) 4′-Phosphopantetheine and coenzyme A biosynthesis in plants. J Biol Chem 278: 38229–38237 [DOI] [PubMed] [Google Scholar]
- Lanz T, Tropf S, Marner FJ, Schröder J, Schröder G. (1991) The role of cysteines in polyketide synthases. Site-directed mutagenesis of resveratrol and chalcone synthases, two key enzymes in different plant-specific pathways. J Biol Chem 266: 9971–9976 [PubMed] [Google Scholar]
- Meyers BC, Tej SS, Vu TH, Haudenschild CD, Agrawal V, Edberg SB, Ghazal H, Decola S. (2004) The use of MPSS for whole-genome transcriptional analysis in Arabidopsis. Genome Res 14: 1641–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore RW, Paran I, Kesseli RV. (1991) Identification of markers linked to disease resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genome regions by using segregating populations. Proc Natl Acad Sci USA 88: 9828–9832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi Y, Shimokawa Y, Wanibuchi K, Noguchi H, Abe I. (2008) Structure function analysis of novel type III polyketide synthases from Arabidopsis thaliana. Biol Pharm Bull 31: 2205–2210 [DOI] [PubMed] [Google Scholar]
- Morant M, Jørgensen K, Schaller H, Pinot F, Møller BL, Werck-Reichhart D, Bak S. (2007) CYP703 is an ancient cytochrome P450 in land plants catalyzing in-chain hydroxylation of lauric acid to provide building blocks for sporopollenin synthesis in pollen. Plant Cell 19: 1473–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli CA, Fahy D, Wang HY, Taylor LP. (1999) white anther: a petunia mutant that abolishes pollen flavonol accumulation, induces male sterility, and is complemented by a chalcone synthase transgene. Plant Physiol 120: 615–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niesbach-Klösgen U, Barzen E, Bernhardt J, Rohde W, Schwartz-Sommer Z, Reif HJ, Wienand U, Saedler H. (1987) Chalcone synthase genes in plants: a tool to study evolutionary relationships. J Mol Evol 26: 213–225 [Google Scholar]
- Niester-Nyveld C, Haubrich A, Kampendonk H, Gubatz S, Tenberge KB, Rittscher M, Wilmesmeier S, Wiermann R. (1997) Immunocytochemical localization of phenolic compounds in pollen walls using antibodies against p-coumaric acid coupled to bovine serum albumin. Protoplasma 197: 148–159 [Google Scholar]
- Nishikawa S, Zinkl GM, Swanson RJ, Maruyama D, Preuss D. (2005) Callose (β-1,3 glucan) is essential for Arabidopsis pollen wall patterning, but not tube growth. BMC Plant Biol 5: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak P, Krofta K, Matousek J. (2006) Chalcone synthase homologues from Humulus lupulus: some enzymatic properties and expression. Biol Plant 50: 48–54 [Google Scholar]
- Obayashi T, Kinoshita K, Nakai K, Shibaoka M, Hayashi S, Saeki M, Shibata D, Saito K, Ohta H. (2007) ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res 35: D863–D869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittscher M, Wiermann R. (1988) Studies on sporopollenin biosynthesis in Tulipa anthers. II. Incorporation of precursors and degradation of the radiolabelled polymer. Sex Plant Reprod 1: 132–139 [Google Scholar]
- Rohde A, Morreel K, Ralph J, Goeminne G, Hostyn V, De Rycke R, Kushnir S, Van Doorsselaere J, Joseleau JP, Vuylsteke M, et al. (2004) Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. Plant Cell 16: 2749–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders PM, Bui AQ, Weterings K, McIntire KN, Hsu YC, Lee PY, Truong MT, Beals TP, Goldberg RB. (1999) Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex Plant Reprod 11: 297–322 [Google Scholar]
- Sankaranarayanan R, Saxena P, Marathe UB, Gokhale RS, Shanmugam VM, Rukmini R. (2004) A novel tunnel in mycobacterial type III polyketide synthase reveals the structural basis for generating diverse metabolites. Nat Struct Mol Biol 11: 894–900 [DOI] [PubMed] [Google Scholar]
- Saxena P, Yadav G, Mohanty D, Gokhale RS. (2003) A new family of type III polyketide synthases in Mycobacterium tuberculosis. J Biol Chem 278: 44780–44790 [DOI] [PubMed] [Google Scholar]
- Schröder J. (1997) A family of plant-specific polyketide synthases: facts and predictions. Trends Plant Sci 2: 373–378 [Google Scholar]
- Schröder J, Raiber S, Berger T, Schmidt A, Schmidt J, Soares-Sello AM, Bardshiri E, Strack D, Simpson TJ, Veit M, et al. (1998) Plant polyketide synthases: a chalcone synthase-type enzyme which performs a condensation reaction with methylmalonyl-CoA in the biosynthesis of C-methylated chalcones. Biochemistry 37: 8417–8425 [DOI] [PubMed] [Google Scholar]
- Schüz R, Heller W, Hahlbrock K. (1983) Substrate specificity of chalcone synthase from Petroselinum hortense: formation of phloroglucinol derivatives from aliphatic substrates. J Biol Chem 258: 6730–6734 [PubMed] [Google Scholar]
- Shaw G. (1971) The chemistry of sporopollenin. Brooks J, Grant PR, Muir M, van Gijzel P, Shaw G, , Sporopollenin. Academic Press, London, pp 305–348 [Google Scholar]
