Summary
The discovery of pathways and regulatory networks whose perturbation contributes to neoplastic transformation remains a fundamental challenge for cancer biology. We show that such pathway perturbations, and the cis-regulatory elements through which they operate, can be efficiently extracted from global gene-expression profiles. Our approach utilizes information-theoretic analysis of expression levels, pathways, and genomic sequences. Analysis across a diverse set of human cancers reveals the majority of previously known cancer pathways. Through de novo motif discovery we associate these pathways with transcription-factor binding sites and miRNA targets, including those of E2F, NF-Y, p53, and let-7. Follow-up experiments confirmed that these predictions correspond to functional in vivo regulatory interactions. Strikingly, the majority of the perturbations, associated with putative cis-regulatory elements, fall outside of known cancer pathways. Our study provides a systems-level dissection of regulatory perturbations in cancer—an essential component of a rational strategy for therapeutic intervention and drug-target discovery.
Keywords: Cancer, regulatory networks, signaling pathways, cancer pathway map, cancer regulatory map, iPAGE, FIRE
INTRODUCTION
Precise molecular definition of pathologic states is an essential component of a rational approach to understanding and treating disease. This is especially true in cancer, where many complex cellular pathways contribute to the initiation and maintenance of the transformation process. Throughout the last decade, microarrays have been widely used for discovering significantly deregulated genes in the tumor samples in order to identify diagnostically and prognostically relevant “molecular signatures” (Rhodes et al., 2004). However, it is becoming increasingly clear that tumor state heterogeneity can often be more accurately described by the behavior of functionally coherent and coordinately regulated sets of genes. Thus, molecular signatures are moving towards pathway-level definitions (Segal et al., 2004; Subramanian et al., 2005). In fact, neoplastic transformation relies on deregulation of diverse oncogenic and tumor-suppressor pathways (Watters and Roberts, 2006), including stimulation of cell growth and proliferation and inhibition of cell cycle arrest and apoptosis (Adjei and Hidalgo, 2005). Global deregulation of these pathways is typically achieved through somatic mutations in key signaling molecules (e.g. Imai et al., 1998), transcription factors (e.g. Gallie, 1994), and post-transcriptional regulators such as microRNAs (e.g. Tavazoie et al., 2008; Wu et al., 2008). Systematic identification of these deregulated pathways and their underlying mutations is a crucial first step in developing a rational strategy for cancer therapy.
In this study, we use an integrated framework to systematically determine deregulated pathways in cancer and identify the transcription factors and other regulators that orchestrate these changes (Figure 1). Our methodology is based on the concept of mutual information (Cover and Thomas, 2006), which provides a general method to detect dependencies between observations, including non-linear correlations and correlations involving continuous (e.g., expression fold-changes) and discrete observations (e.g. expression clusters). Our approach consists of the following steps: first, we identify which known pathways and cellular processes are deregulated in cancer gene expression datasets. This step is based on an information-theoretic pathway analysis called iPAGE, which directly quantifies the mutual information between pathways and expression profiles (see Figure S1A and Suppl. Procedures). Then, we identify promoter and 3′ UTR cis-regulatory elements that best explain gene expression in the same datasets. This is achieved using FIRE, a robust and general information-theoretic framework for cis-regulatory elements discovery from gene expression data (Elemento et al., 2007). The regulators responsible for the observed expression changes are then identified by comparing these uncovered regulatory elements to transcription factor binding sites in JASPAR (Sandelin et al., 2004), TRANSFAC (Matys et al., 2006) and to seed regions of known miRNAs (Griffiths-Jones et al., 2008). Finally, in the last step, we associate the regulatory elements uncovered by FIRE with the deregulated pathways identified by iPAGE. This latter analysis essentially reveals the pathways that are regulated through the discovered putative binding sites by their associated regulatory proteins or RNAs (see Figure S1B and Suppl. Procedures).
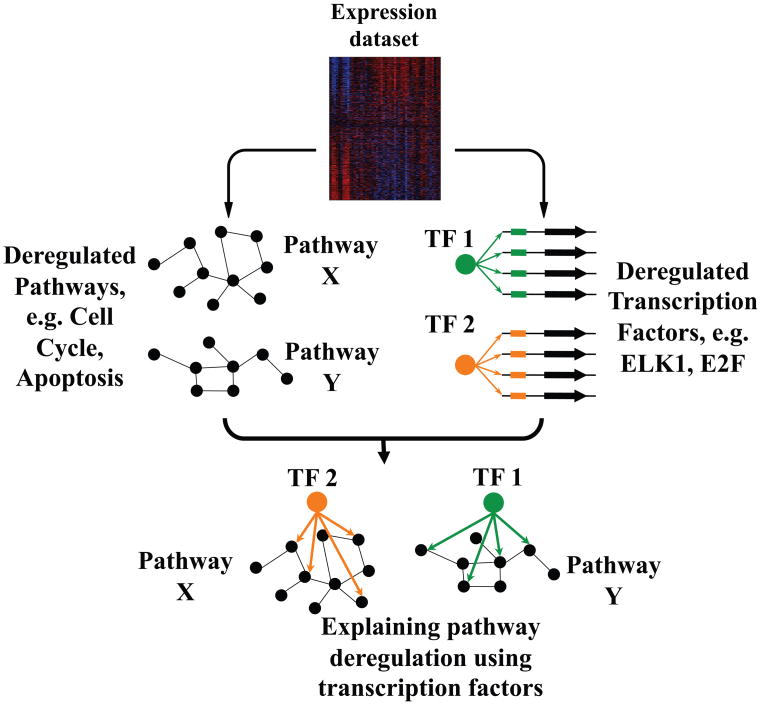
Figure 1. Revealing local regulatory networks from gene expression data.
Perturbed pathways and informative cis-regulatory elements are inferred from cancer-related global gene expression profiles. The discovered pathways are then associated with local DNA and RNA elements in order to reconstruct the underlying regulatory networks (see also Figure S1).
When applied to a large number of cancer gene expression datasets, this data-integrative approach successfully recapitulated the role of many known transcription factors and miRNAs in cancer. This comprehensive analysis of cancer gene expression represents a crucial first step towards achieving a systems-level understanding of regulatory perturbations in cancer. Our discovery of a large repertoire of significant cis-regulatory elements, many of which are RNA binding sites, and their associations with key cellular pathways, highlights our limited current understanding of cancer pathway perturbations.
RESULTS
Discovering deregulated pathways: description of framework and application to bladder cancer
We have created an integrated framework to systematically determine deregulated pathways in cancer and identify the transcription factors and other potential regulators that orchestrate these changes (Figure 1). In what follows, we describe the application of this approach to urinary bladder cancer, the fifth most common malignancy in the US. Using published genome-wide expression profiles of bladder cancer (Dyrskjot et al., 2004) with 41 tumor samples (and 9 normal bladder samples for comparison), we sought to discover the pathways that show significant differential expression in tumor samples compared to their normal controls.
Several methods have been developed to perform this type of analysis, e.g. T-profiler (Boorsma et al., 2005) and GSEA (Subramanian et al., 2005). While undoubtedly powerful, these methods are restricted to continuous gene expression variables, such as fold-changes between cancer and normal samples. More discrete or categorical expression observations, such as co-expression clusters cannot be used as inputs. This is an important limitation, since using co-expression clusters can significantly improve the signal to noise ratio, by taking into account gene expression behavior across different conditions and/or perturbations.
