Abstract
Piwi-interacting RNAs (piRNAs) are essential for silencing of transposable elements in the germline, but their biogenesis is poorly understood. Here we demonstrate that MOV10L1, a germ cell–specific putative RNA helicase, is associated with Piwi proteins. Genetic disruption of the MOV10L1 RNA helicase domain in mice renders both MILI and MIWI2 devoid of piRNAs. Absence of a functional piRNA pathway in Mov10l1 mutant testes causes loss of DNA methylation and subsequent derepression of retrotransposons in germ cells. The Mov10l1 mutant males are sterile owing to complete meiotic arrest. This mouse mutant expresses Piwi proteins but lacks piRNAs, suggesting that MOV10L1 is required for piRNA biogenesis and/or loading to Piwi proteins.
Keywords: meiosis, RNA helicase, transposon silencing
The Piwi clade of Argonaute proteins associates with a class of 26–31-nt germline-specific small RNAs called “piRNAs”. Together they participate in suppression of transposable elements in all animals studied (1–4). In mice, the Piwi clade contains three members: Miwi2, Mili, and Miwi. These three Piwi members exhibit distinct developmental expression patterns. Miwi2 is expressed in perinatal male germ cells (5), whereas Mili is more broadly expressed from embryonic germ cells to postnatal round spermatids (6). Miwi expression begins in pachytene spermatocytes and persists in haploid round spermatids (7). The overlapping temporal expression of Mili with Miwi and Miwi2 points to the pivotal role of MILI in the piRNA pathway, as further supported by the fact that MILI is associated with developmental stage–dependent pools of piRNAs: prenatal, prepachytene, and pachytene piRNAs (5, 8, 9).
The mechanisms of piRNA biogenesis are largely unclear (1–4). One feature of piRNAs in all species is their highly clustered genomic origins. Several of these clusters produce piRNAs only from one strand. This leads to a hypothesized primary processing pathway whereby an unknown nuclease cleaves off mature piRNAs from a long single-stranded precursor transcript. On the other hand, some piRNAs in prenatal and prepachytene pools display signatures indicative of a proposed RNA-mediated amplification loop that uses primary piRNAs to generate secondary piRNAs from precursor transcripts (ping-pong mechanism) (10, 11). Apart from the Piwi proteins themselves, factors directly impacting piRNA production are unknown.
We previously identified Mov10l1 as a gene specifically expressed in mouse germ cells, which encodes a putative RNA helicase of unknown function (12). Whereas the N-terminal half of MOV10L1 is not homologous to any other mouse proteins, its C-terminal RNA helicase domain exhibits low homology (45% amino acid identity) with MOV10. MOV10, the vertebrate homolog of Drosophila Armi, is ubiquitously expressed. In mammalian cells, MOV10 is associated with Argonaute proteins in the RNA-induced silencing complex (RISC) and is functionally required for RNA interference (13, 14). Here we demonstrate that MOV10L1 is an essential factor in the piRNA pathway.
Results
MOV10L1 Is Associated with Piwi Proteins.
To identify potential interaction partners, we isolated MOV10L1-containing protein complexes from testicular extracts by immunoprecipitation. Mass spectrometry analyses of three specific protein bands in the MOV10L1 complex revealed that they corresponded to MOV10L1/TDRD1, MILI, and MIWI (Fig. S1). We and others have also found MOV10L1 in immunoprecipitated MILI, MIWI, and MIWI2 complexes by mass spectrometry (15, 16). Consistent with the mass spectrometry data, coimmunoprecipitations followed by Western blot analysis showed abundant association of MOV10L1 with MILI but less with TDRD1 and MIWI (Fig. 1 A and B). Further coimmunoprecipitation experiments done with cotransfected human 293T cells strongly suggested that MOV10L1 binds to MILI, MIWI, and MIWI2 (Fig. 1C), because mammalian somatic cells lack piRNA pathway components.
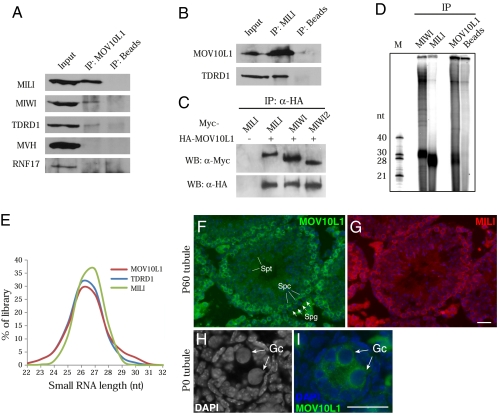
Fig. 1.
