Abstract
In patients with hemochromatosis, cardiac dysfunction may appear years after they have reached a state of iron overload. We hypothesized that cumulative iron-catalyzed oxidant damage to mitochondrial DNA (mtDNA) might explain the cardiomyopathy of chronic iron overload. Mice were given repetitive injections of iron dextran for a total of 4 weeks after which the iron loaded mice had elevated cardiac iron, modest cardiac hypertrophy and cardiac dysfunction. qPCR amplification of near-full-length (~16 kb) mtDNA revealed >50% loss of full length product, whereas amounts of a qPCR product of a nuclear gene (13 kb region of beta globin) were unaffected. Quantitative rtPCR analyses revealed 60–70% loss of mRNA for proteins encoded by mtDNA with no change in mRNA abundance for nuclear encoded respiratory subunits. These changes coincided with proportionate reductions in complex I and IV activities and decreased respiration of isolated cardiac mitochondria. We conclude that chronic iron overload leads to cumulative iron-mediated damage to mtDNA and impaired synthesis of mitochondrial respiratory chain subunits. The resulting respiratory dysfunction may explain the slow development of cardiomyopathy in chronic iron overload and similar accumulation of damage to mtDNA may also explain the mitochondrial dysfunction observed in slowly progressing diseases such as neurodegenerative disorders.
Keywords: mitochondrial DNA, iron overload, cardiomyopathy
Introduction
Patients with iron overload are liable to slowly developing cardiomyopathy characterized by systolic and diastolic dysfunction but the pathophysiologic mechanism(s) underlying this gradual organ damage remain poorly defined. Despite the highly likely conjecture that iron-mediated tissue damage involves interactions between ‘loose’ iron and cellular oxidizing and reducing equivalents, the pathophysiologic events involved in this disorder (as well as hepatic and pancreatic damage seen in chronic iron overload) have not been fully elucidated.
Iron is required for the activity of numerous iron- and heme-containing proteins, but “free” (redox active) iron catalyzes the formation of highly toxic reactive oxygen species (ROS) that damage lipids, proteins and DNA [1]. This damage is assumed to arise from iron-catalyzed hydroxyl radical (•OH) formation or, perhaps more likely, iron-centered radicals such as ferryl and perferryl [2,3]. Iron-driven oxidation events require that the metal interact with cellular reducing and oxidizing equivalents such as superoxide and hydrogen peroxide, a major source of which is 'leak' of electrons from the mitochondrial electron transport chain [4].
We investigated the hypothesis that progressive iron-catalyzed oxidant damage to mitochondrial DNA (mtDNA) might occur in chronic iron overload and that this might lead to loss of respiratory capacity and cardiac dysfunction. For this study, a murine model of iron-overload cardiomyopathy was used. Overall, the results indicate that iron-mediated damage to mtDNA does occur and is associated with decreased expression of mitochondrially-encoded (but not nuclear encoded) mRNA and proteins. These effects are associated with loss of mitochondrial respiratory capacity and quite likely lead directly to cardiac dysfunction.
Materials and Methods
Materials
Iron dextran (batch 018H251711; ferric hydroxide dextran complex, elemental iron content 100 mg/ml containing 0.5% phenol as a preservation), trichloroacetic acid (TCA), Ferene S (3-(2-pyridyl)-5,6-di(2-furyl)-1,2,4-triazine-5',5"-disulfonic acid), Hanks balanced salt solution (HBSS), bovine heart cytochrome c, sodium citrate, potassium phosphate, ethyl acetate, porcine heart isocitrate dehydrogenase, β-NADH, MnCl2, sucrose, potassium phosphate, ethylenediaminetetraacetic acid (EDTA), tris (hydroxymethyl) aminomethane, tris (hydroxymethyl) aminomethane (Tris) and the acid salt tris-HCl, ethylene glycol-bis (β-aminoethyl ether) N,N,N’,N’-tetraacetic acid (EGTA), 3-(N-morpholino)propanesulfoniic acid (MOPS) and ethyl acetate, were purchased from Sigma Chemical Co. (St. Louis, MO). Perchloric acid was purchased from Fisher Scientific (Pittsburgh, PA). The rTth DNA Polymerase XL (eXtra Long) designed for generating long PCR products was obtained from Applied Biosystems (Foster City, CA). GoTaq® Flexi DNA Polymerase and RNasin® ribounclease inhibitor were obtained from Promega (Madison WI). PicoGreen was purchased from Molecular Probes (Eugene, OR).
Animals and treatment
Male B6D2F1 mice aged 6 weeks, average weight 20–25 grams, were purchased from Jackson Laboratory (Bar Harbor, ME). Twenty mice were randomized to receive repetitive intraperitoneal (i.p.) injections of iron dextran (10 mg elemental Fe/day/mouse; 100 ul volume) or saline, 5 days per week for a total of 4 weeks following institutional guidelines and an earlier protocol [5].
Electron microscopy
Myocardial samples were taken from the left ventricles and cut into small pieces (~1-mm3). Tissue samples were fixed in 3% glutaraldehyde in 0.1 mmol/L sodium cacodylate buffer (pH 7.4) for 2 h at 4° C, and post-fixed in 1% osmium tetroxide. The samples were embedded and ultrathin (5 µ) sections were prepared. The sections were then stained with uranyl acetate and lead citrate, and observed under an electron microscope.
