Summary
The intestinal microbiota impacts many facets of human health and is associated with human diseases. Diet impacts microbiota composition, yet mechanisms that link dietary changes to microbiota alterations remain ill-defined. Here we elucidate the basis of Bacteroides proliferation in response to fructans, a class of fructose-based dietary polysaccharides. Structural and genetic analysis disclosed a fructose-binding, hybrid-two-component signaling sensor that controls the fructan utilization locus in Bacteroides thetaiotaomicron. Gene content of this locus differs among Bacteroides species and dictates the specificity and breadth of utilizable fructans. BT1760, an extracellular β2-6 endo-fructanase, distinguishes B. thetaiotaomicron genetically and functionally, and enables the use of the β2-6-linked fructan levan. The genetic and functional differences between Bacteroides species are predictive of in vivo competitiveness in the presence of dietary fructans. Genes that differentiate function serve as potential biomarkers in microbiomic datasets to enable rational manipulation of the microbiota via diet.
Introduction
The trillions of microbial cells that reside within the intestine shape aspects of host metabolism and immune function and extend the physiological definition of humans (Backhed et al., 2005; Hooper, 2009; Louis et al., 2007). While the general composition of the intestinal microbiota is similar in most healthy people, with greater than 90% of the cells belonging to the Firmicutes or Bacteroidetes phyla (Dethlefsen et al., 2008), the species composition is highly personalized (Turnbaugh et al., 2009).
Community membership and function of the microbiota can change due to numerous variables including antibiotic treatment, inflammation, or changes in diet (Dethlefsen et al., 2008; Frank et al., 2007; Jernberg et al., 2007; Ley et al., 2006). Protracted loss of the typical composition has been associated with several disorders including inflammatory bowel diseases (Frank et al., 2007). In addition, changes in composition have been associated with obesity and weight loss; however, factors that cause these changes are not well defined (Duncan et al., 2008; Ley et al., 2006). The alterations in community membership, whether chronic or short-term, are accompanied by changes in the microbiota’s collective genome, or microbiome, and the patterns and metabolic capabilities it specifies (Turnbaugh et al., 2009). Therefore, the mechanisms that link relevant variables, such as changes in diet, to changes in the microbiome, are integral to understanding how environmental factors and behavior influence human biology.
Many complex plant polysaccharides in the human diet are resistant to host-mediated degradation due to either insolubility or lack of human-encoded hydrolytic enzymes (Flint et al., 2008; Louis et al., 2007; Sonnenburg et al., 2005). These carbohydrates are not absorbed in the upper gastrointestinal tract and serve as a major source of carbon and energy for the distal gut microbial community. Polysaccharide degradation is one of the core functions encoded in the microbiome (Lozupone et al., 2008; Turnbaugh et al., 2007). Broad expansion of the genes and operons dedicated to degrading and consuming polysaccharides has occurred within the genomes of microbiota-resident species (Xu et al., 2003; Xu et al., 2007), a logical outcome of the intense competition for these resources. It is, therefore, expected that alterations in the type and quantity of polysaccharides consumed can result in changes in the microbiota community composition and function.
Inulin- and levan-type fructans (homopolymers of β2-1 or β2-6 fructose units, respectively) are common dietary plant polysaccharides that feed the intestinal microbiota (Roberfroid et al., 1993). Multiple bacterial taxa in the gut utilize fructans, including members of Firmicutes, Bacteroides, and Bifidobacterium, (Duncan et al., 2003; Rossi et al., 2005; Van der Meulen et al., 2006), and dietary fructan can result in expansion of Actinobacteria, Firmicutes, or Bacteroides (Kolida et al., 2007; Menne et al., 2000; Ramirez-Farias et al., 2008). Lack of predictability in how the microbiota responds to such dietary interventions reflects our limited understanding of nutrient sensing and utilization by members of the intestinal microbiota.
Bacteroides, a major genera in the human microbiota, have a widely expanded capacity to use diverse types of dietary polysaccharides (Xu et al., 2007). Much of the glycan degrading and import machinery within Bacteroides genomes are encoded within clusters of coregulated genes known as polysaccharide utilization loci (PULs). B. thetaiotaomicron (Bt), a prototypic member of the Bacteroides, possesses 88 PULs, which differ in polysaccharide specificity (Martens et al., 2008). The defining characteristic of a PUL is the presence of a pair of genes homologous to Bt susD and susC, which encode outer membrane proteins that bind and import starch oligosaccharides, respectively (Figure 1A) (Martens et al., 2009; Shipman et al., 2000). The pair of susC and susD homologs is usually associated with genes that encode the machinery necessary to convert extracellular polysaccharides into intracellular monosaccharides, such as glycoside hydrolases (susA, susB, and susG in Figure 1A).
Figure 1. Bt’s use of fructose-containing carbohydrates corresponds to induction of the polysaccharide utilization locus BT1757-1763 and BT1765.
A. Genomic organization of Bt’s Sus locus (top) and putative fructan utilization locus (bottom). Genes of similar function are coded by color; intervening unrelated genes are white; genes without corresponding homologs are grey.
B. Gene expression patterns of differentially regulated susC and susD homologs from Bt grown in rich medium (TYG) at five time points from early log (3.5h) to stationary phase (8.8h) in duplicate.
C. Growth curves of Bt in minimal medium containing indicated carbon source at 0.5% w/v. FOS, fructo-oligosaccharide.
D. RNA abundance for genes relevant to fructan use in cells grown in different carbon sources, relative to growth in minimal medium plus glucose. Standard errors of expression levels from three biological replicate cultures are shown.
In addition to machinery for polysaccharide acquisition, most PULs contain, or are closely linked, to a gene or genes encoding an inner membrane associated sensor-regulator system, including the novel hybrid two-component systems (HTCS) (Sonnenburg et al., 2006). Bt’s genome encodes 32of these HTCS, which may mediate the rapid and specific responses required in the dynamic nutrient environment of the intestine. Here we dissect a Bt PUL required for utilization of fructans to better understand how Bacteroides species acquire and process this common class of dietary carbohydrates. In addition, we provide evidence that the associated HTCS controls the expression of the fructan PUL and that monomeric fructose is the activating signal that binds directly to the periplasmic sensor domain of the regulatory protein. These data provide the first example of a well-defined ligand for a member of this class of novel sensor regulators.
The fructan PUL is conserved to varying extents among Bacteroides species, corresponding to a range of fructan utilization capability across the genus. Using model intestinal microbiotas living within gnotobiotic mice, we demonstrate that dietary fructan can have disparate effects on community composition, depending upon the fructan degrading capacity of members of the microbiota. These studies suggest that within personal microbiomic datasets, we will be able to identify genetic biomarkers of discrete functions. Inference of function from these biomarkers should provide predictive power in determining how an individual’s microbiota will respond to changes in diet and other interventions.
Results
BT1757-BT1763 and BT1765 form a putative polysaccharide utilization locus (PUL) that is transcribed early in Bt’s growth in rich media
BT1757-BT1763 and BT1765 encodes eight open reading frames on the negative strand of the Bt genome, including one susC/susD homolog pair (BT1763 and BT1762), a putative outer membrane lipoprotein (BT1761), a putative inner membrane monosaccharide importer (BT1758), a putative fructokinase (BT1757), and three putative glycoside hydrolases (BT1759, BT1760, BT1765) (Figure 1A). These glycoside hydrolases are members of Glycoside Hydrolase Family 32 (GH32), a family of enzymes specific for fructans (Cantarel et al., 2009). One of these, BT1760, possesses a N-terminal lipidation motif and is predicted to reside on the cell surface; the other two, BT1759 and BT1765, are predicted to be periplasmic and intracellular, respectively (www.cbs.dtu.dk/services/LipoP/ and www.cbs.dtu.dk/services/SignalP/). Directly adjacent to the locus is a putative inner membrane-associated sensor regulator of the HTCS family, BT1754. These data suggest that this PUL encodes the proteins required for Bt’s use of fructans.
