Abstract
Induced pluripotent stem (iPS) cells can be obtained through the introduction of defined factors into somatic cells1. The combination of Oct4, Sox2 and Klf4 (OSK) constitutes the minimal requirement for generating iPS cells from mouse embryonic fibroblasts (MEFs). These cells are thought to resemble embryonic stem cells (ESCs) based on global gene expression analyses; but, few studies have tested their ability and efficiency in contributing to chimerism, colonization of germ tissues, and most importantly, germ-line transmission and life-birth from iPS cells produced with tetraploid complementation. Through the genomic analyses of ESC genes that have roles in pluripotency and fusion-mediated somatic cell reprogramming, we identified Tbx3 as a transcription factor that significantly improves the quality of iPS cells. Induced-PS cells generated with OSK + Tbx3 (OSKT) are superior in both germ cell contribution to the gonads and germ-line transmission frequency. However, global gene expression profiling could not distinguish between OSK and OSKT iPS cells. Genome-wide ChIP-sequencing analysis of Tbx3 binding sites in ESCs suggests that Tbx3 regulates pluripotency-associated and reprogramming factors, in addition to sharing many common downstream regulatory targets with Oct4, Sox2, Nanog and Smad1. This study underscores the intrinsic qualitative differences between iPS cells generated by different methods and highlights the need to rigorously characterize iPS cells beyond in vitro studies.
The pluripotency and self-renewing properties of ESCs are conferred by a set of core factors that helps determine their unique identity. Adult somatic cells can be reprogrammed to resemble ESCs when some of these key transcription factors are introduced1. Induced-PS cells can be obtained by the viral transduction of a few genes in both mouse and human cells, albeit at low efficiency. Supplementations with chemical compounds such as inhibitors to DNA methyltransferase, histone deacetylase, histone methyltransferase, mitogen-activated protein kinase (MAPK) and glycogen synthase kinase-3 (GSK3) have been reported to improve the reprogramming efficiency2-4. Recently, iPS cells have been generated without the use of viral vectors5. While ESC-like iPS cells are routinely obtained with these methods, very few studies have carefully examined their germ-line contribution and transmission frequency6. Although iPS cells have a distinct morphology and express molecular markers similar to ESCs, their ability and degree of contribution to the chimera appear highly varied3,7-9. This suggests that iPS cells do not completely resemble ESCs10, and there is marked disparity in the quality of different iPS cell lines. Hence, we postulated that other factors in addition to the basal requirements of OSK may improve the quality of iPS cells as defined by their capacity for high germ-line competency.
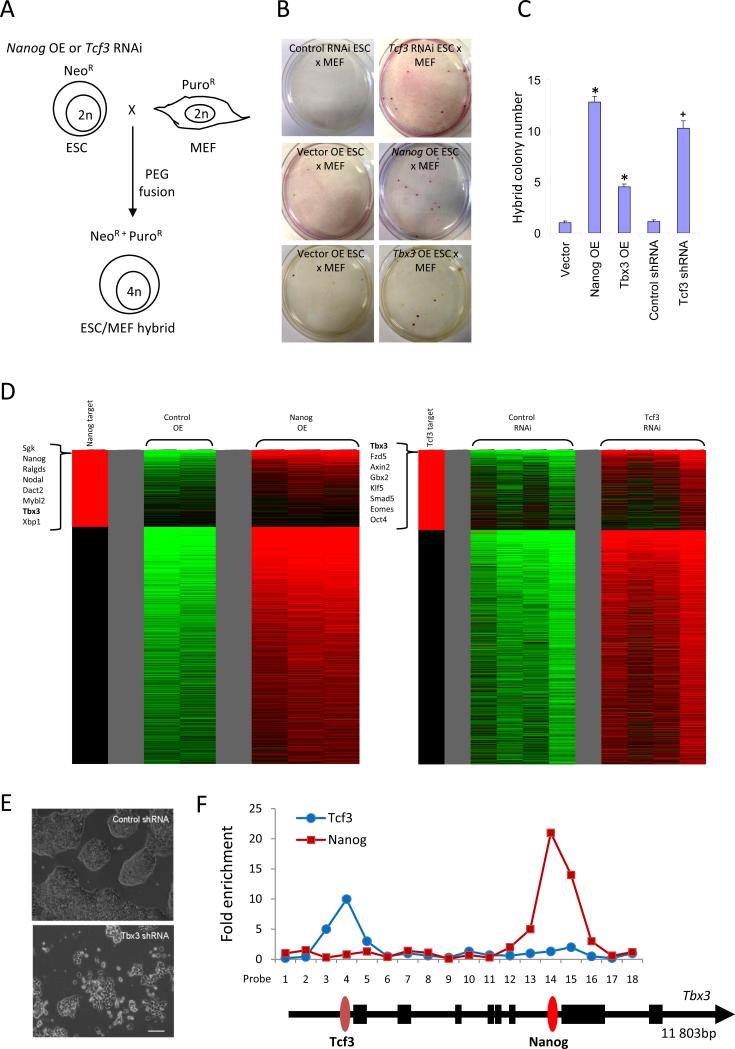