Echocardiography
Transthoracic echocardiography was performed on control and treated animals using a Sonos 5500 Philips echocardiograph with a 15-6L 7–15 MHz multifrequency linear transducer and an S12 5–12 MHz Doppler transducer. Mice were anesthetized with tribromoethanol (Avertin®) (2.5% working stock in isotonic NaCl).
Measurements of arterial blood pressure
Mice were anesthetized as for echocardiography. They were placed on their backs and the trachea and left carotid artery were exposed via an incision in the neck. A cannula was placed in the trachea through an incision below the larynx to insure a patent airway and a catheter (PE-10) was placed in the left carotid artery to measure arterial blood pressure by a transducer (Statham P23ID, Gould, Oxnard, CA).
Measurement of cardiac iron
Cardiac Fe content was measured colorimetrically as previously described [6–8]. Briefly, the tissue (~ 20 mg) was homogenized with 300 µL of Chelex 100 pre-treated phosphate buffered saline (PBS) using a glass homogenizer and precipitated by adding 200 µL of ice-cold 25% TCA (final TCA concentration 10%). Following 30 min incubation on ice, the precipitate was centrifuged at 12,000 × g for 5 min and 400 µL of supernatant was used for assay of "loose" Fe. For measurements of total residual Fe, the remaining precipitate was digested with 100 µL of 50% nitric acid at 56°C for 24 hr. Following the addition of NaOH to neutralize the nitric acid, Fe was assayed as described above [6–8].
Total cellular DNA isolation
Total DNA was isolated using a QIAamp DNA isolation kit (Qiagen, Inc., Valencia, CA) based on the manufacturer’s protocol. Briefly, frozen mouse heart tissue (~20 mg) was disrupted in a Dounce homogenizer in 100 µl of preparation buffer containing 1 mmol/L desferrioxamine equivalents of a high molecular weight hydroxyethyl starch:desferrioxamine conjugate (Biomedical Frontiers Inc., Minneapolis, MN) to remove adventitious iron. After homogenization, an additional 80 µl of preparation buffer and 20 µl proteinase K (20mg/ml) were added. The sample was thoroughly mixed and incubated at 56°C over night. Following this, total genomic DNA was isolated using a QIAamp® DNA isolation kit.
PCR amplification
To assess mtDNA integrity we used total DNA derived from mouse heart tissue as described above. Near full-length mtDNA amplification was done using a GeneAmp® XL PCR kit (Applied Biosystems, Foster City, CA). The PCR products were quantified with PicoGreen® using a plate reading spectrofluorometer at 485 nm excitation and 525 nm emission.
To determine whether iron-mediated DNA damage was specific for mtDNA or perhaps shared by nuclear DNA, we also performed PCR amplification of a long fragment of the nuclear beta-globin gene (13 kb) as well as a short segment of the beta-globin gene (223 bp) (as a control for input of nuclear DNA). Primers for amplification of the 13 kb long fragment and the 223 bp short fragment of the beta-globin gene are listed in the supplementary Table. Amplified products were resolved and quantified as described above [9,10].
Real time quantitative PCR
For quantitative real-time reverse transcript polymerase chain reaction (qRT-PCR) total RNA was extracted using an RNeasy® Mini Kit (Qiagen, Inc., Valencia, CA). qRT-PCR was performed using Power SYBR® Green PCR Master Mix (Applied Biosystems, Foster, CA) on the DNA Engine Opticon II real-time system (Bio-Rad Laboratories, Hercules, CA). qRT-PCR was done for mRNAs encoded by both nuclear and mitochondrial genes. The primers used are listed in the supplementary Table. All tests were performed in triplicate and all experiments were repeated three times. The amplification data were analyzed with Opticon Monitor analysis software. Calculations were based on the ‘Delta-Delta method’ using the equation R (ratio) = 2−[ΔCT sample − ΔCT control] [11]. The integrity of amplified DNA was confirmed by determination of melting temperature. The data were expressed as fold changes of the treatment groups in relation to the controls.
Measurement of mitochondrial oxygen consumption
Mice were anesthetized with Avertin, exsanguinated via aortic puncture and the heart was quickly removed. Heart weight was measured and the heart was immediately placed in 10 ml cold isolation buffer (220 mmol/L mannitol, 70 mmol/L sucrose, 5 mmol/L MOPS, pH 7.4) and washed 5 times with the same buffer. The tissue was homogenized with a Dounce homogenizer on ice in a buffer of 220 mmol/L mannitol, 70 mmol/L sucrose, 5 mmol/L MOPS, 2 mmol/L EGTA and 0.2% bovine serum albumin (BSA), pH 7.4 on ice. The supernatant was collected by passage through cheesecloth and the cheesecloth was washed twice with equal volumes of the buffer. The filtered supernatant was centrifuged at 5,000 × g for 10 min. The pellet was resuspended in 200 µl of cold isolation buffer, and the protein concentration was measured using bicinchoninic acid (Pierce, Rockford, IL, USA) [12]. Oxygen consumption was measured using a Gilson Oxygraph with a Clark electrode (Yellow Springs Instrument Co., Yellow Springs, OH, USA). Results were adjusted for variations in protein to reflect O2 consumed/min/mg protein.
