Abstract
3,8-Divinyl (proto)chlorophyll(ide) a 8-vinyl reductase (DVR) catalyzes the reduction of 8-vinyl group on the tetrapyrrole to an ethyl group, which is indispensable for monovinyl chlorophyll (Chl) synthesis. So far, three 8-vinyl reductase genes (DVR, bciA, and slr1923) have been characterized from Arabidopsis (Arabidopsis thaliana), Chlorobium tepidum, and Synechocystis sp. PCC6803. However, no 8-vinyl reductase gene has yet been identified in monocotyledonous plants. In this study, we isolated a spontaneous mutant, 824ys, in rice (Oryza sativa). The mutant exhibited a yellow-green leaf phenotype, reduced Chl level, arrested chloroplast development, and retarded growth rate. The phenotype of the 824ys mutant was caused by a recessive mutation in a nuclear gene on the short arm of rice chromosome 3. Map-based cloning of this mutant resulted in the identification of a gene (Os03g22780) showing sequence similarity with the Arabidopsis DVR gene (AT5G18660). In the 824ys mutant, nine nucleotides were deleted at residues 952 to 960 in the open reading frame, resulting in a deletion of three amino acid residues in the encoded product. High-performance liquid chromatography analysis of Chls indicated the mutant accumulates only divinyl Chl a and b. A recombinant protein encoded by Os03g22780 was expressed in Escherichia coli and found to catalyze the conversion of divinyl chlorophyll(ide) a to monovinyl chlorophyll(ide) a. Therefore, it has been confirmed that Os03g22780, renamed as OsDVR, encodes a functional DVR in rice. Based upon these results, we succeeded to identify an 8-vinyl reductase gene in monocotyledonous plants and, more importantly, confirmed the DVR activity to convert divinyl Chl a to monovinyl Chl a.
Chlorophyll (Chl) is the main component of the photosynthetic pigments. Chl molecules universally exist in photosynthetic organisms and perform essential processes of harvesting light energy in the antenna systems and by driving electron transfer in the reaction centers (Fromme et al., 2003). In higher plants, there are two Chl species, Chl a and Chl b. The photosynthetic reaction centers contain only Chl a, and the peripheral light-harvesting antenna complexes contain Chl a and Chl b (Grossman et al., 1995). Chl a is synthesized from glutamyl-tRNA, and Chl b is synthesized from Chl a at the last step of Chl biosynthesis (Beale, 1999). So far, genes for all 15 steps in the Chl biosynthetic pathway have been identified in higher plants, at least in angiosperms represented by Arabidopsis (Arabidopsis thaliana; Beale, 2005; Nagata et al., 2005). Analysis of the complete genome of Arabidopsis showed that it has 15 enzymes encoded by 27 genes for Chl biosynthesis from glutamyl-tRNA to Chl b (Nagata et al., 2005). However, only six genes encoding three enzymes involved in Chl biosynthesis have been identified in rice (Oryza sativa). Magnesium chelatase comprises three subunits (ChlH, ChlD, and ChlI) and catalyzes the insertion of Mg2+ into protoporphyrin IX, the last common intermediate precursor in both Chl and heme biosyntheses. Jung et al. (2003) characterized OsCHLH gene for the OsChlH subunit of magnesium chelatase, and Zhang et al. (2006) cloned Chl1 and Chl9 genes encoding the OsChlD and OsChlI subunits of magnesium chelatase. Chl synthase catalyzes esterification of chlorophyllide (Chlide), resulting in the formation of Chl a. Wu et al. (2007) identified the YGL1 gene encoding the Chl synthase. Chl b is synthesized from Chl a by Chl a oxygenase; Lee et al. (2005) identified OsCAO1 and OsCAO2 genes for Chl a oxygenase.
According to the number of vinyl side chains, Chls of oxygenic photosynthetic organisms are classified into two groups: 3,8-divinyl Chl (DV-Chl) and 3-vinyl Chl (monovinyl Chl [MV-Chl]). Almost all of the oxygenic photosynthetic organisms contain MV-Chls, regardless of the variation in their indigenous environments (Porra, 1997). The exceptions are species of Prochlorococcus marinus, marine picophytoplanktons that contain DV-Chls as their photosynthetic pigments (Chisholm et al., 1992).
Chl biosynthetic heterogeneity is assumed to originate mainly in parallel DV- and MV-Chl biosynthetic routes interconnected by 8-vinyl reductases that convert DV-tetrapyrroles to MV-tetrapyrroles by conversion of the vinyl group at position 8 of ring B to the ethyl group (Parham and Rebeiz, 1995; Rebeiz et al., 2003). Most of Chls carry an ethyl group or, less frequently, a vinyl group. For example, Chl a and b occur as the MV-derivatives in green plants, but Chl precursors sometimes accumulate as DV-intermediates, and the ratio between the two forms can vary depending on the species, tissue, and growth conditions (Shioi and Takamiya, 1992; Kim and Rebeiz, 1996). So far, five 8-vinyl reductase activities have been detected at the levels of DV Mg-protoporphyrin IX (Kim and Rebeiz, 1996), Mg-protomonomethyl ester (Kolossov et al., 2006), protochlorophyllide (Pchlide) a (Tripathy and Rebeiz, 1988), Chlide a (Kolossov and Rebeiz, 2001; Nagata et al., 2005), and Chl a (Adra and Rebeiz, 1998). What is not clear at this stage is whether the various 8-vinyl reductase activities are catalyzed by one enzyme of broad specificity or by a family of enzymes of narrow specificity encoded by one gene or multiple genes, as is the case for NADPH Pchlide oxidoreductases (Rebeiz et al., 2003). The issue could be settled by purification of the various putative reductases and comparison of their properties.
Nagata et al. (2005) and Nakanishi et al. (2005) independently identified the AT5G18660 gene of Arabidopsis as a divinyl reductase (DVR) that has sequence similarity to isoflavone reductase. Chew and Bryant (2007) demonstrated that BciA (CT1063), which is an ortholog of the Arabidopsis gene, encodes a DVR of the green sulfur bacterium Chlorobium tepidum TLS. They also considered that BchJ, which had been reported to be a vinyl reductase (Suzuki and Bauer, 1995), is not the enzyme, but it may play an important role in substrate channeling and/or regulation of bacteriochlorophyll biosynthesis. Islam et al. (2008) and Ito et al. (2008) independently identified a novel 8-vinyl reductase gene (Slr1923) in DVR-less cyanobacterium Synechocystis sp. PCC6803. However, no DVR gene has yet been identified in monocotyledonous plants.
