Abstract
Xanthomonas oryzae pv. oryzae causes bacterial blight, a serious disease of rice. Our analysis revealed that the X. oryzae pv. oryzae genome encodes genes responsible for iron uptake through FeoB (homolog of the major bacterial ferrous iron transporter) and a siderophore. A mutation in the X. oryzae pv. oryzae feoB gene causes severe virulence deficiency, growth deficiency in iron-limiting medium, and constitutive production of a siderophore. We identified an iron regulated xss gene cluster, in which xssABCDE (Xanthomonas siderophore synthesis) and xsuA (Xanthomonas siderophore utilization) genes encode proteins involved in biosynthesis and utilization of X. oryzae pv. oryzae siderophore. Mutations in the xssA, xssB, and xssE genes cause siderophore deficiency and growth restriction under iron-limiting conditions but are virulence proficient. An xsuA mutant displayed impairment in utilization of native siderophore, suggesting that XsuA acts as a specific receptor for a ferric-siderophore complex. Histochemical and fluorimetric assays with gusA fusions indicate that, during in planta growth, the feoB gene is expressed and that the xss operon is not expressed. This study represents the first report describing a role for feoB in virulence of any plant-pathogenic bacterium and the first functional characterization of a siderophore-biosynthetic gene cluster in any xanthomonad.
Iron is one of the most important nutrients for bacterial growth. Although iron is abundant in nature, its availability is limiting because of low solubility (9). Bacteria encode multiple iron uptake pathways, which provide specificities and affinities for various forms of environmental or host iron. Under iron-limited conditions bacteria secrete siderophores (low-molecular-weight iron chelators), which facilitate ferric iron acquisition. This ferric-siderophore complex is taken up into bacterial cells by a transporter system comprised of a TonB-ExbBD-dependent outer membrane receptor, a periplasmic binding protein, and an inner membrane ABC permease complex (33, 46). Siderophores can be classified into three major classes based on the ligand that participates in chelating iron and are either catecholates (e.g., enterobactin), hydroxamates (e.g., alcaligin), or α-hydroxy carboxylates (e.g., vibrioferrin). Some siderophores belong to a class of mixed-type siderophores (e.g., aerobactin) (43). A number of siderophores are synthesized as small peptides by multimodular enzymes known as nonribosomal peptide synthetases (NRPSs) (17) while other siderophores are nonpolypeptidic in nature and are synthesized via NRPS-independent pathways (13).
Extensive studies have shown the importance of siderophores in virulence of a number of animal-pathogenic bacteria. For instance, the siderophore pyoverdine is essential for virulence of Pseudomonas aeruginosa, and production of yersiniabactin is required for full virulence of Yersinia pestis and Klebsiella pneumoniae (3, 34, 42). The plant-pathogenic bacterium Erwinia chrysanthemi strain 3937, which causes soft rot disease on a variety of plants, produces two siderophores, namely, achromobactin and chrysobactin, both of which are required for optimal virulence on African violets (Saintpaulia ionantha) (23). Siderophore-deficient mutants of Erwinia amylovora are virulence deficient on apple (22). However, Pseudomonas syringae pv. syringae B301D, a pathogen of stone fruit trees, does not require pyoverdine for virulence (16). P. syringae pv. tomato DC3000, the causal agent of bacterial speck disease of tomato, produces yersiniabactin as well as pyoverdine. Yersiniabactin is produced in planta, and the pchA gene encoding isochorismate synthase is required to synthesize it. However, a pchA mutant of this bacterium does not show any growth defect or altered virulence on tomato and Arabidopsis hosts (30). Also, siderophore-deficient mutants of the plant pathogens Ralstonia solanacearum, Erwinia carotovora subsp. carotovora, and Agrobacterium tumefaciens retain virulence proficiency (5, 11, 38, 58). These findings suggest that there is considerable variation in the contributions of siderophores to plant-bacteria interactions.
In addition to the uptake of ferric iron, many bacteria take up free ferrous iron via the Feo (ferrous iron transporter) system (12). The Feo system encoded by the feoABC operon is well characterized in Escherichia coli. The feoB gene encodes the major bacterial ferrous iron transporter composed of a hydrophilic cytoplasmic domain and an integral membrane domain (31). The cytoplasmic domain of the FeoB protein in E. coli has been shown to act as a GTPase and exhibits sequence similarities to eukaryotic G proteins (40). Binding of GTP/GDP is required for efficient uptake of ferrous iron through the FeoB-dependent transport system. The roles of feoA and feoC are not known, but they are predicted to encode a GTPase-activating protein (GAP) and an Fe-S cluster containing a transcriptional regulator, respectively (12). In animal-pathogenic bacteria such as E. coli, Helicobacter pylori, Legionella pneumophila, Salmonella enterica serovar Typhimurium, Porphyromonas gingivalis, and Campylobacter jejuni, mutation in the feoB gene has been shown to cause deficiency in ferrous iron uptake and virulence (8, 19, 45, 57, 65, 71). There is no report on the role of feoB in virulence of any plant-pathogenic bacterium.
Although iron is essential for bacterial growth, excess intracellular iron is toxic because of the generation of free radicals (70). Hence, the expression of iron acquisition functions must be tightly controlled, and this function is performed by the ferric uptake regulator (Fur) protein. By using ferrous ion as a corepressor, Fur functions as a transcriptional repressor of iron uptake-related genes in bacteria. Fur binds to specific sequences called Fur boxes that are located in the promoter regions of such genes (51).
The Gram-negative bacterial genus Xanthomonas comprises bacteria that cause almost ∼400 different plant diseases (14). In Xanthomonas campestris pv. campestris, which causes black rot of cruciferous plants, a tonB mutant is impaired for ferric ion uptake and exhibits increased extracellular siderophore production and reduced disease symptoms (72, 73). In Xanthomonas oryzae pv. oryzae, causal agent of the serious bacterial blight disease of rice, a fur mutant exhibits a severe in planta growth deficiency that appears to be due to increased oxidative stress (66). In X. campestris pv. phaseoli, which causes common bacterial blight of beans, the Fur protein has been characterized with respect to primary structure (39).
X. oryzae pv. oryzae uses a diverse array of functions to grow within the xylem vessels of rice leaves and cause disease (47). In this study, we have assessed the role of two different iron uptake systems in promoting virulence of X. oryzae pv. oryzae. We have mutated the feoB homolog of this bacterium and demonstrated that this results in a severe virulence deficiency. We have also determined that the xss operon of X. oryzae pv. oryzae is involved in siderophore biosynthesis and transport. Mutations in the xss operon result in siderophore deficiency but do not affect virulence.
MATERIALS AND METHODS
Bacterial strains, plasmids, primers, and culture media.
Bacterial strains and plasmids used in this study are listed in Table 1. Primers and degenerate primers used in this work are listed in Table 2 and Table 3, respectively. Degenerate primers were designed based on sequences of X. campestris pv. campestris strain ATCC 33913 and Xanthomonas axonopodis pv. citri strain 306 using the CODEHOP (consensus-degenerate hybrid oligonucleotide primers) program (59). X. oryzae pv. oryzae strains were grown at 28°C in peptone sucrose (PS) medium, and E. coli strains were grown at 37°C in Luria Bertani (LB) medium as described previously (55). To minimize contamination with exogenous iron, MilliQ water and acid-washed glassware were used in this study. The concentrations of antibiotics used in this study were as follows: rifampin (Rf), 50 μg/ml; ampicillin (Ap), 100 μg/ml; spectinomycin (Sp), 50 μg/ml; kanamycin (Km), 50 μg/ml for E. coli and 15 μg/ml for X. oryzae pv. oryzae; and tetracycline (Tc). 15 μg/ml for E. coli and 5 μg/ml for X. oryzae pv. oryzae.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| E. coli strains | ||
| DH5α | λ− f80dlacZDM15 D(lacZYA-argF)U169 recA1 endA hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | Invitrogen |
| S17-1 | RP4-2 Tc::Mu-Km::Tn7 pro hsdR recA Tra+; used as a mobilizing strain | 64 |
| X. oryzae pv. oryzae strains | ||
| BXO1 | Wild type; Indian isolate | Lab collection |
| BXO43 | rif-2; derivative of BXO1 | Lab collection |
| BXO2201 | feoB1::pK18mob rif-2; Kmr; derived from BXO43 | This work |
| BXO2204 | feoB1::pK18mob rif-2/pHM1; Kmr Spr | This work |
| BXO2206 | feoB1::pK18mob rif-2/pAP3 feoB+; Kmr Spr | This work |
| BXO2211 | xssE1::pK18mob rif-2; Sid− Kmr; derived from BXO43 | This work |
| BXO2214 | xssB1::pK18mob rif-2; Sid− Kmr; derived from BXO43 | This work |
| BXO2217 | xssA1::pK18mob rif-2; Sid− Kmr; derived from BXO43 | This work |
| BXO2220 | mhpE1::pK18mob rif-2; Sid+ Kmr; derived from BXO43 | This work |
| BXO2223 | xssE1::pK18mob rif-2/pAP15; Sid+ Kmr Tcr | This work |
| BXO2224 | xssB1::pK18mob rif-2/pAP15; Sid+ Kmr Tcr | This work |
| BXO2225 | xssA1::pK18mob rif-2/pAP15; Sid+ Kmr Tcr | This work |
| BXO2229 | xssE2::gusA rif-2; Sid− Kmr; derived from BXO43 | This work |
| BXO2230 | feo::gusA rif-2; KmrfeoB+; derived from BXO43 | This work |
| BXO2231 | xsuA1::gusA rif-2; Kmr; derived from BXO43 | This work |
| BXO2233 | xsuA1::gusA rif-2/pAP23; Kmr Tcr | This work |
| BXO2234 | xsuA1::gusA rif-2/pAP15; Kmr Tcr | This work |
| BXO2235 | xsuA2::pK18mob rif-2; Kmr; derived from BXO43c | This work |
| BXO2236 | xsuA3::pK18mob rif-2; Kmr; derived from BXO43b | This work |
| Plasmids | ||
| pCR-Blunt II-TOPO | PCR cloning vector; Kmr | Invitrogen |
| pMOSblue | High-copy-number cloning vector; Apr | Amersham Pharmacia Biotech |
| pK18mob | pUC18 derivative; Mob+ Kmr; does not replicate in X. oryzae pv. oryzae | 62 |
| pHM1 | Broad-host-range cosmid vector (∼13.3 kb); Spr | 28 |
| pBSKS+ | E. coli cloning vector; Apr | Stratagene |
| pUFR034 | IncW; Kmr Mob+mob (P) lacZα+ Par+cos (8.7 kb) | 21 |
| pTc28 | Source of tetracycline cassette; Tcr Apr | 56 |
| pVO155 | pUC119 derivative carrying promoterless gusA; Kmr Apr | 48 |
| pAP1 | pCR-Blunt II-TOPO with 510-bp internal fragment of feoB | This work |
| pAP2 | pK18mob with 510-bp fragment of feoB (from pAP1) cloned in to EcoRI site of pK18mob | This work |
| pAP3 | pHM1 with 2,119-bp fragment containing full length feoB cloned in to HindIII and SacI sites of pHM1 | This work |
| pAP4 | pCR-Blunt II-TOPO with 503-bp internal fragment of xssE | This work |
| pAP5 | pK18mob with 503-bp fragment of xssE (from pAP4) cloned in to EcoRI site of pK18mob | This work |
| pAP6 | pMOSblue with 553-bp internal fragment of xssB | This work |
| pAP7 | pK18mob with 553-bp fragment of xssB (from pAP6) cloned in to EcoRI and HindIII sites of pK18mob | This work |
| pAP8 | pCR-Blunt II-TOPO with 512-bp internal fragment of xssA | This work |
| pAP9 | pK18mob with 512-bp fragment of xssA (from pAP8) cloned in to PstI and HindIII sites of pK18mob | This work |
| pAP10 | pMOSblue with 339-bp internal fragment of mhpE | This work |
| pAP11 | pK18mob with 339-bp fragment of mhpE (from pAP10) cloned in to EcoRI and HindIII sites of pK18mob | This work |
| pAP12 | pUFR034 with 38.2-kb genomic insert of X. oryzae pv. oryzae containing entire xss gene cluster | This work |
| pAP13 | ∼2.4kb tet cassette from pTc28 cloned in to the XbaI and KpnI sites of pMOSblue; Tcr Apr | This work |
| pAP14 | pBSKS with 2.4-kb tet cassette (from pAP13) cloned in to XbaI and SacI sites of pBSKS; Tcr Apr | This work |
| pAP15 | Tcr Kms; derivative of pAP12 (tet cassette from pAP14 cloned in to XhoI site of pAP12) | This work |
| pAP16 | pVO155 with 503-bp fragment of xssE (from pAP4) cloned in to SpeI and XbaI sites of pVO155 | This work |
| pAP17 | pVO155 carrying 343-bp DNA (putative promoter region, +139 to −114) upstream of feoABC operon | This work |
| pAP18 | pCR-Blunt II-TOPO with 505-bp internal fragment of xsuA | This work |
| pAP19 | pVO155 with 505-bp fragment of xsuA (from pAP18) cloned in to SpeI and XhoI sites of pVO155 | This work |
| pAP20 | pK18mob with 505-bp fragment of xsuA (from pAP18) cloned in to EcoRI site of pK18mob; nonpolar version (lacZ promoter of pK18mob and xsuA gene fragment are in the same transcriptional orientation) | This work |
| pAP21 | pK18mob with 505-bp fragment of xsuA (from pAP18) cloned in to EcoRI site of pK18mob; polar version (lacZ promoter of pK18mob and xsuA gene fragment are in opposite transcriptional orientations) | This work |
| pAP22 | A 9.9-kb DNA fragment containing xsuA gene from pAP12 cloned into EcoRI site of pUFR034 | This work |
| pAP23 | Tcr Kms; derivative of pAP22 (tet cassette from pAP14 cloned into XhoI site of pAP22) | This work |
The rif-2 mutation confers resistance to rifampin; Apr, Spr, Tcr, and Kmr indicate resistance to ampicillin, spectinomycin, tetracycline, and kanamycin, respectively. Kms indicates kanamycin sensitivity. Sid+ and Sid− indicate proficiency and deficiency in siderophore production, respectively.