We have developed a principled approach for discovering deregulated pathways from gene expression measurements without the data-type limitation described above. Our framework (called iPAGE) uses the concept of mutual information (Cover and Thomas, 2006) to directly quantify the dependency between expression and known pathways in the Gene Ontology (Ashburner et al., 2000) or in MSigDB (Subramanian et al., 2005). Non-parametric statistical tests are then used to determine whether a pathway is significantly informative about the observed expression measurements. When used on co-expression clusters, enrichment and depletion of pathway components across all clusters contribute to the mutual information; this in turn increases the overall sensitivity and specificity of our approach (see Suppl. Procedures). iPAGE possesses additional advantages over other pathway analysis methods: it can detect non-monotonic pathway association patterns (e.g. pathways with both up-regulated and down-regulated components); it also incorporates a procedure based on the conditional mutual information (Cover and Thomas, 2006) to only return pathways that are independently informative about the expression data being analyzed (Suppl. Procedures).
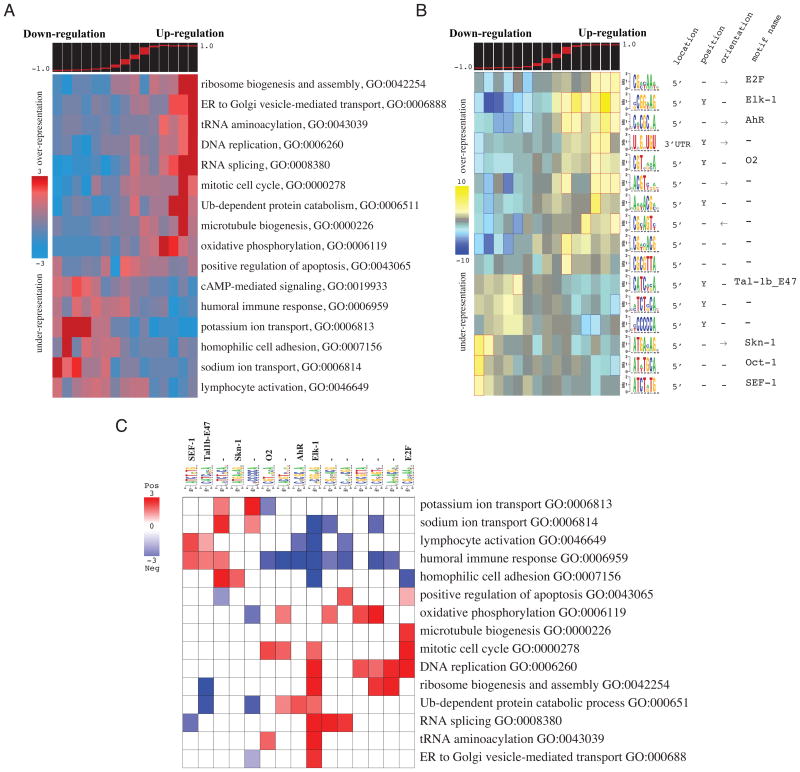
As a first step in the analysis of bladder cancer, we determined the extent to which each gene is differentially expressed between tumors and normal bladders (for details see Suppl. Procedures). We then used iPAGE to search for the pathways (“Biological processes” categories in the Gene Ontology annotations) that are most informative about the observed gene expression differences. As shown in Figure 2A, we found 16 non-redundant pathways with significant deregulation as indicated by the non-random distribution of their components across the spectrum of cancer vs normal expression differences (partitioned into discrete “expression bins”, i.e., contiguous equally populated expression intervals, as described in Suppl. Procedures). These 16 pathways include the up-regulated “mitosis”, “DNA replication” and “oxidative phosphorylation” pathways, which can be explained by the high cell proliferation rate in bladder tumors and the elevated metabolic activity required to sustain it (Arora and Pedersen, 1988; Dyrskjot et al., 2004). The iPAGE analysis also showed that the “lymphocyte activation”, “immune response” and “cell adhesion” pathways are significantly down-regulated. This may indicate suppression of the immune response in these tumors—which can reportedly be overcome by IL2 treatment (Velotti et al., 1991)—in addition to a higher probability of metastasis due to deregulation in cell adhesion components (Cooper and Pienta, 2000).
Figure 2. Pathway and regulatory perturbations in bladder cancer.
(A) Shown are the informative pathways discovered by iPAGE and their patterns of over-representation across the cancer vs. normal expression differences. These differences are partitioned into discrete “expression bins”. Each expression bin includes genes within a specific range of expression values (shown in the top panel). Bins to the left contain genes with lower expression in cancer samples whereas the ones to the right contain genes with higher expression. In the heatmap representation, rows correspond to pathways, and columns to consecutive expression bins. Red entries indicate enrichment of pathway genes in a given expression bin. Enrichment and depletion are measured using hypergeometric p-values (log-transformed) as described in Suppl. Procedures. (B) Shown are the over-representation patterns of the putative cis-regulatory elements discovered by FIRE across the spectrum of cancer vs. normal expression differences. In this heatmap, rows correspond to the discovered motifs and columns to expression bins (see Also Figure S2A and B). Yellow entries in the heatmap indicate motif over-representation (measured by negative log-transformed hypergeometric p-values), while blue entries indicate under-representation (log-transformed p-values). (C) The resulting pathway-regulatory interaction map showing the putative associations between regulatory elements and pathways. Rows correspond to informative iPAGE pathways and columns to informative FIRE motifs. Red entries in this heatmap correspond to a positive association where the genes belonging to a pathway are also enriched in a given motif (measured using log-transformed hypergeometric p-values). Blue entries correspond to significant motif depletions in the upstream sequences (or 3′ UTRs) of genes in a given pathway (see Also Figure S2C).
In the next step, we applied FIRE to identify the cis-regulatory elements that are informative about the same bladder cancer expression changes (Elemento et al., 2007). We identified 16 upstream sequence motifs (including known binding sites for E2F, Elk-1, AhR, SEF-1 and E47) and a single 3′-UTR element (Figure 2B). Approximately two thirds of these motifs are associated with genes that are up-regulated in the tumor state; whereas, the remaining third are enriched in down-regulated genes. Our analysis suggests that the Elk-1 transcription factor, a member of ETS family of ternary complex factors (TCF) and a target of the MAP kinase pathway, plays a central role in bladder cancer. We discovered that many Elk-1 and E2F motifs co-occur within the same promoters (Figure S2A), and that genes with both motifs in their promoters are more likely to be up-regulated in bladder cancer (73%) than genes with either motif considered alone (62% and 65%, respectively). The UNGNUGU element, a 3′ UTR motif, shows essentially a similar pattern of occurrence as the Elk-1 motif (Figure 2B). This motif does not match any of the known miRNA target sites; it may be targeted by an uncharacterized miRNA or by an RNA-binding regulatory protein. Our observation that genes associated with this motif and the Elk-1 motif are more co-expressed than genes associated with each motif considered alone (Figure S2B), suggests a functional cooperation between the factor that binds to this RNA motif and Elk-1.