Cytochrome c oxidase assay
Frozen mouse heart tissue (~20 mg) was homogenized on ice using a Dounce tissue homogenizer at a tissue:buffer ratio of 1:30 (250 mmol/L sucrose, 40 mmol/L potassium chloride, 2 mmol/L EGTA, 0.1% BSA and 20 mmol/L Tris-HCl, pH7.2 @ 37°C). The homogenate was centrifuged at 2,000 × g for 10 min, and the pellet was discarded. The supernatant was used to assay COX activity as reported earlier [13,14]. Results were normalized for protein. In addition, because accurate estimations of COX activity could be compromised by variations in mitochondrial yield and integrity, results were also normalized using a relatively invariant mitochondrial enzyme activity, citrate synthase (see below).
NADH-dehydrogenase
The reaction was followed at 550 nm (ε = 19 mM−1 cm−1) in a solution of 40 µM oxidized cytochrome c and 8 mmol/L NADH, in a buffer consisting of 10 mmol/L Tris (pH 8.0) containing 1 mg/ml BSA and 0.4 mmol/L KCN [14]. The results were normalized for protein.
Citrate synthase
This assay was carried out as described by Williams et al. with minor modifications [15]. The citrate synthase activity was calculated using ε = 13.6 mM−1 cm−1 [16]. The calculated citrate synthase activities were ~54 nmol/min/mg protein and ~52 nmol/min/mg protein in untreated and iron treated groups, respectively.
Results
Cardiac iron concentration
At the end of the four week iron loading, there was a ~50% increase in “loose” (i.e., TCA soluble) cardiac iron (increasing from 0.28 nmol/mg to 0.49 nmol/mg wet weight) in control vs. iron-injected animals, respectively (Table 1). Total cardiac iron (i.e., TCA-soluble and -insoluble) was increased almost 10-fold.
Table 1.
Cardiac iron content in control mice and mice given repetitive injections of iron dextran for a total of 4 weeks. Iron content was measured as described under Materials and Methods. n = 10 in all cases.
| Treatment with iron | Free Iron (nmol/mg tissue) |
Protein Bound Iron (nmol/mg tissue) |
Total Iron (nmol/mg tissue) |
|---|---|---|---|
| Saline Treatment | 0.28 ± 0.11 | 0.45 ± 0.16 | 0.73 ± 0.22 |
| Iron-treatment | 0.49 ± 0.13* | 4.06 ± 0.17* | 4.55 ± 1.75* |
p < 0.01 for saline treated mice vs. iron treated mice (Student’s t test, two-tailed).
Electron microscopy
Hearts of iron-loaded mice had irregularly shaped mitochondria, often with disordered or absent cristae. In addition, electron dense material (possibly within lysosomes) was clearly evident. No such electron dense material was present in the hearts of control mice (Figure 1).
Figure 1.
Thin section (5 µM) electron micrographs of cardiomyocytes from control mice (A) and mice given repetitive injections of iron dextran for 4 weeks (B). M: mitochondria; MF: myofibrils; H: electron dense iron deposits.
Habitus and Cardiac Function
The iron loaded mice became progressively lethargic and the fur became coarse and dull. Unlike the control animals, these mice failed to gain weight after 4 wks of iron loading and actually lost about 10% of their initial body weight (Table 2). Major organs such as heart, liver and spleen of the iron treated mice were larger compared with control animals and generally had a bronze discoloration. Typical echocardiographic results are shown in Table 3 for control vs. iron-loaded mice. In comparison with control mice, LVIDD was significantly decreased in iron treated mice and AWTd, PWTd and RWT were increased. Note that the apparent increase in LVEF in the iron treated mice may be an artifact arising from the cardiac hypertrophy (see Table 2). Perhaps reflecting the decreased LVEDD, arterial blood pressure was found to be lower in iron-loaded animals (83.7 mmHg) than in controls (102.0 mmHg) (p = 0.004; n = 7 and 8 respectively).
Table 2.
Body and organ weights in control mice and mice given repetitive injections of iron dextran for a total of 4 weeks. Iron overload caused modest cardiac hypertrophy and increased hepatic weight. P < 0.01 compared to controls (n = 10 in each group).
| Treatment Group |
Initial Body Weight (g) |
Body Weight at Sacrifice (g) |
Heart Weight (g) |
Heart Weight/ Body Weight |
Liver Weight (g) |
Kidney Weight (g) |
|---|---|---|---|---|---|---|
| Saline Treatment |
26.21 ± 1.52 | 27.74 ± 2.03 | 0.134 ± 0.02 | 0.0049 ± 0.001 | 1.15 ± 0.11 | 0.43 ± 0.05 |
| Iron Treatment |
26.14 ± 0.83 | 23.46 ± 2.20 | 0.144 ± 0.02 | 0.0062 ± 0.001* | 2.30 ± 0.22* | 0.36 ± 0.05 |
p < 0.01 for saline treated mice vs. iron treated mice (Student’s t test, two-tailed).
Table 3.
Cardiac function in control mice and mice given repetitive injections of iron dextran for a total of 4 weeks. Values shown are mean ± 1 standard deviation (n = 10 in each group). In all measures except for heart rate, iron treated mice differ from controls at p<0.001 (Student’s t test, two-tailed).
| Group | Heart Rate | AWTd | PWTd | RWT | LVIDD | LVEF single plane |
|---|---|---|---|---|---|---|
| Control | 512±36.36 | 0.70±0.06 | 0.83±0.07 | 0.43±0.14 | 3.6±0.26 | 0.68±0.06 |
| Iron treated | 472±29.79 | 0.99±0.15 | 0.93±0.01 | 0.67±0.05 | 2.9±0.37 | 0.80±0.07 |
Abbreviations: AWTd: anterior wall thickness in diastole. PWTd: posterior wall thickness in diastole. RWT: relative wall thickness = (AWT + PWT)/LVIDD. LVIDD: left ventricle internal diameter in diastole. LVEF: left ventricular ejection fraction = (EDV - ESV)/ED.