Polar mutation in xsuA.
Nonpolar mutation in xsuA.
TABLE 2.
Oligonucleotide primers used in this study
| Primer name | Nucleotide sequence (5′-3′ [restriction site])a |
|---|---|
| M13F | GTAAAACGACGGCCAGT |
| M13R | GGAAACAGCTATGACCATG |
| T7 | TAATACGACTCACTATAGGG |
| FeoBF | CGGTGGTGATGTTCCTGATCTT |
| FeoBR | ACAGCACCAATCCCTGCTGATT |
| FeoBFF | GACGTGCTGGTCTACGTGA |
| FeoBRR | CGGCCTGCGAAGCTGTAGT |
| FeoAHF | CCCAAGCTTGGGGAGGAAGTCACGGTGCTGGC (HindIII) |
| FeoCSR | CGAGCTCGGACCACGTGCAAGCGCACGA (SacI) |
| 1354F | CAAACTCGATACCGAGCTCGC |
| 1354R | GCAGGTCTGTGCCATGTTCG |
| 1354FF | GCGCATACGTCTACGACCT |
| 1354RR | TGCCAGCACATCCTTGGGT |
| 1357F | CCGGCAGAGTTATGGCCGAGT |
| 1357R | ACCAGTGCAGCACATCGCCGT |
| 1357FF | AACATGGTACGCACGCATGGT |
| 1357RR | GCGCGGGCTGCCATTCGGTAA |
| 1358F | CTATGCCGCCAAGAACAAGGC |
| 1358R | CGATGCTGCGGTAATTGATCT |
| 1358FF | ATCACCTGGCCTATTTCCACG |
| 1358RR | CACGCCCAGGTAATCCTTGTT |
| 1359F | AGCGTTATCTGGTCGACAT |
| 1359R | GTAATGGCGTTGCTGACGTA |
| 1359FF | AGATCGACCGCATCGACAT |
| 1359RR | TGTTATCCAGTTGGATCTGCT |
| 1360F | CGATGAACTTGAACATGCCATC |
| 1360R | ACGGCGAGAATTTCCGGAAG |
| 1360FF | CTCAATGCAGTGCCAAGCGT |
| 1360RR | CAATGCGCGTTGAATCAGTG |
| MF1 | GCTGCCTGGGCGAGCTCGGTATC |
| MR2 | CTGCAGAACACGGTGATCGGTTTC |
| MF3 | CACTCATGCGTTGCAACCATGC |
| MR4 | CGGTGCTTGTGGAGATCAATTAC |
| MF5 | CGGTTTCGTCGAAGATCTTGAT |
| MR6 | CATCGTGCCGATGATCGAAAG |
| FBPFB | CGGGATCCCGCGATCAGTGTCGCATCGAAG (BamHI) |
| FBPFX | GCTCTAGAGCCACCGTGACTTCCTCGCCAG (XbaI) |
| PVO155F | CCGCCTAATGAGCGGGCTT |
| PVO155R | GATCCAGACTGAATGCCCACA |
| FeoAF | GAGGAAGTCACGGTGCTGGC |
| FeoCR | GACCACGTGCAAGCGCACGA |
| FeoBF2 | GGACGATGAAGAAGTGGCG |
| FeoBR1 | CCAGATCGGATGCAGCAAC |
The engineered restriction modification sites are underlined. All of the primers were developed in the course of this study.
TABLE 3.
List of degenerate oligonucleotide primers used in this study
| Primer name | Target genea | Nucleotide sequence (5′-3′)b |
|---|---|---|
| AP2F | xssA | CCTGCCGGGCAAGGAYTGGCARGT |
| AP2R | GGCGTCCCACTGGGCYTCRCAYTC | |
| AP3F | xssB | CCACTACGCCCCGGARTTYGCNCC |
| AP3R | AAGCTCTCGGCCAGGGDATNGCCAT | |
| AP4F | xssD | GCGTGCGCAAGAACGCNTGGTAYGA |
| AP4R | CGCGGGTCGGCCANCCRTC | |
| AP5F | xssE | GGTGTTCCTGCGCATGAAYATHGC |
| AP5R | CGCGGCACGGGTGRTCRTGNCC | |
| AP6F | xsuA | GCGCGTGCGCGCNGARTGGTG |
| AP6R | CGCCGGCGGTCCAYTGYTG | |
| AP7F | xssC | CCCGCGCCCTGGAYTGGACNCA |
| AP7R | CGTCGCCGCGGCKCCACCANGG |
Intragenic fragments were amplified from BXO43 genomic DNA.
The region of degeneracy is indicated by boldface letters. In the sequences, Y is C or T; R is A or G; K is G or T; D is A, G, or T; H is A, C, or T; and N is A, C, G, or T. All of the primers were developed in the course of this study.
Molecular biological and microbiological techniques.
Genomic DNA isolation was performed as described by Leach et al. (35). Plasmid DNA was isolated by the alkaline lysis method (61) or by using a Qiagen plasmid midi-kit. Phusion polymerase (Finnzymes) was used for PCR amplification in applications that required high-fidelity DNA synthesis (such as for complementation analysis) while Taq polymerase was used for all other applications. Restriction digestions were done with enzymes from New England Biolabs (NEB; Beverly, MA). PCR products and restriction enzyme-digested DNA fragments were purified using a QIAquick PCR purification kit and a QIAquick nucleotide removal kit (Qiagen), respectively. Other techniques such as ligation with T4 DNA ligase, agarose gel electrophoresis, and transformation of E. coli were performed as described previously (61). Plasmids were transferred from E. coli to X. oryzae pv. oryzae using either broad-host-range mobilizing strain S17-1 (64) for biparental matings or electroporation (55, 66).
DNA sequencing and bioinformatics tools.
Sequencing was performed with an ABI Prism 3700 automated DNA sequencer (Perkin-Elmer, Foster City, CA). Homology and conserved domain searches were performed in the NCBI database by using the BLAST algorithm (2). Pairwise alignment was carried out using Clustal W (69). The presence of transmembrane helices in proteins was predicted using TMpred and TMHMM, version 2.0, programs at the ExPASy proteomics server. The PSORTb program was used for the prediction of subcellular localization of bacterial proteins (26).
RNA isolation, operon mapping, and expression analysis.
To investigate the transcriptional organization and expression of feoABC genes and the xss gene cluster, reverse transcription-PCR (RT-PCR) was performed with total RNA isolated from X. oryzae pv. oryzae strain BXO43 grown under iron-supplemented and iron-limiting conditions. A culture of X. oryzae pv. oryzae grown in PS medium to an optical density at 600 nm (OD600) of 0.5 was split into two parts. Ferrous sulfate (FeSO4) was added at 100 μM to one part (iron-supplemented conditions), and 2,2′-dipyridyl (DP; an iron chelator) was added at 100 μM to the other part (iron-limiting conditions). Both cultures were further incubated for 6 h, and total RNA from each of the samples was isolated using TRIzol reagent (Invitrogen). Total RNA concentration was determined spectrophotometrically. RNA samples were treated with RNase-free DNase I (Promega) for 1 h at 37°C to remove any DNA contamination. A total of 5 μg of RNA was reverse transcribed using SuperscriptIII RT enzyme (Invitrogen) as per the supplier's instructions. For feo operon mapping, cDNA synthesis was performed with primer FeoCR, complementary to the feoC gene. The resulting cDNAs were used as a template for PCR amplification with Taq polymerase using specific primer pairs for each gene. The following primer pairs were used: FeoCR/FeoBF2 for amplification of feoCB and FeoBR1/FeoAF for the feoBA region. To detect the presence or absence of feoB transcripts, PCR was done with FeoBF and FeoBR primers. To determine the transcriptional organization and iron-mediated regulation of the xss gene cluster, cDNA synthesis was done using RNA isolated from iron-supplemented and iron-limiting conditions, with primer MF1 which is complementary to the xssE gene. Subsequent PCR was done with cDNA as a template. To amplify the xssE-xssD, xssB-xssA, and xsuA-mhpE regions, MF1/MR2, MF3/MR4, and MF5/MR6 primer pairs were used, respectively. RT-PCR products were analyzed on a 1.2% agarose gel along with a 100-bp DNA ladder (NEB). Sequencing of RT-PCR-amplified products was done to confirm their identity.
Construction of feo::gusA and xssE::gusA strains.
Glucuronidase (GUS) reporter gene fusions were created by using the suicide plasmid pVO155 that has the promoterless gusA gene (48). To construct the feo::gusA transcriptional fusion, a 343-bp DNA fragment containing the putative promoter of the feoABC operon (+139 to −224) was amplified by using the FBPFB/FBPFX primers. This promoter fragment was subsequently digested with BamHI and XbaI and directionally cloned upstream of the promoterless gusA gene in to similarly digested pVO155 to create the feo::gusA reporter construct pAP17. To create an xssE::gusA fusion, a 503-bp PCR-amplified internal fragment of the xssE gene was cloned into the pCR-Blunt-II-TOPO vector to obtain the plasmid pAP4. This plasmid was digested with SpeI and XbaI, and the released fragment was directionally cloned upstream of the promoterless gusA gene into the similarly digested pVO155 to create the xssE::gusA reporter construct, pAP16. After confirmation by sequencing, the resulting pAP17 and pAP16 constructs were then introduced in to strain BXO43 by biparental mating using E. coli S17-1. X. oryzae pv. oryzae GUS reporter strains were selected on medium plates containing rifampin (counter-selectable marker) and kanamycin. Since pVO155 cannot replicate in X. oryzae pv. oryzae, kanamycin-resistant colonies are obtained upon chromosomal integration of the plasmid using the cloned DNA sequence as a region of homology. Chromosomal integration of the plasmid was confirmed by PCR using flanking primers (from the chromosomal region of interest) and the vector-specific primers PVO155F/PVO155R and sequencing of the PCR product. The feo::gusA and xssE::gusA transconjugants were named BXO2230 and BXO2229, respectively. In BXO2230, PfeoABC is fused to gusA and leaves an intact functional feoABC operon with its associated native promoter. In BXO2229, the gusA fusion leads to disruption of xssE.
β-Glucuronidase reporter assays.
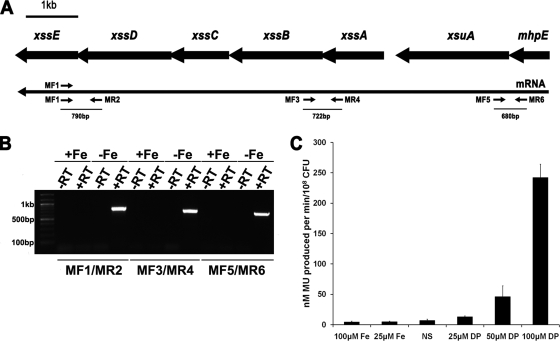
GUS reporter strains were grown to an OD600 of 0.6 in PS medium. Aliquots of 1.1 ml of the cell suspension were taken, and either FeSO4 (at a final concentration of 25 μM or 100 μM to obtain iron supplementation) or DP (at a final concentration of 25 μM, 50 μM, or 100 μM to cause iron limitation) was added. In addition to the above, several 1.1-ml aliquots of the strains were grown without any supplementation. All of these cultures were incubated at 28°C for a further period of 6 h, and 100 μl was removed to determine the number of CFU/ml. The remaining 1 ml of culture was centrifuged, and the obtained pellet was then washed once in sterile MilliQ water to remove the residual supplements. The cell pellets were resuspended in 250 μl of 1 mM MUG (4-methylumbelliferyl β-d-glucuronide) extraction buffer (50 mM sodium dihydrogen phosphate [pH 7.0], 10 mM EDTA, 0.1% Triton X-100, 0.1% sodium lauryl sarcosine, and 10 mM β-mercaptoethanol) (29) and incubated at 37°C. Subsequently, 25-μl aliquots were taken from each reaction mixture, and the reaction was terminated at suitable time intervals by the addition of 225 μl of 0.2 M Na2CO3. Fluorescence was measured on a Spectramax Gemini XS dual scanning microplate spectrofluorimeter (Molecular Devices) with 4-methyl-umbelliferone (MU; Sigma) as the standard. β-Glucuronidase activity was expressed as nanomoles of MU produced/minute/108 CFU.
Generation and complementation of the X. oryzae pv. oryzae feoB mutant.
The X. oryzae pv. oryzae feoB gene was disrupted by homologous suicide plasmid integration using vector pK18mob (62). A 510-bp internal fragment of the feoB gene was amplified by PCR using genomic DNA of X. oryzae pv. oryzae strain BXO43 and the gene-specific FeoBF/FeoBR primer pair and cloned into pCR-Blunt II-TOPO vector (Invitrogen) to generate the plasmid pAP1. The cloned DNA fragment was excised from pAP1 using EcoRI and further cloned into EcoRI-digested pK18mob to create the plasmid pAP2. This construct was then transferred from E. coli strain S17-1 to X. oryzae pv. oryzae via biparental mating, and feoB gene disruptants were selected on kanamycin-containing medium. Gene disruption was confirmed by PCR using flanking gene-specific primers FeoBFF/FeoBRR (which bind in chromosomal regions that flank the insert sequence cloned in pAP2) in combination with vector-specific primers M13F and M13R and sequencing of the PCR products. In pAP2, the transcriptional orientations of the lacZ promoter of pK18mob and of the feoB gene fragment were in the same direction so that mutation caused by pAP2 integration is unlikely to cause any polar effect on downstream genes due to activity of the lacZ promoter (74). One of the confirmed mutants was named BXO2201 and was chosen for further study. For feoB mutant complementation, a 2,119-bp DNA fragment containing the entire feoB coding region with its own ribosomal binding site and extending from 125 bp upstream of the start codon to 128 bp downstream of the stop codon of the feoB gene was amplified by PCR using the total DNA of X. oryzae pv. oryzae and primers FeoAHF/FeoCSR. The amplified fragment was cloned as an HindIII-SacI fragment into broad-host-range vector pHM1 (28) to create recombinant plasmid pAP3. The pAP3 clone and pHM1 vector were individually introduced in to the feoB mutant strain (BXO2201) to obtain strains BXO2206 (feoB/pAP3; for complementation) and BXO2204 (feoB/pHM1; control).
