Abstract
Transcriptional targeting for cardiac gene therapy is limited by the relatively weak activity of most cardiac-specific promoters. We have developed a bidirectional plasmid vector, which uses a two-step transcriptional amplification (TSTA) strategy to enhance the expression of two optical reporter genes, firefly luciferase (fluc) and Renilla luciferase (hrluc), driven by the cardiac troponin T (cTnT) promoter. The vector was characterized in vitro and in living mice using luminometry and bioluminescence imaging to assess its ability to mediate strong, correlated reporter gene expression in a cardiac cell line and the myocardium, while minimizing expression in non-cardiac cell lines and the liver. In vitro, the TSTA system significantly enhanced cTnT-mediated reporter gene expression with moderate preservation of cardiac specificity. After intramyocardial and hydrodynamic tail vein delivery of an hrluc-enhanced variant of the vector, long-term fluc expression was observed in the heart, but not in the liver. In both the cardiac cell line and the myocardium, fluc expression correlated well with hrluc expression. These results show the vector’s ability to effectively amplify and couple transgene expression in a cardiac-specific manner. Further replacement of either reporter gene with a therapeutic gene should allow non-invasive imaging of targeted gene therapy in living subjects.
Keywords: plasmid vector, bidirectional construct, two-step transcriptional amplification, reporter gene expression, cardiac specificity, bioluminescence imaging
Introduction
Somatic cell gene therapy holds tremendous potential for the treatment of various cardiac diseases including myocardial ischemia, heart failure, and focal arrhythmia.1 Its translation into the clinical setting, however, hinges on showing an acceptable level of safety, which has recently been brought into question after genetic correction of metabolic and hematopoietic disorders.2,3 Transcriptional targeting using cardiac-specific promoters has been proposed as a means to improve the safety profile of cardiac gene therapy by minimizing extra-cardiac expression after vector-mediated delivery of a potentially therapeutic gene.4,5 The motivation for developing cardiac tissue-specific expression systems stems from animal studies of cardiac gene therapy showing the possibility of off-target expression leading to untoward side effects such as lower-extremity edema, hemangioma formation, and proliferative diabetic retinopathy, some of which remain controversial.6 Although promising in theory, transcriptional targeting has been difficult to achieve in practice without a corresponding compromise in therapeutic efficacy because of low expression levels from most cardiac-specific promoters in adult hearts.7 Transcriptional amplification has been attempted by appending tandem repeats of cardiac or viral enhancers to a cardiac-specific promoter or multimerizing the promoter itself.8,9 Unfortunately, these approaches lead to either minimal transcriptional amplification or significant reduction in cardiac specificity. Currently, there is not a single strategy that leads to robust expression of therapeutic genes with specificity for cardiac tissue.
We have earlier validated a two-step transcriptional amplification (TSTA) strategy for enhancing the transcriptional activity of a weak tissue-specific promoter.10,11 In this approach, a weak promoter is used to drive the expression of recombinant GAL4–VP2 transcriptional activators, which can bind to multiple Gal4-binding sites placed upstream of an adenovirus E4 TATA minimal promoter to enhance the transcription of a downstream transgene (for example Figure 1; unidirectional TSTA). In a single vector configuration, this system has been shown to increase transgene expression from the prostate-specific antigen promoter by at least 20-fold.10 More recently, in a proof-of-principle study, we showed using a two-plasmid system the potential of adapting the TSTA system to enhance the expression of two linked transgenes in a bidirectional cassette, with one being a therapeutic gene and the other a reporter gene such as firefly luciferase (fluc).12 Firefly luciferase (FLuc) activity, therefore, becomes an indicator of the expression levels of the therapeutic gene and can be indirectly assessed overtime in living animals by bioluminescence imaging (BLI).13 The construction of a single bidirectional TSTA vector would greatly facilitate the monitoring of therapeutic gene expression in cardiac tissue.
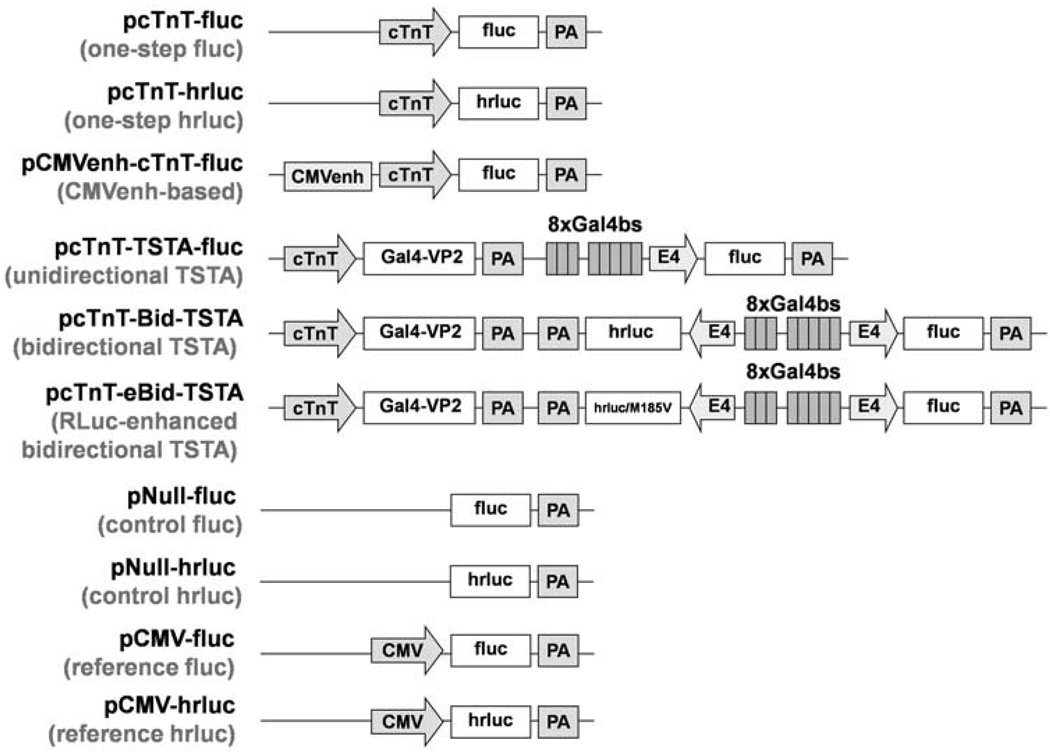
Figure 1.
Diagrams of experimental and control plasmid vectors. Abbreviations: cTnT, rat cardiac troponin T promoter; fluc, firefly luciferase gene; PA, SV40 poly(A) tail; hrluc, synthetic Renilla luciferase gene; CMVenh, cytomegalovirus enhancer; TSTA, two-step transcriptional amplification; Gal4–VP2, fusion gene combining yeast Gal4 and two tandem repeats of herpes simplex virus VP16; 8xGal4bs, 8 tandem repeats of Gal4-binding site; E4, adenovirus E4 minimal promoter; hrluc/M185V, gene expressing the mutant RLuc (RLuc/M185V) with enhanced light output; CMV, cytomegalovirus promoter.
The aim of this study was to develop a versatile bidirectional TSTA vector capable of transcriptional targeting as well as amplifying the coupled expression of two transgenes. For this purpose, we specifically constructed a bidirectional plasmid vector (pcTnT-Bid-TSTA), which uses a weak, truncated cardiac troponin T (cTnT) promoter in conjunction with the TSTA system to drive the expression of two optical reporter genes, fluc and Renilla luciferase (hrluc) (Figure 1; bidirectional TSTA). Here, we show using in vitro reporter enzyme assays and in vivo BLI that the bidirectional TSTA vector and its Renilla luciferase (RLuc)-enhanced variant (pcTnT-eBid-TSTA), which has been optimized for in vivo imaging, can lead to robust, cardiac-specific, and correlated reporter gene expression in cell culture and in living mice, respectively. With further refinement of the vector by replacing one of the reporter genes with a therapeutic gene of interest (for example hypoxia-inducible factor-1α; HIF-1α), the bidirectional TSTA vector should enable non-invasive BLI monitoring of targeted gene therapy in living animal models of human heart disease.
Results
Construction of experimental and control plasmid vectors
Several plasmid vectors were constructed in this study to allow bioluminescence assessment of the bidirectional TSTA vector strategy. These include one-step, cytomegalovirus immediate-early enhancer (CMVenh)-based, unidirectional TSTA, bidirectional TSTA, control, and reference vectors (Figure 1). A one-step vector (pcTnT-fluc or pcTnT-hrluc) uses the cardiac-specific cTnT promoter (−303 to +192 bp14) to drive either fluc expression or hrluc expression. The CMVenh-based vector (pCMVenh-cTnT-fluc) differs from the one-step fluc vector (pcTnT-fluc) in that a CMVenh is placed upstream of cTnT to enhance fluc expression. The unidirectional TSTA vector (pcTnT-TSTA-fluc) uses the cTnT promoter to drive the expression of a Gal4–VP2 fusion gene, whose translated products (GAL4–VP2) can bind to any of the eight tandem repeats of Gal4-binding site (8xGal4bs) to amplify the expression of fluc placed immediately downstream of an adenovirus E4 minimal promoter. The bidirectional TSTA vector (pcTnT-Bid-TSTA) differs from the unidirectional TSTA vector in that an additional E4 promoter is placed on the opposite side of 8xGal4bs from fluc to direct hrluc expression. In such a construct, binding of GAL4–VP2 fusion proteins to 8xGal4bs leads to bidirectional expression of fluc and hrluc. The RLuc-enhanced bidirectional TSTA vector (pcTnT-eBid-TSTA) differs from pcTnT-Bid-TSTA in that it expresses, instead of RLuc, a single mutation variant of RLuc (RLuc/ M185V) that can generate three times more light in the presence of its native substrate (coelenterazine) than RLuc.15 A control vector (pNull-fluc or pNull-hrluc) is promoter-less and contains only fluc or hrluc. A reference vector (pCMV-fluc or pCMV-hrluc) uses the constitutive cytomegalovirus (CMV) promoter to drive either fluc expression or hrluc expression. To test the generalizability of the amplification strategies, two additional sets of experimental vectors, driven by either a truncated myosin light chain 2 promoter (MLC2v; −250 to +18 bp)16 or a truncated α-myosin heavy chain promoter (αMHC; −373 to 16 bp),17 were constructed (Supplementary Figure S1). The RLuc-enhanced versions of the bidirectional TSTA vectors shown in Supplementary Figure S1 (pMLC2v-eBid-TSTA and pαMHC-eBid-TSTA; not shown) were also constructed for comparison with pcTnT-eBid-TSTA.