Expression profiling of Bt in rich medium has revealed the upregulation of several PULs, each of which is confined to a discrete phase of growth (Sonnenburg et al., 2006). Analysis of Bt transcriptional profiles at five timepoints that spanned from early log to stationary phase in vitro in rich medium, compared to basal expression in minimal medium containing glucose as the sole carbohydrate (MM-G), revealed that 14 pairs of susC/susD homologs were induced greater than 20-fold at one or more time points during the growth (Figure 1B) (Gene Expression Omnibus database, www.ncbi.nlm.nih.gov/geo/; accession nos., GSM 40897-40926). The putative fructan PUL, showed upregulation early in Bt’s growth suggesting it is responsive to a high priority substrate accessed early in growth on rich medium (Figure 1B). Genes within this PUL are coexpressed both in vitro in rich medium and in vivo in Bt mono-associated gnotobiotic mice fed a polysaccharide-rich diet (Figure S1A), consistent with the functional relatedness of adjacent genes and operon predictions in Bt (Westover et al., 2005). Bt increases expression of this PUL in vivo while downregulating the vast majority of other PULs when bi-associated in the gnotobiotic mouse intestine with the methanogenic archeon, Methanobrevibacter smithii (Samuel and Gordon, 2006). The upregulation of the putative fructan PUL is concomitant with increased densities of Bt in vivo, suggesting that expression of this locus is associated with growth potentiation of Bt.
Bt upregulates its putative fructan PUL when grown on fructose-containing carbohydrates
We inoculated minimal medium containing specific fructose-based carbohydrates as the only carbon and energy source with Bt to test if the bacterium is competent to grow on fructans. Bt grew on a broad range of fructose-based glycans, including free fructose, sucrose, levan (high MW fructose polymer with predominantly β2-6-linkages), and fructo-oligosaccharides (FOS; short-chain β2-1 polymers of 2-10 fructose units) (Figure 1C; see Figure S2 for carbohydrate structures). However, grew poorly on inulin (β2-1 fructose polymer with an average degree of polymerization of ~25), with growth only apparent three days after inoculation. Doubling times on simple monosaccharides and disaccharide were similar to one another (Table S1). In contrast, growth rates of Bt between the different fructans showed large linkage-dependent differences: β2-6 levan resulted in the fastest doubling time (2.7h), while β2-1 FOS and inulin were significantly slower (doubling times of 5.6h +/− 0.004 and 96.4h +/− 0.05, respectively) (Table S1).
To determine whether these fructose-based substrates induced expression of genes associated with the putative fructan PUL, Bt was grown in either glucose or one of five fructose-containing substrates (fructose, sucrose, levan, FOS, or inulin) as the sole carbohydrate. Cells were harvested at mid-log phase for quantitative RT-PCR (qPCR) analysis, and RNA levels of the 3′ and the 5′ ends of the operon, BT1757 (encoding the fructokinase) and BT1763 (encoding the SusC-like protein), respectively, were used as an indicator of PUL expression (Figure 1D). Both BT1757 and BT1763 were dramatically up regulated in all media containing fructose, whether as a free monosaccharide or in glycosidic linkage. Across all conditions, expression of BT1757, BT1763 and BT1765 showed coordinated increases consistent with the predicted operon structure. However, BT1754 (the PUL-associated putative HTCS) showed no significant induction under all conditions tested. Therefore, the operon that encodes the structural genes of Bt’s putative fructan PUL is transcriptionally responsive to fructose-containing carbohydrates. Published surveys of Bt gene expression in numerous carbohydrates support that up regulation of the fructan PUL is specific to fructose-containing substrates (Martens et al., 2009; Sonnenburg et al., 2005).
Two genes within Bt’s genome that are not physically associated with the putative fructan PUL, a second putative periplasmic GH32 (BT3082) and a second putative fructokinase (BT3305) were likely candidates to be involved in fructan utilization. Analysis of BT3082 and BT3305 expression by qPCR revealed that BT3082 was induced in all fructose-containing media and showed a pattern of induction consistent with those seen for BT1757, BT1763, and BT1765 (Figure 1D); however, BT3305 showed no change in expression or a slightly reduced expression in all conditions (data not shown). These data suggest that the fructosidase, BT3082, but not the putative fructokinase, BT3305, is part of the regulon of the putative fructan PUL.
The hybrid two-component system BT1754 is required for efficient fructan utilization by Bt
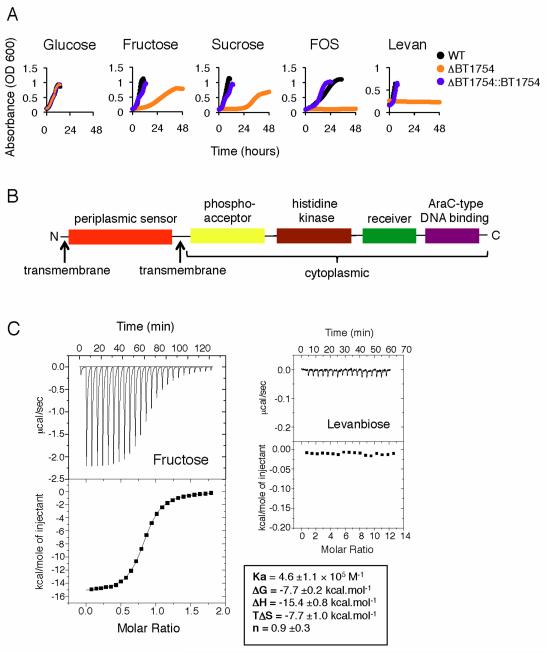
We assessed the ability of an isogenic mutant of Bt lacking the BT1754 gene to grow in a panel of fructose-based minimal media (MM) to test if upregulation of the PUL was dependent upon the HTCS signaling sensor. An in-frame, unmarked deletion of BT1754 was constructed using a standard counter-selectable allele-exchange procedure. Bt-ΔBT1754 exhibited normal colony morphology on solid medium and grew with a similar doubling time to wild type in MM-glucose (2.6h); however, Bt-ΔBT1754 failed to grow in any of the three fructans (FOS, inulin and levan) and showed retarded growth in fructose and sucrose (Figure 2A and Table S1). Additionally, Bt-ΔBT1754 does not exhibit prioritized up regulation of the putative fructan PUL during growth in rich media (Figure S1B). Complementation of this mutant was achieved by introducing the genomic fragment containing BT1754 and its 5′ intergenic upstream promoter region in trans. Growth of the ΔBT1754::BT1754 complemented mutant restored growth in all fructose-based media to levels comparable to wild type (Figure 2A and Table S1). These data demonstrate the HTCS encoded by BT1754 is required for Bt’s use of fructans.
Figure 2. BT1754 HTCS binds fructose and is required for growth on fructose-containing carbohydrates.
A. Growth curves of Bt-ΔBT1754 compared to wild type Bt (WT) and the complemented mutant (ΔBT1754::BT1754) on fructose-based carbon sources.
B. Domain organization of BT1754.
C. Interaction of the N-terminal periplasmic domain of BT1754 with fructose or levanbiose assessed by isothermal calorimetry, showing the raw heats of binding (upper panel) and integrated data (lower panel) fit to a single site binding model (fructose only). Values areaverages and SDs of at least three independent titrations.
