Abstract
Encoded in the genome of most eukaryotes, microRNAs (miRNAs) have been proposed to regulate specifically up to 90% of human genes through a process known as miRNA-guided RNA silencing. The aim of this review is to present this process as the integration of a succession of specialized molecular machines exerting well defined functions. The nuclear microprocessor complex initially recognizes and processes its primary miRNA substrate into a miRNA precursor (pre-miRNA). This structure is then exported to the cytoplasm by the Exportin-5 complex where it is presented to the pre-miRNA processing complex. Following pre-miRNA conversion into a miRNA:miRNA* duplex, this complex is assembled into a miRNA-containing ribonucleoprotein (miRNP) complex, after which the miRNA strand is selected. The degree of complementarity of the miRNA for its messenger RNA (mRNA) target guides the recruitment of the miRNP complex. Initially repressing its translation, the miRNP-silenced mRNA is directed to the P-bodies, where the mRNA is either released from its inhibition upon a cellular signal and/or actively degraded. The potency and specificity of miRNA biogenesis and function rely on the distinct protein·protein, protein·RNA and RNA:RNA interactions found in different complexes, each of which fulfill a specific function in a well orchestrated process.
Keywords: Microprocessor, Drosha, DGCR8, Exportin-5, pre-miRNA processing complex, Dicer, TRBP, PACT, miRNP complex, Ago2, FMRP, P-bodies
2. THE MICRORNA-GUIDED RNA SILENCING PATHWAY
The microRNA (miRNA)-guided RNA silencing pathway is a recently discovered process found to regulate gene expression at the messenger RNA (mRNA) level. Observation of this phenomenon was first reported in the nematode Caenorhabditis elegans, when Lee et al. (1) noted that the lin-4 transcript encoded for a small RNAs of approximately 22 nt, which was found to contain sequences complementary to a repeated sequence element in the 3′ nontranslated region (NTR) of lin-14 mRNA (1). This finding suggested that the lin-14 mRNA could be regulated by RNA:RNA interaction. Evidence of this observation was obtained when the Nobel laureates Fire and Mello reported that a small double-stranded (ds) RNA can induce a more potent genetic interference compared to single-stranded (ss) antisens species (2). Since then, significant advances have been made to elucidate the mechanistic basis of this important regulatory process, mainly through the identification of the components involved in the biogenesis and action of miRNAs.
In human, RNA interference (RNAi) refers to the inhibition of gene expression induced by exogenous small interfering RNA (siRNAs), which take advantage of their similarities with miRNAs to hijack the endogenous miRNA-guided RNA silencing pathway. Both miRNAs and siRNAs are 21- to 23-nt long RNA species that act as guides that mediate downregulation of their specific mRNA target. Either synthetic or episomally expressed in vivo, siRNAs are currently used in several spheres of the research community mainly to study the function of their genes of interest. Expanding our understanding of the endogenous miRNA machinery will further improve the design and therapeutic potential of these potent and specific inhibitors of gene expression. The focus of this review will be mainly on the human miRNA-guided RNA silencing machinery, in which the main components or functional entities seem to be relatively conserved among eukaryotic organisms, such as fungi, plants and animals.
MiRNAs are encoded in the genome of most eukaryotic organisms and are conserved throughout the evolution. Bioinformatic predictions indicate that miRNA genes constitute about 2% of the predicted genes in mammals. They are constitutively or developmentally expressed, and some are tissue-specific or expressed at different levels. Increasing efforts to identify the specific targets of miRNAs lead us to speculate that miRNAs can regulate more than 90% of human genes (3). The importance of an intact synthetic process is mandatory for the regulation of a wide variety of mRNAs implicated in development, cell growth, apoptosis and other cellular processes (for a recent review, see Ouellet et al. (4)). A dysfunctional miRNA-guided RNA silencing pathway can thus lead to important genetic disorders, exemplify by some components of the machinery that have now been linked to specific diseases (for a recent review, see Perron et al. (5)).
The potency and specificity of the miRNA-guided RNA silencing pathway is strikingly correlated to the protein·protein, protein·RNA and RNA:RNA interactions involved. In fact, when considered from a mechanistic point of view, the most notable feature of this process is the orchestrated yet complex, interplay involving specific protein and RNA species. Remarkably, miRNAs can be recognized only by a few proteins through specific structural determinants, so that the machinery in place can generate, process and mediate the action of miRNAs of diverse sequences, their sequences confer all the specificity required for accurate gene regulation through imperfect pairing to their mRNA targets.
3. THE MICROPROCESSOR COMPLEX
MiRNA genes are initially transcribed by RNA polymerase (pol) II, which is responsible for the transcription of protein-coding mRNAs (6). In vitro processing assay using 293 cell extracts had revealed that miRNA genes were expressed as long RNA transcripts, called primary miRNA (pri-miRNAs) that were processed into miRNA precursors (pre-miRNAs) (7). The resulting pri-miRNA forms a long, non-translated RNA harboring a hairpin-loop structure that can be several kilobases long, and possesses the RNA pol II signature, i.e. a 5′ 7-methyl guanylate (m7G) cap and a 3′ poly(A) tail (7–9). Following its transcription, the pri-miRNA is recognized by the nuclear microprocessor complex, which is composed of Drosha and the DiGeorge syndrome critical region gene 8 (DGCR8) protein (10–13).
3.1. Drosha
Initially found to be involved in preribosomal RNA processing and formerly known as the human RNase III (14), Drosha was characterized as the founding member of the class 2 RNase III proteins (14). The RNases III enzyme family regroups endoribonucleases specific for dsRNA substrates. The dsRNA cleavage products of RNAses III bear characteristic termini consisting of a 5′ phosphate group and a 2-nt 3′ overhang hydroxylated end. As recently reviewed in MacRae and Doudna (15), RNases III are divided into 3 classes. Class 1 RNases III, of which bacterial RNase III remains the best documented enzyme, are composed of a single RNase III domain and a C-terminal dsRNA binding domain (dsRBD). Class 2 enzymes, to which Drosha belongs, possess two RNase III domains and a C-terminal dsRBD domain. Class 3 RNases III, represented by Dicer, are the largest enzymes that contain two RNase III domains, a C-terminal dsRBD and a putative N-terminal DExD/H-box helicase domain accompanied by a domain of unknown function (DUF283) and a central PIWI/Ago/Zwille (PAZ) domain.