MOV10L1 is associated with Piwi proteins and piRNAs in testis. (A) Validation of protein associations by coimmunoprecipitation (co-IP) and Western blotting. Note that nuage components MVH and RNF17 are not associated with MOV10L1. (B) Reciprocal IP confirms the association between MOV10L1 and MILI. (C) MOV10L1 interacts with MILI, MIWI, and MIWI2 in cotransfected 293T cells. (D) Analysis of piRNAs in immunoprecipitated MILI, MIWI, and MOV10L1 complexes from adult mouse testes. (E) Length distribution of small RNA reads in three small RNA libraries prepared from adult mouse testes. (F and G) Adjacent testis sections from 2-mo-old (P60) mice were immunostained with anti-MOV10L1 (F, green) and anti-MILI (G, red) antibodies. Strong interstitial signal is due to autofluorescence of Leydig cells. (H and I) Expression of MOV10L1 in gonocytes (Gc) from newborn (P0) testes. In contrast to nuclei of Sertoli cells, gonocyte nuclei contain little heterochromatin (H). Spg, spermatogonium; Spc, spermatocyte; Spt, spermatid. (Scale bar, 25 μm.)
To determine whether MOV10L1 is associated with piRNAs, we immunoprecipitated MOV10L1, MILI, and MIWI from adult mouse testes and assessed the presence of any associated small RNAs (Fig. 1D). As expected, MILI and MIWI were associated with ≈26-nt and ≈30-nt small RNAs, respectively. Consistent with its interaction with Piwi proteins, the MOV10L1 purification revealed the presence of small RNAs in the 26–30-nt size range, with a majority migrating similar to MILI-associated piRNAs (Fig. 1D). It is very likely that MOV10L1 is associated with piRNAs indirectly through its interaction with Piwi proteins.
We then performed Solexa deep sequencing of a small RNA library from MOV10L1 complexes isolated from adult testes. A total of ≈2.2 million reads perfectly mapped to the genome and peaked around 26 to 27 nt (Fig. 1E), similar to those in small RNA libraries prepared from MILI or its interacting partner, TDRD1 (17). A notable fraction (≈8%) of MOV10L1 reads was 29–32 nt in length, a size similar to MIWI-associated piRNAs (18, 19). The MOV10L1 reads display a strong preference (≈80%) for a uridine at position one (U1-bias). Genome annotation of the MOV10L1 library revealed that the majority of reads derive from intergenic (≈56%) unannotated regions, similar to MILI- and TDRD1-associated pachytene piRNAs from adult testes (Fig. S2A) (17, 20). Further analysis of the repeat fractions showed that representation of transposon classes is also similar among these three libraries (Fig. S2B). Pachytene piRNAs derive from ≈250 genomic regions, and this clustered origin is a hallmark of piRNA biogenesis. Reads from all three libraries mapped to the same strand within the same genomic window, confirming that the MOV10L1 reads are MILI-associated primary piRNAs (Fig. S2C). These results show that MOV10L1 is abundantly associated with the MILI–piRNA complex and, to a lesser extent, with the MIWI–piRNA complex in adult testis.
Colocalization of MOV10L1 with MILI in Male Germ Cells.
We found that MOV10L1 localizes to the cytoplasm of germ cells. Mov10l1 is transcribed at a much higher level in spermatocytes than in spermatogonia (21). Consistent with this previous study, the level of MOV10L1 protein in spermatogonia was low (Fig. 1F). MOV10L1 was clearly present in pachytene spermatocytes but absent in postmeiotic spermatids (Fig. 1F). This spatiotemporal localization pattern of MOV10L1 resembles that of MILI in adjacent testis sections (Fig. 1G). In postnatal day 14 (P14) tubules, MOV10L1 colocalized with GASZ, a nuage-associated protein in the piRNA pathway (22). Similar to MILI, MOV10L1 also localized to the cytoplasm of gonocytes from newborn testes (Fig. 1 H and I). These coexpression and colocalization data further support a role for MOV10L1 in the piRNA pathway.
MOV10L1 Is Essential for Male Meiosis and Male Fertility.