In this study, we isolated a spontaneous mutant, 824ys, from indica rice cv 824B. The mutant exhibited a yellow-green leaf phenotype throughout the growth stage, reduced level of Chls, arrested development of chloroplasts, and retarded growth rate. Map-based cloning of the mutant resulted in the identification of the OsDVR gene, showing sequence similarity to the DVR gene of Arabidopsis. In the 824ys mutant, nine nucleotides were deleted at residues 952 to 960 in the open reading frame (ORF), resulting in three amino acid deletion in the encoded protein. HPLC analysis of Chls indicated the mutant accumulates only DV-Chls. Enzymatic analysis demonstrated that the recombinant protein expressed in Escherichia coli is able to catalyze the conversion of DV-Chl(ide) a to MV-Chl(ide) a. Therefore, this study has confirmed that the OsDVR gene encodes a functional DVR in rice.
RESULTS
The 824ys Mutant Showing Reduced Chl Level and Arrested Chloroplast Development
The 824ys mutant was a spontaneous mutant isolated from indica rice cv 824B, which exhibited a yellow-green leaf phenotype throughout development. The mutant and wild type were planted under natural conditions, in which average air temperature per day was 18.6°C to 28.4°C, average sunlight intensity per day was 30 to 580 μmol m−2 s−1, and the maximum sunlight intensity in a day was 524 to 5,948 μmol m−2 s−1. The 824ys mutant grew very slowly. Although its days to heading increased 16 d, its height of plants and number of productive tillers decreased by 25.6% and 64.9% compared with the wild type, respectively (Fig. 1). Besides, its number of spikelets per panicle, seed-setting rate, and 1,000-grain weight also declined by 42.5%, 45.7%, and 26.9%, respectively.
Figure 1.
Plant phenotype of the rice 824ys mutant. A, Four-week-old plants of wild type (left) and 824ys mutant (right). B, Thirteen-week-old plants of wild type (left) and 824ys mutant (right). Bars = 5 cm.
To characterize the yellow-green phenotype of the 824ys mutant, we first measured its Chl contents, and found its contents of Chl a, b, and total Chls to be 41% to 48%, 9% to 22%, and 33% to 42% of that from wild type, respectively (Table I), indicating that the mutant phenotype resulted from reduced Chl level.
Table I. Pigment contents in leaves of wild type and 824ys mutant, in mg g fresh weight−1.
Chl were measured in 95% ethanol extracts from the first, second, and third leaves from the top at indicated growth stages. Values shown are the mean ± sd from five independent determinations.
| Growth Stage | Line | Total Chl | Chl aa | Chl ba | Chl a/b Ratio |
| Seedling stage | Wild type (824B) | 3.32 ± 0.04 | 2.54 ± 0.03 | 0.78 ± 0.01 | 3.26 ± 0.03 |
| 824ys | 1.39 ± 0.19 | 1.22 ± 0.17 | 0.17 ± 0.13 | 7.18 ± 0.34 | |
| Tillering stage | Wild type (824B) | 5.66 ± 0.02 | 4.27 ± 0.01 | 1.39 ± 0.03 | 3.07 ± 0.07 |
| 824ys | 1.89 ± 0.75 | 1.73 ± 0.70 | 0.16 ± 0.06 | 10.81 ± 4.11 | |
| Heading stage | Wild type (824B) | 4.93 ± 0.45 | 3.52 ± 0.36 | 1.41 ± 0.69 | 2.50 ± 0.10 |
| 824ys | 1.67 ± 0.67 | 1.55 ± 0.17 | 0.12 ± 0.02 | 12.92 ± 7.42 |
According to Figure 4, Chl a and b in the 824ys mutant actually were Chl a- and b-like pigment, i.e. DV-Chl a and b, respectively.
Next, transmission electron microscopic analysis was performed to determine if low Chl content in the mutant leaves affects the chloroplast development and morphology. The result revealed that the shape of 824ys mutant chloroplasts was irregular compared with the wild-type chloroplasts. The thylakoid membranes of the mutant chloroplasts were disrupted. The grana stacks in the mutant appeared less dense (Supplemental Fig. S1, C and D). These results indicated that the development of chloroplast thylakoids was suppressed in the mutant.
The 824ys Locus Mapped to a Putative Gene Encoding DVR on Chromosome 3
All F1 plants of the 824ys mutant crossed with four normal green cultivars, i.e. 824B, II-32B, 495R (indica), and 02428 (japonica), displayed normal green leaves. F2 populations from the four crosses all showed a segregation ratio of 3:1 (green:yellow-green plants, χ2 < χ20.05 = 3.84, P > 0.05; Table II). These results suggested that the yellow-green leaf phenotype in the 824ys mutant is controlled by a single recessive nuclear gene.
Table II. Segregation of F2 populations from four crosses.
| Crossa | 824B/824ys | 495R/824ys | II-32B/824ys | 02428/824ys |
| Total nos. of plants | 2,939 | 1,066 | 1,047 | 3,963 |
| Nos. of green plantsb | 2,217 | 816 | 813 | 3,026 |
| Nos. of yellow plants | 722 | 250 | 234 | 937 |
| χ2 | 0.27 | 1.28 | 3.78 | 3.82 |
| Pc | 0.50–0.75 | 0.25–0.50 | 0.05–0.10 | 0.05–0.10 |
The 824ys mutant as the male partner for these crosses.
Green plants and yellow plants were determined by visual inspection.
P > 0.05 considered as significant.
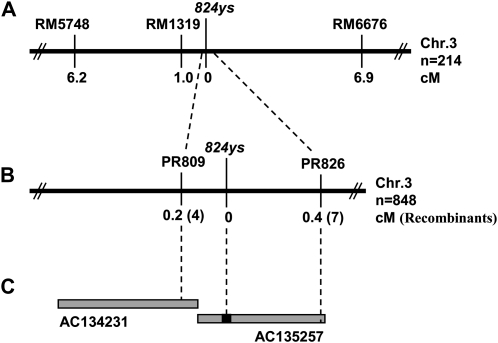
In a previous study, using F2 population from the cross between the 824ys mutant with an indica cultivar, 495R, we preliminarily mapped this mutant gene on the short arm of rice chromosome 3, 5.2 cm from simple sequence repeat (SSR) marker RM282 (Huang et al., 2008). In this investigation, fine mapping of the 824ys locus was performed using F2 population from the cross between the 824ys mutant with a japonica cultivar, 02428. First, three SSR markers (RM5748, RM1319, and RM6676) with parental polymorphism were found, and the 824ys locus was mapped to a region between RM1319 and RM6676 using 214 recessive individuals with the yellow-green leaf phenotype. Genetic distances from the target gene to SSR markers RM1319 and RM6676 were 1.0 and 6.9 cm, respectively (Fig. 2A). Subsequently, two insertion/deletion (InDel) markers (PR809 and PR826) were developed, whose primers were as follows: PR809F, 5′-CTAATCTGTACGCTTCAC-3′; PR809R, 5′-ACACTTCATACCGTCATA-3′; PR826F, 5′-TCCTCTTGCTCCTCTTCT-3′; and PR826R, 5′-ACATTGGGTATGGTGGTC-3′. The 824ys locus was further mapped between the two markers, at genetic distances of 0.2 and 0.4 cm, respectively, with 848 recessive individuals. PR809 and PR826 markers mapped a 142-kb DNA region on two bacterial artificial chromosomes, AC134231 and AC135257 (Fig. 2, B and C).