- Shen JB, Hsu FC. (1992) Brassica anther-specific genes: characterization and in situ localization of expression. Mol Gen Genet 234: 379–389 [DOI] [PubMed] [Google Scholar]
- Shiokawa KI, Inagaki Y, Morita H, Hsu TJ, Iida S, Noguchi H. (2000) The functional expression of the CHS-D and CHS-E genes of the common morning glory (Ipomoea purpurea) in Escherichia coli and characterization of their gene products. Plant Biotechnol 17: 12 [Google Scholar]
- Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM. (1995) Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J 8: 659–671 [DOI] [PubMed] [Google Scholar]
- Shomura Y, Torayama I, Suh DY, Xiang T, Kita A, Sankawa U, Miki K. (2005) Crystal structure of stilbene synthase from Arachis hypogaea. Proteins 60: 803–806 [DOI] [PubMed] [Google Scholar]
- Shulze Osthoff K, Wiermann R. (1987) Phenols as integrated compounds of sporopollenin from Pinus pollen. J Plant Physiol 131: 5–15 [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckigt J, Zenk MH. (1975) Chemical synthesis and properties of hydroxycinnamoylcoenzyme A derivatives. Z Naturforsch [C] 30: 352–358 [DOI] [PubMed] [Google Scholar]
- Tang LK, Chu H, Yip WK, Yeung EC, Lo C. (2008) An anther-specific dihydroflavonol 4-reductase-like gene (DRL1) is essential for male fertility in Arabidopsis. New Phytol 181: 576–587 [DOI] [PubMed] [Google Scholar]
- Taylor LP, Jorgensen R. (1992) Conditional male fertility in chalcone synthase-deficient petunia. J Hered 83: 11–17 [Google Scholar]
- Toufighi K, Brady SM, Austin R, Ly E, Provart NJ. (2005) The Botany Array Resource: e-northerns, expression angling, and promoter analyses. Plant J 43: 153–163 [DOI] [PubMed] [Google Scholar]
- Tropf S, Kärcher B, Schröder G, Schröder J. (1995) Reaction mechanisms of homodimeric plant polyketide synthases (stilbene and chalcone synthases). J Biol Chem 270: 7922–7928 [DOI] [PubMed] [Google Scholar]
- van der Meer IM, Stam ME, van Tunen AJ, Mol JN, Stuitje AR. (1992) Antisense inhibition of flavonoid biosynthesis in petunia anthers results in male sterility. Plant Cell 4: 253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Besser K, Frank AC, Johnson MA, Preuss D. (2006) Arabidopsis HAP2 (GCS1) is a sperm-specific gene required for pollen tube guidance and fertilization. Development 133: 4761–4769 [DOI] [PubMed] [Google Scholar]
- Walden AR, Walter C, Gardner RC. (1999) Genes expressed in Pinus radiata male cones include homologs to anther-specific and pathogenesis response genes. Plant Physiol 121: 1103–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White WH, Gunyuzlu PL, Toyn JH. (2001) Saccharomyces cerevisiae is capable of de novo pantothenic acid biosynthesis involving a novel pathway of beta-alanine production from spermine. J Biol Chem 276: 10794–10800 [DOI] [PubMed] [Google Scholar]
- Wiermann R, Ahlers F, Schmitz-Thom I. (2001) Sporopollenin. Stenbüchel A, Hofrichter M, , Biopolymers, Vol 1. Wiley-VCH Verlag, Weinheim, Germany, pp 209–227 [Google Scholar]
- Wijeratne AJ, Zhang W, Sun Y, Liu W, Albert R, Zheng Z, Oppenheimer DG, Zhao D, Ma H. (2007) Differential gene expression in Arabidopsis wild-type and mutant anthers: insights into anther cell differentiation and regulatory networks. Plant J 52: 14–29 [DOI] [PubMed] [Google Scholar]
- Winkel BSJ. (2004) Metabolic channeling in plants. Annu Rev Plant Biol 55: 85–107 [DOI] [PubMed] [Google Scholar]
- Xing S, Zachgo S. (2008) ROXY1 and ROXY2, two Arabidopsis glutaredoxin genes, are required for anther development. Plant J 53: 790–801 [DOI] [PubMed] [Google Scholar]
- Xu J, Yang C, Yuan Z, Zhang D, Gondwe MY, Ding Z, Liang W, Zhang DB, Wilson ZA. (2010) The ABORTED MICROSPORES regulatory network is required for postmeiotic male reproductive development in Arabidopsis thaliana. Plant Cell 22: 91–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Vizcay-Barrena G, Conner K, Wilson ZA. (2007) MALE STERILITY1 is required for tapetal development and pollen wall biosynthesis. Plant Cell 19: 3530–3548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylstra B, Muskens M, Van Tunen AJ. (1996) Flavonols are not essential for fertilization in Arabidopsis thaliana. Plant Mol Biol 32: 1155–1158 [DOI] [PubMed] [Google Scholar]
- Zhou H, Zhan J, Watanabe K, Xie X, Tang Y. (2008) A polyketide microlactone synthase from the filamentous fungus Gibberella zeae. Proc Natl Acad Sci USA 105: 6251–6254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkl GM, Zwiebel BI, Grier DG, Preuss D. (1999) Pollen-stigma adhesion in Arabidopsis: a species-specific interaction mediated by lipophilic molecules in the pollen exine. Development 126: 5431–5440 [DOI] [PubMed] [Google Scholar]
- Zuurbier KW, Leser J, Berger T, Hofte AJ, Schroder G, Verpoorte R, Schroder J. (1998) 4-Hydroxy-2-pyrone formation by chalcone and stilbene synthase with nonphysiological substrates. Phytochemistry 49: 1945–1951 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.