The periplasmic domain of the hybrid two-component system BT1754 binds to monomeric fructose
One of the key unanswered questions concerning the HTCS family, and many extracellular sensory systems, is the identity of the molecular triggers for signaling events. The predicted inner-membrane localization of Bt’s HTCS family members, including BT1754, suggests that the periplasmic region likely serves as the sensor/receptor, similar to classic two-component systems. Analysis of the sequence of BT1754 revealed a typical HTCS architecture with an N-terminal predicted periplasmic sensor domain flanked by two transmembrane regions and a C-terminal cytoplasmic histidine kinase domain, a phosphoacceptor domain and a response regulator (including a receiver and an HTH_AraC-type DNA binding domain) (Figure 2B and Figure S3). Uniquely within Bt’s HTCS, the sensor domain displays homology to Type I bacterial periplasmic binding proteins (PBPs) (Dwyer and Hellinga, 2004). As PBPs are known to bind small molecules such as sugars, we expressed the periplasmic domain of BT1754 (BT1754-PD; residues 29-343) in a recombinant form and tested for binding to a range of monosaccharides and fructan-derived oligosaccharides to see if direct interaction with a specific carbohydrate is the means of signal perception in BT1754. The isothermal calorimetry data reveal that BT1754-PD binds specifically to fructose, with a Kd of ~2μM and a stoichiometry of 1:1 and does not interact with either β2-1- or β2-6-linked fructooligosaccharides or any other monosaccharides, including glucose and ribose (Figure 2C).
Structure of BT1754 periplasmic sensor domain
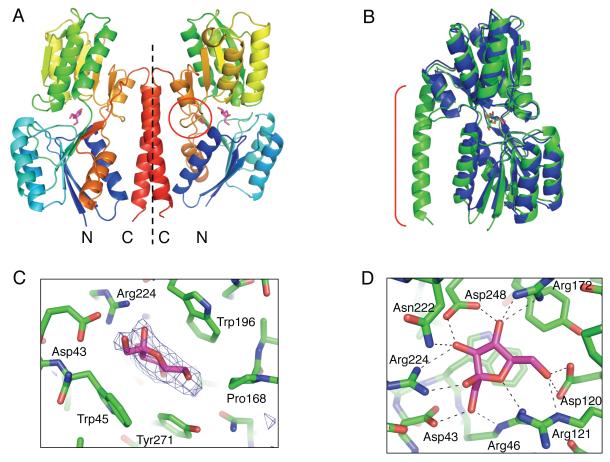
To understand the mechanism of signal perception in more detail, we determined the structure of BT1754-PD in complex with fructose to 2.66Å. The closest homolog with known structure, a ribose-binding PBP from Thermoanaerobacter tengcongensis (TtRBP), PDB 2IOY, was used as a molecular replacement search model. Successful molecular replacement resulted in a dimer in the asymmetric unit. A least squares alignment of the final model with TtRBP gave a root mean square deviation of 1.2 Å for 269 alpha carbons despite the relatively low sequence identity, indicative of the high structural conservation of this family. The BT1754-PD structure comprises a typical two-subdomain PBP-fold, with each subdomain consisting of a core of six β-strands flanked by two or three α-helices (Figure 3A). The polypeptide chain forms a hinge by crossing between the two subdomains three times along one side, the last of these exiting the PBP-fold and then forming a long α-helix, which extends back along the length of the protein to the N-terminal region (Figure 3A).
Figure 3. Structure of BT1754-PD in complex with fructose.
A. Representation of the homodimer of BT1754-PD present in the asymmetric unit, with each monomer separated by a dotted line; molecule of fructose (pink); the flexible hinge between the two subdomains (circle).
B. Overlay of BT1754-PD (green) with TtRBP (blue); the extended C-terminal helix in BT1754-PD (bracket) is unique to BT1754.
C. Side view of the binding site illustrating hydrophobic interactions of BT1754-PD and fructose. Fo-Fc electron density prior to modeling the single molecule of fructose in the β-furanose form is shown (blue mesh contoured at 3σ).
D. Top view of the binding site of BT1754-PD illustrating the numerous H-bonds (dotted black lines) with fructose.
The C-terminal helix of BT1754-PD provides the predominant interface for homo-dimerization and is the main structural difference between classical soluble PBPs such as the TtRBP and BT1754-PD (Figure 3B). Though there are several hydrogen bonds to retain the turn between the PBP-fold and the helix, once the polypeptide has progressed beyond the first residue of the helix (Asn306), the remainder of the contacts, both inter- and intra-molecular, are nonpolar. The dimer, generating a buried surface area of 2640 Å2, appears to be biologically relevant as both the N- and C-termini of each molecule are oriented such that they face in the same direction and, therefore, both molecules are positioned correctly for insertion into the membrane (Figure 3A).
Crystals were grown in the presence of fructose, and electron density indicative of a fructose molecule in the β-furanose form was observed in the cleft between the two subdomains, the typical binding site of PBP family proteins (Dwyer and Hellinga, 2004)(Figure 3A). The sugar ring is sandwiched between two tryptophan residues, one from each subdomain (Trp45 and Trp196), with Tyr-271 and Pro-168 also forming hydrophobic contacts along the C4-C6 edge of the fructose ring (Figure 3C). All remaining interactions with the sugar are polar, with multiple H-bonds formed between side chains, mainly Arg and Asp, and the hyroxyls and ring oxygen of the fructose (Figure 3D). In common with other PBPs, solvent is excluded from the binding site itself. Asp248, which is involved in the interaction with the fructose molecule, is in the disallowed region of the Ramachandran plot and is likely held in an unfavorable conformation by a combination of neighboring amino acids and the sugar itself. The structural and biochemical data for BT1754-PD binding to fructose are consistent with the increased expression of the fructan PUL observed in minimal mediumcontaining only fructose (Figure 1D).
Genetic and biochemical basis of β2-6 fructan specificity of Bt
The lack of linkage recognition by the HTCS sensor suggested that the β2-6-linkage specificity of Bt’s fructan use was encoded within the structural genes of the fructan PUL. We first focused on genes encoding GH32 enzymes, the main family of hydrolases that catalyze the depolymerization of fructans (Cantarel et al., 2009). Three GH32 enzymes (BT1759, BT1760, BT1765) are encoded within the fructan PUL; the other GH32 family member, BT3082, is not encoded within the PUL, but is co-regulated (Figure 1D).
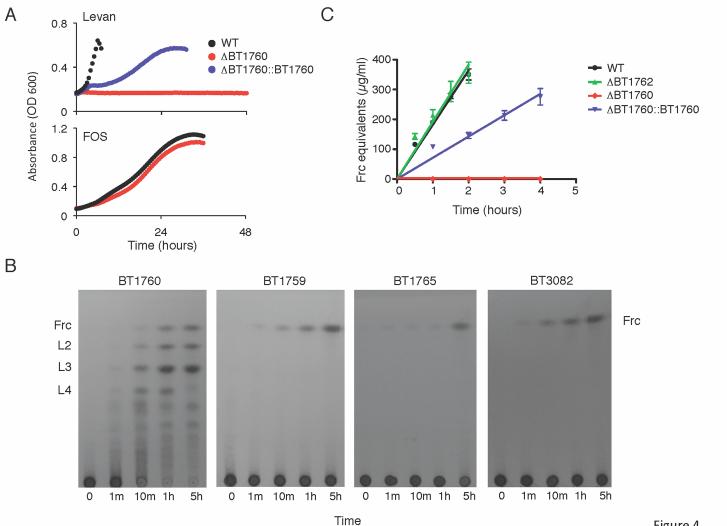
To test whether the only putative cell surface GH32 in Bt, BT1760, is required for levan utilization, an in-frame, unmarked deletion of BT1760 was constructed. Bt-ΔBT1760 exhibited normal colony morphology on solid medium and grew with a normal doubling time in MM-glucose. Bt–ΔBT1760 did not grow on levan, but showed normal growth on all other media tested including β2-1-linked FOS (Figure 4A), with doubling times comparable to wild type in fructose, sucrose, and FOS (Table S1). Complementation in trans of this mutant was achieved by fusing the upstream intergenic promoter region of BT1765 to the 5′ end of the genomic fragment containing BT1760. Levan growth was restored, albeit at a reduced rate, in the complemented Bt-ΔBT1760::BT1760 strain (Figure 4A), confirming the requirement of this gycoside hydrolase for utilization of the β2-6 linked fructan.