Figure 2.
Fine mapping of the 824ys locus. The map was constructed based on the publicly available sequence of rice chromosome 3. Two InDel markers (PR809 and PR826) were developed during this study, while three SSR markers (RM5748, RM1319, and RM6676) were obtained from the public database. A, The 824ys locus was mapped to a region between SSR markers RM1319 and RM6676 on the short arm of rice chromosome 3 (Chr.3) with 214 recessive individuals. B, Fine mapping of the 824ys locus between InDel markers PR809 and PR826 from a segregating population of 848 recessive individuals. C, Two bacterial artificial chromosome contigs (AC134231 and AC135257) covered the 824ys locus, which was narrowed down to a 142-kb DNA region between PR809 and PR826.
Within the PR809 and PR826 region, 20 predicted genes had been annotated in Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/pseudomolecules). According to recombinants between the two InDel markers and the 824ys mutant gene (Fig. 2, B and C), we found one ORF (Os03g22780) encoding a putative isoflavone reductase in the corresponding region, and sequence BLASTX showed it has a significant similarity to DVR (65% identities) in Arabidopsis (Nagata et al., 2005; Nakanishi et al., 2005), which is indispensable for MV-Chl synthesis. Then, we cloned the Os03g22780 gene from the 824ys mutant and the corresponding wild-type parent 824B. DNA sequencing results revealed that 9 bp were deleted at nucleotides 952 to 960 in the ORF in the 824ys mutant, which resulted in the deletion of three amino acid residues (Lys, Val, and Pro; Supplemental Fig. S2). Therefore, the Os03g22780 gene was considered as the candidate gene of 824ys, and designated tentatively as OsDVR gene.
BLAST search in the rice genome database revealed that OsDVR is a single-copy gene with an ORF of 1,218 bp and encodes a 405-amino acid protein with a molecular mass of approximately 43 kD. OsDVR protein contains an apparent chloroplast-targeting sequence of 58 amino acid residues at its N terminus (http://www.cbs.dtu.dk/services/TargetP/; Supplemental Fig. S2).
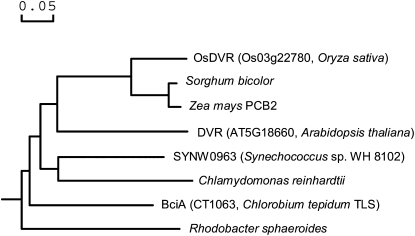
Multiple amino acid sequence alignment indicated that OsDVR has significant similarity to sorghum (Sorghum bicolor) hypothetical protein and maize (Zea mays) PCB2, with identity of 82% and 81%, respectively. OsDVR is also 56% identical to DVR (BciA) in green sulfur bacterium C. tepidum (Chew and Bryant, 2007; Supplemental Fig. S3). We subsequently analyzed the possible phylogenetic relationships between OsDVR and its related proteins from higher plants and other photosynthetic organisms. The result showed that rice OsDVR is more closely related to DVRs of the monocotyledonous plants sorghum and maize than to those of other species (Fig. 3).
Figure 3.
A phylogenetic tree representing alignment of OsDVR protein and its homologs. The rooted neighbor-joining tree using percentage identities was constructed based on a multiple sequence alignment generated with the program DNAMAN. Accession numbers for the respective protein sequences are as follows: rice (OsDVR, Os03g22780, ADE43128); sorghum (XP_002465274); maize (PCB2, NP_001148282); Arabidopsis (DVR, AT5G18660, NP_197367); Synechococcus sp. WH 8102 (SYNW0963, NP_897056); Chlamydomonas reinhardtii (XP_001690168); C. tepidum TLS (BciA [CT1063], NP_661954); and Rhodobacter sphaeroides (ZP_00006667). Scale represents percentage substitution per site.
Exclusive Accumulation of Divinyl Chl a and b in the 824ys Mutant
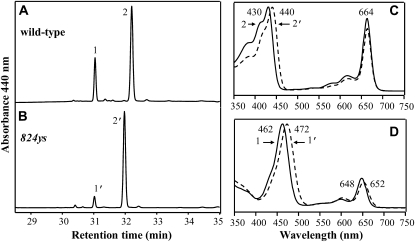
Because both divinyl and monovinyl types of Pchlide and Chlide can serve as substrates for Pchlide oxidoreductase and Chl synthetase, respectively, the final product of Chl biosynthesis should be DV-Chls when the plants have a defect in DVR (Nagata et al., 2005; Nakanishi et al., 2005). So we examined Chl compositions of the 824ys mutant by HPLC according to the described method (Zapata et al., 2000). The result showed that the mutant Chls differed from the wild-type MV-Chls but were identical to DV-Chls (Shedbalkar and Rebeiz, 1992; Zapata et al., 2000; Nagata et al., 2005; Nakanishi et al., 2005). The mutant Chl a-like pigment, whose retention time was about 14 s fewer than that of the wild-type MV-Chl a, had absorbance peaks at 440 and 664 nm. The Soret band peak shifted to the red by 10 nm compared with the MV-Chl a that had absorbance peaks at 430 and 664 nm (Fig. 4). The mutant Chl b-like pigment, whose retention time was close to that of the wild-type MV-Chl b, exhibited absorbance peaks at 472 and 652 nm compared with the MV-Chl b that had absorbance peaks at 462 and 648 nm, respectively (Fig. 4). Based upon these results, we concluded that the 824ys mutant accumulated only DV-Chls, instead of MV-Chls, so the mutant has a defective DVR. Accordingly we concluded the candidate gene underlying the 824ys mutant phenotype is OsDVR.
Figure 4.
HPLC analysis of Chls. The elution profiles of wild type (A) and 824ys mutant (B) were detected at 440 nm. Peak 1, Chl b; peak 2, Chl a; peak 1′, Chl b-like pigment; peak 2′, Chl a-like pigment. The absorption spectra of the peaks 2 and 2′ (C) and the peaks 1 and 1′ (D) in acetone were compared, respectively.