In the last step of our analysis, we evaluate whether the independently discovered pathways and cis-regulatory sequences are mutually informative of each other. This analysis enables us to associate regulators with their target genes, and to reconstruct the local regulatory networks responsible for cancer-related deregulation. In a heat-map built from the resulting information values (Figure 2C; we call this representation pathway-regulatory interaction map), we observed that Elk-1 binding sites are positively associated with several up-regulated pathways, namely “mitosis”, “DNA replication”, “RNA splicing”, “ribosome biogenesis” and “protein degradation”, and negatively associated with several down-regulated ones (e.g. “lymphocyte activation” and “cell adhesion”). The significant depletion of Elk-1 elements from these specific pathways may reflect selective pressure for avoidance of regulatory cross-talk (Elemento et al., 2007). We also observed a significant anti-correlation between the gene expression level of Elk1 and its target genes in “mitosis”, “RNA splicing” and “ribosome biogenesis” (see Figure S2C). The binding site for E2F also showed a significant association with “DNA replication” and “mitosis” (Figure 2C). Indeed, E2F is a known regulator of DNA replication and mitotic events (Ishida et al., 2001). In bladder carcinoma, the expression of TFDP1, an E2F dimerization partner (Chan et al., 2002), shows a significant correlation with the expression of E2F target genes in “mitosis”, “DNA replication” and “microtubule biogenesis” (see Figure S2C). We also predicted a potential association between AhR transcription factor and “ubiquitin-dependent protein degradation”. As shown in Figure S2C, this transcription factor has a lower expression in normal samples and its expression profile is highly correlated with expression profiles of genes involved in protein catabolism (GO:0006511). Prior evidence for this regulation also exists in the literature: in breast cancer cells, it has been shown that AhR down-regulates estrogen receptor α through activation of the proteasome complex (Wormke et al., 2000).
Our analysis of bladder cancer microarray expression data recapitulates many previously known signaling pathway perturbations. In case of E2F and Elk-1 (whose binding sites we identified above), we speculate that mutations in their upstream signaling proteins (e.g., Rb and Erk2 respectively) result in aberrant activities of these transcription factors, which in turn translate into increased cell proliferation. Strikingly, half the regulatory elements uncovered by FIRE do not correspond to known transcription factor binding or miRNA targeting sites, but nonetheless are highly informative of regulatory perturbations in this dataset. The pathway-regulatory interaction map (Figure 2C) is a powerful starting point for exploring the biological role of these elements and their connections to known pathways.
Comparative analysis of cancer sub-types (BL vs. DLBCL)
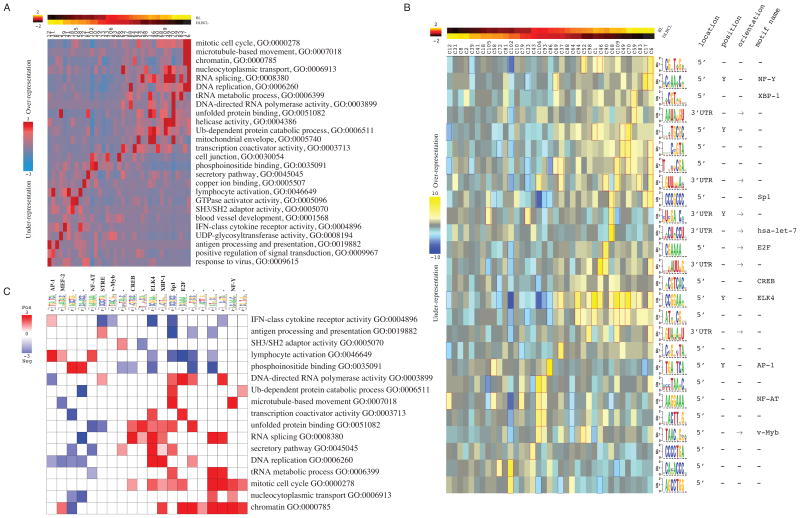
In this section, we demonstrate that our approach can be used to discover deregulated pathways and regulatory networks that distinguish cancer subtypes. We applied this methodology to Burkitt s Lymphoma (BL) and Diffuse Large B-cell Lymphoma (DLBCL), two types of lymphoma that are phenotypically similar but require very different treatment regimens (Frost et al., 2004). We applied iPAGE and FIRE to a microarray analysis of 36 BL and 166 DLBCL samples (Hummel et al., 2006). Based on their expression values across all the samples, we grouped the genes into 110 co-expression clusters (using the k-means clustering algorithm) with each gene uniquely assigned to an index representing a distinct cluster. In contrast with the continuous method used for the bladder cancer dataset, this clustering process increases the sensitivity of our approach by capturing the intra-cancer gene expression heterogeneity, which is usually veiled when averaging expression values across multiple samples of the same tumor type. In this dataset, iPAGE discovered 51 significantly informative and non-redundant pathways. The representative pathways that are associated with the clusters showing differential average expression between BL and DLBCL samples are shown in Figure 3A.
Figure 3. Differential pathway perturbations between Burkitt’s lymphoma (BL) and diffuse large B-cell lymphoma (DLBCL).
(A) Differentially expressed pathways uncovered by iPAGE and their pattern of over-representation across BL/DLBCL co-expression clusters. In this representation, columns represent co-expression clusters while rows correspond to informative pathways. The top panel shows the normalized average expression of each gene cluster in BL and DLBCL samples. (B) A subset of putative cis-regulatory elements discovered by FIRE in BL vs. DLBCL co-expression clusters. (C) The pathway-regulatory interaction map reveals the association between the identified regulatory elements (and their cognate binding factors, when known) and the pathways that are differentially expressed in BL vs DLBCL (see also Figure S3).
Our analysis reveals that several cell cycle-related pathways and processes (e.g., “mitotic cell cycle” and “DNA replication”) are over-represented in co-expression clusters 6 and 17, whose genes show a higher expression level in BL samples (Figure 3A). Along with cell cycle-related genes, protein metabolism pathways such as “protein catabolic process” are also identified as highly informative. These are mostly associated with cluster 109, a cluster of genes with higher expression in BL samples. Moreover, a number of pathways related to immune response, e.g. “cytokine receptor activity” and “antigen processing”, are also significantly deregulated (Figure 3A). These pathways are generally associated with clusters showing lower expression in BL compared to DLBCL (e.g. cluster 8 for “antigen processing” and cluster 39 for “cytokine receptor activity”). The higher expression of lymphocyte-specific pathways in DLBCL has been previously shown by employing immunohistochemical analysis, and revealed the overabundance of B cell activated markers (Gormley et al., 2005).