Figure 4. BT1760 encodes an extracellular endo-levanase required for Bt growth in levan.
A. Growth curves of Bt -ΔBT1760 compared to the complemented mutant (ΔBT1760::BT1760) in levan (top) or FOS (bottom panel).
B. TLC analysis of the products of levan digestion by the Bt GH32 enzymes, BT1760, BT1759, BT1765 and BT3082. Frc, fructose; L2, levanbiose; L3, levantriose; L4, levantetraose.
C. Degradation of levan by Bt cells grown in minimal medium plus fructose. Error bars show the SDs from three independent experiments.
We next assessed whether BT1760 is a β2-6-specific fructanase. Activity of a recombinant form of BT1760 was tested against a range of β2-6 and β2-1 fructan oligo- and polysaccharides. The data show that BT1760 is indeed a β2-6-fructan specific enzyme with no detectable activity against β2-1 fructans or fructooligosaccharides (Table S2). TLC analysis of levan digestion by BT1760 revealed that a mixture of different sized oligosaccharides was produced. Mono-, di-, tri-, and tetra-levanoligosaccharides accumulated as the main products as the reaction proceeded (Figure 4B). These data demonstrate that BT1760 is a β2-6-specific endo-acting fructanase.
To determine whether the β2-6 fructoside hydrolase activity of BT1760 could be detected on the cell surface, we measured the activity of washed whole Bt cells against levan and inulin. Fructose-grown wild-type cells could degrade the β2-6 polymer but had no detectable activity against inulin, mirroring the specificity of recombinant BT1760 (Figure 4C and not shown). This levanase activity was completely lost in the Bt-ΔBT1760 strain and was largely restored in the complemented Bt-ΔBT1760::BT1760 strain (Figure 4C). Moreover, cells grown on glucose displayed ~100-fold lower levan activity, confirming that the levan-specific hydrolysis is inducible by fructose (not shown). Cytoplasmic and periplasmic marker enzyme assays demonstrated that no cell lysis or leakage occurred in the assay conditions used; therefore, the hydrolase activity detected could only be extracellular (data not shown). These data indicate BT1760 is indeed localized on the surface of the bacterium. The localization and activity are consistent with the hydrolase serving as a key step for converting long-chain levan into oligosaccharides for SusC/SusD-homolog-mediated import.
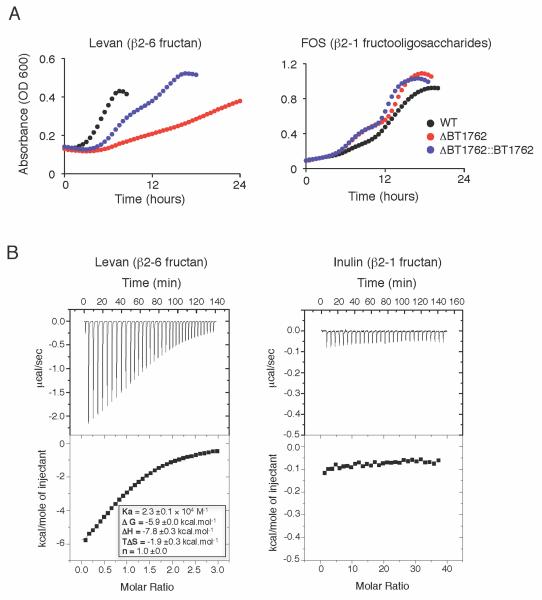
Structural insight into the nature of SusD and a SusD-like protein binding to oligosaccharides suggests that linkage is an important determinant in cell surface structural recognition of oligosaccharides (Koropatkin et al., 2009; Koropatkin et al., 2008). We tested whether BT1760 was the sole specificity determinant in Bt’s efficient use of levan, or whether the SusD homolog within the fructan PUL, BT1762, also exhibited specificity for the β2-6 linkage. We constructed a Bt mutant inwhich BT1762 was deleted, and we tested the ability of this mutant to grow in minimal media in which levan is the sole carbon source. Bt-ΔBT1762 showed significantly retarded growth on levan compared with wild type and growth of this mutant in levan was largely restored upon BT1762 complementation. (Figure 5A and Table S1). Absence of BT1762, however, did not affect extracellular levan degradation, supporting that BT1760 is responsible for cell surface levan degradation (Figure 4C). To determine the specificity of BT1762 directly, the protein was expressed in a recombinant form lacking its signal peptide and lipidation motif, and its interaction with levan and inulin was assessed by isothermal calorimetry (Figure 5B). The data show that BT1762 binds to the β2-6 fructose polymer but displays no affinity for the β2-1 equivalent. BT1762 displays a Kd of ~40μM for levan, similar to the affinity of the prototypic SusD for starch (Koropatkin et al., 2008).
Figure 5. The SusD-homolog encoded by BT1762 is required for efficient Bt utilizaton of levan and binds β2-6 but not β2-1 fructan.
A. Growth curves of wild type Bt, Bt-ΔBT1762, and Bt-ΔBT1762::BT1762 in levan (left) or FOS (right).
B. Interaction of BT1762 with fructans as assessed by isothermal calorimetry. Levan binding data integrated and fit to a single site binding model (bottom left). Values are averages and SDs of at least three independent titrations.
Recent studies have indicated that within many Bacteroides PULs, the gene found downstream of the SusD homolog also encodes a polysaccharide-binding lipoprotein (Martens et al., 2009). Although the products of these ‘susE-positioned’ genes have no obvious homology to one another they appear to be functionally conserved. To explore the role of the susE-positioned gene from the Bt fructan PUL, BT1761, we assessed the ability of a recombinant form of the protein to interact with inulin and levan. The data reveal that BT1761 bound specifically to levan (Figure S4). Reducing sugar and TLC assays with BT1761 and BT1762 against inulin and levan revealed that neither protein had any detectable degradative capacity (not shown). Together, these genetic and biochemical data show that the cell surface components of Bt’s fructan PUL exhibit β2-6 linkage specificity.
Bt has three GH32 enzymes that are not linkage specific
To understand the pattern of fructan degradation in Bt in more detail we biochemically characterized the three other GH32s expressed during growth on fructose-containing media, the predicted periplasmic BT1759 and BT3082 and the predicted intracellular BT1765. The data revealed that all three of these enzymes are exo-acting fructosidases that release fructose from both β2-1 and β2-6 fructans, although some differences in their kinetic characteristics were observed (Table S2 and Figure 4B). BT1759 and BT3082 act equally well on inulin and levan, as well as oligosaccharides of these polymers, although BT3082 appears to be overall a more efficient enzyme with ~2-4 fold higher kcat/KM values than BT1759, driven mainly by its higher turnover rate. Considering the β2-6 fructan preference of Bt, it is interesting that both enzymes display lower KM values (~3-7 fold) for β2-1 oligosaccharides compared to their β2-6 equivalents (Table S2). BT1759 and BT3082 also cleave sucrose, a trait shared with other bacterial fructosidases, although both have a higher KM for the disaccharide than for larger β2-1 kesto- or β2-6 levan-oligosaccharides, indicating a preference for fructose over glucose at the +1 sites of these enzymes. By contrast, BT1765 prefers sucrose over any of the other oligo- or polysaccharides tested, although the enzyme is also able to efficiently degrade levanbiose (Table S2). The predicted cytoplasmic location of BT1765 and substrate specificity suggest that some of the disaccharide products of levan (and possibly FOS) digestion are transported across the inner membrane before they are degraded by the periplasmic fructosidases (see Figure S5).