Conversion of DV-Chl(ide) a to MV-Chl(ide) a by the Recombinant Protein Encoded by OsDVR (Os03g22780)
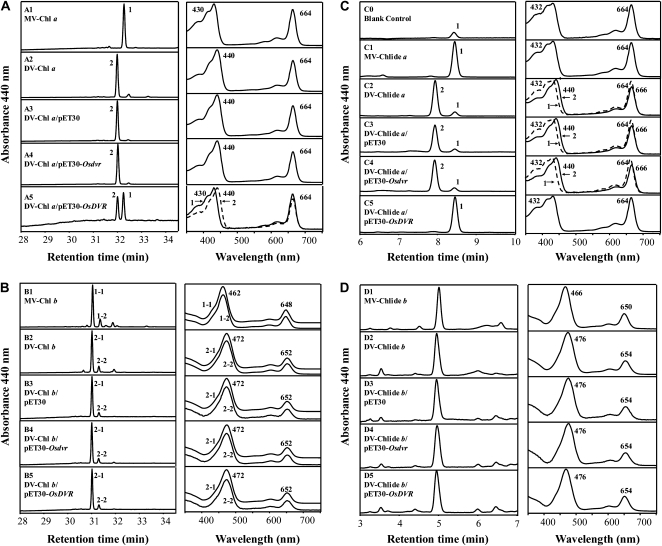
To test the DVR activity of the recombinant OsDVR protein, four DV-tetrapyrroles (DV-Chl a, -Chl b, -Chlide a, and -Chlide b) were used as substrates, in which DV-Chl a and b were prepared from the 824ys mutant of rice, and DV-Chlide a and b were prepared from DV-Chl a and b by the enzymatic reaction of chlorophyllase isolated from Garland chrysanthemum leaves (Holden, 1961; Ito et al., 1993, 1994, 1996). The recombinant OsDVR and Osdvr proteins were expressed in E. coli, and the cell extracts were incubated with the four different DV-tetrapyrroles, respectively, in the presence of NADPH. The results showed that DV-Chl a and -Chlide a were converted to MV-Chl a and -Chlide a, respectively, after 10-min incubation with OsDVR protein (Fig. 5, A5 and C5). However, under the same conditions, DV-Chl a and -Chlide a were not converted to the corresponding MV-compounds with Osdvr mutant protein (Fig. 5, A4 and C4). On the other hand, DV-Chl b and -Chlide b were not converted to corresponding MV-compounds after 10-min incubation with either OsDVR or Osdvr proteins (Fig. 5, B5, B4, D5, and D4). The results confirmed that the OsDVR gene encodes 3,8-divinyl chlorophyll(ide) a 8-vinyl reductase to catalyze the conversion of DV-Chl(ide) a to MV-Chl(ide) a in rice, and that the deletion of three amino acid residues (Lys, Val, and Pro) results in inactivation of OsDVR in the 824ys mutant. We should note that two peaks were detected in the HPLC profiles (Fig. 5B), but the absorption spectra of minor peaks (peaks 1-2 and 2-2) were all identical to those of their corresponding major peaks (peaks 1-1 and 2-1), and there was no difference in the minor peaks of DV-Chl b between before and after incubation with the cell extracts. We speculated that these minor peaks were likely to be isomers of their corresponding major peaks that were formed during separation and purification of MV/DV-Chl b.
Figure 5.
Enzymatic assay of OsDVR. DV-Chl a (A), -Chl b (B), -Chlide a (C), and -Chlide b (D) were incubated with OsDVR expressed in E. coli, and the pigments extracted from reaction mixtures were subjected to HPLC as described in “Materials and Methods,” respectively. The left column is the chromatogram that was detected at 440 nm by HPLC and the right column depicts the spectrum of each peak. A1 and B1 stand for MV-Chl a and b prepared from the wild-type MV-Chls, respectively. A2 and B2 represent DV-Chl a and b prepared from the 824ys mutant DV-Chls, respectively. C1, D1, C2, and D2 stand for MV-Chlide a, MV-Chlide b, DV-Chlide a, and DV-Chlide b prepared from MV-Chl a, MV-Chl b, DV-Chl a, and DV-Chl b by the enzymatic reaction of chlorophyllase isolated from G. chrysanthemum leaves, respectively. A3, B3, C3, and D3 represent products synthesized after incubation with E. coli lysates expressing the empty vector, which were used as negative controls, respectively. A4, B4, C4, and D4 stand for products synthesized after incubation with E. coli lysates expressing Osdvr, which were also used as negative controls, respectively. A5, B5, C5, and D5 represent products synthesized after incubation with E. coli lysates expressing OsDVR, respectively. Each pigment, DV-Chl a (A), -Chl b (B), -Chlide a (C), and -Chlide b (D), was used as a substrate for enzymatic reaction of OsDVR. C0 (blank control) represents MV-Chlide a resulted from incubation only with an acetone powder of G. chrysanthemum leaves, suggesting a small amount of MV-Chl a still remained in the acetone powder after being washed repeatedly, and the minor peak (peak 1) of C2, C3, and C4 was exactly the MV-Chlide a converted from residue MV-Chl a in the acetone powder.
As shown in Figure 5, A5 and C5, a significant difference in conversion efficiency of substrates between DV-Chlide a and DV-Chl a was observed, indicating that OsDVR has higher catalyzing activity in conversion of DV-Chlide a to MV-Chlide a than that in conversion of DV-Chl a to MV-Chl a.
DISCUSSION
Nagata et al. (2005) and Nakanishi et al. (2005) independently identified AT5G18660 gene of Arabidopsis as a DVR gene. Chew and Bryant (2007) demonstrated that BciA (CT1063), which is an ortholog of the Arabidopsis gene, encodes a DVR of the green sulfur bacterium C. tepidum TLS. Islam et al. (2008) and Ito et al. (2008) independently identified a novel 8-vinyl reductase gene (Slr1923) in DVR-less cyanobacterium Synechocystis sp. PCC6803. However, no DVR gene has yet been identified in monocotyledonous plants. In this report, we isolated the rice spontaneous mutant 824ys and cloned OsDVR gene that shows sequence similarity with Arabidopsis DVR gene. In the 824ys mutant, nine nucleotides were deleted in the ORF (Supplemental Fig. S2) and DV-Chls were exclusively accumulated (Fig. 4). A recombinant protein expressed in E. coli was shown to catalyze the conversion of DV-Chl(ide) a to MV-Chl(ide) a (Fig. 5, A and C). Therefore, we succeeded to identify a DVR gene in monocotyledonous plants.