Chronic iron overload leads to mitochondrial DNA damage
The use of long range PCR to estimate the integrity of mtDNA is based on the principle that several types of DNA damage - such as single or double strand breaks or bulky base modifications - will block the polymerase, leading to decreased total PCR product [9,10]. We found that iron overload caused a major loss of full-length mtDNA PCR product (Figure 2A). This DNA damage appears selective for mtDNA inasmuch as there was no change in amount of PCR product from a 13 kb section of a nuclear gene, beta globin (Figure 2B). We should note that this particular assay only indicates the presence of mtDNA damage but not the extent of such damage; diminished long product formation could arise from only a single break or modification or, for that matter, could reflect more extensive damage.
Figure 2.
Decreased PCR-amplifiable full length (16 kb) cardiac mtDNA in mice given repetitive injections of iron dextran for 4 weeks. Total DNA was isolated and amplified by QPCR as described under Materials and Methods. A: Full length mtDNA product was significantly decreased in hearts from iron treated mice while no changes were observed in the short (0.3 kb) PCR product (a control for input of mtDNA). B: No change in the amounts of either a long (13 kb) segment of the beta-globin gene or short (223 bp) segment after iron loading. *p<0.01 (Student's t test, two-tailed; n = 3 in each case).
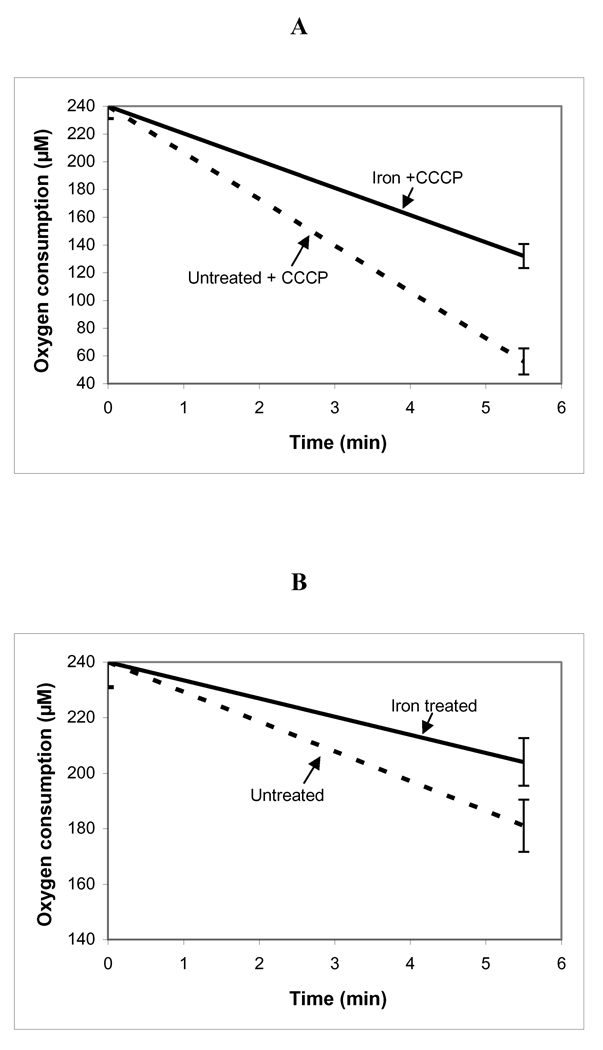
Mitochondrial DNA damage is associated with decreased mitochondrial respiration
Chronic iron overload also caused significantly decreased mitochondrial respiration both in the absence and presence of an uncoupler (CCCP) (Figure 3A,B). Note that in these experiments, the mice were sacrificed individually, the mitochondria were isolated and respiration measured one animal at a time.
Figure 3.
Decreased respiration of mitochondria from hearts of mice given repetitive injections of iron dextran for 4 weeks. A: Maximal oxygen consumption rates in the presence of the uncoupler CCCP were significantly decreased following iron loading. B: Similar iron-mediated decrements in respiration were also observed in the absence of an uncoupler; p<0.01 untreated vs. iron treated mice in both cases (Student's t test, two-tailed; n = 3 separate mitochondrial preparations in each case.
Expression of mRNA for respiratory chain components is decreased in iron overload
As shown in Table 4, very large (60–70%) decreases in mRNA for genes encoded by mtDNA were observed. In contrast, mRNAs for respiratory chain components encoded by the nuclear genome were either unaffected or actually greater than controls. Once again, this implies preferential damage to mtDNA leading to diminished production of mRNA while nuclear DNA appears unaffected.
Table 4.
Decreased mRNA for respiratory chain components encoded by mtDNA but not those encoded by nuclear DNA in mice given repetitive injections of iron dextran for 4 weeks. Real-time rtPCR was used to detect levels of mRNA expression of mitochondrial genes (12S rRNA, COXI, ND4, and ND1) and nuclear genes (SDH, COXVb, NRF1, and GAPDH).
| mRNA | % OF CONTROL | MtDNA ENCODED |
nDNA ENCODED |
|---|---|---|---|
| 16S rRNA | 41 ± 0.74* | + | |
| COXI | 31 ± 0.93* | + | |
| ND4 | 30 ± 0.65* | + | |
| ND1 | 34 ± 1.41* | + | |
| SDH subunit b | 141 ± 0.99 | + | |
| COXVb | 112 ± 1.28 | + | |
| NRF1 | 116 ± 1.10 | + | |
| GAPDH | 106 ± 1.00 | + |
p< 0.001 control (n = 3 in each case).