To uncover the requirement of Mov10l1 for spermatogenesis and the piRNA pathway, we generated a conditional mutant allele (Mov10l1fl) in mice. In the targeted allele, one loxP site was inserted in intron 17 and one in intron 21 (Fig. S3). To disrupt the Mov10l1 gene, Mov10l1fl mice were bred with ACTB-Cre mice, in which Cre recombinase is ubiquitously expressed (23). Deletion of exons 18–21 (encoding amino acids 841–1,018) disrupted the putative RNA helicase domain of MOV10L1. Sequencing of the mutant Mov10l1 transcript amplified from Mov10l1−/− testes showed that splicing occurred precisely from exon 17 to exon 22, maintaining the translation frame. As expected, the internally deleted MOV10L1 protein (1,061 aa; MOV10L1Δ) was expressed in Mov10l1+/− and Mov10l1−/− testes but with reduced abundance (Fig. 2A). The mutant MOV10L1 protein was not readily detectable in Mov10l1−/− testes by immunofluorescence (Fig. S4). Exons 17–27 of the Mov10l1 locus are used to produce a heart-specific alternative transcript (termed Csm/Champ) (24, 25). Despite the lack of this heart-specific transcript, Mov10l1−/− mice were viable and exhibited no overt defects, suggesting that Mov10l1 (Csm/Champ) is not essential for heart development. Interbreeding of heterozygous mice yielded a normal Mendelian ratio of offspring (Mov10l1+/+, Mov10l1+/−, Mov10l1−/−: 56, 129, 63), suggesting that disruption of Mov10l1 does not cause embryonic lethality.
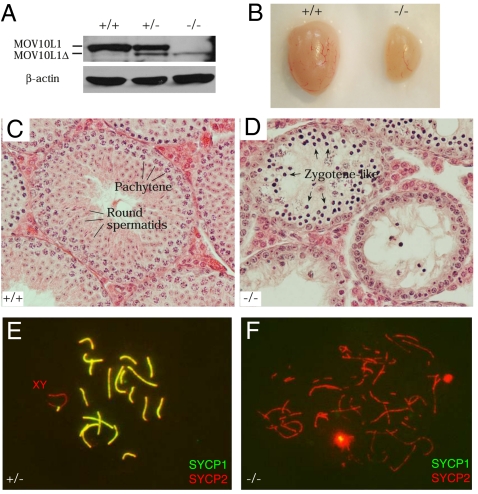
Fig. 2.
Mov10l1 is essential for spermatogenesis and chromosomal synapsis. (A) Western blot analysis of adult wild-type, Mov10l1+/−, and Mov10l1−/− testes. The mutant MOV10L1 protein (1,061 aa; MOV10L1Δ) is indicated. (B) Dramatic size reduction in 7-wk-old Mov10l1−/− testis. (C and D) In contrast to wild-type tubules with full spermatogenesis (C), Mov10l1−/− tubules from adult mice exhibited early meiotic arrest (D). (E) Normal pachytene spermatocyte with 19 pairs of fully synapsed autosomes and the partially synapsed sex chromosomes. (F) Zygotene-like spermatocytes from Mov10l1−/− adult testes.
Mov10l1−/− females displayed normal fertility, but Mov10l1−/− males were sterile. Disruption of Mov10l1 caused a sharp reduction in testis size (Fig. 2B). The weight of Mov10l1−/− testes (48.3 ± 9.1 mg per pair) from 5- to 7-wk-old mice was <37% that of Mov10l1+/− testes (131.1 ± 6.9 mg per pair) (Student's t test, P < 0.0001). In contrast to wild-type seminiferous tubules (Fig. 2C), tubules from adult Mov10l1−/− testes contained only early spermatogenic cells, with a complete lack of postmeiotic germ cells (Fig. 2D). The most advanced germ cells in the mutant were zygotene-like spermatocytes, indicating an early meiotic arrest. The stage of meiotic arrest in Mov10l1−/− mice is similar to that in Mili and Miwi2 mutant mice (6, 26). Thus, MOV10L1 is required for male meiosis and is essential for male fertility.
To further define the meiotic defects in Mov10l1−/− testes, we analyzed the assembly of the synaptonemal complex by immunostaining central and axial elements with anti-SYCP1 and anti-SYCP2 antibodies. In Mov10l1+/− spermatocytes, all chromosomes were fully synapsed except the XY chromosomes, which were only synapsed at the pseudoautosomal region (Fig. 2E). In Mov10l1−/− spermatocytes, synapsis failed to occur, evident from the absence of SYCP1 (Fig. 2F). Thus, disruption of Mov10l1 causes meiotic blockade before the pachytene stage.