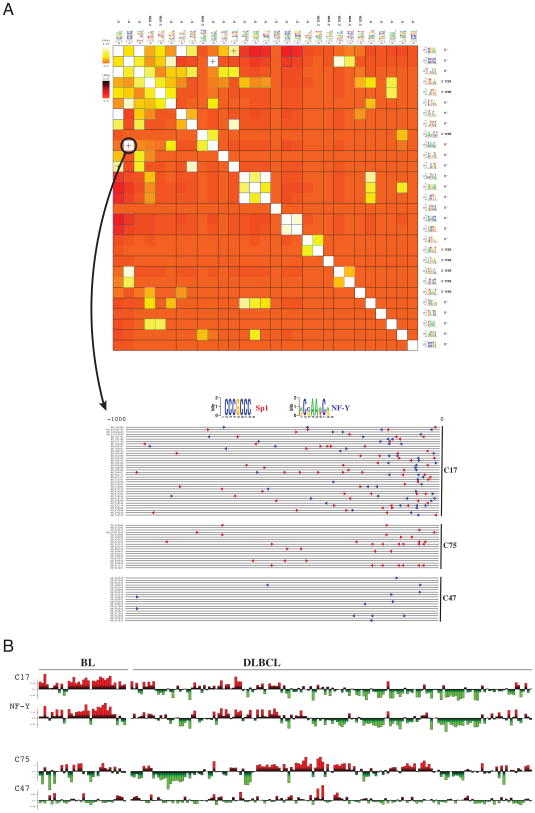
Application of FIRE to the same dataset revealed a collection of informative cis-regulatory elements (both 5′ upstream motifs and 3′ UTR elements) including many known transcription factor binding sites, e.g., E2F, ELK4, NF-Y, NF-AT, MYB, and a microRNA target site for let-7 (see Figure 3B). The let-7 miRNA, whose target genes show significant up-regulation in BL samples, is a known regulator of cell proliferation, and let-7 mutations have been observed in human lung cancers (Johnson et al., 2007). As shown in the pathway-regulatory interaction map, genes with a NF-Y binding site are significantly associated with “mitotic cell cycle” (Figure 3C). NF-Y can activate G1-S cyclins and promote tumorigenesis through cyclin B2 over-expression (Park et al., 2007). The FIRE analysis indicated a strong co-occurrence and co-localization of NF-Y and Sp1 binding sites in cluster 17 (Figure 4A). Cluster 17 genes were highly up-regulated in BL samples (two-tailed t-test, p< 10−10). By comparison, genes in clusters 75 (enriched only in Sp1 motif) and cluster 47 (enriched only in NF-Y motif) show negligible differential expression between BL and DLBCL samples (t-test p-value of 0.5 and 0.3 respectively; Figure 4B); these results suggest a functional interaction between NF-Y and Sp1. One of the shared targets of these two transcription factors with known over-expression in BL is A-myb (Facchinetti et al., 2000) whose binding site (TAACNG reported here as v-Myb) is also captured by FIRE (Figure 3B). The observed correlation between NF-Y mRNA expression and the expression levels of genes in cluster 17 (R=0.73; t-test p<1e-34) further supports the direct role of NF-Y in the regulation of the genes in this cluster (Figure 4B and Figure S3).
Figure 4. Cis-regulatory element interactions and combinatorial regulation.
(A) The FIRE regulatory interaction matrix for the cis-regulatory elements discovered in the BL vs. DLBCL dataset (Figure 3B), and an accompanying motif map showing co-localization of Sp1 and NF-Y sites. In the FIRE interaction matrix, lighter colors (white and yellow) correspond to significant motif co-occurrences. + signs indicate that two motifs tend to co-localize on the DNA or RNA sequences. The NF-Y and Sp1 binding sites show a significant proximal co-occurrence and co-localization in the promoters of their target genes. This co-localization is illustrated by a FIRE motif map, which shows where these two binding sites co-occur in the promoter sequences of genes in cluster 17, in comparison with genes randomly selected from clusters 75 and 47. (B) The average expression profile of genes in co-expression cluster 17, across all BL and DLBCL samples, shows a high correlation with NF-Y mRNA expression. The average expression profiles of the genes in clusters 75 and 47, although enriched in Sp1 and NF-Y putative sites respectively, are not correlated with BL vs. DLBCL classification.
The pathway-regulatory interaction map revealed many known associations but also uncovered previously uncharacterized ones (Figure 3C). For example, the detected association between the AP-1 motif (TGANTCA) and the “lymphocyte activation” and “cytokine receptor activity” pathways correctly recapitulates the prominent role of AP-1 proteins in lymphomas (Vasanwala et al., 2002) and their importance in leukocyte activation and differentiation (Foletta et al., 1998). Figure 3C also clearly highlights the known role of NF-AT in lymphocyte activation (Fisher et al., 2006).
Moreover, our analysis in Figure 3C re-discovered the known association between E2F and mitotic cell cycle and RNA polymerase activity (Ishida et al., 2001). Alongside the mitotic transcription factors, we identified other regulators with potential key roles in defining the biological differences between BL and DLBCL. For example, our results indicate that the binding site for the human X-box binding protein-1 (XBP1), a transcription factor that participates in the unfolded protein response (Calfon et al., 2002), is associated with “unfolded protein binding”. The latter pathway shows a significant up-regulation in BL samples (Figure 3B and C). Although this association has not been observed before in the context of BL, sustaining the activation of the unfolded protein response (UPR) is important for tumor cells due to its cytoprotective action against cytotoxic conditions, e.g. hypoxia and nutrient deprivation, that typically accompany the tumor state.
Global analysis of pathway perturbations across cancers
Our success in revealing regulatory perturbations in cancer vs. normal samples as well as in cancer sub-types motivated us to conduct a more comprehensive meta-analysis of perturbations across diverse human cancers. Our goal was to identify both generic and cancer-type specific deregulations and to reveal the cis-regulatory sequences underlying these changes. To this end, we compiled data from 46 microarray studies of cancer versus normal tissues (see Table S1). In order to capture intra-cancer variation within samples, we employed the same pre-processing step as in the BL vs. DLBCL analysis above; we first clustered the genes based on their expression across normal and tumor samples and then combined the clusters with low average differences into a single background cluster (see Suppl. Procedures for details). We then used iPAGE to find the pathways that best explain the resulting co-expression clusters. We combined the results obtained from all cancer datasets into a cancer pathway heatmap. This map also indicates whether these pathways tend to be up or down-regulated in each cancer type (Figure 5).
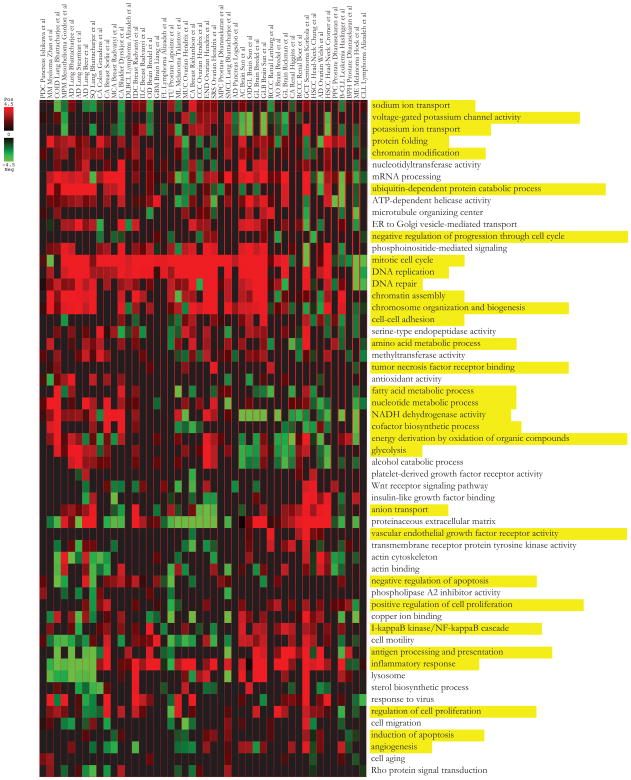
Figure 5. Cancer pathway map.
