SUMMARY
Dramatic pigmentation changes have evolved within most vertebrate groups, including fish and humans. Here we use genetic crosses in sticklebacks to investigate the parallel origin of pigmentation changes in natural populations. High-resolution mapping and expression experiments show that light gills and light ventrums map to a divergent regulatory allele of the Kit ligand (Kitlg) gene. The divergent allele reduces expression in gill and skin tissue, and is shared by multiple derived freshwater populations with reduced pigmentation. In humans, Europeans and East Asians also share derived alleles at the KITLG locus. Strong signatures of selection map to regulatory regions surrounding the gene, and admixture mapping shows that the KITLG genomic region has a significant effect on human skin color. These experiments suggest that regulatory changes in Kitlg contribute to natural variation in vertebrate pigmentation, and that similar genetic mechanisms may underlie rapid evolutionary change in fish and humans.
INTRODUCTION
Many morphological, physiological, and behavioral traits have evolved repeatedly in different populations, species, and phyla. Parallel phenotypic change may arise through different genetic mechanisms even in closely related groups (Wilkens, 1971). However, recent studies in bacteria, plants, insects, birds, fish, and mammals raise the intriguing possibility that particular genes may be used repeatedly to evolve similar phenotypes even in distantly related organisms (Wood et al., 2005). Despite great interest in general mechanisms that may underlie phenotypic change in natural populations, we still have relatively few examples where similar traits have been traced to particular genes in multiple different groups, making it difficult to compare the genetic mechanisms that underlie evolutionary change in different lineages.
Threespine stickleback fish (Gasterosteus aculeatus) have emerged as a powerful new system for studying the genetic basis of parallel evolutionary change in vertebrates (reviewed in Kingsley and Peichel, 2007). Migratory ocean sticklebacks colonized countless new postglacial lakes and streams in coastal regions of North America, Europe, and Asia at the end of the last ice age. Newly established freshwater populations have had approximately 10,000 generations to adapt to new food sources, predators, parasites, salinity, and water temperatures. Today, stickleback populations show striking differences in morphological, physiological, and behavioral traits; many of which have evolved repeatedly in response to similar environmental conditions (reviewed in Bell and Foster, 1994). The reproductive isolation in the wild between most forms can be readily overcome using in vitro fertilization, making it possible to use genetics to map evolved phenotypic differences to particular chromosomes, and to compare the genetic mechanisms that underlie evolution of similar phenotypes in different populations.
Most previous genetic mapping studies in sticklebacks have focused on evolved differences in skeletal structures and body shape. However, pigmentation differences have also evolved repeatedly in different populations, and provide a particularly favorable phenotype for comparative studies across different species and phyla. Pigmentation is known to play an important role in crypsis and sexual display in many different animals (Cott, 1940). Many of the genetic pathways controlling production, migration, and differentiation of pigment-producing cells are well-characterized from studies of both pigmentation disorders in humans, and laboratory mutations in Drosophila, zebrafish, and mice (reviewed in Bennett and Lamoreux, 2003; Wittkopp et al., 2003; Parichy, 2006). Previous studies have identified some coding and regulatory mutations that underlie natural variation in pigmentation of insects, fish, birds, and mammals (Mundy, 2005; Lamason et al., 2005; Prud’homme et al., 2006; Protas et al., 2006; Steiner et al., 2007). While these studies show the promise of pigmentation traits for studying parallel evolution, most examples of pigment change are still incompletely understood in natural populations.
Both sticklebacks and humans have evolved major changes in pigmentation as they migrated out of ancestral environments into new environments around the world. Sticklebacks have evolved either black to mottled or white body colors (reviewed in Bell and Foster, 1994). This variation maximizes contrast of display colors in lakes with different water colors (Moodie, 1972), may provide crypsis or convergence of threat displays with predators (McPhail, 1969), and can contribute to mate choice preferences and speciation (Boughman et al., 2005). In humans, skin color varies with latitude. Significant lightening in northern populations is thought to have arisen relatively recently, and may reflect sexual selection and/or selection for increased vitamin D production in new environments with lower levels of ultraviolet radiation (reviewed in Jablonski, 2004).
While many potential adaptive functions of pigmentation changes have been hypothesized in fish and humans, the underlying genetic basis, and whether similar genetic mechanisms are used in both fish and humans to rapidly evolve changes in pigmentation patterns are largely unknown. Here we use genome-wide linkage mapping in sticklebacks to study the genetic basis of naturally evolved pigmentation changes. We then use admixture mapping in humans to ask whether genetic factors implicated in rapid evolution of pigmentation changes in sticklebacks also contribute to recent evolution of pigmentation differences in humans.
RESULTS
A QTL with a major effect on gill pigmentation
While examining branchial skeletons of marine and derived freshwater threespine sticklebacks, we noted that marine sticklebacks have heavily melanized gills, while derived freshwater Paxton Lake benthic sticklebacks have gills with sparsely distributed melanocytes. F2 progeny from a cross between marine and Paxton Lake benthic fish also showed a range of pigmentation phenotypes (Figure 1). Some F2 fish had few gill melanocytes, giving the gills an overall whitish appearance similar to that seen in Paxton benthic fish (Figure 1A, and see below). Other F2s had many gill melanocytes, giving the gills a dark brownish appearance similar to that seen in marine fish. Other progeny had intermediate phenotypes.
Figure 1. A QTL with a major effect on gill pigmentation.
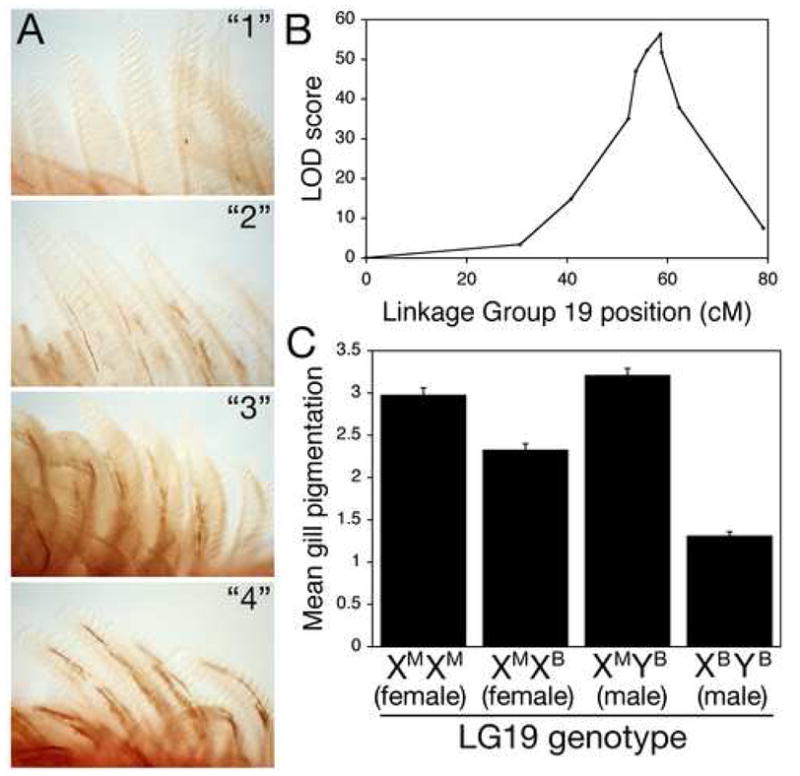
(A) Segregating variation in melanocyte distribution within the gills of Japanese marine × Paxton benthic F2 fish. F2 fish show a range of gill pigmentation, from gills with few melanocytes and overall white appearance (class “1”), to intermediate (classes “2” and “3”), to gills with many melanocytes and overall dark brownish appearance (class “4”).
