Abstract
Objectives
Schizophrenia is associated with dysfunction of glutamatergic neurotransmission, and several studies have suggested glutamatergic abnormalities in bipolar disorder. Recent data suggest involvement of the NMDA receptor-signaling complex, which includes NMDA receptor subunits as well as associated intracellular interacting proteins critical for NMDA receptor assembly, trafficking, and activation; the most well characterized being PSD-93, PDS-95, SAP102, and NF-L. Previously, studies from our laboratories have described changes in glutamate receptor subunit transcript and binding site expression in schizophrenia, and changes in NMDA receptor binding site expression in bipolar disorder in postmortem brain tissue. In the present work, we focus on the expression of these molecules in hippocampus in schizophrenia and bipolar affective disorder I.
Methods
We performed in situ hybridization to assess hippocampal expression of the transcripts encoding NMDA receptor subunits NR1, 2A, 2B, 2C and 2D, and the transcripts for the NMDA receptor associated PSD proteins PSD-95, PSD-93, NF-L and SAP102 in subjects with schizophrenia, bipolar affective disorder I, and a comparison group. We also measured [3H]CGP39653 and [3H]MK-801 binding site expression in the hippocampus in schizophrenia.
Results
There was a significant decrease in the expression of transcripts for NR1 and NR2A subunits and SAP102 in bipolar disorder. We did not detect any changes in these transcripts or in binding site expression in the hippocampus in schizophrenia.
Conclusions
We propose that the NMDA receptor-signaling complex, including the intracellular machinery that is coupled to the NMDA receptor subunits, is abnormal in the hippocampus in bipolar disorder. These data suggest bipolar disorder might be associated with abnormalities of glutamate-linked intracellular signaling and trafficking processes.
Keywords: [3H]CGP39653, [3H]MK-801, binding, in situ hybridization, postmortem, human, glutamate, psychosis, NMDA, PSD, post synaptic density, PDZ domain
Introduction
The glutamate hypothesis of schizophrenia was originally based on the observation that phencyclidine (PCP) can precipitate a schizophreniform psychosis that is indistinguishable from schizophrenia in non-psychiatrically ill persons. Since PCP is an antagonist of the NMDA subtype of the glutamate receptor, it has been postulated that NMDA receptor dysfunction contributes to psychotic pathophysiology. Postmortem studies of persons afflicted with schizophrenia and, to a lesser extent bipolar disorder, have demonstrated region- and subunit-specific alterations in the expression of NMDA receptor subunit transcripts and binding sites, supporting a hypothesis of NMDA receptor dysfunction in these illnesses [1,3,5,17,22,48].
The NMDA receptor is a ligand-gated ion channel formed by an obligate NR1 subunit and combinations of NR2A-D subunits [15]. The NMDA receptor subunits have consensus phosphorylation and protein-protein interaction sites that link the NMDA receptor with intracellular signaling pathways. The development of techniques such as yeast 2-hybrid that permit identification of previously unknown protein-protein interactions has led to the discovery of NMDA receptor-interacting proteins that modulate receptor activity via interactions with the cytoplasmic tails of the NMDA subunits. These protein-protein interactions facilitate clustering and anchoring of the NMDA receptor in the postsynaptic density, leading to the assembly of a signaling complex that involves the receptor, cytostructural proteins, and signal transduction enzymes [30,32]. The NMDA receptor interacting proteins are enriched in the postsynaptic density (PSD), and include PSD95, PSD93, and synapse associated protein 102 (SAP102), which primarily interact with the NR2B subunit, and neurofilament-light (NFL), which interacts with exon 21 containing NR1 subunits [32,41].
Expression of NMDA receptors and NMDA receptor interacting proteins parallel the widespread distribution of glutamatergic synapses throughout the brain. In medial temporal lobe structures, the expression of these molecules is consistent with well-characterized glutamatergic circuitry. This region includes the dentate gyrus, the hippocampal subfields, the subiculum and adjacent cortical regions that are reciprocally interconnected via glutamatergic projections [9]. Previously, changes in the structure and function of the medial temporal lobe in schizophrenia, and to a lesser extent bipolar disorder, have been reported based upon techniques utilizing imaging, histopathology, and measures of gene expression [4,12,37,44,46]. Abnormalities of NMDA receptor expression have been reported in the hippocampus in schizophrenia and bipolar disorder, consistent with the hypothesis of altered glutamate neurotransmission in these illnesses [5,43]. The preponderance of data implicating the hippocampus and related structures in schizophrenia and bipolar disorder, taken together with the robust demonstration of glutamate receptor dysfunction, suggest that alterations of glutamatergic neurotransmission, and in particular the NMDA receptor signaling complex, likely underlie some of these hippocampal abnormalities. Thus, we performed in situ hybridization to assess hippocampal expression of the transcripts encoding NMDA receptor subunits NR1, 2A, 2B, 2C and 2D, and the transcripts for the NMDA receptor associated PSD proteins PSD-95, PSD-93, NF-L and SAP102 in subjects with schizophrenia, bipolar disorder and a comparison group. We also measured NMDA receptor binding site expression in the hippocampus in schizophrenia.
Materials and Methods
Subjects
Twenty-four subjects from the neural tissue repository at the Rebecca L. Cooper Research Laboratories, Victoria, Australia, were studied, comprised of three groups of 8 subjects each with diagnoses of schizophrenia, bipolar disorder, and normal controls. The left hippocampi from each subject were obtained as previously described [6,47]. Blocks of hippocampal tissue at 22mm rostral to the anterior commissure were taken using a standard methodology [42]. Normal controls and subjects with a diagnosis of bipolar affective disorder I (hereafter referred to as bipolar disorder) or schizophrenia were matched for age, pH, PMI and sex. Subjects with bipolar disorder were psychotic at the time of death, and the DSM-IV diagnoses of bipolar affective disorder I and schizophrenia were confirmed by a senior psychologist and a psychiatrist using the Diagnostic Instrument for Brain Studies [20]. Demographic data are shown in Table 1. There were no differences between diagnostic groups for age, sex, PMI or pH. Cases were excluded if more than five hours elapsed between the subject last being seen alive and being found dead. Cryostat prepared (20 um) sections from the left middle hippocampus of each brain were stored at -70°C until used.
Table 1.