Isolation of a cosmid clone containing the entire xss gene cluster from a X. oryzae pv. oryzae genomic library.
A genomic library of X. oryzae pv. oryzae (average insert size of 30 kb) had been constructed in the cosmid vector pUFR034 (21) and maintained in E. coli strain S17-1 (54). A total of 480 clones were screened by PCR using 1354F/1354R (xssE-specific), 1358F/1358R (xssA-specific) and 1360F/1360R (mhpE-specific) primer pairs. Five positive clones were identified, and one of these clones (named pAP12) with an insert size of ∼38.2 kb was characterized further. End sequencing of this clone was performed with M13F and M13R primers, which indicated that the cloned DNA extends from the XOO1344 gene to the XOO1371 gene (as per nomenclature for X. oryzae pv. oryzae strain KACC10331 [36]) while the entire xss gene cluster ranges from XOO1354 to XOO1360.
Construction and complementation analyses of xssA, xssB, xssE, mhpE, and xsuA mutants.
The xssA, xssB, xssE, and mhpE genes were each disrupted by integration of suicide plasmid pK18mob. Three different xsuA mutants were created by integration of either pK18mob or pVO155 (Table 4 lists the primers and plasmids used, steps for mutant construction, and the designation for each mutant).
TABLE 4.
Construction of X. oryzae pv. oryzae xssE, xssB, xssA, mhpE, and xsuA mutants
| Gene | Primer pair for amplification (length of internal gene fragment [bp]) | Plasmid clone (cloning vector) | Restriction enzyme(s) | Plasmid clone used for gene disruption (suicide vector)a | Mutant strain | Primer pair used to confirm gene disruption |
|
|---|---|---|---|---|---|---|---|
| Gene specific | Vector specific | ||||||
| xssE | 1354F/1354R (503) | pAP4 (pCR-Blunt-II-TOPO) | EcoRI | pAP5 (pK18mob)b | BXO2211 | 1354FF/1354RR | M13F/M13R |
| xssB | 1357F/1357R (553) | pAP6 (pMOSblue) | EcoRI-HindIII | pAP7 (pK18mob)b | BXO2214 | 1357FF/1357RR | M13F/M13R |
| xssA | 1358F/1358R (512) | pAP8 (pCR-Blunt-II-TOPO) | PstI-HindIII | pAP9 (pK18mob)b | BXO2217 | 1358FF/1358RR | M13F/M13R |
| mhpE | 1360F/1360R (339) | pAP10 (pMOSblue) | EcoRI-HindIII | pAP11 (pK18mob)b | BXO2220 | 1360FF/1360RR | M13F/M13R |
| xsuA | 1359F/1359R (505) | pAP18 (pCR-Blunt-II-TOPO) | SpeI-XhoI | pAP19 (pVO155) | BXO2231 | 1359FF/1359RR | PVO155F/PVO155R |
| EcoRI | pAP20 (pK18mob)b | BXO2235 | 1359FF/1359RR | M13F/M13R | |||
| EcoRI | pAP21 (pK18mob)c | BXO2236 | |||||
These constructs were transferred from E. coli strain S17-1 to X. oryzae pv. oryzae via biparental matings, and gene disruptants were selected on kanamycin-containing medium.
The transcriptional orientation of the lacZ promoter of pK18mob is in the same direction as the cloned gene fragment. Therefore, integration of the plasmid is likely to result in a nonpolar mutation.
The transcriptional orientation of the lacZ promoter of pK18mob is in the opposite direction of the cloned gene fragment. Therefore, plasmid integration will result in a polar mutation.
A kanamycin resistance determinant is present in the xss mutants (BXO2211, BXO2214, and BXO2217) as well as in the pAP12 cosmid. Therefore, the kanamycin resistance gene of pAP12 was inactivated by insertion of a tetracycline (tet) resistance cassette. To do so, a 2.4-kb tet cassette was excised from the pTc28 vector (56) using XbaI and KpnI and subsequently cloned into the XbaI and KpnI site of pMOSBlue to obtain pAP13. The tet cassette was released from pAP13 by double digestion with XbaI and SacI and then cloned into similarly digested the pBSKS+ (Stratagene) plasmid to obtain pAP14. Subsequently, pAP14 was digested with XhoI to release the tet cassette, which was ligated to XhoI-digested pAP12 to generate pAP15, which was electroporated into BXO2211, BXO2214, and BXO2217 (by selection for tetracycline resistance) to generate complemented strains BXO2223, BXO2224, and BXO2225, respectively. For genetic complementation of BXO2231 (xsuA mutant), a 9.9-kb EcoRI fragment, which includes the full-length mhpE and xsuA genes but not the xss gene, was obtained after digestion of the cosmid pAP12 with EcoRI. This 9.9-kb DNA fragment was cloned into EcoRI-digested pUFR034 to obtain the cosmid pAP22. The kanamycin resistance marker of cosmid pAP22 was then inactivated by insertion of the tet cassette. This was accomplished by digesting pAP14 with XhoI to release the tet cassette, which was ligated to XhoI-digested pAP22 to generate the cosmid pAP23. Cosmids pAP23 and pAP15 were electroporated into BXO2231 by selection for tetracycline resistance to generate complemented strains BXO2233 and BXO2234, respectively.
Siderophore production assay.
Peptone-sucrose agar (PSA)-chrome azurol sulfonate (CAS) siderophore indicator plates (63) were prepared as described previously (15). In order to create iron-limiting conditions, DP (100 μM) was added to PSA-CAS plates. The colonies were then scored for the orange halo phenotype that is indicative of siderophore production. Csaky and Arnow tests (52) were used to test for production of hydroxamate and catechol-type siderophores, respectively. Deferoxamine mesylate ([DFX] a trihydroxamate siderophore; Sigma) and 2,3-dihydroxy benzoic acid ([DHBA] a catechol siderophore; Sigma) were used as positive controls.
Growth experiments.
Bacterial cultures were grown up to saturation in 3 ml of PS medium to obtain a preinoculum. The optical density of preinoculum from each culture was adjusted to an OD600 of 0.6 by dilution with PS medium. The preinoculum (0.2%, vol/vol) was added to PS medium under iron-limited or iron-supplemented conditions and grown at 28°C. For growth under iron-limiting conditions, DP and DFX were added to PS medium at different concentrations. Iron supplementation of PS medium was achieved by adding different concentrations of either FeSO4 or ferric chloride (FeCl3). Growth curves were obtained by monitoring a change in the OD600 over a period of 60 h. To determine growth yields, 0.3% of preinoculum (prepared as indicated above) was added, cultures were grown at 28°C for 48 h, and the number of CFU/ml of culture was determined by dilution plating on PSA medium. After 4 to 5 days, colonies were counted, and the results are expressed as means ± standard deviations (SD) of the log CFU/ml at 0 h and 48 h.
Siderophore supplementation bioassay.
The ability of a siderophore to enhance growth of X. oryzae pv. oryzae strains under iron-limiting conditions was assessed using a siderophore supplementation bioassay. Spent culture supernatant of strain BXO43, grown to stationary phase in PS medium containing 100 μM DP (PS-100 μM DP medium), was used as a source of native siderophore. These culture supernatants were passed through 0.22-μm-pore-size filters (Millipore) to make them cell free. Siderophore activity in the spent supernatant was confirmed in a CAS assay by spotting 100 μl on a PSA-CAS plate and scoring for the appearance of an orange-yellow halo. Bioassay tubes contained 3 ml of PS-100 μM DP medium and ∼106 CFU of selected X. oryzae pv. oryzae strains (i.e., BXO2211, BXO2231, and BXO2233). Tubes were supplemented with the following: 0.5 ml of the supernatant of the BXO43 strain grown in iron-limited or iron-supplemented medium, 0.5 ml of a culture supernatant of BXO2211 (xssE mutant) grown in PS medium, 120 μM FeSO4, 120 μM FeCl3, 50 μM DFX, 50 μM DHBA, and 50 μM ferrichrome (FCR). The growth yield of the X. oryzae pv. oryzae strains under the above-mentioned conditions was determined after 48 h and is expressed as log CFU/ml.
Virulence assay on rice plants.
Forty-day-old greenhouse-grown rice plants of the susceptible rice cultivar Taichung Native-1 (TN-1) were inoculated by clipping leaf tips with sterile scissors dipped in cultures of PS-grown X. oryzae pv. oryzae strains resuspended in sterile double-distilled water (108 CFU/ml) (32). Lesion lengths were measured at regular intervals. In each experiment, 15 leaves were inoculated, and the values are presented as mean lesion lengths and standard deviations.
Reisolation of bacteria from rice leaves and revertant identification.
Infected leaves were surface sterilized by being dipped in 1% (vol/vol) sodium hypochlorite (Loba Chemie) for 1 min and washed three times in distilled water. The leaves were cut at the leading edge of the lesion and dipped in 1 ml of sterile water for 5 min. Bacteria that exuded from the cut edge of the leaf were isolated by plating for individual colonies on PSA. These colonies were patched on PSA plates containing kanamycin, and the feoB+ revertants were identified as kanamycin-sensitive colonies. The kanamycin-sensitive colonies were patched on PSA-CAS plates for assessing the siderophore production phenotype.
Histochemical staining of infected rice leaves.
Rice leaves infected with GUS-marked strains were stained to determine β-glucuronidase activity, 12 days after infection, with 1 mM 5-bromo-4-chloro-3-indolyl β-d-glucuronide (X-Gluc, Sigma) in GUS assay buffer (50 mM sodium dihydrogen phosphate [pH 7.0] 10 mM EDTA, 0.1% Triton X-100, 0.1% sodium lauryl sarcosine, and 10 mM β-mercaptoethanol). Penetration of X-Gluc into leaves was facilitated by vacuum infiltration of leaves in assay buffer containing X-Gluc for 1 h, prior to incubation at 37°C. Destaining was performed by incubating stained leaves in absolute alcohol for 72 h at 37°C. Leaves were then observed under a compound microscope (Leica).
RESULTS
Sequence analysis of X. oryzae pv. oryzae feoABC operon.
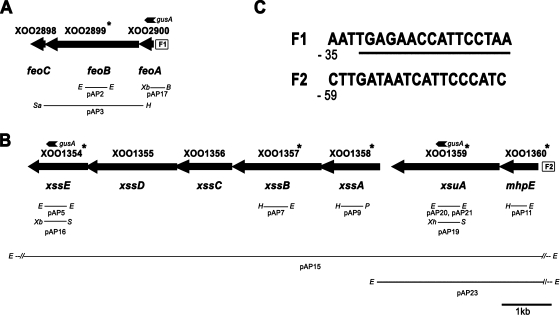
The genome of X. oryzae pv. oryzae KACC10331 strain (36) has a homolog of the E. coli feoABC operon. As in E. coli, the stop codon of the X. oryzae pv. oryzae feoA gene overlaps with the start codon of feoB, and the stop codon of feoB overlaps with the start codon of feoC, suggesting that these genes are co-operonic and translationally coupled (Fig. 1 A). The feoB gene of X. oryzae pv. oryzae encodes a protein of 621 amino acids that has 29% identity and 45% similarity to E. coli FeoB. The N-terminal one-third of X. oryzae pv. oryzae FeoB is hydrophilic while the C terminus is hydrophobic and predicted to have 10 transmembrane α-helices. As in E. coli, the X. oryzae pv. oryzae FeoB protein contains four G protein signature sequences. All four well-defined sequence motifs, G1 (residues 16 to 23), G2 (residues 42 to 44), G3 (residues 63 to 66), and G4 (residues 128 to 131), were found in the N-terminal region of X. oryzae pv. oryzae FeoB. In X. oryzae pv. oryzae, the genes feoA and feoC encode two small proteins of 82 and 84 amino acids, respectively. A putative Fur binding site was identified upstream of the X. oryzae pv. oryzae feoA coding region (Fig. 1C), which contained 10/19 residues conserved in the E. coli Fur-box consensus sequence (GATAATGATAATCATTATC) (51) and 13/19 residues conserved in the X. campestris pv. campestris Fur-box consensus sequence (AATGAGAATCATTCTCATT) (7). This suggests that, as in E. coli, expression of the X. oryzae pv. oryzae feoABC operon is regulated in response to levels of intracellular iron. Interestingly, the Fur binding site overlaps with a Clp (CAP-like protein) binding site. The Clp binding site of X. campestris pv. campestris is TGTGA-N6-TCACA (27). The putative Clp binding site upstream of the feoA coding region (Fig. 1C) has a match of 6/10 with the invariant residues in the X. campestris pv. campestris Clp binding site. Clp is homologous to the cyclic AMP (cAMP) nucleotide receptor protein Crp of E. coli and regulates several important biological functions of X. campestris pv. campestris (27). The feoABC locus is almost identical in all other xanthomonads that have been sequenced to date.
FIG. 1.