In several mutants accumulating DV-Chls, the dvr mutant of Arabidopsis (Nagata et al., 2005) and the slr1923 mutant of Synechocystis sp. PCC6803 (Islam et al., 2008; Ito et al., 2008) were demonstrated to be sensitive to high-light conditions. These mutants were capable of photosynthesizing and growing only under low-light conditions but rapidly died under high-light conditions. Nagata et al. (2005) suggested that DV-Chls could not completely substitute for MV-Chls in the preexisting pigment system and that this substitution led to photodamage under high-light conditions. However, a maize mutant was capable of photosynthetic growth with DV-Chls (Bazzaz, 1981; Bazzaz and Brereton, 1982; Bazzaz et al., 1982). In this study, the 824ys mutant exhibits a yellow-green leaf phenotype, reduced Chl level, and arrested chloroplast development (Fig. 1; Supplemental Fig. S1; Table I). HPLC analysis of Chls showed that the mutant accumulates only DV-Chls (Fig. 4). These results indicate that the rice mutant is able to survive by photosynthetic growth with DV-Chls under natural high-light conditions although it grew slowly (Fig. 1).
The 824ys mutant was a spontaneous mutant, unlike the Arabidopsis dvr mutant isolated from ethyl methanesulfonate mutagenesis (Nagata et al., 2005). Surprisingly, the mutation site of the 824ys mutant was similar to that of the dvr mutant. The dvr point mutation created a single base pair change from C to T at nucleotide 998 in the ORF, which resulted in an amino acid change from Pro to Leu. In the corresponding site of the 824ys mutant, 9 bp were deleted at nucleotides 952 to 960 in the ORF, which resulted in the deletion of three amino acid residues (Lys, Val, and Pro; Supplemental Fig. S2). As a result, the dvr mutant accumulated DV-Chls instead of MV-Chls, but a small amount of MV-Chls (>4% of the total Chls) still remained in the mutant (Nagata et al., 2005), which suggested that the single amino acid exchange from Pro to Leu did not completely abrogate the protein function. However, the 824ys mutant accumulated only DV-Chls and had no MV-Chls (Fig. 4), which suggested that the three amino acid deletion completely abrogated the protein function. Similarly, a single nucleotide substitution at the 85th residue (C to T) in the Arabidopsis pcb2 mutant resulted in an incorporation of an immature stop codon in the ORF of DVR gene, and as a result, DV-Chls was exclusively accumulated (Nakanishi et al., 2005). Based upon these results, we speculate that the three amino acid residues (especially the Pro residue) may play an important role in OsDVR protein function. In addition, Nagata et al. (2005) described that the Arabidopsis DVR possesses one putative α-helix transmembrene domain predicted by the SOSUI program (http://bp.nuap.nagoya-u.ac.jp/sosui/), suggesting that the enzyme likely resided on the membranes. The three amino acid residues deleted in the 824ys mutant were located exactly in the α-helix transmembrene domain, but their detailed functions in enzymatic activity need to be further investigated.
It is unclear whether one or multiple enzymes are responsible for the reduction of the 8-vinyl group of various intermediates of Chl biosynthesis (Rebeiz et al., 2003). 8-Vinyl Chlide a reductase is a membrane-bound enzyme, embedded in etioplast and etiochloroplast membranes (Kolossov and Rebeiz, 2001). Tripathy and Rebeiz (1988) demonstrated that exogenous DV-Pchlide could be partially converted to MV-Pchlide in barley (Hordeum vulgare) plastids, but the conversion of DV-Pchlide to MV-Pchlide was not able to take place in cucumber (Cucumis sativus) plastids under similar incubation conditions. Parham and Rebeiz (1995) demonstrated membrane-bound 8-vinyl Chlide a reductases in etioplast membranes of etiolated cucumber cotyledons, barley, and maize leaves were very active in conversion of exogenous DV-Chlide a, being 50- to 300-fold higher rate than 8-vinyl Pchlide a reductase in conversion of exogenous DV-Pchlide a in barley etioplasts (Tripathy and Rebeiz, 1988). However, solubilization and partial purification of 8-vinyl reductase were not successfully performed from etiolated barley leaves until 2001 (Kolossov and Rebeiz, 2001). Nagata et al. (2007) carried out an enzymatic assay using a recombinant Arabidopsis DVR protein to examine the substrate specificity of this enzyme. Five DV-tetrapyrroles (DV-Pchlide a, -Chlide a, -Chlide b, -Chl a, and -Chl b) were used as substrates. The results indicated that the major substrate of DVR was DV-Chlide a, other substrates were not shown to be converted to corresponding MV-compounds. Nevertheless, Chew and Bryant (2007) demonstrated that recombinant BciA reduced the C-8 vinyl group of DV-Pchlide a in vitro. In this study, we confirmed that the recombinant OsDVR protein not only converted DV-Chlide a to MV-Chlide a (Fig. 5C), but also converted DV-Chl a to MV-Chl a (Fig. 5A) in the presence of NADPH, although it could not convert DV-Chlide b and -Chl b to corresponding MV-compounds (Fig. 5, B and D). These results implied that the DVR proteins derived from various organisms could have distinct DVR activities although their amino acid sequences are considerably similar.
Adra and Rebeiz (1998) reported transient rise and fall of DV-Chl a following a 2.5 ms light-flash treatment of etiolated cucumber cotyledons, and the appearance and disappearance of DV-Chl a was attributed to esterification of DV-Chlide a and conversion of DV-Chl a to MV-Chl a, respectively. They considered that the nascent DV-Chl a was converted to MV-Chl a by 8-vinyl Chlide a reductase, or by a novel 8-vinyl Chl a reductase. In this study, we confirmed that the DV-Chl a can be converted to MV-Chl a by the 8-vinyl chlorophyll(ide) a reductase (OsDVR) in vitro, which is an ortholog of the Arabidopsis DVR. In the current Chl biosynthetic pathway, two different DVR functional activities have been identified (Fig. 6A), i.e. DV-Chlide a is converted to MV-Chlide a by the Arabidopsis DVR (Nagata et al., 2007), and DV-Pchlide a is converted to MV-Pchlide a by the green sulfur bacterium DVR (BciA; Chew and Bryant, 2007). According to this study, we succeeded to demonstrate the third functional activity of DVR protein, i.e. OsDVR catalyzes the conversion reaction of DV-Chl a to MV-Chl a (Fig. 6B). On the other hand, Nagata et al. (2007) presented data showing that DVR protein preferentially reduces DV-Chlide a to MV-Chlide a. Consistently, OsDVR protein was active in conversion reaction of both DV-Chl a and DV-Chlide a, and showed pronounced enzyme activity in DV-Chlide a conversion in our experiment (Fig. 5, A5 and C5). In addition, it was reported that the ratio of DV-Pchlide a to MV-Pchlide a varies from species to species among higher plants (Shioi and Takamiya, 1992). This might be due to the differences in the substrate specificity of the DVR proteins among various organisms and/or to the differences in the accessibility of DVR to DV-Pchlide a (Nagata et al., 2007).