Subject Characteristics
| Diagnosis | Age | Sex | PMI | pH | Cause of death | AS* | MS* |
|---|---|---|---|---|---|---|---|
| CTL | 77 | F | 17 | 6.32 | cardiomyopathy | none | none |
| CTL | 47 | F | 24 | 5.89 | pulmonary embolus | none | none |
| CTL | 57 | M | 27 | 6.43 | ischemic heart disease | none | none |
| CTL | 38 | M | 46 | 6.42 | trauma | none | none |
| CTL | 68 | M | 41 | 6.06 | aortic stenosis | none | none |
| CTL | 39 | F | 65 | 6.38 | mitral valve prolapse | none | none |
| CTL | 56 | F | 24 | 5.88 | cardiac tamponade | none | none |
| CTL | 67 | M | 32 | 6.14 | myocardial infarction | none | none |
| mean/total | 56.1 ± 14 | 4F, 4M | 34.5 ± 15 | 6.2 ± 0.2 | suicide: 0/8 | 0/8 | 0/8 |
| BD | 74 | F | 45 | 6.26 | drug toxicity | FLU | none |
| BD | 58 | F | 41 | 5.68 | ischemic heart disease | none | none |
| BD | 59 | M | 34 | 4.46 | ruptured aorta | none | Li+ |
| BD | 38 | M | 24 | 6.42 | CO poisoning | CPZ | Li+ |
| BD | 66 | M | 17 | 6.41 | aspiration | FLU | none |
| BD | 55 | F | 52 | 6.46 | unknown | CPZ | Li+ |
| BD | 60 | F | 50 | 6.08 | cardiomyopathy | TRI | Li+ |
| BD | 61 | M | 58 | 6.44 | myocardial infarction | FLU, MEL | VPA |
| mean/total | 58.9 ± 10 | 4F, 4M | 39.1 ± 13 | 6.3 ± 0.2 | suicide: 2/8 | 6/8 | 5/8 |
| SCZ | 79 | F | 26 | 6.27 | hypothermia | FLU | none |
| SCZ | 48 | F | 52 | 6.21 | pulmonary embolus | FLU | Li+ |
| SCZ | 57 | M | 24 | 6.06 | coronary atheroma | FLU, CPZ | none |
| SCZ | 38 | M | 40 | 5.52 | mediastinitis | HAL | none |
| SCZ | 65 | M | 41 | 6.57 | ischemic heart disease | FLU | none |
| SCZ | 47 | F | 50 | 6.31 | pneumonia | RIS | none |
| SCZ | 65 | F | 50 | 6.35 | ruptured aorta | FLU | none |
| SCZ | 69 | M | 48 | 6.44 | CO poisoning | HAL | none |
| mean/total | 58.5 ± 14 | 4F, 4M | 41.4 ± 11 | 6.2 ± 0.3 | suicide: 1/8 | 8/8 | 1/8 |
Abbreviations: postmortem interval (PMI), male (M), female (F), control (CTL), bipolar disorder (BD), schizo-phrenia (SCZ), haloperidol (HAL), risperidol (RIS), mellaril (MEL), chlorpromazine (CPZ), fluphenazine (FLU), trifluoperazine (TRI), lithium (Li+), valproic acid (VPA), carbon monoxide (CO). Age and DOI are expressed in years.
Mood stabilizer (MS) or antipsychotic (AS) treatment at time of death. Data expressed as mean+/- SD
In Situ Hybridization
The design of this study was to examine expression of the transcripts encoding the NMDA receptor subunits NR1, NR2A-D, and the NMDA-associated PSD proteins neurofilament light-chain (NF-L), PSD95, PSD93 and synapse-associated protein 102 (SAP102). Subclones for the NMDA receptor subunits NR1 and NR2A-2D were prepared and characterized as previously described [16,17]. Subclones of unique areas of SAP102, NF-L, PSD93, and PSD95 were prepared as previously described [3,16]. Riboprobes were synthesized from linearized plasmid DNA and in situ hybridization was performed as previously described [31,45]. For riboprobe synthesis, 8 μl of [35S]-UTP was dried and 2.0 μl 5X transcription buffer, 1.0 μl 0.1M DTT, 3.0 μl of 10mM ATP, CTP, and GTP, 2.0 μl linearized plasmid DNA, 0.5 μl RNase inhibitor, and 1.5 μl SP6 or T7 RNA polymerase were combined and incubated for 2 hours at 37°C. 1.0 μl DNase (RNase-free) was added and the mixture was incubated for 15 minutes at room temperature (RT). Radiolabeled probe was purified with microspin chromatography columns (Bio-Rad, Hercules, CA). Two slides per subject for each antisense probe were removed from -70°C and placed in 4% (weight:vol) formaldehyde at RT for one hour. The slides were then washed in 2X SSC (300mM NaCl/30 mM sodium citrate, pH 7.2) three times for 5 minutes each. Slides were washed in deionized water for one minute and placed in 0.1M triethanolamine, pH 8.0/acetic anhydride, 400:1 (vol:vol) on a stir plate for ten minutes. A final wash was in 2X SSC for five minutes, followed by dehydration through graded alcohols and air-drying for 30 minutes. A cover slip with 200 μl of riboprobe (2 million cpm, 50% formamide buffer (50% formamide, 10% dextran sulfate, 3 × SSC, 50 mM Na2HPO4 (pH 7.4), 10 mM dithiothreitol, 1 × Denhardt’s solution, 100 ug/ml yeast tRNA), and 0.01M dithiothreitol) was placed on each slide. Slides were placed in a covered tray with a filter paper liner saturated with 50% formamide. After overnight incubation at 55°C, cover slips were removed and the slides were placed in 2X SSC for five minutes, followed by RNase (200 μg/ml in 10 mM Tris-HCl (pH 8.0) and 0.5 M NaCl) at 37°C for thirty minutes and then washed as follows: 2X SSC for 15 minutes at RT; 2X SSC for 15 minutes at RT; 1X SSC for 10 minutes at RT; 0.5X SSC for 5 minutes at RT; 0.1X SSC for 60 minutes at 55°C; 0.1X SSC for 60 minutes at 55°C and 0.5X SSC for 15 minutes at RT. The slides were dehydrated in graded ethanol solutions, air-dried, placed in X-ray cassettes, and apposed to Kodak XAR-5 film for 2-60 days.
Image and Data Analyses
One subject in the bipolar group was not utilized since we were unable to detect any mRNA signal (labeling over the tissue was at background) with any of the probes we utilized for this study, including probes for genes NR1 and NF-L that are highly expressed, suggesting very low RNA quality. Film was developed and used for quantitative computer image analysis (Scion Image, 4.0.2), as previously described [34]. Areas of the hippocampus evaluated include CA1, CA2, CA3, CA4 and the dentate gyrus (DG). We analysed the entirety of each subfield for each section studied, with conservative margins where the subfields interface, and the collection of imaging data was performed blind to diagnosis. The subiculum (Sub) was grouped with the hippocampal subfields and the DG for statistical analysis. Tissue background readings were subtracted from gray scale values collected for each specific region of interest for each tissue section. Two tissue sections were studied for each subject. These background corrected values were averaged, providing one mean value per region, per subject, for each probe. Background-adjusted, averaged gray scale values were converted into optical density. Optical density values were converted to units of bound radiation from a standard curve generated from a [14C] microscale standard (Amersham Life Sciences, Amersham UK) placed on each film [10,50]. Transcript concentration, in turn, was determined from the amount of bound radiation, the specific activity of each batch of [35S]-uridine triphosphate, and the number of uracil residues contained in each probe sequence [14,16,19,26,33,35,36]. All data are expressed as femtomoles mRNA per gram tissue (fmol/g).
