Abstract
Cellular responses to environmental stimuli require conserved signal transduction pathways. In budding yeast (Saccharomyces cerevisiae), nutrient limitation induces morphological changes that depend on the protein kinase A (PKA) pathway and the Kss1 mitogen-activated protein kinase (MAPK) pathway. It was unclear to what extent and at what level there is synergy between these two distinct signaling modalities. We took a systematic genetic approach to clarify the relationship between these inputs. We performed comprehensive epistasis analysis of mutants lacking different combinations of all relevant pathway components. We found that these two pathways contribute additively to nutrient limitation-induced haploid invasive growth. Moreover, full derepression of either pathway rendered it individually sufficient for invasive growth and thus, normally, both are required only because neither is maximally active. Furthermore, in haploids, the MAPK pathway contributes more strongly than the PKA pathway to cell elongation and adhesion, whereas nutrient limitation-induced unipolar budding is independent of both pathways. In contrast, in diploids, upon nutrient limitation the MAPK pathway regulates cell elongation, the PKA pathway regulates unipolar budding, and both regulate cell adhesion. Thus, although there are similarities between haploids and diploids, cell type-specific differences clearly alter the balance of the signaling inputs required to elicit the various nutrient limitation-evoked cellular behaviors.
EUKARYOTIC cells respond to a wide variety of environmental conditions by using a comparatively limited number of conserved signal transduction mechanisms. Potentially, there are diverse ways in which such pathways can interact with and regulate each other to integrate stimuli, generate positive or negative feedback, and coordinate, antagonize, or potentiate subprocesses, thereby generating by their combinatorial action specific cellular behaviors.
External nutrient supply affects the division pattern, adhesivity, and morphology of cells and the colonies of the budding yeast Saccharomyces cerevisiae (Gancedo 2001; Gagiano et al. 2002; Palecek et al. 2002; Schneper et al. 2004; Truckses et al. 2004; Verstrepen and Klis 2006; Chen and Thorner 2007; Granek and Magwene 2010). During conditions of nutrient sufficiency, cells are ovoid, and haploid cells bud to form daughter cells at the cell pole corresponding to their own birth site (an “axial” budding pattern), whereas diploid cells bud alternately from their birth end and from the opposite cell pole (a “bipolar” budding pattern). When nutrients are plentiful, the combination of a rounder cell shape, decreased adherence, and division near existing cells may facilitate denser and more rapid population of the current niche. By contrast, during conditions of nutrient limitation, cells exhibit increased cell–cell adhesion, cell–substratum adhesion, and substratum penetration, as well as an elongated morphology and a “unipolar” pattern of proliferation, in which daughter cells arise only at the cell pole opposite their mother's birth end. This combination of adhesion, substratum invasion, cell elongation, and directional growth (division away from, as opposed to next to, existing cells) is thought to permit a colony of individually non-motile yeast cells to spread out, facilitating the exploration of its surroundings for areas of greater nutritional abundance.
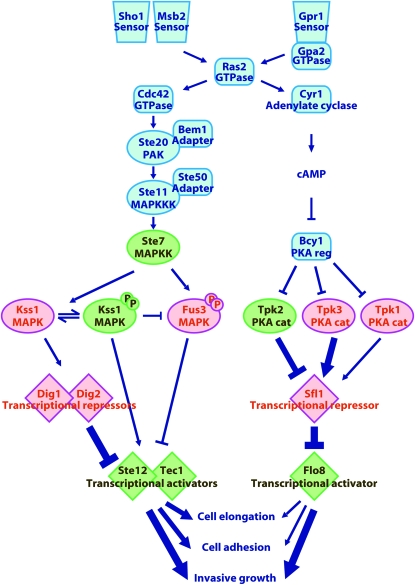
The changes in growth characteristics elicited by nutrient limitation are termed “pseudohyphal growth” in diploids and “invasive growth” in haploids or, generally, “filamentous growth” (FG). Pseudohyphal growth under nitrogen source limitation and invasive growth under carbon source limitation involve many of the molecular factors and signaling pathways (for an overview of these pathway components, please refer to the schematic summary of the results of this study shown in Figure 7). In particular, pseudohyphal growth and invasive growth require two classical signaling modalities: (i) a mitogen-activated protein kinase (MAPK) cascade and (ii) 3′, 5′-cyclic adenosine monophosphate (cAMP) and cAMP-dependent protein kinase A (PKA) (Lengeler et al. 2000; Pan et al. 2000; Truckses et al. 2004). The MAPK cascade that promotes FG consists of Ste20 (PAK/MAPKKKK), Ste11 (MAPKKK), Ste7 (MAPKK), and Kss1 (MAPK). Fus3, another MAPK activated by Ste7 in response to a different stimulus (mating pheromone), inhibits FG (Cook et al. 1997; Madhani et al. 1997) at least in part by targeting a transcription factor (Tec1) required for FG gene expression for ubiquitin- and proteasome-mediated destruction (Bao et al. 2004; Chou et al. 2004). Of the three isoforms of the PKA catalytic subunit encoded in the S. cerevisiae genome, Tpk2 activates filamentous growth, Tpk3 inhibits it, and the role of Tpk1 has been unclear (Robertson and Fink 1998; Pan and Heitman 1999).
Figure 7.—
Signaling circuitry involved in the response of haploid cells to nutrient limitation. This study focused on the relative contributions of distal components in these pathways that have either positive (green) or negative (red) roles. Arrows, activation; T-bars, inhibition, inactivation, or repression. Line thickness indicates the strength of the observed effect: the thicker the line, the stronger the effect. The various effects depicted contribute in an additive (not all-or-none) fashion.
One node at which the MAPK and PKA pathways are known to converge is on the promoters of certain genes, such as FLO11 (Rupp et al. 1999), which encodes a surface glycoprotein important for cell–substratum adhesion. Tpk2-mediated phosphorylation displaces repressor Sfl1, thereby permitting binding of transcriptional activator Flo8 (Pan and Heitman 2002). Similarly, Kss1 catalyzes phosphorylation and displacement of the related repressors Dig1 and Dig2 from transactivator Ste12 (Cook et al. 1996; Tedford et al. 1997; Bardwell et al. 1998b), allowing its hetero-dimerization with Tec1 to induce FG genes (Cook et al. 1996; Madhani and Fink 1997; Tedford et al. 1997; Rupp et al. 1999; Zeitlinger et al. 2003; Chou et al. 2006).
The MAPK and PKA pathways also share a common upstream activator, the GTPase Ras2 (Mösch et al. 1999), a homolog of mammalian H-Ras. Ras2 activates adenylate cyclase (Toda et al. 1985), which stimulates cAMP production; cAMP binding to the regulatory (inhibitory) subunit (Bcy1) of PKA releases the active catalytic subunits. Separately, Ras2 action stimulates formation of the GTP-bound form of another small GTPase, Cdc42 (Mosch et al. 1996), which both stimulates Ste20 and tethers nearby its immediate substrate, Ste11 (via an adaptor protein, Ste50) (Truckses et al. 2006), thereby triggering the cascade that activates Kss1.
Because FG requires both MAPK and PKA activity, it seemed reasonable that these inputs might exhibit coordinate and synergistic regulation, such that activation of one pathway might promote activation of the other. Evidence obtained in some studies suggests instead that the MAPK and PKA pathways function independently, at least for regulation of diploid pseudohyphal growth (Pan and Heitman 1999). However, overexpression in haploids of any of the three PKA isoforms stimulated expression of a filamentous growth reporter construct [FRE(Ty1))∷lacZ], and this stimulation was completely dependent on Ste12 and Tec1, on which basis it was proposed that a component of the cAMP pathway at or downstream of the PKAs regulates a component of the MAPK pathway at or upstream of the transcriptional regulators (Mösch et al. 1999). However, it has also been reported that haploid cells lacking either Fus3 or Kss1 exhibit substantially altered cAMP levels, suggesting regulation of the PKA pathway by the MAPK pathway (Cherkasova et al. 2003). In light of these ambiguities about whether the MAPK regulates the PKA pathway or PKA regulates the MAPK pathway or the two function independently, we applied a comprehensive and systematic genetic approach to analyze whether cross-regulation between these two pathways is essential for nutrient limitation-evoked phenotypic responses.
