Abstract
LMP1 induces the expression of two members of the family of Id proteins, Id1 and Id3, and affects cell cycle regulation by decreasing the expression of the cyclin dependent kinase inhibitor, p27, and increasing levels and phosphorylation of cdk2 and Rb. In the present study, the contribution of the Id proteins to LMP1-mediated transformation was determined. Although LMP1 effectively inhibited p27 expression, the Id proteins alone did not affect expression of p27, cdk2, and Rb. Neither Id1 nor Id3 was sufficient to transform Rat-1 cells and inhibition of Id1 expression did not affect LMP1-induced morphologic transformation of Rat-1 cells or reduction of p27. However, reduced Id expression resulted in smaller foci and impaired the growth rate of Rat-1 cells. These data indicate that overexpression of the Id proteins is not sufficient for the effects of LMP1 on the cell cycle but that inhibition of Id expression does affect the growth of LMP1-transformed and parental Rat1 cells.
Keywords: EBV, LMP1, Id1, p27, Transformation
Introduction
Epstein–Barr virus (EBV) is a DNA tumor virus that efficiently transforms B-lymphocytes in vitro and is associated with a number of epithelial and lymphoid malignancies (Kieff, 2007; Raab-Traub, 2002; Young et al., 1989; Young and Rickinson, 2004). Latently infected B-cells initially express a number of EBV proteins and factors important to ensure the survival of the latently infected cell. The latently infected cells express different subsets of latent proteins and these gene expression patterns are characteristic of different EBV-associated disease states. In nasopharyngeal carcinoma (NPC) and some types of Hodgkin disease, latent membrane protein 1 (LMP1), latent membrane protein 2 (LMP2A, LMP2B), EBV nuclear antigen 1 (EBNA1), and the EBER and BamHI-A transcripts are expressed (Kieff, 2007).
Many of the EBV proteins expressed during latency have profound effects on gene regulation, control cell growth, and several key pathways that are affected have been identified. The nuclear proteins, EBNA2, EBNA3A, B, and C impinge on Notch signaling (Grossman et al., 1994; Henkel et al., 1994; Robertson et al., 1996a,b) and LMP2A inhibits B-cell receptor signaling (Miller et al., 1995, 1994, 1993) and activates phosphatidylinositol 3-kinase (PI3K) (Morrison et al., 2003; Scholle et al., 2000; Swart et al., 2000). LMP1 is a constitutively active member of the tumor necrosis factor receptor family and activates NFκB, PI3K, mitogen-activated protein kinases (MAPK), and c-Jun N-terminal kinase (JNK) signaling (Dawson et al., 2000; Eliopoulos and Young, 1998; Izumi and Kieff, 1997; Mainou et al., 2005; Miller et al., 1997; Paine et al., 1995; Roberts and Cooper, 1998; Thornburg et al., 2006). Recently viral microRNAs have been identified as being processed from the BamHI-A transcripts (Cai et al., 2006).
LMP1 is expressed in most cancers associated with EBVand is considered the EBV oncogene as it can transform rodent fibroblasts by conferring anchorage-independent growth and loss of contact inhibition (Baichwal and Sugden, 1988; Everly et al., 2004; Kaye et al., 1993; Wang et al., 1985). Fibroblasts expressing LMP1 form tumors in nude mice and can grow in reduced serum conditions (Wang et al., 1985). The LMP1 protein has a short cytoplasmic amino-terminus and a six-pass integral membrane domain. The transmembrane domain mediates ligand-independent self-association (Devergne et al., 1996; Franken et al., 1996). Oligomerization in the membrane initiates signaling from two carboxyl-terminal activating regions (CTARs), CTAR1 and CTAR2, by recruiting adapter molecules including tumor necrosis receptor associated factors (TRAFs). CTAR1 can bind TRAF1, 2, 3, or 5, while CTAR2 recruits TRAF2 and 6 through binding of other factors like tumor necrosis factor receptor associated death domain (TRADD) protein, RIP and BS69 (Devergne et al., 1996; Izumi et al., 1999, 1997; Izumi and Kieff, 1997; Miller et al., 1998; Mosialos et al., 1995; Wan et al., 2006). Signaling induced by LMP1 is analogous to constitutively activated tumor necrosis factor receptor (TNFR) signaling and LMP1 is constitutively present in the cholesterol-rich lipid raft domains (Ardila-Osorio et al., 2005; Higuchi et al., 2001; Pioche-Durieu et al., 2005; Yasui et al., 2004). Experiments using rodent fibroblasts have determined that LMP1 via CTAR1 induces two members of the family of Id proteins, Id1 and Id3, and regulates the cell cycle by down-regulating the expression of cyclin dependent kinase inhibitor, p27KIP, and increasing levels and phosphorylation of cdk2 and Rb (Everly et al., 2004).
The Id proteins are a family of proteins named both for their ability to inhibit DNA binding and inhibit differentiation (Sikder et al., 2003). There are four members of the family of Id proteins, Id1, Id2, Id3, and Id4. The Id proteins do not dimerize with each other but do contain a helix–loop–helix domain that allows them to heterodimerize and negatively regulate helix–loop–helix transcription factor family members (E-box proteins). Id proteins lack the basic DNA-binding domains present in the E-box proteins and are thought to act as dominant-negative transcription factors by altering the subnuclear localization of the E-box proteins, from chromatin-associated to freely diffusing (O’Toole et al., 2003). They are critical regulators of cell growth as they inhibit differentiation and promote cell cycle progression (Barone et al., 1994; Hara et al., 1994; Peverali et al., 1994). The expression of the Id proteins is deregulated and increased in many cancers and knockout mouse models containing deletions of the Id proteins have developmental defects (Perk et al., 2005). Important targets of the Id proteins are the cyclin dependent kinase inhibitors (cdki) that are critical regulators of the cell cycle. Expression of two cdkis is controlled in part by the repressive and activating activity of the Id and E-box proteins, respectively (Ohtani et al., 2001; Prabhu et al., 1997; Zheng et al., 2004). Id1 and several E-box proteins, E47 and Ets1/2, regulate the p16INK4 promoter (Ohtani et al., 2001; Zheng et al., 2004). Another E-box protein, E2A, regulates cdki p21WAF1 in conjunction with Id1 and controls cell growth (Prabhu et al., 1997).
In the present study, the contribution of the Id proteins to LMP1-mediated effects on the cell cycle proteins and transformation was determined. Although LMP1 effectively inhibited p27 expression, the Id proteins did not affect expression of p27 or other regulators of the cell cycle. Neither Id1 nor Id3 was sufficient to transform Rat-1 cells. Silencing Id1 expression did not block LMP1-mediated transformation, however, the growth of both control and LMP1 expressing Rat-1 cells was impaired. The data indicate that the Id proteins contribute to the growth of LMP1-transformed cells but are not required for the effects of LMP1 on cell cycle markers.
Results
LMP1 induces the expression of two members of the Id family of proteins, Id1 and Id3, in epithelial cells and during rodent fibroblast transformation (Everly et al., 2004). Id proteins have been shown to bind to helix–loop–helix transcription factor family members, inhibit differentiation, and promote proliferation (Barone et al., 1994; Hara et al., 1994; Peverali et al., 1994; Sikder et al., 2003). The overexpression of Id1 in primary epithelial cells delays senescence and in some cases leads to their immortalization (Alani et al., 1999; Nickoloff et al., 2000). In addition, Id proteins are often deregulated and overexpressed in many cancers (Perk et al., 2005). In this study, the contribution of the Id proteins to LMP1-mediated rodent fibroblast transformation was determined.
Id1 and Id3 are insufficient for transformation of Rat-1 cells
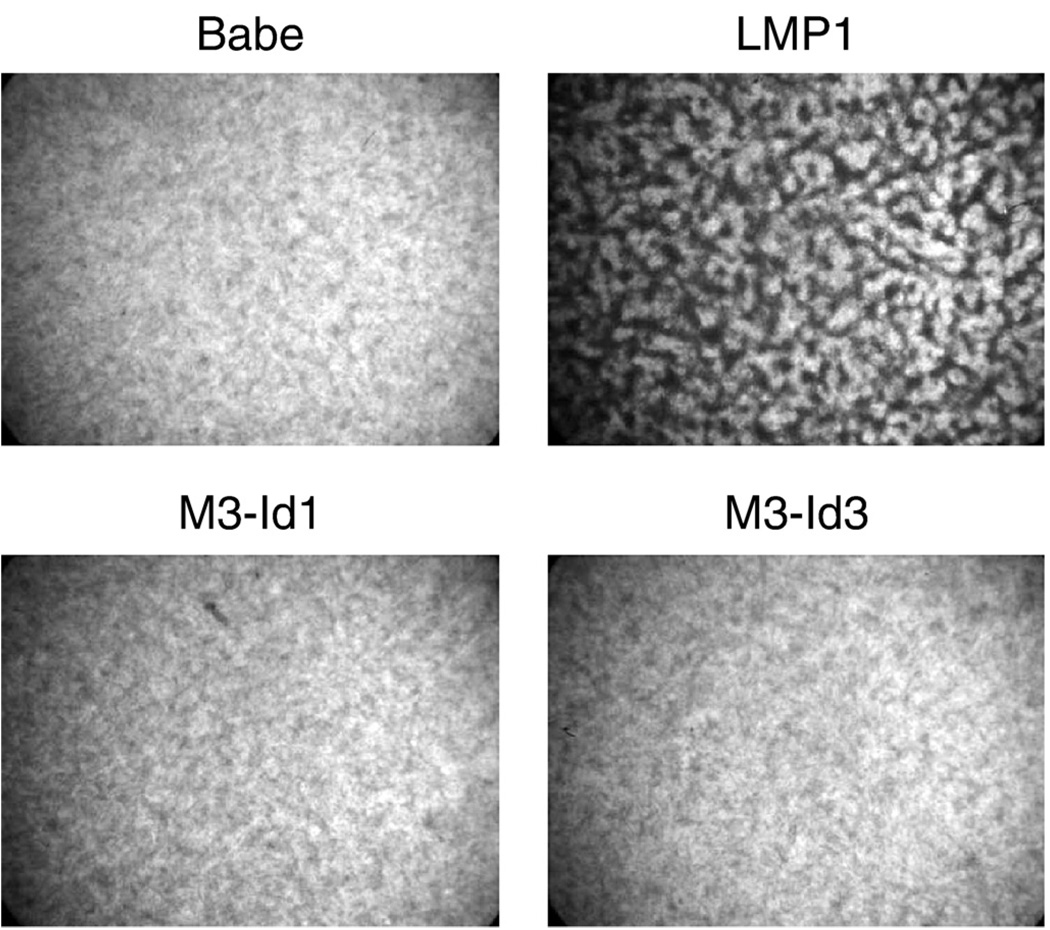
To determine if the Id proteins have transforming properties, Rat-1 cells were transduced with the vector control, pBabe, or retroviruses expressing LMP1, Id1, or Id3, and selected with puromycin for stable cell lines. Stable cell lines were plated and incubated at confluency for 10–14 days and tested for ability to overcome contact-inhibition of growth (Fig. 1). The pBabe control cells were contact inhibited and stopped growing once the monolayer reached confluency. In contrast, the LMP1 expressing cells were transformed, overcame contact-inhibition, and continued to grow. Neither the cells expressing Id1 nor the cells expressing Id3 were able to transform Rat-1 cells, as they were contact inhibited and did not continue to grow once the cells reached confluency (bottom panels). Expression of LMP1 and expression of myc-tagged Id proteins were confirmed by western blotting for LMP1 and the myc-epitope tag, respectively (Fig. 2). Detection of the tagged Id constructs was also confirmed using Id-specific antibodies and indicated that the levels of expression of the tagged Id proteins were similar to that induced by LMP1 (data not shown). The cell lines stably expressing Id1 or Id3 were also transduced with retrovirus expressing Id3 or Id1, respectively, and analyzed for focus formation. No foci were detected in either the Id1 stable cells with Id3 retrovirus or Id3 stable cells with Id1 retrovirus. These data indicate that the induced expression of the Id proteins is not sufficient to transform fibroblasts and that other properties of LMP1, possibly in concert with the induction of the Id proteins, are required for transformation.