We speculated that iPS cell-reprogramming factors may share common characteristics with pluripotency-associated genes whose perturbed levels in ESCs confer resistance to differentiation. Previous studies have shown that mouse ESCs over-expressing Nanog are resistant to differentiation11, express higher levels of pluripotency-associated genes, and are more effective at reprogramming somatic cells through cell fusion12. Another transcription factor Tcf3 when depleted in ESCs limits their differentiation ability, and upregulates the expression of pluripotency markers that includes Oct4, Sox2, Nanog and Sall413. As both Nanog and Tcf3 regulate each other, and are core features of the ESC transcriptional network, we speculated that similar to Nanog over-expressing ESCs, the loss of Tcf3 may enhance fusion-mediated reprogramming of somatic hybrid cells. To test this, we used polyethyleneglycol (PEG) to generate duo drug-resistant fusion hybrids between Nanog over-expressing (OE) or Tcf3 RNAi ESCs that were neomycin-resistant (NeoR) and primary MEFs that were puromycin-resistant (PuroR) (Figure 1A). Consistent with previous observations, Nanog OE ESCs showed enhanced reprogramming efficiency (Figure 1B & C). Using Tcf3-deficient ESC lines, a significant increase in the number of hybrid clones was also observed (Figure 1B & C). Karyotype analysis confirmed that these were tetraploid (Figure S1). The hybrids possess properties similar to the parental modified ESC lines, including their response to the lack of leukemia inhibitory factor (LIF) and epigenetic reprogramming of the Nanog promoter (Figure S2 & S3). We eliminated the possibility that improvements in reprogramming frequency was attributed to increased cell fusion events12 (Figure S4).
Figure 1.
Global gene expression profiling reveals Tbx3 aids cell fusion-mediated reprogramming. (A) Modified ESCs with Nanog over-expression (OE) or Tcf3 RNAi were fused with MEFs to generate tetraploid ESC/MEF hybrids resistant to neomycin and puromycin. (B) Nanog OE, Tbx3 OE and Tcf3 RNAi enhanced cell fusion-mediated reprogramming of MEFs. Representative examples illustrate the emergence of ESC/MEF hybrid colonies. Control ESC fusion with MEFs resulted in an average of one per experiment whereas Tcf3 RNAi, Nanog OE or Tbx3 OE ESCs produced numerous hybrid clones. (C) Nanog OE ESCs were efficient in reprogramming MEFs, generating 13 colonies, followed by Tcf3 RNAi (10) and Tbx3 OE (4.5). The numbers represent the average of four independent fusion experiments. * denotes significantly different from vector, + denotes significantly different from control shRNA; error bars represent s.e.m. (D) The heat-map shows all genes which were increased in treated ESCs compared to controls. Tbx3 was among the most highly up-regulated genes in Nanog OE and Tcf3 RNAi ESCs. The left-most column in red indicates direct gene targets of Nanog or Tcf3 based on the ChIP-PET21 and ChIP-chip13 databases respectively. (E) RNAi knockdown of Tbx3 in ESCs led to a loss of self-renewal and induced differentiation. Scale bar = 100 μm. (F) Enrichment of Tcf3 and Nanog occupancy on the Tbx3 gene, as measured by ChIP-qPCR.