Comparison of TSTA and CMVenh-based strategies for transcriptional amplification
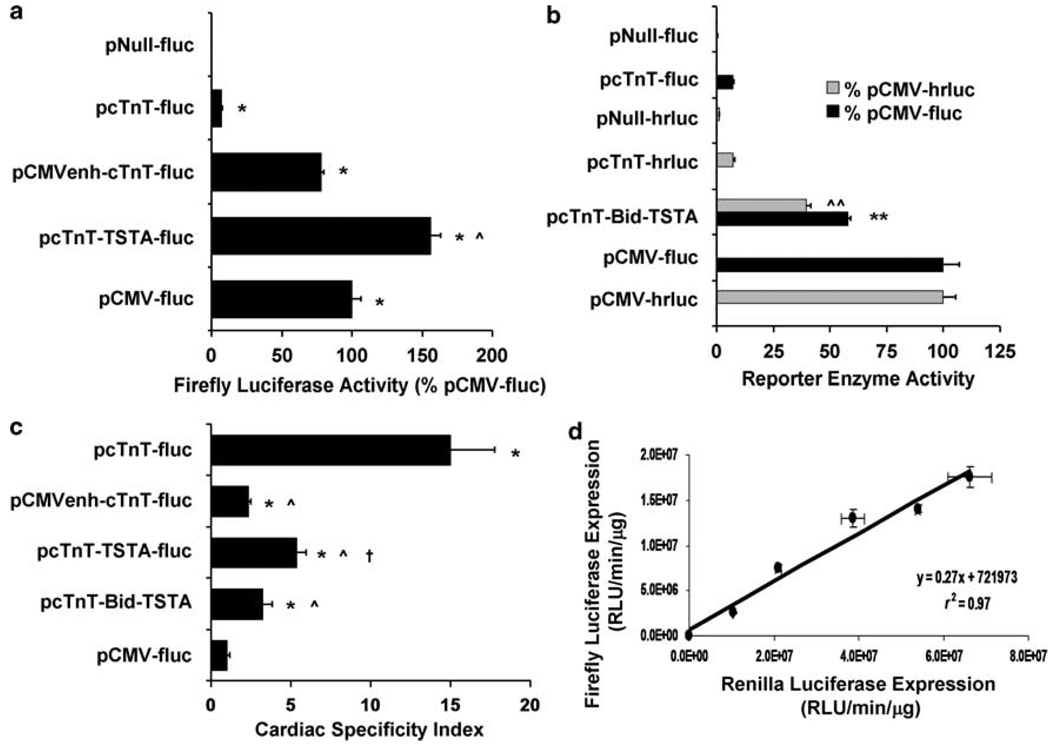
To evaluate TSTA’s ability to enhance the transcriptional activity of a weak cardiac-specific promoter, we tested the performance of TSTA against another promising transcriptional amplification approach, the CMVenh-based strategy, in cell culture assays. Specifically, we transfected HL-1 cardiac cells separately with control (pNull-fluc), one-step (pcTnT-fluc), CMVenh-based (pCMVenh-cTnT-fluc), unidirectional TSTA (pcTnT-TSTA-fluc), and reference (pCMV-fluc) vectors, with each vector co-delivered with a normalization vector (pCMV-β-gal), which expresses β-galactosidase (β-GAL) under the regulation of a CMV promoter. Twenty-four hours later, the cell lysates were assayed for FLuc, RLuc, and β-GAL activities using in vitro reporter enzyme assays. In comparison with pCMV-fluc, the FLuc activity of pcTnT-fluc was found to be notably low (7.3±0.2%), confirming the weak transcriptional capacity of cTnT (Figure 2a). With CMVenh cloned upstream of cTnT, the FLuc activity was significantly enhanced for pCMVenh-cTnT-fluc (78.3±2.0%; P<0.001). Transcriptional amplification was even greater with TSTA such that pcTnT-TSTA-fluc led to a greater FLuc activity than pCMV-fluc (156.3±6.7%; P < 0.004 compared with pCMV-fluc and P < 0.001 compared with pCMVenh-cTnT-fluc). The total fold amplification was 10.8±0.7-fold for the CMVenh-based strategy and 21.5±1.8-fold for the TSTA strategy. pNull-fluc led to only a minimal level of FLuc activity (0.2±0.0%). When the same experiments were performed using similar vectors that carry different cardiac-specific promoters (MLC2v and αMHC) (Supplementary Figure S1), it was found that both strategies could enhance transcription by at least 10-fold, irrespective of the promoter used (Supplementary Table S1). The CMVenh-based strategy could amplify the strength of MLC2v (116.1±12.1-fold) significantly better (P<0.03) than the TSTA strategy (60.5±8.8-fold), whereas the latter strategy could enhance the activity of αMHC (27.5±3.5-fold) significantly more (P<0.05) than the former (10.6±0.4-fold). The level of FLuc activity achieved by the unidirectional TSTA vectors qualitatively tracked with the strength of their respective promoters (pcTnT-TSTA-fluc >pαMHC-TSTA-fluc > pMLC2v-TSTA-fluc and pcTnT-fluc > pαMHC-fluc > pMLC2v-fluc; P<0.02 for all comparisons except for that between pcTnT-TSTA-fluc and pαMHC-TSTA-fluc (P = NS)).
Figure 2.
In vitro characterizations of the TSTA-based strategies in terms of transcriptional amplification, cardiac specificity, and correlated reporter gene expression. Cardiac (HL-1) and non-cardiac cells were separately transfected with control (pNull-fluc, pNull-hrluc), one-step (pcTnT-fluc, pcTnT-hrluc), CMVenh-based (pCMVenh-cTnT-fluc), unidirectional TSTA (pcTnT-TSTA-fluc), bidirectional TSTA (pcTnT-Bid-TSTA), and reference (pCMV-fluc, pCMV-hrluc) vectors, with each vector co-delivered with a normalization vector (pCMV-β-gal). The cell lysates were assayed 24 h later for FLuc, RLuc, and β-GAL activities. The FLuc and RLuc activities were normalized to total protein, corrected for transfection efficiency (β-GAL activity), and, respectively, expressed as a percentage of FLuc activity of pCMV-fluc and RLuc activity of pCMV-hrluc. (a) The FLuc activity is shown for all fluc-containing vectors except for pcTnT-Bid-TSTA, whose FLuc (black solid bar) and RLuc (gray solid bar) activities are shown in (b) and compared against those of fluc- and hrluc-containing control, one-step, and reference vectors. *P< 0.005 compared with pNull-fluc; ^P< 0.001 compared with pCMVenh-cTnT-fluc; **P< 0.001 compared with pcTnT-fluc; ^^P< 0.002 compared with pcTnT-hrluc. (c) The cardiac-specificity index (CSI), derived from FLuc activity, is shown for each vector except for pNull-fluc, whose CSI could not be fairly assessed mathematically because of its near-zero expression in both cardiac and non-cardiac cell lines. *P< 0.04 compared with pCMV-fluc; ^P< 0.03 compared with pcTnT-fluc; †P< 0.01 compared with pCMVenh-cTnT-fluc. (d) HL-1 cells transfected with increasing doses of pcTnT-Bid-TSTA were assayed 24 h later for both FLuc and RLuc activities, which were normalized by total protein and plotted here against each other for each plasmid dose used. For all figures, the error bars represent s.e.m. for triplicate determinations.
To investigate whether TSTA used in a bidirectional vector can significantly enhance the expression of two transgenes, we performed the same experiments on HL-1 cardiac cells using the bidirectional TSTA vector, pcTnT-Bid-TSTA, as well as additional hrluc-expressing control (pNull-hrluc), one-step (pcTnT-hrluc), and reference (pCMV-hrluc) vectors for comparison of hrluc expression in terms of RLuc activity. The FLuc activity of pcTnT-Bid-TSTA was greater than that of pcTnT-fluc (7.3±0.2%; P<0.001), reaching 57.9±1.2% of pCMV-fluc. Similarly, the RLuc activity of pcTnT-Bid-TSTA (39.7±1.9% of pCMV-hrluc) was significantly greater than that of pcTnT-hrluc (7.2±0.9%; P<0.0002) (Figure 2b). The FLuc activity of pcTnT-Bid-TSTA was significantly less than that of its unidirectional counterpart, pcTnT-TSTA-fluc (156.3±6.7%; P<0.0002). This qualitative relationship was also observed between unidirectional and bidirectional TSTA vectors containing other cardiac-specific promoters tested (P<0.03) (Supplementary Table S1). In the absence of the cTnT promoter, the bidirectional TSTA vector (pNull-Bid-TSTA; not shown) led to only negligible FLuc (1.56±0.35% of pCMV-fluc) and RLuc (0.56±0.04% of pCMV-hrluc) activities when tested under the same experimental conditions (data not shown).