Fig. 1.
Rat-1 transformation by LMP1, Id1, and Id3. Rat-1 cells were transduced with the control retrovirus, pBabe, or retrovirus expressing LMP1, or myc-tagged Id1 or Id3, M3-Id1 or M3-Id3, respectively. Stable cell lines were selected with puromycin, plated in 6-well plates, and allowed to grow for 7 to 10 days to assess effects on contact inhibition. Monolayers were fixed, stained with crystal violet, and photographed by light microscopy using a dissecting microscope. Representative monolayer fields are presented.
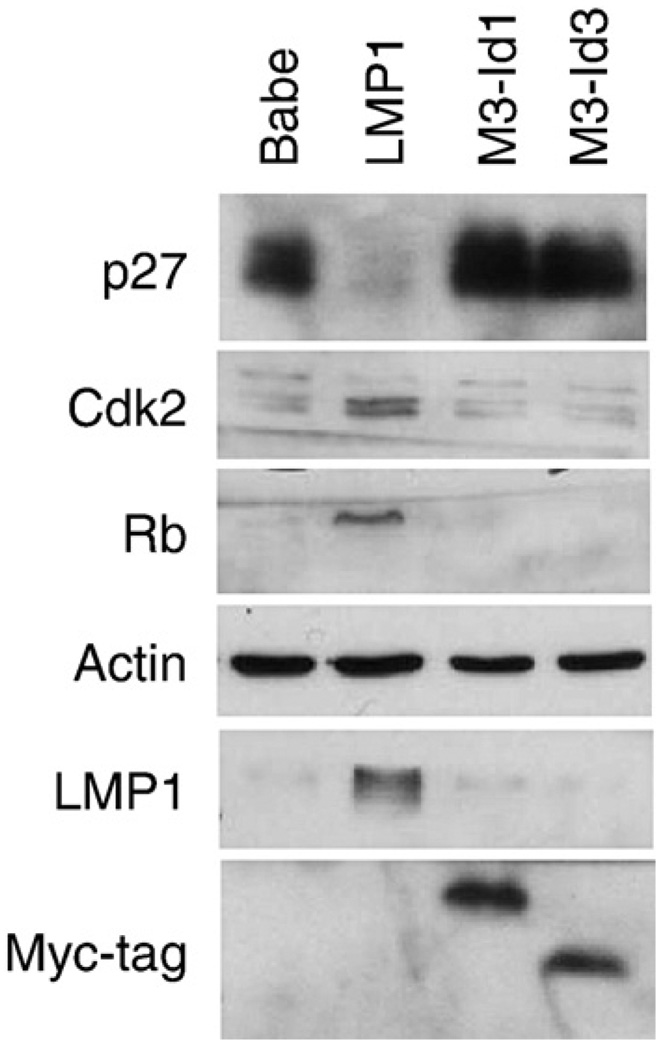
Fig. 2.
Regulation of cell cycle markers in Rat-1 stable cells. Rat-1 cell lines stably expressing LMP1, Id1, or Id3 were analyzed by western blotting. Total cell lysates from control (pBabe), LMP1, myc-tagged Id1 (M3-Id1), or myc-tagged Id3 (M3-Id3) were analyzed for levels of p27, Cdk2 and Rb protein. Actin was used as a loading control and expression of constructs was confirmed by blotting with LMP1-specific and myc-tag antibodies. Fifty micrograms of protein were loaded per lane.
Effects of Id proteins on cell cycle markers
Although the overexpression of the Id proteins was not sufficient to transform Rat-1 cells, the effects of the Id proteins on the cell cycle markers that are affected by LMP1 during transformation were determined. The Id proteins have been shown to regulate expression of the cyclin dependent kinase inhibitors, p16INK and p21WAF. Rat-1 fibroblasts do not express p16 or p21 and only express the p27 cdki enabling assessment of the effects of the Id proteins on p27. As previously shown, expression of the cyclin dependent kinase inhibitor, p27, was markedly decreased in the LMP1-expressing cells (Fig. 2). However, in the cell lines stably expressing Id1 or Id3 expression of p27 were not affected. Cell lines that stably expressed both Id1 and Id3 also failed to downregulate p27 (data not shown).
The effects of the Id proteins on other aspects of the cell cycle that are altered during LMP1-mediated transformation were also determined. In Rat-1 cells transformed with LMP1, the amount of cyclin dependent kinase 2 (cdk2) and hyperpho-sphorylated Rb and total Rb was increased as previously described (Fig. 2). However, in the cells expressing Id1 or Id3, the amounts of p27, cdk2, and Rb were not affected (Fig. 2). These findings indicate that the cell cycle proteins that are affected by LMP1 during transformation are not affected by overexpression of the Id proteins in the absence of LMP1.
LMP1 regulates p27KIP transcription
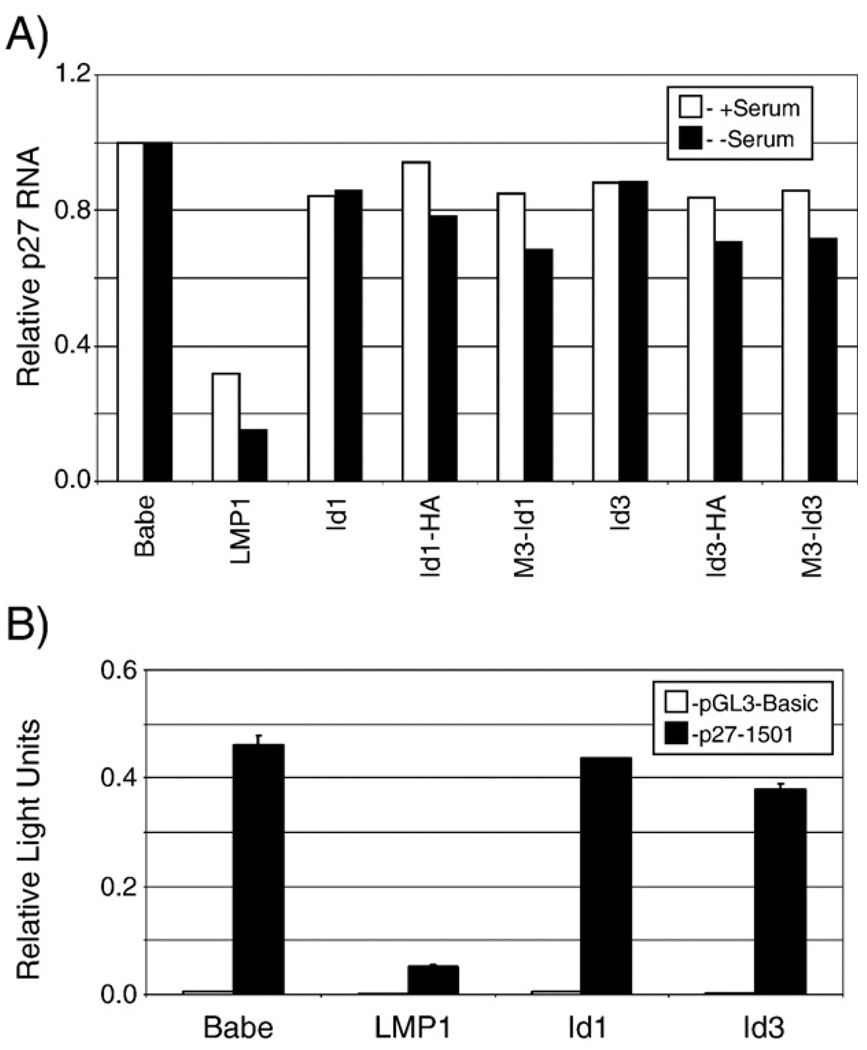
Both transcriptional and post-transcriptional regulation of p27 has been described (Auerkari, 2006; Chandramohan et al., 2004). To determine if LMP1 or the Id proteins affected p27 transcription, Rnase protection assays (RPA) were performed using extracts from the stable cell lines expressing LMP1 or the Id proteins. The Id proteins are serum responsive proteins, therefore RNA was harvested from cells in full serum or after serum-starving (0.2%) for 16 h. The RNA was annealed with rat p27 probe, digested, and resolved on polyacrylamide gels. Protected p27 fragments were quantitated using a phosphorimager and plotted relative to GAPDH RNA levels (Fig. 3, panel A). In the presence or absence of serum, LMP1 decreased p27 RNA levels approximately 70–80%, similarly to its effects on the protein levels (Fig. 2). The Id proteins did not affect p27 RNA levels.