Binding Studies
The binding densities of the radioligands [3H]MK-801 and [3H]CGP39653 were carried out as previously described [43]. For [3H]MK-801 binding, sections were pre-incubated in assay buffer (50 mM Tris-acetate (pH 7.4) containing 100 μM glutamate, 50 μM glycine and 50 μM spermidine) at room temperature (RT) for 30 minutes. The specific binding of [3H]MK-801 (20 nM) was defined as the difference in radiolabel binding in the absence (TB) and presence (NS) of MK-801 (100μM) following a 60 minute incubation, in assay buffer, at RT [18]. The density of [3H]CGP39653 binding was determined following a 45 minute pre-incubation in assay buffer (50 mM Tris-HCl (pH 8)) at RT. Sections were incubated in assay buffer with [3H]CGP39653 (20 nM) in the absence (TB) or presence (NS) of L-glutamic acid (1 mM) for 45 minutes at RT [16]. Following their incubations with the respective radioligands, all sections were washed twice in ice-cold assay buffer and then dipped in distilled water, to remove salts, before being dried in a stream of cool air. Sections used for the determination of [3H]MK-801 were apposed to Hyperfilm-[3H] with [3H]microscales until an image suitable for analysis was obtained. The film was then developed in Kodak D19 developer and fixed in Hypam fixative. The images were analysed from film using MCID M1 image analysis system. The density of specific binding was calculated as total binding of the radioligand (TB, three sections) minus the binding of the radioligand in the presence of a non-radioactive drug that binds to the same binding site. The sections used for the determination of [3H]CGP39653 were partially fixed overnight in paraformaldehyde fumes at RT before being apposed to BAS-TR2025 phosphoimage plates with [3H]microscales until an image suitable for analysis was obtained. The phosphoimage plates were then scanned using a BAS5000 high-resolution phosphoimager and the images analysed using AIS image analysis software. For both [3H]CGP39653 and [3H]MK-801 the density of the images was converted to dpm/mg estimated tissue equivalent (ETE) by comparison with the standard curve generated using the relative densities of the [3H]microscales. Using the specific activities of the appropriate radioligand, the values were converted to fmole/mg ETE.
Statistics
Multiple linear regression was performed to test for associations between gene expression and postmortem interval, age, and tissue pH. When significant associations were detected, we utilized analysis of covariance (ANCOVA) as our primary statistic. When ANCOVA was utilized, regressed values were used for post-hoc analysis. Otherwise, our dependent measures were analyzed by factorial analysis of variance with diagnosis and hippocampal subregion as independent variables. Where appropriate, posthoc analyses were performed with Tukey’s honestly significant difference test. For all statistical applications we used the Statistica (Statsoft, Tulsa, OK) software package for Windows 2000. For all tests, α = 0.05. The presence or absence of antipsychotic or mood stabilizer (lithium and valproic acid) treatment was secondarily analyzed for an association with transcript expression.
Results
Sense and antisense probes were prepared for NR1, NR2A, NR2B, NR2C, NR2D, PSD95, PSD93, SAP102, and NF-L. Specific labeling was only observed for sections incubated with antisense riboprobe. We detected NR1, NR2A, NR2B, NR2C, NR2D, PSD95, PSD93, SAP102, and NF-L transcript expression in the hippocampal subfields CA1, CA2, CA3, and CA4, the dentate gyrus (DG), and the subiculum (Sub) (Figures 1 and 2).
Figure 1.
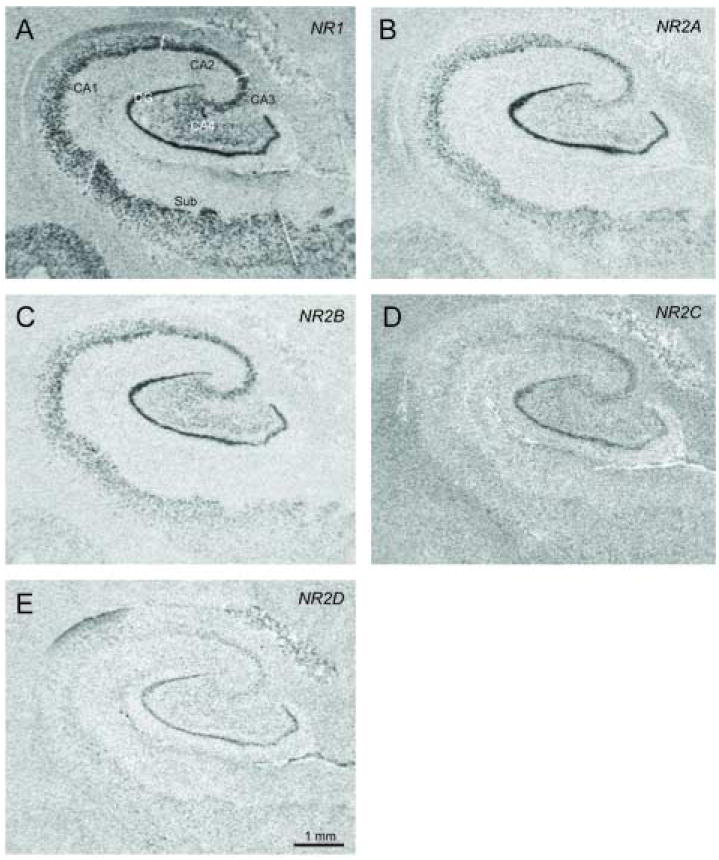
In situ hybridization using [35S] labeled antisense riboprobes for NMDA receptor subunit transcripts in the hippocampal subfields CA1, CA2, CA3, and CA4, the dentate gyrus (DG), and the subiculum (Sub).
Figure 2.
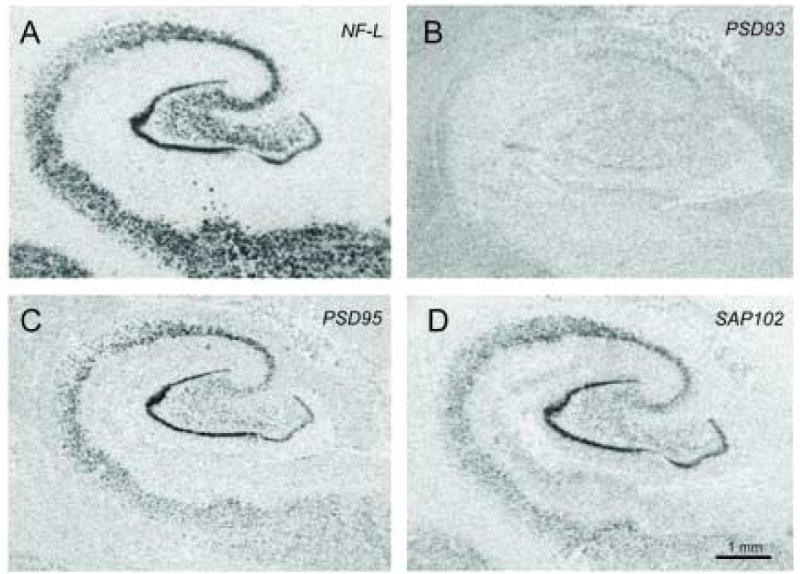
In situ hybridization using [35S] labeled antisense riboprobes for neurofilament light (NF-L) (A), postsynaptic density protein 93 (PSD93) (B), PSD95 (C), and synapse associated protein 102 (SAP102) (D) in hippocampal subfields, the dentate gyrus, and the subiculum.
In the hippocampal subfields, DG, and Sub, we detected an association between NR1, NR2A, and NR2C, but not NR2B or NR2D, mRNA expression and age (NR1: R = 0.36 p = 0.003, NR2A: R = 0.28, p = 0.017, NR2C: R = 0.31, p = 0.037). Multiple regression analyses are summarized in Table 2. We also detected an association between NR1, NR2A, and NR2D, but not NR2B or NR2C, mRNA expression in this region and pH (NR1: R = 0.36, p = 0.0002, NR2A: R = 0.28, p = 0.005, NR2D: R = 0.24, p = 0.01)(Table 2). We did not detect any associations between NMDA receptor subunit expression and PMI. Using ANCOVA, we detected a main effect for diagnosis for NR1 (F (2, 105) = 3.91, p = 0.02) and NR2A (F (2, 106) = 4.37, p = 0.02), but not for NR2B, NR2C, or NR2D, transcript expression in the hippocampus (Figure 3). Post-hoc analysis revealed significantly lower levels of mRNA expression for NR1 (p = 0.02) and NR2A (p = 0.049) in bipolar disorder compared to controls. We also detected a main effect for region for NR1, NR2A, NR2B, NR2C and NR2D mRNA expression (p < 0.001 for each gene for region). Post-hoc analysis revealed significantly (p < 0.05) higher levels of mRNA expression for all of the NMDA subunits in the DG and CA2 versus CA1, CA3, CA4 and the Sub. We did not detect any diagnosis by region interactions for NR1 or NR2A-D in the hippocampus.