Assessment of the effects of TSTA and CMVenh-based strategies on cardiac specificity
To study the influence of TSTA and CMVenh-based strategies on the cardiac specificity conferred by the cTnT promoter, we transfected four different cell lines (HL-1 cardiac cells, C2C12 skeletal myoblasts, Hepa1-6 hepatoma, and NIH3T3 embryonic fibroblasts) separately with control (pNull-fluc), one-step (pcTnT-fluc), CMVenh-based (pCMVenh-cTnT-fluc), unidirectional TSTA (pcTnT-TSTA-fluc), bidirectional TSTA (pcTnT-Bid-TSTA), and reference (pCMV-fluc) vectors, with each vector co-delivered with a normalization vector (pCMV-β-gal). The cell lysates were assayed 24 h later for FLuc, RLuc, and β-GAL activities using in vitro reporter enzyme assays. A cardiac-specificity index (CSI) was calculated for every vector to assess its ability to specifically express fluc in cardiac cells relative to non-cardiac cells, compared with pCMV-fluc.
As expected, the CSI was found to be the greatest for pcTnT-fluc (15.0±2.8) and the least for pCMV-fluc (1.0±0.2) (Figure 2c), confirming the cardiac specificity of cTnT and the constitutive nature of the CMV immediate-early promoter. All vectors containing cTnT had a greater CSI than pCMV-fluc (P<0.04). Compared with the expression from cTnT when there is no amplification, the CSI was modest when amplified using either the unidirectional TSTA strategy (pcTnT-TSTA-fluc; 5.4±0.6; P<0.03) or the bidirectional TSTA strategy (pcTnT-Bid-TSTA; 3.3±0.6; P<0.02), and worse when amplified using the CMVenh-based strategy (pCMVenh-cTnT-fluc; 2.4±0.2; P<0.05). The CSI of pcTnT-TSTA-fluc was significantly greater than that of pCMVenh-cTnT-fluc (P<0.01), but not statistically different from that of pcTnT-Bid-TSTA (P = NS). Qualitatively, the CSI results also extended to bidirectional TSTA vectors containing other cardiac-specific promoters, namely MLC2v and αMHC (Supplementary Table S2). There was a trend for the cardiac specificity of a given promoter to decrease when amplified using either TSTA strategy and to drop further when enhanced using the CMVenh-based strategy. The bidirectional TSTA strategy could in some cases significantly reduce the cardiac specificity of a promoter more than its unidirectional counterpart (P<0.04 for MLC2v- and αMHC-based vectors).
Correlation of reporter gene expression in HL-1 cardiac cells
To study whether the bidirectional TSTA vector (pcTnT-Bid-TSTA) can couple the expression of fluc and hrluc, we transfected HL-1 cells with increasing doses of the vector and assayed for both FLuc and RLuc activities 24 h later. A high degree of linear correlation was observed between the enzymatic activities of FLuc and RLuc (r2 = 0.97; P<0.001; Figure 2d).
BLI of pcTnT-eBid-TSTA-mediated cardiac reporter gene expression after intramyocardial vector delivery
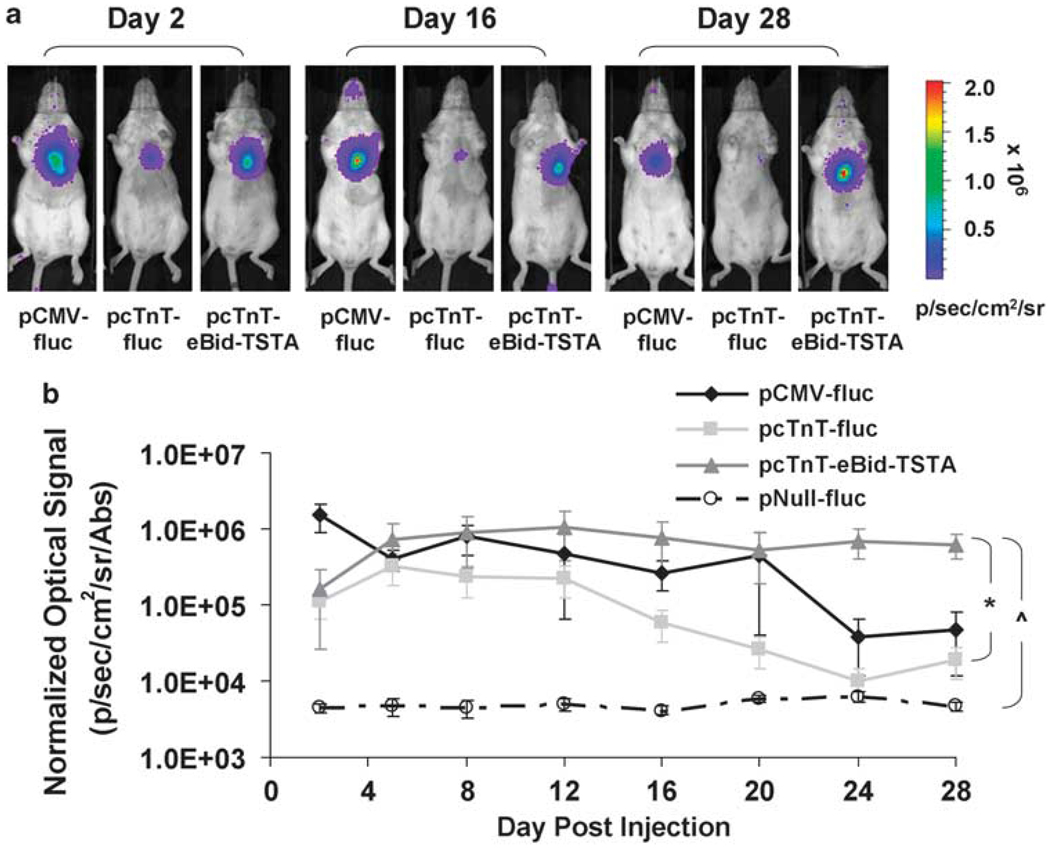
To assess the degree of transcriptional amplification achievable with the bidirectional TSTA strategy in living subjects, we first mutated the hrluc gene of our bidirectional vector to enhance the RLuc activity, so it can be more easily imaged in living mice. The mutant RLuc (RLuc/M185V) has three times greater light-generating capacity than the wild-type RLuc.15 To characterize the RLuc-enhanced bidirectional TSTA vector (pcTnT-eBid-TSTA) in vivo, we performed serial BLI for 4 weeks (days 2, 5, 8, 12, 16, 20, 24, and 28) on five cohorts of mice (n = 5 per group), which had undergone intramyocardial injection of (1) pCMV-fluc, (2) pcTnT-fluc, (3) pcTnT-eBid-TSTA, (4) pNull-fluc, or (5) pcTnT-TSTA-fluc (all at 80 µg per mouse; 16 µg per mouse of pCMV-β-gal was co-injected with each vector to correct for in vivo transfection efficiency). pNull-fluc served as negative control for the experiment. BLI revealed a strong FLuc-mediated heart signal for pCMV-fluc on day 2 (1.5±0.6×106 photons s−1 cm−2 sr−1 per absorbance (Abs)), compared with cTnT-eBid-TSTA (1.6±1.4×105 photons s−1 cm−2 sr−1 per Abs) and pcTnT-fluc (1.1±0.5×105 photons s−1 cm−2 sr−1 per Abs) (Figures 3a and b). pNull-fluc showed only background signal on day 2 (4.4±0.7×103 photons s−1 cm−2 sr−1 per Abs) and for the remainder of the study period. The average heart signal declined gradually over the next 4 weeks for both pCMV-fluc and pcTnT-fluc, but less so for pcTnT-eBid-TSTA. By day 28, the signal for pcTnT-eBid-TSTA (6.3±2.3×105 photons s−1 cm−2 sr−1 per Abs) became the greatest at 27.9±13.9-fold of pcTnT-fluc (1.9±0.9×104 photons s−1 cm−2 sr−1 per Abs) (P<0.02). Considering the entire study period, a significant difference was observed between pcTnT-eBid-TSTA and pcTnT-fluc (P<0.012), but not between pcTnT-eBid-TSTA and pCMV-fluc (P = NS) or between pcTnT-eBid-TSTA and pcTnT-TSTA-fluc (P = NS; Supplementary Figure S2a). All vectors led to an average signal significantly greater than pNull-fluc (P<0.007).
Figure 3.
Bioluminescence imaging of pcTnT-eBid-TSTA-mediated cardiac reporter gene expression in living mice. Serial BLI was performed on four mouse cohorts, which had separately undergone intramyocardial co-injections of (1) pCMV-fluc+pCMV-β-gal, (2) pcTnT-fluc+pCMV-β-gal, (3) pcTnT-eBid-TSTA+pCMV-β-gal, or (4) pNull-fluc+pCMV-β-gal. (a) The BLI images of three representative mice that received pCMV-fluc (left), pcTnT-fluc (middle), and pcTnT-eBid-TSTA (right), respectively, are shown for only days 2, 16, and 28. Each image is displayed on a rainbow scale in units of photons s−1 cm−2 sr−1) and overlaid onto a grayscale reference image of the corresponding mice. (b) The average heart signal on the post-operative days indicated was corrected for transfection efficiency [β-GAL activity in units of absorbance (Abs)] and plotted for each mouse cohort: pCMV-fluc (solid black diamond), pcTnT-fluc (light gray square), pcTnT-eBid-TSTA (dark gray triangle), and pNull-fluc (empty circle). Note that the signal is displayed on a log scale. The error bars represent s.e.m. for five mice. When considering the entire study period, *P< 0.012 between pcTnT-eBid-TSTA and pcTnT-fluc; ^P< 0.007 between pcTnT-eBid-TSTA and pNull-fluc.