MATERIALS AND METHODS
Yeast strains and growth conditions:
S. cerevisiae strains used in this study are listed in Table 1. In the strains generated for this study, deletion alleles represent precise deletions of the corresponding open reading frames, except where otherwise noted. dig1Δ represents the same genomic deletion as that of the dig1-Δ1∷HIS3 allele in JCY501; kss1Δ represents the same genomic deletion as that of the kss1Δ∷hisG allele in JCY110; ste12Δ represents the same genomic deletion as that of the ste12Δ∷LEU2 allele in JCY512. The disruption markers KanMX4, HIS3MX6, CgHIS3, CgLEU2, CgTRP1, KlURA3, hphNT1, and natNT2 were amplified from pFA6a-KanMX4 (Wach et al. 1994), pFA6a-HIS3MX6 (Wach et al. 1997), pCgH, pCgL, pCgW, pKlU, pRS306H, and pRS306N (Taxis and Knop 2006), respectively. Unless otherwise indicated, strains were cultivated at 30° in standard rich (YP) or defined (SC) media (Burke et al. 2000) containing 2% glucose (Glc)/dextrose (D). When necessary, selection for plasmids was maintained by the addition of G418 (0.2 g/liter) or the omission of appropriate nutrients. Standard yeast genetic techniques were according to Burke et al. (2000).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| JCY100 | MATahis3Δ∷hisG leu2Δ∷hisG trp1Δ∷hisG ura3-52 | Cook et al. (1997) |
| JCY101 | MATα otherwise isogenic to JCY100 | Bardwell et al. (1998b) |
| JCY102 | MATa/MATα (JCY100 × JCY101) | Bardwell et al. (1998b) |
| JCY107 | JCY100 ste7Δ∷ura3 | Cook et al. (1997) |
| JCY110 | JCY100 kss1Δ∷hisG | Cook et al. (1997) |
| JCY117 | JCY100 kss1Δ∷hisG ste7Δ∷ura3 | Cook et al. (1997) |
| JCY120 | JCY100 fus3Δ∷TRP1 | Cook et al. (1997) |
| JCY127 | JCY100 fus3Δ∷TRP1 ste7Δ∷HIS3 | Cook et al. (1997) |
| JCY130 | JCY100 kss1Δ∷hisG fus3Δ∷TRP1 | Cook et al. (1997) |
| JCY137 | JCY100 kss1Δ∷hisG fus3Δ∷TRP1 ste7Δ∷ura3 | Cook et al. (1997) |
| JCY501 | JCY100 dig1-Δ1∷HIS3 dig2-Δ1∷ura3 | Bardwell et al. (1998b) |
| JCY512 | JCY100 dig1-Δ1∷HIS3 dig2-Δ1∷ura3 ste12Δ∷LEU2 | Bardwell et al. (1998b) |
| JCY600 | JCY100 ste12Δ∷LEU2 | Bardwell et al. (1998b) |
| RCY006 | JCY100 tpk1Δ∷KanMX4 | This study |
| RCY007 | JCY100 tpk2Δ∷KanMX4 | This study |
| RCY008 | JCY100 tpk3Δ∷KanMX4 | This study |
| RCY009 | JCY100 kss1Δ∷hisG tpk1Δ∷KanMX4 | This study |
| RCY010 | JCY100 kss1Δ∷hisG tpk3Δ∷KanMX4 | This study |
| RCY012 | JCY100 kss1Δ∷hisG tpk2Δ∷KanMX4 | This study |
| RCY015 | JCY100 kss1Δ∷hisG tpk1Δ∷KanMX4 tpk3Δ∷HIS3MX6 | This study |
| RCY016 | JCY100 tpk1Δ∷KanMX4 tpk2Δ∷HIS3MX6 | This study |
| RCY017 | JCY100 tpk1Δ∷KanMX4 tpk3Δ∷HIS3MX6 | This study |
| RCY018 | JCY100 tpk2Δ∷HIS3MX6 tpk3Δ∷KanMX4 | This study |
| RCY020 | JCY100 ste7Δ∷ura3 tpk1Δ∷KanMX4 | This study |
| RCY021 | JCY100 ste7Δ∷ura3 tpk2Δ∷KanMX4 | This study |
| RCY022 | JCY100 ste7Δ∷ura3 tpk3Δ∷KanMX4 | This study |
| RCY023 | JCY100 kss1Δ∷hisG ste7Δ∷ura3 tpk1Δ∷KanMX4 | This study |
| RCY024 | JCY100 kss1Δ∷hisG ste7Δ∷ura3 tpk2Δ∷KanMX4 | This study |
| RCY025 | JCY100 kss1Δ∷hisG ste7Δ∷ura3 tpk3Δ∷KanMX4 | This study |
| RCY026 | JCY100 fus3Δ∷TRP1 ste7Δ∷HIS3 tpk1Δ∷KanMX4 | This study |
| RCY027 | JCY100 fus3Δ∷TRP1 ste7Δ∷HIS3 tpk2Δ∷KanMX4 | This study |
| RCY028 | JCY100 fus3Δ∷TRP1 ste7Δ∷HIS3 tpk3Δ∷KanMX4 | This study |
| RCY029 | JCY100 kss1Δ∷hisG fus3Δ∷TRP1 ste7Δ∷ura3 tpk1Δ∷KanMX4 | This study |
| RCY030 | JCY100 kss1Δ∷hisG fus3Δ∷TRP1 ste7Δ∷ura3 tpk2Δ∷KanMX4 | This study |
| RCY031 | JCY100 kss1Δ∷hisG fus3Δ∷TRP1 ste7Δ∷ura3 tpk3Δ∷KanMX4 | This study |
| RCY033 | JCY100 flo8Δ∷HIS3MX6 sfl1Δ∷KanMX4 | This study |
| RCY104 | JCY100 flo8Δ∷HIS3MX6 | This study |
| RCY108 | JCY100 tec1Δ∷HIS3MX6 | This study |
| RCY109 | JCY100 sfl1Δ∷KanMX4 | This study |
| RCY121 | JCY100 fus3Δ∷TRP1 tpk1Δ∷KanMX4 | This study |
| RCY122 | JCY100 fus3Δ∷TRP1 tpk2Δ∷KanMX4 | This study |
| RCY123 | JCY100 fus3Δ∷TRP1 tpk3Δ∷KanMX4 | This study |
| RCY131 | JCY100 kss1Δ∷hisG fus3Δ∷TRP1 tpk1Δ∷KanMX4 | This study |
| RCY132 | JCY100 kss1Δ∷hisG fus3Δ∷TRP1 tpk2Δ∷KanMX4 | This study |
| RCY133 | JCY100 kss1Δ∷hisG fus3Δ∷TRP1 tpk3Δ∷KanMX4 | This study |
| RCY608 | JCY100 ste12Δ∷LEU2 tec1Δ∷HIS3MX6 | This study |
| RCY609 | JCY100 sfl1Δ∷HIS3MX6 ste12Δ∷LEU2 | This study |
| RCY640 | JCY100 ste12Δ∷LEU2 tpk1Δ∷KanMX4 | This study |
| RCY660 | JCY100 ste12Δ∷LEU2 tpk3Δ∷KanMX4 | This study |
| RCY664 | JCY100 flo8Δ∷HIS3MX6 ste12Δ∷LEU2 tpk3Δ∷KanMX4 | This study |
| RCY668 | JCY100 ste12Δ∷LEU2 tec1Δ∷HIS3MX6 tpk3Δ∷KanMX4 | This study |
| RCY698 | JCY100 sfl1Δ∷KanMX4 ste12Δ∷LEU2 tec1Δ∷HIS3MX6 | This study |
| RCY9041 | JCY102 FUS3/fus3Δ∷KlURA3 KSS1/kss1Δ∷CgTRP1 TPK1/tpk1Δ∷KanMX4 TPK2/tpk2Δ∷HIS3MX6 TPK3/tpk3Δ∷CgLEU2 | This study |
| RCY9043 | JCY102 DIG1/dig1Δ∷CgLEU2 DIG2/dig2Δ∷KlURA3 KSS1/kss1Δ∷CgTRP1 SFL1/sfl1Δ∷KanMX4 TPK2/tpk2Δ∷HIS3MX6 | This study |
| RCY9103 | JCY102 FLO8/flo8Δ∷CgTRP1 FUS3/fus3Δ∷KlURA3 STE12/ste12Δ∷CgHIS3 TPK1/tpk1Δ∷KanMX4 TPK3/tpk3Δ∷CgLEU2 | This study |
| RCY9107 | JCY102 DIG1/dig1Δ∷CgLEU2 DIG2/dig2Δ∷KlURA3 FLO8/flo8Δ∷CgTRP1 SFL1/sfl1Δ∷KanMX4 STE12/ste12Δ∷CgHIS3 | This study |
| RCY9113 | JCY100 kss1Δ∷CgTRP1 | This study |
| RCY9123 | JCY100 fus3Δ∷KlURA3 kss1Δ∷CgTRP1 | This study |
| RCY9140 | JCY100 dig1Δ∷CgLEU2 dig2Δ∷KlURA3 | This study |
| RCY9150 | JCY100 tpk3Δ∷CgLEU2 | This study |
| RCY9153 | JCY100 ste12Δ∷CgHIS3 | This study |
| RCY9155 | JCY100 ste12Δ∷CgHIS3 tpk3Δ∷CgLEU2 | This study |
| RCY9161 | JCY100 flo8Δ∷CgTRP1 | This study |
| RCY9163 | JCY100 flo8Δ∷CgTRP1 tpk3Δ∷CgLEU2 | This study |
| RCY9169 | JCY100 flo8Δ∷CgTRP1 ste12Δ∷CgHIS3 | This study |
| RCY9171 | JCY100 flo8Δ∷CgTRP1 ste12Δ∷CgHIS3 tpk3Δ∷CgLEU2 | This study |
| RCY9179 | JCY100 sfl1Δ∷KanMX4 | This study |
| RCY9182 | JCY100 dig1Δ∷CgLEU2 dig2Δ∷KlURA3 sfl1Δ∷KanMX4 | This study |
| RCY9184 | JCY100 dig1Δ∷CgLEU2 dig2Δ∷KlURA3 ste12Δ∷CgHIS3 | This study |
| RCY9185 | JCY100 sfl1Δ∷KanMX4 ste12Δ∷CgHIS3 | This study |
| RCY9188 | JCY100 dig1Δ∷CgLEU2 dig2Δ∷KlURA3 sfl1Δ∷KanMX4 ste12Δ∷CgHIS3 | This study |
| RCY9191 | JCY100 dig1Δ∷CgLEU2 dig2Δ∷KlURA3 flo8Δ∷CgTRP1 | This study |
| RCY9192 | JCY100 flo8Δ∷CgTRP1 sfl1Δ∷KanMX4 | This study |
| RCY9195 | JCY100 dig1Δ∷CgLEU2 dig2Δ∷KlURA3 flo8Δ∷CgTRP1 sfl1Δ∷KanMX4 | This study |
| RCY9198 | JCY100 dig1Δ∷CgLEU2 dig2Δ∷KlURA3 flo8Δ∷CgTRP1 ste12Δ∷CgHIS3 | This study |
| RCY9199 | JCY100 flo8Δ∷CgTRP1 sfl1Δ∷KanMX4 ste12Δ∷CgHIS3 | This study |
| RCY9202 | JCY100 dig1Δ∷CgLEU2 dig2Δ∷KlURA3 flo8Δ∷CgTRP1 sfl1Δ∷KanMX4 ste12Δ∷CgHIS3 | This study |
| RCY9327 | JCY102 dig1Δ∷CgLEU2/dig1Δ∷CgLEU2 DIG2/dig2Δ∷KlURA3 KSS1/kss1Δ∷CgTRP1 STE12/ste12Δ∷CgHIS3 | This study |
Plasmids and recombinant DNA methods:
Plasmids (Table 2) were constructed and propagated in Escherichia coli using standard recombinant DNA methods. pRC136, pRC137, and pRC138 were generated by site-directed mutagenesis of YCpU-KSS1 and were verified by nucleotide sequence analysis.