Genetic organization of X. oryzae pv. oryzae loci encoding functions involved in iron acquisition. Solid arrows represent open reading frames. Gene numbers are as specified by Lee et al. (36). (A) The X. oryzae pv. oryzae feoABC operon and cloned fragments. F1 represents a putative Fur binding site (FBS). (B) Organization of the X. oryzae pv. oryzae xss gene cluster involved in biosynthesis, export, and utilization of siderophore, including xss (Xanthomonas siderophore synthesis) genes, xsuA (Xanthomonas siderophore utilization) gene, and the mhpE gene encoding 4-hydroxy 2-oxovalerate aldolase. F2 represents an FBS. (C) Sequences of F1 and F2. The numbers indicate position of the FBS relative to translational start site, and the underlined sequence in F1 represents a putative Clp (CAP-like protein) regulator binding site. Thin lines represent cloned genomic regions, internal fragments of genes, and promoter regions that were used for complementation analysis, construction of mutations, and chromosomal GUS reporter fusions. Small arrowheads marked with gusA represent the positions of GUS reporter fusions. Gene numbers with asterisks indicate that the mutations in the corresponding genes were generated in this study. Restriction sites are abbreviated as follows: E, EcoRI; H, HindIII; Xb, XbaI; B, BamHI; Sa, SacI; P, PstI; S, SpeI; Xh, XhoI.
Transcriptional organization of feoABC genes.
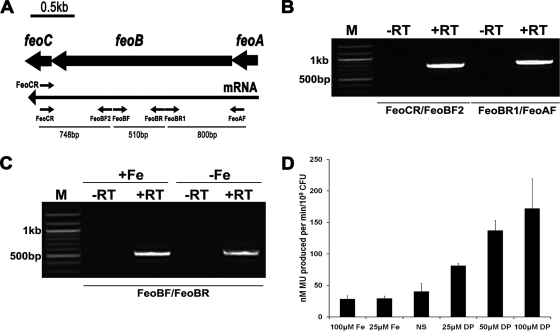
RT-PCR demonstrated that X. oryzae pv. oryzae feoA, feoB, and feoC genes are cotranscribed under iron-supplemented and iron-limiting conditions (Fig. 2A to C). A chromosomal transcriptional fusion strain (BXO2230; feo::gusA) was constructed such that the promoter of the feoABC operon is fused to a gusA reporter while retaining a functional copy of feoABC with its native promoter. When this strain was assayed for β-glucuronidase activity under iron-limiting conditions imposed by the addition of either 25 μM, 50 μM, or 100 μM 2,2′-dipyridyl ([DP] an iron chelator) to the PS medium, a gradual increase in β-glucuronidase activity was observed, indicating the induction of expression of the feoABC operon following iron depletion (Fig. 2D). The level of expression of the feo::gusA fusion is almost the same in either PS medium or PS medium supplemented with 25 μM or 100 μM ferrous sulfate. Overall, the RT-PCR data and GUS reporter assays indicate that, although expression of feoABC operon is induced by iron limitation, a basal level is observed even under high-iron conditions.
FIG. 2.
Operon mapping and expression analysis of X. oryzae pv. oryzae feo operon. (A) Solid arrows represent the feoA, feoB, and feoC ORFs. The long arrow represents the feoABC mRNA. The small arrows denote the positions and directions of primers used in RT-PCR. FeoCR primer was used in cDNA first-strand synthesis while FeoCR as well as other primers were used in subsequent PCRs. Lengths of expected PCR products are also shown for every primer pair. (B) RT-PCR analysis indicates that feoABC genes are cotranscribed as a single mRNA. The FeoBF2/FeoCR and the FeoAF/FeoBR1 primer pairs yield the expected 748-bp and 800-bp fragments, respectively. (C) The feoB transcript is present under iron-supplemented (100 μM FeSO4; +Fe) and iron-limited (100 μM DP, an iron chelator; −Fe) growth conditions. The expected 510-bp fragment is amplified by the FeoBF/FeoBR primer pair under both conditions. Lanes: −RT, without reverse transcriptase; +RT, with reverse transcriptase; M, 100-bp DNA molecular markers. (D) Expression of feo::gusA transcriptional fusion. The feo::gusA strain (BXO2230; FeoB+) was grown under either iron-supplemented (different concentrations of FeSO4; Fe), or iron-limited (different concentrations of DP) conditions or with no supplementation (NS). β-Glucuronidase activity is represented as nanomoles of methyl-umbelliferone (MU) produced/minute/108 CFU of bacteria. Data represent means ± standard deviations of values from three independent experiments.
X. oryzae pv. oryzae feoB mutant overproduces siderophores and exhibits growth deficiency under iron-limiting conditions.
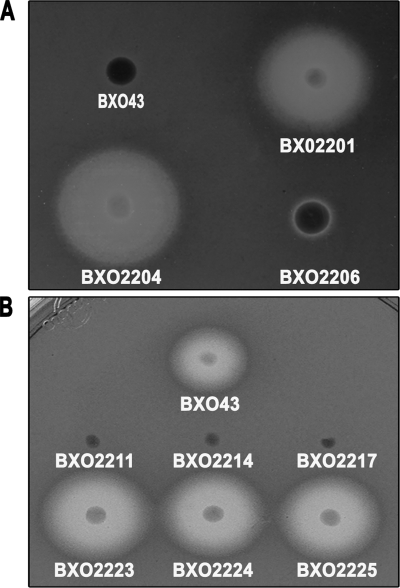
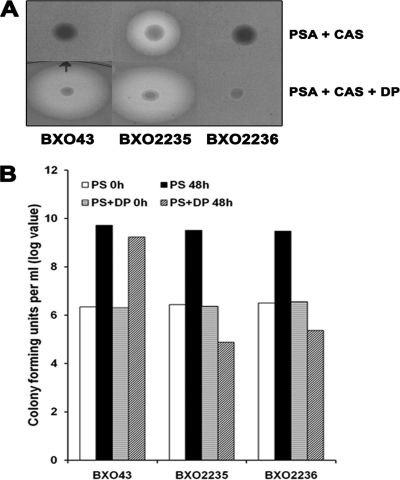
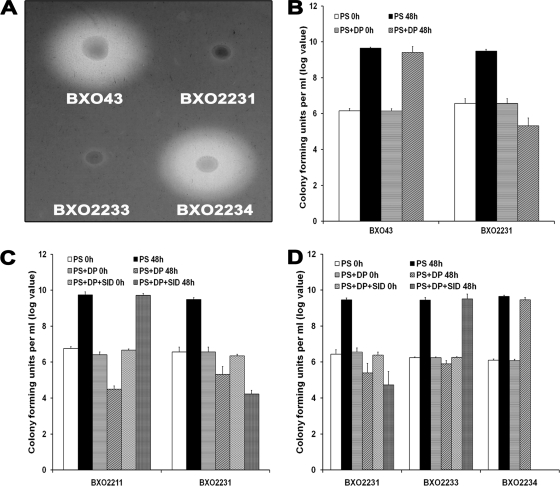
The X. oryzae pv. oryzae feoB mutant (strain BXO2201) overproduces siderophores on peptone-sucrose agar and chrome azurol S (PSA-CAS) medium. The wild-type strain BXO43 does not produce siderophores under these conditions (Fig. 3A). Introduction of the feoB gene on a complementing plasmid (BXO2206), but not the empty vector (BXO2204), abolishes the siderophore overproduction phenotype of the feoB mutant. The overproduction of siderophores by the feoB mutant might be a compensatory measure to overcome the loss of feoB function.
FIG. 3.
Siderophore production phenotype of X. oryzae pv. oryzae strains with mutations in genes involved in iron acquisition. Siderophore production is assayed as the presence of a halo around colonies growing on PSA-CAS medium. (A) An X. oryzae pv. oryzae feoB mutant overproduces siderophores. The X. oryzae pv. oryzae strains used are BXO43 (wild-type strain), BXO2201 (feoB1::pK18mob), BXO2204 (BXO2201/pHM1; vector control), and BXO2206 (BXO2201/pAP3; complemented strain). The pAP3 plasmid carries the cloned X. oryzae pv. oryzae feoB gene. (B) Mutations in xss genes result in a deficiency for siderophore production. Siderophore production was assessed by growth on PSA-CAS medium containing 100 μM DP. The X. oryzae pv. oryzae strains used are BXO43 (wild-type strain), BXO2211 (xssE1::pK18mob), BXO2214 (xssB1::pK18mob), BXO2217 (xssA1::pK18mob), BXO2223 (BXO2211/pAP15;, complemented strain), BXO2224 (BXO2214/pAP15; complemented strain), and BXO2225 (BXO2217/pAP15; complemented strain). The pAP15 cosmid contains the entire xss gene cluster of X. oryzae pv. oryzae. Plasmid pK18mob has an outreading lacZ promoter for transcribing downstream genes.
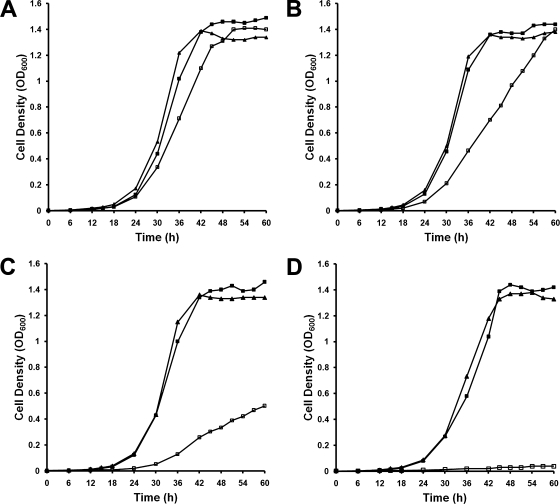
BXO43, BXO2201, and BXO2206 were grown in PS medium, with and without addition of DP. All three strains grew equally well in PS medium (Fig. 4A). There was no significant effect of the addition of DP (25 μM or 50 μM) on growth of either BXO43 or BXO2206. In contrast, BXO2201 displayed a significant decrease in growth as the concentration of DP increased (Fig. 4B and C). At 100 μM DP, growth of BXO2201 was almost negligible (Fig. 4D). The feoB mutant also exhibited a growth deficiency in PS medium containing deferoxamine mesylate (a specific ferric iron chelator) (data not shown). Taken together, these data suggested that FeoB is required for growth of X. oryzae pv. oryzae in low-iron medium.
FIG. 4.
A feoB mutant of X. oryzae pv. oryzae is deficient for growth under conditions of iron limitation. Wild-type BXO43 (filled squares), feoB mutant (feoB1::pK18mob; open squares), and complemented strain BXO2206 (BXO2201/pAP3; filled triangles) were grown in PS medium (A) PS supplemented with 25 μM DP (B) PS medium with 50 μM DP (C), and PS medium with 100 μM DP (D). Bacterial growth was monitored by determining the OD600. The results were reproducible in an independent experiment.
Identification of a siderophore-biosynthetic gene cluster in X. oryzae pv. oryzae.
By employing the Arnow and Csaky tests (52), we were able to determine that the siderophore produced by X. oryzae pv. oryzae is not of either the catecholate or hydroxamate type (data not shown). This suggested that the X. oryzae pv. oryzae siderophore might be of the α-hydroxy carboxylate type. We found that X. campestris pv. campestris strain ATCC 33913 and X. axonopodis pv. citri (a pathogen of citrus) strain 306 (20) have a homolog of the Vibrio parahaemolyticus pvs locus, which encodes five genes involved in the biosynthesis of vibrioferrin, an α-hydroxy carboxylate-type of siderophore, and the pvuA gene, which is involved in vibrioferrin uptake (67). Degenerate oligonucleotide primers (Table 3) were used to amplify internal fragments of all six genes from the genome of X. oryzae pv. oryzae strain BXO43, and sequencing confirmed the identity of these genes. During the course of this work, the genome sequence of X. oryzae pv. oryzae strain KACC10331 (36) was released, and we found that the genes XOO1354-XOO1359 are homologous to pvsABCDE and pvuA genes. In addition, we observed that the stop codon of XOO1360 overlaps with the start codon of XOO1359. We named this gene cluster xss (Xanthomonas siderophore synthesis) (Fig. 1B), and we designated open reading frames (ORFs) XOO1354-XOO1359 xssE, xssD, xssC, xssB, xssA, and xsuA while ORF XOO1360 was named mhpE (encodes 4-hydroxy 2-oxovalerate aldolase) based on its homology. The similarity/identity of XOO1354-XOO1360 to the genes involved in biosynthesis/uptake of siderophores in other organisms is shown in Table 5.
TABLE 5.