To dissect the commonalities between Nanog and Tcf3 pathways, we examined the repertoire of genes elevated in Nanog OE and Tcf3 RNAi ESCs that could suggest shared downstream mediators. The intersected expression profiles revealed a handful of genes such as Dazl, Fzd10, Hal, 4930502E18Rik, and Erf that were upregulated in both conditions (Table S1 & S2). Strikingly, Tbx3, a transcription factor previously reported to sustain pluripotency14 was strongly elevated (Figure 1D; Figure S5A). RNAi knockdown of Tbx3 in ESCs induced differentiation (Figure 1E), with concomitant downregulation of pluripotency-associated genes (Table S3). Tbx3 is also directly bound by both Nanog and Tcf3 (Figure 1F). In reprogrammed ESC/MEF hybrids, Tbx3 levels remained highly elevated (Figure S5B; Table S4 & S5). To test the role of Tbx3 in cell fusion-mediated reprogramming, we fused NeoR Tbx3 over-expressing ESC lines with MEFs (Figure S6). Indeed, there was an increase in the number of hybrids (Figure 1B & C), that was not due to enhanced cell fusion (Figure S4).
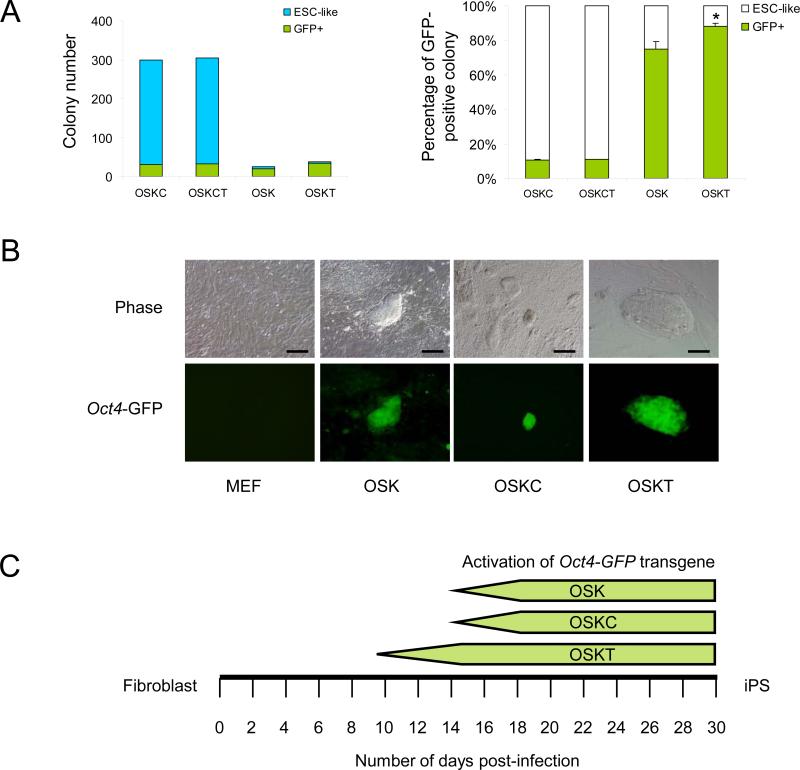
We postulated that Tbx3 may also improve the reprogramming efficiency or the quality of iPS cells. Retroviral infection of MEFs bearing the Oct4-GFP transgene with OSKC induced ~300 ESC-like colonies per 5 × 104 starting cells (Figure 2A). However, only ~10% of these showed activation of the transgene. The addition of Tbx3 (OSKCT) did not increase the frequency of GFP+ colony numbers (Figure 2A). With three factors (OSK), the total number of ESC-like colonies was dramatically reduced but 74% of OSK colonies expressed GFP (Figure 2A). The addition of Tbx3 (OSKT) improved the colony count (~38 on average) when compared with OSK (~26), and the percentage of GFP+ colonies also significantly increased to 89% (Figure 2A). Other qualitative differences between iPS cells obtained with different factor combinations were observed. OSKC iPS cell colonies were difficult to distinguish amongst transformed and partially reprogrammed cells that did not show GFP expression. OSK and OSKT iPS cell colonies were morphologically similar, with uniform GFP expression within individual colonies (Figure 2B). However, while the activation of Oct4 typically required 14 days post-infection with OSK and OSKC, the use of OSKT took 9-10 days, suggesting that Tbx3 accelerated the reprogramming process (Figure 2C). The efficiency of isolating stable iPS cell lines from GFP+ colonies was similar between OSK and OSKT transduction, but almost two-fold higher than OSKC and OSKCT (Figure S7). For all the iPS cell lines obtained, we performed PCR analyses and confirmed genomic integration of the respective transduced genes (Figure S8). Molecular characterization of iPS cells from OSKT confirmed these were alkaline phosphatase-positive, expressed Nanog, Sox2 and SSEA1 (Figure S9). They also form teratomas comprised of multiple differentiated cell types when xenografted into SCID mice (Figure S10).