Fig. 3.
Regulation of p27KIP transcription. Regulation of the p27KIP promoter was determined by ribonuclease protection assays (A) and promoter reporter assays (B). Ribonuclease protection assays (A) were performed using total RNA from stable cell lines. Fragments protected by rat p27KIP RNA probe relative to control (GAPDH) RNA probe were quantitated using a Phosphorimager and p27 expression is depicted relative to control (Babe) stables set arbitrarily as one. Stable cell lines included cells expressing LMP1, untagged Id proteins, Id1 and Id3, C-terminally HA-tagged Id proteins, Id1-HA and Id3-HA, and N-terminally triple myc-tagged Id proteins, M3-Id1 and M3-Id3. The experiment was performed three times and a representative experiment is presented. Promoter reporter assays (B) were performed by transfection of 293T cells with control pRL-SV40, pGL3-Basic (empty reporter) or p27-1501 (containing 1500 base pairs of the Rattus novegicus p27KIP promoter) reporter plasmids, and vector control (Babe) or plasmids expressing LMP1, or untagged Id1 or Id3. Forty hours post-transfection cells were harvested, and dual-luciferase assays were performed. Relative luciferase activity was determined by the firefly luciferase activity of the reporter constructs relative to the control renilla luciferase activity. Error bars represent the standard deviation from the mean in triplicate wells.
To ensure that the failure to downregulate p27 was not the result of the triple myc-tag at the N-terminus, the Id proteins were subcloned with a carboxyl-terminal HA tag or untagged. Neither the N-terminally myc-tagged nor C-terminally HA-tagged versions of the Id proteins affected p27 RNA levels in either high or low serum (Fig. 3). In the cell lines stably expressing untagged Id1 and Id3, p27 RNA levels were also approximately equivalent or slightly higher than the control cell levels. Expression of the differentially tagged Id proteins in stable cell lines was confirmed by western blotting. The expression levels of the tagged Id proteins in the stable lines were also assessed using antibodies to Id1 and Id3. These assays revealed that expression of Id1 and Id3 was approximately two to three times greater than control cell lines and was comparable to the levels induced by LMP1 ((Everly et al., 2004) and data not shown).
To assess the ability of LMP1 or the Id proteins to regulate the p27KIP promoter, the control region (1500 base pairs) of the p27KIP promoter was cloned into a promoter reporter constructs for reporter assays. 293T cells were cotransfected with the empty reporter vector (pGL3-Basic) or p27 reporter (p27-1501), transfection control renilla luciferase, and effector plasmids, encoding LMP1, Id1 or Id3. p27 promoter activity was plotted relative to the internal control renilla luciferase activity (Fig. 3B). LMP1 decreased the activity of the p27 promoter reporter plasmid indicating that LMP1 regulates the p27KIP1 promoter. However, confirming the western blotting and RPA assays, the Id proteins did not negatively regulate the p27 promoter reporter. These findings indicate that LMP1 negatively regulates p27 expression transcriptionally but that the expression of Id1 or Id3, in the absence of LMP1, is not sufficient for this effect.
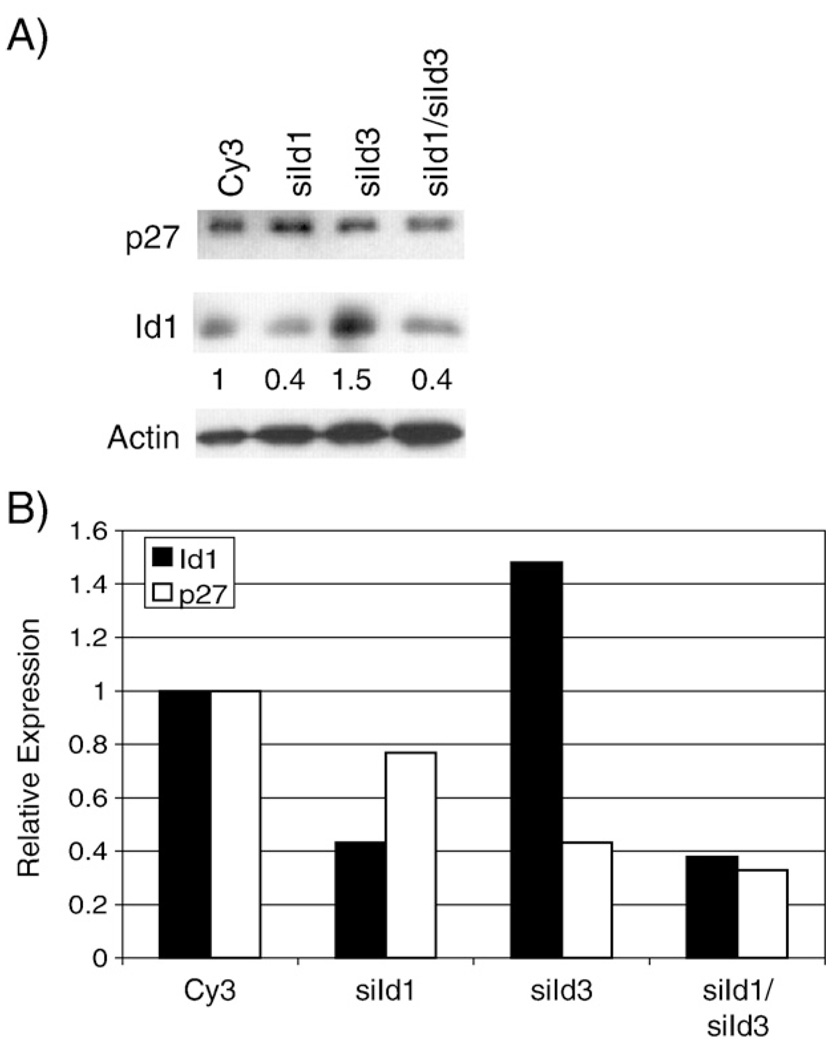
To determine if inhibition of Id expression affects the ability of LMP1 to decrease p27 expression, siRNA for a negative control siRNA (Cy3), Id1, Id3, or for both Id1 and Id3 was transfected into Rat1 cells which were then transduced with LMP1. Expression of Id1 was assessed using an Id1 specific antibody (Fig. 4A). The levels of expression were normalized to actin and quantitated (Fig. 4B). The Id1 siRNA reduced Id1 expression approximately 60%. The effect of the Id3 siRNA could not be determined due to the poor quality of the Id3 antibody. As shown, in some experiments, the Id3 siRNA increased expression of Id1, however, this unexpected effect did indicate successful transfection of the Id3 siRNA. When both Id1 and Id3 siRNAs were transfected, the level of Id1 was again reduced by 60%. Despite successful transfection of the siRNAs and effective reduction of Id1 expression, the levels of p27 did not increase as anticipated but rather were further decreased (Fig. 4B). These data suggest that the LMP1-induced decrease of p27 is not mediated by its ability to increase Id1 and Id3.
Fig. 4.
Effects of siId1 and siId3 on LMP1-mediated decrease of p27. Rat-1 cells were transfected with negative control siRNA (Cy3), siId1, siId3, or siId1 and siId3 and transduced with LMP1-expressing retrovirus, HSCG-LMP1. Expression of Id1 and p27 was determined by immunoblotting (panel A). Expression of Id1 and p27 was quantitated relative to actin expression (numbers below Id1 blot in panel A and black and white bars, respectively, in panel B) using ImageJ densitometry software.
Inhibition of Id1 expression in Rat-1 cells
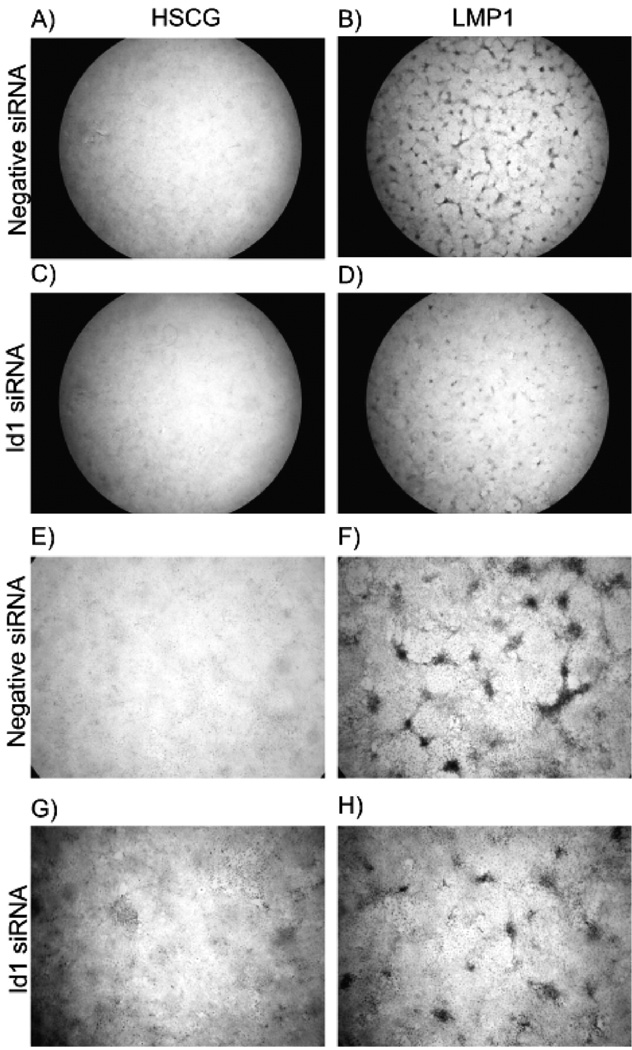
To assess the possible contribution of Id expression to LMP1-mediated transformation, expression of Id1 was inhibited with siRNA and transformation assays were performed. Rat-1 cells were transiently transfected with Cy3-labeled negative control or Id1 siRNA and then transduced with GFP-expressing retroviruses, pHSCG (control) or pHSCG-LMP1. The transfected and transduced cells were then allowed to grow for 7 days to assess transformation. Monolayers were examined at the end of the experiment by phase contrast and fluorescence microscopy to analyze transfection and transduction. Efficiency of siRNA transfection was assessed by fluorescence of the cells transfected with the Cy3-labelled control siRNA. Greater than 95% of the cells were transfected indicating efficient transfection of siRNAs (data not shown). Co-expression of green fluorescent protein was used as a control for retrovirus transduction efficiency and differences were not detected in the numbers of green cells between control and LMP1 expressing retrovirus in the presence or absence of Id1 siRNA. Monolayers were stained with crystal violet and examined for focus formation (Fig. 5). In the presence of the nonspecific siRNA, LMP1 transformed Rat-1 cells and formed foci (Fig. 5B and F). However in the presence of Id1 siRNA, the foci were fewer and smaller (Fig. 5D and H). As shown in Fig. 4, Id1 was effectively targeted by the silencing sequence.