Iron overload leads to decrements in the activities of complexes I and IV
Greatly decreased activities of both complex I (NADH dehydrogenase) and complex IV (cytochrome c oxidase) were found in mice following iron overload (Figure 4). Since both have subunits encoded by mtDNA, it is very likely that these decreases are secondary to the mtDNA damage and decreased expression of mRNAs for respiratory chain subunits described above.
Figure 4.
Cardiac NADH-DH and COX activities in iron treated mice and untreated treated mice. The results are shown as a ratio of NADH-DH/citrate synthase activities (A) and as a ratio of COX/citrate synthase activities (B). In both cases, *p<0.01 (Student’s t test, two-tailed) (n = 5 and 7, respectively).
Discussion
Damage caused by chronic iron overload affects a wide range of tissues. Especially in patients with secondary (transfusion-mediated) iron overload, heart failure is a prominent cause of death [17,18]. The fact that the cardiomyopathy associated with iron overload may only appear years after patients have been iron overloaded suggests a sort of biologic ‘memory’. Iron-mediated oxidation of proteins and lipids should be repaired rather quickly, so the putative 'memory' might involve damage to another cellular component. We have investigated the possibility that iron causes progressive damage to mtDNA, leading to loss of intact mtDNA and, pari pasu, decreased expression of respiratory chain subunits encoded by mtDNA followed by respiratory insufficiency and cardiac dysfunction. Overall, our results support this chain of events.
The present results mirror those we recently reported in a cultured cardiac myocyte cell line exposed to increased iron (in the form of ferric ammonium citrate) [19]. In this earlier work, we similarly found progressive loss of intact mtDNA, decreased mRNAs and protein products for mitochondrially (but not nuclear) encoded genes, loss of activity for complexes I, III and IV and decreased mitochondrial respiration. These changes may have occurred more rapidly (i.e., within 7 days) than those reported here but the results with intact animals only represent a single time point. Nonetheless, the overall process of iron-mediated damage is quite likely the same.
Chronic iron overload leads to heart, liver and pancreatic dysfunction. Interestingly, the liver and heart have high steady-state production of superoxide and hydrogen peroxide [20], largely derived from mitochondrial activity [21]. The pancreatic beta cell is also rich in mitochondria and appears to be highly sensitive to oxidant-generating xenobiotics (such as alloxan and streptozotocin) and exogenous oxidants [22,23]. Histopathologic and ultrastructural studies of livers from patients with iron overload revealed swollen mitochondria having an electron dense matrix and ruptured mitochondrial membranes [24]. In the heart, the early accumulation of iron is seen predominantly in the epicardium [25]. As overload progresses, the stainable iron tends to have a sarcoplasmic localization but, to the best of our knowledge, the precise subcellular distribution of iron has not been well defined. Diastolic dysfunction appears early in the course of iron overload while systolic dysfunction occurs very late [18].
Early in the course of transfusion-mediated iron overload, numerous homeostatic mechanisms likely prevent damage from the accumulating iron. The induction of ferritin can, to an extent, limit the amounts of redox active iron and suppress iron-catalyzed oxidant damage to cells [26,27]. However, there is probably a limit to the amounts of ferritin which can be accumulated and ultimately this homeostatic mechanism may fail. Similarly, antioxidant enzymes may be induced when cells are exposed to the oxidant challenge of 'free' iron coupled with endogenous redox agents. However, increases in ant-ioxidants and/or anti-oxidant enzymes may be of minor importance in protection against oxidant damage which is promoted by transition metals such as iron [28]. In part, this may derive from the paradoxical involvement of intracellular reducing species such as reduced glutathione (GSH) in promotion of iron-mediated oxidative reactions (by facilitating the cyclic reduction of ferric to ferrous iron) [29]. In fact, Giulivi and Candenas found that H2O2-mediated damage to the DNA of isolated mitochondria is actually decreased by prior GSH depletion [30].
Numerous reports indicate that, in addition to synergizing the oxidation of polyunsaturated fatty acids and proteins, “loose” intracellular iron also promotes DNA damage. Cochrane and colleagues reported that oxidant-induced damage to naked DNA and intracellular DNA is greatly enhanced by iron [31,32]. In the absence of transition metals such as iron and copper, DNA is quite unreactive with oxidants such as H2O2 whereas, in the presence of iron, oxidative DNA scission occurs readily [33,34]. The products of reactions involving DNA, iron and oxidants are not yet fully known but do include strand breaks, oxidatively modified bases, DNA-protein cross-links [35,36] and lipid peroxidation products which form covalent adducts with DNA [37]. 32P-postlabelling of hepatic DNA from patients with hereditary hemochromatosis has revealed increased amounts of bulky DNA lesions (of unknown structures) in hepatic DNA from patients with hereditary hemochromatosis [38]. The relative importance of all these products in long-term or irreversible oxidative DNA damage is not yet known.