BLI of hepatic reporter gene expression after hydrodynamic tail vein injection of pcTnT-eBid-TSTA
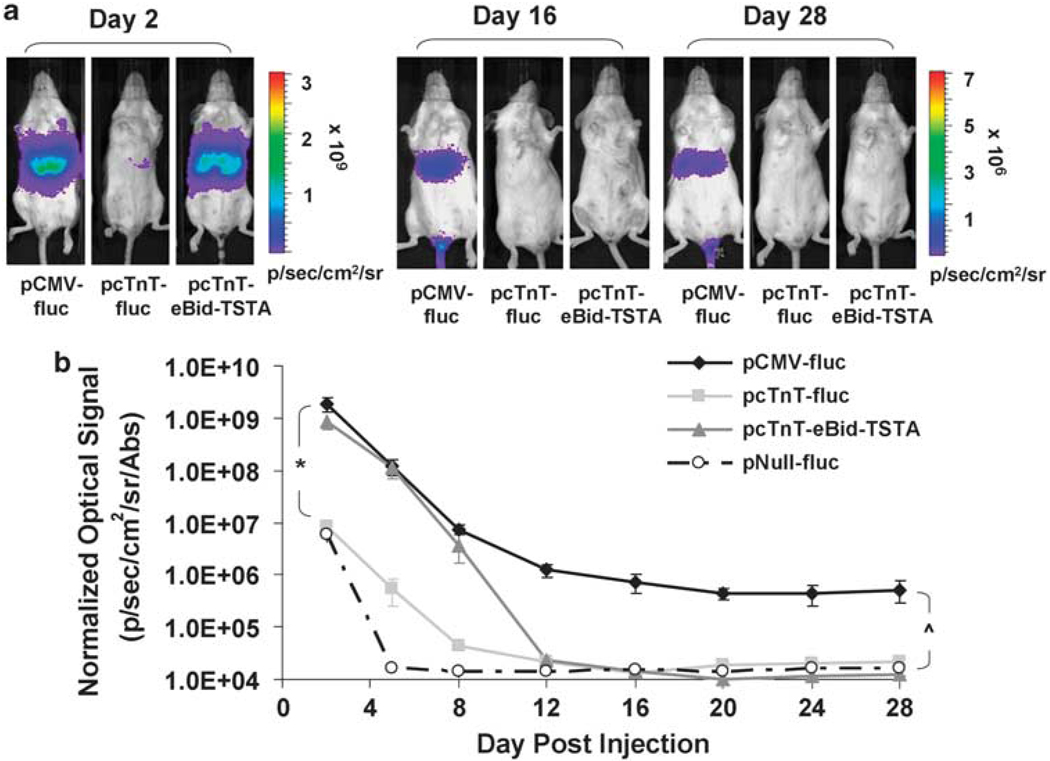
To assess whether pcTnT-eBid-TSTA can effectively minimize extra-cardiac transgene expression, we performed serial BLI (days 2, 5, 8, 12, 16, 20, 24, and 28) on four mouse cohorts (n = 5 per group) that received hydro-dynamic tail vein (HTV) injection of (1) pCMV-fluc, (2) pcTnT-fluc, (3) pcTnT-eBid-TSTA, or (4) pNull-fluc. pCMV-β-gal (4 µg per mouse) was co-delivered with each vector to correct for in vivo transfection efficiency. HTV injection is an established technique for transfecting hepatocytes in vivo18 and was used here to test the hypothesis that cTnT, a cardiac-specific promoter, would lead to minimal transgene expression in the liver. BLI revealed strong FLuc-mediated hepatic signal for both pCMV-fluc (1.9±0.6×109 photons s−1 cm−2 sr−1 per Abs) and pcTnT-eBid-TSTA (8.4±2.4×108 photons s−1 cm−2 sr−1 per Abs) initially on day 2, compared with much less signal for pcTnT-fluc (8.2±1.5×106 photons s−1 cm−2 sr−1 per Abs) and pNull-fluc (5.8±1.5×106 photons s−1 cm−2 sr−1 per Abs) (Figures 4a and b). In all groups, the hepatic signal dropped drastically over the first 12 days until it stabilized or fell below the detection threshold. Before day 12, no statistical difference (P = NS) was found between pCMV-fluc and pcTnT-eBid-TSTA, or between pcTnT-fluc and pNull-fluc, whereas a significant difference was observed between the former and latter pairs (P<0.0001). After day 12, significant hepatic signal was only associated with pCMV-fluc (P<0.0001), but not with other groups, which were not significantly different from each other (P = NS).
Figure 4.
Bioluminescence imaging of pcTnT-eBid-TSTA-mediated hepatic reporter gene expression in living mice. Serial BLI was performed on four mouse cohorts, which had separately undergone hydrodynamic tail vein co-injections of (1) pCMV-fluc+pCMV-β-gal, (2) pcTnT-fluc+pCMV-β-gal, (3) pcTnT-eBid-TSTA+pCMV-β-gal, or (4) pNull-fluc+pCMV-β-gal. (a) The BLI images of three representative mice, one from each of the groups that received pCMV-fluc (left), pcTnT-fluc (middle), and pcTnT-eBid-TSTA (right), are shown for days 2, 16, and 28. Each image is displayed on a rainbow scale in units of photons s−1 cm−2 sr−1 and overlaid onto a grayscale reference image of the corresponding mouse. (b) The average hepatic signal on the post-operative days shown was normalized for transfection efficiency (β-GAL activity) and plotted for the different mouse groups: pCMV-fluc (solid black diamond), pcTnT-fluc (light gray square), pcTnT-eBid-TSTA (dark gray triangle), and pNull-fluc (empty circle). Note that the signal is displayed on a log scale. The error bars represent s.e.m. for five mice. *P< 0.0001 between pcTnT-eBid-TSTA (or pCMV-fluc) and pcTnT-fluc (or pNull-fluc) before day 12. ^P< 0.0001 between pCMV-fluc and all other vectors after day 12.
BLI performed on mice injected with (1) pcTnT-TSTA-fluc, (2) pαMHC-eBid-TSTA, or (3) pMLC2v-eBid-TSTA showed similar patterns of decline in hepatic signal after HTV vector administration. Notably, pcTnT-TSTA-fluc led to an initially greater (P<0.02) signal on day 2 than that of pcTnT-eBid-TSTA, but this difference became statistically insignificant (P = NS) by day 16, when the signal reached background levels (Supplementary Figure S2b). Compared with pcTnT-eBid-TSTA, both pαMHC-eBid-TSTA and pMLC2v-eBid-TSTA led to significant hepatic signal initially which then decreased to background levels by day 20, beyond which persistent signal was only observed for pCMV-fluc (P < 0.0001 compared with all other groups) (Supplementary Figure S3a).
Assessment of HTV-induced hepatic cTnT expression by quantitative reverse transcription–polymerase chain reaction analysis
To study whether the HTV procedure itself can acutely activate non-specific gene transcription in hepatocytes, we subjected mice to HTV injection of phosphate-buffered saline (PBS) and assessed the hepatic mRNA levels of cTnT and β-actin 48 h later using ex vivo quantitative reverse transcription–polymerase chain reaction (qRT–PCR). The cTnT and β-actin mRNA levels (as inferred from the average fluorescence intensity) were found to be greater (P<0.05) in the liver samples of mice injected with PBS, compared with those of untreated mice (Supplementary Figure S3b), after 26 and 44 PCR cycles, respectively. In both PBS-treated and untreated mouse groups, the average fluorescence intensity associated with β-actin mRNA saturates after cycle number 33.
Correlation of reporter gene expression in the mouse myocardium
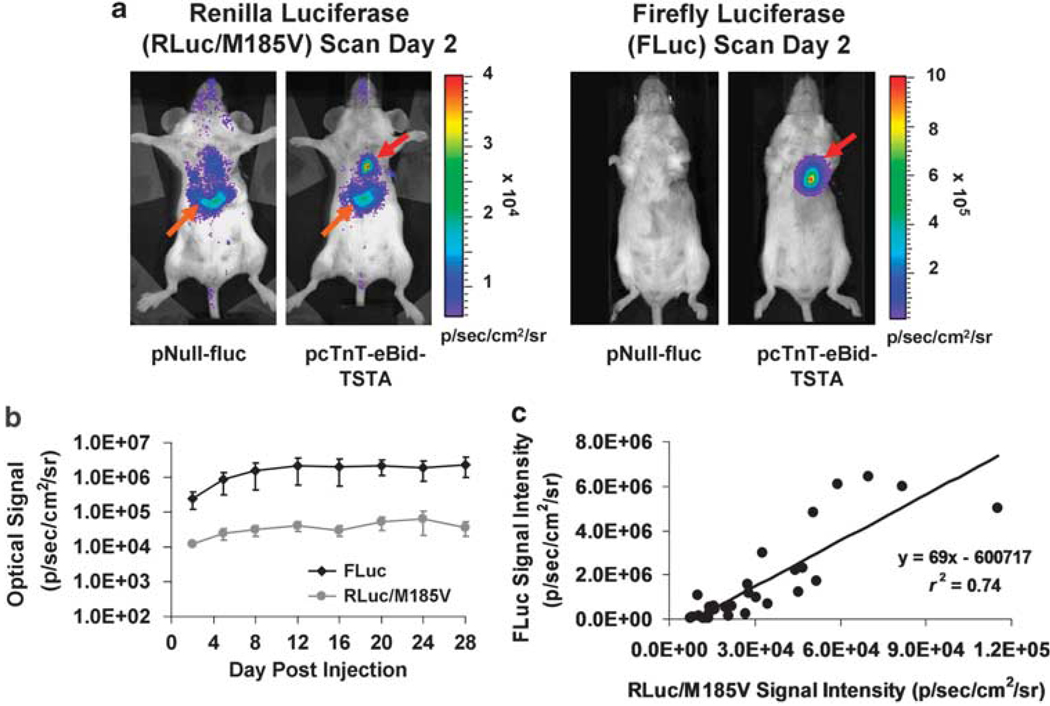
Longitudinal BLI was performed on mice that received pcTnT-eBid-TSTA to assess the long-term fluc and hrluc/M185Vexpression in the mouse myocardium. As early as 2 days after intramyocardial gene delivery, strong FLuc-and RLuc/M185V-mediated heart signals were observed in the experimental mice, compared with minimal levels of signal in the control mice that received pNull-fluc (Figure 5a). Both the average FLuc signal and the average RLuc/M185V signal increased gradually over a week and stabilized at high levels for the remainder of the study period (Figure 5b), during which the RLuc/M185V and FLuc-mediated signals correlated well with each other (Figure 5c; r2 = 0.74; P<0.001). The ex vivo FLuc and RLuc/M185V activities of the explanted mouse hearts at various time points (days 2, 16, and 28) after pcTnT-eBid-TSTA injection also correlated strongly with each other (Supplementary Figure S4; r2 = 0.93; P<0.0005).