Fig. 5.
Transient silencing of Id1 in transformation assays. Rat-1 cells were transfected with negative control (panels A, B, E, F) or Id1 siRNA (panels C, D, G, and H) in twelve-well plates. The following day cells were transduced with control HSCG retrovirus (panels A, C, E, G), or LMP1-expressing retrovirus, HSCG-LMP1 (panels B, D, F, H) and focus formation assays were performed for 7 days. Monolayers were fixed, stained with crystal violet, and examined for focus formation by light microscopy using a dissecting microscope. Representative micrographs are depicted at 10X (panels A, B, C, D) or 40X (panels E, F, G, H).
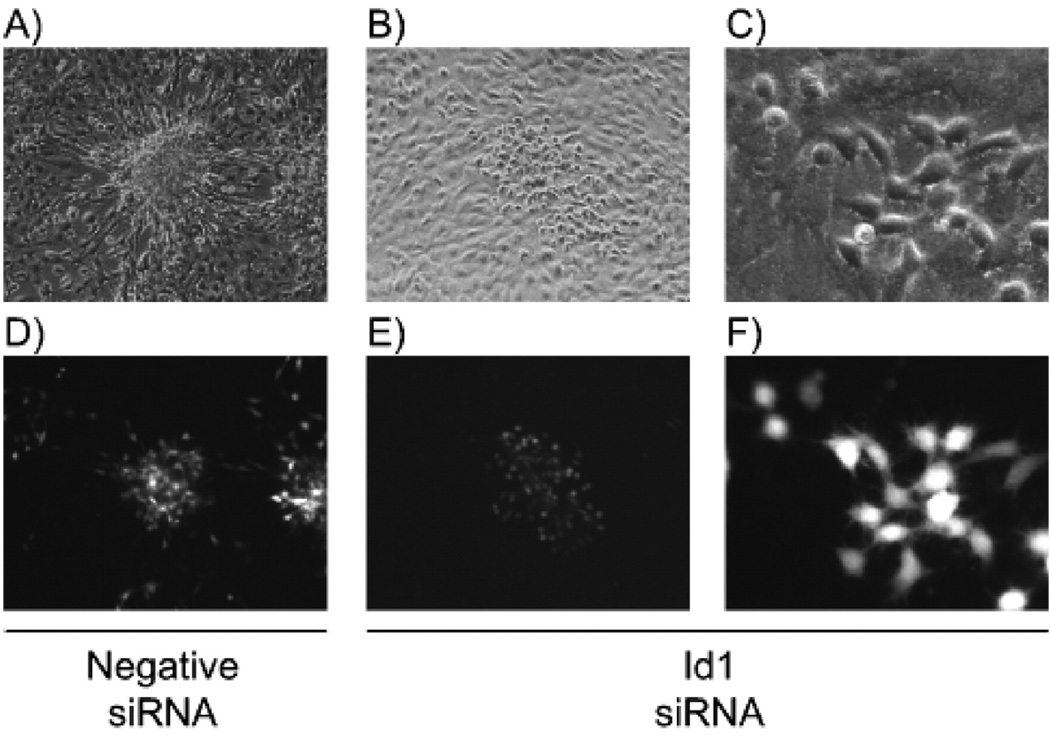
Fluorescence microscopy (Fig. 6) revealed that in the cells transfected with control siRNA, LMP1 induced brightly green and transformed foci (Fig. 6A and D, respectively). In the cells transfected with siRNA to Id1, patches of green LMP1 transduced cells were detected. The cells were not as large as the control LMP1 expressing cells but appeared morphologically different than the surrounding cells (Fig. 6B and E, respectively). At high power the siId1 transfected cells that were transduced with LMP1 (green) were rounded and refractile, suggestive of transformation (Fig. 6C and F, respectively). The cells adjacent to the foci were normal and control cells expressing only GFP had normal morphology identical to surrounding untransduced cells. These observations indicate that the siId1 RNA was not toxic. The smaller foci observed in the Id1 siRNA transfected cells could be due to decreased silencing after 7 days, which is considered to be the limit of the efficacy of transfected siRNA, or that inhibition of Id1 expression slowed cell growth but did not completely block transformation.
Fig. 6.
Morphology of LMP1 transformed cells. Rat-1 cells were transfected with negative control (panels A and D) or Id1 siRNA (panels B, C, E, and F) in twelve-well plates. The following day cells were transduced with LMP1-expressing retrovirus and focus formation assays were performed for 7 days. Monolayers were examined by phase contrast (panels A–C) and fluorescence microscopy (panels D–F) for focus formation at 10× low power (A, B, D, and E) and 40× high power (C and F). Representative micrographs are depicted.
Effects of stably silencing Id1 expression in LMP1 transformed cells
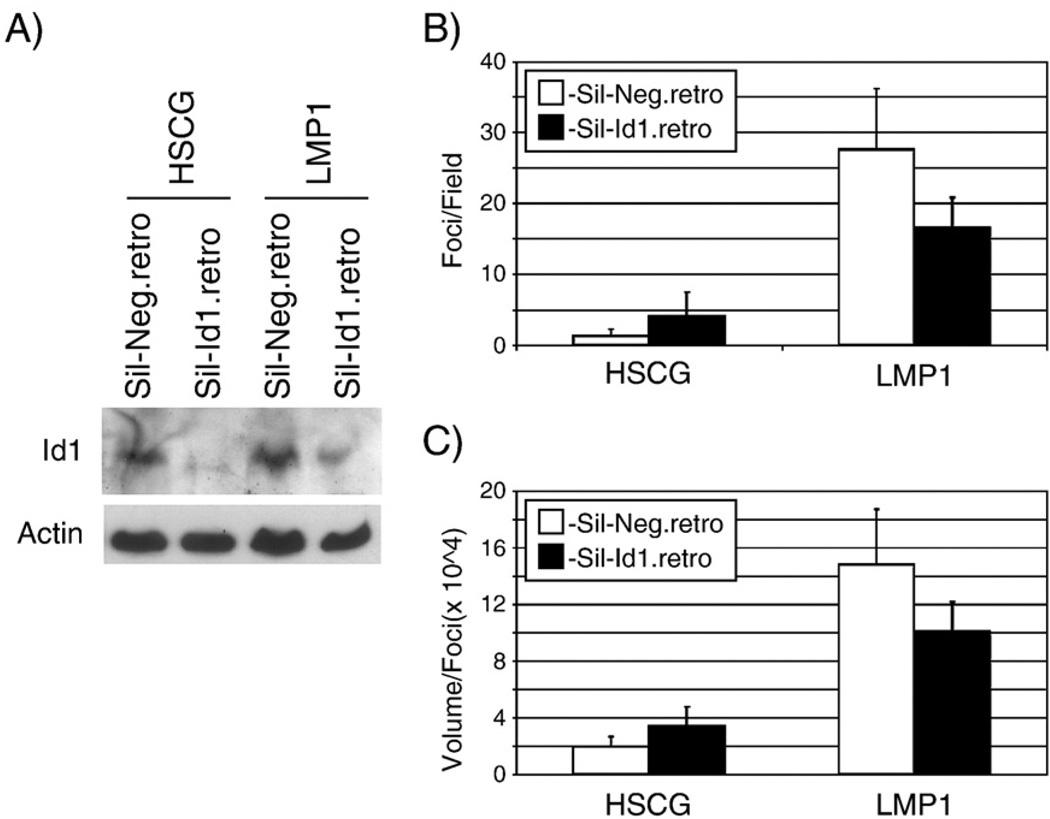
To ensure that Id1 expression was effectively decreased throughout the 7- to 10-day transformation assay, the siRNA Id1 targeting sequence and a control irrelevant sequence were cloned into a retrovirus. Rat-1 cells were transduced with control and Id1 siRNA expressing retroviruses and stable cell lines were selected with puromycin. Cells were then transduced with HSCG or HSCG-LMP1 retrovirus and cells were tested in transformation assays for focus formation and colony formation. In parallel, cells were harvested to confirm knockdown of Id1. In cells transduced with HSCG or HSCG-LMP1, the cells expressing the Id1 siRNA had lower Id1 protein expression, confirming knockdown of Id1 expression (Fig. 7A). The effects on LMP1 induced transformation in cells stably expressing Id1 siRNA were similar to those observed with the transiently transfected siRNA with the formation of fewer and smaller foci (Fig. 5). The number and size of the crystal violet stained foci were quantitated using ImageQuant software in five separate fields for each condition and plotted (Fig. 7B and C, respectively). The cell lines expressing the Id1 siRNA reduced the number of the foci by approximately 50% and size of the foci by about a third in the presence of LMP1. The finding that foci were produced despite decreased Id1 expression suggests that in the absence of Id1 LMP1 is still able to transform cells. However, the decreased number and size of foci suggests that LMP1 induction of Id1 contributes to cell growth.