Homologous proteins for functions encoded in the xss gene cluster of X. oryzae pv. oryzae
|
X. oryzae pv. oryzae protein |
Putative homologue |
E valuea | I/Sb | ||
|---|---|---|---|---|---|
| Name (length [aa]) | Function | Name and/or function (% match in length) | Bacterium | ||
| XssE (426) | Diaminopimelate decarboxylase/PLP-dependent decarboxylasec | PvsE; vibrioferrin biosynthesis (86) | Vibrio parahaemolyticus | 2e-111 | 53/67 |
| AcsE; achromobactin biosynthesis (87) | Erwinia chrysanthemi | 3e-80 | 45/58 | ||
| Siderophore synthesis decarboxylase (87) | Ralstonia solanacearum | 2e-85 | 45/58 | ||
| SbnH; staphyloferrin B biosynthesis (88) | Staphylococcus aureus | 3e-85 | 40/58 | ||
| XssD (591) | Type A siderophore synthetase | PvsD; vibrioferrin biosynthesis (94) | Vibrio parahaemolyticus | 5e-126 | 43/57 |
| Siderophore biosynthetic protein (73) | Ralstonia solanacearum | e-40 | 31/46 | ||
| AcsD; achromobactin biosynthesis (97) | Erwinia chrysanthemi | 7e-53 | 29/44 | ||
| RhbC; rhizobactin biosynthesis (77) | Sinorhizobium meliloti | 8e-37 | 28/41 | ||
| SbnE; staphyloferrin B biosynthesis (81) | Staphylococcus aureus | 2e-40 | 27/43 | ||
| IucA; aerobactin biosynthesis (82) | Escherichia coli | 3e-30 | 25/39 | ||
| XssC (398) | Major facilitator superfamily multidrug efflux pump | PvsC; multidrug efflux pump homologue (100) | Vibrio parahaemolyticus | 8e-73 | 43/58 |
| XssB (600) | Type B siderophore synthetase | PvsB; vibrioferrin biosynthesis (95) | Vibrio parahaemolyticus | 6e-120 | 44/58 |
| AcsA; achromobactin biosynthesis (66) | Erwinia chrysanthemi | 3e-28 | 31/45 | ||
| LbtA; legiobactin biosynthesis (54) | Legionella pneumophila | 7e-23 | 29/47 | ||
| SbnC; staphyloferrin B biosynthesis (64) | Staphylococcus aureus | 3e-22 | 24/44 | ||
| IucC; aerobactin biosynthesis (50) | Escherichia coli | e-12 | 24/39 | ||
| XssA (408) | d-Alanine-d-Alanine ligase/ATP-dependent amino acid ligase | PvsA; vibrioferrin biosynthesis (95) | Vibrio parahaemolyticus | e-92 | 45/60 |
| XsuA (713) | TonB-dependent outer membrane receptor | PvuA; ferric vibrioferrin receptor (93) | Vibrio parahaemolyticus | 2e-156 | 43/61 |
| FecA; ferric citrate outer membrane receptor (92) | Escherichia coli | 2e-47 | 31/47 | ||
| MhpE (253) | 4-Hydroxy 2-oxovalerate aldolase | AcsB; achromobactin biosynthesis (93) | Erwinia chrysanthemi | e-32 | 36/53 |
| SbnG; staphyloferrin B biosynthesis (93) | Staphylococcus aureus | 7e-32 | 35/52 | ||
E values were obtained by using the BLAST algorithm.
I/S, % identity/% similarity.
PLP, pyridoxal 5′-phosphate.
The xssE gene product is closely related to pyridoxal 5′-phosphate (PLP)-dependent decarboxylase family enzymes and also to diaminopimelate decarboxylases (LysA) which are predicted to be involved in siderophore biosynthesis. The protein products encoded by xssD and xssB belong to type A and B NRPS-independent siderophore (NIS) synthetases and showed 25% and 24% identity (39% and 39% similarity) to IucA and IucC proteins of E. coli, respectively, which catalyze amide bond formation during aerobactin biosynthesis in E. coli (13, 41). The xssA gene product is homologous to a family of proteins which include d-Ala-d-Ala ligase enzymes. The mhpE gene encodes a 4-hydroxy 2-oxovalerate aldolase protein which belongs to the HpcH/HpaI aldolase/citrate lyase family. This enzyme acts on 4-hydroxy 2-oxovalerate to release pyruvate and acetaldehyde (37). The xssC gene is predicted to encode a cytoplasmic membrane protein (as determined by PSORTb) having a membrane spanning region comprised of 12 α-helices and is homologous to multidrug efflux pumps of the major facilitator superfamily (MFS) of proteins. MFS proteins facilitate the transport of a variety of substrates across the cytoplasmic membrane (53). XssC is the X. oryzae pv. oryzae homolog of PvsC, which functions as an exporter of vibrioferrin (68). The xsuA gene is predicted to encode a 713-amino-acid-long outer membrane-located (as determined by PSORTb) protein that is homologous to PvuA, the ferric vibrioferrin receptor (24). These similarities suggest that the xss gene cluster is involved in biosynthesis, export, and uptake of a vibrioferrin kind of siderophore. Overlapping start and start codons were present between all genes of the xss cluster except xssA and xsuA, which are separated by 235 bp (Fig. 1B). This suggested that this gene cluster of ∼10.5 kb might be transcriptionally organized as an operon. A putative Fur binding site was identified upstream of the X. oryzae pv. oryzae mhpE coding region (Fig. 1C), which contained 12/19 residues conserved in E. coli Fur-box consensus sequence (51) and 14/19 residues conserved in X. campestris pv. campestris Fur-box consensus sequence (7).
Although the xss gene cluster is present in the genomes of all other xanthomonads that have been sequenced to date, this gene cluster is not present in the genomes of the closely related Xylella fastidiosa (6) and Stenotrophomonas maltophilia (18). In X. oryzae pv. oryzae and other xanthomonads, the xss gene cluster is adjacent to the vitamin B12-biosynthetic gene cluster. The XOO1362 (a cation-proton antiporter) and XOO1337 (clpB) genes flank this genomic region. Interestingly, BLAST N analysis reveals that the S. maltophilia genes Smlt3726 (homolog of XOO1362) and Smlt3732 (homolog of XOO1337) are present near each other with a few Stenotrophomonas-specific genes and the metB gene (encodes for cystathionine gamma-synthase) being located between them. The colinearity at this locus between the Xanthomonas and Stenotrophomonas genomes suggests that the xss gene cluster was either introduced into this locus at an early stage in the Xanthomonas lineage or that it was deleted from the Stenotrophomonas lineage. However, S. maltophilia has a gene cluster that is homologous to a gene cluster involved in biosynthesis of enterobactin, a catecholate-type of siderophore produced by E. coli (60).
The xss genes are part of an operon which is induced by iron limitation.
RT-PCR demonstrated that xssA, xssB, xssC, xssD, xssE, xsuA, and mhpE genes are cotranscribed as a polycistronic mRNA whose expression is controlled in response to iron availability (Fig. 5A and B). The expression of an xssE::gusA transcriptional fusion (BXO2229) was analyzed under iron-supplemented and iron-limiting conditions. High levels of β-glucuronidase activities were observed under iron-depleted conditions (addition of different concentrations of DP) (Fig. 5C). A very low basal level of expression was observed for the xssE::gusA fusion during growth in either PS medium or under iron-supplemented conditions.
FIG. 5.
Operon mapping and expression analysis of xss gene cluster of X. oryzae pv. oryzae. (A) Solid arrows represent the xssABCDE, xsuA, and mhpE ORFs. The long arrow represents the mRNA transcript. The small arrows denote the positions and directions of primers used in RT-PCR. MF1 primer was used in cDNA first-strand synthesis, and MF1 as well as other primers were used in subsequent PCRs. Lengths of expected PCR products are also shown for every primer pair. (B) Lanes contain RT-PCR products amplified from total RNA isolated from X. oryzae pv. oryzae cultures grown under iron-supplemented (+Fe; 100 μM FeSO4) and iron-limiting (-Fe; 100 μM DP, an iron chelator) conditions as described in Materials and Methods. The primer pairs MF1/MR2, MF3/MR4, and MF5/MR6 yielded the expected DNA fragments of 790 bp, 722 bp, and 680 bp, respectively, exclusively from mRNA templates under iron-limiting conditions. No amplification was observed from mRNA templates expressed under iron-supplemented conditions. Lanes −RT and +RT, without and with reverse transcriptase, respectively; lane M, 100-bp DNA molecular marker. (C) Transcriptional analysis of xssE gene involved in siderophore biosynthesis carried out with GUS reporter strain BXO2229 (xssE2::gusA). BXO2229 was grown under either iron-supplemented (different concentrations of FeSO4; Fe) or iron-limited (different concentrations of [DP]) conditions or with no supplementation (NS). β-Glucuronidase activity is represented as the nanomoles of methyl-umbelliferone produced/minute/108 CFU of bacteria. Data represent mean ± standard deviation of values from three independent experiments.
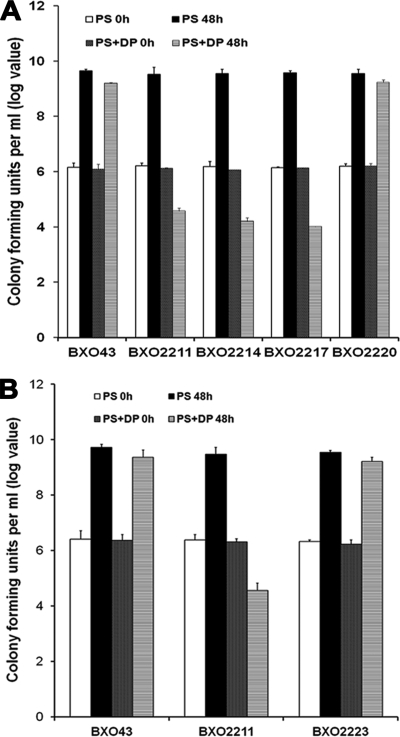
In order to exclude the possibility that there is a promoter between the xssA and xsuA genes, a nonpolar mutant (BXO2235; xsuA2::pK18mob) and a polar mutant (BXO2236; xsuA3::pK18mob) of the xsuA gene were constructed as described in Materials and Methods. A polar mutation in the xsuA gene blocked siderophore biosynthesis while a nonpolar mutation did not affect the production of siderophore on PSA-CAS-DP medium (see Fig. 8A). Moreover, BXO2235 produces siderophore on PS medium, a condition in which BXO43 does not produce siderophore, indicating that the lacZ promoter of the integrated vector pK18mob is driving the transcription of xssABCDE genes in this strain. These data suggest that there is no relevant promoter in the intergenic region between xssA and xsuA and that a promoter present upstream of mhpE drives transcription of the xss operon.
FIG. 8.
Siderophore production and growth phenotype of xsuA mutants of X. oryzae pv. oryzae constructed by integration of suicide plasmid pK18mob. (A) Strains depicted are BXO43 (wild-type strain), BXO2235 (xsuA2::pK18mob; a nonpolar mutation), and BXO2236 (xsuA3::pK18mob; a polar mutation). X. oryzae pv. oryzae strains were grown either on PSA-CAS medium or on PSA-CAS medium supplemented with 100 μM DP. Siderophore production is evident as a halo around the colonies. (B) BXO2235 and BXO2236 are deficient for growth under iron-limiting conditions. Cell numbers were estimated in X. oryzae pv. oryzae cultures grown in PS medium and PS-100 μM DP medium at 0 h and after 48 h of growth.
xssA, xssB, and xssE mutants are siderophore deficient and exhibit growth deficiency under low-iron conditions.
Nonpolar mutations were created by plasmid integration in the xssE, xssB, xssA, and mhpE genes as described in Materials and Methods, and the generated mutants were named as BXO2211, BXO2214, BXO2217, and BXO2220, respectively. The ability of these mutants to produce siderophores was assessed on PSA-CAS-DP plates. In contrast to BXO43, mutants BXO2211, BXO2214, and BXO2217 failed to produce a halo on PSA-CAS-DP medium (Fig. 3B), indicating that these mutants are impaired in siderophore production. We introduced a cosmid clone (pAP15) containing the entire xss gene cluster to these mutants to get complemented strains BXO2223 (for the xssE mutant), BXO2224 (for the xssB mutant), and BXO2225 (for the xssA mutant). These complemented strains produced wild-type levels of siderophore on PSA-CAS-DP medium (Fig. 3B). Overall, these results indicate that mutation of the xssA, xssB, and xssE genes affects the ability of X. oryzae pv. oryzae to produce siderophore, a phenotype which can be restored by complementation in trans. However, although it is less likely, there is a possibility that the siderophore-deficient phenotype of the xssA and xssB mutants is not due to gene inactivation but to overexpression of downstream genes caused by the nonpolar mutation. The mhpE mutant (BXO2220) showed no defect in siderophore production on PSA-CAS-DP medium (data not shown), indicating that this gene is not essential for siderophore biosynthesis in X. oryzae pv. oryzae.
In PS medium, no significant difference in growth yields was observed between the wild-type and the BXO2211, BXO2214, BXO2217, and BXO2220 strains. When these mutants were cultured in PS-100 μM DP medium (iron-limiting conditions), the siderophore-deficient mutants (BXO2211, BXO2214, and BXO2217) exhibited a severe deficiency for growth (Fig. 6A). The siderophore-proficient strains, BXO43, BXO2220, and BXO2223, grew quite well under these conditions (Fig. 6). The siderophore-deficient mutants (BXO2211, BXO2214, and BXO2217) also exhibited a growth deficiency in PS medium containing deferoxamine mesylate (a specific ferric iron chelator) (data not shown). These results indicate that the xssA, xssB, and xssE genes are essential for siderophore biosynthesis and growth under low-iron conditions.
FIG. 6.
Mutations in xss genes affect growth of X. oryzae pv. oryzae in iron-limited medium. Cell numbers were estimated in X. oryzae pv. oryzae cultures grown in PS medium and PS medium containing 100 μM DP (an iron chelator) at 0 h and after 48 h of growth. Data are the mean log CFU values ± standard deviations from three independent experiments. (A) Mutations in xss genes, but not the mhpE gene, result in a severe growth deficiency under iron-limiting conditions. Strains are BXO43 (wild-type strain), BXO2211 (xssE1::pK18mob), BXO2214 (xssB1::pK18mob), BXO2217 (xssA1::pK18mob), and BXO2220 (mhpE1::pK18mob). (B) Genetic complementation restores growth of a xssE1::pK18mob mutant under iron-limiting medium. Strains are BXO43 (wild-type strain), BXO2211 (xssE1::pK18mob), and BXO2223 (BXO2211/pAP15). The pAP15 cosmid contains the entire xss gene cluster of X. oryzae pv. oryzae. The pAP15 cosmid also corrects the growth deficiency of the xssB and xssA mutants on iron-limited medium (data not shown). Plasmid pK18mob has an outreading lacZ promoter for transcribing downstream genes.
Siderophore restores the growth of the xssE mutant in iron-deficient medium.