Figure 2.
Generation of iPS cells with Oct4, Sox2, Klf4 and Tbx3 retroviral transduction. (A) OSKC and OSKCT induced ~300 ESC-like colonies with ~10% expressing GFP, whereas OSKT induced an average of 38 compared to 26 ESC-like colonies from OSK, with 89% and 74% GFP activation respectively. Colonies were assessed and counted on day 16. Data represents the average of three independent transduction experiments. * denotes significantly different from OSK; p<0.05; error bars represent s.e.m. (B) OSK and OSKT iPS cell clones showed tight, domed-shaped ESC-like morphology and uniform GFP expression throughout the colony, whereas OSKC clones appeared as flattened, transformed cells with sparse GFP expression. Scale bar = 100 μm. (C) OSKT induced the activation of Oct4-GFP transgene in iPS derived from primary MEFs at 9-10 days post-infection while OSK and OSKC combinations required 14 days observed in four independent transduction experiment.
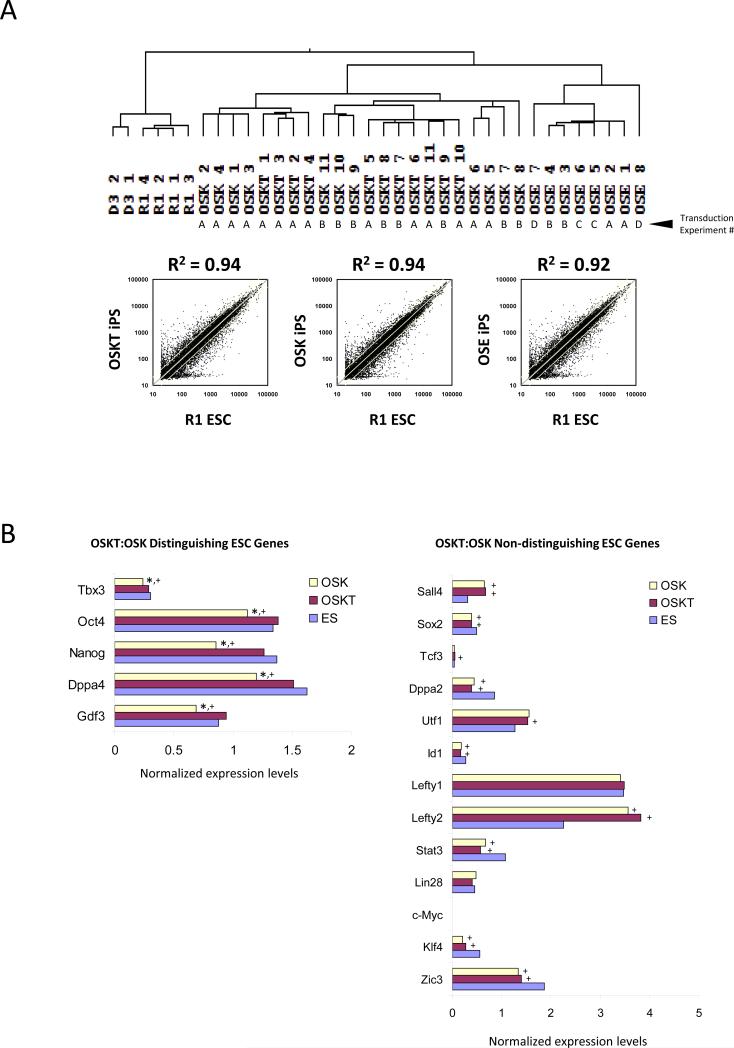
Next we sought to examine the differences in global transcriptome profiles between iPS cells generated using different factor combinations. To eliminate variations that could have arisen from handling techniques, and to ensure the reproducibility of properties inherent to iPS cell lines generated with different combinations, clones were isolated from two or more independent transduction experiments. Hierarchical clustering revealed that the recently reported iPS cells generated with OS + Esrrb (OSE)7 were most dissimilar to wild-type R1 and D3 ESCs with a correlation coefficient (R2) of 0.92 (Figure 3A). Both OSK and OSKT iPS cells bore closer resemblance to ESCs but were indistinguishable from each other; R2 = 0.94. Hence, global expression profiling was not sufficiently sensitive in detecting differences between these clones. Closer examination of specific gene