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Source or reference |
|---|---|---|
| pRC136 | YCplac33 PKSS1-kss1 (S303A) | This study |
| pRC137 | YCplac33 PKSS1-kss1 (S303D) | This study |
| pRC138 | YCplac33 PKSS1-kss1 (S303E) | This study |
| YCplac33 | CEN URA3 | Gietz and Sugino (1988) |
| YCpU-KSS1 | YCplac33 PKSS1-KSS1 | Bardwell et al. (1998a) |
| YCpU-kss1(K42R Q45P) | YCplac33 PKSS1-kss1 (K42R Q45P) | Cook et al. (1997) |
| YCpU-kss1(T183A Y185F) | YCplac33 PKSS1-kss1 (T183A Y185F) | Cook et al. (1997) |
Random spore analysis:
Strains were sporulated at 26° in 0.03 m potassium acetate, 0.02% raffinose. Unsporulated diploids were eliminated by adding 2 vol of ethyl ether and vortexing for 30 sec. After a 20-min room temperature incubation, the aqueous phase was plated onto YPD. Colonies arising were genotyped by their auxotrophies and abilities to induce growth arrest halos on lawns of MATa or MATα cells. In parallel, the colonies were grown on YPD plates and assessed for invasive growth.
Invasive growth assay:
Cells were grown on a solid medium and assessed for the extent to which they remained adhered to the plate following exposure to a gentle stream of tap water. For the experiments summarized in Table 3, random spore analysis was conducted for >800 spore colonies resulting from the sporulation of strains RCY9041, RCY9043, RCY9103, and RCY9107. The invasive growth of each colony was scored blind to its genotype, which was determined in parallel. In this experiment, the phenotypic range permitted a graded scoring of the degree of invasive growth, as described in the legend to Table 3, which lists the average invasive growth of all colonies of each indicated genotype. No significant differences in invasive growth were observed between otherwise isogenic MATa and MATα colonies.
TABLE 3.
Invasive growth of MAPK and PKA pathway mutants
| TPK2 | tpk2Δ | FLO8 | flo8Δ | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | KSS1 | kss1Δ | tpk2Δ | kss1Δ | STE12 | ste12Δ | flo8Δ | ste12Δ | ||
| FUS3 | TPK3 | TPK1 | + | − | − | x | + | –/x | x | x |
| tpk1Δ | +/++ | +∼ | − | x | +/++ | −∼ | x | x | ||
| tpk3Δ | +/++ | + | − | x | +/++ | +/++ | x | x | ||
| tpk3Δ | tpk1Δ | +/++ | + | ND | ND | +/++ | +/++ | x | x | |
| fus3Δ | +/++ | + | ∼ | − | +/++ | − | –/x | x | ||
| fus3Δ | tpk1Δ | +/++ | + | ∼ | − | +/++ | ∼/+∼ | –/x | x | |
| fus3Δ | tpk3Δ | +/++ | + | ∼ | − | +/++ | +/++ | –/x | x | |
| fus3Δ | tpk3Δ | tpk1Δ | +/++ | + | ND | ND | +/++ | +/++ | – | x |
| SFL1 | DIG1 | DIG2 | + | − | − | x | + | –/x | x | x |
| dig2Δ | −/−∼ | − | –/x | x | −/−∼ | –/x | x | x | ||
| dig1Δ | +/++ | ++ | ++ | ++ | +/++ | − | − | x | ||
| dig1Δ | dig2Δ | ++ | ++ | ++ | ++ | ++ | –/x | + | x | |
| sfl1Δ | ++ | ++ | ++ | ++ | ++ | ++ | − | − | ||
| sfl1Δ | dig2Δ | ++ | ++ | ++ | ++ | ++ | ++ | − | –/x | |
| sfl1Δ | dig1Δ | ++ | ++ | ++ | ++ | ++ | ++ | +/++ | − | |
| sfl1Δ | dig1Δ | dig2Δ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | –/x |
Invasive growth was scored as follows: ++, no erosion of growth patch by water stream. +, only slight erosion of growth patch by water stream (defined as that displayed by wild-type strain JCY100). +∼, some significant erosion, but majority of growth patch remained. ∼, borderline cases not unambiguously closer to either “+∼” or “−∼”. −∼, non-invasive, but after wash-off by water stream some residual material remained. –, non-invasive, but after wash-off by water stream, patch imprint readily detectable. x, non-invasive, and after wash-off by water stream, patch imprint difficult to detect. ND, not determined; no spores of these genotypes were obtained because tpk1Δ tpk2Δ tpk3Δ mutants are inviable (Toda et al. 1987).
Flocculation assay:
Liquid cultures were thoroughly mixed, transferred to spectrometry cuvettes, thoroughly mixed again, and immediately checked for absorbance at 600 nm (time 0). The cuvettes were then allowed to sit without agitation, and A600 values were read at various times up to 4 hr later. For all strains, the A600 values exhibited exponential decay over time, and the times required for A600 values to halve [t0.5 (A600)] were calculated on the basis of fitting to a first-order exponential decay curve.
Preparation of cell extracts and immunoblotting:
Cell pellets were resuspended in buffer containing 25 mm Tris–Cl (pH 7.4), 150 mm NaCl, 10% glycerol, 1 mm DTT, 2 mm EDTA, 5% SDS, 0.1% Triton X-100, protease inhibitors (Roche Complete EDTA-free protease inhibitor tablets), and phosphatase inhibitors (25 mm NaF, 0.25 mm NaVO3, 0.25 mm Na3VO4, 1 mm sodium pyrophosphate, 25 mm β-glycerol phosphate). Glass beads were added and samples were vortexed, boiled, and clarified by centrifugation. Samples were resolved by SDS–PAGE, transferred to nitrocellulose filters, incubated with appropriate primary antibodies—i.e., anti-Fus3 (sc-6773, Santa Cruz Biotechnology), anti-Kss1 (sc-6775, Santa Cruz Biotechnology), or anti-phospho-MAPK (#9101, Cell Signaling Technology #9101)—and then with appropriate infrared dye-conjugated secondary antibodies, and visualized by using an infrared imaging system (Odyssey; Li-Cor).
RESULTS
A PKA consensus sequence in Kss1 is not required for its function in invasive growth:
The optimal consensus motif for phosphorylation by PKA is well characterized and consists of two basic residues (most often Arg) at the −3 and −2 positions relative to the Ser (or Thr) phosphoacceptor residue, followed at +1 by a hydrophobic residue: -(R/K)(R/K)×(S/T)Hpo- (Pearson and Kemp 1991). We noted that Kss1 contains such a motif (-Lys-Arg-Ile-Ser303-Ala-) and considered the possibility that PKA could influence Kss1 activity and/or affect its interaction with binding partners and/or substrates by modifying this site. If so, then mutating Ser303 should disrupt coordination between PKA and MAPK function in FG responses.
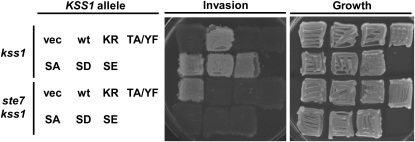
Consequently, we generated both a nonphosphorylatable (S303A) and two phospho-mimetic (S303D and S303E) alleles and tested their ability to support a diagnostic hallmark of haploid FG. In wild-type cells, Kss1 activity is required to elicit invasive growth (Cook et al. 1997); a kss1Δ strain carrying an empty vector or the same plasmid expressing either an unactivatable [Kss1(T183A Y185F)] or a catalytically inactive [Kss1(K42R Q45P)] mutant cannot invade the agar, whereas invasiveness is restored when the plasmid expresses wild-type Kss1 (Figure 1, top rows). Kss1 in its inactive state (e.g., in the absence of its activator Ste7) represses invasive growth (Cook et al. 1997) by remaining bound to Ste12 (Bardwell et al. 1998a); a kss1Δ ste7Δ double mutant is able to invade agar, and introduction of a Kss1-expressing plasmid, but not the empty vector, abolishes the invasiveness of the kss1Δ ste7Δ cells (Figure 1, bottom rows). In either context, the Kss1(S303A), Kss1(S303D), and Kss1(S303E) mutants all behaved indistinguishably from wild-type Kss1. Thus, elimination of Ser303 does not affect either the stimulatory or the inhibitory functions of Kss1 in invasive growth. Therefore, modification of this residue by PKA (or any other protein kinase) is unlikely to be of any importance for modulating Kss1 function in haploid FG.
Figure 1.—
Mutation of a PKA consensus phosphorylation motif in Kss1 does not affect either its positive or its negative functions in invasive growth. Strains JCY110 (kss1Δ) and JCY117 (kss1Δ ste7Δ) were transformed with an empty vector (vec; YCplac33) or with plasmids encoding Kss1 (wt; YCpU-KSS1), Kss1(K42R Q45P) [KR; YCpU-kss1(K42R Q45P)], Kss1(T183A Y185F) [TA/YF; YCpU-kss1(T183A Y185F)], Kss1(S303A) (SA; pRC136), Kss1(S303D) (SD; pRC137), or Kss1(S303E) (SE; pRC138). Transformants were grown on SCD–Ura plates, replicated to YPD, and assessed for invasive growth.