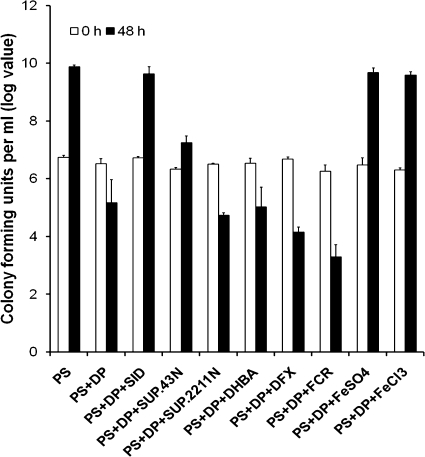
We examined the ability of siderophore to promote the growth of BXO2211 (xssE mutant) on PS-100 μM DP medium by supplementation with culture supernatants (taken as source of native siderophore) of a BXO43 strain grown PS-100 μM DP medium. The presence of siderophore in the supernatant of BXO43 grown in PS-100 μM DP medium was confirmed by spotting supernatants on PSA-CAS plates. The addition of culture supernatant of the BXO43 strain enhanced the growth of BXO2211, resulting in a level of cell density similar to that observed in PS medium. Interestingly, cell-free supernatant of BXO43 culture grown in PS medium also promoted (∼10-fold) the growth of BXO2211. This suggests that during growth in PS medium, BXO43 produces a slight amount of siderophore which can minimally enhance growth of the BXO2211 strain. However, this level of siderophore production was not detectable on PSA-CAS plates. In addition, supplementation with either FeSO4 or FeCl3 was equally efficient in restoring the growth of BXO2211 (Fig. 7). Restoration of growth of BXO2211 to the level that is observed in iron-sufficient medium by siderophore supplementation again confirmed that siderophore production is required for growth on low-iron medium. Importantly, supplementation with the culture supernatant of BXO2211 grown in PS medium did not promote growth of BXO2211.
FIG. 7.
Siderophore supplementation bioassays. Cell numbers (log value) were estimated at 0 h and after 48 h of growth in cultures of a siderophore-deficient X. oryzae pv. oryzae mutant (strain BXO2211; xssE1::pK18mob) grown in PS medium, PS-100 μM DP medium, and PS-100 μM DP medium with supplements. The different supplements used are as follows: SID, cell-free supernatant of a saturated culture of wild-type X. oryzae pv. oryzae strain BXO43 grown in iron-deficient medium (PS-100 μM DP medium; SUP.43N, cell-free supernatant of a saturated culture of BXO43 grown in PS medium; SUP.2211N, cell-free supernatant of a saturated culture of the siderophore-deficient mutant BXO2211 grown in PS medium; 50 μM DHBA; 50 μM DFX; 50 μM FCR; 120 μM FeSO4; and 120 μM FeCl3. The culture volume was 3 ml, of which 0.5 ml was cell-free culture supernatant in applicable experiments. The results from three independent experiments are expressed as means ± standard deviations.
As bacteria often can utilize exogenous siderophores that cannot be biosynthesized by them, the ability of BXO2211 to utilize three exogenous siderophores was tested. We used 2,3-dihydroxy benzoic acid, a catecholate type of siderophore, as well as ferrichrome and deferoxamine mesylate, two hydroxamate siderophores. These siderophores did not exhibit a growth-promoting activity on BXO2211 (Fig. 7).
The xsuA gene appears to be involved in siderophore uptake.
In addition to the two xsuA mutants described above, a polar mutation in the xsuA gene was generated (as described in Materials and Methods) by integration of plasmid pVO155 to create the strain BXO2231 (xsuA1::pVO155). The siderophore production phenotype of the xsuA mutants was assessed on PSA-CAS-DP. BXO2231 and BXO2236 were found to be defective in siderophore production while BXO2235 was able to produce siderophores (Fig. 8A and Fig. 9A). The ability of the BXO2235 (xsuA2::pK18mob) mutant to produce siderophores indicated that xsuA is not essential for siderophore production and that the mutation in this strain is not polar on expression of downstream xss genes. The siderophore-deficient phenotype of the BXO2231 and BXO2236 strains indicated that, in these strains, the insertion mutation in the xsuA gene is polar on expression of downstream xss genes. In the BXO2231 background, a cosmid (pAP23) containing the xsuA gene, but devoid of xss genes, and another cosmid having the entire xss gene cluster including xsuA (pAP15) were mobilized to get complemented strains, BXO2233 and BXO2234, respectively. As expected, BXO2234 was able to produce siderophores, but BXO2233 was still defective for siderophore production (Fig. 9A).
FIG. 9.
The phenotype of an xsuA X. oryzae pv. oryzae mutant is suggestive of deficiency in siderophore uptake. (A) Siderophore production phenotype of an xsuA mutant of X. oryzae pv. oryzae. Strains depicted are BXO43 (wild-type strain), BXO2231 (xsuA1::gusA; a polar mutation), BXO2233 (BXO2231/pAP23; this cosmid contains the xsuA gene but does not contain any intact xss genes), and BXO2234 (BXO2231/pAP15; this cosmid contains the entire xss gene cluster including the xsuA gene). X. oryzae pv. oryzae strains were grown on PSA-CAS medium and 100 μM DP. Siderophore production is evident as a halo around the colonies. (B). An xsuA mutant of X. oryzae pv. oryzae is deficient for growth under iron-limiting conditions. Cell numbers (log values) were estimated in X. oryzae pv. oryzae cultures grown in PS medium and PS-100 μM DP medium at 0 h and after 48 h of growth. (C) An xsuA mutant is unable to use exogenously supplied siderophore. Strains are BXO2211 (xssE1::pK18mob) and BXO2231 (xsuA1::gusA). (D) Genetic complementation restores the ability of an X. oryzae pv. oryzae xsuA mutant to utilize exogenously supplied siderophore. Strains are BXO2231 (xsuA1::gusA), BXO2233 (BXO2231/pAP23), and BXO2234 (BXO2231/pAP15). Cell numbers were estimated in 3 ml of X. oryzae pv. oryzae cultures grown in PS medium, PS-100 μM DP medium, and PS-100 μM DP supplemented with SID (cell-free supernatant of a saturated culture of wild-type X. oryzae pv. oryzae strain BXO43 grown in iron-deficient medium [PS-100 μM DP]) at 0 h and after 48 h of growth. The results from three independent experiments are expressed as means ± standard deviations.
Growth yield assays were performed for xsuA mutants under iron-limiting conditions imposed by the addition of 100 μM DP. Under these conditions all three mutants, BXO2231, BXO2235, and BXO2236, were severely deficient for growth compared with wild-type BXO43. These strains grew as well as BXO43 in PS medium (Fig. 8B and 9B). The growth deficiency of the BXO2235 mutant on low-iron medium, in spite of the mutant's proficient siderophore production, is consistent with the possibility that xsuA encodes a siderophore receptor. To investigate the ability of xsuA mutants to utilize native siderophore, supplementation was carried out with spent culture supernatants of a BXO43 strain grown on PS—100 μM DP medium. Upon supplementation with the culture supernatant of BXO43, a BXO2211 (xssE mutant) strain was able to grow in PS—100 μM DP medium, but the BXO2231 was unable to grow under these conditions (Fig. 9C). This suggested that BXO2231 is unable to utilize the siderophore present in the culture medium, a phenotype that is indicative of a deficiency in siderophore uptake. Introduction of the plasmid pAP23 (which carries only the xsuA gene) restores the ability of the BXO2231 strain to grow on PS—100 μM DP medium supplemented with culture supernatant of BXO43 (Fig. 9D, BXO2233). As expected, introduction of the cosmid pAP15 (which carries the entire xss gene cluster including xsuA) into BXO2231 also restores the ability of this strain to grow on PS—100 μM DP medium (Fig. 9D, BXO2234).
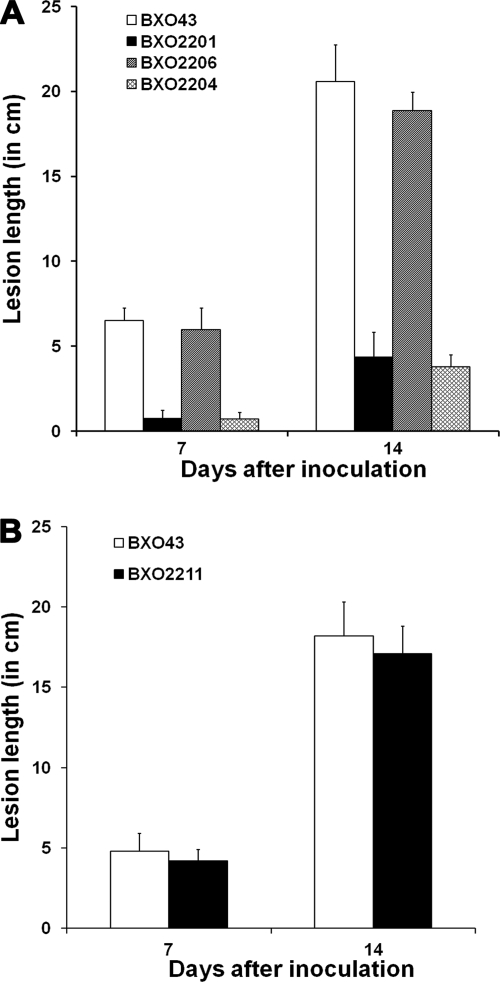
feoB mutant is virulence-deficient while siderophore-deficient mutants are virulence proficient.
The virulence phenotypes of BXO43 (wild-type strain), BXO2201 (feoB mutant), BXO2204 (BXO2201/pHM1; control), and BXO2206 (BXO2201/pAP3, which carries feoB for complementation) were determined by assessing lesion lengths in rice 7 and 14 days after inoculation (as described in Materials and Methods). At both time points, the BXO2201 and BXO2204 strains showed significantly reduced lesions compared to lesions produced by BXO43 and BXO2206 (Fig. 10A). However, we observed that the BXO2201 and BXO2204 strains produced slightly elongated lesions at 14 days after inoculation compared to lesions observed at 7 days after inoculation. To determine whether this increase in lesion length was caused by the presence of any feoB+ revertants in planta, X. oryzae pv. oryzae cells were reisolated (as described in Materials and Methods) from leaves infected with BXO2201. Reisolated bacteria were found to exhibit the wild-type phenotype with respect to siderophore production; i.e., they did not produce siderophore on PSA medium but produced siderophore on PSA-DP medium (data not shown). Also, these bacteria were kanamycin susceptible, suggesting that they arose after loss of the integrated plasmid. Taken together, these results remarkably illustrate the importance of FeoB for virulence of X. oryzae pv. oryzae on rice. In contrast, an xssE mutant (BXO2211) was found to be as virulence proficient as the wild-type strain BXO43 (Fig. 10B). An xssB mutant (BXO2214), xssA mutant (BXO2217), and xsuA mutant (BXO2231) were also as virulence proficient as BXO43 (data not shown). These observations indicate that siderophore-mediated iron acquisition is not essential for in planta growth of X. oryzae pv. oryzae. Virulence proficiency of BXO2220 (data not shown) also indicated that the mhpE gene does not have a role in virulence of X. oryzae pv. oryzae.
FIG. 10.
Virulence phenotype of X. oryzae pv. oryzae strains with mutations in genes involved in iron acquisition. Inoculations were performed on leaves of 40-day-old greenhouse-grown plants of susceptible rice cultivar Taichung native-1 (TN-1). Lesion lengths were measured 7 and 14 days after inoculation. Means and standard deviations of 15 replicate measurements are given. Results from one experiment are presented. Similar results were obtained in independent experiments. (A) An X. oryzae pv. oryzae feoB mutant is severely virulence deficient. Strains are BXO43 (wild-type strain), BXO2201 (feoB1::pK18mob), BXO2206 (BXO2201/pAP3; complemented strain; pAP3 is pHM1-feoB), and BXO2204 (BXO2201/pHM1; vector control). (B) A siderophore-deficient mutant of X. oryzae pv. oryzae is virulence proficient. Strains are BXO43 and BXO2211 (xssE1::pK18mob). The values obtained for BXO2211 were not significantly different (P < 0.05, for 7 days and 14 days) from those for BXO43 in a Student's two-tailed t test for independent means. Similar results (data not shown) were obtained with BXO2214 (xssB1::pK18mob), BXO2217 (xssA1::pK18mob), BXO2220 (mhpE1::pK18mob), and BXO2231 (xsuA1::gusA) strains.
feoB gene of X. oryzae pv. oryzae is expressed during in planta growth.
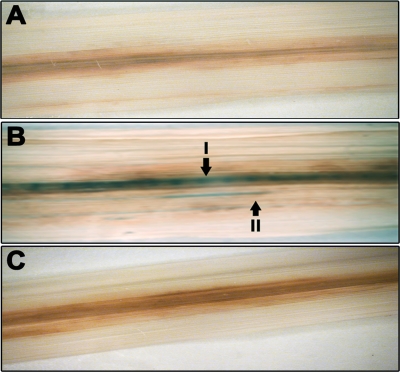
Rice leaves were inoculated with either BXO2230 (feo::gusA) or BXO2229 (xssE::gusA), and the leaves were stained for GUS expression 12 days after infection. As described above, the feo::gus fusion retains a wild-type copy of the feo operon under the control of its native promoter and is, hence, proficient for virulence. GUS activity was assessed by staining the infected leaves with the chromogenic substrate X-Gluc, as described in Materials and Methods. GUS staining (blue-green) was not detected in xylem vessels from leaves infected with either BXO43 (as a negative control) or BXO2229 (Fig. 11A and C), whereas GUS activity was detected in major longitudinal veins and minor longitudinal veins of rice leaves infected with BXO2230 (Fig. 11B, I and II, respectively). Bacteria were reisolated from rice leaves infected with either BXO2229 or BXO2230, and all of the reisolated bacteria (100/100 tested) retained the kanamycin resistance marker that is associated with the gus fusion. These reisolated bacteria also retained GUS expression in vitro under iron-limiting conditions. This indicated that the lack of GUS staining in leaves inoculated with BXO2229 is not due to loss of the gus fusion. In addition, the number of CFU/cm2 of rice leaves infected with these strains also did not differ, indicating that variations in the bacterial density are not the cause of differences in GUS activity (data not shown). Furthermore, GUS expression in bacterial exudates from cut ends of rice leaf pieces infected with BXO2229 and BXO2230 was determined. GUS activity for feo::gusA and xssE::gusA fusions in leaf exudates was 64 ± 7.95 nM MU/minute/108 CFU and 5.2 ± 0.98 nM MU/minute/108 CFU, respectively. This again indicates that the feo operon is expressed at a much higher level than the xss operon during in planta growth.