(B) Mapping results on linkage group 19. Genetic distance in centiMorgans (cM) is on the x-axis, and LOD score on the y-axis. Significance threshold is 4.5 (van Ooijen, 1999). Markers from left to right are Stn303, Stn185, Stn186, Stn187, Stn193, Stn194, Stn191, Stn398, Stn399, Stn400.
(C) Gill pigmentation phenotypes of F2 fish according to genotype at Stn191, the peak QTL marker (mean scores +/− SEM). Since the grandparents were a Japanese marine female and Paxton benthic male, F2 fish can have any of four LG19 genotypes. Females can either be homozygous for marine chromosomes (XMXM) or heterozygous for marine and Paxton benthic chromosomes (XMXB). Males all have the benthic Y chromosome (YB), and can have a marine or freshwater X chromosome. Differences between genotypic classes are highly significant for both males (p = 9 × 10−40) and females (p = 2 × 10−8).
To examine the genetic control of gill melanization, we compared the segregation of pigmentation phenotypes in 360 F2 progeny with genotypes of a genome-wide set of genetic markers previously developed for linkage mapping in sticklebacks (Peichel et al., 2001). A highly significant quantitative trait locus (QTL) on linkage group (LG) 19 controlled over half of the variation in pigmentation scores (Figure 1B; genotype at peak marker Stn191: 56.1 percent variance explained, log likelihood ratio of linkage (LOD) of 56.4).
This QTL controlling gill pigmentation maps near the sex-determining region (Peichel et al., 2004). However, pigmentation scores show significant linkage to the same region within each sex, ruling out sexual dimorphism as the basis of the trait (Figure 1C). Instead, the pigmentation difference is conferred by genetic variation between marine and freshwater X chromosomes. In males, X chromosome genotype at the peak marker explains an even larger percentage of the variance in pigmentation scores (73.9%).
Fine mapping the pigmentation QTL
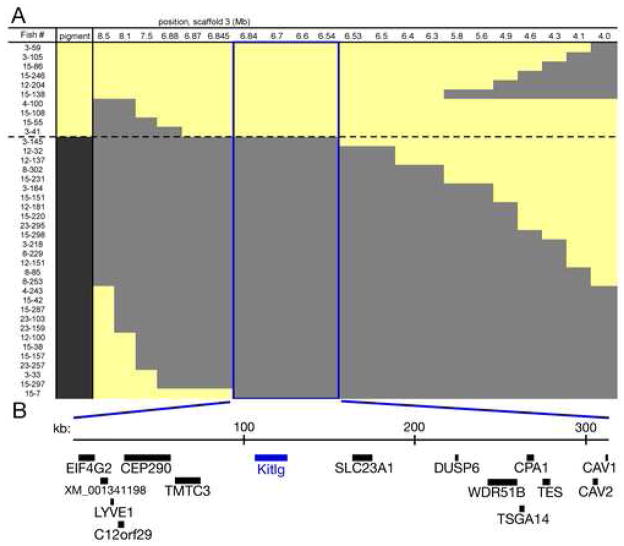
Stn194 and Stn398, the genetic markers flanking the peak marker Stn191 on LG19, have LOD scores over four units lower than the score at Stn191, indicating that the pigmentation QTL maps within the 2.9 cM region between these two markers (Figure 1B). To fine map the pigmentation QTL, we genotyped and phenotyped more F2 progeny, focusing on male fish with extreme pigmentation scores, and testing for concordance with genotype at additional LG19 markers (Figures 2A and S1). Genotyping 1182 F2 fish identified a total of 38 informative male progeny with recombination events within the Stn194-Stn398 interval. Inspection of a draft stickleback genome assembly showed this interval to be 4.5 megabases (Broad Institute, 2006). We designed 17 new microsatellite markers within this region, and compared the genotypes at each new marker with gill phenotypes. In the interval between Stn405 and Stn409 (blue box in Figure 2A), the presence of marine or freshwater alleles was completely concordant with the presence of dark or light gills.
Figure 2. Fine mapping the LG19 pigmentation QTL.
(A) X chromosome genotypes and gill pigmentation phenotypes of recombinant F2 males. Each row is an informative recombinant F2 male. Light (yellow) or dark (gray) gill score is shown in the first column, followed by genotype at markers along the X chromosome. Positions at top correspond to location in megabases of scaffold 3 from the stickleback genome assembly (Broad Institute, 2006). The first and last marker columns are Stn194 and Stn398, the markers flanking the QTL peak in Fig 1B. Microsatellites are listed in Table S1. Genotypes are coded yellow for Paxton benthic, gray for marine. The 315 kb minimal region concordant with gill pigmentation phenotype is boxed in blue.
(B) Schematic of the 315 kb QTL interval, which contains 15 genes including Kit ligand (Kitlg). Genes are listed in Table S2.
This minimal region defined by high resolution mapping spans 315 kb in the stickleback genome assembly, and contains 15 genes (Figure 2B, and Table S2). Fourteen of these genes have no known role in pigmentation. The remaining gene (Kit ligand or Kitlg) is an outstanding candidate for a gene controlling pigmentation phenotypes, based upon its major role in melanocyte patterning in both laboratory mice and zebrafish (reviewed in Wehrle-Haller, 2003; Hultman et al., 2007).
The LG19 pigmentation QTL also controls melanocyte distribution in ventral skin
Kitlg is expressed in the skin of mice, and controls the proliferation, migration, differentiation, and survival of Kit receptor-expressing melanocytes (reviewed in Wehrle-Haller, 2003). Homozygous loss of Kitlg produces white mice. In contrast, heterozygous reduction of Kitlg typically causes selective loss of pigment in ventral skin, which is located furthest from the embryonic origin of melanocytes in the dorsal neural tube.
To test whether the stickleback pigmentation locus also affects melanocyte distribution outside the gills, we scored melanocyte patterns in different skin regions of F2 fish from the same marine by Paxton benthic cross. Dramatic external pigmentation differences segregate in the cross, particularly in posterior ventral regions where F2 fish can be heavily or sparsely melanized (Figure 3B,C). Melanization scores in the ventral region depend on genotype at the LG19 pigment QTL (Figure 3D). Similar to gill pigmentation phenotypes, we find a more pronounced difference in F2 males than F2 females, though effects are significant in both sexes. In contrast, we find no significant pigmentation differences controlled by this chromosome region on the dorsal head region. Melanocyte numbers in a defined area of ventral skin are significantly lower in animals inheriting Paxton benthic X alleles, suggesting that lightening of ventral skin arises in part from changes in melanocyte number (Figure S2; XMYB vs. XBYB males, p = 8 × 10−6; XMXM vs. XMXB females, p = 0.03). Importantly, the ventral melanocyte phenotype cosegregates with the gill pigmentation phenotype in the recombinant males used for fine mapping in Figure 2, indicating that the same 315 kb region controls both gill and ventral skin pigmentation.
Figure 3. LG19 pigmentation QTL controls melanization of ventral skin.
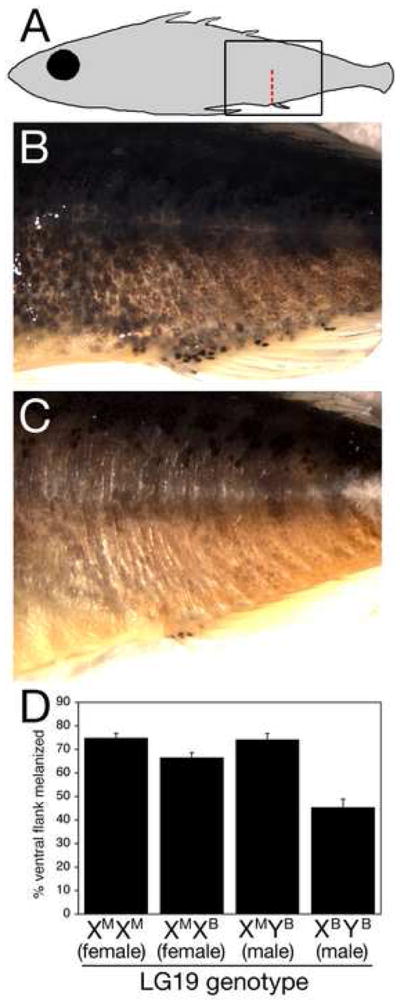
(A) Schematic of region shown in B and C (box). The red dashed line between the posterior lateral line and the cloaca shows where melanization was scored in (D).