Fig. 7.
Effects of decreased Id1 on focus formation. Rat-1 stable cell lines were analyzed by western blotting for Id1 expression (panel A). Total cell lysates from cell lines stably expressing control siRNA (Sil-Neg.retro) or Id1 siRNA (Sil-Id1.retro) and transduced with control (HSCG) or LMP1-expressing retrovirus were analyzed for Id1 levels compared to Actin loading control. Fifty micrograms of protein were loaded per lane. Doubly transduced cells were also seeded in 12-well plates for focus formation assays for 10 days. Monolayers were fixed and stained with crystal violet and five separate fields were photographed per condition using a dissecting microscope. The foci were quantitated using ImageQuant software and the mean number and size of foci were plotted (panels B and C, respectively). Error bars represent the standard deviation in the five separate fields.
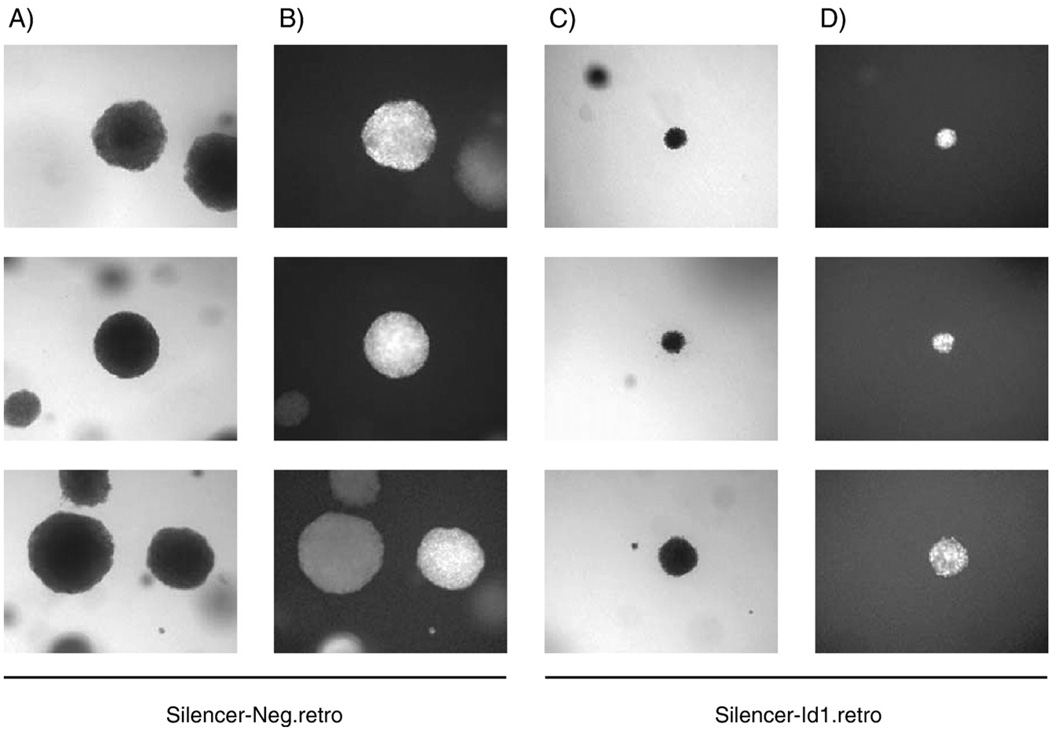
To further assess effects on transformation, the cell lines with decreased Id1 expression were tested for their ability to grow in an anchorage-independent fashion and form colonies in soft agar. Equal numbers of siRNA expressing cells were transduced with control and LMP1 expressing retrovirus overnight and then an equal number of cells were seeded into soft agar and maintained under puromycin selection to maintain siRNA expression, for ten to 14 days. Cells were examined by phase contrast and fluorescence microscopy for colony formation. Cells stably expressing the siRNAs that were transduced with control retrovirus, HSCG, did not form colonies in soft agar (data not shown). However, cell lines expressing either nonspecific or Id1 siRNA with LMP1 were transformed and grew in soft agar (Fig. 8 columns A and C, respectively). The cells were fluorescent green indicating that they were effectively transduced with the LMP1 expressing retrovirus (Fig. 8 columns B and D). Similarly to the focus formation assays, the colonies expressing Id1 siRNA were smaller compared to cells expressing the negative control siRNA. These data confirm that Id1 silencing does not inhibit LMP1-induced transformation to anchorage-independence. However, the slower growth of the cells expressing Id1 siRNA suggests that the Id proteins contribute to enhanced cell growth.
Fig. 8.
Effects of reduced Id1 expression in soft agar assays. Equal numbers of Rat-1 stable cell lines expressing control siRNA (Sil-Neg.retro, columns A and B) or Id1 siRNA (Sil-Id1.retro, columns C and D) and transduced with LMP1-expressing retrovirus were seeded into soft agar assays and allowed to grow for 10 to 14 days. Wells were examined by phase contrast (columns A and C) and fluorescence microscopy (columns B and D) for colony formation at 10×low power. Randomly selected fields containing one or more colonies are depicted.
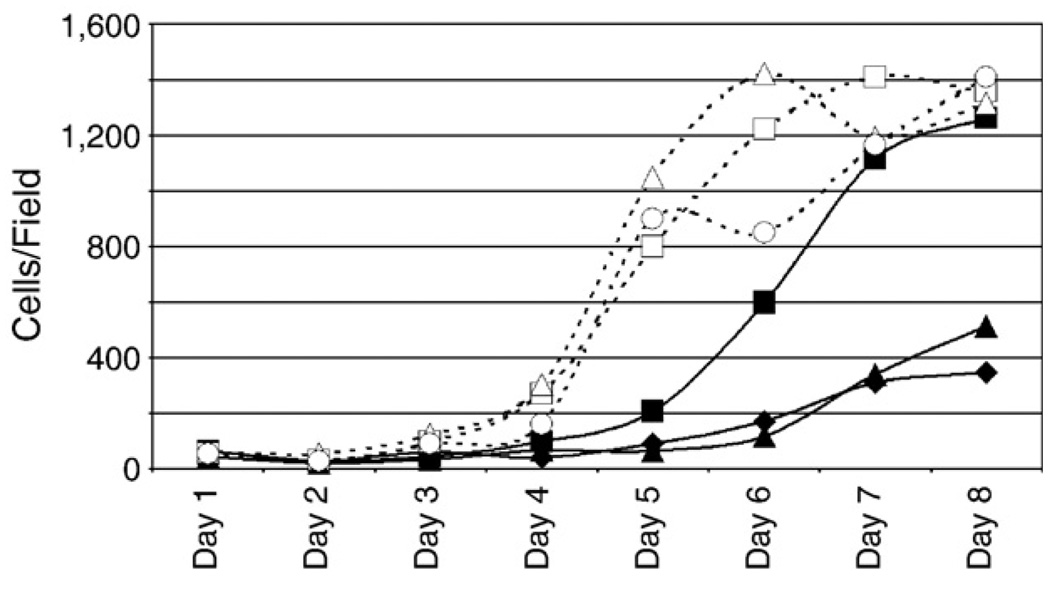
To assess the effects of silencing Id1 expression on the growth of Rat-1 cells, equal numbers of three different pools of separate retrovirus transductions of control and Id1 siRNA expressing Rat-1 cells were plated in duplicate in eight 12-well plates and cells were allowed to grow from one to 8 days. Cells were fixed and stained with DAPI, photographed by fluorescence microscopy, and DAPI stained nuclei were counted using ImageJ using the ITCN plugin. Four different fields were counted for each duplicate per pool per day and the average number of cells per field is plotted for each day (Fig. 9). All three pools expressing the negative control siRNA grew to confluence by day six or seven at approximately the same rate (open symbols and dashed lines). Two of Id1 siRNA (closed triangles and closed diamonds), grew much slower and only reached about one third of the confluency of the negative pools by day eight. The third Id1 pool (closed squares) grew to an intermediate level, reaching half confluency by about day six and similar density to the negatives by day eight. These data indicate that inhibition of Id1 expression in Rat-1 cells impairs cell growth as the cells grow more slowly and to a lower density than the control cells.
Fig. 9.
Effects of Id1 silencing on cell growth. Three separate pools of Rat-1 cells were transduced with different retrovirus aliquots expressing control siRNA (open symbols and dashed lines) or Id1 siRNA (closed symbols and solid lines) and selected with puromycin for 48 h. Equal numbers of stable cells from each pool were seeded in duplicate into eight 12-well plates. The next day and each day afterwards, one plate (containing two wells per pool) was fixed and stained with DAPI. Four randomly selected fields for each well were photographed using a fluorescent microscope and DAPI stained nuclei were counted using the ITCN plugin of ImageJ. The average number of cells per field for each pool on each day is graphed.
Discussion
The data presented here assess the contribution of the up-regulation of the Id proteins by LMP1 to LMP1 transformation of Rat1 cells. Expression of the Id proteins alone was not sufficient to transform rodent fibroblast and did not affect the expression of the cell cycle regulators, p27, cdk2, or Rb. However, inhibition of Id1 expression impaired the growth of Rat-1 cells but did not inhibit LMP1-mediated transformation. These results suggest that LMP1 mediated upregulation of the Id proteins contributes to enhanced cellular growth independent of potential effects on the expression of cyclin dependent kinase inhibitors.
The Id proteins are known regulators of cyclin dependent kinase inhibitors including p16INK4 and p21WAF1, however, our data demonstrate that the Id proteins are clearly not sufficient to downregulate p27KIP expression and that the distinct cdkis are not regulated in common by the Id proteins. Distinct mechanisms of regulation of cdkis during cancer development have also been identified. The loss of expression of p16 in cancer cells frequently involves genetic mechanisms including mutation and deletion (Auerkari, 2006). In contrast, p27 is frequently regulated at the post-transcriptional level during cancer development and in cancer cells (Viglietto et al., 2002). These differences in regulation likely reflect the different properties of the different cdkis.