FIG. 11.
The feoB gene of X. oryzae pv. oryzae is expressed during in planta growth. Inoculations were performed on leaves of 40-day-old greenhouse-grown plants of susceptible rice cultivar TN-1 with BXO43 (GUS negative) (A) and Gus reporter strains BXO2230 (feo::gusA) (B) and BXO2229 (xssE2::gusA) (C). Twelve days after inoculation, the leaves were stained for GUS activity as described in Materials and Methods. Arrows indicate major (I) and minor (II) longitudinal veins. Blue-green staining in the veins in panel indicates GUS expression.
DISCUSSION
In this study, we have characterized two different X. oryzae pv. oryzae iron uptake systems. The X. oryzae pv. oryzae feoABC genes are homologues of well-characterized ferrous transporters in other bacteria wherein the feoB gene encodes the major bacterial ferrous ion transporter. The xss operon is homologous to siderophore-biosynthetic genes in other bacteria and is essential for siderophore production by X. oryzae pv. oryzae. We have studied the regulation of these X. oryzae pv. oryzae iron uptake systems, demonstrated that they are required for growth on low-iron media, and assessed their roles in virulence. This is the first report on the role of specific iron uptake systems in virulence of any member of the important xanthomonad group of plant pathogens.
Iron supplementation of PS medium suppresses siderophore production by the X. oryzae pv. oryzae feoB mutant (data not shown), indicating that the strain is experiencing iron limitation on PS medium and that it is overproducing siderophore as a compensatory measure. Quantitative GUS assays showed that, in wild-type cells, the expression of the feo operon is induced under iron-limiting conditions (i.e., in the presence of DP), but a basal level of expression is retained even under iron-replete conditions (i.e., medium supplemented with FeSO4). A similar expression pattern of the feo operon has been reported for X. fastidiosa, in which the levels of feo transcripts were not affected under iron-replete conditions while an increase in the amounts of feo transcripts was detected under iron-limiting conditions (76). Altogether, it seems that the X. oryzae pv. oryzae feoABC operon is expressed at a basal level even under high-iron conditions and that iron uptake via FeoB may form a constitutive pathway that is utilized by this bacterium under all growth conditions.
Other than the mhpE gene, homologs of the remaining six genes in the xss operon are present within the pvs gene cluster which is involved in biosynthesis of vibrioferrin, an α-hydroxy carboxylate siderophore of V. parahaemolyticus (67). Homologs of the X. oryzae pv. oryzae mhpE gene, named acsB and sbnG, are present in gene clusters involved in the biosynthesis of the α-hydroxy carboxylate-type of siderophores, achromobactin of E. chrysanthemi and staphyloferrin B of Staphylococcus aureus, respectively (4, 23). Mutations have not been isolated in the acsB and sbnG genes. It has been suggested (23) that MhpE-like proteins might maintain a pool of certain carbohydrates that are essential for the formation of siderophore. However, at least in X. oryzae pv. oryzae, MhpE is not essential for siderophore production.
Bacterial siderophore biosynthetic gene clusters generally include genes that encode functions involved in export of siderophore and uptake of the ferric-siderophore complex. Most characterized siderophore exporters are members of the MFS family, including proteins such as exporter of enterobactin in E. coli (EntS) (25), vibrioferrin in V. parahaemolyticus (PvsC) (68), achromobactin in E. chrysanthemi (YhcA) (23), alcaligin in Bordetella spp. (AlcS) (10), bacillibactin in Bacillus subtilis (YmfE) (44), legiobactin in L. pneumophila (LbtB) (1), and protochelin in Azotobacter vinelandii (CbsX) (49). The observation that XssC is an MFS family protein and its homology to PvsC suggest that xssC might be encoding an exporter involved in siderophore secretion. The homology of XsuA protein to PvuA, which acts as a ferric-vibrioferrin receptor in V. parahaemolyticus (24), and the observation that xsuA mutants are unable to utilize native siderophore suggest that XsuA encodes a siderophore receptor protein.
Several studies have established a role for ferrous iron acquisition via FeoB in the virulence of animal-pathogenic bacteria (12). Our observation that the feoB mutant of X. oryzae pv. oryzae is severely virulence deficient represents the first demonstration of the role of the feoB gene in virulence of any plant-pathogenic bacterium. It is likely that this virulence deficiency is due to a defect in iron uptake although the possibility that FeoB is also importing something else other than iron or that it is serving as an exporter cannot be excluded. In contrast, X. oryzae pv. oryzae mutants defective for siderophore production/utilization were as virulent as the wild-type strain. Our data indicate that, under iron-deficient conditions in the laboratory, the FeoB protein as well as siderophore are involved in iron uptake as a mutation in either one of these systems affects ability growth on low-iron medium.
It is generally agreed that iron exists as Fe3+-citrate complex in xylem vessels of plants (50). However, recently, speciation analysis of xylem sap of rice demonstrated that iron is present in Fe2+ and Fe3+ forms and that the concentration of iron in the xylem sap is in the micromolar range (75). Bacterial requirement for iron is generally in the micromolar range (9), suggesting that there is an ample availability of iron for X. oryzae pv. oryzae in the rice xylem vessels within which it grows. The lack of expression of the xss operon during in planta growth is also consistent with the possibility that X. oryzae pv. oryzae does not ordinarily experience a limitation for iron during in planta growth. In this context, it is worth noting that R. solanacearum also does not produce the staphyloferrin B siderophore in tomato xylem sap (which contains >5 μM iron) (5). It is unclear why siderophores are important for virulence in some plant-pathogenic bacteria and are apparently disposable for virulence in others. The levels of available iron within plant tissues and the presence of alternate iron uptake systems might be contributing factors in determining the virulence phenotype of siderophore-deficient mutants of plant-pathogenic bacteria. Although siderophore is not required for growth of X. oryzae pv. oryzae within the host, it is possible that siderophore may have an important role in other phases of the life cycle of this pathogen wherein it has to survive outside the host.
X. oryzae pv. oryzae mutants for rpfF (regulation of pathogenicity factor; a global regulator) are deficient for virulence and growth under low-iron conditions and are siderophore overproducers (15). Exogenous iron supplementation has been shown to promote in planta growth of X. oryzae pv. oryzae rpfF mutants. It appears that the rpfF gene promotes virulence of X. oryzae pv. oryzae by facilitating iron uptake. A possibility that needs to be tested is that rpfF promotes iron acquisition by X. oryzae pv. oryzae through control of feoB expression. In summary, we have demonstrated that X. oryzae pv. oryzae possesses an FeoB-mediated and siderophore-mediated iron uptake system. We have further shown that FeoB protein is required for X. oryzae pv. oryzae to grow inside the rice plant. Another possibility is that X. oryzae pv. oryzae FeoB can be used as a target for developing new molecules that can be used for application on rice fields against bacterial blight disease and possibly diseases caused by other xanthomonads.
Acknowledgments
We thank Matthieu Arlat for providing the plasmid pVO155.
A.P. was supported by a Senior Research Fellowship from the Council of Scientific and Industrial Research (CSIR), Government of India. This work was supported, in part, by a grant to R.V.S. from the Department of Biotechnology, Government of India.
Footnotes
Published ahead of print on 9 April 2010.
REFERENCES
- 1.Allard, K. A., V. K. Viswanathan, and N. P. Cianciotto. 2006. lbtA and lbtB are required for production of the Legionella pneumophila siderophore legiobactin. J. Bacteriol. 188:1351-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Bearden, S. W., J. D. Fetherston, and R. D. Perry. 1997. Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis. Infect. Immun. 65:1659-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beasley, F. C., E. D. Vines, J. C. Grigg, Q. Zheng, S. Liu, G. A. Lajoie, M. E. Murphy, and D. E. Heinrichs. 2009. Characterization of staphyloferrin A biosynthetic and transport mutants in Staphylococcus aureus. Mol. Microbiol. 72:947-963. [DOI] [PubMed] [Google Scholar]
- 5.Bhatt, G., and T. P. Denny. 2004. Ralstonia solanacearum iron scavenging by the siderophore staphyloferrin B is controlled by PhcA, the global virulence regulator. J. Bacteriol. 186:7896-7904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhattacharyya, A., S. Stilwagen, N. Ivanova, M. D'Souza, A. Bernal, A. Lykidis, V. Kapatral, I. Anderson, N. Larsen, T. Los, G. Reznik, E. Selkov Jr., T. L. Walunas, H. Feil, W. S. Feil, A. Purcell, J. L. Lassez, T. L. Hawkins, R. Haselkorn, R. Overbeek, P. F. Predki, and N. C. Kyrpides. 2002. Whole-genome comparative analysis of three phytopathogenic Xylella fastidiosa strains. Proc. Natl. Acad. Sci. U. S. A. 99:12403-12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanvillain, S., D. Meyer, A. Boulanger, M. Lautier, C. Guynet, N. Denance, J. Vasse, E. Lauber, and M. Arlat. 2007. Plant carbohydrate scavenging through TonB-dependent receptors: a feature shared by phytopathogenic and aquatic bacteria. PLoS One 2:e224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyer, E., I. Bergevin, D. Malo, P. Gros, and M. F. Cellier. 2002. Acquisition of Mn(II) in addition to Fe(II) is required for full virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 70:6032-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braun, V., and H. Killmann. 1999. Bacterial solutions to the iron-supply problem. Trends Biochem. Sci. 24:104-109. [DOI] [PubMed] [Google Scholar]
- 10.Brickman, T. J., and S. K. Armstrong. 2005. Bordetella AlcS transporter functions in alcaligin siderophore export and is central to inducer sensing in positive regulation of alcaligin system gene expression. J. Bacteriol. 187:3650-3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bull, C. T., S. R. Carnegie, and J. E. Loper. 1996. Pathogenicity of mutants of Erwinia carotovora subsp. carotovora deficient in aerobactin and catecholate siderophore production. Phytopathology 86:260-266. [Google Scholar]
- 12.Cartron, M. L., S. Maddocks, P. Gillingham, C. J. Craven, and S. C. Andrews. 2006. Feo—transport of ferrous iron into bacteria. Biometals 19:143-157. [DOI] [PubMed] [Google Scholar]
- 13.Challis, G. L. 2005. A widely distributed bacterial pathway for siderophore biosynthesis independent of nonribosomal peptide synthetases. Chembiochem 6:601-611. [DOI] [PubMed] [Google Scholar]
- 14.Chan, J. W., and P. H. Goodwin. 1999. The molecular genetics of virulence of Xanthomonas campestris. Biotechnol. Adv. 17:489-508. [DOI] [PubMed] [Google Scholar]
- 15.Chatterjee, S., and R. V. Sonti. 2002. rpfF mutants of Xanthomonas oryzae pv. oryzae are deficient for virulence and growth under low iron conditions. Mol. Plant Microbe Interact. 15:463-471. [DOI] [PubMed] [Google Scholar]
- 16.Cody, Y. S., and D. C. Gross. 1987. Outer membrane protein mediating iron uptake via pyoverdinpss, the fluorescent siderophore produced by Pseudomonas syringae pv. syringae. J. Bacteriol. 169:2207-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crosa, J. H., and C. T. Walsh. 2002. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol. Mol. Biol. Rev. 66:223-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crossman, L. C., V. C. Gould, J. M. Dow, G. S. Vernikos, A. Okazaki, M. Sebaihia, D. Saunders, C. Arrowsmith, T. Carver, N. Peters, E. Adlem, A. Kerhornou, A. Lord, L. Murphy, K. Seeger, R. Squares, S. Rutter, M. A. Quail, M. A. Rajandream, D. Harris, C. Churcher, S. D. Bentley, J. Parkhill, N. R. Thomson, and M. B. Avison. 2008. The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol. 9:R74. http://genomebiology.com/2008/9/4/R74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dashper, S. G., C. A. Butler, J. P. Lissel, R. A. Paolini, B. Hoffmann, P. D. Veith, N. M. O'Brien-Simpson, S. L. Snelgrove, J. T. Tsiros, and E. C. Reynolds. 2005. A novel Porphyromonas gingivalis FeoB plays a role in manganese accumulation. J. Biol. Chem. 280:28095-28102. [DOI] [PubMed] [Google Scholar]