In a study aimed to identify the proteins involved in miRNA biogenesis, the RNase III Drosha was identified as the core nuclease that executes the initiation step of miRNA processing in the nucleus (8). Drosha was shown to bind to the pri-miRNA and excise the ~70-nt pre-miRNA. Downreglation of Drosha expression by RNAi indicate of its requirement for miRNA biogenesis, as its absence was correlated with a diminution of mature miRNA production (8). A structural model of the protein proposed that Drosha functions as a monomer, with its RNase III domains forming an internal dimer structure able to recognize its substrate and cleave it into a pre-miRNA (12). Surprisingly, purified recombinant Drosha can not cleave the pri-miRNA with precision (11), indicating that the specificity of substrate recognition and proper positioning of Drosha RNase III catalytic center must be conferred by DGCR8, a Drosha binding partner (11, 15). In fact, Drosha was found to be part of two protein complexes, the smaller one with DGCR8, and a larger one with multiple classes of RNA-associated proteins, including RNA helicases, dsRNA binding proteins, novel heterogeneous nuclear ribonucleoproteins (RNPs) and the Ewing’s sarcoma family of proteins (11). Accessory domains present in the different RNase III proteins are determinants of substrate selectivity, which in turn governs their specific biological function. The accessory domains in Drosha are a proline-rich region (PRR) and an arginine/serine (RS)-rich region in its N-terminal region. DGCR8 possesses a WW domain that may be responsible of its interaction with the PRR domain of Drosha (11). Other interacting proteins may confer distinct functionalities to Drosha in the nucleus (15).
3.2 DiGeorge syndrome critical region gene 8 (DGCR8) protein
The DGCR8 gene is present in a common monoallelic deleted genomic region containing ~30 genes located in the q11.2 region of the human chromosome 22 (16). Heterozygous deletion of this locus leads to the most common human genetic deletion syndrome known as the DiGeorge syndrome. The patients carrying this deletion demonstrate various conditions, ranging from congenital heart defect and characteristic facial appearance to immunodeficiency and behavioral problems (17).
The DGCR8 protein was identified in Flag immunoprecipitates prepared from HEK293 cells stably expressing the Flag-Drosha fusion protein (11). In similar experiments using cells stably expressing Flag-DGCR8, Drosha was found in Flag immunoprecipitates, which were also found to convert pri-miRNAs into pre-miRNAs (11). Individual knockdown of Drosha or DGCR8 by RNAi decreased the level of mature miRNAs, indicating that both proteins are essential for miRNA biogenesis (11). These findings were confirmed in DGCR8 knockout embryonic stem (ES) mouse cell (18).
DGCR8 possesses two dsRBDs that are complemented by a WW domain. Since Drosha lacks specificity in substrate processing in its absence, DGCR8 has been proposed to guide Drosha by functioning as a molecular anchor that mesures the distance from the dsRNA-ssRNA junction in the pri-miRNA (11, 12). By creating and testing deletion mutants of pri-miR30a by in vitro processing assay, Lee et al. (8) demonstrated that the sequence flanking the miRNA-to-become duplex is important for efficient miRNA biogenesis. More specifically, the sequences ~20-nt upstream and ~25-nt downstream of the expected cleaving site are important for the recognition by the microprocessor complex (8).
Recently, Han et al. (19) proposed a new model for substrate recognition by the Drosha·DGCR8 complex. Based on the fact that Drosha is the only enzyme that can cleave a wide variety of pri-miRNAs in the cell, they estimated that the Drosha·DGCR8 complex must recognize common structural features in these RNA species. To establish this model, they divided the structure of a pri-miRNA into four parts: the terminal loop, the upper stem, the lower stem and the basal segments. By combining computational and biochemical approches, the authors determined that the terminal loop does not seem to be essential for the cleavage by the microprocessor complex, while the flanking ssRNA segments appear critical. As for the cleavage site, it is determined mainly by the distance (~11 bp) from the stem-ssRNA junction. They also observed that the immunopurified Flag-DGCR8, but not Flag-Drosha, interacts with pri-miRNAs both directly and specifically, and that the flanking ssRNA segments are required for this binding to occur (19). Crucial in defining the miRNA sequence-to-be, the Drosha·DGCR8 complex is responsible for generating one extremity of the miRNA duplex, whereas the opposite end will be created upon cleavage by the other RNase III of the miRNA pathway, Dicer.
4. THE EXPORTIN-5 COMPLEX
After its production in the nucleus, the pre-miRNA is subsequently exported to the cytoplasm, through a mechanism known as a rate-limiting step in the miRNA-guided RNA silencing pathway (20). Pre-miRNA export is achieved by the transporter Exportin-5 (Exp-5) which was shown to bind specifically a typical pre-miRNA substrate in the presence of RanGTP by gel-shifted retardation assay (21). Exp-5 is a member of the nuclear karyopherin β transporter family, which can mediate nuclear export of dsRNA binding proteins (22). The initial model stipulates that Exp-5 and RanGTP associate with a dsRBD-containing protein in the nucleus and that this export complex translocates through the nuclear pore complex (NPC) to the cytoplasm (22). Then, RanGTP hydrolysis releases in the cytoplasm the dsRBD-containing protein that will allow it to interact with regulatory elements of its mRNA target. Further release or degradation of the mRNA target would permit import of the dsRBD-containing protein back to the nucleus (22).
Experiment using RNAi showed that Exp-5 is required for efficient inhibition of gene expression induced by a pre-miRNA or a pre-miRNA mimetic short hairpin RNA (shRNA), but not by a siRNA, in dual luciferase reporter gene assay in 293T cells (21), suggesting that the pre-miRNA or shRNA export precedes their processing. Entering in the RNAi pathway downstream of Exp-5, siRNAs do not required Exp-5 for their activity. Moreover, miRNA biogenesis is dependent on the presence of Exp-5 in HeLa cells (23). Indeed, upon depletion of Exp-5 by RNAi for 48 to 72 hours, the levels of mature miRNAs was reduced by 40 to 60% (23). On the other hand, episomal overexpression of Exp-5 enhances the capacity for RNAi induced either by miRNAs or shRNAs, but not siRNAs (20). The expression of endogenous pre-miRNAs and mature miRNAs is also enhanced by Exp-5 overexpression, indicating that the presence of Exp-5 is a rate-limiting step in miRNA biogenesis. Achilles heel of the miRNA pathway, the Exp-5 transporter is saturated by structural VA1 RNA overexpressed by adenoviruses (24).
5. THE PRE-MICRORNA PROCESSING COMPLEX
First protein component of the miRNA pathway to be characterized in its recombinant form (25, 26), Dicer is known to be part of the pre-miRNA processing complex. Acting together with TAR RNA binding protein (TRBP), Dicer accepts the pre-miRNA product issued from the microprocessor complex and converts it into a miRNA duplex of ~21 to 23 pb. The complex is then incorporated into an effector miRNA-containing ribonucleoprotein (miRNP) complex.