MILI and TDRD1 Are Lost in Mov10l1−/− Spermatocytes but Retained in Spermatogonia.
We next investigated the consequence of loss of MOV10L1 function on the expression of piRNA pathway components. Normally, MILI and TDRD1 are expressed in both spermatogonia and spermatocytes (Fig. S5). However, both MILI and TDRD1 were detectable in spermatogonia but not in spermatocytes in Mov10l1−/− testis (Fig. S5 B and D). In contrast, MVH, MAEL, and GASZ were still expressed in both spermatogonia and spermatocytes in Mov10l1−/− testis (Fig. S5F) (22, 27). The expression pattern of MILI and TDRD1 in P14 Mov10l1−/− testes was the same as in the adult (P60) mutant testes. Thus, disruption of MOV10L1 impacted the abundance of MILI and TDRD1 most dramatically. Decreased abundance of nuage proteins has also been observed in Gasz mouse mutant and a number of Drosophila nuage mutants (22, 28, 29).
Binary Derepression of LINE1 and IAP Retrotransposons in Postnatal Mov10l1−/− Testes.
piRNAs are required for silencing of transposable elements in the germline in various species (1, 2, 30). We examined the expression of LINE1 and IAP retrotransposons in Mov10l1−/− testes. P10 testes contain predominantly spermatogonia and also preleptotene and leptotene spermatocytes. P14 testes also contain more advanced spermatocytes, such as zygotene and pachytene cells. Quantitative RT-PCR analyses showed that the abundance of both LINE1 and IAP transcripts increased sharply in Mov10l1−/− testes (Fig. 3A). We confirmed these findings by Northern blot analyses (Fig. 3B). Western blot analyses showed that LINE1 ORF1p abundance increased significantly in P10 Mov10l1−/− testes and more dramatically in P14 mutant testes (Fig. 3C). IAP protein abundance increased modestly in P10 and P14 Mov10l1−/− testes (Fig. 3C). Thus, disruption of Mov10l1 results in derepression of LINE1 and IAP retrotransposons.
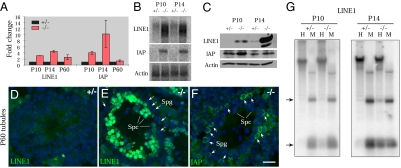
Fig. 3.
Binary derepression of LINE1 and IAP retrotransposons in mitotic vs. meiotic germ cells. (A) Quantitative RT-PCR analysis. (B) Northern blot analysis. (C) Western blotting analysis. (D and E) Immuofluorescence analysis of LINE1 ORF1p in 2-mo-old (P60) Mov10l1+/− and Mov10l1−/− testes. (F) Immunofluorescence analysis of IAP in Mov10l1−/− testes. Spc, spermatocytes; Spg, spermatogonia. (G) Methylation-sensitive Southern blot analysis of testis genomic DNA. Genomic DNA was digested with methylation sensitive (HpaII, H) and methylation insensitive (MspI, M) enzymes. Arrows indicate the position of methylation-sensitive (H) restriction products in the Mov10l1−/− testes. (Scale bar in F, 25 μm for D–F.)
We next identified the cell types that express retrotransposon-encoded proteins by immunofluorescence analyses on adult testis sections (P60), in which spermatogonia and spermatocytes can be unequivocally identified. In wild-type tubules, LINE1 and IAP were barely detectable (Fig. 3D). Mov10l1−/− testes, however, exhibited highly abundant expression of LINE1 in spermatocytes but not spermatogonia (Fig. 3E). In contrast, IAP protein was readily visualized in spermatogonia but not in spermatocytes of mutant testes (Fig. 3F). This binary derepression pattern of LINE1 and IAP was also observed in P14 Mov10l1−/− testes. These data suggest that these two classes of retrotransposons (LINE1 and IAP) are silenced in a MOV10L1-dependent manner but are regulated differently in spermatogonia and spermatocytes.
Cytosine DNA methylation of retrotransposon regulatory regions in mouse causes transcriptional silencing (31). Methylation-sensitive Southern blotting showed that demethylation of LINE1 elements was discernible in Mov10l1−/− testes at P10 and clearly detectable in P14 mutant testes (Fig. 3G). Quantification of LINE1 demethylation by bisulfite sequencing showed methylation of 84% and 85% of CpGs in P10 and P14 Mov10l1+/− testes, respectively. Consistent with our Southern result, 72% of LINE1 CpGs were methylated in P10 Mov10l1−/− testes, but only 54% were methylated in P14 mutant testes. These data showed that disruption of Mov10l1 results in loss of DNA methylation and thus derepression of LINE1 and IAP retrotransposons.