- 20.da Silva, A. C., J. A. Ferro, F. C. Reinach, C. S. Farah, L. R. Furlan, R. B. Quaggio, C. B. Monteiro-Vitorello, M. A. Van Sluys, N. F. Almeida, L. M. Alves, A. M. do Amaral, M. C. Bertolini, L. E. Camargo, G. Camarotte, F. Cannavan, J. Cardozo, F. Chambergo, L. P. Ciapina, R. M. Cicarelli, L. L. Coutinho, J. R. Cursino-Santos, H. El-Dorry, J. B. Faria, A. J. Ferreira, R. C. Ferreira, M. I. Ferro, E. F. Formighieri, M. C. Franco, C. C. Greggio, A. Gruber, A. M. Katsuyama, L. T. Kishi, R. P. Leite, E. G. Lemos, M. V. Lemos, E. C. Locali, M. A. Machado, A. M. Madeira, N. M. Martinez-Rossi, E. C. Martins, J. Meidanis, C. F. Menck, C. Y. Miyaki, D. H. Moon, L. M. Moreira, M. T. Novo, V. K. Okura, M. C. Oliveira, V. R. Oliveira, H. A. Pereira, A. Rossi, J. A. Sena, C. Silva, R. F. De Souza, L. A. Spinola, M. A. Takita, R. E. Tamura, E. C. Teixeira, R. I. Tezza, M. Trindade dos Santos, D. Truffi, S. M. Tsai, F. F. White, J. C. Setubal, and J. P. Kitajima. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417:459-463. [DOI] [PubMed] [Google Scholar]
- 21.DeFeyter, R., C. I. Kado, and D. W. Gabriel. 1990. Small, stable shuttle vectors for use in Xanthomonas. Gene 88:65-72. [DOI] [PubMed] [Google Scholar]
- 22.Dellagi, A., M. N. Brisset, J. P. Paulin, and D. Expert. 1998. Dual role of desferrioxamine in Erwinia amylovora pathogenicity. Mol. Plant Microbe Interact. 11:734-742. [DOI] [PubMed] [Google Scholar]
- 23.Franza, T., B. Mahe, and D. Expert. 2005. Erwinia chrysanthemi requires a second iron transport route dependent of the siderophore achromobactin for extracellular growth and plant infection. Mol. Microbiol. 55:261-275. [DOI] [PubMed] [Google Scholar]
- 24.Funahashi, T., K. Moriya, S. Uemura, S. Miyoshi, S. Shinoda, S. Narimatsu, and S. Yamamoto. 2002. Identification and characterization of pvuA, a gene encoding the ferric vibrioferrin receptor protein in Vibrio parahaemolyticus. J. Bacteriol. 184:936-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furrer, J. L., D. N. Sanders, I. G. Hook-Barnard, and M. A. McIntosh. 2002. Export of the siderophore enterobactin in Escherichia coli: involvement of a 43 kDa membrane exporter. Mol. Microbiol. 44:1225-1234. [DOI] [PubMed] [Google Scholar]
- 26.Gardy, J. L., M. R. Laird, F. Chen, S. Rey, C. J. Walsh, M. Ester, and F. S. Brinkman. 2005. PSORTb v. 2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics 21:617-623. [DOI] [PubMed] [Google Scholar]
- 27.He, Y. W., A. Y. Ng, M. Xu, K. Lin, L. H. Wang, Y. H. Dong, and L. H. Zhang. 2007. Xanthomonas campestris cell-cell communication involves a putative nucleotide receptor protein Clp and a hierarchical signalling network. Mol. Microbiol. 64:281-292. [DOI] [PubMed] [Google Scholar]
- 28.Innes, R. W., M. A. Hirose, and P. L. Kuempel. 1988. Induction of nitrogen-fixing nodules on clover requires only 32 kilobase pairs of DNA from the Rhizobium trifolii symbiosis plasmid. J. Bacteriol. 170:3793-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jefferson, R. A., T. A. Kavanagh, and M. W. Bevan. 1987. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6:3901-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones, A. M., S. E. Lindow, and M. C. Wildermuth. 2007. Salicylic acid, yersiniabactin, and pyoverdin production by the model phytopathogen Pseudomonas syringae pv. tomato DC3000: synthesis, regulation, and impact on tomato and Arabidopsis host plants. J. Bacteriol. 189:6773-6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kammler, M., C. Schon, and K. Hantke. 1993. Characterization of the ferrous iron uptake system of Escherichia coli. J. Bacteriol. 175:6212-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kauffman, H. E., A. P. K. Reddy, S. P. Y. Hsieh, and S. D. Merca. 1973. An improved technique for evaluation of resistance of rice varieties to Xanthomonas oryzae. Plant Dis. Rep. 57:537-541. [Google Scholar]
- 33.Krewulak, K. D., and H. J. Vogel. 2008. Structural biology of bacterial iron uptake. Biochim. Biophys. Acta 1778:1781-1804. [DOI] [PubMed] [Google Scholar]
- 34.Lawlor, M. S., C. O'connor, and V. L. Miller. 2007. Yersiniabactin is a virulence factor for Klebsiella pneumoniae during pulmonary infection. Infect. Immun. 75:1463-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leach, J. E., F. F. White, M. L. Rhoads, and H. Leung. 1990. A repetitive DNA sequence differentiates Xanthomonas campestris pv. oryzae from other pathovars of X. campestris. Mol. Plant Microbe Interact. 3:238-246. [Google Scholar]
- 36.Lee, B. M., Y. J. Park, D. S. Park, H. W. Kang, J. G. Kim, E. S. Song, I. C. Park, U. H. Yoon, J. H. Hahn, B. S. Koo, G. B. Lee, H. Kim, H. S. Park, K. O. Yoon, J. H. Kim, C. H. Jung, N. H. Koh, J. S. Seo, and S. J. Go. 2005. The genome sequence of Xanthomonas oryzae pathovar oryzae KACC10331, the bacterial blight pathogen of rice. Nucleic Acids Res. 33:577-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee, S. J., J. H. Ko, H. Y. Kang, and Y. Lee. 2006. Coupled expression of MhpE aldolase and MhpF dehydrogenase in Escherichia coli. Biochem. Biophys. Res. Commun. 346:1009-1015. [DOI] [PubMed] [Google Scholar]
- 38.Leong, S. A., and J. B. Neilands. 1981. Relationship of siderophore-mediated iron assimilation to virulence in crown gall disease. J. Bacteriol. 147:482-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loprasert, S., R. Sallabhan, S. Atichartpongkul, and S. Mongkolsuk. 1999. Characterization of a ferric uptake regulator (fur) gene from Xanthomonas campestris pv. phaseoli with unusual primary structure, genome organization, and expression patterns. Gene. 239:251-258. [DOI] [PubMed] [Google Scholar]
- 40.Marlovits, T. C., W. Haase, C. Herrmann, S. G. Aller, and V. M. Unger. 2002. The membrane protein FeoB contains an intramolecular G protein essential for Fe(II) uptake in bacteria. Proc. Natl. Acad. Sci. U. S. A. 99:16243-16248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinez, J. L., M. Herrero, and V. de Lorenzo. 1994. The organization of intercistronic regions of the aerobactin operon of pColV-K30 may account for the differential expression of the iucABCD iutA genes. J. Mol. Biol. 238:288-293. [DOI] [PubMed] [Google Scholar]
- 42.Meyer, J. M., A. Neely, A. Stintzi, C. Georges, and I. A. Holder. 1996. Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect. Immun. 64:518-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miethke, M., and M. A. Marahiel. 2007. Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 71:413-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miethke, M., S. Schmidt, and M. A. Marahiel. 2008. The major facilitator superfamily-type transporter YmfE and the multidrug-efflux activator Mta mediate bacillibactin secretion in Bacillus subtilis. J. Bacteriol. 190:5143-5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Naikare, H., K. Palyada, R. Panciera, D. Marlow, and A. Stintzi. 2006. Major role for FeoB in Campylobacter jejuni ferrous iron acquisition, gut colonization, and intracellular survival. Infect. Immun. 74:5433-5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neilands, J. B. 1995. Siderophores: structure and function of microbial iron transport compounds. J. Biol. Chem. 270:26723-26726. [DOI] [PubMed] [Google Scholar]
- 47.Nino-Liu, D. O., P. C. Ronald, and A. J. Bogdanove. 2006. Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol. Plant Pathol. 7:303-324. [DOI] [PubMed] [Google Scholar]
- 48.Oke, V., and S. R. Long. 1999. Bacterial genes induced within the nodule during the Rhizobium-legume symbiosis. Mol. Microbiol. 32:837-849. [DOI] [PubMed] [Google Scholar]
- 49.Page, W. J., E. Kwon, A. S. Cornish, and A. E. Tindale. 2003. The csbX gene of Azotobacter vinelandii encodes an MFS efflux pump required for catecholate siderophore export. FEMS Microbiol. Lett. 228:211-216. [DOI] [PubMed] [Google Scholar]
- 50.Palmer, C. M., and M. L. Guerinot. 2009. Facing the challenges of Cu, Fe and Zn homeostasis in plants. Nat. Chem. Biol. 5:333-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Panina, E. M., A. A. Mironov, and M. S. Gelfand. 2001. Comparative analysis of FUR regulons in gamma-proteobacteria. Nucleic Acids Res. 29:5195-5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Payne, S. M. 1994. Detection, isolation, and characterization of siderophores. Methods Enzymol. 235:329-344. [DOI] [PubMed] [Google Scholar]
- 53.Putman, M., H. W. van Veen, and W. N. Konings. 2000. Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 64:672-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rajagopal, L., S. Dharmapuri, A. T. Sayeepriyadarshini, and R. V. Sonti. 1999. A genomic library of Xanthomonas oryzae pv. oryzae in the broad host range mobilizing Escherichia coli strain S17-1. Int. Rice Res. Notes 24:20-21. [Google Scholar]
- 55.Ray, S. K., R. Rajeshwari, and R. V. Sonti. 2000. Mutants of Xanthomonas oryzae pv. oryzae deficient in general secretory pathway are virulence deficient and unable to secrete xylanase. Mol. Plant Microbe Interact. 13:394-401. [DOI] [PubMed] [Google Scholar]
- 56.Regha, K., A. K. Satapathy, and M. K. Ray. 2005. RecD plays an essential function during growth at low temperature in the Antarctic bacterium Pseudomonas syringae Lz4W. Genetics 170:1473-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robey, M., and N. P. Cianciotto. 2002. Legionella pneumophila feoAB promotes ferrous iron uptake and intracellular infection. Infect. Immun. 70:5659-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rondon, M. R., K. S. Ballering, and M. G. Thomas. 2004. Identification and analysis of a siderophore biosynthetic gene cluster from Agrobacterium tumefaciens C58. Microbiology 150:3857-3866. [DOI] [PubMed] [Google Scholar]
- 59.Rose, T. M., E. R. Schultz, J. G. Henikoff, S. Pietrokovski, C. M. McCallum, and S. Henikoff. 1998. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res. 26:1628-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ryan, R. P., S. Monchy, M. Cardinale, S. Taghavi, L. Crossman, M. B. Avison, G. Berg, D. van der Lelie, and J. M. Dow. 2009. The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat. Rev. Microbiol. 7:514-525. [DOI] [PubMed] [Google Scholar]
- 61.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 62.Schafer, A., A. Tauch, W. Jager, J. Kalinowski, G. Thierbach, and A. Puhler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 63.Schwyn, B., and J. B. Neilands. 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160:47-56. [DOI] [PubMed] [Google Scholar]
- 64.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1:784-791. [Google Scholar]
- 65.Stojiljkovic, I., M. Cobeljic, and K. Hantke. 1993. Escherichia coli K-12 ferrous iron uptake mutants are impaired in their ability to colonize the mouse intestine. FEMS Microbiol. Lett. 108:111-115. [DOI] [PubMed] [Google Scholar]
- 66.Subramoni, S., and R. V. Sonti. 2005. Growth deficiency of a Xanthomonas oryzae pv. oryzae fur mutant in rice leaves is rescued by ascorbic acid supplementation. Mol. Plant Microbe Interact. 18:644-651. [DOI] [PubMed] [Google Scholar]
- 67.Tanabe, T., T. Funahashi, H. Nakao, S. Miyoshi, S. Shinoda, and S. Yamamoto. 2003. Identification and characterization of genes required for biosynthesis and transport of the siderophore vibrioferrin in Vibrio parahaemolyticus. J. Bacteriol. 185:6938-6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tanabe, T., H. Nakao, T. Kuroda, T. Tsuchiya, and S. Yamamoto. 2006. Involvement of the Vibrio parahaemolyticus pvsC gene in export of the siderophore vibrioferrin. Microbiol. Immunol. 50:871-876. [PubMed] [Google Scholar]
- 69.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Touati, D. 2000. Iron and oxidative stress in bacteria. Arch. Biochem. Biophys. 373:1-6. [DOI] [PubMed] [Google Scholar]
- 71.Velayudhan, J., N. J. Hughes, A. A. McColm, J. Bagshaw, C. L. Clayton, S. C. Andrews, and D. J. Kelly. 2000. Iron acquisition and virulence in Helicobacter pylori: a major role for FeoB, a high-affinity ferrous iron transporter. Mol. Microbiol. 37:274-286. [DOI] [PubMed] [Google Scholar]
- 72.Wiggerich, H. G., B. Klauke, R. Koplin, U. B. Priefer, and A. Puhler. 1997. Unusual structure of the tonB-exb DNA region of Xanthomonas campestris pv. campestris: tonB, exbB, and exbD1 are essential for ferric iron uptake, but exbD2 is not. J. Bacteriol. 179:7103-7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wiggerich, H. G., and A. Puhler. 2000. The exbD2 gene as well as the iron-uptake genes tonB, exbB and exbD1 of Xanthomonas campestris pv. campestris are essential for the induction of a hypersensitive response on pepper (Capsicum annuum). Microbiology 146:1053-1060. [DOI] [PubMed] [Google Scholar]
- 74.Windgassen, M., A. Urban, and K. E. Jaeger. 2000. Rapid gene inactivation in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 193:201-205. [DOI] [PubMed] [Google Scholar]
- 75.Yokosho, K., N. Yamaji, D. Ueno, N. Mitani, and J. F. Ma. 2009. OsFRDL1 is a citrate transporter required for efficient translocation of iron in rice. Plant Physiol. 149:297-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zaini, P. A., A. C. Fogaca, F. G. Lupo, H. I. Nakaya, R. Z. Vencio, and A. M. da Silva. 2008. The iron stimulon of Xylella fastidiosa includes genes for type IV pilus and colicin V-like bacteriocins. J. Bacteriol. 190:2368-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]