5.1. Dicer
The enzyme responsible for the generation of ~21–23 nt RNA guide sequences from dsRNA substrates that initiate RNAi in Drosophila S2 cells was identified as Dicer (27). In fact, miRNA duplexes bear the signature of RNases III, i.e. a 5′ phosphate and a 3′ hydroxylated end with 2-nt overhang (28). Dicer has been reported to be localized in the cytoplasm (29) and/or associated with the endoplasmic reticulum (ER) (25). Although its localization remains a matter of debate its positioning at the ER would be strategic in term of proximity to the Exp-5 complex and the translational machinery. Dicer thus acts as the core nuclease within the pre-miRNA processing complex, converting pre-miRNAs into miRNA:miRNA* duplexes.
In mammals, the presence of Dicer is essential, as Dicer-deficient mice die at the embryonic stage, suggesting that Dicer is required for mammalian development (30, 31). Dicer-deficient mouse ES cells are defective in differentiation and centromeric silencing. The role of miRNAs in ES cell differentiation has been recently studied by generating a Dicer knockout model (32). Analysis of Dicer-null ES cells revealed an impairement in miRNA biogenesis and a severe defect in differentiation both in vivo and in vitro. Epigenetic silencing of centromeric repeat sequences and concomittant expression of homologous small dsRNAs were also markedly reduced. Noticeably, the phenotype was rescued by the re-expression of Dicer in these cells (32). Their results suggest that Dicer participates in multiple, fundamental biological processes in mammals, ranging from stem cell differentiation to the maintenance of centromeric heterochromatin structure and centromeric silencing (32). Deregulation of Dicer expression has also been observed in cases of cancer. A reduced expression of Dicer in non-small cell lung cancer (33) and an overexpression in prostate adenocarcinoma and in precursor lesions of lung adenocarcinoma (34, 35) have been reported, indicating that an adequate level of Dicer expression is required for maintaining normal cell functions.
A functional model of Dicer catalytic activity proposed by Zhang et al. (36), based on an X-ray structure of the Aquifex aeolicus RNase III and biochemical data obtained from recombinant human Dicer (26), stipulates that Dicer contains a single catalytic center. In this model, Dicer would function by the intramolecular dimerization of its two RNase III domains, like Drosha, assisted by the PAZ and dsRBD. Its central PAZ domain specifically recognizes the 2-nt 3′ overhang present in the pre-miRNA and generated upon cleavage by Drosha. The PAZ and RNase IIIa domains may then act as a caliper in generating similarly-sized miRNAs. Each RNase III domain is responsible of the cleavage of a single strand of the miRNA duplex after approximately two turns of a-helices, at the opposite end of the extremity produced by Drosha. The distance from the PAZ domain to the RNase III domains determines the length of the miRNA produced by Dicer. Precious structural information emanated from the elucidation of the crystal structure of Dicer from Giardia intestinalis, which is of special interest because this Giardial enzyme is considered as a “minimal” Dicer. The structure prediction of this Dicer form containing only the PAZ and tandem RNase III domains is thus transposable to the C-terminal half of human Dicer (37). When viewed from the front, the molecule resembles the shape of a hatchet, with the RNase III domains making up the blade and the PAZ domain at the base of the handle. The PAZ and RNase III domains are connected by a long helix that runs along the length of the handle. The N-terminal residues of the protein encompass the connector helix, forming a flat platform on the face of the molecule. For Giardia Dicer, the length of the connector helix seems to be longer than human Dicer, which can explained that dsRNAs produced by Giardia Dicer are ~25 to 27 nt in length as compared to only ~21 to 23 nt long for human Dicer (15, 37).
5.2. TAR RNA binding protein (TRBP)
Initially discovered in 1991, TRBP was found to be a cellular factor acting in synergy with the viral TAT protein in the transactivation of the long terminal repeat (LTR) of human immunodeficiency virus type 1 (HIV-1), a process that results in the transcription of viral genes (38). TRBP possesses three dsRBD and is found in two isoforms in the cell, as TRBP2 is 21 amino acids longer than the TRBP1 isoform (39–41). TRBP is also known to interact with the protein Tax encoded by the human T-cell leukemia virus type 1 (HTLV-1) and inhibits its function of activation of the transcription from the long terminal repetition via the association with host cellular factors (42). TRBP exerts other functions, including inhibition of the interferon (IFN)-induced dsRNA regulated protein kinase R (PKR) (40), as well as a growth promoting role with oncogenic potential activity (43). Acting as a tumor suppressor, Merlin regulates the oncogenic activities of TRBP through a direct interaction (44).
TRBP was identified by mass spectroscopy analysis of Dicer complexes prepared by anti-Flag immunoprecipitation of HEK293 cells stably expressing Flag-Dicer (45). Conversely, a cell line expressing Flag-TRBP allowed to find Dicer in Flag immunoprecipitates. The region in TRBP that was found to interact with Dicer comprises the third dsRBD domain located at the C-terminal end (46). Both recombinant Dicer and TRBP proteins interact with each other, as evidenced by their cosedimentation in gel filtration assay. Furthermore, the TRBP·Dicer complex interacts with Argonaute 2 (Ago2) in a sequence of events in which TRBP is required for the recruitment of Ago2 to the siRNA bound by Dicer (45). Depletion studies have shown that TRBP is required for optimal RNA silencing mediated by siRNAs and endogenous miRNAs (46). Despite its important role in miRNA biogenesis and function, its exact role in miRNA-guided RNA silencing pathway is not completely understood (47, 48). Whether the interaction between Dicer and TRBP is reminiscent to that occurring between Drosha and DGCR8 remains to be investigated.
5.3. PACT
Recently, the PKR-activating protein (PACT) has been reported to be associated with a complex containing Dicer, Ago2, and TRBP. PACT interacts with Dicer, and the interaction involves the third dsRBD domain of PACT and the N-terminal region of Dicer containing the helicase motif (49). The depletion of PACT strongly affects the accumulation of mature miRNAs in vivo and moderately reduces the efficiency of siRNA-induced RNAi (49). An independant group confirmed these findings in shRNA-induced RNAi, but not when using siRNAs, suggesting that TRBP and PACT function primarily in siRNA production (50). Moreover, the presence of both TRBP and PACT increased the ability of Dicer to cleave a 566-bp long dsRNA in vitro, as compared to TRBP and PACT added individually (50).