level alterations, however, revealed key differences (Figure 3B; Figure S11). We compared the pluripotency-associated gene levels among OSK, OSKT iPS and ES cells. The majority of these genes such as Sall4, Tcf3, Sox2, Zfx, Lin28, Utf1 and Zic3 were non-distinguishing between OSKT and OSK cells as their levels were similar. Surprisingly, a small subset of distinguishing features appeared to define OSKT from OSK cells. Oct4, Nanog, Gdf3, Dppa4 and Tbx3 levels in OSKT cells were equivalent to ESCs, but significantly reduced in OSK cells. This suggests that exogenous Tbx3 may be crucial for assisting in re-establishing proper levels of certain ESC factors critical for the induction of pluripotency that cannot be completely achieved with OSK alone.
Figure 3.
Transcriptome of iPS cell clones generated with different combinations of reprogramming factors. (A) Hierarchical clustering of global expression profiling showed that OSKT and OSK clones are more similar to wild-type ESCs than OSE, but indistinguishable from each other based on correlation coefficient. R2 value was obtained from the average individual gene signal intensity of all iPS clones in each combination and compared against R1 ESCs. The independent transduction experiments where the clones were isolated are denoted as A, B, C, and D. (B) Analysis of individual ESC-associated gene profiles revealed a subset that could distinguish OSKT from OSK-derived iPS cells. ‘Distinguishing’ ESC genes were expressed at levels similar between OSKT and ESCs but significantly lower in OSK. The majority of other ESC-associated genes were ‘non-distinguishing’ and present at levels similar between both OSKT and OSK. * denotes significantly different from OSKT; + denotes significantly different from ESC; p<0.05. Changes in gene expression based on microarray were confirmed with qPCR (Figure S11).