MILI Is Depleted of piRNAs in Postnatal Mov10l1−/− Testes.
Given the association of MOV10L1 with the MILI piRNA ribonucleoprotein particles (piRNPs), we examined the impact of loss of Mov10l1 on piRNA populations. In testes from P10 and P14 mice, MILI binds to prepachytene and pachytene piRNAs, respectively (8, 20). Although the abundance of MILI was reduced in Mov10l1−/− testes, a substantial amount of MILI was immunoprecipitated from P10 or P14 mutant testes but was found to be devoid of piRNAs in Mov10l1−/− testes (Fig. 4A). To rule out the possibility that the observed loss of MILI-bound piRNAs is due to the detection limit of immunoprecipitation and 5′-end radiolabeling assay, we repeated the experiment and, in parallel, performed immunoprecipitations with serial dilutions of P10 wild-type testis extract. MILI-associated piRNAs were readily detected in wild-type controls, even when MILI protein was not detectable by Western blotting (Fig. 4B, lanes 5 and 6), demonstrating the high sensitivity of this method. In conclusion, MILI is unloaded in the P10 and P14 Mov10l1−/− testes.
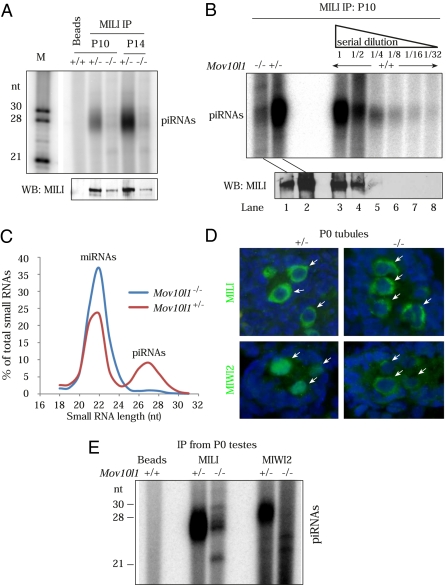
Fig. 4.
Biogenesis blockade of both prepachytene and perinatal piRNAs in Mov10l1−/− testes. (A) MILI is unloaded in P10 and P14 Mov10l1−/− testes. One tenth of the immunoprecipitated material was used for detection of associated RNAs, whereas the remaining was used for Western blotting (WB) to detect MILI. (B) MILI immunoprecipitations with P10 Mov10l1−/−, Mov10l1+/−, and serial dilutions (1:2) of P10 wild-type testicular extracts. (C) Profile of read lengths in total small RNA (18–32 nt) libraries from P10 Mov10l1+/− and Mov10l1−/− testes. (D) MIWI2 is localized to the cytoplasm in Mov10l1−/− perinatal (P0) gonocytes (arrows). (E) MILI and MIWI2 are devoid of piRNAs in Mov10l1−/− newborn (P0) testes.
It is possible that, in Mov10l1 mutant testes, piRNAs are produced but fail to get incorporated into MILI. To test this, we prepared and sequenced 18–32-nt total small RNA libraries from testes of P10 Mov10l1+/− and Mov10l1−/− mice. Analysis of read-lengths (Fig. 4C) revealed a loss of ≈26–28-nt sequences (presumably piRNAs) in Mov10l1−/− testes. The ≈21-nt reads, the bulk of which are annotated miRNAs, are still present in the Mov10l1−/− testes (Fig. 4C). These data demonstrate that MOV10L1 is required for biogenesis of piRNAs but not miRNAs.
Both MILI and MIWI2 Are Depleted of piRNAs in Mov10l1−/− Perinatal Germ Cells.