Table 2.
Summary of Regression Analysis
| Gene Studied | Multiple Regression Coefficient (R) | Demographic | Beta | p-value |
|---|---|---|---|---|
| NR1 | 0.36 | age | -0.31 | 0.0033 |
| pH | 0.35 | 0.00025 | ||
| NR2A | 0.28 | age | -0.25 | 0.016 |
| pH | 0.27 | 0.0046 | ||
| NR2B | 0.19 | age | -0.16 | 0.15 |
| pH | 0.20 | 0.54 | ||
| NR2C | 0.31 | age | -0.22 | 0.037 |
| pH | -0.15 | 0.10 | ||
| NR2D | 0.24 | age | 0.039 | 0.71 |
| pH | -0.25 | 0.0099 | ||
| NF-L | 0.36 | age | -0.12 | 0.23 |
| pH | 0.39 | 0.000061 | ||
| PSD 93 | 0.25 | age | -0.019 | 0.87 |
| pH | 0.14 | 0.19 | ||
| PSD 95 | 0.43 | age | -0.14 | 0.14 |
| pH | 0.46 | 0.000001 | ||
| SAP102 | 0.41 | age | -0.21 | 0.045 |
| pH | 0.45 | 0.000003 |
Figure 3.
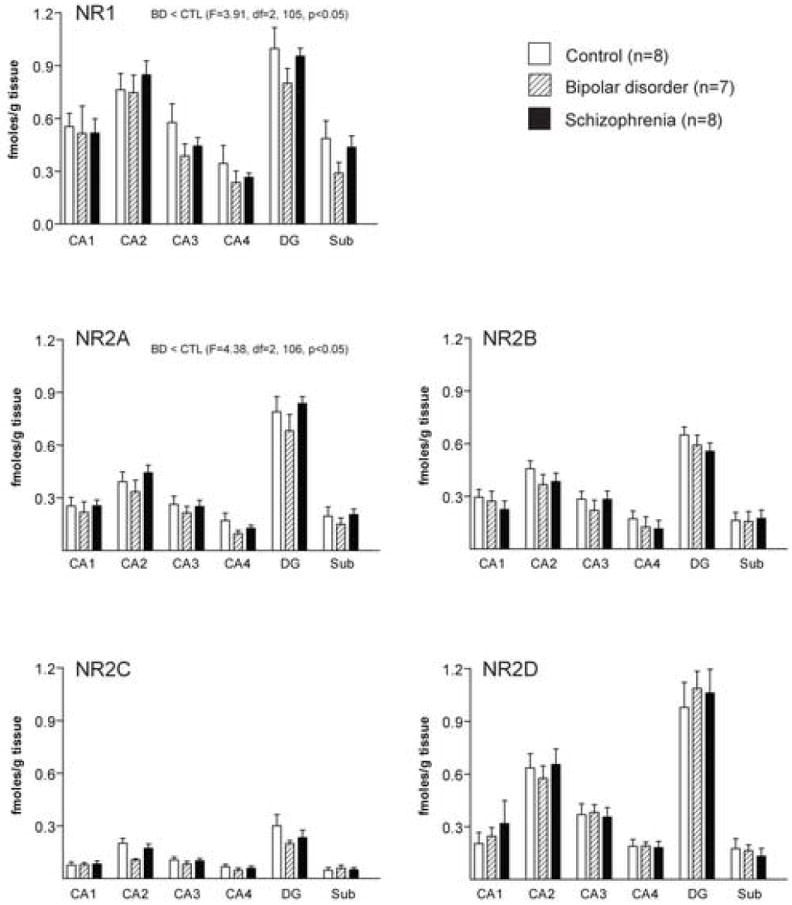
Expression of NMDA receptor subunit transcripts in hippocampal regions CA1, CA2, CA3, CA4, dentate gyrus (DG) and subiculum (Sub) from patients with bipolar disorder (BD), schizophrenia and a control group (CTL). Data expressed as fmol mRNA/g tissue (mean ± SEM).
In the hippocampal subfields, DG, and Sub, we detected an association between NF-L, SAP102, and PSD95, but not PSD93, mRNA expression and pH (NF-L: R = 0.36, p = 0.000061, SAP102: R = 0.41, p = 0.000003, PSD95: R = 0.43, p = 0.000001)(Table 2). We also detected an association between SAP102 transcript expression in this region and age (R = 0.41, p = 0.045)(Table 2). We did not detect any associations between NMDA receptor interacting protein transcript expression and PMI. We detected a main effect for diagnosis for SAP102 (F (5, 104) = 3.61, p = 0.03), PSD93 (F (5, 95) = 3.83, p = 0.03), and NF-L (F (5, 105) = 9.57, p = 0.0002) transcript expression in the hippocampus (Figure 4). We did not detect a main effect for diagnosis for PSD95 mRNA expression. Post-hoc analysis revealed significantly lower levels of mRNA expression for SAP102 (p = 0.048) in bipolar disorder compared to controls. We also detected significantly reduced levels of transcripts for PSD93 (p = 0.02), and NF-L (p = 0.0009) in the hippocampus in bipolar disorder compared to schizophrenic subjects. We detected a main effect for region for SAP102, PSD95, PSD93, and NF-L mRNA expression (p < 0.001 for each gene for region). Post-hoc analysis revealed significantly (p < 0.05) higher levels of mRNA expression for SAP102, PSD95, PSD93 or NF-L in the DG and CA2 versus CA1, CA3, CA4 and the Sub. We did not detect any diagnosis by region interactions for SAP102, PSD95, PSD93 or NF-L in the hippocampus.
Figure 4.
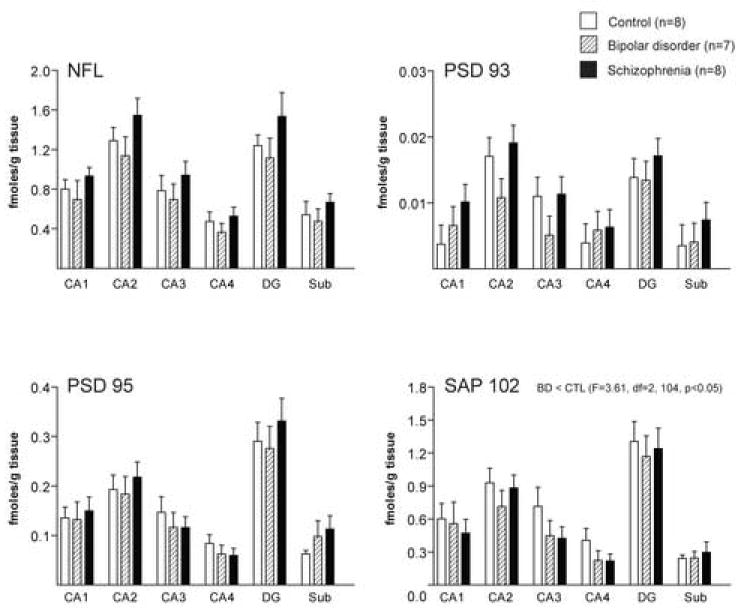
Expression of transcripts for NMDA receptor interacting molecules in hippocampal regions CA1, CA2, CA3, CA4, dentate gyrus (DG) and subiculum (Sub) from patients with bipolar disorder (BD), schizophrenia and a control group (CTL). Data expressed as fmol mRNA/g tissue (mean ± SEM).