Shown are the 58 non-redundant iPAGE-discovered pathways with significant patterns of deregulation across 46 cancer vs. normal samples. Each entry in this heatmap represents the most significant over-representation of a given pathway across all non-background co-expression clusters for a given cancer. Over-representation is measured using log-transformed hypergeometric p-values. The colors indicate whether the genes in a given pathway are up-regulated (red) or down-regulated (green) in the tumor samples. The pathways discussed in the text are highlighted in yellow.
As expected, our analysis reveals that multiple pathways are deregulated in many cancers; some of these deregulated pathways are well-known core cancer pathways while others, to the best of our knowledge, have not been previously associated with the tumor state. As expected, our results show that pathways responsible for growth and proliferation are consistently up-regulated in tumor samples as compared to normal controls. This includes “mitotic cell cycle”, “DNA replication” and “chromatin assembly” genes (Figure 5). Metabolic pathways such as “glycolysis” and “organic compounds oxidation” are also up-regulated in many tumors (Arora and Pedersen, 1988); on the other hand, stress responses that lead to cell cycle arrest such as “negative regulation of progression through cell cycle” are often down-regulated. Among the signal transduction pathways, the expression of “NF-κB pathway” components is significantly increased in many cancers, recapitulating the broad oncogenic role of this signaling pathway. Increased levels of NF-κB, a negative regulator of apoptosis, have indeed been reported in many solid and hematopoietic primary tumors and tumor cell lines (see Rayet and Gelinas, 1999 for review).
Our results also suggest an important role for ion transport pathways in oncogenesis and/or tumor maintenance. For example, sodium and potassium transport activities are deregulated in many types of cancer (Figure 5). This is consistent with previous reports that showed active avoidance of sodium transport in some tumors (e.g. Morgan et al., 1986). We also observed a general increase in the expression of the genes encoding anion transporters (especially phosphate transporters) in most of the tumor cells compared to their corresponding normal samples. The cytoplasmic Pi concentration has been suggested to play a critical role in metabolic control in animal cells; a measurable decline in cytoplasmic Pi is accompanied by a decrease in glycolytic or respiratory rates (Geck and Bereiter-Hahn, 1991). The degree to which limited Pi uptake restricts glycolysis, respiration, or cell growth in normal or malignant tissues has been studied extensively (e.g. Wehrle and Pedersen, 1982).
Alongside broadly deregulated pathways, we also identified informative pathways that are only associated with a single or a small number of tumor types. For example, “TNF receptor binding” pathway is most prominent in serous ovarian cancer. The molecular mechanisms of tumor survival in this cancer are not well-understood; however, a recent study has reported that the over-expression of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is correlated with prolonged survival in advanced ovarian cancers (Lancaster et al., 2003). VEGF receptor activity is another pathway that our analysis finds to be up-regulated primarily in renal cancers. Accordingly, inhibitors of VEGF receptor have been widely considered as potential treatment for this type of cancer (Duncan et al., 2008).
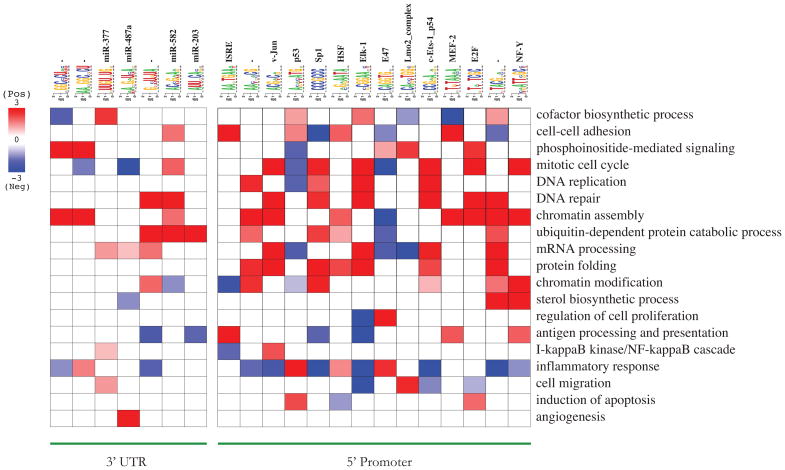
One of the main advantages of our analysis is that it does not only detect deregulated cancer pathways but also reveals the mechanisms by which the observed perturbations may come about. In order to map regulatory networks onto these deregulated pathways, we systematically searched for informative cis-regulatory elements in each cancer gene expression dataset. Combining the resulting motifs into a non-redundant list, we generated a “cancer regulatory map” in which the up- and down-regulation of the genes associated with each motif is captured across all cancers (see Figure S4A and B). Subsequent assessment of the relationship between the deregulated pathways and informative motifs suggests potential key roles for previously known regulatory elements as well as for many previously uncharacterized motifs (Figure S4C). Figure 6, which shows a subset of these relationships, indicates that our approach successfully assigns p53 to “induction of apoptosis” (Stiewe, 2007), Jun, Elk-1 and E2F to “mitotic cell cycle” (Gurzov et al., 2008; Ishida et al., 2001; Smith et al., 2004) and HSF to “protein folding” (Mosser et al., 1993). We hypothesize that the observed deregulations at the transcription level root from perturbations in the upstream signaling pathways leading to the activation or inactivation of key regulators. For example, the IFN-stimulated response element (ISRE) is associated with “antigen processing and presentation”. It is indeed known that interferon β increases gene expression at the transcriptional level through binding of factors to the ISRE upstream of interferon-inducible genes, such as HLA class I (Lefebvre et al., 2001).
Figure 6. Cancer pathway-regulatory interaction map.
Shown is a subset of the cis-regulatory motif-pathway associations from the cancer pathway-regulatory interaction map in Figure S4C. As in Figure 2C, red entries represent positive associations between pathways and regulatory elements.
In another case, genes harboring the MEF-2 motif show significant changes in expression level across different tumor types (Figure S4A). These perturbations are, by and large, comparable across similar cancers, e.g. MEF-2 target genes tend to be up-regulated in most lung cancer samples (Figure S4A). MEF-2, which in our study is associated with “cell-cell adhesion” (Figure 6), is a known regulator of αT-catenin promoter (Vanpoucke et al., 2004). The loss of α-catenin expression, a cadherin-associated protein, results in the disruption of cell-cell adhesion and is associated with an increase in tumor malignancy (Shimoyama et al., 1992).
In addition to promoter motifs, there are also a large number of predicted 3′ UTR elements associated with key pathways. Recent studies have highlighted the role of miRNAs in tumorigenesis and metastasis (e.g., Tavazoie et al., 2008). Among them is miR-203 which was found to be down-regulated in metastatic cells from breast-cancer tumors (see Figure 1a in Tavazoie et al., 2008). Interestingly, our approach associates this miRNA with “ubiquitin-dependent protein catabolic process” (Figure 6). Similarly, we have also associated three known miRNA target sites, miR-377, miR487a and miR-582, with “cofactor biosynthesis”, “angiogenesis”, and “protein degradation”, respectively (Figure 6).