Compared to nuclear DNA, mtDNA is much more sensitive to oxidant damage [39,40]. It has been reported that at very low fluxes of exogenously generated hydrogen peroxide, only mtDNA damage occurs and nuclear DNA is unscathed [41]. Even in the absence of oxidative stress, the steady-state level of oxidized bases in mtDNA has been reported to be 10–15 times higher than in nuclear DNA [41–43] (although Hegler et al. argue that many estimates of steady-state mtDNA oxidative modifications have been exaggerated [44]). There may be several reasons that mtDNA is so vulnerable. These include the fact that mitochrondria generate reactive oxygen species. Furthermore, mitochondria - the site of final synthesis of heme and iron-sulfur complexes - are intrinsically rich in iron, some of it easily released by oxidants from, e.g., iron-sulfur centers. In addition, mtDNA lacks histones which may help protect nuclear DNA against oxidant damage. Finally, repair of damage to mtDNA is less effective (depending on the types of damage involved) [45]. In the present instance, we are unsure of the exact amounts of intramitochondrial iron present in our iron-loaded mice because technical difficulties precluded precise measures of this parameter.
Why will damage to mtDNA lead to dysfunction of organs such as the heart? In this regard, the pertinent experiments of nature may be inherited mitochondrial diseases (such as myoclonic epilepsy and ragged red fibers [MERRF] and mitochondrial myopathy, encephalomyopathy, lactic acidosis and stroke-like episodes [MELAS]. In these cases, the mitochondrial defects (caused by point mutations interrupting the function of single mitochondrial genes) are heteroplasmic (i.e., present on some but not all copies of mtDNA). In some instances, individuals in whom 50% of mtDNA carries one of these mutations may have no symptoms whatsoever. However, when the frequency of a dysfunctional mitochondrial gene rises to 70–80%, the disease complex - often involving both the nervous system and muscles such as the heart - may suddenly appear [46]. This level of homoplasmy presumably represents the point beyond which metabolic compensation is impossible.
In humans with iron overload, iron-mediated damage to mtDNA in an organ such as the heart may take a period of years but the consequences can become manifest precipitously. In fact, Liu and Olivieri remark on the frequent observation in hemochromatotic patients of a rapid deterioration of systolic function, leading to heart failure and death, which may happen within a few days (with an ejection fraction of >50% falling to <30% over this brief period) [18]. There could, of course, be several interpretations of such a cataclysmic event but one possibility is acute 'mitochondrial failure' arising from years of damage to the mtDNA (similar to the drift towards homoplasmy in disorders such as MERRF and MELAS mentioned above). In the specific case of chronic iron overload, there is the additional possibility that mitochondria with sufficiently damaged genomes may become factories for the production of increased amounts of activated oxygen species which, in turn, further accelerate organ damage. We should note that, at present, we do not know the extent to which the mtDNA damage might be reversed by effective chelator therapy.
The present results strongly support the idea that damage to mtDNA is central to the development of cardiac and, perhaps, hepatic and pancreatic, dysfunction. The consequent, presumably slow, development of respiratory insufficiency leads ultimately to death from cardiac failure. Improved knowledge of the precise pathophysiologic events may prove important in the development of new iron chelators for the treatment of chronic iron overload.
Supplementary Material
Acknowledgments
This work was supported by NIH RO1 DK073586, RO1AA018016 and by the Commonwealth of Kentucky Research Challenge Trust Fund (J.W.E.).
List of Abbreviations
- BSA
bovine serum albumin
- CCCP
carbonyl cyanide m-chlorophenylhydrazone
- EDTA
ethylenediaminetetraacetic Acid
- Ferene S
(3-(2-pyridyl)-5,6-di(2-furyl)-1,2,4-triazine-5',5"-disulfonic acid)
- HBSS
Hanks balanced salt solution
- MOPS
3-(N-morpholino)propanesulfonic acid
- mtDNA
mitochondrial DNA
- ROS
reactive oxygen species
- TCA
trichloroacetic Acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Quinlan GJ, Evans TW, Gutteridge JM. Iron and redox status of the lungs. Free Radic. Biol. Med. 2002;33:1306–1313. doi: 10.1016/s0891-5849(02)00903-6. [DOI] [PubMed] [Google Scholar]
- 2.Ernst SH, Stuart L. Formation, prevention, and repair of DNA damage by iron/hydrogen peroxide. J. Biol. Chem. 1997;272:19095–19098. doi: 10.1074/jbc.272.31.19095. [DOI] [PubMed] [Google Scholar]