The data presented here indicate that LMP1 transcriptionally regulates p27 expression as evidenced by repression of a p27 promoter reporter construct and decreased p27 protein and RNA. However expression of Id1 or Id3 is not sufficient for this effect. It is possible that LMP1 affects p27 transcription through regulation of other pathways independent of the Id proteins or possibly through the concerted effects of the Id proteins with other regulators of p27 transcription. One likely target of LMP1 is FOXO3A, which is known to regulate p27 transcription (Chandramohan et al., 2004). FOXO3A transcriptional activity is negatively regulated by the kinase Akt. Akt is activated by LMP1 and the PI3K/Akt pathway is required for LMP1-mediated transformation (Mainou et al., 2005). Activation of the PI3K/Akt pathway by LMP1 could phosphorylate and inactivate FOXO3A leading to the transcriptional downregulation of p27. It is also possible that LMP1 also post-transcriptionally regulates p27 expression. The further study of the effects of LMP1 on p27 expression and the functional consequences is likely to reveal new mechanisms of LMP1-mediated cell control.
The Id proteins have been shown to delay the senescence of primary cell lines and are frequently highly expressed in a number of cancers including the EBV-associated tumor, nasopharyngeal carcinoma (Wang et al., 2002). A recent study in prostate cancer cells defined distinct properties of the different Id proteins by specifically inhibiting the expression of the Id proteins individually with targeted siRNAs (Asirvatham et al., 2006). This study indicated that loss of Id1 and Id3 resulted in loss of proliferation, while loss of Id2 did not affect proliferation but resulted in increased apoptosis. Similarly in breast cancer cells, inhibition of Id1 expression with siRNA or antisense RNA resulted in a growth defect similar to inhibition of myc expression (Swarbrick et al., 2005). In the current study, inhibition of Id expression similarly affected proliferation of Rat-1 cells. This inhibition did not block transformation as the LMP1 transformed cells still had altered morphology, formed foci under contact-inhibiting conditions, and produced colonies in an anchorage-independent fashion. However, the impairment of cell growth suggests that Id1 and Id3 induced by LMP1 contribute to the enhanced growth of LMP1 expressing cells. Identification of the target genes and pathways affected by Id induction in LMP1 transformed cells may lead to the identification of new targets of LMP1 that contribute to enhanced cellular growth and vitality.
Materials and methods
Plasmids
The cloning of wild type LMP1 into retrovirus packaging vector pBabe was previously described (Everly et al., 2004). LMP1 was subcloned into a retrovirus-packaging vector, pHSCG (a kind gift of Lishan Su, University of North Carolina, Chapel Hill), which co-expresses a gene of interest under control of the LTR and GFP under the CMV promoter in order to identify transduced cells by green fluorescence. HA-tagged LMP1 was amplified by pcr with Platinum Pfx (Invitrogen) according to the manufacturer’s instructions with HA-EcoRI5′ (gccgaattcatggcttacccatacgatgttccag) and LMP1-SalI3′ (gcggtccagaatgtggcttttcagcctagac), digested with EcoRI and SalI and ligated into pHSCG digested with EcoRI and XhoI by compatible ends.
Retrovirus expressing Id1 siRNA, pSilencer-Id1, was constructed by inserting Id1 specific sequences into pSilencer™ 5.1-U6 retro (Ambion) according to the manufacturer’s directions. Id1 primers siId13805′ (gatccgtcgcgaccgccggaggccttcaagagaggcctccggcggtcgcgacttttttggaaa) and siId13803′ (agcttttccaaaaaagtcgcgaccgccggaggcctctcttgaaggcctccggcggtcgcgacg) were mixed at 1 mg/ml in 1x annealing solution (Ambion), boiled, slowly cooled, and ligated into pSilencer™ 5.1-U6 retro digested with BamHI and HindIII by compatible ends. Mature siRNA produced in transduced cells should target the sequence ttgtcgcgaccgccggaggcc from 380 to 400 bp in the Rattus novegicus Id1 message (GenBank accession NM_012797).
Retroviruses expressing untagged and myc-tagged Id1 and Id3 were constructed by cloning into pBabe and pM3-Babe, respectively, described previously (Mainou et al., 2007). Human Id1 (GenBank accession NM_002165) was subcloned from pLXSN-Id1 (kindly provided by Pierre-Yves Desprez, California Pacific Medical Research Institute) by pcr amplification as described above with Id1KZ5′ (cgacggatccctgccaccatgaaagtcgccagtggcagc) and EcoId13′ (atcacgaggaattccgcttcagcgacacaagatgc) with BamHI and EcoRI. Id3 (GenBank accession NM_013058) was cloned from cDNA generated from total Rat-1 RNA by reverse transcription with SuperScript™ III Rnase H-Reverse Transcriptase (Invitrogen) according to the manufacturer’s instructions with RNId3E3′ (atcacgaggaattcaggacgaccgggtcagtggc), followed by pcr with RNId3E3′ and Id3KZ5′ (cgacggatccctgccaccatgaaggcgctgagcccggtgcgcggc) and cloning using BamHI and EcoRI. Resulting constructs express untagged or amino-terminal triple myc-tagged protein Id1 and Id3 protein. Caborxyl-terminal HA-tagged Id1 and Id3 pBabe vectors were produced by amplification of Id1 and Id3 by pcr with the same 5′ primers, Id1KZ5′ and Id3KZ5′, respectively, with Id1C-HA′ (ggcgaattcctaaccggtacgagcgtaatctggaacatcgtatgggtaagctccggtgcgacacaagatgcgatcgtc) and Id3C-HA′ (ggcgaattcctaaccggtacgagcgtaatctggaacatcgtatgggtaagctccggtgtggcaaaaactcctcttgtcc), respectively, as described above.
A promoter reporter construct containing upstream regulatory sequence of p27Kip1/cyclin-dependent kinase inhibitor 1B of Rattus novegicus was constructed. Control sequences for the p27 promoter were amplified from Rat-1 genomic DNA using p27RN-5′ (gacagatctgaggaaggaaggagctgttgtagttggcg) and p27RN-3′ (gacagatctcttcctccccgggcgggtgtgg) as described above and cloned into the BglII site of pGL3-Basic (Promega). The resulting promoter reporter plasmid contains sequences 1140 base pairs upstream of the transcription start site and the 360 base pair 5′ untranslated region up to but not including the translation start site for a total of 1500 base pairs.
The Rat p27Kip1/cyclin-dependent kinase inhibitor 1B gene was cloned for generation of rat-specific ribonuclease protection probes. The gene for p27Kip1 (GenBank accession NM_031762) was amplified from genomic DNA as described above using p27RN5′ (cgacggatccatatgtcaaacgtgagagtgtctaacggg) and p27RN3′ (atcacgaggaattcggagctgtttacgtctggcg) and cloned into the BamHI and EcoRI sites of pcDNA3 (Invitrogen). Digestion with BamHI followed by in vitro transcription with SP6 RNA polymerase will yield a probe that will protect exon 1 and exon 2 of the rat p27Kip1 gene in ribonuclease protection assays.
Transfection and retrovirus transduction
Cells were transfected with FuGENE 6 (Boehringer Mannheim) according to the manufacturer’s directions. Recombinant retroviruses were generated as previously described (Scholle et al., 2000) by transfection of 293T cells with pHSCG, pHSCGLMP1, pBABE, pBABE-HA-LMP1, pBABE-Id1, pBABE-Id3, pSilencer-Neg (nonspecific siRNA expression vector, Ambion), or pSilencer-Id1, and VSV-G (pG1-VSV-G) and gag-pol (pGPZ9) expressing plasmids. After 24 h, transfection media was removed and replaced with fresh media, and cells were incubated at 33 °C overnight. The following day culture supernatants were harvested and centrifuged 1000×g for 10 min to remove any cells or cellular debris. Rat-1 cells were transduced with clarified 293T culture supernatants overnight with 8 µg/ml polybrene.
Cell culture and stable cell lines
Cell lines were maintained in Dulbecco modified Eagle medium (Gibco) supplemented with antibiotic/antimycotic mixture and 10% (v/v) heat-inactivated fetal bovine serum (FBS). Rat-1, rodent fibroblast, stable cell lines were established by transduction with retrovirus and selection with puromycin (1 µg/ ml, Sigma). For serum starvation, selected cells were grown to confluence and then changed to media with 0.2% serum for 24 h.
Id1 silencing
Transient silencing of Id1 was accomplished by transfection with small interfering RNAs. Transfection was optimized using siRNATransfection Optimization Kit from Ambion according to the manufacturer’s directions using predesigned Cy3-labelled negative-control and GAPDH silencer siRNA, Ambion. Id1 siRNA as selected from Ambion’s predesigned siRNA database and transfected according to the optimized conditions with siPORT™ NeoFX™ transfection agent (Ambion). Knockdown of targets was confirmed by western blotting and ribonuclease protection assays.
Ribonuclease protection assay
Ribonuceotide protection assays (RPA) were performed using a Direct Protect™ Lysate RPA kit (Ambion) according to the manufacturer’s directions. Briefly, RPA probes were annealed with RNA overnight in lysis buffer. The following day samples were digested with RNase and protease and precipitated. Protected probes were resolved on a 5% polyacrylamide gel, dried, and imaged using a PhosphorImager (Molecular Dynamics). Bands were quantitated using ImageQuant 5.1 (Molecular Dynamics) and p27 expression was determined relative to GAPDH levels.