(B) F2 male with marine X chromosome and heavily melanized ventrum. A nearly continuous field of melanocytes covers the ventrum.
(C) F2 male with Paxton benthic X chromosome and sparsely melanized ventrum.
(D) Mean percentage of ventral flank melanization for F2s of each possible LG19 QTL genotype. Each genotypic class contains from 24 to 31 F2 fish non-recombinant for the Stn194-Stn398 interval. Shown are the means +/− the SEM. Differences between classes are highly significant (p = 1 × 10−7 for one-way ANOVA) and LG19 genotype controls flank melanization in both males and females (XMYB vs. XBYB males, p = 4 × 10−9; XMXM vs. XMXB females, p = 0.017).
Parallel evolution of gill and skin phenotypes
The gill and skin pigmentation differences could be due to coding and/or regulatory changes between marine and Paxton benthic fish. We sequenced Kitlg from both grandparental populations used for the cross, and found two nonsynonomous changes. Neither change is predicted to have a strong functional effect using the SIFT algorithm (Ng and Henikoff, 2003), and both occur at nonconserved positions that differ between sticklebacks and zebrafish (Figure S3). To test for possible changes in promoter usage or splicing patterns that could lead to more severe alterations in protein structure, we used 5′-RACE and RT-PCR to compare Kitlg transcript size and structure in marine and freshwater fish. A single transcription start and a single splicing pattern across the entire protein coding region of the Kitlg gene was seen in both populations, suggesting that major differences in transcript splicing or protein structure are unlikely to contribute to reduced pigmentation in gill tissue.
The Paxton benthic Kitlg allele shows numerous noncoding differences from marine Kitlg sequences (Figure 4C and S3). Comparative sequencing showed that the Little Campbell River marine population from Canada has sequences that closely resemble those in Japanese marine fish. In contrast, sticklebacks from two geographically distant freshwater populations have Kitlg sequences that closely resemble those seen in Paxton benthic fish (Fishtrap Creek in Washington State, and G. williamsoni from Southern California). Bottom-dwelling (benthic) sticklebacks exist as a species pair with morphologically distinct open water (limnetic) sticklebacks in Paxton lake (McPhail, 1992). Interestingly, although they exist in the same lake, the Paxton limnetic fish have marine-like, rather than Paxton benthic-like Kitlg sequences (Figure 4B,C). Phylogenetic analysis shows strong bootstrap support for two separate clades at the Kitlg locus, one made up of marine and Paxton limnetic fish, and a separate group made up of a variant Kitlg found in Paxton benthic, Fishtrap Creek and williamsoni fish (Figure 4B).
Figure 4. The freshwater Kit ligand variant is found in other populations with sparsely melanized gills and skin.
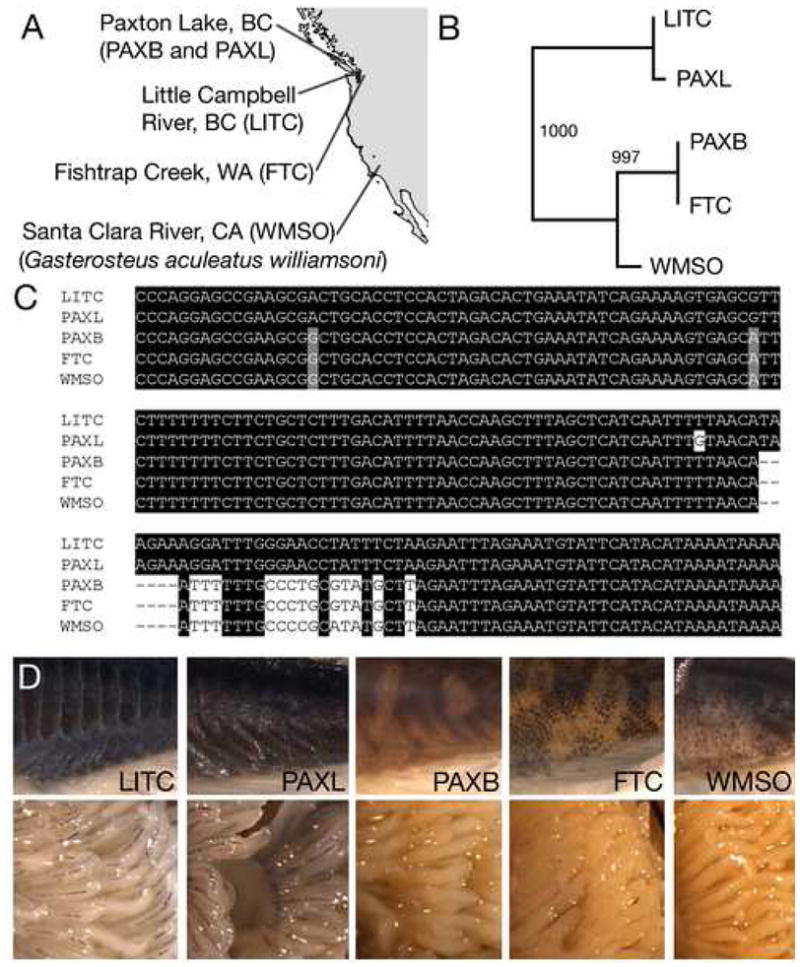
(A) Map of west coast of North America showing locations of marine fish (Little Campbell River, BC; LITC) and derived freshwater populations (Paxton benthic, PAXB; Paxton limnetic, PAXL; Fishtrap Creek, FTC; and G. williamsoni, WMSO).
(B) Neighbor-joining phylogenetic tree showing that FTC and WMSO have PAXB-like Kitlg alleles. In contrast, PAXL, which occurs as a species pair with PAXB, has marine-like alleles.
(C) Nucleotide alignment of part of exon 8 and intron 8, showing that PAXB, FTC, and WMSO share a closely related Kitlg haplotype.
(D) Ventrums (top) and gills (bottom) from populations shown in A-C. Ventrums are views like in Figure 3B,C. LITC and PAXL have heavily melanized ventrums and gills, while PAXB, FTC, and WMSO all have sparsely melanized ventrums and gills.
The presence of Paxton benthic-like Kitlg alleles in other freshwater fish suggests that similar gill and pigment changes may have evolved in other stickleback populations. We verified this prediction by examining gill and skin pigmentation in the same fish populations used for sequencing (Figure 4D). Fishtrap Creek and willamsoni fish have extremely white gills and light ventrums, like those seen in Paxton benthic fish. In contrast, fish from both marine populations and Paxton limnetic fish have more heavily melanized gills and dark ventrums.
Recent studies of armor plate patterning in multiple freshwater stickleback populations have shown that parallel evolution of low armor phenotypes occurs by repeated selection on a variant Ectodysplasin (Eda) allele that is found at low frequency in migratory marine fish (Colosimo et al., 2005). To determine if the variant freshwater Kitlg allele is also present at low frequency in marine sticklebacks, we genotyped fully-plated fish from the ocean outlet of the Navarro River in California. Paxton benthic-like alleles were found at a frequency of 12% in 107 fully-plated fish from the Navarro site. The variant Eda allele is found in 3.8% of fish from the same location (Colosimo et al., 2005). None of the carriers of the variant Kitlg allele were carriers of the Eda allele, suggesting that rare carriers are not simply F1 hybrids between marine and freshwater fish.
cis-regulatory changes in Kit ligand gene expression
The absence of clear functional changes in the predicted Kitlg amino acid sequence of Paxton benthic fish raises the question of whether the numerous noncoding changes in the variant freshwater Kitlg allele produce regulatory changes in gene expression. In situ hybridization showed lower overall Kitlg expression in gill tissue of Fishtrap Creek fish compared to marine fish (Figure 5). As expected from the known role of Kitlg as a secreted signal that has a cell non-autonomous effect on melanocyte development (reviewed in Wehrle-Haller, 2003), expression in both populations was observed in gill tissue surrounding melanocytes, rather than melanocytes themselves.