Binding data for the bipolar group from these experiments was published previously [43]. We did not detect any associations between NMDA receptor binding site expression and PMI, pH, or age. We measured [3H]CGP39653 and [3H]MK-801 binding in the hippocampal subfields, DG, and Sub (Figure 5). We did not detect any changes in [3H]CGP39653 or [3H]MK-801 binding in the hippocampus in schizophrenia (Figure 6). Finally, we performed correlation analyses for the effects of treatment with psychotropic medications. We did not detect a significant association between NR1, NR2A, or SAP102 mRNA expression and treatment with a mood stabilizer (lithium or valproic acid), for the entire group of 24 subjects or within the bipolar cohort (data not shown).
Figure 5.
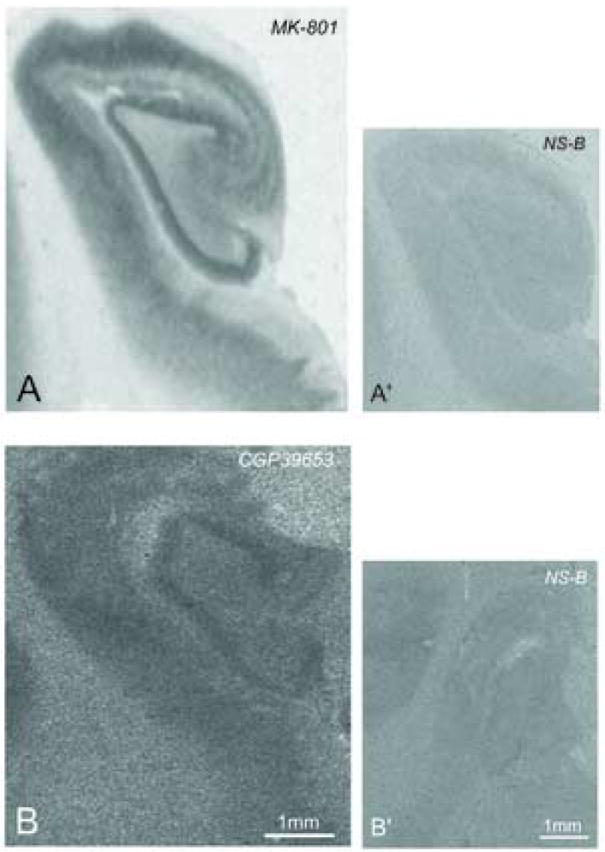
[3H]MK-801 (A) and [3H]CGP39653 (B) binding site expression in hippocampal regions CA1, CA2, CA3, dentate gyrus (DG) and subiculum (Sub). Panel A’ is [3H]MK-801 binding in the presence of unlabeled MK-801, Panel B’ is [3H]CGP39653 binding in the presence of unlabeled L-glutamic acid.
Figure 6.
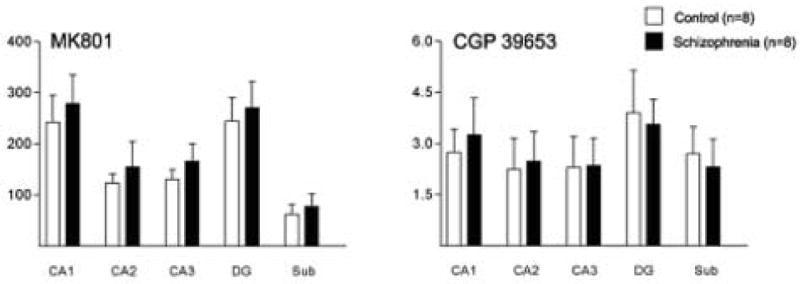
[3H]CGP39653 and [3H]MK-801 binding site expression in hippocampal regions CA1, CA2, CA3, CA4, dentate gyrus (DG) and subiculum (Sub) from patients with schizophrenia and a control group. Data expressed as fmol/mg estimated tissue equivalent.
Summary of results
We found decreased NR1, NR2A and SAP102 transcript expression in bipolar disorder, but not schizophrenia. The largest decreases in NR1 mRNA expression were detected in CA3 (33%) and the Sub (41%), for NR2A in CA4 (44%), and for SAP102 in CA2 (24%), CA3 (37%), and CA4 (45%). We did not detect any significant changes in NR2B, NR2C, NR2D, PSD93, PSD95, or NF-L expression in the hippocampus in bipolar disorder or schizophrenia. We did not detect any changes in [3H]CGP39653 or [3H]MK-801 binding in schizophrenia.
Discussion
We found significant decreases in NR1 and NR2A transcript expression in the hippocampus in bipolar disorder (Figure 3). These findings are consistent with a previous report of decreased [3H]CGP39653 and [3H]MK-801 binding site expression in bipolar disorder in the same subjects. [3H]CGP39653 binds to the glycine coagonist site which is on the NR1 subunit, while [3H]MK-801 binds to a site accessible only when the ion channel is open and preferentially binds NR2A- or NR2B-containing receptor complexes [38]. Thus, our findings of decreased NR1 and NR2A transcripts and the previously published binding data from the same cohort are consistent with one another, and they suggest a decrease in the total number of NMDA receptors as well as a decrease in the number of open ion channels in bipolar disorder [43]. Decreased NR1 mRNA has also been reported in the dentate gyrus in a different cohort of subjects with bipolar disorder, while another study did not detect changes in NR1 immunoreactivity in the hippocampus in this illness [22,48]. Taken together, the balance of these studies suggests that NMDA receptor expression is altered in the hippocampus in bipolar disorder. Since NMDA receptors mediate molecular correlates of learning and memory, such as long term potentiation, changes in NMDA receptor expression may have a profound effect on brain function in this illness.
In bipolar subjects, we found changes in NR1 and NR2A, but not NR2B, transcript expression. While expression of NMDA receptor subunits may be coordinately regulated, previous studies have found that NMDA receptor subunit transcript expression may be divergent in human disease states [27-29]. Our data suggest that NMDA receptor subunit mRNAs, in particular NR1 and NR2B, may not be co-regulated at the transcript level in bipolar disorder.
We also detected decreased expression of SAP102 transcripts in the hippocampus in bipolar disorder. This finding represents the first such report of an altered NMDA receptor interacting protein in the hippocampus in this illness. SAP102 mediates NMDA receptor trafficking by interacting with the cytoplasmic tail of the NR2B subunit and the intracellular trafficking molecule Sec8 [41]. SAP102 also binds signaling molecules, including protein kinase C alpha, that contribute to the NMDA receptor signaling complex in the PSD [23]. Thus, a decrease in SAP102 expression, in the context of no changes in NR2B expression, might lead to a decrease in the number of NR2B-containing NMDA receptors delivered to the postsynaptic density (PSD), and alterations in the composition of NMDA receptor signaling complexes in the PSD. Such changes could have a significant impact on synaptic plasticity, since electrophysiological processes in the hippocampus, such as long term potentiation, are mediated by NMDA receptor activation [25].