Sharing 44% of identity, both PACT and TRBP possess three similar dsRBD domains and bind to PKR, although exerting opposing effects, i.e. as PACT activates PKR, TRBP inhibits it (51–53). PKR is a dsRNA-dependent serine/threonine protein kinase, which phosphorylates the translation initiation factor eIF2 to cause a general reduction of protein synthesis (54). The third dsRBD of PACT and TRBP, which is devoid of any detectable dsRNA binding activity (53), mediates their interaction with Dicer as well as their regulation of PKR (45, 46, 53). The C-terminal domain of PACT can also mediate homomultimerization (55). Interestingly, a recent study demonstrated that PACT directly interacts with TRBP and that this complex associates with Dicer to facilitate siRNAs production (50). PACT and TRBP may establish an interesting, but intriguing redundant link between the miRNA-guided RNA silencing pathway and the PKR signaling pathway.
6. THE MICRORNA-CONTAINING RIBONUCLEOPROTEIN (MIRNP) COMPLEX
The miRNP complex is the effector complex of the miRNA-guided RNA silencing pathway. Guided by a miRNA of imperfect complementarity, the miRNP recognizes a specific mRNA target, inhibiting its translation (56). However, if the miRNA bears a perfectly complementary with its mRNA target, degradation will be induced. For instance, miR-196 induces degradation of HOXB8 mRNA, to which it is perfectly complementary (except for the presence of a G:U wobble base pairing) (57). Loaded with a siRNA (siRNP), the RNA-induced silencing complex (RISC) is the best characterized RNP that also induces cleavage of its mRNA target (58).
MiRNAs usually exhibit an imperfect complementarity with their natural mRNA targets. Elucidation of the architecture of the well-studied interaction between let-7 and lin41 in C. elegans have allowed a better understanding of the pairing determinants of a miRNA with its mRNA target (59, 60). The let-7 miRNA forms an imperfect duplex with six complementary sites present in the lin-41 3′ NTR, and the occupancy of only two of the sites, separated by a 27-nt sequence, are sufficient to induce silencing of lin-41. Perfect pairing of the miRNA seed, referred to as nucleotides 2 to 8 from the 5′ end, to a sequence usually located in the 3′ NTR region of an mRNA is extremely important in target recognition (60). The 27-nt sequence between the two binding sites for let-7 also seems to be important, as mutational deletion or nucleotides substitution in the sequence prevent lin-41 silencing (60). However, the characteristics of an experimentally validated miRNA:target pair are not sufficient by themselves to establish the rules governing mRNA recognition by miRNAs, which may require more elaborated studies of several different miRNA:mRNA combinations.
A large multiprotein RNP effector complex of ~150 kDa to 500 kDa was isolated from Drosophila cells and found to have a sequence specific nuclease activity: this RNP was named RISC (61–63). In 2002, a miRNP complex was identified and found to share several characteristics with the RISC (64). The miRNP complex was composed of a wide variety of miRNAs forming a complex with three major proteins: Gemin3, Gemin4 and EIF2C2 (Ago2). Gemin3 is a DEAD-box putative helicase which is probably involved in the unwinding of the miRNA:miRNA* duplex, releasing the miRNA for its binding to its binding site on the mRNA. Three studies published in 2005 in the same issue of Cell provided additional insight on the composition and function of the human RISC (65–67) through the identification of Dicer, TRBP, and Ago2 as protein constituants of the human RISC (65). Recently a role in that complex for PACT (49), the Fragile X mental retardation protein (FMRP) (68, 69) and the RNA helicase A (RHA) (70) has been proposed.
The functional role of the miRNP complex in the miRNA-guided RNA silencing pathway is to receive the miRNA:miRNA* duplex, select the miRNA guide based the relative stability of the duplex extremities, destroy the miRNA* or passenger strand, recognize the mRNA target with its loaded miRNA guide, exert its inhibitory effects on mRNA translation, and possibly carry it into the P-bodies.
6.1. Argonaute 2 (Ago2)
Since the pre-miRNA processing complex is connected to the downstream effector complex, it is not surprising to find proteins involved in these two steps of miRNA biogenesis, such as Dicer and TRBP. In fact, both proteins were found to bind to Ago2 (45), thereby linking the initiation and effector steps of the miRNA pathway. Additionnal Ago2-binding proteins include PACT (49) and RHA, which was recently found to interact with the human RISC and to function in RISC loading (70). Robb and Rana (70) showed that RHA interacts with siRNA, Ago2, TRBP, and Dicer in human cells. In functional assays, RNAi was reduced in RHA-depleted cells as a consequence of decreased levels of active, Ago2-containing RISC assembled with the guide-strand RNA (70).
Ago2 is a member of the PAZ and PIWI domain (PPD) protein family expressed in metazoans and fungi, but not apparently in the budding yeast Saccharomyces cerevisiae (71, 72). Among the eight members of the family present in humans, Ago isoforms 1 to 4 are the most closely related and bind to siRNAs and miRNAs, although only Ago2 is found within miRNP complexes (73, 74). In fact, coincubation of recombinant Ago2 protein with an siRNA is sufficient to form a minimal RISC that accurately cleaves target RNAs (75). The crystal structure of the Pyrococcus furiosus Ago protein (76) have led to the discovery that human Ago2 possesses the slicer activity of the RISC (74).
Ago2 is essential for mouse development, and cells lacking this protein are unable to mount an experimental response to siRNAs (74). Ago2 possesses a central PAZ domain and a C-terminal PIWI domain. The PIWI domain and the RNase H share a common catalytic DDE motif and, like RNase H, the catalytic activity of Ago2 requires divalent cations such as Mg2+ or Mn2+ (74, 76, 77). Mutations of the DDE motif within the PIWI domain of Ago2, as identified by comparison with the structure of P. furiosus protein, inactivate RISC activity, indicating that this activity resides within the PIWI domain of the protein (74). The PIWI domain consists of a 5-stranded β-sheets surrounded by three helices (76) and mediates siRNA binding (77, 78). The crystal structure of the Archaeoglobus fulgidus PIWI domain, in complex with an siRNA duplex, indicates that this domain may function as the receptor site for the obligatory 5′ phosphate of siRNAs and specify the cleavage site on the target mRNA (79). The target mRNA is cleaved at the position corresponding to the phosphodiester bond linking nt 10 and 11 from the 5′ end of the siRNA located in close proximity to the PIWI catalytic site (79).