We also discovered a large number of previously uncharacterized motifs that are associated with deregulated pathways (Figure S4C and Table S2). For example, AA[CT]N[AC]CG is a putative upstream binding element which our analysis associates with genes involved in “chromatin assembly” (Figure 6). Genes with the TACGN[AC] motif in their promoters, on the other hand, tend to be involved in “DNA repair”, “mRNA processing” and “protein folding” (Figure 6). Besides these upstream elements, we also discovered many associations involving predicted RNA motifs from 3′ UTRs. For example, GN[CU]U[GU]UA is associated with “DNA repair”, GGC[CU]CU[AU] with “chromatin assembly and AANGGCNCU with “PI3-K signaling” (Figure 6). Our discovery of a large number of RNA motifs suggests an important role for as yet unknown miRNAs or RNA binding regulatory proteins in cancer.
Experimental validation of predicted regulatory interactions
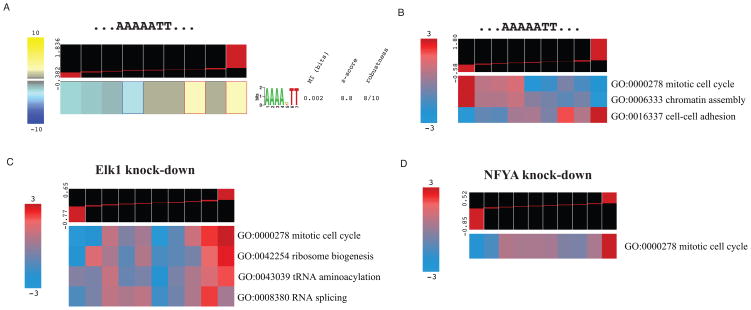
All known and putative cis-regulatory elements presented in Figure S4A are strongly associated with multiple cancer datasets. These elements are therefore very likely to be functional regulatory sequences, e.g. binding sites for transcription factors or RNA-binding proteins, with a broad impact on gene expression. Nevertheless, they are computational predictions that ought to be validated experimentally. In order to test these predictions, we used an oligonucleotide decoy transfection strategy, where the presence of double-stranded DNA titrates away the cognate TF from its genomic target sites and causes a measurable change in their expression (Cutroneo and Ehrlich, 2006; Sinha et al., 2008). We chose to test the upstream sequence motif AAAA[AGT]TT which is independently discovered in more than 15 cancer datasets in Figure S4A. We transfected double-stranded decoy oligonucleotides containing this motif into MDA-MB-231 cells, using a shuffled version of each sequence as a control (see Suppl. Procedures). We then performed expression profiling 72 hours post-transfection. The genes harboring AAAA[AGT]TT motif showed a significant non-random distribution across the expression profile with a significant enrichment in the up-regulated genes (Figure 7A). These experimentally obtained results thus show that the computationally predicted AAAA[AGT]TT motif is capable of influencing the expression of many genes in human cells. In addition, we observed that in our pathway-regulatory interaction map, AAAA[AGT]TT is significantly associated with chromatin assembly and cell-cell adhesion pathways. Consistently, iPAGE discovers these pathways to be significantly deregulated across the profile (Figure 7B). Interestingly, mitotic genes are also notably deregulated in MDA-MB-231 cells, which may indicate a key tumorigenic role for the unidentified protein that binds to this element (Figure 7B).
Figure 7. Experimental validation of the discovered associations.
(A) Genes harboring the AAAA[AGT]TT motif are up-regulated upon transfection of decoy oligonucleotides matching that sequence. (B) Transfection of AAAA[AGT]TT oligos deregulates the expression of “mitotic cell cycle”, “chromatin assembly”, and “cell-cell adhesion” genes (see also Figure S5A). (C) Knocking down Elk1 mRNA up-regulates genes in several pathways associated with the binding site of this transcription factor (see also Figure S5B). (D) Knocking down NFYA is accompanied by up-regulation in the mitotic cell cycle genes (see also Figure S5C).
We then sought to perform experiments to test our ability to identify motif-pathway associations using siRNA knockdowns of selected transcription factors in MDA-MB-231 cancer cell lines, followed by gene expression profiling (see Suppl. Procedures). Our analyses predicted that Elk1-regulated genes in primary tumors are also components of several pathways including mitotic cell cycle, DNA replication, ribosome biogenesis, protein catabolism, and RNA splicing (Figure 2B). The gene expression profile of MDA-MB-231 cells upon Elk1 knock-down shows a significant deregulation in four of these pathways (Figure 7C). The anti-correlation between Elk1 and the “mitotic cell cycle”, “ribosome biogenesis” and “RNA splicing” genes, which was previously discovered in the bladder carcinoma dataset (Figure S2C), was also observed here. Our analysis of the BL vs. DLBCL dataset (Figure 3C) also predicted that NFYA-regulated genes are often involved in mitotic cell cycle, microtubule-based movement and chromatin structure. The iPAGE analysis of the gene expression profile of MDA-MB-231 cells upon NFYA knock-down indeed revealed the broad deregulation of “mitotic cell cycle” (Figure 7D). The expression profiles for the TF knock-downs and decoy vs scrambles experiments can be accessed from GEO (GSE18874) and the processed data along with detailed results are also available online at http://tavazoie.princeton.edu/iPAGE. Altogether, these experimental results clearly demonstrate that the iPAGE/FIRE computational predictions correspond to true and functional in vivo regulatory interactions.
DISCUSSION
The identification of regulatory pathways whose perturbations are causal to the initiation and maintenance of the tumor state is one of the major challenges in cancer biology. In this study, we have introduced a computational framework for simultaneous extraction of perturbed cellular pathways and their underlying regulatory programs from cancer gene expression datasets. Our results clearly show a general over-expression of mitotic pathways and down-regulation of immune response pathways in tumors compared to normal tissues; however, we did not detect any other “universal” tumor pathway signature. The diversity of the perturbed pathways, and their association with specific cancers, as represented in the cancer pathway map, highlights the broad heterogeneity underlying the tumor cellular state. Despite this heterogeneity, pathway-level analysis of cancer gene expression can be employed for classification purposes in the sense that the tumors with similar phenotypes (in terms of which pathways are deregulated and to what extent) can be identified (one such analysis is described in detail in Suppl. Results and clearly shows that similar cancers tend to cluster together when compared on the basis of their deregulated pathways).
In addition to uncovering the deregulated pathways, we have employed a de novo and systematic cis-regulatory element discovery strategy in order to identify the regulators (transcription factors, miRNAs or RNA-binding proteins) through which the perturbations in the cellular pathways come about. The regulators that we identify using our approach are often downstream effectors of signaling pathways with long-established roles in tumorigenesis, and we uncover a substantial fraction of them. Our approach predicts the involvement of many known transcriptional or post-transcriptional regulators in cancer-associated pathways, thus revealing putative oncogenes and tumor suppressors, and yielding potential drug target candidates. We have validated some of the predicted associations using siRNA-based knock-down of transcription factors followed by gene expression profiling in cancer cell lines.