Figure 5. Reduced Kit ligand expression in gills of Fishtrap Creek fish.
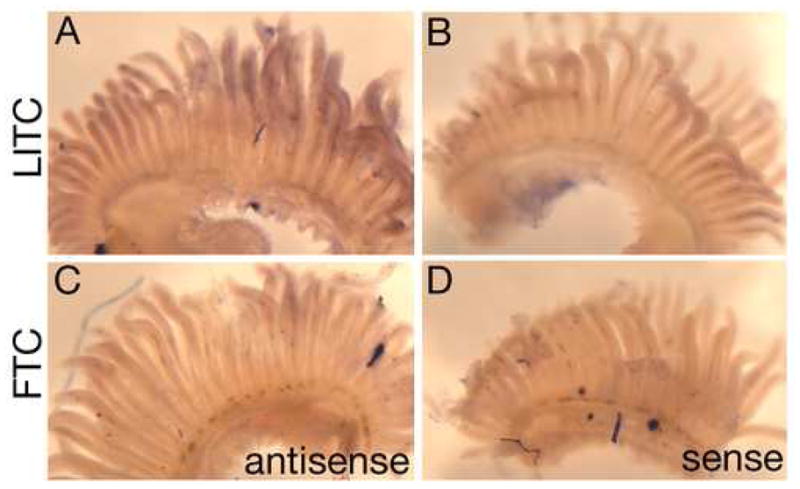
(A-D) In situ hybridization of gill arches from marine fish from Little Campbell River (LITC; A,B) and derived freshwater population Fishtrap Creek (FTC; C,D). (A,C) Antisense probe showing overall reduced Kitlg expression levels in FTC relative to LITC. (B,D) Sense probe controls showing staining observed is specific.
The reduced expression could be due either to cis-acting regulatory changes at the Kitlg locus, or trans-acting changes in other linked or unlinked genes that affect Kitlg expression. To distinguish these possibilities, we generated F1 hybrids heterozygous for marine and derived Kitlg alleles, and tested whether allele-specific expression differences were still seen in the otherwise identical trans-acting environment of F1 hybrid fish (Figure 6).
Figure 6. cis-regulatory changes in Kit ligand expression.
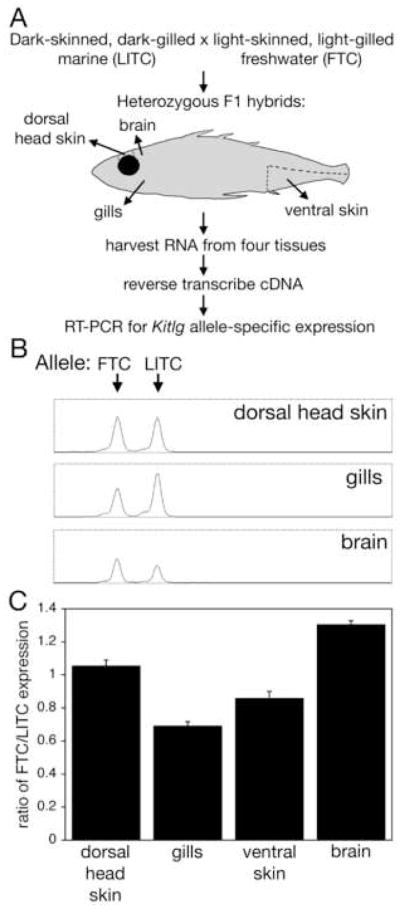
(A) Schematic of allele-specific expression experiments.
(B) Chromatogram showing representative results for size separation of Kitlg RT-PCR products from three different F1 hybrid tissues. Size is on the x-axis and fluorescence intensity on the y-axis. In dorsal head skin, near equal levels of each allele is amplified. In gills, the LITC allele is overexpressed relative to the FTC allele. In brains, the FTC allele is more abundant.
(C) Ratio of freshwater to marine Kitlg expression in dorsal head skin, gills, ventral skin, and brain. Shown are means +/− SEM. For each tissue, at least eight F1 hybrids were analyzed. Expression in different tissues is significantly different (p = 1 × 10−14 by one-way ANOVA); as are all pairwise comparisons (p < 0.03 by Tuky’s HSD post hoc pairwise comparisons).
Expression of the two different alleles was examined in four different tissues of F1 hybrids, using RT-PCR and size differences in the 5′ UTR to quantify the level of expression of each allele (Figure 6B). In some tissues, such as dorsal head skin, we observed near equivalent levels of expression of marine and freshwater Kitlg alleles. However, in both ventral skin and gill tissue, we found the freshwater Kitlg allele to be expressed at significantly lower levels than the marine allele. Conversely, in brain tissue we find the freshwater Kitlg variant to be upregulated, showing that reduced levels of gene expression in gills and ventral skin are not due to inherent bias in ability to amplify the two alleles (Figure 6B,C). The quantitative changes in gene expression were confirmed in at least eight animals (Figure 6C), and essentially identical results were seen with another method using single nucleotide polymorphisms (SNPs) in a non-overlapping region of Kitlg (Figure S4). These results demonstrate that the marine and freshwater Kitlg alleles are expressed at different levels in multiple tissues, and that the differences in gene expression are due to cis-acting changes at the Kitlg locus, rather than differences in the trans-acting environment of marine and freshwater fish, or differences in the number of particular cell types in marine and freshwater tissues.
Human KITLG region controls skin pigmentation
The stickleback results suggest that regulatory changes at the Kitlg locus have been repeatedly selected in natural environments. A variety of recent molecular tests also reveal strong signatures of positive selection in a large genomic region surrounding the KITLG gene in humans (The Chimpanzee Sequencing and Analysis Consortium, 2005; Lao et al., 2007; Williamson et al., 2007). Reexamination of the region using Haplotter (Voight et al., 2006) reveals scores in the top one-thousandth of the genome based on Fay and Wu’s H score (Figure 7A), which detects an excess of high frequency derived alleles, a signal of positive selection (Fay and Wu, 2000). The major peaks are found in people of European ancestry and East Asians, and map to the large intergenic regions flanking KITLG. Many adjacent SNPs in the KITLG intergenic regions show extreme frequency differences among human populations, with a predominance of ancestral alleles in West Africans, and derived alleles in Europeans and East Asians (The International HapMap Consortium, 2005).
Figure 7. Effect of KITLG genotype on skin pigmentation in African-Americans.
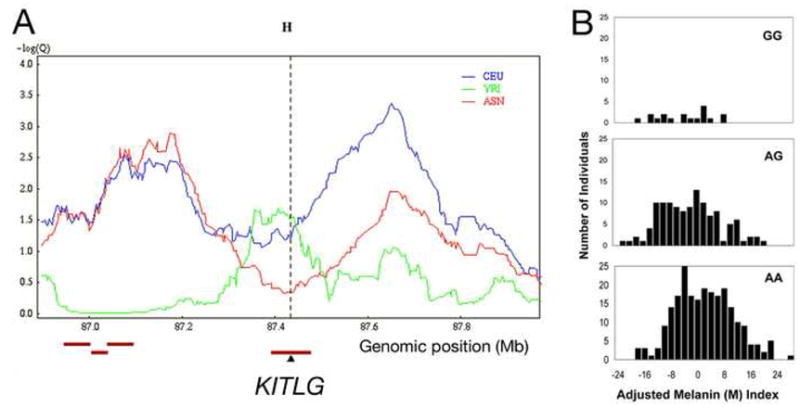
(A) Fay and Wu’s H from Haplotter (Voight et al., 2006) for a 1 Mb window centered on human KITLG. Strong signals of selection are detected 5′ (right) and 3′ (left) of KITLG in a population sample of European descent (CEU: CEPH, Utah residents with ancestry from northern and western Europe) and in a population sample from East Asia (ASN: Han Chinese in Beijing and Japanese in Tokyo). YRI: West Africans (Yoruba in Ibadan, Nigeria) (The International HapMap Consortium, 2005). A -log(Q) value of 3.0 indicates a value in the top one-thousandth of all scores in the human genome.