Figure 6.
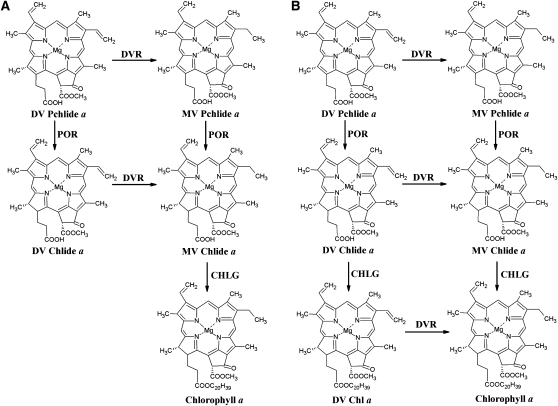
Activity of DVR. DVR activities described previously (A) and proposed in this study (B). In current Chl biosynthetic pathway, there are the two DVR activities that have been confirmed (A), i.e. DV-Chlide a is converted to MV-Chlide a by the Arabidopsis DVR (Nagata et al., 2007) and DV-Pchlide a is converted to MV-Pchlide a by the green sulfur bacterium DVR (BciA; Chew and Bryant, 2007). Here we succeeded to demonstrate the third DVR activity, i.e. DV-Chl a is converted to MV-Chl a by OsDVR (B).CHLG, Chl synthase; POR, Pchlide oxidoreductase.
Interestingly, CvrA (Slr1923) encodes a novel DVR or a subunit essential for MV-Chl synthesis in DVR-less cyanobacterium Synechocystis sp. PCC680 (Islam et al., 2008; Ito et al., 2008). Though the deduced amino acid sequence of Slr1923 has no homology to that of both DVR (AT5G18660) and OsDVR (Os03g22780), the homologs of Slr1923 are found in Arabidopsis (AT1G04620) and rice (Os04g25400) genome. If both enzymes (DVR and Slr1923 homologs) are functioning in the chloroplast, inactivation of dvr alone will not suffer from incapability of the conversion from DV-Chlide to MV-Chlide. However, the DVR-inactivated mutant accumulated only DV-Chls and no MV-Chls (Nakanishi et al., 2005). Several possible explanations for this discrepancy were proposed (Islam et al., 2008; Ito et al., 2008). Beale (2005) considered the mutation of DVR (AT5G18660), resulting in the complete abolition of MV-Chls (Nagata et al., 2005; Nakanishi et al., 2005), implied that Arabidopsis contains only one functional DVR. In this article, the OsDVR (Os03g22780)-inactivated 824ys mutant also accumulated only DV-Chls and had no MV-Chls. The deduced amino acid sequence of Os04g25400 gene product is 58% identical to that of the Slr1923 gene product, and putative function of hypothetical protein of Os04g25400 gene is coenzyme F420 hydrogenase/dehydrogenase β-subunit (http://rice.plantbiology.msu.edu). Here we assumed that the product of Os04g25400 gene might not be 8-vinyl reductase in rice, but it could not be excluded as a regulatory factor. In any case, analysis of the enzymatic activity is necessary by expressed protein.
MATERIALS AND METHODS
Plant Materials and Mapping Population
The 824ys mutant was a spontaneous yellow-green leaf mutant isolated from indica cv 824B of rice (Oryza sativa). The F2 mapping population was generated by crossing the 824ys mutant with normal green japonica cv 02428 of rice. Plant materials were planted under natural conditions in April to August, in Wenjiang District (latitude 30°42′N, longitude 103°50′E, and altitude 539.3 m), Chengdu City, Sichuan Province. Air temperature and sunlight (300–3,000 nm of wavelength) intensity were automatically measured using dry ball thermometer (X3420031; Vaisala) and pyranometer (TBQ-2-B), respectively. The related meteorological data were provided by Wenjiang weather station, which is less than 100 m near the experimental field, of Chengdu Meteorological Bureau.
Marker Development
The SSR markers were obtained from Gramene (http://www.gramene.org/microsat/) based on the SSR linkage map constructed by McCouch et al. (2002). To narrow down the region of the target locus, the BLAST search in the National Center for Biotechnology Information database was conducted to find InDel sequence divergence between the japonica rice cv Nipponbare and the indica rice cv 9311 around this locus (http://www.ncbi.nlm.nih.gov/BLAST/), and InDel markers were designed around the sequence divergence including 15 to 100 bp InDel using the Primer 5.0 software.
Sequence Analysis
The full-length DNA and protein sequences of OsDVR and its homologs were retrieved from GenBank (http://www.ncbi.nlm.nih.gov). The chloroplast signal peptide was predicted with http://www.cbs.dtu.dk/services/TargetP (Emanuelsson et al., 2000). Multiple sequence alignment and phylogenetic analysis were conducted using DNAMAN version 6.0 (Lynnon Biosoft).
Transmission Electron Microscopy Analysis
Rice leaf samples of 824ys and 824B were harvested from 4-week-old seedlings grown in natural conditions. Leaf sections were fixed in a solution of 3% glutaraldehyde and post fixed with 1% osmium tetroxide. The tissues were dehydrated in a gradient acetone series and embedded in Epon812 medium prior to thin sectioning. The samples were stained with uranyl acetate and Reynolds' lead citrate, and observed under a transmission electron microscope (H-600IV; Hitachi).
Analysis of Pigments
Chls were extracted from 0.2 g fresh leaves of wild-type 824B and the 824ys mutant with 95% ethanol, and the Chl content were determined with UV-1700 UV-visible spectrophotometer (Shimadzu) according to the method of Lichtenthaler (1987).
Chls used for HPLC analysis were extracted from fresh rice leaf tissue of 4-week-old 824B and 824ys plants with 100% acetone, the extract was centrifuged at 12,857g (5804R; Eppendorf) for 15 min, and the supernatants were subjected to HPLC on a C8 column (4.6 mm i.d. × 150 mm long; 3.5 μm; Agilent) according to the method of Zapata et al. (2000). Elution profiles were monitored by A440, and Chl a and b standards (Sigma) are used as control.
Preparation of Chl(ide) a and b
DV-Chl a and b were prepared from the mutant 824ys of rice. MV-Chl a and b were prepared from the wild-type 824B of rice. Chls were extracted from fresh rice leaf tissue with 100% acetone, the extract was centrifuged at 12,857g (5804R; Eppendorf) for 15 min, and the supernatants were dried under N2 gas and redissolved with 100% acetone. DV/MV-Chl a and b were separated from each other by HPLC on a C18 column (4.6 mm i.d. × 150 mm long; 5 μm; Agilent), and eluted with solvent (methanol:acetonitrile:acetone = 1:3:1) at a flow rate of 1.0 mL min−1 at 40°C (as described by Nakanishi et al. [2005]). Elution profiles were monitored by A660.