We did not detect changes in NMDA receptor subunit transcript expression in the hippocampus in schizophrenia (Figure 3). This finding is consistent with the absence of changes in glutamate and phencyclidine (PCP) binding site expression in the hippocampus in this illness in two previous reports [11,21], while a third study only found changes in the PCP binding site in CA3 [5]. However, other studies have detected changes in NR1 and NR2B transcript expression in the hippocampus in schizophrenia, including one study with a cohort of different subjects from the same brain bank as the subjects utilized in the present study [11,22].
Several factors may account for these divergent mRNA expression findings. Ours was the only cohort studied that was comprised of subjects who were actively psychotic at the time of death, and there may be significant differences in hippocampal gene expression in psychotic versus nonpsychotic states. We measured gene expression in the middle hippocampus, but not the dorsal or ventral areas. Hippocampal gene expression is not homogenous, and we only sampled one level of this structure, possibly accounting for the disparate findings of NMDA receptor expression in the present study compared to previous reports [13]. Another possibility is that our study was underpowered to detect changes in NR1 or NR2B transcript expression, since our cohort has fewer subjects than the previous studies reporting differences in schizophrenia. This is unlikely given that we were able to detect a moderate (10-15%) change in NR1 expression in the hippocampus in bipolar disorder. Finally, the use of more sensitive methods such as quantitative PCR might have found differences in hippocampal gene expression in schizophrenia. Nevertheless, while our transcript studies are not consistent with other studies of NMDA subunit mRNA expression in the hippocampus, there is a consensus of no detectable changes in studies that have measured NMDA receptor binding sites in the hippocampus in schizophrenia.
Dissimilar to our results with bipolar disorder, we did not detect any changes in NMDA receptor interacting protein expression in the hippocampus in schizophrenia (Figure 4). This result was unexpected given our previous findings of altered PSD95 and NF-L transcript expression in the thalamus [3] and prefrontal cortex [2], as well as a report of decreased SAP102 protein expression in the hippocampus in this illness [49]. Taken together, these findings suggest that the hippocampus may be spared with regard to abnormalities of NMDA receptor interacting molecule expression in schizophrenia. Supporting this hypothesis, one study reported differences in PSD95 mRNA expression in the prefrontal cortex, but not in the hippocampus, in schizophrenia [40].
Our finding of decreased NR1, NR2A, and SAP102 mRNA expression in bipolar disorder is potentially confounded by the presence of mood stabilizer treatment. Valproic acid (VPA) and lithium have been shown to modulate glutamatergic neurotransmission in the rat brain [7,8]. In addition, chronic lithium treatment has been shown to inhibit NMDA receptor mediated calcium currents and suppress phosphorylation of molecules in the NMDA receptor signaling complex [24,39]. These results indicate that mood stabilizers may regulate NMDA receptor expression and function. Since five out of eight of the subjects with bipolar disorder were taking lithium (4 subjects) or valproic acid (1 subject) at the time of death, we performed a secondary analysis to determine if there was an association between mood stabilizer treatment and mRNA expression. We did not detect a significant association between NR1, NR2A, or SAP102 mRNA expression and treatment with a mood stabilizer (lithium or valproic acid), for the entire group of 24 subjects or within the bipolar cohort. This result is difficult to interpret because of the inherent selection bias of a psychiatric population where the patients who are the most ill, with possibly the most glutamatergic dysfunction, are frequently taking a mood stabilizer. Due to the small sample size of our cohort and the correspondingly low analytic power to detect a medication effect, it remains a possibility that our findings of altered NR1, NR2A, and SAP102 expression in bipolar disorder might be secondary to regulation by a mood stabilizer.
In addition, since we only measured mRNA expression for the NMDA interacting proteins, our findings do not indicate whether or not there was a change in total protein expression. Another confounding issue is the very low expression levels of NR2C, raising the possibility that the lack of an effect for expression of this transcript was due to a floor effect. However, NR2C expression is generally low in the HPC and for each subject we did detect a grey scale value above background. Finally, changes in mRNA or binding site expression do not necessarily indicate a change in receptor function. There may also be changes in receptor surface expression, localization to the proper region of the plasma membrane, or linkage to intracellular signaling machinery.
Conclusions
Our findings of decreased expression of NMDA receptor subunits and an NMDA receptor interacting protein in the hippocampus in bipolar disorder suggest that NMDA receptor activity may be abnormal in this illness. Decreases in NMDA receptor subunit and binding site expression suggest that the overall level of NMDA receptor expression may be decreased in bipolar disorder. The decrease in SAP102 mRNA expression suggests that there may be a deficit in NMDA receptor trafficking. Our data suggest that the pathophysiology of bipolar disorder includes abnormalities of NMDA receptor expression, and support modulation of glutamate transmission as a target for pharmacological intervention in this devastating illness.
Acknowledgments
This work was supported by a Pfizer Postdoctoral Fellowship (REM) and MH53327 (JMW), Stanley Research Centre Grant 03-RC-002 (BD) and in part by a project grant from the National Health and Medical Research Council of Australia #14253 (BD). The authors would like to thank Geoff Pavey for technical assistance and as curator of the brain bank and Robyn Bradbury for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Akbarian S, Sucher NJ, Bradley D, Tafazzoli A, Trinh D, Hetrick WP, Potkin SG, Sandman CA, Bunney WE, Jr, Jones EG. Selective alterations in gene expression for NMDA receptor subunits in prefrontal cortex of schizophrenics. J Neurosci. 1996;16:19–30. doi: 10.1523/JNEUROSCI.16-01-00019.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beneyto M, Vandervliet JD, Velasquez GR, Meador-Woodruff JH. Cell and Laminar-specific abnormalities in AMPA and NMDA associated postsynaptic protein expression in prefrontal cortex in schizophrenia and mood disorders. Society for Neuroscience Annual Meeting. 2004;1022.4 [Google Scholar]