Prior studies that addressed the problem of uncovering regulatory networks perturbed in cancer have largely relied on known cis-regulatory elements or genome-wide binding data (ChIP-chip), e.g., Lemmens et al. (2006) and Sinha et al. (2008). However, on average, only 10% (~32/292) of our discovered motifs correspond to previously known binding sites, even though a majority of them are conserved when evaluating conservation using the network-level approach described in Elemento et al. (2005) (see Figure S4B). This underscores both the complexity and our relatively primitive understanding of the tumor state. For example, we discovered 11 putative regulatory elements with significant positive associations with DNA repair (p < 10−3). Besides, many regulatory elements are highly informative about groups of coordinately regulated genes in cancer versus normal tissues but are not associated with any known pathways. We hypothesize that these putative regulatory elements predict previously uncharacterized cancer-associated pathways. Similarly, only a minority of the 3′ UTR elements we discovered (~10%) match known miRNA target sites. These findings point to a largely unexplored role for post-transcriptional regulation (involving both miRNAs and RNA-binding proteins) in cancer. These cis-regulatory element predictions provide molecular anchors into the sequence, allowing subsequent identification of their cognate trans-factors and the upstream signaling pathways using techniques such as (Freckleton et al., 2009).
To conclude, we have introduced a powerful framework for revealing regulatory perturbations in cancer. We anticipate that this framework, freely available at http://tavazoie.princeton.edu/iPAGE, will enable the rapid and comprehensive analysis of cancer expression data by experts and non-experts alike. As a final note, we stress that although our analyses here have been focused on gene expression perturbations in cancer, our framework is general in concept and can be utilized to study regulatory perturbations across other human diseases.
Highlights.
A computational framework reveals deregulated pathways across cancer expression data.
Deregulated pathways are mapped to known and predicted regulatory elements.
The approach rediscovers known cancer pathways and predicts many new ones.
In vivo experiments validate the function of predicted regulatory interactions.
Supplementary Material
Acknowledgments
We thank Bambi Tsui for developing the FIRE/iPAGE web servers. We are also grateful to Jia Min Loo, Kim Png, Hien Tran and Sohail Tavazoie for their technical support with the experimental validations. S.T. was supported by grants from the NHGRI (R01HG003219), NIGMS (P50 GM071508), and the NIH Director s Pioneer Award (1DP10D003787-01).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adjei AA, Hidalgo M. Intracellular signal transduction pathway proteins as targets for cancer therapy. J Clin Oncol. 2005;23:5386–5403. doi: 10.1200/JCO.2005.23.648. [DOI] [PubMed] [Google Scholar]
- Arora KK, Pedersen PL. Functional significance of mitochondrial bound hexokinase in tumor cell metabolism. Evidence for preferential phosphorylation of glucose by intramitochondrially generated ATP. J Biol Chem. 1988;263:17422–17428. [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boorsma A, Foat BC, Vis D, Klis F, Bussemaker HJ. T-profiler: scoring the activity of predefined groups of genes using gene expression data. Nucleic Acids Res. 2005;33:W592–595. doi: 10.1093/nar/gki484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- Chan JA, Olvera M, Lai R, Naing W, Rezk SA, Brynes RK. Immunohistochemical expression of the transcription factor DP-1 and its heterodimeric partner E2F-1 in non-Hodgkin lymphoma. Appl Immunohistochem Mol Morphol. 2002;10:322–326. doi: 10.1097/00129039-200212000-00006. [DOI] [PubMed] [Google Scholar]
- Cooper CR, Pienta KJ. Cell adhesion and chemotaxis in prostate cancer metastasis to bone: a minireview. Prostate Cancer Prostatic Dis. 2000;3:6–12. doi: 10.1038/sj.pcan.4500387. [DOI] [PubMed] [Google Scholar]
- Cover T, Thomas J. Elements of Information Theory. 2. Hoboken, NJ: Wiley-Interscience; 2006. [Google Scholar]
- Cutroneo KR, Ehrlich H. Silencing or knocking out eukaryotic gene expression by oligodeoxynucleotide decoys. Crit Rev Eukaryot Gene Expr. 2006;16:23–30. doi: 10.1615/critreveukargeneexpr.v16.i1.20. [DOI] [PubMed] [Google Scholar]
- Duncan TJ, Al-Attar A, Rolland P, Scott IV, Deen S, Liu DT, Spendlove I, Durrant LG. Vascular endothelial growth factor expression in ovarian cancer: a model for targeted use of novel therapies? Clin Cancer Res. 2008;14:3030–3035. doi: 10.1158/1078-0432.CCR-07-1888. [DOI] [PubMed] [Google Scholar]
- Dyrskjot L, Kruhoffer M, Thykjaer T, Marcussen N, Jensen JL, Moller K, Orntoft TF. Gene expression in the urinary bladder: a common carcinoma in situ gene expression signature exists disregarding histopathological classification. Cancer Res. 2004;64:4040–4048. doi: 10.1158/0008-5472.CAN-03-3620. [DOI] [PubMed] [Google Scholar]
- Elemento O, Slonim N, Tavazoie S. A universal framework for regulatory element discovery across all genomes and data types. Mol Cell. 2007;28:337–350. doi: 10.1016/j.molcel.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elemento O, Tavazoie S. Fast and systematic genome-wide discovery of conserved regulatory elements using a non-alignment based approach. Genome Biol. 2005;6:R18. doi: 10.1186/gb-2005-6-2-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchinetti V, Lopa R, Spreafico F, Bolognese F, Mantovani R, Tavner F, Watson R, Introna M, Golay J. Isolation and characterization of the human A-myb promoter: regulation by NF-Y and Sp1. Oncogene. 2000;19:3931–3940. doi: 10.1038/sj.onc.1203730. [DOI] [PubMed] [Google Scholar]
- Fisher WG, Yang PC, Medikonduri RK, Jafri MS. NFAT and NFkappaB activation in T lymphocytes: a model of differential activation of gene expression. Ann Biomed Eng. 2006;34:1712–1728. doi: 10.1007/s10439-006-9179-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foletta VC, Segal DH, Cohen DR. Transcriptional regulation in the immune system: all roads lead to AP-1. J Leukoc Biol. 1998;63:139–152. doi: 10.1002/jlb.63.2.139. [DOI] [PubMed] [Google Scholar]
- Freckleton G, Lippman SI, Broach JR, Tavazoie S. Microarray profiling of phage-display selections for rapid mapping of transcription factor-DNA interactions. PLoS Genet. 2009;5:e1000449. doi: 10.1371/journal.pgen.1000449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost M, Newell J, Lones MA, Tripp SR, Cairo MS, Perkins SL. Comparative immunohistochemical analysis of pediatric Burkitt lymphoma and diffuse large B-cell lymphoma. Am J Clin Pathol. 2004;121:384–392. doi: 10.1309/8WYN-VUTG-V9RP-HUQH. [DOI] [PubMed] [Google Scholar]