The structure of the D. melanogaster Ago1 PAZ domain has been solved by nuclear magnetic resonance (80, 81). The determined structure consists of a left-handed, six-stranded β-barrel capped at one end by two α-helices and wrapped on one side by a distinctive appendage comprised of a long β-hairpin and a short a-helix. The PAZ domain contributes to the specific recognition of siRNAs by providing a binding pocket for their characteristic 2-nt 3′ overhangs (76, 80–82). Thus, the 3′ end of the guide siRNA strand is anchored in the PAZ domain, whereas the PIWI domain, acting in concert with the PAZ, cleaves the mRNA strand between the siRNA nt 10 and 11, as discussed above. An active RISC can then be regenerated and initiate, armed with the same siRNA, a new round of mRNA cleavage (83). When a miRNA perfectly complementary to its mRNA target is loaded into a miRNP, the cleavage occurs at precisely the same site as that seen for siRNA-guided cleavage (84). After cleavage of the mRNA, the miRNA remains intact and can guide the recognition and destruction of additional mRNAs (84).
Pertaining to the translational repression induced by miRNAs, the most attractive hypothesis relates to the presence of a bulge in the miRNA:target duplex that prevents the cleavage by Ago2 by moving the nts complementary to nt 10 and 11 of the miRNA away from the catalytic site. The mechanism underlying translational inhibition likely involves multiple binding sites for the same or different miRNAs in the mRNA 3′ NTR and cooperation among the different miRNPs attached to these binding sites in the targeted message. These miRNPs may prevent initiation of translation or block active translation through combinatorial control and steric hindrance of the ribosomal machinery (85, 86). This scenario is supported by the relatively high degree of conservation among the different miRNAs recognizing the same target throughout the evolution. Some computational approaches do exploit this characteristic in order to identify potential miRNA targeted genes.
6.2 Fragile X mental retardation protein (FMRP)
FMRP, encoded by the FMR1 gene, has been identified in a miRNP complex containing Dicer and Ago2 proteins in mammalian cells in vivo (87). In human, loss of expression of the FMR1 gene product is the etiologic factor of the fragile X syndrome, the most frequent cause of inherited mental retardation (88, 89). FMRP is an RNA-binding protein, containing two K-homology (KH) domains and an RGG box, known to be involved in the regulation of mRNA translation, mRNA transfer and local modulation of synaptic mRNA translation (90–94). FMRP has been reported to behave as a negative regulator of translation both in vitro and in vivo (90–94).
Recently, the fact that human FMRP can act as a miRNA acceptor protein for the RNase III Dicer and facilitate assembly of miRNAs on specific target RNA sequences has been demonstrated (68). The requirement of FMRP for efficient RNAi in vivo was unveiled by reporter gene silencing assays using various small RNA inducers, which also supports its involvement in an siRNP effector complex in mammalian cells. From these results, a model has been proposed in which FMRP could facilitate miRNA assembly on target mRNAs. Functioning within a duplex miRNP, FMRP may also mediate mRNA targeting through a strand exchange mechanism, in which the miRNA* of the duplex is swapped for the mRNA (69). Furthermore, FMRP may contribute to the relief of miRNA-guided mRNA repression through a reverse strand exchange reaction, possibly initiated by a specific cellular signal, that would liberate the mRNA for translation (69). Although the intracellular sites hosting these events remain to be determined, we cannot exclude the possible involvement of the P-bodies.
7. THE P-BODIES
Characterized only recently (95, 96), the P-bodies are specific cytoplasmic foci of aggregated mRNA-containing RNP (mRNP) complexes associated with the translation repression and mRNA decay machinery. P-bodies are referred to as the GW182-containing bodies, because they contain the GW182 RNA-binding protein as well as other proteins implicated in protein degradation (97, 98). Recently, a link between these bodies and the miRNA-guided RNA silencing pathway has been established. In fact, Ago proteins have been localized to these protein complexes (95) and found to interact directly with GW182 (99). Indeed, silencing of GW182 delocalizes resident P-body proteins and impairs silencing of a miRNA reporter system. Moreover, mutations that prevent Ago proteins from localizing to P-bodies also prevent translational repression of mRNAs (99).
MiRNAs are present in P-bodies, as the formation of this structure seems to be a consequence, rather than be the cause, of miRNA genesis (100). The authors reported that endogenous let-7 miRNA co-precipitates with a GW182 protein complex. In addition, knockdown of two proteins, Drosha and DGCR8, which are essential for the generation of mature miRNAs, results in a loss of the P-bodies (100). P-bodies thus represent the cellular site where repressed mRNAs accumulate and are ultimately either degraded or rescued and redirected to the translational machinery.
MiRNA-guided mRNA repression appears to be a reversible process. Recently, Bhattacharyya et al. (101) observed that upon a specific stress, a repressed mRNA can be released and translated back into proteins. The authors showed that cationic amino acid transporter 1 (CAT-1) mRNA and reporter genes bearing its 3′ NTR can be relieved from miR-122-induced inhibition in human hepatocarcinoma cells subjected to various stress conditions. The derepression of CAT-1 mRNA is accompanied by its release from cytoplasmic processing bodies and its recruitment to polysomes. mRNA derepression seems to require binding of HuR, an AU-rich element (ARE) binding protein, to the 3′ NTR of CAT-1 mRNA. They proposed that proteins interacting with the 3′ NTR will generally act as modifiers, altering the potential of miRNAs to repress gene expression (101).
8. CONCLUSION
As reflected from the nature of its main steps and components, the miRNA-guided RNA silencing pathway is a highly organized process well orchestrated by a variety of protein·protein, RNA·protein and RNA:RNA interactions. Although the main steps and components have been identified and the complexity of this gene regulatory process being unreveiled, our comprehension of the mechanism underlying miRNA biogenesis as well as the translational repression of specific mRNAs guided by miRNAs remains incomplete. A better understanding of this process will help identify of miRNA-regulated mRNAs and determine importance of this regulation in health and diseases.
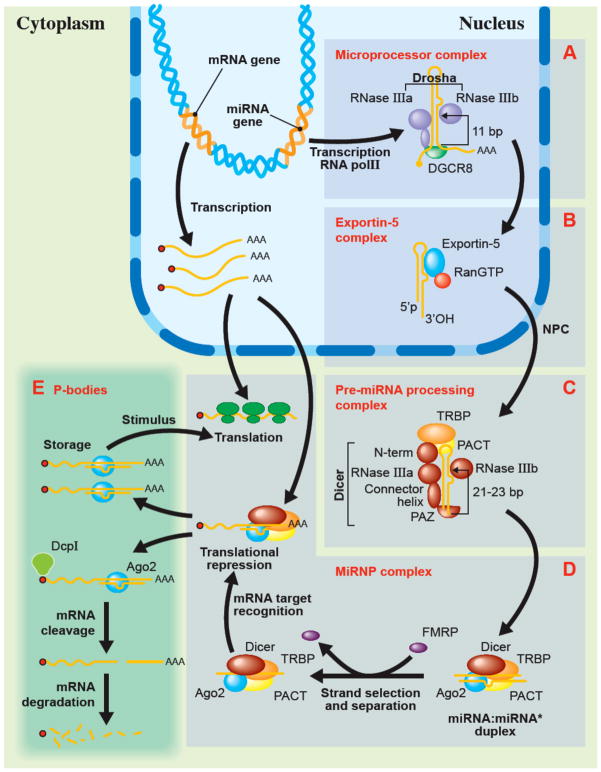
Figure 1.