Expression and activation of MAPKs Kss1 and Fus3 are independent of PKA:
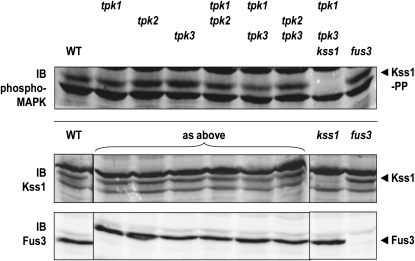
Although Kss1 MAPK does not appear to be a direct target of PKA, it is possible that PKA action potentiates the activity of this MAPK via an indirect mechanism. For example, in lymphocytes, PKA phosphorylates a MAPK-inactivating phosphatase, causing a reduction in its binding affinity for the MAPK, thereby extending the duration of MAPK activation (Saxena et al. 1999). Among the known MAPK-targeted phosphatases in yeast, Ptp3 contains two consensus PKA motifs (-KRAT75A- and -RKNT675M-), raising the possibility that PKA action might potentiate Kss1 function in FG by preventing its efficient Ptp3-mediated dephosphorylation. Furthermore, it is claimed that, in cerebellar granule cells, elevated cAMP promotes (via PKA) the activation of Ras, thereby stimulating the MAPKKK (Raf) of the cascade that activates the mammalian Kss1 ortholog (Erk2) (Obara et al. 2007). To test whether PKA activity affects Kss1 activation in any way, we assessed the extent of MAPK phosphorylation in extracts from cells lacking Tpk1, Tpk2, or Tpk3 (or combinations of these gene products) (Figure 2). Only modest effects on the level of activated (dually phosphorylated) Kss1 were observed, and they varied somewhat from trial to trial. Therefore, we conclude that the absence of any single PKA isoform or even pairs of PKA isoforms [cells lacking all three are inviable (Toda et al. 1987)] does not reproducibly alter Kss1 activation under FG conditions. Likewise, Fus3 (which in wild-type cells is unphosphorylated in the absence of pheromone stimulation) did not become detectably phosphorylated in any of the PKA mutant strains. Furthermore, the total levels of Kss1 and Fus3 were unaffected by loss of PKA (Figure 2). Thus, PKA does not regulate the amount or activation state of Kss1 or Fus3.
Figure 2.—
Absence of PKA does not affect levels or activation of Fus3 or Kss1. Whole-cell extracts from exponentially growing cultures of strains JCY100, JCY110, JCY120, RCY006, RCY007, RCY008, RCY015, RCY016, RCY017, and RCY018 were immunoblotted to assess levels of phospho-MAPK, Kss1, and Fus3. Strains lacking Kss1 or Fus3 serve to identify the appropriate bands. The bottom two images were spliced to reorder lanes for ease of comparison.
The function of PKA during invasive growth does not require Kss1 MAPK:
The preceding results indicated that Kss1 MAPK is not regulated by PKA, either directly as a substrate or indirectly in terms of its expression or activation level. Therefore, we investigated more generally whether any functional interrelationships between these two kinases exist that contribute to their mutual regulation of the biological process of invasive growth.
Of the MAPKs involved in invasive growth, Kss1 is a positive regulator because mutation of Kss1 reduces the invasiveness of wild-type cells, and Fus3 is a negative regulator because mutation of Fus3 restores invasiveness to kss1Δ cells (Cook et al. 1997). Among the PKA isoforms, Tpk2 is a positive regulator because tpk2Δ cells are non-invasive, and Tpk3 is a negative regulator because tpk3 cells are hyper-invasive (consistent with this, tpk2/tpk2 diploids are defective for pseudohyphal growth and tpk3/tpk3 diploids display enhanced pseudohyphal growth) (Robertson and Fink 1998). One group reported that tpk1Δ haploids and tpk1/tpk1 diploids displayed indistinguishable invasive growth and pseudohyphal growth, respectively, relative to wild-type cells, suggesting that Tpk1 has no role in these processes (Robertson and Fink 1998); on the other hand, another group reported that tpk1/tpk1 diploids exhibited enhanced pseudohyphal growth, suggesting that Tpk1 is a negative regulator of FG, at least in diploids (tpk1 haploids were not tested) (Pan and Heitman 1999). This ambiguity about the role of Tpk1 may stem from the fact that the Σ1278b backgrounds used in the two studies cited derive from different laboratories and have undergone different manipulations and, thus, are likely not strictly isogenic. In any event, Kss1 and Tpk2 positively regulate haploid invasive growth, Fus3 and Tpk3 negatively regulate this phenotype, and it was unclear whether Tpk1 is either neutral or, perhaps, a negative regulator of this behavior in haploids.
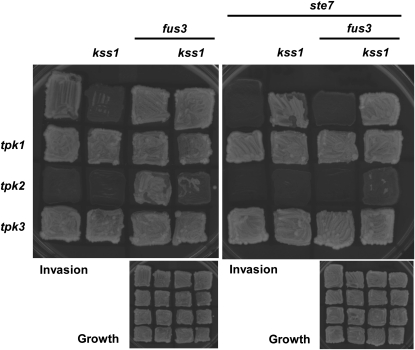
Previous genetic analyses of invasive growth have generally been performed either on MAPK pathway components only or on PKA pathway components only. To examine interpathway regulatory relationships, we combined tpk1, tpk2, and tpk3 null mutations with kss1 and/or fus3 null mutations. Interestingly, like removal of Fus3, absence of either Tpk1 or Tpk3 restored invasiveness to a kss1Δ strain (Figure 3). Similarly, absence of Fus3 substantially increased the invasiveness of cells lacking Tpk2 (Figure 3). These results demonstrate, first, that Tpk1 indeed performs a negative function during invasive growth, corroborating the conclusions reached for its role in diploid pseudohyphal growth (Pan and Heitman 1999). Second, our findings indicate that the stimulatory roles of Kss1 and Tpk2 are weaker than (or farther upstream) the inhibitory roles of Fus3, Tpk1, and Tpk3. Third, these data show that the requirement for Kss1 can be bypassed by removal of any of several different negatively acting factors and that these factors contribute independently and act additively, suggesting that Fus3, Tpk1, and Tpk3 do not act together in a linear pathway whose inhibitory effect is counteracted by Kss1 action. Consistent with this conclusion, the invasiveness of the tpk2Δ fus3Δ strain was largely lost in cells lacking Kss1 (Figure 3). Thus, even in the absence of the inhibitory factor Fus3, either Tpk2 or Kss1 is needed in a positive sense to elicit any response. The implications of these results will be discussed in a larger context below.
Figure 3.—
In the absence of Tpk1 or Tpk3, the MAPK cascade is not necessary for invasive growth, and in the absence of the MAPK cascade, Tpk2 is still required for invasive growth. Strains JCY100, JCY107, JCY110, JCY117, JCY120, JCY127, JCY130, JCY137, RCY006, RCY007, RCY008, RCY009, RCY010, RCY012, RCY020, RCY021, RCY022, RCY023, RCY024, RCY025, RCY026, RCY027, RCY028, RCY029, RCY030, RCY031, RCY121, RCY122, RCY123, RCY131, RCY132, and RCY133 were assessed for invasive growth.
A strain lacking Ste7 (the MAPKK that activates both Kss1 and Fus3) is non-invasive (Roberts and Fink 1994). Just as absence of Tpk1 or Tpk3 restored invasiveness to a kss1Δ strain (Figure 3), absence of either Tpk1 or Tpk3 also restored invasiveness to a ste7Δ strain (Figure 3). Active Kss1 (as in STE7 strains) has an activating function, whereas inactive Kss1 (as in ste7Δ strains) acts as a repressor (Cook et al. 1997; Madhani et al. 1997; Bardwell et al. 1998a,b). For this reason, elimination of Kss1 restores invasiveness to a ste7Δ strain (Cook et al. 1997). However, importantly, we found that the invasiveness of ste7Δ kss1Δ, kss1Δ fus3Δ, and ste7Δ kss1Δ fus3Δ strains still required Tpk2 (Figure 3), indicating that Tpk2 performs an essential function in invasive growth that cannot be mediated by the MAPK cascade.
Together, these results indicate that both promotion of invasive growth by the PKA isoform Tpk2 and inhibition of invasive growth by the PKA isoforms Tpk1 and Tpk3 are independent of the action of the MAPKs Kss1 and Fus3. In other words, these two signaling modules are not connected at the level of their effector kinases.
Systematic analysis of distal components of the MAPK and PKA pathways in invasive growth:
The kinases in both the MAPK and PKA pathways directly interact with, phosphorylate, and modify the activity of transcriptional repressors and activators. Downstream of Kss1 are Dig1 (repressor), Dig2 (repressor), and Ste12 (activator) (Cook et al. 1997; Madhani et al. 1997; Tedford et al. 1997; Bardwell et al. 1998a,b). Downstream of Tpk2 are Sfl1 (repressor) and Flo8 (activator) (Robertson and Fink 1998; Pan and Heitman 1999, 2002). To determine whether any regulatory interrelationships exist among these transcriptional modulators, we assessed invasive growth in strains lacking combinations of these pathway components. Toward this end, four diploid strains were constructed. Two were heterozygous for the activating kinases (KSS1/kss1 TPK2/tpk2), and two of the strains were heterozygous for the transcriptional activators (STE12/ste12 FLO8/flo8). From each strain, derivatives that were heterozygous for either the inhibitory kinases (TPK1/tpk1 TPK3/tpk3 FUS3/fus3) or the transcriptional repressors (DIG1/dig1 DIG2/dig2 SFL1/sfl1) were generated. Such quintuply heterozygous diploid strains were allowed to sporulate, generating haploid spores containing various combinations of PKA and MAPK pathway mutations, each identifiable by a distinct associated marker. These spores were plated and grown into colonies, and colonies were assessed for their invasive growth phenotype blind to its genotype, which was determined subsequently. Table 3 catalogs the average invasive growth phenotype of all colonies of each genotype (otherwise isogenic MATa and MATα colonies exhibited very similar properties).
Typically, determination of the invasive growth phenotype is binary—i.e., strains are scored as invasive or non-invasive. Due to the very large number of colonies analyzed in parallel (>800) in our experiments, we were able to distinguish a wider phenotypic range and assign finer gradations to the strength of the invasive growth response (see legend to Table 3). We found that strains of different genotypes reproducibly exhibited distinct degrees of invasiveness over a rather broad range (Table 3). Thus, our systematic analysis revealed that invasive growth is not an all-or-none response, but rather a quantitative or at least semiquantitative trait.