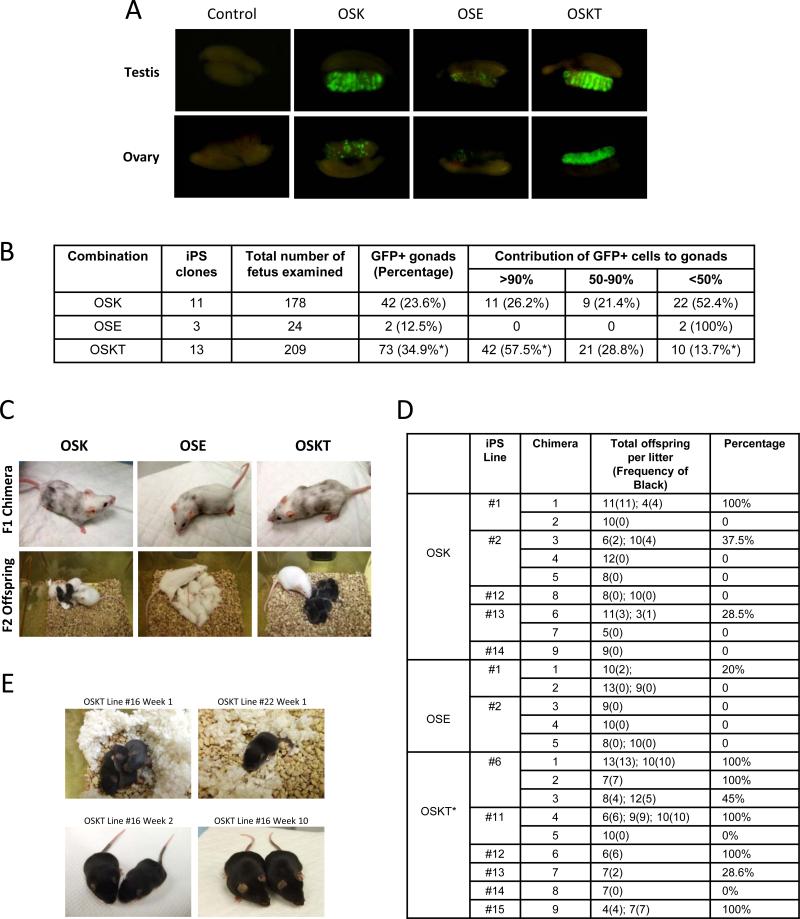
We then investigated whether OSKT cells were of higher quality than OSK and OSE cells (Figure S12; summarized in Table S7). We selected numerous iPS cell lines generated from each combination which showed homogeneous activation of Oct4-GFP within each colony: OSKT #1,2,4,6,11-22; OSK #1-3,12-14,16-22; OSE #1,2,9. These lines were derived from four independent transduction experiments performed at different times to eliminate any biasness in clonal selection that may arise from the stochastic behavior of individual clones. Induced-PS cells were then injected into 4-8 cell embryos and cultured in vitro to blastocysts (Figure S13). There was no difference in the maturation efficiency (~95%) between the three iPS cell types (data not shown). All blastocysts transplanted into the surrogate female mice initially had shown robust contribution of GFP+ cells to the inner cell mass (ICM) (Figure S13) and live chimeras were obtained. As evaluated by coat color, all OSKT and OSK chimeras showed coats with varying density of black fur, signifying iPS cell contribution; whereas OSE chimeras clearly had lesser black coat (Figure 4C, top panels), indicating OSE cells tend to contribute poorly to chimerism.
Figure 4.
OSKT iPS cells show enhanced germ-line contribution and transmission. The iPS clone ID and the types of analyses that were performed is summarized in Table S7. (A) Representative photos showing the quantitative contribution and spatial distribution of GFP+ cells generated with different reprogramming factor combinations in the gonads of chimeric embryos. (B) Tabulation comparing the contribution of GFP+ iPS cell-derived germ cells to the gonads of chimeric fetuses and their spatial distribution with different combinations of factors. OSKT iPS cells were most effective at colonizing the gonads, compared to OSK and OSE iPS cells. * denotes significantly different from OSK, p<0.01. At least eleven independent sets of microinjections were performed for OSK versus OSKT comparison. (C) Representative photos showing the contribution of iPS cells to chimeric coat and the production of F2 offspring after crossing with albino mice. Black offspring indicates germ-line transmission of iPS cells. (D) Table summarizing the germ-line transmission frequency for iPS cells generated with the different combination of factors. For each combination, between two to six iPS cell lines were used to obtain live chimeras, of which, at least one male from each combination were crossed with an albino female to determine the frequency of black F2. * denotes significantly different from OSK and OSE, p<0.005 and p<0.001 respectively. (E) Live birth animals obtained from two OSKT iPS cell lines tested by tetraploid complementation.