Figure 5.
Correlated cardiac reporter gene expression from pcTnT-eBid-TSTA after intramyocardial vector delivery. (a) Two representative mice were intramyocardially injected with either pcTnT-eBid-TSTA (experimental) or pNull-fluc (control) and scanned for both hrluc/M185V and fluc expression 2 days later using BLI. Both RLuc/M185V (left two images) and FLuc (right two images) images (rainbow scale and grayscale for luminescence and reference images, respectively) show a robust heart signal (red arrow) for the experimental pcTnT-eBid-TSTA mouse and minimal to no signal for the pNull-fluc control mouse. Note that for the dose of coelenterazine used in this study (50 µg), a background hepatic signal (orange arrows) can always be seen on the RLuc/M185V images of both control and experimental mice. (b) Serial BLI of the entire mouse cohort injected with cTnT-eBid-TSTA revealed the kinetics of hrluc/M185V and fluc expression over 28 days. The average RLuc/M185V- and FLuc-mediated signals are displayed on a log scale. The error bars represent s.e.m. for five mice. (c) The FLuc signal at each time point is plotted against the corresponding RLuc/M185V signal. All signal measures are expressed in units of photons s−1 cm−2 sr−1.
Discussion
In this study, we used a systematic approach to characterize a bidirectional TSTA plasmid vector (pcTnT-Bid-TSTA) capable of amplifying the expression of two transgenes (fluc and hrluc) under the regulation of a weak cardiac-specific promoter (cTnT). We first verified in vitro the superiority of the TSTA strategy in both mediating robust cardiac-specific transcriptional amplification and minimizing transgene expression in non-cardiac cell lines, by comparing it with another promising strategy that is CMVenh based. We then showed in cell culture the ability of this vector to achieve transcriptional amplification by as much as 7.3-fold in a cardiac-specific manner. After modification of the RLuc moiety, the optimized vector (pcTnT-eBid-TSTA) was found to exhibit excellent long-term transcriptional targeting and transcriptional amplification after delivery into living mice, as well as the ability to lead to highly correlated reporter gene expression. The development of such a versatile vector, with further optimization, should set the groundwork for future attempts to monitor targeted cardiac gene therapy in living subjects using a reporter surrogate.
To our knowledge, this is the first study in which various cardiac-specific promoters were tested for their compatibility with the TSTA system. Our results here illustrate that the TSTA system is fairly generalizable and works the best with cTnT, followed by αMHC and then MLC2v. In most cases, TSTA can enhance the transcription of a single gene to levels beyond that can be achieved with the strong CMV promoter. Consistent with earlier studies showing 10–25-fold amplification with TSTA,10,19 the unidirectional TSTA vectors used in this study achieved comparable degrees of amplification for cTnT and αMHC (22-fold), and an even greater amplification for MLC2v (61-fold), the weakest promoter tested, suggesting the propensity of TSTA to better amplify a weaker promoter. For practical concerns, however, the strongest possible promoter should still be used as our results showed that stronger promoters tend to mediate greater overall transgene expression. Although proven to be better in performance than the CMVenh-based approach, the TSTA strategy is more complex in design because of the inclusion of multiple elements besides the promoter and the reporter gene (Gal4VP2 gene, 8xGal4bs, E4 promoter). The large size of the TSTA construct (7.1 kb for pcTnT-TSTA-fluc versus 5.3 kb for pCMVenh-cTnT-fluc) may become an issue to consider when an additional transgene is included, as in the case for bidirectional vectors (for example, 8.6 kb for pcTnT-Bid-TSTA), making potential incorporation of the construct into a viral vector more difficult because of the insert size limit of most common viral vectors (for example, 8 kb for adenovirus, 4 kb for adeno-associated virus). A potential solution to this is the use of viral backbones that can accept large transgene inserts (for example, up to 30 kb for ‘gutless’ adenovirus).
The development of a bidirectional TSTA vector was based on the premise that a reporter gene linked to a therapeutic gene would allow non-invasive monitoring of the therapeutic gene expression.20,21 Two optical reporter genes were included in our bidirectional construct so that BLI could be better used to serially examine the in vivo correlation between the two transgene expression. Compared with its unidirectional counterpart, our bidirectional vector, pcTnT-Bid-TSTA, achieved significantly less (2.7-fold) transcriptional amplification for each transgene. Nevertheless, the in vitro fluc and hrluc expression achieved by the vector were still 58% and 40% of pCMV-hrluc and pCMV-fluc, respectively, more than sufficient for in vivo imaging with BLI. The use of the bidirectional TSTA strategy, however, was associated with a moderate reduction in cardiac specificity (4.6-fold), compared with the case without amplification. This drop was likely overestimated because the CSI analysis was performed on three non-cardiac cells lines, one of which (C2C12 skeletal myoblasts) could have a transcriptional profile for the muscle proteins (MLC2v, cTnT, αMHC) similar to that of HL-1 cells. A CSI analysis excluding C2C12 cells would have led to a greater CSI and an even lesser reduction in cardiac specificity (3.5-fold; data not shown). This level of cardiac specificity, however, would imply a potentially 30% off-target transgene expression in vivo. For eventual gene therapy applications, additional strategies to improve the overall cardiac specificity of the bidirectional TSTA vector should be used, and these may include (1) using a promoter more cardiac specific than pcTnT (for example, the cardiac sodium-calcium exchanger promoter; NCX1)22 to drive the TSTA system or (2) placing a ‘pause signal’23 upstream of the cTnT promoter to minimize transcriptional read-through and unintentional transcription of both Gal4-VP16 and fluc in non-cardiac cells. Ultimately, the bidirectional TSTA transcriptional targeting strategy presented herein can be combined with various existing transductional targeting strategies (for example, redirecting adenoviral tropism)24 to achieve the utmost cardiac specificity in vivo after intramyocardial vector administration. Nevertheless, considering a six- to eightfold gain in the transcriptional amplification of each transgene, the net benefit of choosing the bidirectional TSTA construct over the one-step vector is evident. Given the fact that in a true therapy application there may not be an imaging probe available for ‘directly’ imaging transgene expression, the benefit of being able to ‘indirectly’ monitor the expression of the therapeutic gene through a surrogate reporter gene should outweigh the compromise in transcriptional amplification when choosing a bidirectional TSTA design over its one-step or unidirectional counterpart. Although not suitable for ‘indirect’ imaging of therapeutic gene expression, the robust unidirectional TSTA vector strategy can be useful for studying the downstream effects of gene therapy. For instance, when replaced with a vascular endothelial growth factor promoter and expressed in transgenic mice, such a unidirectional TSTA strategy can be used to non-invasively image the kinetics of stimulated myocardial angiogenesis (vascular endothelial growth factor upregulation) after angiogenic gene delivery.25
The transcriptional amplification of the RLuc-enhanced bidirectional TSTA vector (pcTnT-eBid-TSTA) was excellent in vivo, reaching ∼28-fold of the one-step vector (pcTnT-fluc) at 28 days post-injection, statistically comparable with that achieved by the unidirectional TSTA vector. After HTV injection of the vector, the hepatic transgene expression, although initially high, declined drastically over 2 weeks to background levels by day 12, leading to an excellent long-term tissue specificity comparable with that of the one-step vector, pcTnT-fluc. The unexpectedly high level of hepatic expression initially is not necessarily reflective of poor tissue specificity and can be partially attributed to the HTV procedure itself, which is known to both upregulate activator protein-1 transcriptional factor and lead to significant activation of many promoters that contain activator protein-1 or activator protein-1-like-binding sites (for example, human elongation factor 1),26 including the cTnT promoter.27 This is supported by our qRT–PCR experiments showing acute upregulation of cTnT mRNA, in addition to β-actin mRNA, 48 h after HTV injection of PBS. The fact that significantly elevated fluc expression was also seen shortly after HTV injection of bidirectional TSTA vectors driven by other cardiacspecific promoters (αMHC and MLC2v) further confirms the non-specific nature of promoter activation after the harsh HTV procedure. Thus, only long-term tissue specificity could be fairly assessed when HTV is used for gene delivery. More studies, however, will be needed to determine the optimal time point (for example, >12 or 16 days) beyond which the interpretation of tissue specificity is least confounded by the HTV procedure.