- 3.Graf E, Mahoney JR, Bryant RG, Eaton JW. Iron-catalyzed hydroxyl radical formation. Stringent requirement for free iron coordination site. J. Biol. Chem. 1984;259:3620–3624. [PubMed] [Google Scholar]
- 4.Loschen G, Azzi A. Proceedings: Formation of oxygen radicals and hydrogen peroxide in mitochondrial membranes. Hoppe Seylers Z Physiol Chem. 1974;355:1226. [PubMed] [Google Scholar]
- 5.Bartfay WJ, Bartfay E. Iron-overload cardiomyopathy: evidence for a free radical--mediated mechanism of injury and dysfunction in a murine model. Biol. Res. Nurs. 2000;2:49–59. doi: 10.1177/109980040000200106. [DOI] [PubMed] [Google Scholar]
- 6.Qian MW, Eaton JW. Tobacc-borne siderophoric activity. Arch. Biochem. Biophys. 1989;275:280–288. doi: 10.1016/0003-9861(89)90374-3. [DOI] [PubMed] [Google Scholar]
- 7.Eskelinen S, Haikonen M, Raisanen S. Ferene-S as the chromogen for serum iron determinations. Scand. J. Clin. Lab. Invest. 1983;43:453–455. [PubMed] [Google Scholar]
- 8.Artiss JD, Vinogradov S, Zak B. Spectrophotometric study of several sensitive reagents for serum iron. Clin. Biochem. 1981;14:311–315. doi: 10.1016/s0009-9120(81)91065-1. [DOI] [PubMed] [Google Scholar]
- 9.Ayala-Torres S, Chen Y, Svoboda T, Rosenblatt J, Van Houten B. Analysis of gene-specific DNA damage and repair using quantitative polymerase chain reaction. Methods. 2000;22:135–147. doi: 10.1006/meth.2000.1054. [DOI] [PubMed] [Google Scholar]
- 10.Yakes FM, Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc. Natl. Acad. Sci. USA. 1997;94:514–519. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 12.Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 13.Jarreta D, Orus J, Barrientos A, Miro O, Roig E, Heras M, Moraes CT, Cardellach F, Casademont J. Mitochondrial function in heart muscle from patients with idiopathic dilated cardiomyopathy. Cardiovasc, Res. 2000;45:860–865. doi: 10.1016/s0008-6363(99)00388-0. [DOI] [PubMed] [Google Scholar]
- 14.Kruidering M, Van de WB, de Heer E, Mulder GJ, Nagelkerke JF. Cisplatin-induced nephrotoxicity in porcine proximal tubular cells: mitochondrial dysfunction by inhibition of complexes I to IV of the respiratory chain. J. Pharmacol. Exp. Ther. 1997;280:638–649. [PubMed] [Google Scholar]
- 15.Williams AJ, Coakley J, Christodoulou J. Automated analysis of mitochondrial enzymes in cultured skin fibroblasts. Anal. Biochem. 1998;259:176–180. doi: 10.1006/abio.1998.2624. [DOI] [PubMed] [Google Scholar]
- 16.Augustinsson K-B, Eriksson H. The effects of two disulphides on cholinesterase activity in the spectrophotometric assay. Biochem. J. 1974;139:123–127. doi: 10.1042/bj1390123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aldouri MA, Wonke B, Hoffbrand AV, Flynn DM, Ward SE, Agnew JE, Hilson AJ. High incidence of cardiomyopathy in beta-thalassaemia patients receiving regular transfusion and iron chelation: reversal by intensified chelation. Acta Haematol. 1990;84:113–117. doi: 10.1159/000205046. [DOI] [PubMed] [Google Scholar]
- 18.Liu P, Olivieri N. Iron overload cardiomyopathies: new insights into an old disease. Cardiovasc Drugs Ther. 1994;8:101–110. doi: 10.1007/BF00877096. [DOI] [PubMed] [Google Scholar]
- 19.Gao X, Campian JL, Qian M, Sun XF, Eaton JW. Mitochondrial DNA damage in iron overload. J. Biol. Chem. 2009;284:4767–4775. doi: 10.1074/jbc.M806235200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chance B, Sies H, Boveris A. Hydroperoxide metabolism in mammalian organs. Physiol Rev. 1979;59:527–605. doi: 10.1152/physrev.1979.59.3.527. [DOI] [PubMed] [Google Scholar]
- 21.Harman D. Free radical theory of aging: Consequences of mitochondrial aging. Age. 1983;6:86–94. [Google Scholar]
- 22.Olejnicka BT, Ollinger K, Brunk UT. A short exposure to a high-glucose milieu stabilizes the acidic vacuolar apparatus of insulinoma cells in culture to ensuing oxidative stress. APMIS. 1997;105:689–698. doi: 10.1111/j.1699-0463.1997.tb05072.x. [DOI] [PubMed] [Google Scholar]
- 23.Zhang H, Ollinger K, Brunk U. Insulinoma cells in culture show pronounced sensitivity to alloxan-induced oxidative stress. Diabetologia. 1995;38:635–641. doi: 10.1007/BF00401832. [DOI] [PubMed] [Google Scholar]
- 24.Thakerngpol K, Fucharoen S, Boonyaphipat P, Srisook K, Sahaphong S, Vathanophas V, Stitnimankarn T. Liver injury due to iron overload in thalassemia: histopathologic and ultrastructural studies. Biometals. 1996;9:177–183. doi: 10.1007/BF00144623. [DOI] [PubMed] [Google Scholar]
- 25.Olson LJ, Edwards WD, McCall JT, Ilstrup DM, Gersh BJ. Cardiac iron deposition in idiopathic hemochromatosis: histologic and analytic assessment of 14 hearts from autopsy. J. Am. Coll. Cardiol. 1987;10:1239–1243. doi: 10.1016/s0735-1097(87)80124-9. [DOI] [PubMed] [Google Scholar]
- 26.Balla G, Jacob HS, Balla J, Rosenberg M, Nath K, Apple F, Eaton JW, Vercellotti GM. Ferritin: a cytoprotective antioxidant stratagem of endothelium. J. Biol. Chem. 1992;267:18148–18153. [PubMed] [Google Scholar]