Cell harvesting and western blotting
Cell lines were grown in 100-mm tissue culture plates to confluency and harvested. Cells were washed with ice cold phosphate buffered saline (PBS, Gibco) and lysed with 100– 250 µl RIPA buffer (10 mM Tris–HCl, pH 8.0, 140 mM NaCl, 1% Triton X-100, 0.1% SDS, 1% deoxycholic acid, protease and phosphatase inhibitors (Sigma)). Cell lysates were clarified by centrifugation and quantitated by Bio-Rad DC protein assay system (Bio-Rad). Samples were then boiled in SDS sample buffer and indicated amounts of proteins were separated using 10 or 15% acrylamide SDS-PAGE, and transferred to Optitran membranes (Whatman) for western blotting analysis. Primary antibodies used included β-Actin (I-19), c-myc (9E10), Id1 (C-20 and Z-8), and Id3 (H-70) (Santa Cruz), p27 (Calbiochem), Cdk2 (55) and Rb (G3-245) (BD Biosciences), and CS1-4 (anti-LMP1) (Dako). Bound proteins were detected with horseradish peroxidase-conjugated secondary antibodies (Amersham Pharmacia and Dako) and Pierce Supersignal West Pico System (Pierce) followed by exposure to film. Relative protein expression was quantitated using ImageJ software, http://rsb.info.nih.gov/ij/ (Rasband, 1997–2007).
Rat-1 foci and soft agar assays
Rat-1 cells or Rat-1 stable cell lines were plated 1:5 in 6- or 12-well plates and infected the following day with retroviruses with 8 µg/ml polybrene. The media was changed the following day and every 2 to 3 days thereafter, and foci were allowed to form for 10 to 15 days. Cells were examined by phase contrast and fluorescence microscopy or transduction and transformation and stained with 1% crystal violet in 50% ethanol and imaged with stereo microscopy. The number and size of foci were quantified using ImageQuant-TL v2005 (GE Healthcare) using the colony counting functions.
Soft agar assays were performed as described previously (Scholle et al., 2000). Briefly, Bacto agar medium (5% Bacto agar diluted in DMEM with 10% FBS to a final concentration of 0.5%) was poured into 12-well plates and allowed to solidify. 1–2×105 cells were suspended in Bacto agar medium and overlaid the solidified wells. Once solidified, the cell-agar suspensions were feed every few days for 14–21 days with normal media containing puromycin to maintain the expression of the Id1 siRNA. Colonies were imaged with phase contrast and fluorescence microscopy.
p27 promoter luciferase assays
Cells were plated 1:5 into 12-well plates 1 day prior to transfection. Cells were transfected with 0.2 µg of pRL-SV40 (Promega), 0.2 µg of pGL3-Basic or p27-1501 promoter plasmids, and 0.2 µg of pBABE or pBABE-expression clones. The following day the media was changed, and 40 h post-transfection cells were harvested and luciferase activity was assayed using the Dual-Luciferase® Reporter Assay System (Promega) according to the manufacturer’s directions. Relative luciferase activity was determined by dividing the firefly luciferase activity of the p27 promoter constructs by the internal control Renilla luciferase activity. Each condition was done in triplicate and replicated in different experiments.
Acknowledgments
The authors thank Stephanie Mazzuca for technical assistance. This work was supported by NIH grants CA 32979 and CA 19014 to N.R.T and NIH T32 CA09156 to D.N.E.
References
- Alani RM, Hasskarl J, Grace M, Hernandez MC, Israel MA, Munger K. Immortalization of primary human keratinocytes by the helix–loop–helix protein, Id-1. Proc. Natl. Acad. Sci. U. S. A. 1999;96(17):9637–9641. doi: 10.1073/pnas.96.17.9637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardila-Osorio H, Pioche-Durieu C, Puvion-Dutilleul F, Clausse B, Wiels J, Miller W, Raab-Traub N, Busson P. TRAF interactions with raft-like buoyant complexes, better than TRAF rates of degradation, differentiate signaling by CD40 and EBV latent membrane protein 1. Int. J. Cancer. 2005;113(2):267–275. doi: 10.1002/ijc.20503. [DOI] [PubMed] [Google Scholar]
- Asirvatham AJ, Schmidt MA, Chaudhary J. Non-redundant inhibitor of differentiation (Id) gene expression and function in human prostate epithelial cells. Prostate. 2006;66(9):921–935. doi: 10.1002/pros.20366. [DOI] [PubMed] [Google Scholar]
- Auerkari EI. Methylation of tumor suppressor genes p16(INK4a), p27 (Kip1) and E-cadherin in carcinogenesis. Oral Oncol. 2006;42(1):5–13. doi: 10.1016/j.oraloncology.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Baichwal VR, Sugden B. Transformation of Balb 3T3 cells by the BNLF-1 gene of Epstein-Barr virus. Oncogene. 1988;2(5):461–467. [PubMed] [Google Scholar]
- Barone MV, Pepperkok R, Peverali FA, Philipson L. Id proteins control growth induction in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 1994;91(11):4985–4988. doi: 10.1073/pnas.91.11.4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Schafer A, Lu S, Bilello JP, Desrosiers RC, Edwards R, Raab-Traub N, Cullen BR. Epstein–Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2006;2(3):e23. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandramohan V, Jeay S, Pianetti S, Sonenshein GE. Reciprocal control of Forkhead box O 3a and c-Myc via the phosphatidylinositol 3-kinase pathway coordinately regulates p27Kip1 levels. J. Immunol. 2004;172(9):5522–5527. doi: 10.4049/jimmunol.172.9.5522. [DOI] [PubMed] [Google Scholar]
- Dawson CW, Eliopoulos AG, Blake SM, Barker R, Young LS. Identification of functional differences between prototype Epstein-Barr virus-encoded LMP1 and a nasopharyngeal carcinoma-derived LMP1 in human epithelial cells. Virology. 2000;272(1):204–217. doi: 10.1006/viro.2000.0344. [DOI] [PubMed] [Google Scholar]
- Devergne O, Hatzivassiliou E, Izumi KM, Kaye KM, Kleijnen MF, Kieff E, Mosialos G. Association of TRAF1, TRAF2, and TRAF3 with an Epstein-Barr virus LMP1 domain important for B-lymphocyte transformation: role in NF-kappaB activation. Mol. Cell. Biol. 1996;16(12):7098–7108. doi: 10.1128/mcb.16.12.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliopoulos AG, Young LS. Activation of the cJun N-terminal kinase (JNK) pathway by the Epstein-Barr virus-encoded latent membrane protein 1 (LMP1) Oncogene. 1998;16(13):1731–1742. doi: 10.1038/sj.onc.1201694. [DOI] [PubMed] [Google Scholar]
- Everly DN, Jr, Mainou BA, Raab-Traub N. Induction of Id1 and Id3 by latent membrane protein 1 of Epstein-Barr virus and regulation of p27/ Kip and cyclin-dependent kinase 2 in rodent fibroblast transformation. J. Virol. 2004;78(24):13470–13478. doi: 10.1128/JVI.78.24.13470-13478.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franken M, Devergne O, Rosenzweig M, Annis B, Kieff E, Wang F. Comparative analysis identifies conserved tumor necrosis factor receptor-associated factor 3 binding sites in the human and simian Epstein–Barr virus oncogene LMP1. J. Virol. 1996;70(11):7819–7826. doi: 10.1128/jvi.70.11.7819-7826.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman SR, Johannsen E, Tong X, Yalamanchili R, Kieff E. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the J kappa recombination signal binding protein. Proc. Natl. Acad. Sci. U. S. A. 1994;91(16):7568–7572. doi: 10.1073/pnas.91.16.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara E, Yamaguchi T, Nojima H, Ide T, Campisi J, Okayama H, Oda K. Id-related genes encoding helix–loop–helix proteins are required for G1 progression and are repressed in senescent human fibroblasts. J. Biol. Chem. 1994;269(3):2139–2145. [PubMed] [Google Scholar]
- Henkel T, Ling PD, Hayward SD, Peterson MG. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science. 1994;265(5168):92–95. doi: 10.1126/science.8016657. [DOI] [PubMed] [Google Scholar]
- Higuchi M, Izumi KM, Kieff E. Epstein-Barr virus latent-infection membrane proteins are palmitoylated and raft-associated: protein 1 binds to the cytoskeleton through TNF receptor cytoplasmic factors. Proc. Natl. Acad. Sci. U. S. A. 2001;98(8):4675–4680. doi: 10.1073/pnas.081075298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi KM, Kieff ED. The Epstein-Barr virus oncogene product latent membrane protein 1 engages the tumor necrosis factor receptor-associated death domain protein to mediate B lymphocyte growth transformation and activate NF-kappaB. Proc. Natl. Acad. Sci. U. S. A. 1997;94(23):12592–12597. doi: 10.1073/pnas.94.23.12592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi KM, Kaye KM, Kieff ED. The Epstein-Barr virus LMP1 amino acid sequence that engages tumor necrosis factor receptor associated factors is critical for primary B lymphocyte growth transformation. Proc. Natl. Acad. Sci. U. S. A. 1997;94(4):1447–1452. doi: 10.1073/pnas.94.4.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi KM, Cahir McFarland ED, Ting AT, Riley EA, Seed B, Kieff ED. The Epstein-Barr virus oncoprotein latent membrane protein 1 engages the tumor necrosis factor receptor-associated proteins TRADD and receptor-interacting protein (RIP) but does not induce apoptosis or require RIP for NF-kappaB activation. Mol. Cell. Biol. 1999;19(8):5759–5767. doi: 10.1128/mcb.19.8.5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye KM, Izumi KM, Kieff E. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc. Natl. Acad. Sci. U. S. A. 1993;90(19):9150–9154. doi: 10.1073/pnas.90.19.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieff ED, Rickinson AB. Epstein–Barr Virus and Its Replication. In: Knipe DM, PMH, editors. Fields Virology. Fifth ed. vol. II. Lippincott Williams and Wilkins; 2007. pp. 2603–2654. [Google Scholar]