The MAPK and PKA pathways contribute additively to promote invasive growth:
Cells lacking all three PKA catalytic subunits are inviable (Toda et al. 1987). The inviability of a tpk1Δ tpk2Δ tpk3Δ triple mutant can be rescued, however, by removal of another protein kinase, Yak1 (Garrett and Broach 1989), suggesting that the essential function of PKA is to antagonize the action of Yak1. Consistent with this conclusion, mass spectrometry analysis has shown that Yak1 is phosphorylated in vivo at its five PKA consensus sites (Zappacosta et al. 2002). In our analysis (Table 3), no strains were obtained in which all three PKA catalytic subunits were deleted, regardless of the presence or absence of one or both of the MAPKs, indicating that neither Kss1 nor Fus3 has a relationship to PKA similar to that of Yak1.
Scoring of the invasive growth phenotype on a finer scale revealed some previously unrecognized interpathway relationships. For example, Tpk1 can clearly be seen to be a negative regulator of invasive growth not only because its loss restores invasiveness to a non-invasive kss1Δ mutant (Figure 3), but also because its absence enhanced the invasiveness of even an otherwise wild-type strain (Table 3).
More generally, our analysis could provide insight into the interconnections among the positively acting and negatively acting factors, as follows. Suppose X and Y are two signal transduction proteins whose corresponding mutations, denoted x and y, give rise to opposing phenotypes. If the phenotype of the double-mutant x y is identical to the phenotype of one of the single mutants, for example, y, then this suggests that Y acts downstream of X in a single pathway. For example, as shown above (Figure 3), because a kss1Δ fus3Δ strain displays the same phenotype as a fus3Δ strain (invasive), but not a kss1Δ strain (non-invasive) (Cook et al. 1997), this suggests that Fus3 acts, in a genetic sense, downstream of Kss1 in its regulation of invasive growth. Formally, this relationship means that the sole role of Kss1 is to counteract the effects of Fus3. If, however, the phenotype of x y is intermediate between those of x and y, then X and Y are likely to contribute independently to the net phenotype.
The invasive growth behavior of strains lacking various combinations of the kinases is shown in Table 3 (note that Table 3 includes all of the STE7 genotypes depicted in Figure 3 and also others). The extent of invasiveness corresponds to genotype in a graded, as opposed to abrupt, manner. Overall, our scheme permitted us to deduce certain rankings. For example, Tpk2 is a more potent activator of invasive growth than Kss1, and Fus3 is a more potent repressor than Tpk3, which is a more potent repressor than Tpk1. In no case did we discern that loss of Tpk1 or Tpk3 caused any increase in the invasiveness of a strain already lacking Tpk2, indicating that the inhibitory functions of Tpk1 and Tpk3 are exerted on Tpk2; i.e., they likely function in the same pathway. With the exception of these relationships among the PKA isoforms, invasiveness increases diagonally from the upper right corner of the quadrant to the lower left, because it can be enhanced either by adding another activatory kinase or by deleting another inhibitory kinase, suggesting that the effects of PKA and the MAPKs Kss1 and Fus3 contribute independently and additively to invasive growth. Indeed, combining the mutations in an invasive strain with the mutations in a non-invasive strain generally caused the resultant strain to exhibit intermediate invasiveness. Importantly, all strains lacking the activators Tpk2 and Kss1 are less invasive than the otherwise identical strains lacking only one of these factors; this would not be the case if one was upstream of the other in a single pathway. In agreement with the data presented in earlier sections indicating that PKA isoforms do not function upstream of MAPK isoforms during invasive growth, the results compiled in Table 3 further broaden and support the conclusion that the PKA and MAPK kinases act independently and additively to facilitate the FG response.
Analysis of mutants containing combinations of the activating kinases (Kss1 and Tpk2) and the transcriptional repressors (Dig1, Dig2, and Sfl1) is shown in Table 3. As previously observed, cells lacking Kss1, Dig1, and Dig2 were slow growing (Breitkreutz et al. 2003), which is most likely due to the toxicity of hyperactive Ste12 (Dolan 1996), and quickly gave rise to faster-growing variants. Indeed, tetrad analysis of a dig1Δ∷CgLEU2/dig1Δ∷CgLEU2 DIG2/dig2Δ∷KlURA3 KSS1/kss1Δ∷CgTRP1 STE12/ste12Δ∷ CgHIS3 strain (RCY9327) revealed that the growth defect of a kss1Δ dig1Δ dig2Δ mutant was completely rescued by loss of Ste12. It has been reported that deletion of DIG2 enhances the invasiveness of a dig1Δ strain, reduces the invasiveness of fus3Δ and fus3Δ kss1Δ strains, and does not affect the invasiveness of otherwise wild-type strains (Breitkreutz et al. 2003). Consistent with the apparent dual nature of the Dig2 function, in our analysis, deletion of DIG2 increased the invasiveness of a dig1Δ strain and decreased the invasiveness of a tpk2Δ strain (Table 3). However, in our hands, loss of Dig2 in an otherwise wild-type strain detectably diminished invasive growth. In contrast to Dig2, the loss of Dig1 (and/or Sfl1) caused hyper-invasiveness even in the absence of both Kss1 and Tpk2 (Table 3). This functional difference between Dig1 and Dig2 is consistent with reports that Dig1 and Dig2 interact with separate regions of Ste12 (Olson et al. 2000) and that Dig1 and Dig2 incorporate differentially into transcription factor complexes (Ste12-Dig1-Dig2 and Tec1-Ste12-Dig1) (Chou et al. 2006), which is consistent with distinct mechanisms of action.
We observed that kss1Δ tpk2Δ dig1Δ and kss1Δ tpk2Δ sfl1Δ strains are more strongly hyper-invasive than fus3Δ tpk1Δ tpk3Δ strains (Table 3). These results suggest, first, that Fus3, Tpk1, and Tpk3 do not inhibit transcription as strongly as Dig1 and Sfl1 do. Second, this finding reveals that derepression of the transcriptional apparatus downstream of either the MAPK or the PKA pathway is sufficient to bypass the need for the upstream signaling kinases in either pathway. Thus, the contributions of the two pathways are not specialized with respect to invasive growth, and the sole essential reason for the combined function of Kss1 MAPK and PKA during filamentous growth is to elicit a sufficiently potent level of transcriptional induction from a common set of genes. There is evidence that optimal invasive growth may require cap-independent translation of certain mRNAs (Gilbert et al. 2007). Our observations suggest either that cap-independent translation during invasive growth does not require either Kss1 or Tpk2 or that cap-independent translation becomes dispensable when Dig1- or Sfl1-regulated transcription targets become highly derepressed. Additionally, it has been reported that glucose depletion triggers rapid PKA-dependent inhibition of translation (Ashe et al. 2000; Uesono et al. 2004). Our results suggest that this acute response is not a critical component of the much longer process of achieving efficacious invasive growth.
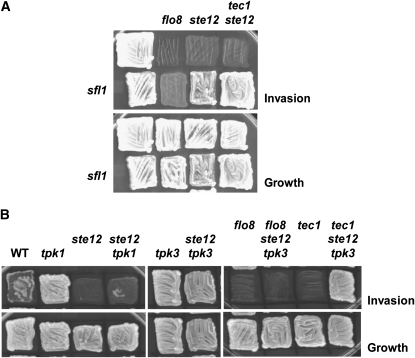
The phenotypes of mutants lacking the transcriptional activators Ste12 or Flo8, or both, combined with the loss of the inhibitory kinases Tpk1, Tpk3, and/or Fus3 or with the absence of the transcriptional repressors Dig1, Dig2, and Sfl1, are shown in Table 3. Strains lacking both Flo8 and Ste12 were non-invasive even in the absence of the inhibitory kinases or the repressors, indicating that the combined effect of these negative regulators is exerted solely via their regulation on Flo8 and Ste12. Because loss of Sfl1 caused a ste12Δ flo8Δ strain to display a slight degree of invasiveness, it is possible that Sfl1 negatively regulates one or more FG genes that are not Ste12 and/or Flo8 targets. Most strikingly, however, absence of Sfl1 (the Flo8 repressor) restored invasiveness to ste12Δ and ste12Δ tec1Δ strains (but not to a flo8Δ strain) (Table 3 and Figure 4A) and, reciprocally, absence of Dig1 and Dig2 (the Ste12 repressors) restored invasiveness to a flo8Δ strain (but not to a ste12Δ strain), in further agreement with the view that the PKA and MAPK inputs act separately and additively.
Figure 4.—
Lack of the PKA isoform Tpk3 or the transcriptional repressor Sfl1 renders the transcriptional activator Ste12 unnecessary for invasive growth. (A) Strains JCY100, JCY600, RCY033, RCY104, RCY109, RCY608, RCY609, and RCY698 were assessed for invasive growth. (B) Strains JCY100, JCY600, RCY006, RCY008, RCY104, RCY108, RCY640, RCY660, RCY664, and RCY668 were assessed for invasive growth.
Although invasiveness could be fully restored to a flo8Δ strain by loss of Dig1 and Dig2 (or Dig1 and Sfl1), it was only slightly increased by loss of Fus3, Tpk1, and Tpk3, suggesting that, in comparison to the repressors, inhibition of Ste12 by these kinases is rather weak. In contrast, invasiveness was restored to ste12Δ and ste12Δ tec1Δ strains not only by loss of Sfl1, but also by loss of Tpk3 (Table 3 and Figure 4B), suggesting that this upstream kinase is, operationally, as potent an inhibitor of Flo8 activity as its directly interacting transcriptional repressor. Although invasiveness was restored to a kss1Δ strain by removing either Tpk1 or Tpk3, invasiveness could be restored to a ste12Δ strain only by removing Tpk3, and not by eliminating Tpk1, which further highlights that these two PKA isoforms have different inhibitory strengths.
In summary, complete derepression of the transcriptional activator controlled by the MAPK pathway, or of the transcriptional activator controlled by the PKA pathway, is sufficient for invasive growth, even in the complete absence of the other pathway. Thus, each of these two inputs does not contribute unique activities or specialized functions that must be integrated with the other in some combinatorial fashion to elicit the desired response; rather, each signal impinges separately and additively to achieve the required level of total signal strength.