The fructan PUL is variably conserved in sequenced Bacteroides, which have differing capacity to utilize fructan
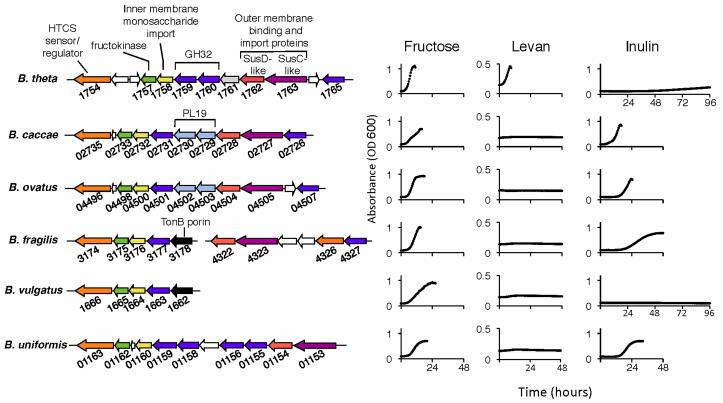
We performed a comparative genomic analysis focused on Bt’s fructan utilization locus between five sequenced species of Bacteroides to gain further insight into the mechanism of fructan use for this major group of gut resident microbes. Using the N-terminal fructose-binding domain of the HTCS BT1754 to query a BLAST database consisting of the Bacteroides species B. caccae, B. vulgatus, B. uniformis, B. fragilis, and B. ovatus, we have identified a single orthologous HTCS in each species, with the exception of B. fragilis, which harbors two BT1754-like genes. Sequence identity between the periplasmic sensor domains of the BT1754 orthologs was high for all but one, ranging from 93% for the B. ovatus protein to 58% for the B. vulgatus domain. Furthermore, the residues involved in fructose binding in BT1754 are almost completely conserved among orthologs, consistent with conservation of the the ligand sensed by each HTCS (Figure S3). The periplasmic domain of one of the two B. fragilis orthologs (BF4326) displayed only 36% identity with BT1754-PD, and this domain was unique in its lack of fully conserved fructose binding residues (Figure S3). Regions adjacent to the HTCS in each genome were analyzed and found to display local synteny with the Bt locus (Figure 6, left panel), including the presence of open reading frames that are predicted to play a role in utilization of fructose-containing carbohydrates. In all six Bacteroides species, the HTCS is adjacent to a predicted fructokinase, a putative inner membrane monosaccharide importer, and GH32-family glycoside hydrolases. In each genome, except that of B. vulgatus, the syntenic regions also contain a susC/susD homologous pair.
Figure 6. Comparative genomic and functional analysis of fructan utilization among Bacteroides species.
Fructan-utilization loci from Bacteroides species (left). Common predicted functions are color coded, intervening unrelated genes are white. PL19, polysaccharide lyase family 19; GH32, glycoside hydrolase family 32. Growth curves (right) of each Bacteroides species in fructose-based carbohydrates.
The presence of an apparent fructan PUL in multiple Bacteroides species suggested that fructan utilization is shared between members of this genus. Testing for growth on fructose-based glycans revealed that all six species are competent for growth on fructose (Figure 6, right panel), sucrose and FOS (Table S1). All Bacteroides species tested, except B. vulgatus, were able to grow efficiently using one of the long-chain fructans, inulin or levan. The inability of B. vulgatus to grow on long-chain fructans is consistent with the absence of a susC/susD-like pair within its locus. B. caccae, B. ovatus, B. fragilis and B. uniformis can utilize inulin with efficiency similar to their use of glucose. This contrasts with Bt inulin use, which is only observed after three days (Figure 6).
Bt was the only species tested able to use levan, which was particularly striking when considering the overall similarity in PUL structure between Bt, B. caccae, and B. ovatus. However, examination of PUL gene content of the two inulin-utilizing species revealed genes encoding PL19 enzymes, a family that is known to include members capable of degrading the β2-1 fructan. Additionally, Bt’s extracellular β2-6-specific GH32, BT1760, does not possess an orthologous gene in the other species (Figure S6). Notably, two other sequenced Bt strains utilize levan more efficiently than inulin in vitro (not shown), similar to the type strain. Both of these strains possess orthologs to the type strain’s BT1760 (Figure S6). Together these data demonstrate that differences in fructanspecificity of Bacteroides species correspond to differences in the gene content of their respective fructan PULs.
Genomic content of Bacteroides species predicts changes in microbiota composition induced by an inulin-based diet
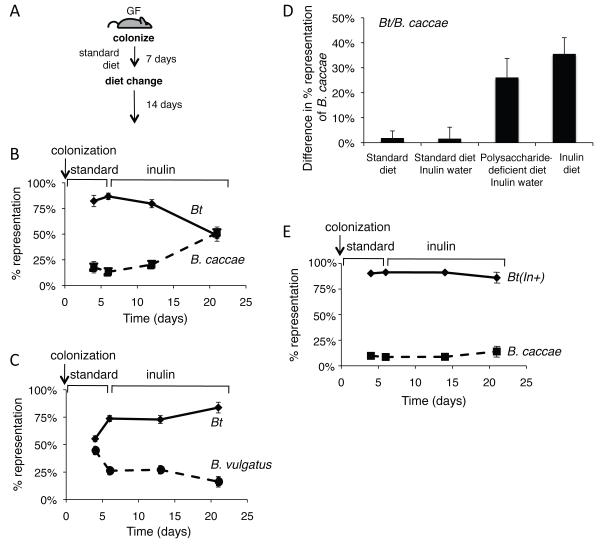
The differences in ability to utilize fructans between the Bacteroides species implies that the relative success of a species within a gut ecosystem may be determined, in part, by the abundance and type of fructan in the host diet. Furthermore, the comparison of genomic sequences and differences in fructan use between species suggests that personalized predictions of microbiota response to specific dietary polysaccharides may be made based on metagenomic microbiome sequence data. We constructed defined two-member communities of Bacteroides species within the intestines of gnotobiotic mice to test how model microbiotas respond in vivo to dietary inulin, which, unlike levan, is available in pure form in quantities sufficient to conduct such a study. Our in vivo experiment aimed to test how differing functionalities embedded within the genomes of two different two-species model microbiotas influence inulin-induced changes in community composition.
Due to B. caccae’s superior ability to use inulin compared to Bt, we tested whether B. caccae would become dominant over Bt within the intestines of mice fed an inulin-supplemented diet. Conversely, Bt’s poor growth on inulin is better than B. vulgatus, which is unable to utilize inulin, suggesting that Bt might benefit from inulin when colonized with B. vulgatus. Two groups of 8-12-week-old, germ-free mice were colonized with equivalent quantities (108 colony forming units, CFU) of Bt and B. caccae or Bt and B. vulgatus. Each mouse was maintained on a standard polysaccharide-rich diet for the first 7 days of colonization and then switched to a diet in which the sole polysaccharide was inulin (10% w/w) for an additional 14 days (Figure 7A). Mice were individually housed throughout the experiment to ensure no cross inoculation could occur and bedding was changed every two days. Total bacterial colonization density was determined by assessing the CFUs in feces over 21 days. The change in each species’ relative abundance before and after dietary inulin supplementation was assessed using species-specific primers in a quantitative PCR assay.