Chlorophyllase (an acetone powder of Garland chrysanthemum leaves) was prepared according to the method of Ito et al. (1994). A total of 200 μg of purified DV/MV Chl a and b were dissolved in 4 mL of a solution containing 25 mm Tris-HCl (pH 7.5) and 40% acetone, and incubated with 200 mg of an acetone powder of G. chrysanthemum leaves for 2 h at 28°C (Holden,1961; Ito et al., 1996), respectively. After incubation of the mixture in the dark, DV/MV Chlide a and b were purified as described by Ito et al. (1993).
Recombinant Enzymes Activity Assays
Full-length OsDVR and Osdvr genomic DNA were amplified by PCR from wild-type and 824ys genomic DNA, respectively, using primer F (5′-CAGGATCCATGGCTGCCCTCCTCCTCT-3′) and primer R (5′-GAAGAATTCCGAGGCCTAGAAGATGGT-3′). The primers incorporated a BamHI site at the N-terminal end and a EcoRI site at the C-terminal end of the ORF. PCR products were inserted into the pMD18-T vector (TaKaRa) and sequenced to obtain the correct clones, pMD-OsDVR and pMD-Osdvr. The pMD-OsDVR and pMD-Osdvr plasmids were then digested by BamHI and EcoRI enzymes, and cloned into the corresponding site of the bacterial expression vector pET30a(+) (Novagen) to generate pET30-OsDVR and pET30-Osdvr, sequenced to confirm OsDVR and Osdvr sequences, respectively, and finally introduced into Escherichia coli BL21 for protein expression. The cells transformed with an empty vector were used as a negative control.
Protein expression and recombinant enzyme activity assays were performed according to the methods as described by Nagata et al. (2005, 2007) with a slight modification. Cultures of E. coli strain (BL21) containing pET30-OsDVR, pET30-Osdvr, or pET30 plasmid alone were grown overnight in 2 mL of Luria-Bertani broth containing 50 μg mL−1 of kanamycin. Overnight cultures were subsequently used to inoculate 50 mL of Luria-Bertani broth containing 50 μg mL−1 of kanamycin. To maximize protein production, isopropylthio-β-galactoside was added to the culture at a final concentration of 0.5 mm after 30 min at 30°C. After 7 h of incubation, the culture was harvested and resuspended with 2.4 mL of solution containing 6.7 μg mL−1 of lysozyme and 3.3 μg mL−1 of DNaseI in 50 mm Tris-HCl, pH 8.0. The culture lysate was stored at −20°C until use. The culture lysate corresponding to 300 μg of proteins was used as the source of the enzyme for every experiment. DVR activity was determined in 500 μL of incubation buffer (40 mm citric acid, 80 mm K2HPO4, 0.5 mm NADPH, 20% acetone, pH 7.0) at 30°C. The incubation buffer was preincubated with the culture lysate for 5 min before the initiation of the reaction. The reaction was started by adding 20 μm of a substrate that was dissolved in 10 μL of acetone, and incubated for 10 min. The reaction was stopped by adding 500 μL of acetone. The pigments were transferred from acetone to diethylether and dried under N2 gas. These pigments were dissolved in a small volume of acetone and analyzed by HPLC on a C8 column (4.6 mm i.d. × 150 mm long; 3.5 μm; Agilent) according to the method of Zapata et al. (2000). Elution profiles were monitored by A440.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number GU733918.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Electron microscopic analysis of the rice 824ys mutant.
Supplemental Figure S2. Nucleotide sequence and deduced amino acid sequence of OsDVR.
Supplemental Figure S3. Alignment of the deduced amino acid sequence of OsDVR and its homologs.
Supplementary Material
Acknowledgments
We thank Mr. Kai Jiang from Sichuan Agricultural University for helping with HPLC manipulation. We also thank Prof. Yongsheng Liu from Sichuan University for help with revision on the manuscript.
References
- Adra AN, Rebeiz CA. (1998) Chloroplast biogenesis 81: transient formation of divinyl chlorophyll a following a 2.5 ms light flash treatment of etiolated cucumber cotyledons. Photochem Photobiol 68: 852–856 [Google Scholar]
- Bazzaz MB. (1981) New chlorophyll chromophores isolate from a chlorophyll deficient mutant of maize. Photobiochem Photobiophys 2: 199–207 [Google Scholar]
- Bazzaz MB, Bradley CV, Brereton RG. (1982) 4-Vinyl-4-desethyl chlorophyll a: characterization of a new naturally occurring chlorophyll using fast atom bombardment, field desorption and “in beam” electron impact mass spectroscopy. Tetrahedron Lett 23: 1211–1214 [Google Scholar]
- Bazzaz MB, Brereton RG. (1982) 4-Vinyl-4-desethyl chlorophyll a: a new naturally occurring chlorophyll. FEBS Lett 138: 104–108 [Google Scholar]
- Beale SI. (1999) Enzymes of chlorophyll biosynthesis. Photosynth Res 60: 47–73 [Google Scholar]
- Beale SI. (2005) Green genes gleaned. Trends Plant Sci 10: 309–312 [DOI] [PubMed] [Google Scholar]
- Chew AGM, Bryant DA. (2007) Characterization of a plant-like protochlorophyllide a divinyl reductase in green sulfur bacteria. J Biol Chem 282: 2967–2975 [DOI] [PubMed] [Google Scholar]
- Chisholm SW, Frankel SL, Goericke R, Olson RJ, Palenik B, Waterbury JB, West-Johnsrud L, Zettler ER. (1992) Prochlorococcus marinus nov. gen. nov. sp.: a marine prokaryote containing divinylchlorophyll a and b. Arch Microbiol 157: 297–300 [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, Heijne GV. (2000) Predicting subcellular localization of protein based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Fromme P, Melkozernov A, Jordan P, Krauss N. (2003) Structure and function of photosystem I: interaction with its soluble electron carriers and external antenna systems. FEBS Lett 555: 40–44 [DOI] [PubMed] [Google Scholar]
- Grossman AR, Bhaya D, Apt KE, Kehoe DM. (1995) Light-harvesting complexes in oxygenic photosynthesis: diversity, control, and evolution. Annu Rev Genet 29: 231–288 [DOI] [PubMed] [Google Scholar]