(B) Histograms for melanin index (M) after adjustment for individual genetic ancestry in individuals with two European (GG), one European and one Afrrican (AG), or two African (AA) alleles at rs642742 upstream of the KITLG gene. Mean (SD) adjusted melanin indexes: for GG, −4.7 (7.7), n=18; for AG, −2.9 (8.8), n=121; for AA, 1.3 (9.1), n=232.
What phenotypes may be associated with selection of KITLG variants in different human populations? Although the pleiotropic functions of KITLG suggest many possibilities, one of the most obvious differences between Africans, Europeans, and East Asians is skin color. To evaluate whether the KITLG gene has measurable effects on human skin pigmentation, we used admixture mapping in African Americans, a population with a combination of ancestry from both West Africa and Europe (Figure 7B). We genotyped an ancestry-informative SNP (rs642742A>G) located 326 kb upstream of the KITLG transcription start site in a sample of 374 African Americans. Gene frequency differences at rs642742 are dramatic, including frequencies of 92% or more for the ancestral allele in West Africans switching to 86% or more for the derived allele in both Europeans and East Asians (The International HapMap Consortium, 2005). The derived G allele alters a noncoding sequence that is highly conserved in mammals, and therefore represents a candidate mutation that could affect KITLG expression. The same African American individuals genotyped at rs642742 have previously been measured for skin pigmentation using narrow-band reflectometry, and tested for individual genetic ancestry using 29 other ancestry informative markers distributed at other locations around the genome (Shriver et al., 2003; Bonilla et al., 2005).
ANCOVA analyses show that, after controlling for sex and ancestry, there is a significant association between rs642742 genotypes and skin pigmentation as measured by the melanin (M) index (P<0.001). Individuals with two African alleles (AA), one African and one European allele (AG) or two European alleles (GG) show mean M index scores of 55.2, 50.9, and 48.8 respectively. The regression coefficient, β, representing the average amount the M index decreases when one A allele is added to the model, is 3.8 melanin index units. Although these data suggest a model for the mode of inheritance that is between an additive model and a dominant model, a much larger sample size is needed to have sufficient statistical power to contrast these models.
We also performed Bayesian admixture mapping using the ADMIXMAP 3.7 software to test for linkage between the KITLG gene region and skin pigmentation. This program provides a score test of gene effect that controls for individual admixture and evaluates dependence of pigmentation on the proportion of alleles of West African ancestry at rs642742. The results with ADMIXMAP are also significant (p=0.0048). The effect of rs642742 on the M index is estimated at 2.8 melanin units, slightly lower than using ANCOVA.
The combined results suggest that replacement of two West African alleles with two European alleles at the KITLG locus lightens a person’s color by an average of 6 to 7 melanin units. This compares with an overall skin reflectance difference of approximately 30 melanin units between West Africans and Europeans (Shriver et al., 2003).
DISCUSSION
The current results represent the first example of a pigmentation difference between stickleback populations that has been traced to changes in a corresponding gene. As previously seen for some skeletal traits (Shapiro et al., 2004; Colosimo et al., 2004; Cresko et al., 2004; Kimmel et al., 2005), pigmentation differences map to a major locus that controls much of the variation between marine and freshwater fish. The large phenotypic effect of the LG19 QTL made it possible to fine map the pigmentation difference to a small region containing the gene for a major developmental signaling molecule variously known as Kit ligand, Steel factor, Mast cell Growth Factor, or Stem Cell Factor.
Previous studies have shown that skin expression of Kitlg stimulates melanocyte migration, proliferation, differentiation, and survival, and is required for ongoing maintenance and survival of normal melanocyte numbers in adults (reviewed in Wehrle-Haller, 2003). The mapping and expression results in sticklebacks suggest that melanocyte numbers in adult ventral skin are also controlled by cis-acting changes in Kitlg expression, thus linking quantitative variation in Kitlg expression to significant phenotypic variation in natural populations.
Evolution through pleiotropic master regulators
Three of the major effect QTLs now identified in sticklebacks map to major developmental control genes that are essential for formation of several different cell types or tissues (Shapiro et al., 2004; Colosimo et al., 2005; this study). Steel mutations in mice were first recognized because of pleiotropic effects on melanocytes, hematopoietic stem cells, and germ cells. Subsequent studies found Kitlg to also be required for normal development of mast cells and pacemaker cells of the enteric nervous system (reviewed in Wehrle-Haller, 2003). Furthermore, Kitlg plays a physiological role in the adult mammalian brain. Kitlg mutant mice have defects in spatial learning, a phenotype proposed to be due to the Kitlg expression domain in the adult hippocampus (Motro et al., 1991; 1996). These spatial learning defects in Kitlg mutant mice are intriguing in light of our results, which show that in the brain, the divergent Paxton-benthic-like Kitlg variant is expressed at higher levels than a marine allele. Previous studies have shown that benthic sticklebacks have increased spatial learning abilities relative to fish living in open water environments (Girvan and Braithwaite, 1998). Interestingly, in a second lake with a benthic-limnetic species pair, Priest Lake, the bottom-dwelling species has largely fixed the same divergent Kitlg haplotype found in Paxton benthic fish, while the open water (limnetic) species in this lake shows the same haplotype seen in both marine and Paxton limnetic fish (Miller, Schluter, and Kingsley, unpublished data). Parallel fixation of similar alleles in similar ecological settings suggests the divergent Paxton benthic haplotype may be advantageous in bottom-dwelling environments, perhaps because of changes in the cryptic value of countershading with water depth (Johnsen, 2002), or because of changes in spatial learning or other neurological phenotypes in fish that forage in more complex, structured environments. These and other possibilities can now be tested by additional phenotypic comparisons, and by monitoring long-term changes in the frequency of the divergent Kitlg allele when appropriate stickleback populations are transplanted to contrasting environments.
The diverse roles of Kitlg may make it seem like an unlikely candidate for evolving phenotypic change in natural populations. Previously known Kitlg mutations usually produce significant defects in fertility, hematopoeisis and viability (Charlier et al., 1996; Rajaraman et al., 2002). However, genetic analysis in mice shows that some of the different functions of Kitlg can be separated by large-scale chromosome rearrangements that map over a hundred kilobases 5′ of Kitlg coding exons (Bedell et al., 1995, 1996). The large size of Kitlg regulatory regions has made it difficult to identify specific sequences responsible for expression in different tissues. Nonetheless, the complex regulatory architecture clearly makes it possible to recover mutations that disrupt only a subset of normal Kitlg functions, including mutations with much less severe viability and sterility problems than typically seen with coding region mutations.
Additional studies are required to determine if the two Kitlg amino acid differences identified in this study contribute to functional differences in marine and freshwater sticklebacks. However, it is already clear that there are significant regulatory differences between the marine and freshwater Kitlg genes, and that these changes are due to cis-acting changes at the Kitlg locus (Figure 6). We do not yet know what specific sequence changes are responsible for the cis-acting regulatory alterations in gills, ventral skin and brain. Although regulatory mutations are much more difficult to identify than coding region mutations, recent transgenic studies have begun to characterize specific enhancer elements responsible for phenotypic change in Drosophila (Marcellini and Simpson, 2006; Prud’homme et al., 2006; McGregor et al., 2007). Similar enhancer surveys of the divergent Kitlg alleles in sticklebacks will help determine whether cis-regulatory differences we observe in several different tissues are due to single or multiple mutations in stickleback Kitlg enhancers.