Glucose limitation-induced unipolar budding pattern is independent of Ste12 and Flo8:
In addition to agar invasion and increased cell–cell adhesion (see next section), haploids undergoing FG also exhibit a slightly elongated morphology and a unipolar budding pattern. To investigate the roles of the MAPK and PKA signaling pathways in regulating these other phenotypic behaviors associated with FG, we analyzed the mutants generated for our epistasis analysis that yielded the most informative and dramatic insights about how the PKA and MAPK pathways contribute to agar invasiveness (Table 3).
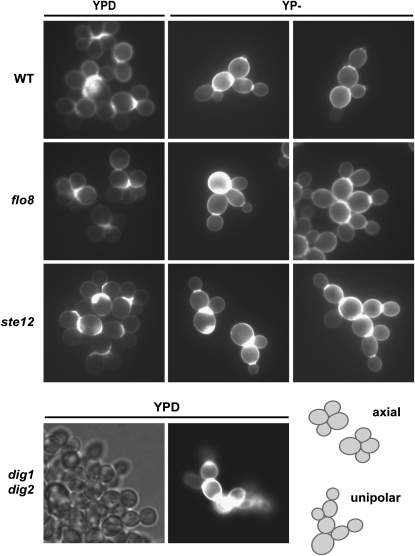
Cells lacking Ste12 were generally rounder (less ovoid) than STE12 cells when grown in a medium containing excess glucose (YPD); cells lacking both Ste12 and Flo8 were generally rounder than STE12 FLO8 cells when grown in a glucose-limiting (YP−) medium (Table 4). All of the strains examined exhibited axial budding in rich medium, with the exception of STE12 strains lacking both Dig1 and Dig2, which exhibited unipolar budding (and more elongated cells) under the same conditions (Table 4 and Figure 5). Upon a shift to a glucose-limiting medium, all of the strains exhibited unipolar budding (Table 4). Thus, the switch in cell division pattern that occurs upon glucose limitation is independent of both Ste12 and Flo8.
TABLE 4.
Cell morphology, cell–cell adhesion, and budding pattern of MAPK and PKA pathway mutants
| Genotype | + | flo8Δ | ste12Δ | flo8 ste12Δ |
|---|---|---|---|---|
| + | ||||
| tpk3Δ | A1, a1 | |||
| sfl1Δ | A1, a1; b | A1 | ||
| dig1Δ dig2Δ | A2, a1; B, b | A2, a1; B, b | ||
| dig1Δ dig2Δ sfl1Δ | A2, a2; B, b | A2, a2; B, b | a1 |
Cultures of strains JCY100, RCY9140, RCY9150, RCY9153, RCY9155, RCY9161, RCY9163, RCY9169, RCY9171, RCY9179, RCY9182, RCY9184, RCY9185, RCY9188, RCY9191, RCY9192, RCY9195, RCY9198, RCY9199, and RCY9202 were examined by microscopy. YPD: cultures were grown to exponential phase in YPD. YP−: overnight YPD cultures were diluted 10× directly into YP and incubated for 16 hr. Unless otherwise indicated, cell clusters were small in both YPD and YP−, and cells exhibited axial budding in YPD and unipolar budding in YP−. ste12Δ strains tended to be rounder (less ovoid) than STE12 strains in YPD. ste12Δ, flo8Δ and ste12Δ flo8Δ strains tended to be rounder than STE12 FLO8 strains in YP−. A1: medium-sized clusters in YPD; a1: medium-sized clusters in YP−; A2: very large, tight cell clusters in YPD; a2: very large, tight cell clusters in YP−; B: unipolar budding and markedly elongated cells in YPD; b: elongated cells in YP−.
Figure 5.—
Glucose limitation-induced unipolar budding pattern is independent of Ste12 and Flo8. Wild type (WT; JCY100), flo8Δ (RCY104), ste12Δ (JCY600), and dig1Δ dig2Δ (JCY501) cells were examined by microscopy. To facilitate assessment of budding patterns, cell walls and cell division junctions were stained with calcofluor white (0.1 g/liter) just before visualization. YPD: cultures were grown to exponential phase in YPD. YP−: overnight YPD cultures were diluted 10× directly into YP and incubated for 16 hr.
Ste12 is a more potent inducer of cell–cell adhesion than Flo8:
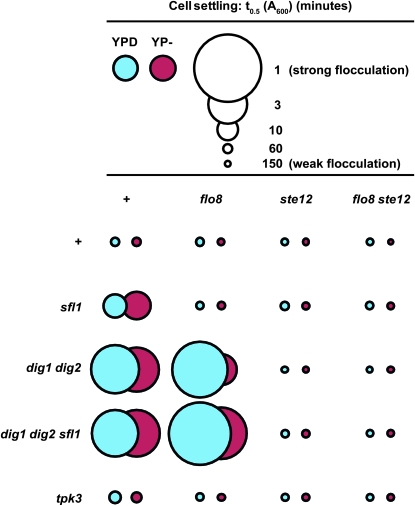
To assess cell–cell adhesion (flocculation), we measured the rate at which cells settled out of liquid cultures (Figure 6) and also examined by microscopy the extent of cell clustering in liquid cultures (Table 4). Surprisingly, glucose limitation, which is known to trigger invasive growth and unipolar budding (Cullen and Sprague 2000), had relatively little effect on flocculation and cell clustering, at least as compared to the markedly more pronounced effects caused by mutations (Figure 6 and Table 4). Absence of Tpk3 from otherwise wild-type cells caused an increase in flocculation (shorter time of cell settling), but the most highly flocculent strains were those lacking the transcriptional repressors Dig1 and Dig2, or Sfl1 (Figure 6). Interestingly, the hyper-flocculation of the sfl1Δ and tpk3Δ strains required the presence of both Flo8 and Ste12, whereas the dig1Δ dig2Δ strains were hyper-flocculent even in the absence of Flo8. The flocculation behavior of the mutants tested correlated well with the size of the cell clusters observed by microscopy; specifically, increased flocculation corresponded to larger average cell cluster size (Table 4). In toto, these results indicate that, similar to their functions in invasive growth, the adhesion-inducing functions of Flo8 and Ste12 are not maximally activated in wild-type cells even under nutrient-limiting conditions. Furthermore, it is clear that Ste12 is a much more potent inducer of cell–cell adhesion than Flo8, since derepression of Ste12 permits enhanced adhesion even in the absence of Flo8, whereas adhesion is not significantly increased by deletion of repressors in strains lacking Ste12.
Figure 6.—
Ste12 is a more potent activator of flocculation than Flo8. Strains of the indicated genotypes were assessed for flocculation. Circle areas depict the strength of flocculation on the basis of the rate of settling: the larger the circle, the stronger the flocculation. YPD (blue): cultures were grown to exponential phase in YPD; YP− (red): overnight YPD cultures were diluted 10× directly into YP and incubated for 16 hr. Strains used were JCY100, RCY9123, RCY9140, RCY9150, RCY9153, RCY9155, RCY9161, RCY9163, RCY9169, RCY9171, RCY9179, RCY9182, RCY9184, RCY9185, RCY9188, RCY9191, RCY9192, RCY9195, RCY9198, RCY9199, and RCY9202.
DISCUSSION
The MAPK Kss1 activates the transcription factor Ste12 by relieving repression imposed by Dig1 and Dig2 (Bardwell et al. 1998a,b). The PKA Tpk2 activates the transcription factor Flo8 by relieving repression imposed by Sfl1 (Robertson and Fink 1998; Pan and Heitman 2002). Both of these kinase-regulated transcription circuits are necessary for nitrogen limitation-induced pseudohyphal growth in diploids and glucose limitation-induced invasive growth in haploids. These nutrient limitation-induced responses are accompanied by cell–cell and cell–substratum adhesion, cell elongation, and a unipolar budding pattern. Here we analyzed the contributions of the MAPK and PKA pathways to these behaviors by using a comprehensive and systematic genetic approach and examined the extent to which agar invasion is representative of the other FG-associated phenotypes in haploids. A summary of our conclusions is shown in schematic form in Figure 7.
Functions of the MAPK and PKA pathways for cellular response to nutrient limitation:
It has been observed that, in diploids growing on nitrogen-limited media, Ste12 is necessary for cell elongation and normal (linear) cell–cell junctions, whereas Tpk2 is necessary for normal cell–cell junctions and the switch from bipolar to unipolar budding (Pan and Heitman 1999). Thus, it was concluded that, in diploids, the MAPK pathway specifically regulates cell elongation, the PKA pathway specifically regulates unipolar budding, and both pathways regulate cell adhesion (Pan and Heitman 1999).
As we observed in this study, in haploids, Ste12 regulates cell elongation, but Flo8 also contributes during glucose limitation (Table 4). However, these differences were rather subtle because the nutrient limitation-induced elongation of haploid cells is much less pronounced than in diploids (Galitski et al. 1999). Cell–cell adhesion was relatively independent of glucose levels, but much more strongly dependent on Ste12 than on Flo8 (Figure 6 and Table 4). The switch from axial to unipolar budding upon glucose limitation in haploids was, by contrast, independent of both Ste12 and Flo8 (Table 4 and Figure 5), as well as independent of Tpk2 (Chen 2008). It is possible that this Ste12- and Flo8-independent effect may be mediated by the action of a third class of protein kinase, Snf1 (AMP-activated protein kinase), which becomes activated upon glucose limitation (Hardie et al. 1998; Hedbacker and Carlson 2008) and contributes to FG by antagonizing two zinc-finger repressors, Nrg1 and Nrg2 (Vyas et al. 2001, 2003; Kuchin et al. 2002). Thus, in haploids, cell elongation and especially cell–cell adhesion are primarily regulated by the MAPK pathway, with a more limited contribution by the PKA pathway, whereas glucose limitation-induced unipolar budding is independent of both of these pathways, which is somewhat different from the regulatory hierarchy required for eliciting similar responses in diploids. Indeed, it has been noted that expression of Flo11, a cell-surface glycoprotein required for FG in both haploids and diploids, is ∼10× higher in haploids than in diploids (Mösch et al. 1999). Taken together with our results, it is clear that, despite the similarities in cellular behaviors that accompany FG in diploids and haploids, and despite the fact that the same kinase pathways are involved, cell type-specific differences impose different regulatory dependencies and outcomes in the face of similar inputs. It will be interesting to compare the detailed regulatory arrangements in other yeasts and pathogenic fungi, whose filamentous behaviors generally require components orthologous to the MAPK and PKA signaling pathways (and the downstream transcription factors) first described in S. cerevisiae (Bruno et al. 1996; Banuett 1998; Martinez-Espinoza et al. 2004; Verstrepen and Klis 2006; Klosterman et al. 2007; Rispail et al. 2009; Hoi and Dumas 2010).