The protein complexes of the microRNA (miRNA)-guided RNA silencing pathway. Both miRNA and messenger RNA (mRNA) encoding genes are transcribed by RNA polymerase II (RNA pol II). Whereas the latter yields a translatable mRNA transcript, the miRNA genes generate a non-translated RNA transcript known as the primary miRNA (pri-miRNA), which is recognized by the microprocessor complex composed of the ribonuclease III (RNase III) Drosha and the accessory DiGeorge syndrome critical region gene 8 (DGCR8) protein. DGCR8 binds to the dsRNA-ssRNA junction at the base of the pri-miRNA and acts as a molecular caliper that mesures the distance of 11 bp, where the two RNase III domains of Drosha form an intramolecular structure with each domain cleaving one strand to produce the miRNA precursor (pre-miRNA). This structure is then exported into the cytoplasm by the Exportin-5 complex in a sequence of events involving RanGTP hydrolysis. The pre-miRNA processing complex, which is composed of Dicer, TRBP and PACT, recognizes and processes the pre-miRNA substrate into a miRNA:miRNA* duplex. The PAZ domain of Dicer has been proposed to act as an anchor for the 3′ hydroxylated 2-nt overhang extremity. The connector helix is responsible of the measurement of the 21–23 bp length from the extremity and determines the positioning at the cleavage site of its two RNase III domains that form, as in Drosha, an intramolecular dimer with a unique catalytic site (36). The resulting miRNA:miRNA* loaded complex then joins the miRNA-containing ribonucleoprotein (miRNP) complex that contains Ago2, after which the miRNA strand is selected. The complementarity of the miRNA to its target brings the miRNP complex on a specific mRNA to negatively regulate it, and ultimately direct it to the P-bodies. These punctuate foci in the cell contain silenced mRNAs, which can be either released from their inhibition upon a cellular signal, or simply degraded.
Acknowledgments
We are grateful to Gilles Chabot for graphic design. M. P. P. was supported by a doctoral studentship from Natural Sciences and Engineering Research Council of Canada (NSERC). P. P. is a New Investigator of the Canadian Institutes of Health Research and Junior 2 Scholar from the Fonds de la Recherche en Santé du Québec. This work was financially supported by a Discovery grant from NSERC (262938-03).
Abbreviations
- 3′ NTR
3′ nontranslated region
- Ago
Argonaute
- DGCR8
DiGeorge syndrome critical region gene 8
- dsRNA
double-stranded RNA
- dsRBD
dsRNA binding domain
- embryonic stem
ES
- Exp-5
Exportin-5
- FMRP
Fragile X mental retardation protein
- mRNA
messenger RNA
- miRNA
microRNA
- pre-miRNA
miRNA precursor
- pri-miRNA
primary miRNA
- miRNP
miRNA-containing ribonucleoprotein
- nt
nucleotide
- PACT
protein kinase R (PKR)-activating protein
- RISC
RNA-induced silencing complex
- RNAi
RNA interference
- RNase
ribonuclease
- shRNA
short hairpin RNA
- siRNA
small interfering RNA
- siRNP
siRNA-containing ribonucleoprotein
- ssRNA
single-stranded RNA
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 3.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–17. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 4.Ouellet DL, Perron MP, Gobeil LA, Plante P, Provost P. MicroRNAs in Gene Regulation: When the Smallest Governs It All. J Biomed Biotechnol. 2006;2006:69616. doi: 10.1155/JBB/2006/69616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perron MP, Boissonneault V, Gobeil LA, Ouellet DL, Provost P. Regulatory RNAs: future perspectives in diagnosis, prognosis, and individualized therapy. Methods Mol Biol. 2007;361:311–26. doi: 10.1385/1-59745-208-4:311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. Embo J. 2004;23:4051–60. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. Embo J. 2002;21:4663–70. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 9.Basyuk E, Suavet F, Doglio A, Bordonne R, Bertrand E. Human let-7 stem-loop precursors harbor features of RNase III cleavage products. Nucleic Acids Res. 2003;31:6593–7. doi: 10.1093/nar/gkg855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–5. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 11.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–40. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 12.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–27. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14:2162–7. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 14.Wu H, Xu H, Miraglia LJ, Crooke ST. Human RNase III is a 160-kDa protein involved in preribosomal RNA processing. J Biol Chem. 2000;275:36957–65. doi: 10.1074/jbc.M005494200. [DOI] [PubMed] [Google Scholar]
- 15.MacRae IJ, Doudna JA. Ribonuclease revisited: structural insights into ribonuclease III family enzymes. Curr Opin Struct Biol. 2007;17:138–45. doi: 10.1016/j.sbi.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 16.Yamagishi H, Srivastava D. Unraveling the genetic and developmental mysteries of 22q11 deletion syndrome. Trends Mol Med. 2003;9:383–9. doi: 10.1016/s1471-4914(03)00141-2. [DOI] [PubMed] [Google Scholar]
- 17.Shiohama A, Sasaki T, Noda S, Minoshima S, Shimizu N. Molecular cloning and expression analysis of a novel gene DGCR8 located in the DiGeorge syndrome chromosomal region. Biochem Biophys Res Commun. 2003;304:184–90. doi: 10.1016/s0006-291x(03)00554-0. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–5. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 20.Yi R, Doehle BP, Qin Y, Macara IG, Cullen BR. Overexpression of Exportin 5 enhances RNA interference mediated by short hairpin RNAs and microRNAs. Rna. 2005;11:220–6. doi: 10.1261/rna.7233305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–6. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brownawell AM, Macara IG. Exportin-5, a novel karyopherin, mediates nuclear export of double-stranded RNA binding proteins. J Cell Biol. 2002;156:53–64. doi: 10.1083/jcb.200110082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–8. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 24.Gwizdek C, Ossareh-Nazari B, Brownawell AM, Doglio A, Bertrand E, Macara IG, Dargemont C. Exportin-5 mediates nuclear export of minihelix-containing RNAs. J Biol Chem. 2003;278:5505–8. doi: 10.1074/jbc.C200668200. [DOI] [PubMed] [Google Scholar]