Parallel evolution from preexisting mutations
The large number of different stickleback populations provides an excellent opportunity to test whether similar phenotypic changes evolve using similar or different genetic mechanisms. Previous studies have shown that parallel loss of armor plates in most freshwater sticklebacks occurs by repeated use of alternative alleles at the Ectodysplasin (Eda) locus (Colosimo et al., 2005). The pigmentation changes studied here are not as prevalent in freshwater fish as armor plate reduction. However, at least three freshwater populations have evolved light pigmentation of gills and ventral skin, and all three populations have closely related Kitlg alleles. These populations are not close geographically. In addition, previous phylogenetic analysis shows that Paxton benthic and williamsoni fish fall in separate clades when analyzed with random nuclear markers, showing that these fish do not share a recent demographic origin from a single freshwater source population (Colosimo et al., 2005). For both armor and pigmentation phenotypes, the alleles for alternative freshwater morphology can be found at low frequencies in fully-plated marine sticklebacks, suggesting that repeated evolution of light gill and ventrum pigmentation phenotypes in multiple freshwater populations may occur by selection from standing variation in migratory marine populations.
It remains to be seen whether the majority of common freshwater stickleback traits evolve from variant alleles that are present at low frequency in marine populations. However, the regular spawning of marine fish in fresh water brings migratory populations into annual contact with stream-resident fish that have already evolved a series of freshwater adaptations. This life history may enhance the probability that alleles for alternative phenotypes are present at low frequency in marine fish.
Pigmentation evolution in fish and humans
Like sticklebacks, modern humans have recently migrated out of an ancestral environment and colonized many new locations around the world. Some of the strongest signatures of positive selection in the human genome map to the region of the KITLG gene (The Chimpanzee Sequencing and Analysis Consortium, 2005; Lao et al., 2007; Williamson et al., 2007; Figure 7A). Based on the location of these signatures in the large 5′ flanking regions of the gene, Williamson et al. have suggested that regulatory differences in KITLG have undergone selective sweeps in both European-American and Asian populations. The current results show that genotype in the KITLG upstream region is associated with significant differences in human skin color, one of the most obvious superficial differences between human populations (reviewed in Jablonski, 2004).
Previous histological studies suggest that human skin color differences arise from the number, size, color, and aggregation of pigment granules, rather than from differences in the number of pigment-producing cells (reviewed in Barsh, 2003). Many of the genes previously associated with human skin color are genes that influence pigment production within melanocytes, including MC1R, TYRP1, OCA2, MATP and SLC24A5 (reviewed in McEvoy et al., 2006). These previous results do not conflict with an additional important role of KITLG as one of several significant factors influencing human skin color. For example, recent work on the golden gene in fish and humans pointed out that changes in SLC24A5 were only found at high frequency in European populations, and other variants must contribute to skin color changes in East Asia, Europe, or both populations (Lamason et al., 2005). Interestingly, strong signatures of selection in the KITLG region are shared in both Europeans and East Asians (Figure 7A). In addition, injection of KITLG into human skin increases the size and dendricity of melanocytes (Grichnik et al., 1998), and previous skin culture experiments show that some of the differences in skin color between human groups are controlled by keratinocytes (Minwalla et al., 2001), the same major epithelial cells of the skin that express KITLG (reviewed in Wehrle-Haller, 2003). Although no amino acid differences are known in the KITLG protein of different human groups, KITLG is expressed at significantly higher levels in skin keratinocytes from Africans than Europeans (Yoshida et al., 2007). These and the current results strongly support the idea that regulatory changes in Kitlg expression contribute to pigmentation differences in both sticklebacks and humans.
Recent studies have revealed several examples of the same genes controlling morphological evolution in distantly related species. Dorsal trichomes have been lost repeatedly in Drosophila through regulatory changes in ovo (Sucena et al., 2003); wing spot patterns have evolved repeatedly through regulatory changes in yellow (Prud’homme et al., 2006); pigment loss has evolved in cavefish through coding region changes in OCA2 (Protas et al., 2006); both increases and decreases of pigmentation have evolved repeatedly in birds and mammals through coding region changes in MC1R (reviewed in Mundy, 2005; Hoekstra, 2006); and pelvic reduction in threespine sticklebacks, ninespine sticklebacks, and manatees may all occur through changes in Pitx1 (Shapiro et al., 2006).
Given the enormous evolutionary distance between sticklebacks and humans, why might similar genes be involved in controlling very recent evolutionary changes in both groups? Rapid phenotypic evolution in both humans and sticklebacks may occur most easily through genes that have large phenotypic effects, and through mutations that have some detectable phenotypic effects in heterozygotes. Only a subset of genes may meet these criteria, including key developmental or structural genes required for formation and differentiation of particular tissues. If these genes are only expressed in particular tissues, coding region mutations may generate major phenotypic change without producing deleterious side effects (e.g. MC1R, OCA2, and SLC24A5). In contrast, if these genes are normally expressed in many different tissues, the only mutations that may be compatible with viability and fitness may be regulatory mutations. Only some genes have complex, highly modular enhancers; and regulatory changes in such genes may also underlie repeated evolutionary changes in natural populations (Kitlg, ovo, Pitx1).
Genetic analysis in both sticklebacks and humans has reached the point where it is possible to carry out genome-wide searches for factors contributing to recent evolutionary change in different environments around the world. Given the increasing number of examples of parallel evolution through similar genes, it will be interesting to see if other morphological, physiological, or behavioral differences have also evolved through related genetic mechanisms in both organisms.
EXPERIMENTAL PROCEDURES
Mapping
Branchial skeletons were dissected from Alizarin stained F2 progeny from a marine by Paxton benthic cross (Colosimo et al., 2004). Three new microsatellites, Stn398-Stn400, were added to the LG19 meiotic map (Peichel et al., 2004) using JoinMap 3.0 as described (Peichel et al., 2001). Gill pigmentation was assigned a score from 1 to 4 (Figure 1A) and QTL mapping was done using MapQTL 4.0 (van Ooijen et al., 2002) as described (Peichel et al., 2001).
In situ hybridization
Gill arches were dissected from adult LITC and FTC fish, and whole mount in situ hybridization was done essentially as described (Shapiro et al., 2004) using hydrolyzed Kitlg riboprobe (Supplemental Experimental Procedures).
Allele-specific expression
A Little Campbell River marine female was crossed to a Fishtrap Creek male using in vitro fertilization. Adult F1 females were sacrificed, and tissue was collected from four sites. For gill tissue, gills were trimmed away from the branchial bones. For brain tissue, the entire brain was removed. For ventral skin tissue, incisions were made along the posterior lateral line, caudal peduncle, the ventral midline, and dorsally from the cloaca, and posterior ventral skin removed. For dorsal head skin, a square of head skin tissue was removed by two interorbital incisions.
For these and other RT-PCR experiments, RNA was isolated using RNAwiz (Ambion) and cDNA was reverse transcribed with random hexamers using Invitrogen’s Superscript III following the manufacturers recommendations. Kitlg was amplified using a 5′ 6FAM fluorescently labeled primer 5′-TTGATTTCACGTTATTTGCAG-3′ and unlabeled primer 5′-CAGCAGTATATGGACACAGACG-3′, which flank a small 5′ UTR size polymorphism, and PCR profile of: 94 ºC for 3min, 31 cycles of 94 ºC for 15s, 58 ºC for 15s, 72 ºC for 20s followed by a 3 min extension at 72 ºC. PCR products were size separated on an ABI3730xl, and the intensity of the amplicon from each allele was quantified using peak height in ABI’s Genemapper software.