- Holden M. (1961) The breakdown of chlorophyll by chlorophyllase. Biochem J 78: 359–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XQ, Wang PR, Zhao HX, Deng XJ. (2008) Genetic analysis and molecular mapping of a novel chlorophyll-deficit mutant gene in rice. Rice Sci 15: 7–12 [Google Scholar]
- Islam MR, Aikawa S, Midorikawa T, Kashino Y, Satoh K, Koike H. (2008) slr1923 of Synechocystis sp. PCC6803 is essential for conversion of 3,8-divinyl(proto)chlorophyll(ide) to 3-monovinyl (proto) chlorophyll(ide). Plant Physiol 148: 1068–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Ohtsuka T, Tanaka A. (1996) Conversion of chlorophyll b to chlorophyll a via 7-hydroxymethyl chlorophyll. J Biol Chem 271: 1475–1479 [DOI] [PubMed] [Google Scholar]
- Ito H, Takaichi S, Tsuji H, Tanaka A. (1994) Properties of synthesis of chlorophyll a from chlorophyll b in cucumber etioplasts. J Biol Chem 269: 22034–22038 [PubMed] [Google Scholar]
- Ito H, Tanaka Y, Tsuji H, Tanaka A. (1993) Conversion of chlorophyll b to chlorophyll a by isolated cucumber etioplasts. Arch Biochem Biophys 306: 148–151 [DOI] [PubMed] [Google Scholar]
- Ito H, Yokono M, Tanaka R, Tanaka A. (2008) Identification of a novel vinyl reductase gene essential for the biosynthesis of monovinyl chlorophyll in Synechocystis sp. PCC6803. J Biol Chem 283: 9002–9011 [DOI] [PubMed] [Google Scholar]
- Jung KH, Hur J, Ryu CH, Choi Y, Chung YY, Miyao A, Hirochika H, An G. (2003) Characterization of a rice chlorophyll-deficient mutant using the T-DNA gene-trap system. Plant Cell Physiol 44: 463–472 [DOI] [PubMed] [Google Scholar]
- Kim JS, Rebeiz CA. (1996) Origin of the chlorophyll a biosynthetic heterogeneity in higher plants. J Biochem Mol Biol 29: 327–334 [Google Scholar]
- Kolossov VL, Bohnert HJ, Rebeiz CA. (2006) Chloroplast biogenesis 92: in situ screening for divinyl chlorophyll(ide) a reductase mutants by spectrofluorometry. Anal Biochem 348: 192–197 [DOI] [PubMed] [Google Scholar]
- Kolossov VL, Rebeiz CA. (2001) Chloroplast biogenesis 84: solubilization and partial purification of membrane-bound [4-vinyl] chlorophyllide a reductase from etiolated barley leaves. Anal Biochem 295: 214–219 [DOI] [PubMed] [Google Scholar]
- Lee S, Kim JH, Yoo ES, Lee CH, Hirochika H, An G. (2005) Differential regulation of chlorophyll a oxygenase genes in rice. Plant Mol Biol 57: 805–818 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK. (1987) Chlorophylls and carotenoids: pigments of photosynthetic membranes. Methods Enzymol 148: 350–382 [Google Scholar]
- McCouch SR, Teytelman L, Xu YB, Lobos KB, Clare K, Walton M, Fu BY, Maghirang R, Li ZK, Xing YZ, et al. (2002) Development and mapping of 2240 new SSR markers for rice (Oryza sativa L). DNA Res 9: 199–207 [DOI] [PubMed] [Google Scholar]
- Nagata N, Tanaka R, Satoh S, Tanaka A. (2005) Identification of a vinyl reductase gene for chlorophyll synthesis in Arabidopsis thaliana and implications for the evolution of Prochlorococcus species. Plant Cell 17: 233–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata N, Tanaka R, Tanaka A. (2007) The major route for chlorophyll synthesis includes [3,8-divinyl]-chlorophyllide a reduction in Arabidopsis thaliana. Plant Cell Physiol 48: 1803–1808 [DOI] [PubMed] [Google Scholar]
- Nakanishi H, Nozue H, Suzuki K, Kaneko Y, Taguchi G, Hayashida N. (2005) Characterization of the Arabidopsis thaliana mutant pcb2 which accumulates divinyl chlorophylls. Plant Cell Physiol 46: 467–473 [DOI] [PubMed] [Google Scholar]
- Parham R, Rebeiz CA. (1995) Chloroplast biogenesis 72: a [4-vinyl] chlorophyllide a reductase assay using divinyl chlorophyllide a as an exogenous substrate. Anal Biochem 231: 164–169 [DOI] [PubMed] [Google Scholar]
- Porra RJ. (1997) Recent progress in porphyrin and chlorophyll biosynthesis. Photochem Photobiol 63: 492–516 [Google Scholar]
- Rebeiz CA, Kolossov VL, Briskin D, Gawienowski M. (2003) Chloroplast biogenesis: chlorophyll biosynthetic heterogeneity, multiple biosynthetic routes, and biological spin-offs. Nalwa HS, , Handbook of Photochemistry and Photobiology, Vol 4 American Scientific, Los Angeles, pp 183–248 [Google Scholar]
- Shedbalkar VP, Rebeiz CA. (1992) Chloroplast biogenesis: determination of the molar extinction coefficients of divinyl chlorophyll a and b and their pheophytins. Anal Biochem 207: 261–266 [DOI] [PubMed] [Google Scholar]
- Shioi Y, Takamiya K. (1992) Monovinyl and divinyl protochlorophyllide pools in etiolated tissues of higher plants. Plant Physiol 100: 1291–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki JY, Bauer CE. (1995) Altered monovinyl and divinyl protochlorophyllide pools in bchJ mutants of Rhodobacter capsulatus. J Biol Chem 270: 3732–3740 [PubMed] [Google Scholar]
- Tripathy BC, Rebeiz CA. (1988) Chloroplast biogenesis 60: conversion of divinyl protochlorophyllide to monovinyl protochlorophyllide in green(ing) barley, a dark monovinyl/light divinyl plant species. Plant Physiol 87: 89–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu ZM, Zhang X, He B, Diao LP, Sheng SL, Wang JL, Guo XP, Su N, Wang LF, Jiang L, et al. (2007) A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis. Plant Physiol 145: 29–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapata M, Rodríguez F, Garrido JL. (2000) Separation of chlorophylls and carotenoids from marine phytoplankton: a new HPLC method using a reversed phase C8 column and pyridine containing mobile phases. Mar Ecol Prog Ser 195: 29–45 [Google Scholar]
- Zhang H, Li J, Yoo JH, Yoo SC, Cho SH, Koh HJ, Seo HS, Paek NC. (2006) Rice Chlorina-1 and Chlorina-9 encode ChlD and ChlI subunits of Mg-chelatase, a key enzyme for chlorophyll synthesis and chloroplast development. Plant Mol Biol 62: 325–337 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.