Given that MOV10L1 is expressed in perinatal gonocytes (Fig. 1I), we wondered whether the piRNA pathway is affected in the early germ cells in Mov10l1 mutant. We first examined the expression of MILI and MIWI2 in perinatal Mov10l1−/− germ cells. MIWI2 localized predominantly to the nucleus in P0 Mov10l1+/− gonocytes, but it was excluded from the nuclei and accumulated in the cytoplasm of Mov10l1−/− gonocytes (Fig. 4D). A nuclear-to-cytoplasmic redistribution of MIWI2 was also observed in both Mili and Tdrd1 mouse mutants (5, 16, 17). The cytoplasmic localization of MILI was maintained in Mov10l1−/− gonocytes (Fig. 4D). Furthermore, LINE1 elements were strongly activated, and IAP was moderately derepressed in Mov10l1−/− gonocytes (Fig. S6). These data strongly indicate that the piRNA pathway is defective in Mov10l1−/− perinatal gonocytes.
In embryonic germ cells, MILI and MIWI2 are associated with repeat-rich prenatal piRNAs (5, 9). To ascertain the piRNA-association status of MILI and MIWI2 in Mov10l1−/− perinatal germ cells, we immunoprecipitated both proteins from P0 testes. In testes from Mov10l1+/− pups, MILI and MIWI2 were loaded with their respective ≈26- and ≈28-nt small RNAs. In contrast, both MILI and MIWI2 were completely devoid of piRNAs in Mov10l1−/− testes (Fig. 4E). Taken together with our findings in P10 and P14 mutant testes, we conclude that MOV10L1 is required for biogenesis and/or stability of both perinatal (MILI- or MIWI2-bound) and prepachytene (MILI-bound) piRNAs.
Discussion
Here we report a mouse mutant in which MILI-associated piRNAs are absent, whereas the MILI protein is still detectable. Our studies point to a central role for MOV10L1 in the biogenesis and/or stability of MILI-, MIWI2-, and possibly MIWI-bound piRNAs. First, we have demonstrated an essential role for MOV10L1 in the biogenesis of MILI-bound piRNAs in both perinatal (P0) and prepachytene (P10) stages. Second, MOV10L1 may also be directly required for the loading of presumably secondary piRNAs onto MIWI2 in perinatal gonocytes, because these two proteins bind to each other and MIWI2 is unloaded in the Mov10l1 mutant. Alternatively, in the Mov10l1 mutant, the “empty” MILI (lack of bound primary piRNAs) may fail to guide the biogenesis of secondary piRNAs that would bind to MIWI2. Such a possibility is further supported by the unloaded status of MIWI2 in the Mili mutant (5). Finally, the fact that MOV10L1 is associated with both MILI and MIWI in adult mouse testes also implicates it in the pachytene piRNA biogenesis. However, this is not readily testable, because disruption of Mov10l1 causes meiotic block before the pachytene stage.
MOV10L1 exhibits limited homology with MOV10, which is the vertebrate homolog of Drosophila Armi. Armi is a sequence homolog of SDE3, which is required for posttranscriptional gene silencing in Arabidopsis (32). Armi is required for RISC maturation and oskar mRNA silencing (33, 34). Armi is also important for piRNA biogenesis and silencing of the Stellate repeat locus and retrotransposons (28, 35). Therefore, Armi functions in both RNAi and piRNA pathways in Drosophila. Our data demonstrate that the vertebrate-specific MOV10L1 is specialized in the piRNA pathway.
Members of the RNA helicase superfamily are required for all biological processes involving RNA molecules, such as ribosome biogenesis, splicing, translation, and RNA interference (36). MOV10L1 is a putative RNA helicase that contains all of the conserved helicase motifs, including ATPase and unwindase domains. Our findings that the truncated mutant protein lacks only the RNA helicase domain and is defective in piRNA biogenesis demonstrate that this domain is required for MOV10L1 functions. Biochemical studies of Drosophila Armi suggest that Armi facilitates ATP-dependent incorporation of single-stranded siRNA into RISC (34). Analogous to the role of Armi in RISC maturation, MOV10L1 may facilitate loading of piRNAs into piRNP complexes. One explanation for the loss of 25–32-nt small RNAs (piRNAs) in the total small RNA library from P10 Mov10l1 mutant testes is that piRNAs are generated but fail to incorporate into MILI and thus are degraded. Alternatively, MOV10L1 might be required for the primary processing in the biogenesis of piRNAs.
Materials and Methods
Antibodies, Western Blot Analyses, and DNA Constructs.
Two GST-MOV10L1 (amino acids 1–101 and 53–253) fusion proteins were expressed in Escherichia coli. Purified recombinant proteins were used to immunize rabbits. Other antibodies used were MILI (Abcam), MIWI (Abcam), MIWI2 (J. Martinez and G. Hannon), TDRD1, RNF17, MVH (T. Noce), LINE1 ORF1p (S. L. Martin), IAP GAG (B. R. Cullen), MAEL (Abcam), GASZ (M. M. Matzuk), β-actin (Sigma-Aldrich), HA (Santa Cruz Biotechnology), and Myc (EMBL MACF).