Figure 7. Effect of dietary fructans on Bacteorides competition within the intestine.
A. Experimental design for in vivo experiments. GF, germ-free.
B. Average relative fecal proportion (% total bacteria) of Bt and B. caccae at 4, 6, 14, and 21 days after colonization; n=7 mice.
C. Average relative fecal proportion (% total bacteria) of Bt and B. vulgatus at 4, 6, 14, and 21 days after colonization; n=3 mice.
D. Increase in proportion (%) of B. caccae over Bt from day 6 (1 day prior to diet change) to day 21 (14 days after diet change). All groups received a standard diet on days 1-7; type of diet and whether the mice received inulin in their water on days 7-21 is indicated; n=3-7 individually housed mice.
E. Average relative fecal proportion (% total bacteria) of inulin-utilizing Bt(In+) and B. caccae at 4, 6, 14, and 21 days after colonization; n=7 individually housed mice.
Our results disclosed that total fecal bacterial densities over the course of the experiment did not differ significantly upon dietary shift (total densities ranged from 1010-1011 bacteria/ml of fecal material). Relative densities were determined on days 4 and 6 (standard diet) and on days 13 and 21 (6 and 14 days after dietary switch). In the Bt/B. caccae, bi-associated mice, before the diet switch (day 6 post-colonization in mice fed a standard diet), Bt comprised 87±3% of the community, indicating that Bt is better adapted than B. caccae to these in vivo conditions. Six days after a change to the inulin-based diet, Bt levels dropped to 80±4%, and B. caccae increased to 20±4%. After two weeks consuming the inulin diet, the relative proportion of the two species showed a more drastic shift in favor of B. caccae:Bt representation decreased to approximately 49±6% vs. 51±6% B. caccae (p = 8 × 10−5, day 21 versus day 6; n=7 mice; Student’s t-test) (Figure 7B). In contrast, the Bt/B. vulgatus bi-associated mice did not exhibit any significant trend in changed community composition after 6 days on an inulin-based diet, but Bt increased in abundance from 74±3% on day 6 to 84±5% on day 21 (p=0.1; n=3 mice) on the inulin-enriched diet (Figure 7C). The delayed and modest effect of diet influencing the composition of the Bt/B. vulgatus bi-association is consistent with poor inulin use by Bt and no inulin use by B. vulgatus (Figure 6). Together these data are consistent with dietary polysaccharide-induced changes in the microbiota composition that are predictable based on the resident species’ ability to use that polysaccharide.
In the previous experiment, inulin was the sole polysaccharide in the diet. We wondered whether we would observe the same inulin-induced increase in B. caccae relative to Bt if other polysaccharides were also present in the diet. To test this, gnotobiotic mice were co-colonized with Bt and B. caccae and maintained on the standard diet with inulin supplementation in the water (1% w/v). Over the 14 days the mice ingested an average of 117±6 mg of inulin daily via the water (compared to 355±7mg/day with the inulin diet). Fecal samples were tested by qPCR over the course of the 21-day experiment for relative levels of Bt or B. caccae. These data revealed no statistical difference in the change in relative colonization between mice fed inulin-supplemented water compared to controls that received the same standard diet for 21 days, but received no inulin (Figure 7D). These data suggest that when mice were fed a diet rich in carbohydrates, the presence of inulin did not provide enough of an advantage to B. caccae to allow it to out-compete Bt; however, the amount of inulin supplied in the water (117mg/day average) was less than the amount derived from the inulin diet (355mg/day average) potentially contributing to the lack of the B. caccae response.
We decided to feed mice a custom diet deficient in all polysaccharides and supplement inulin in the water to determine whether a lower dose of inulin in the absence of other polysaccharides was sufficient to provide B. caccae a competitive advantage over Bt in vivo. Under this experimental paradigm the mice consumed an average of 97mg of inulin per day. After 14 days on inulin-water supplementation, the proportion of B. caccae increased by 26±8% (Figure 7D). While not as robust an increase as observed in the inulin-only diet experiment (which showed a 36±7% increase in B. caccae), these data demonstrate that reduced inulin consumption in the absence of competing polysaccharides, offers a significant competitive advantage to inulin-utilizing B. caccae, consistent with the flexible nutrient foraging the Bacteroides species exhibit. The wide range of polysaccharides present in the standard diet allows Bt to compete effectively with B. caccae even in the presence of inulin.
We finally demonstrate the importance of inulin utilization for conferring a competitive advantage in hosts fed an inulin-rich diet using a genetic proof of this effect. The region of the B. caccae fructan-utilization locus from the susC-like gene through the GH32-encoding gene (BC02727-BC02731) was cloned and expressed in a strain of Bt that is compromised in its ability to utilize levan (Bt-ΔBT1763) under the control of the BT1763 promoter (not shown). The resulting strain, Bt(In+), exhibits efficient growth in minimal medium containing inulin, similar to B. caccae (Figure S7). Repeating our original in vivo competition experiment with Bt(In+) revealed that conferring inulin use ability upon Bt eliminates the ability of B. caccae to become dominant in the presence of an inulin-based diet (Figure 7E). This result confirms that the specificity of dietary polysaccharide use is the key functionality that dictates the alterations in the model microbiota that we observe. These results support our hypothesis that changes in microbiota community membership brought on by dietary change can be inferred based on genomic and functional knowledge of resident microbial populations. They also suggest that diet can be a dominant determinant in dictating changes in microbiota composition.
Discussion
Inulin (β2-1 fructan) and levan (β2-6 fructan) are polysaccharides that are abundant in the human diet, but are resistant to host-mediated digestion in the upper gastrointestinal tract. These glycans instead serve as a carbon and energy source for the bacteria that reside in the distal intestine. Bacteroides thetaiotaomicron, a resident of the human GI tract, encodes a fructan utilization locus, BT1757-63 and BT1765, the gene products of which enable efficient acquisition and use of levan-type carbohydrates.
The fructan PUL is adjacent to a hybrid two-component system sensor-regulator, BT1754, which binds only to monomeric fructose, a signal sufficient to induce transcription of the locus. While the upregulation of polysaccharide utilization machinery in response to a monosaccharide may seem unexpected, this signal is a likely consequence of the environment in which Bt resides. Within the natural habitat of the large bowel, free fructose and simple disaccharides, such as sucrose,do not occur at appreciable levels as the host absorbs such sugars within the small intestine. Therefore, the regulation of this locus evolved in the absence of selective pressure to discriminate free monosaccharide from polysaccharides. In addition, unlike many other monosaccharides, fructose is found in only a single class of polysaccharide, namely homopolymeric fructans. Bt appears to use the liberated fructose as a proxy (i.e., indicator) for fructan, which results in upregulation of the machinery to utilize the polysaccharide. This is consistent with previous data that demonstrate Bt’s constitutive, low-level expression of signaling sensors and glycoside hydrolases in conditions lacking the relevant substrates (Martens et al., 2009; Sonnenburg et al., 2005), as well as the low level cell surface levanase activity we observe with whole cells grown in glucose. The constitutive expression suggests that Bt employs a strategy of being prepared to degrade multiple polysaccharides immediately upon their arrival into the distal gut environment. Specific liberated carbohydrates that result from the degradation serve as signalsthat augment expression of the appropriate PUL via a specific sensor-regulator such as a HTCS.