More interestingly, obvious differences were discerned between OSKT and OSK cells in their ability to colonize the germ tissues. At E13.5, gonads were obtained from the F1 chimeric embryos. 34.9% of the gonads from OSKT embryos showed contribution of iPS cells with GFP expression contrasting with 23.6% from OSK and 12.5% from OSE embryos (Figure 4A & B; Figure S14; p<0.01). Further assessment based on GFP distribution within these gonads revealed extensive contribution by OSKT cells as 57.5% of the chimeric gonads were >90% GFP+, compared to 26.2% of those from OSK embryos (Figure 4B; p<0.01). This indicates OSKT cells were more efficient in colonizing the germ tissues. Employing one of the most stringent criteria for demonstrating the quality of iPS cell clones, we tested their frequency for germ-line transmission and production of viable F2 offspring. Once again, we used several iPS cell lines from each combination. Induced-PS cells generated with OSE had very poor capacity for germ-line transmission (Figure 4C, bottom panels; and Figure 4D). Using five chimeras from two lines which were bred with albino mice, a total of seven litters were obtained. Only one litter contained two of 10 offspring that had iPS cell-derived black coat. With OSK, nine chimeras from five lines were used for breeding. 13 litters were produced, of which only two had 100% black offspring, and another four had an average of ~33%. Strikingly, with the nine OSKT chimeras obtained from six lines, nine of 14 litters had 100% black offspring, three had 41%, 50% and 28% each, and only two with none (Figure 4D; p<0.005).
Previous reports had implicated improper retroviral silencing and the frequency of retroviral integrations in altering the efficiency of generating iPS cells and their resultant properties15. To exclude these possibilities, we confirmed that exogenous expression from the retroviral plasmids was silenced (Figure S15). Southern blot analyses showed that retroviral integration of Oct4, Sox2 and Klf4 transgenes into either OSK or OSKT cells were comparable (Figure S16). Importantly, even with the additional integration of exogenous Tbx3, these cells were consistently of higher quality, thus ruling out the effects of DNA damage. We then speculated that the higher quality OSKT iPS cells could be used to generate viable mice composed entirely of the engineered cells through tetraploid complementation16,17. This has not been previously shown, perhaps owing to the difficulty of generating high quality iPS cells. Five lines of each OSK and OSKT were tested. In two lines of each, surrogate females were sacrificed at E19 to check for embryos. Only one embryo was obtained out of 66 implanted tetraploid aggregates for OSK cells, whereas four embryos were obtained out of 59 implants for OSKT cells. With the remaining three lines from each, implanted females were followed to term. With OSK lines, only one produced live birth, with a cumulative yield of three births from 105 implants whereas two of the OSKT lines yielded 11 live births out of 107 implants, and two animals continued to thrive beyond ten weeks (Figure 4E).
To better understand how Tbx3 may contribute to improving iPS cell quality, we performed Solexa ChIP-sequencing to uncover the direct regulatory targets of Tbx3 in ESCs (Table S6). Hierarchical clustering of Tbx3 with the previously mapped ESC factors18 revealed that it shares a large number of common binding sites with the classic pluripotency-associated transcription factors Oct4, Sox2, Nanog and Smad1 (Figure S17). Tbx3 is also found to target ESC factors Oct4, Sox2, Sall4, Lefty1, Lefty2, Zfp42, as well as reprogramming factors Klf2, Klf4, Klf5, n-Myc and c-Myc (Figure S18).
Taken altogether, our study highlights the success of OSKT combination in generating high quality iPS cells capable of germ-line transmission at high efficiency. Previous studies have primarily employed morphological assessments and global gene expression analyses as indicators of iPS cell pluripotency. However, in vitro analyses are not sufficient to distinguish bona fide iPS cells with true ESC properties from poor quality iPS cells which do not possess germ-line competency. The ability of OSKT cells in generating high frequency live birth mice through tetraploid complementation further strengthens the qualitative impact of Tbx3. While Tbx3 retroviral transduction in conjunction with OSK significantly improves iPS cell quality, we noted that not all OSKT clones would manifest robust germ-line transmission. Two reports of more uniform platforms for reprogramming could help address this issue further. Firstly, an inducible system in homogenous starting fibroblast population19 that includes Tbx3 for reprogramming may allow for derivation and evaluation of iPS clones with more consistent biological properties. Secondly, the dual inhibition of MAPK signaling pathway and GSK3, which greatly enhances the conversion of pre-iPS to fully pluripotent iPS cells4 may also lead to the production of iPS clones with more uniform properties.