Coding sequences for Miwi, Miwi2, and Mili were cloned into the pcDNA3 vector with an N-terminal 3× Myc tag. The Mov10l1 ORF was inserted into the pCIneo vector with an N-terminal HA tag. Plasmids were cotransfected into HEK 293T cells, and lysates were prepared after 48 h for immunoprecipitations.
Immunoprecipitation, Mass Spectrometry, and Detection of Small RNAs.
Eight pairs of 18- to 20-d testes (≈300 mg) were used for immunoprecipitation with affinity-purified anti-MOV10L1 antibody followed by SDS/PAGE. Gel bands of interest were cut and sent for protein identification by mass spectrometry at the PENN Proteomics Core facility.
Mouse testicular extract preparation, immunoprecipitations, purification of MILI complexes for complex identification, and 5′ end-labeling of piRNAs were performed as described previously (17). Equal numbers of age-matched Mov10l1+/− or Mov10l1−/− testes were used in experiments describing small RNA association with MILI and MIWI2 (Fig. 4).
Targeted Inactivation of the Mov10l1 Gene.
To generate the Mov10l1 targeting construct, DNA fragments were amplified by high-fidelity PCR using a Mov10l1 BAC clone (RP23-269F24) as template. V6.5 ES cells were electroporated with linearized Mov10l1 targeting construct (pKe16-1/ClaI). Screening of ES cells was described previously (37). Two ES cell clones (A1 and B10) harboring the Mov10l1fl allele were injected into B6C3F1 (Taconic) blastocysts. The Mov10l1fl allele was transmitted through the germline in chimeric mice derived from both clones. All offspring were genotyped by PCR. Wild-type (398 bp) and floxed (592 bp) alleles were assayed by PCR with the primers GGCCTATGGGTTGAATGTGT and CAGGAAGAGCAGGTGAAGTG. The Mov10l1 knockout (461 bp) allele was assayed by PCR with the primers GGGTCGTGGATCTGGGATAT and CAGGAAGAGCAGGTGAAGTG.
Histological, Surface-Spread, and Immunofluorescence Analyses.
For histology, testes were fixed in Bouin's solution overnight, dehydrated in ethanol, embedded in paraffin, sectioned, and stained with hematoxylin and eosin. Surface-spread analysis of spermatocyte nuclei and immunofluorescence analyses of testis sections were described previously (37).
Southern and Northern Blot Analyses.
Methylation-sensitive Southern blot, Northern blot, and bisulfite analyses were described previously (17).
Small RNA Library Construction, Sequencing, and Bioinformatic Analyses.
MOV10L1-associated small RNAs from adult mouse testes extract and 18–32-nt total small RNAs from Mov10l1+/− and Mov10l1−/− P10 testes were used for preparation of libraries for deep sequencing by Solexa technology (Illumina). MILI and TDRD1 libraries were previously described (17). Sequencing reads that perfectly mapped to the mouse genome were considered for analyses: MOV10L1 (≈2.2 million), Mov10l1−/− (≈500,000), and Mov10l1+/− (≈125,000). Small RNA sequencing data used in this study are deposited in the GEO database under the accession number GSE21763.
Supplementary Material
Acknowledgments
We thank the following for gifts of antibodies: J. Martinez (Institute of Molecular Biotechnology GmbH, Vienna) and G. Hannon (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) for MIWI2 antibody, B. R. Cullen (Duke University Medical Center, Durham, NC) for IAP antibody, S. L. Martin (University of Colorado School of Medicine, Denver) for L1 ORF1p antibody, M. M. Matzuk (Baylor College of Medicine, Houston) for GASZ antibody, T. Noce (Shiga University of Medical Science, Shiga, Japan) for MVH antibody, and C. Höög (Karolinska Institutet, Stockholm) for SYCP1 antibody; we also thank C. Yuan for mass spectrometry and Stephanie Eckhardt for imaging analysis; the European Molecular Biology Laboratory (EMBL) Protein Expression and Gene Core facilities for antibody production and Solexa sequencing; and D. O'Carroll, F. Yang, A. Verdel, and members of the Pillai group for comments on the manuscript. This work was supported by EMBL (R.S.P.), Agence National de la Recherche Jeune Chercheur program (piRmachines) (R.S.P), and National Institutes of Health/National Institute of General Medical Sciences Grant RO1GM076327 (to P.J.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Solexa deep-sequencing data have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE21763).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1003953107/-/DCSupplemental.