Admixture Mapping
rs642742A>G was genotyped by PCR-restriction fragment length polymorphism (Supplemental Experimental Procedures) in 374 African Americans previously characterized for skin pigmentation using narrow-band reflectometry and measured for individual genetic ancestry (Shriver et al., 2003; Bonilla et al., 2005).
Association between rs642742A>G and skin pigmentation (quantified by the melanin (M) index, Shriver et al., 2003) was tested using both analysis of covariance (ANCOVA) and Bayesian admixture mapping. Since, as in many admixed populations, individual genetic ancestry shows correlations with skin reflectance in this sample of African Americans (Parra et al., 2004), and KITLG, like several other pigmentation candidate genes (McEvoy et al., 2006) shows substantial differences in allele frequencies across these parental populations, it is important to statistically adjust for the effects of genetic ancestry on skin pigmentation before testing for genetic effects to avoid the risk of false positive associations (Shriver et al., 2003). Both the ANCOVA and Bayesian admixture mapping methods accomplish this adjustment (Supplemental Experimental Procedures).
Supplementary Material
Acknowledgments
We thank Esteban Parra, Jorge Rocha, Greg Barsh, Katie Peichel, Mike Shapiro, Paul Khavari, Amy Adams, Abe Bassan and members of the Kingsley laboratory for useful discussions. This work was supported in part by the Jane Coffin Childs Memorial Fund (CTM), the Portuguese Foundation for Science and Technology (SB), the Natural Sciences and Engineering Research Council of Canada and the Canada Foundation for Innovation (DS), and the National Institutes of Health (DMK, MDS). DS is a Canada Research Chair, and DMK is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barsh GS. What controls variation in human skin color? PLoS Biol. 2003;1:E27. doi: 10.1371/journal.pbio.0000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedell MA, Brannan CI, Evans EP, Copeland NG, Jenkins NA, Donovan PJ. DNA rearrangements located over 100 kb 5′ of the Steel (Sl)-coding region in Steel-panda and Steel-contrasted mice deregulate Sl expression and cause female sterility by disrupting ovarian follicle development. Genes Dev. 1995;9:455–470. doi: 10.1101/gad.9.4.455. [DOI] [PubMed] [Google Scholar]
- Bedell MA, Cleveland LS, O’Sullivan TN, Copeland NG, Jenkins NA. Deletion and interallelic complementation analysis of Steel mutant mice. Genetics. 1996;142:935–944. doi: 10.1093/genetics/142.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell MA, Foster SA. The Evolution of the Threespine Stickleback. Oxford University Press; Oxford: 1994. [Google Scholar]
- Bennett DC, Lamoreux ML. The color loci of mice--a genetic century. Pigment Cell Res. 2003;16:333–344. doi: 10.1034/j.1600-0749.2003.00067.x. [DOI] [PubMed] [Google Scholar]
- Bonilla C, Boxill LA, Donald SA, Williams T, Sylvester N, Parra EJ, Dios S, Norton HL, Shriver MD, Kittles RA. The 8818G allele of the agouti signaling protein (ASIP) gene is ancestral and is associated with darker skin color in African Americans. Hum Genet. 2005;116:402–406. doi: 10.1007/s00439-004-1251-2. [DOI] [PubMed] [Google Scholar]
- Boughman JW, Rundle HD, Schluter D. Parallel evolution of sexual isolation in sticklebacks. Evolution. 2005;59:361–373. [PubMed] [Google Scholar]
- Broad Institute. A draft of the stickleback genome. 2006 http://www.ensembl.org/Gasterosteus_aculeatus/index.html.
- Charlier C, Denys B, Belanche JI, Coppieters W, Grobet L, Mni M, Womack J, Hanset R, Georges M. Microsatellite mapping of the bovine roan locus: a major determinant of White Heifer disease. Mamm Genome. 1996;7:138–142. doi: 10.1007/s003359900034. [DOI] [PubMed] [Google Scholar]
- Colosimo PF, Peichel CL, Nereng K, Blackman BK, Shapiro MD, Schluter D, Kingsley DM. The genetic architecture of parallel armor plate reduction in threespine sticklebacks. PLoS Biol. 2004;2:E109. doi: 10.1371/journal.pbio.0020109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colosimo PF, Hosemann KE, Balabhadra S, Villarreal G, Jr, Dickson M, Grimwood J, Schmutz J, Myers RM, Schluter D, Kingsley DM. Widespread parallel evolution in sticklebacks by repeated fixation of Ectodysplasin alleles. Science. 2005;307:1928–1933. doi: 10.1126/science.1107239. [DOI] [PubMed] [Google Scholar]
- Cott HB. Adaptive Colouration in Animals. Methuen & Co. Ltd; London: 1940. [Google Scholar]
- Cresko WA, Amores A, Wilson C, Murphy J, Currey M, Phillips P, Bell MA, Kimmel CB, Postlethwait JH. Parallel genetic basis for repeated evolution of armor loss in Alaskan threespine stickleback populations. Proc Natl Acad Sci USA. 2004;101:6050–6055. doi: 10.1073/pnas.0308479101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Wu CI. Hitchhiking under positive Darwinian selection. Genetics. 2000;155:1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girvan JR, Braithwaite VA. Population differences in spatial learning in three-spined sticklebacks. Proc R Soc Lond B. 1998;265:913–918. [Google Scholar]
- Grichnik JM, Burch JA, Burchette J, Shea CR. The SCF/KIT pathway plays a critical role in the control of normal human melanocyte homeostasis. J Invest Dermatol. 1998;111:233–238. doi: 10.1046/j.1523-1747.1998.00272.x. [DOI] [PubMed] [Google Scholar]
- Hoekstra HE. Genetics, development, and evolution of adaptive pigmentation in vertebrates. Heredity. 2006;97:222–234. doi: 10.1038/sj.hdy.6800861. [DOI] [PubMed] [Google Scholar]
- Hultman KA, Bahary N, Zon LI, Johnson SL. Gene duplication of the zebrafish kit ligand and partitioning of melanocyte development functions to kit ligand a. PLoS Genet. 2007;3:e17. doi: 10.1371/journal.pgen.0030017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonski NG. The evolution of human skin and skin color. Ann Rev Anthrop. 2004;33:585–623. [Google Scholar]
- Johnsen S. Cryptic and conspicuous coloration in the pelagic environment. Proc R Soc Lond B. 2002;269:243–256. doi: 10.1098/rspb.2001.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ullmann B, Walker C, Wilson C, Currey M, Phillips PC, Bell MA, Postlethwait JH, Cresko WA. Evolution and development of facial bone morphology in threespine sticklebacks. Proc Natl Acad Sci USA. 2005;102:5791–5796. doi: 10.1073/pnas.0408533102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley DM, Peichel CL. The molecular genetics of evolutionary change in sticklebacks. In: Ostlund-Nilsson S, Mayer I, Huntingford FA, editors. Biology of the Three-spined Stickleback. CRC Press; 2007. pp. 41–81. [Google Scholar]
- Lamason RL, Mohideen MA, Mest JR, Wong AC, Norton HL, Aros MC, Jurynec MJ, Mao X, Humphreville VR, Humbert JE, et al. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science. 2005;310:1782–1786. doi: 10.1126/science.1116238. [DOI] [PubMed] [Google Scholar]
- Lao O, de Gruijter JM, van Duijn K, Navarro A, Kayser M. Signatures of positive selection in genes associated with human skin pigmentation as revealed from analyses of single nucleotide polymorphisms. Ann Hum Genet. 2007;71:354–369. doi: 10.1111/j.1469-1809.2006.00341.x. [DOI] [PubMed] [Google Scholar]