- 3.Clinton SM, Haroutunian V, Davis KL, Meador-Woodruff JH. Altered Transcript Expression of NMDA Receptor-Associated Postsynaptic Proteins in the Thalamus of Subjects With Schizophrenia. Am J Psychiatry. 2003;160:1100–9. doi: 10.1176/appi.ajp.160.6.1100. [DOI] [PubMed] [Google Scholar]
- 4.Csernansky JG, Wang L, Jones D, Rastogi-Cruz D, Posener JA, Heydebrand G, Miller JP, Miller MI. Hippocampal deformities in schizophrenia characterized by high dimensional brain mapping. Am J Psychiatry. 2002;159:2000–6. doi: 10.1176/appi.ajp.159.12.2000. [DOI] [PubMed] [Google Scholar]
- 5.Dean B, Scarr E, Bradbury R, Copolov D. Decreased hippocampal (CA3) NMDA receptors in schizophrenia. Synapse. 1999;32:67–9. doi: 10.1002/(SICI)1098-2396(199904)32:1<67::AID-SYN9>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 6.Dean B, Scarr E, Pavey G, Copolov D. Studies on serotonergic markers in the human hippocampus: changes in subjects with bipolar disorder. J Affect Disord. 2003;75:65–9. doi: 10.1016/s0165-0327(02)00021-6. [DOI] [PubMed] [Google Scholar]
- 7.Dixon JF, Hokin LE. The antibipolar drug valproate mimics lithium in stimulating glutamate release and inositol 1,4,5-trisphosphate accumulation in brain cortex slices but not accumulation of inositol monophosphates and bisphosphates. Proc Natl Acad Sci U S A. 1997;94:4757–60. doi: 10.1073/pnas.94.9.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dixon JF, Hokin LE. Lithium acutely inhibits and chronically up-regulates and stabilizes glutamate uptake by presynaptic nerve endings in mouse cerebral cortex. Proc Natl Acad Sci U S A. 1998;95:8363–8. doi: 10.1073/pnas.95.14.8363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douglas R, Martin KAC. Neocortex. Third edition. Oxford University Press; New York: 1990. pp. 389–438. [Google Scholar]
- 10.Downs AM, Williams MA. An improved approach to the analysis of autoradiographs containing isolated sources of simple shape: method, theoretical basis and reference data. J Microsc. 1984;136:1–22. doi: 10.1111/j.1365-2818.1984.tb02542.x. [DOI] [PubMed] [Google Scholar]
- 11.Gao XM, Sakai K, Roberts RC, Conley RR, Dean B, Tamminga CA. Ionotropic glutamate receptors and expression of N-methyl-D-aspartate receptor subunits in subregions of human hippocampus: effects of schizophrenia. Am J Psychiatry. 2000;157:1141–9. doi: 10.1176/appi.ajp.157.7.1141. [DOI] [PubMed] [Google Scholar]
- 12.Harrison PJ, Eastwood SL. Neuropathological studies of synaptic connectivity in the hippocampal formation in schizophrenia. Hippocampus. 2001;11:508–19. doi: 10.1002/hipo.1067. [DOI] [PubMed] [Google Scholar]
- 13.Harrison PJ, Law AJ, Eastwood SL. Glutamate receptors and transporters in the hippocampus in schizophrenia. Ann N Y Acad Sci. 2003;1003:94–101. doi: 10.1196/annals.1300.006. [DOI] [PubMed] [Google Scholar]
- 14.Healy DJ, Meador-Woodruff JH. Glutamatergic modulation of subcortical motor and limbic circuits. Ann N Y Acad Sci. 1999;877:684–7. doi: 10.1111/j.1749-6632.1999.tb09301.x. [DOI] [PubMed] [Google Scholar]
- 15.Hollmann M, Heinemann S. Cloned glutamate receptors. Annu Rev Neurosci. 1994;17:31–108. doi: 10.1146/annurev.ne.17.030194.000335. [DOI] [PubMed] [Google Scholar]
- 16.Ibrahim HM, Healy DJ, Hogg AJ, Jr, Meador-Woodruff JH. Nucleus-specific expression of ionotropic glutamate receptor subunit mRNAs and binding sites in primate thalamus. Brain Res Mol Brain Res. 2000;79:1–17. doi: 10.1016/s0169-328x(00)00072-3. [DOI] [PubMed] [Google Scholar]
- 17.Ibrahim HM, Hogg AJ, Healy DJ, Haroutunian V, Davis KL, Meador-Woodruff JH. Ionotropic glutamate receptor binding and subunit mRNA expression in thalamic nuclei in schizophrenia. Am J Psychiatry. 2000;157:1811–1823. doi: 10.1176/appi.ajp.157.11.1811. [DOI] [PubMed] [Google Scholar]
- 18.Johnson M, Perry EK, Ince PG, Shaw PJ, Perry RH. Autoradiographic comparison of the distribution of [3H]MK801 and [3H]CNQX in the human cerebellum during development and aging. Brain Res. 1993;615:259–66. doi: 10.1016/0006-8993(93)90036-m. [DOI] [PubMed] [Google Scholar]
- 19.Julius D, Huang KN, Livelli TJ, Axel R, Jessell TM. The 5HT2 receptor defines a family of structurally distinct but functionally conserved serotonin receptors. Proc Natl Acad Sci U S A. 1990;87:928–32. doi: 10.1073/pnas.87.3.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keks N, Hill C, Opeskin K, Copolov DL, Dean B. Psychiatric diagnosis after death: the problems of accurate diagnosis. In: Gordon J, editor. The Use of CNS Autopsy Tissue in Psychiatric Research: A Practical Guide. Breach Science Publishers; Sydney: 1999. pp. 19–37. [Google Scholar]
- 21.Kornhuber J, Mack-Burkhardt F, Riederer P, Hebenstreit GF, Reynolds GP, Andrews HB, Beckmann H. [3H]MK-801 binding sites in postmortem brain regions of schizophrenic patients. J Neural Transm. 1989;77:231–6. doi: 10.1007/BF01248936. [DOI] [PubMed] [Google Scholar]
- 22.Law AJ, Deakin JF. Asymmetrical reductions of hippocampal NMDAR1 glutamate receptor mRNA in the psychoses. Neuroreport. 2001;12:2971–4. doi: 10.1097/00001756-200109170-00043. [DOI] [PubMed] [Google Scholar]
- 23.Lim IA, Hall DD, Hell JW. Selectivity and promiscuity of the first and second PDZ domains of PSD-95 and synapse-associated protein 102. J Biol Chem. 2002;277:21697–711. doi: 10.1074/jbc.M112339200. [DOI] [PubMed] [Google Scholar]
- 24.Ma J, Zhang GY, Liu Y, Yan JZ, Hao ZB. Lithium suppressed Tyr-402 phosphorylation of proline-rich tyrosine kinase (Pyk2) and interactions of Pyk2 and PSD-95 with NR2A in rat hippocampus following cerebral ischemia. Neurosci Res. 2004;49:357–62. doi: 10.1016/j.neures.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 25.Malenka RC, Nicoll RA. Long-term potentiation--a decade of progress? Science. 1999;285:1870–4. doi: 10.1126/science.285.5435.1870. [DOI] [PubMed] [Google Scholar]
- 26.Mansour A, Thompson RC, Akil H, Watson SJ. Delta opioid receptor mRNA distribution in the brain: comparison to delta receptor binding and proenkephalin mRNA. J Chem Neuroanat. 1993;6:351–62. doi: 10.1016/0891-0618(93)90010-2. [DOI] [PubMed] [Google Scholar]