- 25.Provost P, Dishart D, Doucet J, Frendewey D, Samuelsson B, Radmark O. Ribonuclease activity and RNA binding of recombinant human Dicer. Embo J. 2002;21:5864–74. doi: 10.1093/emboj/cdf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. Embo J. 2002;21:5875–85. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–6. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 28.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Billy E, Brondani V, Zhang H, Muller U, Filipowicz W. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc Natl Acad Sci U S A. 2001;98:14428–33. doi: 10.1073/pnas.261562698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–7. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 31.Yang WJ, Yang D, Na S, Sandusky G, Zhang Q, Zhao G. Dicer is required for embryonic angiogenesis during mouse development. J Biol Chem. 2004 doi: 10.1074/jbc.M413394200. [DOI] [PubMed] [Google Scholar]
- 32.Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karube Y, Tanaka H, Osada H, Tomida S, Tatematsu Y, Yanagisawa K, Yatabe Y, Takamizawa J, Miyoshi S, Mitsudomi T, Takahashi T. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–5. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chiosea S, Jelezcova E, Chandran U, Acquafondata M, McHale T, Sobol RW, Dhir R. Up-regulation of dicer, a component of the MicroRNA machinery, in prostate adenocarcinoma. Am J Pathol. 2006;169:1812–20. doi: 10.2353/ajpath.2006.060480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiosea S, Jelezcova E, Chandran U, Luo J, Mantha G, Sobol RW, Dacic S. Overexpression of Dicer in precursor lesions of lung adenocarcinoma. Cancer Res. 2007;67:2345–50. doi: 10.1158/0008-5472.CAN-06-3533. [DOI] [PubMed] [Google Scholar]
- 36.Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 37.Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–8. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 38.Gatignol A, Buckler-White A, Berkhout B, Jeang KT. Characterization of a human TAR RNA-binding protein that activates the HIV-1 LTR. Science. 1991;251:1597–600. doi: 10.1126/science.2011739. [DOI] [PubMed] [Google Scholar]
- 39.Duarte M, Graham K, Daher A, Battisti PL, Bannwarth S, Segeral E, Jeang KT, Gatignol A. Characterization of TRBP1 and TRBP2. Stable stem-loop structure at the 5′ end of TRBP2 mRNA resembles HIV-1 TAR and is not found in its processed pseudogene. J Biomed Sci. 2000;7:494–506. doi: 10.1159/000025485. [DOI] [PubMed] [Google Scholar]
- 40.Daher A, Longuet M, Dorin D, Bois F, Segeral E, Bannwarth S, Battisti PL, Purcell DF, Benarous R, Vaquero C, Meurs EF, Gatignol A. Two dimerization domains in the trans-activation response RNA-binding protein (TRBP) individually reverse the protein kinase R inhibition of HIV-1 long terminal repeat expression. J Biol Chem. 2001;276:33899–905. doi: 10.1074/jbc.M103584200. [DOI] [PubMed] [Google Scholar]
- 41.Gatignol A, Duarte M, Daviet L, Chang YN, Jeang KT. Sequential steps in Tat trans-activation of HIV-1 mediated through cellular DNA, RNA, and protein binding factors. Gene Expr. 1996;5:217–28. [PMC free article] [PubMed] [Google Scholar]
- 42.Donzeau M, Winnacker EL, Meisterernst M. Specific repression of Tax trans-activation by TAR RNA-binding protein TRBP. J Virol. 1997;71:2628–35. doi: 10.1128/jvi.71.4.2628-2635.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benkirane M, Neuveut C, Chun RF, Smith SM, Samuel CE, Gatignol A, Jeang KT. Oncogenic potential of TAR RNA binding protein TRBP and its regulatory interaction with RNA-dependent protein kinase PKR. EMBO J. 1997;16:611–24. doi: 10.1093/emboj/16.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee JY, Kim H, Ryu CH, Kim JY, Choi BH, Lim Y, Huh PW, Kim YH, Lee KH, Jun TY, Rha HK, Kang JK, Choi CR. Merlin, a tumor suppressor, interacts with transactivation-responsive RNA-binding protein and inhibits its oncogenic activity. J Biol Chem. 2004;279:30265–73. doi: 10.1074/jbc.M312083200. [DOI] [PubMed] [Google Scholar]
- 45.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–4. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–7. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gatignol A, Laine S, Clerzius G. Dual role of TRBP in HIV replication and RNA interference: viral diversion of a cellular pathway or evasion from antiviral immunity? Retrovirology. 2005;2:65. doi: 10.1186/1742-4690-2-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rossi JJ. Mammalian Dicer finds a partner. EMBO Rep. 2005;6:927–9. doi: 10.1038/sj.embor.7400531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–32. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kok KH, Ng MH, Ching YP, Jin DY. Human TRBP and PACT Directly Interact with Each Other and Associate with Dicer to Facilitate the Production of Small Interfering RNA. J Biol Chem. 2007;282:17649–57. doi: 10.1074/jbc.M611768200. [DOI] [PubMed] [Google Scholar]
- 51.Patel RC, Sen GC. PACT, a protein activator of the interferon-induced protein kinase, PKR. EMBO J. 1998;17:4379–90. doi: 10.1093/emboj/17.15.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peters GA, Hartmann R, Qin J, Sen GC. Modular structure of PACT: distinct domains for binding and activating PKR. Mol Cell Biol. 2001;21:1908–20. doi: 10.1128/MCB.21.6.1908-1920.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gupta V, Huang X, Patel RC. The carboxy-terminal, M3 motifs of PACT and TRBP have opposite effects on PKR activity. Virology. 2003;315:283–91. doi: 10.1016/s0042-6822(03)00589-0. [DOI] [PubMed] [Google Scholar]
- 54.Taylor SS, Haste NM, Ghosh G. PKR and eIF2alpha: integration of kinase dimerization, activation, and substrate docking. Cell. 2005;122:823–5. doi: 10.1016/j.cell.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 55.Hitti EG, Sallacz NB, Schoft VK, Jantsch MF. Oligomerization activity of a double-stranded RNA-binding domain. FEBS Lett. 2004;574:25–30. doi: 10.1016/j.febslet.2004.07.080. [DOI] [PubMed] [Google Scholar]
- 56.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 57.Yekta S, I, Shih H, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–6. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 58.Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–74. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 59.Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′ UTR. Genes Dev. 2004;18:132–7. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vella MC, Reinert K, Slack FJ. Architecture of a validated microRNA::target interaction. Chem Biol. 2004;11:1619–23. doi: 10.1016/j.chembiol.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 61.Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–50. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 62.Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–6. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 63.Nykanen A, Haley B, Zamore PD. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–21. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- 64.Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, Rappsilber J, Mann M, Dreyfuss G. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16:720–8. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–40. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 66.