- Marcellini S, Simpson P. Two or four bristles: functional evolution of an enhancer of scute in Drosophilidae. PLoS Biol. 2006;4:e386. doi: 10.1371/journal.pbio.0040386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEvoy B, Beleza S, Shriver MD. The genetic architecture of normal variation in human pigmentation: an evolutionary perspective and model. Hum Mol Genet. 2006;15(Spec No 2):R176–181. doi: 10.1093/hmg/ddl217. [DOI] [PubMed] [Google Scholar]
- McGregor AP, Orgogozo V, Delon I, Zanet J, Srinivasan DG, Payre F, Stern DL. Morphological evolution through multiple cis-regulatory mutations at a single gene. Nature. 2007;448:587–590. doi: 10.1038/nature05988. [DOI] [PubMed] [Google Scholar]
- McPhail JD. Predation and the evolution of a stickleback (Gasterosteus) J Fish Res Brd Canada. 1969;26:3183–3208. [Google Scholar]
- McPhail JD. Ecology and evolution of sympatric sticklebacks (Gasterosteus): evidence for a species pair in Paxton Lake, Texada Island, British Columbia. Canadian J Zool. 1992;70:361–369. [Google Scholar]
- Minwalla L, Zhao Y, Le Poole IC, Wickett RR, Boissy RE. Keratinocytes play a role in regulating distribution patterns of recipient melanosomes in vitro. J Invest Dermatol. 2001;117:341–347. doi: 10.1046/j.0022-202x.2001.01411.x. [DOI] [PubMed] [Google Scholar]
- Moodie GEE. Predation, natural selection and adaptation in an unusual threespine stickleback. Heredity. 1972;28:155–167. [Google Scholar]
- Motro B, van der Kooy D, Rossant J, Reith A, Bernstein A. Contiguous patterns of c-kit and steel expression: analysis of mutations at the W and Sl loci. Development. 1991;113:1207–1221. doi: 10.1242/dev.113.4.1207. [DOI] [PubMed] [Google Scholar]
- Motro B, Wojtowicz JM, Bernstein A, van der Kooy D. Steel mutant mice are deficient in hippocampal learning but not long-term potentiation. Proc Natl Acad Sci USA. 1996;93:1808–1813. doi: 10.1073/pnas.93.5.1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy NI. A window on the genetics of evolution: MC1R and plumage colouration in birds. Proc R Soc Lond B. 2005;272:1633–1640. doi: 10.1098/rspb.2005.3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parichy DM. Evolution of danio pigment pattern development. Heredity. 2006;97:200–210. doi: 10.1038/sj.hdy.6800867. [DOI] [PubMed] [Google Scholar]
- Parra EJ, Kittles RA, Shriver MD. Implications of correlations between skin color and genetic ancestry for biomedical research. Nat Genet. 2004;36:S54–60. doi: 10.1038/ng1440. [DOI] [PubMed] [Google Scholar]
- Peichel CL, Nereng KS, Ohgi KA, Cole BL, Colosimo PF, Buerkle CA, Schluter D, Kingsley DM. The genetic architecture of divergence between threespine stickleback species. Nature. 2001;414:901–905. doi: 10.1038/414901a. [DOI] [PubMed] [Google Scholar]
- Peichel CL, Ross JA, Matson CK, Dickson M, Grimwood J, Schmutz J, Myers RM, Mori S, Schluter D, Kingsley DM. The master sex-determination locus in threespine sticklebacks is on a nascent Y chromosome. Curr Biol. 2004;14:1416–1424. doi: 10.1016/j.cub.2004.08.030. [DOI] [PubMed] [Google Scholar]
- Protas ME, Hersey C, Kochanek D, Zhou Y, Wilkens H, Jeffery WR, Zon LI, Borowsky R, Tabin CJ. Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism. Nat Genet. 2006;38:107–111. doi: 10.1038/ng1700. [DOI] [PubMed] [Google Scholar]
- Prud’homme B, Gompel N, Rokas A, Kassner VA, Williams TM, Yeh SD, True JR, Carroll SB. Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene. Nature. 2006;440:1050–1053. doi: 10.1038/nature04597. [DOI] [PubMed] [Google Scholar]
- Rajaraman S, Davis WS, Mahakali-Zama A, Evans HK, Russell LB, Bedell MA. An allelic series of mutations in the Kit ligand gene of mice. II. Effects of ethylnitrosourea-induced Kitl point mutations on survival and peripheral blood cells of Kitl(Steel) mice. Genetics. 2002;162:341–353. doi: 10.1093/genetics/162.1.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro MD, Marks ME, Peichel CL, Blackman BK, Nereng KS, Jonsson B, Schluter D, Kingsley DM. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature. 2004;428:717–723. doi: 10.1038/nature02415. [DOI] [PubMed] [Google Scholar]
- Shapiro MD, Bell MA, Kingsley DM. Parallel genetic origins of pelvic reduction in vertebrates. Proc Natl Acad Sci USA. 2006;103:13753–13758. doi: 10.1073/pnas.0604706103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriver MD, Parra EJ, Dios S, Bonilla C, Norton H, Jovel C, Pfaff C, Jones C, Massac A, Cameron N, et al. Skin pigmentation, biogeographical ancestry and admixture mapping. Hum Genet. 2003;112:387–399. doi: 10.1007/s00439-002-0896-y. [DOI] [PubMed] [Google Scholar]
- Steiner CC, Weber JN, Hoekstra HE. Adaptive variation in beach mice produced by two interacting pigmentation genes. PLoS Biol. 2007;5(9):e219. doi: 10.1371/journal.pbio.0050219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucena E, Delon I, Jones I, Payre F, Stern DL. Regulatory evolution of shavenbaby/ovo underlies multiple cases of morphological parallelism. Nature. 2003;424:935–938. doi: 10.1038/nature01768. [DOI] [PubMed] [Google Scholar]
- The Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooijen JW. LOD significance thresholds for QTL analysis in experimental populations of diploid species. Heredity. 1999;83:613–624. doi: 10.1038/sj.hdy.6886230. [DOI] [PubMed] [Google Scholar]
- van Ooijen JW, Boer MP, Jansen RC, Maliepaard C. MapQTL 4.0: Software for the calculation of QTL positions on genetic maps. Wageningen (the Netherlands): Plant Research International; 2002. [Google Scholar]
- Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrle-Haller B. The role of Kit-ligand in melanocyte development and epidermal homeostasis. Pigment Cell Res. 2003;16:287–296. doi: 10.1034/j.1600-0749.2003.00055.x. [DOI] [PubMed] [Google Scholar]
- Wilkens H. Genetic interpretation of regressive evolutionary processes: studies on hybrid eyes of two Astyanax cave populations (Characidae pisces) Evolution. 1971;25:530–544. doi: 10.1111/j.1558-5646.1971.tb01913.x. [DOI] [PubMed] [Google Scholar]
- Williamson SH, Hubisz MJ, Clark AG, Payseur BA, Bustamante CD, Nielsen R. Localizing recent adaptive evolution in the human genome. PLoS Genet. 2007;3:e90. doi: 10.1371/journal.pgen.0030090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Carroll SB, Kopp A. Evolution in black and white: genetic control of pigment patterns in Drosophila. Trends Genet. 2003;19:495–504. doi: 10.1016/S0168-9525(03)00194-X. [DOI] [PubMed] [Google Scholar]
- Wood TE, Burke JM, Rieseberg LH. Parallel genotypic adaptation: when evolution repeats itself. Genetica. 2005;123:157–170. doi: 10.1007/s10709-003-2738-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Hachiya A, Sriwiriyanont P, Ohuchi A, Kitahara T, Takema Y, Visscher MO, Boissy RE. Functional analysis of keratinocytes in skin color using a human skin substitute model composed of cells derived from different skin pigmentation types. Faseb J. 2007;21:1–11. doi: 10.1096/fj.06-6845com. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.