BLI assessment of RLuc-enhanced bidirectional TSTA vector in the heart was made possible partially by the improved light-generating ability of RLuc/M185V, compared with RLuc.15 To date, no other studies have reported successful imaging of hrluc expression in the heart of living rodents with the use of BLI, mainly because the suboptimal, blue-wavelength photons emitted by RLuc when coelenterazine is used as its substrate (λpeak = 480 nm) are strongly attenuated by the chest wall.28 By comparison, fluc expression can be more easily imaged because the red-wavelength photons generated by FLuc (λpeak = 612 nm) are less attenuated by tissue.28 Another impediment to imaging hrluc expression in the heart is the high hepatic background signal (likely related to albumin-mediated auto-oxidation of coelenterazine)29 that often makes the quantification of heart RLuc signal difficult. In this study, we circumvented this problem by injecting our vector near the base of the heart, far from the liver, so that RLuc/ M185V signal could be better distinguished from the background liver signal. For a therapeutic application in which injections may need to be made more apically into the infarct zone, we could use a red-shifted hrluc (λpeak = 547 nm), optionally coupled with a red-shifted coelenterazine analog (λpeak = 588 nm),30 so that hrluc expression can be efficiently imaged at lower doses of substrate that lead to much less hepatic signal.
The main limitations of this study concern the assessment of cardiac-specific gene expression both in cell culture and in living mice. Specifically, the in vitro CSI-based assessment of various vectors was limited by the use of only one cardiac cell line (HL-1), as the other publicly available cardiac cell line, H9c2 rat embryonic cardiomyoblast, exhibits additional skeletal muscle phenotypes, making it less ‘pure’ for cardiac-specificity testing. Neonatal cardiomyocytes were not used in this study because of their poor transfection efficiency, making simultaneous testing of multiple cell lines of significantly greater transfection efficiency difficult, especially when an assay with a poor dynamic range (that is β-gal assay) is used to correct for transfection efficiency. The use of viral vector-mediated gene transfer as the next step should greatly facilitate the testing of cardiac specificity in neonatal cardiomyocytes. The in vivo assessment of cardiac specificity was limited by the use of HTV injection for highly efficient hepatic transfection, which would be otherwise difficult to achieve with intramyocardial administration alone because of the limited half-life of plasmid DNA in blood (<5 min).31 Biodistribution studies that are typically performed for viral vector-mediated gene delivery are not applicable here because of inadequate uptake and expression of plasmids in most non-cardiac tissues.32 Therefore, the in vivo results presented here do not comprise a complete assessment of tissue specificity, but do support the notion that the bidirectional TSTA strategy can effectively minimize extra-cardiac expression.
In conclusion, we have validated a generalizable TSTA vector capable of achieving effective transcriptional targeting and amplification. The ability of such a vector to couple the expression of two transgenes through a bidirectional cassette further makes it useful for correlative assessment of therapeutic gene expression. Future incorporation of the construct into a high capacity adenoviral vector should enable a better delivery of the therapeutic construct, while allowing for a more complete BLI characterization of transcriptional targeting in small living animals. Eventual replacement of the optical reporter genes with a PET reporter gene (for example HSV1-sr39tk20) and a therapeutic gene of choice should allow non-invasive monitoring of transcriptionally targeted gene therapy in large animals and human beings.
Materials and methods
Cloning reagents and starting plasmids
Starting plasmids PGL3-Basic (referred herein as pNull-fluc) and pCMV-hRL (referred herein as pCMV-hrluc) were obtained from Promega (Madison, WI, USA), whereas pcDNA3.1(+) was purchased from Invitrogen (Carlsbad, CA, USA). The plasmid vector PBCVP2-G5L from an earlier study was used as the starting vector for constructing all TSTA-based vectors.11 The site-directed mutagenesis kit was obtained from Strategene (La Jolla, CA, USA).
Construction of experimental and control vectors
Rat genomic DNA was extracted from rat peripheral blood using DNAzol (BD Biosciences, San Jose, CA, USA) as described earlier.33 A truncated cTnT (−303 to +192 bp14) promoter was generated by PCR amplification of rat genomic DNA and subcloned into the MluI/NheI restriction enzyme sites of PGL3-Basic (in a 5′ to 3′ orientation with respect to fluc) to generate pcTnT-fluc. pCMV-fluc was similarly constructed by subcloning the MluI/NheI fragment excised from pcDNA3.1(+) into the MluI/NheI sites of PGL3-Basic. The CMV enhancer (CMVenh) obtained by PCR amplification of pcDNA3.1(+) (312–631 bp) was subcloned into the SacI/MluI sites of pcTnT-fluc (in a 5′ to 3′ orientation with respect to fluc) to produce pCMVenh-cTnT-fluc. The cTnT promoter fragment obtained earlier was further amplified and subcloned into the BglII/NheI sites of pCMV-hrluc (in a 5′ to 3′ orientation with respect to hrluc) to generate pcTnT-hrluc.
The cTnT promoter fragment earlier generated by PCR was further subcloned into the MluI/NheI sites of PBCVP2-G5L to produce an intermediate unidirectional TSTA vector (pcTnT-TSTA(G5)-fluc), which contains on the anti-sense strand from 5′ to 3′ the cTnT promoter, a Gal4–VP2 fusion gene, and an SV40 polyA, as well as on the sense strand from 5′ to 3′ 5 tandem repeats of 17-bp Gal4-binding site, an adenovirus E4 minimal promoter, an fluc gene, and an SV40 poly(A). Three tandem repeats of Gal4-binding site (G3) synthesized by annealing primers flanked by KpnI were inserted into the intermediate vector to generate pcTnT-TSTA-fluc. The hrluc fragment from pCMV-hrluc was PCR amplified and subcloned into the BglII/SalI sites of pcTnT-TSTA-fluc (in a 5′ to 3′ orientation with respect to the Gal4-binding sites) to form its hrluc counterpart (pcTnT-TSTA-hrluc). Subsequent PCR amplification of a fragment containing (from 5′ to 3′ end) an adenovirus E4 promoter, an hrluc, and an SV40 polyA, followed by insertion of the fragment into pcTnT-TSTA(G5), further generated the intermediate vector pcTnT-hrluc-TSTA(G5)-fluc. Subcloning of earlier made G3 into the KpnI site of this vector led to the final bidirectional TSTA vector, pcTnT-Bid-TSTA. pcTnT-eBid-TSTA was made from pcTnT-Bid-TSTA by mutating hrluc using the Strategene site-directed mutagenesis kit and primers described earlier.15 A sealing fragment of the sequence 5′-GC GCG-3′ was made by annealing sense and anti-sense primers containing BglII/NheI, respectively, and subcloned into the BglII/NheI sites of pCMV-hrluc to generate pNull-hrluc.
A truncated myosin light chain 2 promoter (MLC2v; −250 to +18 bp)16 and a truncated α-myosin heavy chain promoter (αMHC; −373 to 16 bp)17 were generated by PCR amplification of rat genomic DNA as described earlier for the cloning of cTnT. All one-step, unidirectional TSTA, CMVenh-based, bidirectional TSTA, and RLuc-enhanced bidirectional TSTA vectors containing either promoter were constructed in the same manner as was performed for cTnT using the same backbone vectors and restriction enzyme sites.
Cell culture and transfection
HL-1 mouse atrial cardiomyocytes were a gift from Dr Gregory Kovacs (Stanford University, Stanford, CA, USA). C2C12 mouse skeletal myoblasts, Hepa1-6 mouse hepatoma, and NIH3T3 mouse embryonic fibroblasts were purchased from American Type Culture Collection (Manassas, VA, USA). HL-1 cells were maintained in flasks coated with 0.00125% fibronectin and 0.02% gelatin (both from Sigma-Aldrich, St Louis, MO, USA) and grown in Claycomb medium (JRH Biosciences, Lenexa, KS, USA) supplemented with 0.1 mM norepinephrine (Sigma-Aldrich) and 2 mM L-glutamine (Invitrogen) for routine passaging. Other cell lines were cultured in Dulbecco’s Modified Eagle’s Medium (Invitrogen). All culture media were supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin solution (both from Invitrogen).
The in vitro testing of all vectors (Figure 1; Supplementary Figure S1) in four different cell lines was performed in a single transfection experiment. Specifically, triplicates of cells plated in 12-well plates for 24 h were co-transfected with a given experimental or control plasmid vectors (1.5 µg) and a normalization plasmid vector (pCMV-β-gal; 0.3 µg) using lipofectamine2000 (3 µl per 1.5 µg DNA; Invitrogen). pCMV-β-gal expresses β-GAL under the regulation of a CMV promoter. Twenty-four hours later, cells were harvested, washed three times with PBS, and stored in −80 °C for in vitro reporter enzyme assays on the following day. For the correlation study, pcDNA3.1(+) (0–2.5 µg) was used to control for total plasmid. pCMV-β-gal was neither needed nor used for normalization of transfection efficiency.
In vitro protein and reporter enzyme assays
Cells in −80 °C were thawed on ice, lysed in 1X Passive Lysis Buffer (Promega), and centrifuged to obtain protein, whose concentration was determined using the Bio-Rad protein assay (Bio-Rad Laboratories, Hercules, CA, USA) as described earlier.34 fluc and hrluc expression were determined by measuring FLuc and RLuc enzymatic activities in relative light units using a TD 20/ 20n luminometer (Turner Designs, Sunnyvale, CA, USA) as described earlier.34 β-gal expression, in term of β-GAL enzymatic activity, was assessed using the β-GAL Enzyme Assay (Promega) according to the manufacturer’s protocol, with the exception that 15 µg of lysate was used per reaction and the substrate incubation time was extended from 30 min to 1 h. Both FLuc and RLuc activities were normalized to total protein and β-GAL activity (Abs at 420 nm), and expressed in units of relative light units per Abs.