- 27.Balla J, Jacob HS, Balla G, Nath K, Eaton JW, Vercellotti GM. Endothelial cell heme uptake from heme proteins: induction of sensitization and desensitization to oxidant damage. Proc. Natl. Acad. Sci. USA. 1993;90:9285–9289. doi: 10.1073/pnas.90.20.9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scott MD, Eaton JW. Markers of free radical-mediated tissue injury: tales of caution and woe. In: Wallace KB, editor. Free Radical Toxicology. Washington, DC: Taylor & Francis; 1997. pp. 401–420. [Google Scholar]
- 29.Scott MD, Eaton JW. Thalassaemic erythrocytes: cellular suicide arising from iron and glutathione-dependent oxidation reactions? Br J Haematol. 1995;91:811–819. doi: 10.1111/j.1365-2141.1995.tb05394.x. [DOI] [PubMed] [Google Scholar]
- 30.Giulivi C, Cadenas E. The role of mitochondrial glutathione in DNA base oxidation. Biochem. Biophys. Acta. 1998;1366:265–274. doi: 10.1016/s0005-2728(98)00125-x. [DOI] [PubMed] [Google Scholar]
- 31.Jackson JH, Schraufstatter IU, Hyslop PA, Vosbeck K, Sauerheber R, Weizman SA, Cochrane CG. Role of oxidants in DNA damage: hydroxyl radical mediates the synergistic DNA damaging effects of asbestos and cigarette smoke. J. Clin. Invest. 1987;80:1090–1095. doi: 10.1172/JCI113165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schraufstatter I, Hyslop PA, Jackson JH, Cochrane CG. Oxidant-induced DNA damage of target cells. J, Clin, Invest. 1988;82:1040–1050. doi: 10.1172/JCI113660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Enright H, Miller WJ, Hebbel RP. Nucleosomal histone protein protects DNA from iron-mediated damage. Nucleic Acids Res. 1992;20:3341–3346. doi: 10.1093/nar/20.13.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Enright H, Hebbel RP, Nath KA. Internucleosomal cleavage of DNA as the sole criterion for apoptosis may be artifactual. J. Lab. Clin. Med. 1994;124:63–68. [PubMed] [Google Scholar]
- 35.Zastawny TH, Altman SA, Randers-Eichhorn L, Madurawe R, Lumpkin JA, Dizdaroglu M, Rao G. DNA base modifications and membrane damage in cultured mammalian cells treated with iron ions. Free Radic. Biol. Med. 1995;18:1013–1022. doi: 10.1016/0891-5849(94)00241-b. [DOI] [PubMed] [Google Scholar]
- 36.Altman SA, Zastawny TH, Randers-Eichhorn L, Cacciuttolo MA, Akman SA, Dizdaroglu M, Rao G. Formation of DNA-protein cross-links in cultured mammalian cells upon treatment with iron ions. Free Radic, Biol, Med. 1995;19:897–902. doi: 10.1016/0891-5849(95)00095-f. [DOI] [PubMed] [Google Scholar]
- 37.Vaca CE, Harms-Ringdahl M. Interaction of lipid peroxidation products with nuclear macromolecules. Biochim. Biophys. Acta. 1989;23(1001):35–43. doi: 10.1016/0005-2760(89)90304-4. [DOI] [PubMed] [Google Scholar]
- 38.Carmichael PL, Hewer A, Osborne MR, Strain AJ, Phillips DH. Detection of bulky DNA lesions in the liver of patients with Wilson's disease and primary haemochromatosis. Mutat. Res. 1995;326:235–343. doi: 10.1016/0027-5107(94)00177-7. [DOI] [PubMed] [Google Scholar]
- 39.Yakes FM, Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc. Natl. Acad, Sci. USA. 1997;94:514–519. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salazar JJ, Van Houten B. Preferential mitochondrial DNA injury caused by glucose oxidase as a steady generator of hydrogen peroxide in human fibroblasts. Mutat. Res. 1997;385:139–149. doi: 10.1016/s0921-8777(97)00047-5. [DOI] [PubMed] [Google Scholar]
- 41.Richter C, Park JW, Ames BN. Normal oxidative damage to mitochondrial and nuclear DNA is extensive. Proc. Natl. Acad. Sci. USA. 1988;85:6465–6467. doi: 10.1073/pnas.85.17.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shigenaga MK, Hagen TM, Ames BN. Oxidative damage and mitochondrial decay in aging. Proc. Natl. Acad. Sci. USA. 1994;91:10771–10778. doi: 10.1073/pnas.91.23.10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zastawny TH, Dabrowska M, Jaskolski T, Klimarczyk M, Kulinski L, Koszela A, Szczesniewicz M, Sliwinska M, Witkowski P, Olinski R. Comparison of oxidative base damage in mitochondrial and nuclear DNA. Free Radic. Biol. Med. 1998;15:722–725. doi: 10.1016/s0891-5849(97)00331-6. [DOI] [PubMed] [Google Scholar]
- 44.Hegler J, Bittner D, Boiteux S, Epe B. Quantification of oxidative DNA modifications in mitochondria. Carcinogenesis. 1993;14:2309–2312. doi: 10.1093/carcin/14.11.2309. [DOI] [PubMed] [Google Scholar]
- 45.LeDoux SP, Wilson GL, Beecham EJ, Stevnsner T, Wassermann K, Bohr VA. Repair of mitochondrial DNA after various types of DNA damage in Chinese hamster ovary cells. Carcinogenesis. 1992;13:1967–1973. doi: 10.1093/carcin/13.11.1967. [DOI] [PubMed] [Google Scholar]
- 46.Wallace DC. Mitochondrial DNA sequence variation in human evolution and disease. Proc. Natl. Acad. Sci. USA. 1994;91:8739–8746. doi: 10.1073/pnas.91.19.8739. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.