- Gallie BL. Retinoblastoma gene mutations in human cancer. N Engl J Med. 1994;330:786–787. doi: 10.1056/NEJM199403173301112. [DOI] [PubMed] [Google Scholar]
- Geck P, Bereiter-Hahn J. The role of electrolytes in early stages of cell proliferation. Cell Biol Rev. 1991;25:85–104. [PubMed] [Google Scholar]
- Gormley RP, Madan R, Dulau AE, Xu D, Tamas EF, Bhattacharyya PK, LeValley A, Xue X, Kumar P, Sparano J, et al. Germinal center and activated b-cell profiles separate Burkitt lymphoma and diffuse large B-cell lymphoma in AIDS and non-AIDS cases. Am J Clin Pathol. 2005;124:790–798. doi: 10.1309/7CEA-WV0D-NLLU-WQTF. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurzov EN, Bakiri L, Alfaro JM, Wagner EF, Izquierdo M. Targeting c-Jun and JunB proteins as potential anticancer cell therapy. Oncogene. 2008;27:641–652. doi: 10.1038/sj.onc.1210690. [DOI] [PubMed] [Google Scholar]
- Hummel M, Bentink S, Berger H, Klapper W, Wessendorf S, Barth TF, Bernd HW, Cogliatti SB, Dierlamm J, Feller AC, et al. A biologic definition of Burkitt’s lymphoma from transcriptional and genomic profiling. N Engl J Med. 2006;354:2419–2430. doi: 10.1056/NEJMoa055351. [DOI] [PubMed] [Google Scholar]
- Imai Y, Tsurutani N, Oda H, Inoue T, Ishikawa T. Genetic instability and mutation of the TGF-beta-receptor-II gene in ampullary carcinomas. Int J Cancer. 1998;76:407–411. doi: 10.1002/(sici)1097-0215(19980504)76:3<407::aid-ijc19>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Ishida S, Huang E, Zuzan H, Spang R, Leone G, West M, Nevins JR. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol Cell Biol. 2001;21:4684–4699. doi: 10.1128/MCB.21.14.4684-4699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- Lancaster JM, Sayer R, Blanchette C, Calingaert B, Whitaker R, Schildkraut J, Marks J, Berchuck A. High expression of tumor necrosis factor-related apoptosis-inducing ligand is associated with favorable ovarian cancer survival. Clin Cancer Res. 2003;9:762–766. [PubMed] [Google Scholar]
- Lefebvre S, Berrih-Aknin S, Adrian F, Moreau P, Poea S, Gourand L, Dausset J, Carosella ED, Paul P. A specific interferon (IFN)-stimulated response element of the distal HLA-G promoter binds IFN-regulatory factor 1 and mediates enhancement of this nonclassical class I gene by IFN-beta. J Biol Chem. 2001;276:6133–6139. doi: 10.1074/jbc.M008496200. [DOI] [PubMed] [Google Scholar]
- Lemmens K, Dhollander T, De Bie T, Monsieurs P, Engelen K, Smets B, Winderickx J, De Moor B, Marchal K. Inferring transcriptional modules from ChIP-chip, motif and microarray data. Genome Biol. 2006;7:R37. doi: 10.1186/gb-2006-7-5-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan K, Spurlock G, Brown RC, Mir MA. Release of a sodium transport inhibitor (inhibitin) from cultured human cancer cells. Cancer Res. 1986;46:6095–6100. [PubMed] [Google Scholar]
- Mosser DD, Duchaine J, Massie B. The DNA-binding activity of the human heat shock transcription factor is regulated in vivo by hsp70. Mol Cell Biol. 1993;13:5427–5438. doi: 10.1128/mcb.13.9.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Yu GR, Kim WH, Moon WS, Kim JH, Kim DG. NF-Y-dependent cyclin B2 expression in colorectal adenocarcinoma. Clin Cancer Res. 2007;13:858–867. doi: 10.1158/1078-0432.CCR-06-1461. [DOI] [PubMed] [Google Scholar]
- Rayet B, Gelinas C. Aberrant rel/nfkb genes and activity in human cancer. Oncogene. 1999;18:6938–6947. doi: 10.1038/sj.onc.1203221. [DOI] [PubMed] [Google Scholar]
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Friedman N, Koller D, Regev A. A module map showing conditional activity of expression modules in cancer. Nat Genet. 2004;36:1090–1098. doi: 10.1038/ng1434. [DOI] [PubMed] [Google Scholar]
- Shimoyama Y, Nagafuchi A, Fujita S, Gotoh M, Takeichi M, Tsukita S, Hirohashi S. Cadherin dysfunction in a human cancer cell line: possible involvement of loss of alpha-catenin expression in reduced cell-cell adhesiveness. Cancer Res. 1992;52:5770–5774. [PubMed] [Google Scholar]
- Sinha S, Adler AS, Field Y, Chang HY, Segal E. Systematic functional characterization of cis-regulatory motifs in human core promoters. Genome Res. 2008;18:477–488. doi: 10.1101/gr.6828808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ER, Smedberg JL, Rula ME, Xu XX. Regulation of Ras-MAPK pathway mitogenic activity by restricting nuclear entry of activated MAPK in endoderm differentiation of embryonic carcinoma and stem cells. J Cell Biol. 2004;164:689–699. doi: 10.1083/jcb.200312028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiewe T. The p53 family in differentiation and tumorigenesis. Nat Rev Cancer. 2007;7:165–168. doi: 10.1038/nrc2072. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massague J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanpoucke G, Goossens S, De Craene B, Gilbert B, van Roy F, Berx G. GATA-4 and MEF2C transcription factors control the tissue-specific expression of the alphaT-catenin gene CTNNA3. Nucleic Acids Res. 2004;32:4155–4165. doi: 10.1093/nar/gkh727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasanwala FH, Kusam S, Toney LM, Dent AL. Repression of AP-1 function: a mechanism for the regulation of Blimp-1 expression and B lymphocyte differentiation by the B cell lymphoma-6 protooncogene. J Immunol. 2002;169:1922–1929. doi: 10.4049/jimmunol.169.4.1922. [DOI] [PubMed] [Google Scholar]
- Velotti F, Stoppacciaro A, Ruco L, Tubaro A, Pettinato A, Morrone S, Napolitano T, Bossola PC, Franks CR, Palmer P, et al. Local activation of immune response in bladder cancer patients treated with intraarterial infusion of recombinant interleukin-2. Cancer Res. 1991;51:2456–2462. [PubMed] [Google Scholar]
- Watters JW, Roberts CJ. Developing gene expression signatures of pathway deregulation in tumors. Mol Cancer Ther. 2006;5:2444–2449. doi: 10.1158/1535-7163.MCT-06-0340. [DOI] [PubMed] [Google Scholar]
- Wehrle JP, Pedersen PL. Characteristics of phosphate uptake by Ehrlich ascites tumor cells. J Biol Chem. 1982;257:9698–9703. [PubMed] [Google Scholar]
- Wormke M, Stoner M, Saville B, Safe S. Crosstalk between estrogen receptor alpha and the aryl hydrocarbon receptor in breast cancer cells involves unidirectional activation of proteasomes. FEBS Lett. 2000;478:109–112. doi: 10.1016/s0014-5793(00)01830-5. [DOI] [PubMed] [Google Scholar]
- Wu M, Jolicoeur N, Li Z, Zhang L, Fortin Y, Denis LA, Yue Z, Shen S. Genetic variations of microRNAs in human cancer and their effects on the expression of miRNAs. Carcinogenesis. 2008 doi: 10.1093/carcin/bgn073. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.