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–20. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 67.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–9. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 68.Plante I, Davidovic L, Ouellet DL, Gobeil LA, Tremblay S, Khandjian EW, Provost P. Dicer-Derived MicroRNAs Are Utilized by the Fragile X Mental Retardation Protein for Assembly on Target RNAs. J Biomed Biotechnol. 2006;2006:64347. doi: 10.1155/JBB/2006/64347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Plante I, Provost P. Hypothesis: A Role for Fragile X Mental Retardation Protein in Mediating and Relieving MicroRNA-Guided Translational Repression? J Biomed Biotechnol. 2006;2006:16806. doi: 10.1155/JBB/2006/16806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Robb GB, Rana TM. RNA helicase A interacts with RISC in human cells and functions in RISC loading. Mol Cell. 2007;26:523–37. doi: 10.1016/j.molcel.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 71.Kataoka Y, Takeichi M, Uemura T. Developmental roles and molecular characterization of a Drosophila homologue of Arabidopsis Argonaute1, the founder of a novel gene superfamily. Genes Cells. 2001;6:313–25. doi: 10.1046/j.1365-2443.2001.00427.x. [DOI] [PubMed] [Google Scholar]
- 72.Cerutti L, Mian N, Bateman A. Domains in gene silencing and cell differentiation proteins: the novel PAZ domain and redefinition of the Piwi domain. Trends Biochem Sci. 2000;25:481–2. doi: 10.1016/s0968-0004(00)01641-8. [DOI] [PubMed] [Google Scholar]
- 73.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–97. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 74.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–41. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 75.Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol. 2005;12:340–9. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- 76.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–7. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 77.Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–70. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Parker JS, Roe SM, Barford D. Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. Embo J. 2004;23:4727–37. doi: 10.1038/sj.emboj.7600488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Parker JS, Roe SM, Barford D. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature. 2005;434:663–6. doi: 10.1038/nature03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yan KS, Yan S, Farooq A, Han A, Zeng L, Zhou MM. Structure and conserved RNA binding of the PAZ domain. Nature. 2003;426:468–74. doi: 10.1038/nature02129. [DOI] [PubMed] [Google Scholar]
- 81.Lingel A, Simon B, Izaurralde E, Sattler M. Nucleic acid 3′-end recognition by the Argonaute2 PAZ domain. Nat Struct Mol Biol. 2004;11:576–7. doi: 10.1038/nsmb777. [DOI] [PubMed] [Google Scholar]
- 82.Lingel A, Simon B, Izaurralde E, Sattler M. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature. 2003;426:465–9. doi: 10.1038/nature02123. [DOI] [PubMed] [Google Scholar]
- 83.Yuan YR, Pei Y, Ma JB, Kuryavyi V, Zhadina M, Meister G, Chen HY, Dauter Z, Tuschl T, Patel DJ. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol Cell. 2005;19:405–19. doi: 10.1016/j.molcel.2005.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–60. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 85.Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–42. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–11. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jin P, Zarnescu DC, Ceman S, Nakamoto M, Mowrey J, Jongens TA, Nelson DL, Moses K, Warren ST. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat Neurosci. 2004;7:113–7. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- 88.O’Donnell WT, Warren ST. A decade of molecular studies of fragile X syndrome. Annu Rev Neurosci. 2002;25:315–38. doi: 10.1146/annurev.neuro.25.112701.142909. [DOI] [PubMed] [Google Scholar]
- 89.Bardoni B, Mandel JL. Advances in understanding of fragile X pathogenesis and FMRP function, and in identification of X linked mental retardation genes. Curr Opin Genet Dev. 2002;12:284–93. doi: 10.1016/s0959-437x(02)00300-3. [DOI] [PubMed] [Google Scholar]
- 90.Laggerbauer B, Ostareck D, Keidel EM, Ostareck-Lederer A, Fischer U. Evidence that fragile X mental retardation protein is a negative regulator of translation. Hum Mol Genet. 2001;10:329–38. doi: 10.1093/hmg/10.4.329. [DOI] [PubMed] [Google Scholar]
- 91.Li Z, Zhang Y, Ku L, Wilkinson KD, Warren ST, Feng Y. The fragile X mental retardation protein inhibits translation via interacting with mRNA. Nucleic Acids Res. 2001;29:2276–83. doi: 10.1093/nar/29.11.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mazroui R, Huot ME, Tremblay S, Filion C, Labelle Y, Khandjian EW. Trapping of messenger RNA by Fragile X Mental Retardation protein into cytoplasmic granules induces translation repression. Hum Mol Genet. 2002;11:3007–17. doi: 10.1093/hmg/11.24.3007. [DOI] [PubMed] [Google Scholar]
- 93.Costa A, Wang Y, Dockendorff TC, Erdjument-Bromage H, Tempst P, Schedl P, Jongens TA. The Drosophila fragile X protein functions as a negative regulator in the orb autoregulatory pathway. Dev Cell. 2005;8:331–42. doi: 10.1016/j.devcel.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 94.Zhang YQ, Bailey AM, Matthies HJ, Renden RB, Smith MA, Speese SD, Rubin GM, Broadie K. Drosophila fragile X-related gene regulates the MAP1B homolog Futsch to control synaptic structure and function. Cell. 2001;107:591–603. doi: 10.1016/s0092-8674(01)00589-x. [DOI] [PubMed] [Google Scholar]
- 95.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7:719–23. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–82. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Eystathioy T, Chan EK, Tenenbaum SA, Keene JD, Griffith K, Fritzler MJ. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol Biol Cell. 2002;13:1338–51. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Eystathioy T, Jakymiw A, Chan EK, Seraphin B, Cougot N, Fritzler MJ. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA. 2003;9:1171–3. doi: 10.1261/rna.5810203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu J, Rivas FV, Wohlschlegel J, Yates JR, 3rd, Parker R, Hannon GJ. A role for the P-body component GW182 in microRNA function. Nat Cell Biol. 2005;7:1261–6. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pauley KM, Eystathioy T, Jakymiw A, Hamel JC, Fritzler MJ, Chan EK. Formation of GW bodies is a consequence of microRNA genesis. EMBO Rep. 2006;7:904–10. doi: 10.1038/sj.embor.7400783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–24. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]