The binding of BT1754 to monomeric fructose also results in a failure of the sensor to differentiate β2-1 and β2-6 linkages despite Bt being much more efficient in use of the levan-type fructans. Specificity of signal is instead derived from the cell surface structural components of the PUL, which serve as the “gateway” for substrates crossing the outer membrane. The cell surface SusD homologue, BT1762, the susE-positioned gene product, BT1761, and the endo-levanase, BT1760, all contribute to the specific import of β2-6 fructans into Bt’s periplasm. BT1754 relies upon the specificity of the cell surface polysaccharide degradation and binding machinery to provide fructose derived from β2-6 fructan to the periplasm where the sensor is sequestered.
Despite Bt’s inability to utilize inulin efficiently it is able to grow well on FOS, a short chain β2-1 fructan. Notably, the fructan PUL of Bt is up regulated during growth in vitro in minimal medium containing FOS or inulin. Bt’s ability to grow in FOS at a rate that is significantly faster than inulin is likely due to the difference in degree of polymerization between the two substrates. Whether small oligosaccharides from these substrates undergo passive diffusion into the periplasm or are accessed via another mechanism requires further investigation.
Among the Bacteroides species tested, Bt appears to be unique in its ability to utilize levan, whereas other species are adept at utilizing polymeric β2-1 fructans. Such phenotypic differences, combined with dietary variation between individuals, could provide the basis for the striking person-to-person variability observed for Bacteroidetes in human microbiota enumeration studies (Eckburg et al., 2005). Our in vivo studies illustrate that species well-adapted to use inulin gain a competitive advantage when hosts are fed an inulin-based diet. Although a genetic loss-of function experiment, in which inulin use is compromised, could be used to test whether the observed changes in species abundance are due to inulin use, we have used a gain-of-function experiment, in which inulin use is conferred upon Bt, to illustrate this point unequivocally. These results suggest that some aspects of diet-induced changes in microbiota composition may be predetermined based on the intrinsic capacity of an individual species to use the substrates that are being consumed by the host. We speculate that diets enriched in different polysaccharides, or polysaccharide-deficient diets, could result in microbiotas of very different species composition. Future studies that follow species and gene composition of the human intestinal microbiota during consumption of levan- or inulin-based diets will provide insight into the rapidity with which members of a complex community adapt at a functional, compositional, and genetic level. How such niche specialization occurs over the course of evolution and the role that diet plays in determining a species’ glycan utilization repertoire remain important yet difficult questions to address.
Perspective
As the age of personal genomes approaches, some aspects of diet and medical therapies will be customized based on genotype. Diet can also be personalized to optimize microbiota function and interaction with the host based on the metagenomic analysis of an individual’s microbiota. A prerequisite for incorporating vast amounts of microbial genomic data into personalized, preventative medicine is to attain a mechanistic understanding of the most dominant aspects of microbiota function. Here we present a case study of how understanding the mechanisms that link the microbome to microbiota function may enable individualized predictions of microbiota response to perturbations. We have taken two-species model microbiotas that collectively possess close to 10,000 genes and predicted how they will respond to a specific dietary cue based on a functional understanding of the ~20 relevant genes. A similar distillation of full microbiomic datasets that contain >106 genes, to a relevant subset, will be required to make microbiota management tractable. With an ever-increasing understanding of how the biology of host and microbiota integrate, we may soon be able to use genomic and microbiomic sequence data to intentionally program or re-program the emergent properties of the host-microbial superorganism.
Materials and Methods
Culturing bacteria
Bacteria were cultured in TYG and MM as described previously (Martens et al., 2008; Sonnenburg et al., 2005). The following bacteria were used: Bt (VPI-5482), B. caccae (ATCC-43185), B. ovatus (ATCC-8483), B. fragilis (NCTC-9343), B. uniformis (ATCC-8492), and B. vulgatus (ATCC-8482). Growth curves in MM were obtained using a Powerwave (Biotek) reading OD600 every 30 min from anaerobic cultures at 37°C.
Quantitative RT-PCR analysis
Quantitative RT-PCR was performed using gene-specific primers as described previously (Table S3) with SYBR Green (ABgene) in a MX3000P thermocycler (Strategene) (Martens et al., 2008).
Gene deletion and complementation in Bt
In-frame (non-polar) gene deletions for mutants were generated using counter-selectable allele exchange (Martens et al., 2008). PCR amplified genes for complementation were ligated into the pNBU2-tetQb vector and conjugated into Bt via E. coli S17.1 λ-pir (Martens et al., 2008). Resulting clones were screened by PCR and sequenced to confirm isolates.
Gene cloning
Genes for expression were amplified from Bt genomic DNA using the primers stated in Table S3 and cloned into pRSETA (Invitrogen) or pET22b (Novagen).
Protein expression and purification
Recombinant proteins were expressed in E. coli C41 or BL21 cells and purified in a single step using metal affinity chromatography as described previously (Bolam et al., 2004).
Sources and preparation of carbohydrates
Monosaccharides, sucrose, and chicory inulin for enzymatic and binding assays were obtained from Sigma. Growth of Bacteroides strains, qRT-PCR, and mouse experiments used inulin, FOS (Orafti-Beneo group; OraftiHP, OraftiP95, respectively) and levan (Sigma; 66674). Kestooligosaccharides were from Megazyme. Levanoligosaccharides were produced by partial acid hydrolysis (1M HCl at 25°C for 20 min-1 h) of levan (Montana Polysaccharides). NaOH-neutralized samples were separated on BioGel P2 (BioRad) size exclusion resin.
Isothermal titration calorimetry
Measurements were carried out essentially as described previously (Bolam et al., 2004), except that a Microcal VP-ITC machine was used, and proteins were dialyzed into 20mM Tris-HCl, pH8.0. The assumption that n=1 for BT1762 binding to levan was based on the structure of the starch binding SusD (Koropatkin et al., 2008).
Thin layer chromatography
Samples were spotted onto foil backed silica plates and placed in a glass tank equilibrated with butanol:acetic acid:H2O (2:2:1). Sugars were visualized using orcinol-sulphuric acid (sulphuric acid:ethanol:water 3:70:20 v/v, orcinol 1% w/v), 90°C for 5-10 min.
Enzyme assays
All assays were carried out at 37°C in 20mM Tris-HCl, pH8.0. Activity of BT1760 was determined by quantifying the amount of reducing sugar released using the DNSA assay (Miller, 1959). Free fructose was determined using a modified fructose detection kit (Megazyme International). Kinetic parameters were determined by fitting initial rates vs. substrate concentration (measured at six substrate concentrations that spanned the KM) to a non-linear model of the Michaelis-Menten equation (Graphpad Prism, v5.0).
Enzyme localization studies
Cultures grown on 0.5% (w/v) fructose or glucose were harvested by centrifugation (OD600 ~1.0). PBS-washed cells and 0.5% levan or inulin in 20mM Tris-HCl, pH8.0, were incubated at 37°C. Reducing sugar present was quantified using DNSA reagent (Miller, 1959). Activities of the periplasmic marker alkaline phosphatase and cytoplasmic marker glucose-6-phophate dehydrogenase were compared to lysed cells to ensure no cell lysis/leakage occurred.
Bacterial colonization and density determination of germ-free mice
Germ-free Swiss-Webster mice were maintained in gnotobiotic isolators and fed an autoclaved standard diet (Purina LabDiet 5K67) or custom diet (Bio-Serv, http://bio-serv.com/), in accordance with A-PLAC, the Stanford IACUC. Mice were bi-associated using oral gavage (108 CFU of each bacterial species). Relative densities of bacteria were determined by qPCR using strain-specific primers (Table S3) (Martens et al., 2008).