Definition and calculation of CSI
A CSI was defined and calculated for every vector using the following formula:
where flucHL-1, flucC2C12, flucHepa1-6, and flucNIH3T3 are the fluc expression mediated by a given vector in HL-1, C2C12, Hepa1-6, and NIH3T3 cells, respectively, whereas flucCMV, HL-1, flucCMV, C2C12, flucCMV, Hepa1-6, and flucCMV, NIH3T3 are pCMV-fluc-mediated fluc expression in HL-1, C2C12, Hepa1-6, and NIH3T3 cell lines, respectively. For a given vector, the above definition of cardiac specificity compares the fluc expression in cardiac cells to that of three different non-cardiac cell lines (C2C12, Hepa1-6, NIH3T3). The denominator accounts for the differential fluc expression of pCMV-fluc, a non-tissue-specific vector, in different cell lines. The CSI index measures the cardiac specificity of a vector relative to pCMV-fluc.
Animal groups and study design
All animal studies were approved by the Stanford Institutional Animal Care and Use Committee. Fourteen groups of 7–8-week-old female Balb/c mice (total n = 146) were purchased from the Stanford In-house Breeding Colony. Groups 1–5 (n = 22 for group 3, n = 10 for all other groups) received intramyocardial co-injections of (1) pCMV-fluc+pCMV-β-gal, (2) pcTnT-fluc+pCMV-β-gal, (3) pcTnT-eBid-TSTA+pCMV-β-gal, (4) pNull-fluc+pCMV-β-gal, and (5) pcTnT-TSTA-fluc+pCMV-β-gal, respectively (16 µg per mouse for pCMV-β-gal and 80 µg per mouse for all other plasmids). Groups 6–10 (n = 10 per group) received the same plasmids as groups 1–5, respectively, except through HTV administration (4 µg per mouse for pCMV-β-gal and 20 µg per mouse for all other plasmids). Groups 11 and 12 (n = 10 per group) received HTV co-injections of (1) pMLC2v-eBid-TSTA+pCMV-β-gal and (2) pαMHC-eBid-TSTA+pCMV-β-gal, respectively. Group 13 (n = 7) received HTV injection of PBS, whereas group 14 (n = 7) received no treatment. For groups 1–12, five mice from each group underwent serial BLI (on days 2, 5, 8, 12, 16, 20, 24, and 28). The remaining five mice were killed on day 2, with their hearts (groups 1–5) or livers (groups 6–12) harvested and assayed for β-gal expression to correct for in vivo transfection efficiency. Twelve mice from group 3 were killed at different time points (days 2, 16, and 28; n = 4 per time point), with their hearts explanted and assayed for both FLuc and RLuc activities. All mice in groups 13 and 14 were killed on day 2, with their explanted livers assayed for cTnT and β-actin mRNAs using qRT–PCR.
Animal surgery and plasmid vector delivery
For intramyocardial administration of plasmid vectors, mice were induced with 3% isoflurane, orotracheally intubated and ventilated with 2% isoflorane using a Harvard rodent ventilator (Harvard Apparatus, Holliston, MA, USA). After a left thoracotomy, two injections of 15 µl each were made at one site near the base of the left ventricle. Afterwards, the chest was closed in layers and pneumothorax evaculated by suction using a 16G angiocath (BD Biosciences). Animals received subcutaneous buprenorphene (0.1 mg kg−1) for post-operative analgesia and recovered in 100% oxygen. For HTV administration, mice under anesthesia (3 and 2% isoflurane for induction and maintenance, respectively) were injected with 2 ml of plasmid diluted in PBS (with a total volume equal to 9% of the body weight) into the mouse tail vein at a rate of 0.5 ml s−1, using a 3-ml syringe capped with a 26 1/2 gauge needle (both from BD Biosciences). After injection, mice were recovered in 100% oxygen.
Tissue harvest and ex vivo protein and reporter enzyme assays
After mice were killed by carbon dioxide asphyxiation, their explanted hearts or livers were washed with PBS, immersed in 1 ml of 1X Passive Lysis Buffer (Promega), and homogenized using a Kontes Duall tissue grinder (Fisher Scientific, Pittsburg, PA, USA). Supernatants were obtained from tissue homogenates by centrifugation, quantified for protein content using the Bio-Rad protein assay, and assayed for FLuc, RLuc, or β-GAL enzyme activity as described earlier for cell lysates, except with minor modifications to the β-GAL assay protocol: 75 µg of supernatant was used per reaction, and the substrate incubation time was extended to 60 min for the heart supernatant and 15 min for the liver supernatant.
BLI of living mice and data analysis
Mice were anesthetized with 2% isoflurane, injected with the appropriate imaging substrate (375 mg kg−1 D-luciferin (Biosynth International, Naperville, IL, USA) injected intraperitoneally for FLuc scan; 2.5 mg kg−1 coelenterazine (NanoLight Technology, Pinetop, AZ, USA) delivered intravenously through tail vein injection for RLuc/M185V scan), imaged using the In Vivo Imaging System (IVIS 100; a Xenogen product from Caliper Life Sciences, Alameda, CA, USA), and recovered with 100% oxygen through nose cone. The imaging protocol for assessing fluc expression consisted of 3 to 5 5-min scans until the imaging signal peaked and was identical for all groups, except for groups that received pCMV-fluc, pcTnT-eBid-TSTA, pcTnT-TSTA-fluc, pαMHC-eBid-TSTA, or pMLC2v-eBid-TSTA by tail vein, for which the scan times were shortened only on day 2 to 2 s and 5 s, respectively, to avoid saturation of the detector. For the same reason, the lens aperture was reduced to an f-stop value of 4 for these groups and kept the same for other groups that received HTV injections. For imaging hrluc/M185V expression, a single 4-min acquisition was performed immediately after intravenous substrate injection. Image analysis was performed using Living Image 2.50 (Caliper Life Sciences). FLuc images were analyzed by placing a fixed size, default region of interest over the target region (heart for groups 1–5 and liver for groups 6–12) and measuring the maximum radiance in units of photons s−1 cm−2 sr−1). Only the peak heart signal after intraperitoneal injection was used for analysis. The RLuc/M185V images were similarly analyzed, except that the region of interest was carefully placed over the heart to exclude confounding liver signal.
For comparison of optical signals mediated by different plasmid vectors co-delivered with pCMV-β-gal, the average myocardial and hepatic optical signals were normalized by the ex vivo myocardial and hepatic β-GAL activities, respectively, 2 days post-injection to account for the differential in vivo transfection efficiency of different plasmid vectors. The normalized optical signal is expressed in units of photons s−1 cm−2 sr−1 per Abs.
Ex vivo qRT–PCR
Total RNAs (250 ng) extracted from mouse liver samples (∼100 mg) using Qiagen RNA extraction kit (Qiagen, Valencia, CA, USA) were used to generate corresponding cDNAs using random hexamers and Superscript III reverse transcriptase (Invitrogen). Real-time PCR reactions were performed on 20-µl mixtures, with each containing cDNA (50 ng RNA equivalent), 2X SYBR green qPCR master mix (Applied Biosystems, Carlsbad, CA, USA), and either of the following sets of primers: cTnT forward and reverse primers (5′ -gccaaagatgctgaa-gaagg-3′ and 5′-ttctcgaagtgagcctcgat-3′, respectively); β-actin forward and reverse primers (5′-gctctggctcctagcac cat-3′ and 5′-gccaccgatccacacagagt-3′, respectively). PCR reactions were performed using an Eppendorf Realplex thermal cycler (Eppendorf, Germany) under the following cycling conditions: 30 s at 94 °C, 30 s at 55 °C, and 30 s at 68 °C for 60 cycles. Real-time quantification of amplified DNA was based on fluorescence intensity in arbitrary units.
Statistical analysis
All data are presented as mean ± s.e.m. The unpaired, two-tailed Student’s t-test was used for comparison between two measurements. For comparison of two groups of measurements over time, two-way fixed-effects repeated measures ANOVA was performed on the log-transformed measurements, with factors of group and time. Greenhouse–Geisser corrections for lack of sphericity were used. Bonferroni-corrected-specific contrasts were also tested. Linear regression analysis was performed to assess the linearity between two variables. The strength of correlation was quantified in terms of the square of Pearson product moment coefficient (r2). The significance of correlation was calculated by performing the Student’s t-test against the null hypothesis that the correlation coefficient (r) is zero. P-values <0.05 were considered statistically significant.
Supplementary Material
Acknowledgements
We thank Dr Jarrett Rosenberg for his kind assistance with statistical analysis. This work was supported in part by NHLBI 5R01HL078632 (SSG), NCI ICMIC P50 CA114747 (SSG), NCI SAIRP, American Heart Association Pre-doctoral Fellowship (IYC), Stanford Bio-X Graduate Student Fellowship (IYC), K99-R00 HL88048 (MRP), HL095571 (JCW), and HL093172 (JCW).