Subthreshold signaling through the MAPK and PKA pathways combines additively to promote agar invasion:
Prior to our study, the existence and direction of cross-regulation between the Kss1 MAPK and PKA pathways in this developmental process have been unclear. Some evidence indicated that the MAPK and PKA pathways function independently, at least in the regulation of diploid pseudohyphal growth (Pan and Heitman 1999), whereas other findings suggested that, for haploid invasive growth, the PKA pathway regulates the MAPK pathway (Mösch et al. 1999). However, another study concluded the converse, namely that the MAPK pathway regulates the PKA pathway in haploids (Cherkasova et al. 2003).
As documented here, to clarify the relationship between the MAPK and PKA pathway inputs in haploid FG, we performed a systematic genetic analysis using cross-combinations of pathway mutants. Each of these pathways contains multiple terminal kinase isoforms. Among the overarching conclusions of this work are that certain isoforms are positive regulators and others are negative regulators and, within each class, regulators that exert control in the same direction nonetheless do so with distinctly different strength. Most revealing, we found that positively acting kinases are individually necessary, but only to relieve the repression imposed by the negatively acting kinases. Furthermore, each signaling pathway regulates a discrete transcriptional activation apparatus, including distinct repressors that act on different activators. Both transcriptional activators of each pathway are normally required for haploid invasive growth, but only because neither activator is maximally active, even under nutrient-limiting conditions, and thus input from both signaling pathways is required to reach an adequate level of expression to elicit the appropriate cellular behavior (Figure 7). In other words, the contributions of the Kss1 MAPK pathway and the PKA pathway are additive and overlapping (nearly redundant) in function because full derepression of either transcriptional activator is sufficient for invasive growth, even in the total absence of input from the other pathway.
Differential control of agar invasion, flocculence, cell elongation, and unipolar budding:
We found that agar invasion, cell elongation, and flocculation are strongly dependent on Ste12. Nonetheless, we were able to distinguish different degrees of dependence of flocculation, cell elongation, unipolar budding, and agar invasion on the MAPK pathway, the PKA pathway, and glucose limitation. Thus, in a certain sense, these processes are not tightly coupled and are to some extent separable. For example, strains that exhibited the most robust agar invasion were not uniformly those that were the most flocculent, and the glucose withdrawal-induced switch to a unipolar budding pattern was not accompanied by a notable increase in flocculence. Thus, although both sfl1Δ ste12Δ (Flo8 on, Ste12 off) and dig1Δ dig2Δ flo8Δ (Flo8 off, Ste12 on) strains display agar invasion, they are not identical. In any event, the requirement in wild-type cells for both the MAPK and PKA inputs for optimal haploid FG is to achieve sufficient signal strength, not to yield the sum of any specific subprocesses uniquely evoked by either pathway.
Physiological rationale for combining multiple partially derepressed pathways:
Although both Ste12 and Flo8 are necessary for optimal invasive growth, our results suggest that neither is maximally active. What advantages might there be in using two partially active pathways instead of one fully active one?
A common interpretation for such a situation is that requiring two pathways permits the integration of upstream signals. The extent to which that occurs in this particular system is currently unclear, as it has not been shown that haploid invasive growth requires simultaneous stimulation by multiple independently sensed stimuli. In any case, the existence of a shared factor, Ras2, upstream of both the MAPK and PKA pathways (Mösch et al. 1999) suggests that signal integration could in principle occur at that node without the necessity for rebranching downstream of Ras2 and reintegrating at target gene promoters.
A different advantage of using two partially active pathways is that it avoids strong activation of either pathway, which may lead to negative side effects due to processes for which there are unique consequences. For example, as revealed by the properties of dig1Δ dig2Δ cells, partially unregulated Ste12 activity leads to cell elongation, greater cell clumping, grossly misshapen morphology, and extraordinary flocculence, which may be quite deleterious in the wild. Removal of all of the negative controls known to impinge directly on Ste12 (kss1Δ dig1Δ dig2Δ cells) resulted in a pronounced growth defect that was completely rescued by also eliminating Ste12. Thus, full derepression of Ste12 in response to any signal would presumably negatively impact cell proliferation. In fact, the need to hold Ste12 in check may explain why it is subject to at least three different negative regulators and cannot be fully derepressed under nutrient-limiting conditions if the growth needed for FG is going to ensue, which takes place over the course of many hours, and even days. Indeed, and in contrast, upon exposure to mating pheromone, haploid cells undergo an acute cell cycle arrest on the 20- to 30-min time scale that is due, in part, to Ste12-dependent gene regulation (Dolan 1996). Thus, costimulatory input from the PKA pathway acting via Flo8 avoids the necessity of full-bore activation of Ste12 via the MAPK pathway. This use of partial activation via two distinct pathways with a shared function during the regulation of haploid invasive growth may reflect the necessity to avoid hyper-activation of certain pathway-specific side processes that may be counterproductive to the appropriate biological outcome.
If two discrete, largely parallel, signaling routes are activated by the same stimulus, but under somewhat different threshold conditions (in this case, at different levels as the glucose concentration falls), another potential advantage of such synergy is that it allows for a reasonable degree of homeostasis in biological response as the conditions change. Yet another potential advantage of controlling cell behavior via two parallel inputs—each operating at an intermediate level rather than via one pathway functioning at its maximum level—is that it may help the cell avoid irreversible commitment to a fate from which it may not be able to recover easily. For example, the ability to rapidly switch from filamentous to yeast-form growth and back, and/or from invasive to non-invasive growth and back, might provide distinct advantages to the cell in competition with other species or mutants that cannot so readily adapt. All of these regulatory strategies may also occur in the control of other developmental processes in more complex organisms.
Acknowledgments
We thank all members of the Thorner laboratory for advice and constructive suggestions and especially Michael McMurray and Françoise Roelants for their insights and critical reading of the manuscript. This work was supported by National Institutes of Health (NIH) Predoctoral Traineeship GM07232 (to R.E.C.) and by NIH Research GM21841 (to J.T.).
References
- Ashe, M. P., S. K. De Long and A. B. Sachs, 2000. Glucose depletion rapidly inhibits translation initiation in yeast. Mol. Biol. Cell 11 833–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banuett, F., 1998. Signalling in the yeasts: an informational cascade with links to the filamentous fungi. Microbiol. Mol. Biol. Rev. 62 249–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao, M. Z., M. A. Schwartz, G. T. Cantin, J. R. Yates, III, and H. D. Madhani, 2004. Pheromone-dependent destruction of the Tec1 transcription factor is required for MAP kinase signaling specificity in yeast. Cell 119 991–1000. [DOI] [PubMed] [Google Scholar]
- Bardwell, L., J. G. Cook, D. Voora, D. M. Baggott, A. R. Martinez et al., 1998. a Repression of yeast Ste12 transcription factor by direct binding of unphosphorylated Kss1 MAPK and its regulation by the Ste7 MEK. Genes Dev. 12 2887–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell, L., J. G. Cook, J. X. Zhu-Shimoni, D. Voora and J. Thorner, 1998. b Differential regulation of transcription: repression by unactivated mitogen-activated protein kinase Kss1 requires the Dig1 and Dig2 proteins. Proc. Natl. Acad. Sci. USA 95 15400–15405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkreutz, A., L. Boucher, B. J. Breitkreutz, M. Sultan, I. Jurisica et al., 2003. Phenotypic and transcriptional plasticity directed by a yeast mitogen-activated protein kinase network. Genetics 165 997–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno, K. S., R. Aramayo, P. F. Minke, R. L. Metzenberg and M. Plamann, 1996. Loss of growth polarity and mislocalization of septa in a Neurospora mutant altered in the regulatory subunit of cAMP-dependent protein kinase. EMBO J. 15 5772–5782. [PMC free article] [PubMed] [Google Scholar]
- Burke, D., D. Dawson and T. Stearns, 2000. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Chen, R. E., 2008. Function and regulation of mitogen-activated protein kinases in the yeast Saccharomyces cerevisiae. Ph.D. Thesis, University of California, Berkeley.