References
- 1.Ghildiyal M, Zamore PD. Small silencing RNAs: An expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 4.Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: Progress and prospect. Annu Rev Cell Dev Biol. 2009;25:355–376. doi: 10.1146/annurev.cellbio.24.110707.175327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aravin AA, et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuramochi-Miyagawa S, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- 7.Deng W, Lin H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell. 2002;2:819–830. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]
- 8.Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- 9.Kuramochi-Miyagawa S, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gunawardane LS, et al. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 11.Brennecke J, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 12.Wang PJ, McCarrey JR, Yang F, Page DC. An abundance of X-linked genes expressed in spermatogonia. Nat Genet. 2001;27:422–426. doi: 10.1038/86927. [DOI] [PubMed] [Google Scholar]
- 13.Meister G, et al. Identification of novel argonaute-associated proteins. Curr Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- 14.Chendrimada TP, et al. MicroRNA silencing through RISC recruitment of eIF6. Nature. 2007;447:823–828. doi: 10.1038/nature05841. [DOI] [PubMed] [Google Scholar]
- 15.Chen C, et al. Mouse piwi interactome identifies binding mechanism of tdrkh tudor domain to arginine methylated miwi. Proc Natl Acad Sci USA. 2009;106:20336–20341. doi: 10.1073/pnas.0911640106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vagin VV, et al. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev. 2009;23:1749–1762. doi: 10.1101/gad.1814809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reuter M, et al. Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat Struct Mol Biol. 2009;16:639–646. doi: 10.1038/nsmb.1615. [DOI] [PubMed] [Google Scholar]
- 18.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 19.Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aravin A, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 21.Wang PJ, Page DC, McCarrey JR. Differential expression of sex-linked and autosomal germ-cell-specific genes during spermatogenesis in the mouse. Hum Mol Genet. 2005;14:2911–2918. doi: 10.1093/hmg/ddi322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma L, et al. GASZ is essential for male meiosis and suppression of retrotransposon expression in the male germline. PLoS Genet. 2009;5:e1000635. doi: 10.1371/journal.pgen.1000635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewandoski M, Meyers EN, Martin GR. Analysis of Fgf8 gene function in vertebrate development. Cold Spring Harb Symp Quant Biol. 1997;62:159–168. [PubMed] [Google Scholar]
- 24.Ueyama T, Kasahara H, Ishiwata T, Yamasaki N, Izumo S. Csm, a cardiac-specific isoform of the RNA helicase Mov10l1, is regulated by Nkx2.5 in embryonic heart. J Biol Chem. 2003;278:28750–28757. doi: 10.1074/jbc.M300014200. [DOI] [PubMed] [Google Scholar]
- 25.Liu ZP, Olson EN. Suppression of proliferation and cardiomyocyte hypertrophy by CHAMP, a cardiac-specific RNA helicase. Proc Natl Acad Sci USA. 2002;99:2043–2048. doi: 10.1073/pnas.261708699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carmell MA, et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 27.Soper SF, et al. Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev Cell. 2008;15:285–297. doi: 10.1016/j.devcel.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malone CD, et al. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li C, et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137:509–521. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 31.Bourc'his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 32.Dalmay T, Horsefield R, Braunstein TH, Baulcombe DC. SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J. 2001;20:2069–2078. doi: 10.1093/emboj/20.8.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cook HA, Koppetsch BS, Wu J, Theurkauf WE. The Drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell. 2004;116:817–829. doi: 10.1016/s0092-8674(04)00250-8. [DOI] [PubMed] [Google Scholar]
- 34.Tomari Y, et al. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell. 2004;116:831–841. doi: 10.1016/s0092-8674(04)00218-1. [DOI] [PubMed] [Google Scholar]
- 35.Vagin VV, et al. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 36.Tanner NK, Linder P. DExD/H box RNA helicases: From generic motors to specific dissociation functions. Mol Cell. 2001;8:251–262. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- 37.Yang F, et al. Meiotic failure in male mice lacking an X-linked factor. Genes Dev. 2008;22:682–691. doi: 10.1101/gad.1613608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.