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on Gene Therapy website (http://www.nature.com/gt)
References
- 1.Rissanen TT, Yla-Herttuala S. Current status of cardiovascular gene therapy. Mol Ther. 2007;15:1233–1247. doi: 10.1038/sj.mt.6300175. [DOI] [PubMed] [Google Scholar]
- 2.Raper SE, Chirmule N, Lee FS, Wivel NA, Bagg A, Gao GP, et al. Fatal systemic inflammatory response syndrome in a ornithine transcarbamylase deficient patient following adenoviral gene transfer. Mol Genet Metab. 2003;80:148–158. doi: 10.1016/j.ymgme.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 3.Gaspar HB, Thrasher AJ. Gene therapy for severe combined immunodeficiencies. Expert Opin Biol Ther. 2005;5:1175–1182. doi: 10.1517/14712598.5.9.1175. [DOI] [PubMed] [Google Scholar]
- 4.Franz WM, Rothmann T, Frey N, Katus HA. Analysis of tissue-specific gene delivery by recombinant adenoviruses containing cardiac-specific promoters. Cardiovasc Res. 1997;35:560–566. doi: 10.1016/s0008-6363(97)00154-5. [DOI] [PubMed] [Google Scholar]
- 5.Huard J, Lochmüller H, Acsadi G, Jani A, Massie B, Karpati G. The route of administration is a major determinant of the transduction efficiency of rat tissues by adenoviral recombinants. Gene Ther. 1995;2:107–115. [PubMed] [Google Scholar]
- 6.Isner JM, Vale PR, Symes JF, Losordo DW. Assessment of risks associated with cardiovascular gene therapy in human subjects. Circ Res. 2001;89:389–400. doi: 10.1161/hh1701.096259. [DOI] [PubMed] [Google Scholar]
- 7.Ma H, Sumbilla CM, Farrance IK, Klein MG, Inesi G. Cell-specific expression of SERCA, the exogenous Ca2+ transport ATPase, in cardiac myocytes. Am J Physiol Cell Physiol. 2004;286:C556–C564. doi: 10.1152/ajpcell.00328.2003. [DOI] [PubMed] [Google Scholar]
- 8.Jin Y, Pasumarthi KB, Bock ME, Chen Y, Kardami E, Cattini PA. Effect of ‘enhancer’ sequences on ventricular myosin light chain-2 promoter activity in heart muscle and nonmuscle cells. Biochem Biophys Res Commun. 1995;210:260–266. doi: 10.1006/bbrc.1995.1655. [DOI] [PubMed] [Google Scholar]
- 9.Boecker W, Bernecker OY, Wu JC, Zhu X, Sawa T, Grazette L, et al. Cardiac-specific gene expression facilitated by an enhanced myosin light chain promoter. Mol Imaging. 2004;3:69–75. doi: 10.1162/15353500200404103. [DOI] [PubMed] [Google Scholar]
- 10.Zhang L, Adams JY, Billick E, Ilagan R, Iyer M, Le K, et al. Molecular engineering of a two-step transcription amplification (TSTA) system for transgene delivery in prostate cancer. Mol Ther. 2002;5:223–232. doi: 10.1006/mthe.2002.0551. [DOI] [PubMed] [Google Scholar]
- 11.Iyer M, Wu L, Carey M, Wang Y, Smallwood A, Gambhir SS. Two-step transcriptional amplification as a method for imaging reporter gene expression using weak promoters. Proc Natl Acad Sci USA. 2001;98:14595–14600. doi: 10.1073/pnas.251551098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ray S, Paulmurugan R, Hildebrandt I, Iyer M, Wu L, Carey M, et al. Novel bidirectional vector strategy for amplification of therapeutic and reporter gene expression. Hum Gene Ther. 2004;15:681–690. doi: 10.1089/1043034041361271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Contag CH, Spilman SD, Contag PR, Oshiro M, Eames B, Dennery P, et al. Visualizing gene expression in living mammals using a bioluminescent reporter. Photochem Photobiol. 1997;66:523–531. doi: 10.1111/j.1751-1097.1997.tb03184.x. [DOI] [PubMed] [Google Scholar]
- 14.Wang G, Yeh HI, Lin JJ. Characterization of cis-regulating elements and trans-activating factors of the rat cardiac troponin T gene. J Biol Chem. 1994;269:30595–30603. [PubMed] [Google Scholar]
- 15.Loening AM, Fenn TD, Wu AM, Gambhir SS. Consensus guided mutagenesis of Renilla luciferase yields enhanced stability and light output. Protein Eng Des Sel. 2006;19:391–400. doi: 10.1093/protein/gzl023. [DOI] [PubMed] [Google Scholar]
- 16.Henderson SA, Spencer M, Sen A, Kumar C, Siddiqui MA, Chien KR. Structure, organization, and expression of the rat cardiac myosin light chain-2 gene. Identification of a 250-base pair fragment which confers cardiac-specific expression. J Biol Chem. 1989;264:18142–18148. [PubMed] [Google Scholar]
- 17.Molkentin JD, Jobe SM, Markham BE. Alpha-myosin heavy chain gene regulation: delineation and characterization of the cardiac muscle-specific enhancer and muscle-specific promoter. J Mol Cell Cardiol. 1996;28:1211–1225. doi: 10.1006/jmcc.1996.0112. [DOI] [PubMed] [Google Scholar]
- 18.Liu F, Song Y, Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6:1258–1266. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- 19.Iyer M, Salazar FB, Wu L, Carey M, Gambhir SS. Bioluminescence imaging of systemic tumor targeting using a prostate-specific lentiviral vector. Hum Gene Ther. 2006;17:125–132. doi: 10.1089/hum.2006.17.125. [DOI] [PubMed] [Google Scholar]
- 20.Wu JC, Chen IY, Wang Y, Tseng JR, Chhabra A, Salek M, et al. Molecular imaging of the kinetics of vascular endothelial growth factor gene expression in ischemic myocardium. Circulation. 2004;110:685–691. doi: 10.1161/01.CIR.000013815302213.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen IY, Wu JC, Min JJ, Sundaresan G, Lewis X, Liang Q, et al. Micro-positron emission tomography imaging of cardiac gene expression in rats using bicistronic adenoviral vector-mediated gene delivery. Circulation. 2004;109:1415–1420. doi: 10.1161/01.CIR.0000121727.59564.5B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barth AS, Kizana E, Smith RR, Terrovitis J, Dong P, Leppo MK, et al. Lentiviral vectors bearing the cardiac promoter of the Na+-Ca2+ exchanger report cardiogenic differentiation in stem cells. Mol Ther. 2008;16:957–964. doi: 10.1038/mt.2008.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eggermont J, Proudfoot NJ. Poly(A) signals and transcriptional pause sites combine to prevent interference between RNA polymerase II promoters. EMBO J. 1993;12:2539–2548. doi: 10.1002/j.1460-2075.1993.tb05909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Douglas JT, Curiel DT. Strategies to accomplish targeted gene delivery to muscle cells employing tropism-modified adenoviral vectors. Neuromuscul Disord. 1997;7:284–298. doi: 10.1016/s0960-8966(97)00053-9. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Iyer M, Annala A, Wu L, Carey M, Gambhir SS. Noninvasive indirect imaging of vascular endothelial growth factor gene expression using bioluminescence imaging in living transgenic mice. Physiol Genomics. 2006;24:173–180. doi: 10.1152/physiolgenomics.00308.2004. [DOI] [PubMed] [Google Scholar]
- 26.Nishikawa M, Nakayama A, Takahashi Y, Fukuhara Y, Takakura Y. Reactivation of silenced transgene expression in mouse liver by rapid, large-volume injection of isotonic solution. Hum Gene Ther. 2008;19:1009–1020. doi: 10.1089/hum.2008.020. [DOI] [PubMed] [Google Scholar]
- 27.Schubert W, Yang XY, Yang TT, Factor SM, Lisanti MP, Molkentin JD, et al. Requirement of transcription factor NFAT in developing atrial myocardium. J Cell Biol. 2003;161:861–874. doi: 10.1083/jcb.200301058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao H, Doyle TC, Coquoz O, Kalish F, Rice BW, Contag CH. Emission spectra of bioluminescent reporters and interaction with mammalian tissue determine the sensitivity of detection in vivo. J Biomed Opt. 2005;10:41210. doi: 10.1117/1.2032388. [DOI] [PubMed] [Google Scholar]
- 29.Campbell AK, Patel A, Houston WA, Scolding NJ, Frith S, Morgan BP, et al. Photoproteins as indicators of intracellular free Ca2+ J Biolumin Chemilumin. 1989;4:463–474. doi: 10.1002/bio.1170040161. [DOI] [PubMed] [Google Scholar]
- 30.Loening AM, Wu AM, Gambhir SS. Red-shifted Renilla reniformis luciferase variants for imaging in living subjects. Nat Methods. 2007;4:641–643. doi: 10.1038/nmeth1070. [DOI] [PubMed] [Google Scholar]
- 31.Lew D, Parker SE, Latimer T, Abai AM, Kuwahara-Rundell A, Doh SG, et al. Cancer gene therapy using plasmid DNA: pharmacokinetic study of DNA following injection in mice. Hum Gene Ther. 1995;6:553–564. doi: 10.1089/hum.1995.6.5-553. [DOI] [PubMed] [Google Scholar]
- 32.Son MK, Choi JH, Lee DS, Kim CY, Choi SM, Kang KK, et al. Pharmacokinetics and biodistribution of a pGT2-VEGF plasmid DNA after administration in rats. J Cardiovasc Pharmacol. 2005;46:577–584. doi: 10.1097/01.fjc.0000179625.89331.41. [DOI] [PubMed] [Google Scholar]
- 33.Chomczynski P, Mackey K, Drews R, Wilfinger W. DNAzol: a reagent for the rapid isolation of genomic DNA. Biotechniques. 1997;22:550–553. doi: 10.2144/97223pf01. [DOI] [PubMed] [Google Scholar]
- 34.Bhaumik S, Lewis XZ, Gambhir SS. Optical imaging of Renilla luciferase, synthetic Renilla luciferase, and firefly luciferase reporter gene expression in living mice. J Biomed Opt. 2004;9:578–586. doi: 10.1117/1.1647546. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.