- 27.Mathern GW, Mendoza D, Lozada A, Pretorius JK, Dehnes Y, Danbolt NC, Nelson N, Leite JP, Chimelli L, Born DE, Sakamoto AC, Assirati JA, Fried I, Peacock WJ, Ojemann GA, Adelson PD. Hippocampal GABA and glutamate transporter immunoreactivity in patients with temporal lobe epilepsy. Neurology. 1999;52:453–72. doi: 10.1212/wnl.52.3.453. [DOI] [PubMed] [Google Scholar]
- 28.Mathern GW, Pretorius JK, Kornblum HI, Mendoza D, Lozada A, Leite JP, Chimelli L, Born DE, Fried I, Sakamoto AC, Assirati JA, Peacock WJ, Ojemann GA, Adelson PD. Altered hippocampal kainate-receptor mRNA levels in temporal lobe epilepsy patients. Neurobiol Dis. 1998;5:151–76. doi: 10.1006/nbdi.1998.0200. [DOI] [PubMed] [Google Scholar]
- 29.Mathern GW, Pretorius JK, Mendoza D, Leite JP, Chimelli L, Born DE, Fried I, Assirati JA, Ojemann GA, Adelson PD, Cahan LD, Kornblum HI. Hippocampal N-methyl-D-aspartate receptor subunit mRNA levels in temporal lobe epilepsy patients. Ann Neurol. 1999;46:343–58. doi: 10.1002/1531-8249(199909)46:3<343::aid-ana10>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 30.McCullumsmith RE, Clinton SM, Meador-Woodruff JH. Schizophrenia as a disorder of neuroplasticity. Int Rev Neurobiol. 2004;59:19–45. doi: 10.1016/S0074-7742(04)59002-5. [DOI] [PubMed] [Google Scholar]
- 31.McCullumsmith RE, Meador-Woodruff JH. Striatal excitatory amino acid transporter transcript expression in schizophrenia, bipolar disorder, and major depressive disorder. Neuropsychopharmacology. 2002;26:368–75. doi: 10.1016/S0893-133X(01)00370-0. [DOI] [PubMed] [Google Scholar]
- 32.Meador-Woodruff JH, Clinton SM, Beneyto M, McCullumsmith RE. Molecular abnormalities of the glutamate synapse in the thalamus in schizophrenia. Ann N Y Acad Sci. 2003;1003:75–93. doi: 10.1196/annals.1300.005. [DOI] [PubMed] [Google Scholar]
- 33.Meador-Woodruff JH, Damask SP, Watson SJ., Jr Differential expression of autoreceptors in the ascending dopamine systems of the human brain. Proc Natl Acad Sci U S A. 1994;91:8297–301. doi: 10.1073/pnas.91.17.8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meador-Woodruff JH, Haroutunian V, Powchik P, Davidson M, Davis KL, Watson SJ. Dopamine receptor transcript expression in striatum and prefrontal and occipital cortex. Focal abnormalities in orbitofrontal cortex in schizophrenia. Arch Gen Psychiatry. 1997;54:1089–95. doi: 10.1001/archpsyc.1997.01830240045007. [DOI] [PubMed] [Google Scholar]
- 35.Meador-Woodruff JH, Mansour A, Civelli O, Watson SJ. Distribution of D2 dopamine receptor mRNA in the primate brain. Prog Neuropsychopharmacol Biol Psychiatry. 1991;15:885–93. doi: 10.1016/0278-5846(91)90016-t. [DOI] [PubMed] [Google Scholar]
- 36.Meador-Woodruff JH, Mansour A, Grandy DK, Damask SP, Civelli O, Watson SJ., Jr Distribution of D5 dopamine receptor mRNA in rat brain. Neurosci Lett. 1992;145:209–12. doi: 10.1016/0304-3940(92)90024-2. [DOI] [PubMed] [Google Scholar]
- 37.Medoff DR, Holcomb HH, Lahti AC, Tamminga CA. Probing the human hippocampus using rCBF: contrasts in schizophrenia. Hippocampus. 2001;11:543–50. doi: 10.1002/hipo.1070. [DOI] [PubMed] [Google Scholar]
- 38.Nankai M, Klarica M, Fage D, Carter C. The pharmacology of native N-methyl-D-aspartate receptor subtypes: different receptors control the release of different striatal and spinal transmitters. Prog Neuropsychopharmacol Biol Psychiatry. 1998;22:35–64. doi: 10.1016/s0278-5846(97)00180-2. [DOI] [PubMed] [Google Scholar]
- 39.Nonaka S, Hough CJ, Chuang DM. Chronic lithium treatment robustly protects neurons in the central nervous system against excitotoxicity by inhibiting N-methyl-D-aspartate receptor-mediated calcium influx. Proc Natl Acad Sci U S A. 1998;95:2642–7. doi: 10.1073/pnas.95.5.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohnuma T, Kato H, Arai H, Faull RL, McKenna PJ, Emson PC. Gene expression of PSD95 in prefrontal cortex and hippocampus in schizophrenia. Neuroreport. 2000;11:3133–7. doi: 10.1097/00001756-200009280-00019. [DOI] [PubMed] [Google Scholar]
- 41.Sans N, Prybylowski K, Petralia RS, Chang K, Wang YX, Racca C, Vicini S, Wenthold RJ. NMDA receptor trafficking through an interaction between PDZ proteins and the exocyst complex. Nat Cell Biol. 2003;5:520–30. doi: 10.1038/ncb990. [DOI] [PubMed] [Google Scholar]
- 42.Scarr E, Pavey G, Copolov D, Dean B. Hippocampal 5-hydroxytryptamine receptors: abnormalities in postmortem brain from schizophrenic subjects. Schizophr Res. 2004;71:383–92. doi: 10.1016/j.schres.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 43.Scarr E, Pavey G, Sundram S, MacKinnon A, Dean B. Decreased hippocampal NMDA, but not kainate or AMPA receptors in bipolar disorder. Bipolar Disord. 2003;5:257–64. doi: 10.1034/j.1399-5618.2003.00024.x. [DOI] [PubMed] [Google Scholar]
- 44.Seidman LJ, Faraone SV, Goldstein JM, Kremen WS, Horton NJ, Makris N, Toomey R, Kennedy D, Caviness VS, Tsuang MT. Left hippocampal volume as a vulnerability indicator for schizophrenia: a magnetic resonance imaging morphometric study of nonpsychotic first-degree relatives. Arch Gen Psychiatry. 2002;59:839–49. doi: 10.1001/archpsyc.59.9.839. [DOI] [PubMed] [Google Scholar]
- 45.Smith RE, Haroutunian V, Davis KL, Meador-Woodruff JH. Vesicular glutamate transporter transcript expression in the thalamus in schizophrenia. Neuroreport. 2001;12:2885–7. doi: 10.1097/00001756-200109170-00026. [DOI] [PubMed] [Google Scholar]
- 46.Takamori S, Rhee JS, Rosenmund C, Jahn R. Identification of a vesicular glutamate transporter that defines a glutamatergic phenotype in neurons. Nature. 2000;407:189–94. doi: 10.1038/35025070. [DOI] [PubMed] [Google Scholar]
- 47.Thomas EA, Dean B, Scarr E, Copolov D, Sutcliffe JG. Differences in neuroanatomical sites of apoD elevation discriminate between schizophrenia and bipolar disorder. Mol Psychiatry. 2003;8:167–75. doi: 10.1038/sj.mp.4001223. [DOI] [PubMed] [Google Scholar]
- 48.Thompson PM, Egbufoama S, Vawter MP. SNAP-25 reduction in the hippocampus of patients with schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry. 2003;27:411–7. doi: 10.1016/S0278-5846(03)00027-7. [DOI] [PubMed] [Google Scholar]
- 49.Toyooka K, Iritani S, Makifuchi T, Shirakawa O, Kitamura N, Maeda K, Nakamura R, Niizato K, Watanabe M, Kakita A, Takahashi H, Someya T, Nawa H. Selective reduction of a PDZ protein, SAP-97, in the prefrontal cortex of patients with chronic schizophrenia. J Neurochem. 2002;83:797–806. doi: 10.1046/j.1471-4159.2002.01181.x. [DOI] [PubMed] [Google Scholar]
- 50.Williams MA. Autoradiography: its methodology at the present time. J Microsc. 1982;128:79–94. doi: 10.1111/j.1365-2818.1982.tb00439.x. [DOI] [PubMed] [Google Scholar]