- Chen, R. E., and J. Thorner, 2007. Function and regulation in MAPK signaling pathways: lessons learned from the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1773 1311–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkasova, V. A., R. McCully, Y. Wang, A. Hinnebusch and E. A. Elion, 2003. A novel functional link between MAP kinase cascades and the Ras/cAMP pathway that regulates survival. Curr. Biol. 13 1220–1226. [DOI] [PubMed] [Google Scholar]
- Chou, S., L. Huang and H. Liu, 2004. Fus3-regulated Tec1 degradation through SCFCdc4 determines MAPK signaling specificity during mating in yeast. Cell 119 981–990. [DOI] [PubMed] [Google Scholar]
- Chou, S., S. Lane and H. Liu, 2006. Regulation of mating and filamentation genes by two distinct Ste12 complexes in Saccharomyces cerevisiae. Mol. Cell. Biol. 26 4794–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, J. G., L. Bardwell, S. J. Kron and J. Thorner, 1996. Two novel targets of the MAP kinase Kss1 are negative regulators of invasive growth in the yeast Saccharomyces cerevisiae. Genes Dev. 10 2831–2848. [DOI] [PubMed] [Google Scholar]
- Cook, J. G., L. Bardwell and J. Thorner, 1997. Inhibitory and activating functions for MAPK Kss1 in the S. cerevisiae filamentous-growth signalling pathway. Nature 390 85–88. [DOI] [PubMed] [Google Scholar]
- Cullen, P. J., and G. F. Sprague, Jr., 2000. Glucose depletion causes haploid invasive growth in yeast. Proc. Natl. Acad. Sci. USA 97 13619–13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan, J. W., 1996. Novel aspects of pheromone-induced cell-cycle arrest in yeast. Curr. Genet. 30 469–475. [DOI] [PubMed] [Google Scholar]
- Gagiano, M., F. F. Bauer and I. S. Pretorius, 2002. The sensing of nutritional status and the relationship to filamentous growth in Saccharomyces cerevisiae. FEMS Yeast Res. 2 433–470. [DOI] [PubMed] [Google Scholar]
- Galitski, T., A. J. Saldanha, C. A. Styles, E. S. Lander and G. R. Fink, 1999. Ploidy regulation of gene expression. Science 285 251–254. [DOI] [PubMed] [Google Scholar]
- Gancedo, J. M., 2001. Control of pseudohyphae formation in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25 107–123. [DOI] [PubMed] [Google Scholar]
- Garrett, S., and J. Broach, 1989. Loss of Ras activity in Saccharomyces cerevisiae is suppressed by disruptions of a new kinase gene, YAKI, whose product may act downstream of the cAMP-dependent protein kinase. Genes Dev. 3 1336–1348. [DOI] [PubMed] [Google Scholar]
- Gietz, R. D., and A. Sugino, 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74 527–534. [DOI] [PubMed] [Google Scholar]
- Gilbert, W. V., K. Zhou, T. K. Butler and J. A. Doudna, 2007. Cap-independent translation is required for starvation-induced differentiation in yeast. Science 317 1224–1227. [DOI] [PubMed] [Google Scholar]
- Granek, J. A., and P. M. Magwene, 2010. Environmental and genetic determinants of colony morphology in yeast. PLoS Genet. 6 e1000823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie, D. G., D. Carling and M. Carlson, 1998. The AMP-activated/SNF1 protein kinase subfamily: Metabolic sensors of the eukaryotic cell? Annu. Rev. Biochem. 67 821–855. [DOI] [PubMed] [Google Scholar]
- Hedbacker, K., and M. Carlson, 2008. SNF1/AMPK pathways in yeast. Front. Biosci. 13 2408–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoi, J. W., and B. Dumas, 2010. STE12 and STE12-like proteins: fungal transcription factors regulating development and pathogenicity. Eukaryot. Cell 9 480–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klosterman, S. J., M. H. Perlin, M. Garcia-Pedrajas, S. F. Covert and S. E. Gold, 2007. Genetics of morphogenesis and pathogenic development of Ustilago maydis. Adv. Genet. 57 1–47. [DOI] [PubMed] [Google Scholar]
- Kuchin, S., V. K. Vyas and M. Carlson, 2002. Snf1 protein kinase and the repressors Nrg1 and Nrg2 regulate FLO11, haploid invasive growth, and diploid pseudohyphal differentiation. Mol. Cell. Biol. 22 3994–4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengeler, K. B., R. C. Davidson, C. D'Souza, T. Harashima, W. C. Shen et al., 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64 746–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani, H. D., and G. R. Fink, 1997. Combinatorial control required for the specificity of yeast MAPK signaling. Science 275 1314–1317. [DOI] [PubMed] [Google Scholar]
- Madhani, H. D., C. A. Styles and G. R. Fink, 1997. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell 91 673–684. [DOI] [PubMed] [Google Scholar]
- Martinez-Espinoza, A. D., J. Ruiz-Herrera, C. G. Leon-Ramirez and S. E. Gold, 2004. MAP kinase and cAMP signaling pathways modulate the pH-induced yeast-to-mycelium dimorphic transition in the corn smut fungus Ustilago maydis. Curr. Microbiol. 49 274–281. [DOI] [PubMed] [Google Scholar]
- Mösch, H. U., R. L. Roberts and G. R. Fink, 1996. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93 5352–5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mösch, H. U., E. Kubler, S. Krappmann, G. R. Fink and G. H. Braus, 1999. Crosstalk between the Ras2p-controlled mitogen-activated protein kinase and cAMP pathways during invasive growth of Saccharomyces cerevisiae. Mol. Biol. Cell 10 1325–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obara, Y., A. M. Horgan and P. J. Stork, 2007. The requirement of Ras and Rap1 for the activation of ERKs by cAMP, PACAP, and KCl in cerebellar granule cells. J. Neurochem. 101 470–482. [DOI] [PubMed] [Google Scholar]
- Olson, K. A., C. Nelson, G. Tai, W. Hung, C. Yong et al., 2000. Two regulators of Ste12p inhibit pheromone-responsive transcription by separate mechanisms. Mol. Cell. Biol. 20 4199–4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palecek, S. P., A. S. Parikh and S. J. Kron, 2002. Sensing, signalling and integrating physical processes during Saccharomyces cerevisiae invasive and filamentous growth. Microbiology 148 893–907. [DOI] [PubMed] [Google Scholar]
- Pan, X., and J. Heitman, 1999. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol. Cell. Biol. 19 4874–4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, X., and J. Heitman, 2002. Protein kinase A operates a molecular switch that governs yeast pseudohyphal differentiation. Mol. Cell. Biol. 22 3981–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, X., T. Harashima and J. Heitman, 2000. Signal transduction cascades regulating pseudohyphal differentiation of Saccharomyces cerevisiae. Curr. Opin. Microbiol. 3 567–572. [DOI] [PubMed] [Google Scholar]
- Pearson, R. B., and B. E. Kemp, 1991. Protein kinase phosphorylation site sequences and consensus specificity motifs: tabulations. Methods Enzymol. 200 62–81. [DOI] [PubMed] [Google Scholar]
- Rispail, N., D. M. Soanes, C. Ant, R. Czajkowski, A. Grunler et al., 2009. Comparative genomics of MAP kinase and calcium-calcineurin signalling components in plant and human pathogenic fungi. Fungal Genet. Biol. 46 287–298. [DOI] [PubMed] [Google Scholar]
- Roberts, R. L., and G. R. Fink, 1994. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 8 2974–2985. [DOI] [PubMed] [Google Scholar]
- Robertson, L. S., and G. R. Fink, 1998. The three yeast A kinases have specific signaling functions in pseudohyphal growth. Proc. Natl. Acad. Sci. USA 95 13783–13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp, S., E. Summers, H. J. Lo, H. Madhani and G. Fink, 1999. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J. 18 1257–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena, M., S. Williams, K. Tasken and T. Mustelin, 1999. Crosstalk between cAMP-dependent kinase and MAP kinase through a protein tyrosine phosphatase. Nat. Cell Biol. 1 305–311. [DOI] [PubMed] [Google Scholar]
- Schneper, L., K. Duvel and J. R. Broach, 2004. Sense and sensibility: nutritional response and signal integration in yeast. Curr. Opin. Microbiol. 7 624–630. [DOI] [PubMed] [Google Scholar]
- Taxis, C., and M. Knop, 2006. System of centromeric, episomal, and integrative vectors based on drug resistance markers for Saccharomyces cerevisiae. Biotechniques 40 73–78. [DOI] [PubMed] [Google Scholar]
- Tedford, K., S. Kim, D. Sa, K. Stevens and M. Tyers, 1997. Regulation of the mating pheromone and invasive growth responses in yeast by two MAP kinase substrates. Curr. Biol. 7 228–238. [DOI] [PubMed] [Google Scholar]
- Toda, T., I. Uno, T. Ishikawa, S. Powers, T. Kataoka et al., 1985. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell 40 27–36. [DOI] [PubMed] [Google Scholar]
- Toda, T., S. Cameron, P. Sass, M. Zoller and M. Wigler, 1987. Three different genes in S. cerevisiae encode the catalytic subunits of the cAMP-dependent protein kinase. Cell 50 277–287. [DOI] [PubMed] [Google Scholar]
- Truckses, D. M., J. E. Bloomekatz and J. Thorner, 2006. The RA domain of Ste50 adaptor protein is required for delivery of Ste11 to the plasma membrane in the filamentous growth signaling pathway of the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 26 912–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truckses, D. M., L. S. Garrenton and J. Thorner, 2004. Jekyll and Hyde in the microbial world. Science 306 1509–1511. [DOI] [PubMed] [Google Scholar]
- Uesono, Y., M. P. Ashe and E. A. Toh, 2004. Simultaneous yet independent regulation of actin cytoskeletal organization and translation initiation by glucose in Saccharomyces cerevisiae. Mol. Biol. Cell 15 1544–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstrepen, K. J., and F. M. Klis, 2006. Flocculation, adhesion and biofilm formation in yeasts. Mol. Microbiol. 60 5–15. [DOI] [PubMed] [Google Scholar]
- Vyas, V. K., S. Kuchin and M. Carlson, 2001. Interaction of the repressors Nrg1 and Nrg2 with the Snf1 protein kinase in Saccharomyces cerevisiae. Genetics 158 563–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas, V. K., S. Kuchin, C. D. Berkey and M. Carlson, 2003. Snf1 kinases with different beta-subunit isoforms play distinct roles in regulating haploid invasive growth. Mol. Cell. Biol. 23 1341–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach, A., A. Brachat, R. Pohlmann and P. Philippsen, 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10 1793–1808. [DOI] [PubMed] [Google Scholar]
- Wach, A., A. Brachat, C. Alberti-Segui, C. Rebischung and P. Philippsen, 1997. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast 13 1065–1075. [DOI] [PubMed] [Google Scholar]
- Zappacosta, F., M. J. Huddleston, R. L. Karcher, V. I. Gelfand, S. A. Carr et al., 2002. Improved sensitivity for phosphopeptide mapping using capillary column HPLC and microionspray mass spectrometry: comparative phosphorylation site mapping from gel-derived proteins. Anal. Chem. 74 3221–3231. [DOI] [PubMed] [Google Scholar]
- Zeitlinger, J., I. Simon, C. T. Harbison, N. M. Hannett, T. L. Volkert et al., 2003. Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell 113 395–404. [DOI] [PubMed] [Google Scholar]