Crystallization, structure determination and refinement of BT1754-PD
Crystals formed in 0.7 M K/Na phosphate, 0.1 M HEPES pH 8.0 (protein at 8 mg/ml with 5 mM fructose). Diffraction data, collected at Diamond Light Source (Oxford, UK) on a tiled ADSC Q315 CCD detector were processed with MOSFLM (Leslie, 1992). Scaling of data, search model generation, molecular replacement and structure refinement were carried out using SCALA, CHAINSAW, MOLREP and REFMAC (Collaborative Computational Project;1994) respectively with model rebuilding in COOT (Emsley and Cowtan, 2004).
Supplementary Material
Acknowledgements
We thank Karla Kirkegaard and Stanley Falkow for valuable comments and Sara Fisher for editing the manuscript. Inulin and FOS for mouse experiments were a kind gift from Beneo-orafti. Levan was a kind gift from Montana Polysaccharides. We thank Jeffrey Gordon and members of the Gordon Lab for valuable advice, Carl Morland for excellent technical assistance and Eric Martens and Andrew Goodman for development of genetic tools used in this paper. Some Bacteroides genomic data were produced by The Genome Center at Washington University School of Medicine in St. Louis (genome.wustl.edu). This work was funded in part by grants from National Institutes of Health through the NIH Director’s New Innovator Award Program (DP2-OD006515) the NIDDK (K01-DK077053), the Stanford Digestive Disease Center (PO3-DK56339) and BBSRC (BBF0141631).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science (New York, NY. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- Bolam DN, Xie H, Pell G, Hogg D, Galbraith G, Henrissat B, Gilbert HJ. X4 modules represent a new family of carbohydrate-binding modules that display novel properties. The Journal of biological chemistry. 2004;279:22953–22963. doi: 10.1074/jbc.M313317200. [DOI] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic acids research. 2009;37:D233–238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project The CCP4 suite: programs for protein crystallography. Acta crystallographica. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan SH, Lobley GE, Holtrop G, Ince J, Johnstone AM, Louis P, Flint HJ. Human colonic microbiota associated with diet, obesity and weight loss. International journal of obesity (2005) 2008;32:1720–1724. doi: 10.1038/ijo.2008.155. [DOI] [PubMed] [Google Scholar]
- Duncan SH, Scott KP, Ramsay AG, Harmsen HJ, Welling GW, Stewart CS, Flint HJ. Effects of alternative dietary substrates on competition between human colonic bacteria in an anaerobic fermentor system. Applied and environmental microbiology. 2003;69:1136–1142. doi: 10.1128/AEM.69.2.1136-1142.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer MA, Hellinga HW. Periplasmic binding proteins: a versatile superfamily for protein engineering. Current opinion in structural biology. 2004;14:495–504. doi: 10.1016/j.sbi.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science (New York, NY. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta crystallographica. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nature reviews. 2008;6:121–131. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV. Do symbiotic bacteria subvert host immunity? Nature reviews. 2009;7:367–374. doi: 10.1038/nrmicro2114. [DOI] [PubMed] [Google Scholar]
- Jernberg C, Lofmark S, Edlund C, Jansson JK. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. The ISME journal. 2007;1:56–66. doi: 10.1038/ismej.2007.3. [DOI] [PubMed] [Google Scholar]
- Kolida S, Meyer D, Gibson GR. A double-blind placebo-controlled study to establish the bifidogenic dose of inulin in healthy humans. European journal of clinical nutrition. 2007;61:1189–1195. doi: 10.1038/sj.ejcn.1602636. [DOI] [PubMed] [Google Scholar]
- Koropatkin N, Martens EC, Gordon JI, Smith TJ. Structure of a SusD homologue, BT1043, involved in mucin O-glycan utilization in a prominent human gut symbiont. Biochemistry. 2009;48:1532–1542. doi: 10.1021/bi801942a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koropatkin NM, Martens EC, Gordon JI, Smith TJ. Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure. 2008;16:1105–1115. doi: 10.1016/j.str.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography. 1992;(No. 26) [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Louis P, Scott KP, Duncan SH, Flint HJ. Understanding the effects of diet on bacterial metabolism in the large intestine. Journal of applied microbiology. 2007;102:1197–1208. doi: 10.1111/j.1365-2672.2007.03322.x. [DOI] [PubMed] [Google Scholar]
- Lozupone CA, Hamady M, Cantarel BL, Coutinho PM, Henrissat B, Gordon JI, Knight R. The convergence of carbohydrate active gene repertoires in human gut microbes. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:15076–15081. doi: 10.1073/pnas.0807339105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell host & microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Koropatkin NM, Smith TJ, Gordon JI. Complex glycan catabolism by the human gut microbiota: The bacteroidetes Sus-like paradigm. The Journal of biological chemistry. 2009 doi: 10.1074/jbc.R109.022848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menne E, Guggenbuhl N, Roberfroid M. Fn-type chicory inulin hydrolysate has a prebiotic effect in humans. The Journal of nutrition. 2000;130:1197–1199. doi: 10.1093/jn/130.5.1197. [DOI] [PubMed] [Google Scholar]
- Miller GL. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Analytical Chemistry. 1959;31:426–428. [Google Scholar]
- Ramirez-Farias C, Slezak K, Fuller Z, Duncan A, Holtrop G, Louis P. Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. The British journal of nutrition. 2008:1–10. doi: 10.1017/S0007114508019880. [DOI] [PubMed] [Google Scholar]
- Roberfroid M, Gibson GR, Delzenne N. The biochemistry of oligofructose, a nondigestible fiber: an approach to calculate its caloric value. Nutrition reviews. 1993;51:137–146. doi: 10.1111/j.1753-4887.1993.tb03090.x. [DOI] [PubMed] [Google Scholar]
- Rossi M, Corradini C, Amaretti A, Nicolini M, Pompei A, Zanoni S, Matteuzzi D. Fermentation of fructooligosaccharides and inulin by bifidobacteria: a comparative study of pure and fecal cultures. Applied and environmental microbiology. 2005;71:6150–6158. doi: 10.1128/AEM.71.10.6150-6158.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel BS, Gordon JI. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10011–10016. doi: 10.1073/pnas.0602187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipman JA, Berleman JE, Salyers AA. Characterization of four outer membrane proteins involved in binding starch to the cell surface of Bacteroides thetaiotaomicron. Journal of bacteriology. 2000;182:5365–5372. doi: 10.1128/jb.182.19.5365-5372.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg ED, Sonnenburg JL, Manchester JK, Hansen EE, Chiang HC, Gordon JI. A hybrid two-component system protein of a prominent human gut symbiont couples glycan sensing in vivo to carbohydrate metabolism. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:8834–8839. doi: 10.1073/pnas.0603249103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science (New York, NY. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Meulen R, Makras L, Verbrugghe K, Adriany T, De Vuyst L. In vitro kinetic analysis of oligofructose consumption by Bacteroides and Bifidobacterium spp. indicates different degradation mechanisms. Applied and environmental microbiology. 2006;72:1006–1012. doi: 10.1128/AEM.72.2.1006-1012.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westover BP, Buhler JD, Sonnenburg JL, Gordon JI. Operon prediction without a training set. Bioinformatics (Oxford, England) 2005;21:880–888. doi: 10.1093/bioinformatics/bti123. [DOI] [PubMed] [Google Scholar]
- Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, Hooper LV, Gordon JI. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science (New York, NY. 2003;299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- Xu J, Mahowald MA, Ley RE, Lozupone CA, Hamady M, Martens EC, Henrissat B, Coutinho PM, Minx P, Latreille P, et al. Evolution of Symbiotic Bacteria in the Distal Human Intestine. PLoS Biol. 2007;5:e156. doi: 10.1371/journal.pbio.0050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.