- Mainou BA, Everly DN, Jr, Raab-Traub N. Epstein-Barr virus latent membrane protein 1 CTAR1 mediates rodent and human fibroblast transformation through activation of PI3K. Oncogene. 2005;24(46):6917–6924. doi: 10.1038/sj.onc.1208846. [DOI] [PubMed] [Google Scholar]
- Mainou BA, Everly DN, Jr, Raab-Traub N. Unique signaling properties of CTAR1 in LMP1-mediated transformation. J. Virol. 2007;81(18):9680–9692. doi: 10.1128/JVI.01001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CL, Longnecker R, Kieff E. Epstein-Barr virus latent membrane protein 2A blocks calcium mobilization in B lymphocytes. J. Virol. 1993;67(6):3087–3094. doi: 10.1128/jvi.67.6.3087-3094.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CL, Lee JH, Kieff E, Longnecker R. An integral membrane protein (LMP2) blocks reactivation of Epstein-Barr virus from latency following surface immunoglobulin crosslinking. Proc. Natl. Acad. Sci. U.S.A. 1994;91(2):772–776. doi: 10.1073/pnas.91.2.772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CL, Burkhardt AL, Lee JH, Stealey B, Longnecker R, Bolen JB, Kieff E. Integral membrane protein 2 of Epstein-Barr virus regulates reactivation from latency through dominant negative effects on protein- tyrosine kinases. Immunity. 1995;2(2):155–166. doi: 10.1016/s1074-7613(95)80040-9. [DOI] [PubMed] [Google Scholar]
- Miller WE, Mosialos G, Kieff E, Raab-Traub N. Epstein-Barr virus LMP1 induction of the epidermal growth factor receptor is mediated through a TRAF signaling pathway distinct from NF kappaB activation. J. Virol. 1997;71(1):586–594. doi: 10.1128/jvi.71.1.586-594.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WE, Cheshire JL, Raab-Traub N. Interaction of tumor necrosis factor receptor-associated factor signaling proteins with the latent membrane protein 1 PXQXT motif is essential for induction of epidermal growth factor receptor expression. Mol. Cell. Biol. 1998;18(5):2835–2844. doi: 10.1128/mcb.18.5.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison JA, Klingelhutz AJ, Raab-Traub N. Epstein-Barr virus latent membrane protein 2A activates beta-catenin signaling in epithelial cells. J. Virol. 2003;77(22):12276–12284. doi: 10.1128/JVI.77.22.12276-12284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosialos G, Birkenbach M, Yalamanchili R, VanArsdale T, Ware C, Kieff E. The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell. 1995;80(3):389–399. doi: 10.1016/0092-8674(95)90489-1. [DOI] [PubMed] [Google Scholar]
- Nickoloff BJ, Chaturvedi V, Bacon P, Qin JZ, Denning MF, Diaz MO. Id-1 delays senescence but does not immortalize keratinocytes. J. Biol. Chem. 2000;275(36):27501–27504. doi: 10.1074/jbc.C000311200. [DOI] [PubMed] [Google Scholar]
- O’Toole PJ, Inoue T, Emerson L, Morrison IE, Mackie AR, Cherry RJ, Norton JD. Id proteins negatively regulate basic helix-loop-helix transcription factor function by disrupting subnuclear compartmentalization. J. Biol. Chem. 2003;278(46):45770–45776. doi: 10.1074/jbc.M306056200. [DOI] [PubMed] [Google Scholar]
- Ohtani N, Zebedee Z, Huot TJ, Stinson JA, Sugimoto M, Ohashi Y, Sharrocks AD, Peters G, Hara E. Opposing effects of Ets and Id proteins on p16INK4a expression during cellular senescence. Nature. 2001;409(6823):1067–1070. doi: 10.1038/35059131. [DOI] [PubMed] [Google Scholar]
- Paine E, Scheinman RI, Baldwin AS, Jr, Raab-Traub N. Expression of LMP1 in epithelial cells leads to the activation of a select subset of NF-kappa B/Rel family proteins. J. Virol. 1995;69(7):4572–4576. doi: 10.1128/jvi.69.7.4572-4576.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perk J, Iavarone A, Benezra R. Id family of helix-loop-helix proteins in cancer. Nat. Rev., Cancer. 2005;5(8):603–614. doi: 10.1038/nrc1673. [DOI] [PubMed] [Google Scholar]
- Peverali FA, Ramqvist T, Saffrich R, Pepperkok R, Barone MV, Philipson L. Regulation of G1 progression by E2A and Id helix- loop-helix proteins. EMBO J. 1994;13(18):4291–4301. doi: 10.1002/j.1460-2075.1994.tb06749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pioche-Durieu C, Keryer C, Souquere S, Bosq J, Faigle W, Loew D, Hirashima M, Nishi N, Middeldorp J, Busson P. In nasopharyngeal carcinoma cells, Epstein-Barr virus LMP1 interacts with galectin 9 in membrane raft elements resistant to simvastatin. J. Virol. 2005;79(21):13326–13337. doi: 10.1128/JVI.79.21.13326-13337.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhu S, Ignatova A, Park ST, Sun XH. Regulation of the expression of cyclin-dependent kinase inhibitor p21 by E2A and Id proteins. Mol. Cell. Biol. 1997;17(10):5888–5896. doi: 10.1128/mcb.17.10.5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab-Traub N. Epstein-Barr virus in the pathogenesis of NPC. Semin. Cancer Biol. 2002;12(6):431–441. doi: 10.1016/s1044579x0200086x. [DOI] [PubMed] [Google Scholar]
- Rasband WS. Image J. Bethesda, Maryland, USA: U.S. National Institutes of Health; 1997–2007. [Google Scholar]
- Roberts ML, Cooper NR. Activation of a ras-MAPK-dependent pathway by Epstein-Barr virus latent membrane protein 1 is essential for cellular transformation. Virology. 1998;240(1):93–99. doi: 10.1006/viro.1997.8901. [DOI] [PubMed] [Google Scholar]
- Robertson ES, Lin J, Kieff E. The amino-terminal domains of Epstein-Barr virus nuclear proteins 3A, 3B, and 3C interact with RBPJ (kappa) J. Virol. 1996a;70(5):3068–3074. doi: 10.1128/jvi.70.5.3068-3074.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson ES, Ooka T, Kieff ED. Epstein-Barr virus vectors for gene delivery to B lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 1996b;93(21):11334–11340. doi: 10.1073/pnas.93.21.11334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholle F, Bendt KM, Raab-Traub N. Epstein-Barr virus LMP2A transforms epithelial cells, inhibits cell differentiation, and activates Akt. J. Virol. 2000;74(22):10681–10689. doi: 10.1128/jvi.74.22.10681-10689.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikder HA, Devlin MK, Dunlap S, Ryu B, Alani RM. Id proteins in cell growth and tumorigenesis. Cancer Cell. 2003;3(6):525–530. doi: 10.1016/s1535-6108(03)00141-7. [DOI] [PubMed] [Google Scholar]
- Swarbrick A, Akerfeldt MC, Lee CS, Sergio CM, Caldon CE, Hunter LJ, Sutherland RL, Musgrove EA. Regulation of cyclin expression and cell cycle progression in breast epithelial cells by the helix loop helix protein Id1. Oncogene. 2005;24(3):381–389. doi: 10.1038/sj.onc.1208188. [DOI] [PubMed] [Google Scholar]
- Swart R, Ruf IK, Sample J, Longnecker R. Latent membrane protein 2A-mediated effects on the phosphatidylinositol 3-kinase/Akt pathway. J. Virol. 2000;74(22):10838–10845. doi: 10.1128/jvi.74.22.10838-10845.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornburg NJ, Kulwichit W, Edwards RH, Shair KH, Bendt KM, Raab-Traub N. LMP1 signaling and activation of NF-kappaB in LMP1 transgenic mice. Oncogene. 2006;25(2):288–297. doi: 10.1038/sj.onc.1209023. [DOI] [PubMed] [Google Scholar]
- Viglietto G, Motti ML, Fusco A. Understanding p27(kip1) deregulation in cancer: down-regulation or mislocalization. Cell Cycle. 2002;1(6):394–400. doi: 10.4161/cc.1.6.263. [DOI] [PubMed] [Google Scholar]
- Wan J, Zhang W, Wu L, Bai T, Zhang M, Lo KW, Chui YL, Cui Y, Tao Q, Yamamoto M, Akira S, Wu Z. BS69, a specific adaptor in the latent membrane protein 1-mediated c-Jun N-terminal kinase pathway. Mol. Cell. Biol. 2006;26(2):448–456. doi: 10.1128/MCB.26.2.448-456.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Liebowitz D, Kieff E. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell. 1985;43(3 Pt 2):831–840. doi: 10.1016/0092-8674(85)90256-9. [DOI] [PubMed] [Google Scholar]
- Wang X, Xu K, Ling MT, Wong YC, Feng HC, Nicholls J, Tsao SW. Evidence of increased Id-1 expression and its role in cell proliferation in nasopharyngeal carcinoma cells. Mol. Carcinog. 2002;35(1):42–49. doi: 10.1002/mc.10072. [DOI] [PubMed] [Google Scholar]
- Yasui T, Luftig M, Soni V, Kieff E. Latent infection membrane protein transmembrane FWLY is critical for intermolecular interaction, raft localization, and signaling. Proc. Natl. Acad. Sci. U. S. A. 2004;101(1):278–283. doi: 10.1073/pnas.2237224100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat. Rev., Cancer. 2004;4(10):757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- Young L, Alfieri C, Hennessy K, Evans H, O’Hara C, Anderson KC, Ritz J, Shapiro RS, Rickinson A, Kieff E, et al. Expression of Epstein–Barr virus transformation-associated genes in tissues of patients with EBV lymphoproliferative disease. N. Engl. J. Med. 1989;321(16):1080–1085. doi: 10.1056/NEJM198910193211604. [DOI] [PubMed] [Google Scholar]
- Zheng W, Wang H, Xue L, Zhang Z, Tong T. Regulation of cellular senescence and p16(INK4a) expression by Id1 and E47 proteins in human diploid fibroblast. J. Biol. Chem. 2004;279(30):31524–31532. doi: 10.1074/jbc.M400365200. [DOI] [PubMed] [Google Scholar]