With the emergence of modified genetic and chemical methods, and an emphasis on the use of minimal reprogramming factors to derive iPS cells, some benchmark as exemplified here by OSKT-derived iPS cells should be employed to evaluate their qualitative and biological properties (Figure S19). The precise mechanistic role for Tbx3 in vastly improving iPS cell quality needs more clarification. The initial ChIP-sequencing data suggests that Tbx3 may be important for the effective re-establishment of the ESC circuitry during the onset of reprogramming, and its subsequent maintenance. The presence of exogenous Tbx3 during the initiation of reprogramming may ensure proper titration of pluripotency-associated and reprogramming factors which are reactivated by OSK to the optimal level. We propose a model whereby the re-establishment of pluripotency from a somatic state is achieved in an increasing probabilistic step-wise manner (Figure S19). The use of different factor combinations results in the generation of iPS populations and clones with markedly varied developmental potentials centered upon attaining progressive “landmarks” of pluripotency. The addition of Tbx3 to a particular combination increases the probabilistic frequency of iPS cells that attain a pluripotent state equivalent or closest to ESCs within the entire population of reprogrammed cells.
Methods Summary
Tbx3 was cloned into pLenti6-UBC and pMXs vectors; Oct4, Sox2, Klf4, c-Myc plamids were obtained from Addgene; Esrrb was obtained from Feng et al7. Retrovirus and lentivirus were generated as previously described1,20. Equal amounts of virus encoding different combination of factors were applied to 5 × 104 plated Oct4-GFP transgenic MEFs in 10 cm2 dishes. After 24 h, inactivated feeder cells were added, and the culture was then maintained for up to 21 days. To generate chimera, albino embryos were isolated at 2-cell stage, matured to 4-8-cell stage and microinjected with iPS cells using the Piezo Micro Manipulator. Injected embryos were cultured to the blastocyst stage and transferred to the uterine horns of E2.5 pseudopregnant F1 (CBA × C57BL/6J) females. Chimeric embryos were harvested at E13.5 for analyses of GFP expression in the gonads. For tetraploid complementation, tetraploid (4n) embryos were generated using 2-cell electro-fusion and incubated in KSOM till 4-8 cell stage prior to aggregation with iPS cells. The zona pellucida of the tetraploid 4-8 cell stage embryos were removed by brief exposure to acid Tyrode's solution. Three tetraploid embryos and ~40 iPS cells were aggregated in a single well, and incubated in medium for 24 h to form a morula or blastocyst16,17. Approximately 10-14 embryos were transferred into the uterus or oviducts of pseudopregnant mice.
Materials and Methods
Cell culture and transfection
All cell cultures were maintained at 37°C with 5% CO2. The culture of mouse R1 and D3 ESCs was described previously22. HEK293T cells were maintained in DMEM supplemented with 10% FBS and penicillin/streptomycin. Transfection of plasmids into mouse ESCs and HEK293 cells was performed using Lipofectamine 2000 (Invitrogen). Plat-E packaging cells (Cell Biolabs, INC), which were used to produce retrovirus, were maintained according to the manufacturer's guide.
Plasmid construction, viral packaging and infection
Coding sequences of Tbx3 were amplified from mouse ESCs by RT-PCR and cloned into pLenti6-UBC (Invitrogen) and pMXs vectors; Oct4, Sox2, Klf4, c-Myc in pMXs were obtained from Addgene; Esrrb was obtained from Feng et al7. The retrovirus and lentivirus were generated as previously described1,20. For the generation of iPS cells, equal amounts of virus encoding different combination of factors were applied to 5 × 104 plated MEFs in 10 cm2 dishes in 10% FBS DMEM media containing 8 ng ml-1 polybrene. After 24 h, inactivated feeder cells and fresh media were added, and the culture was then maintained for up to 21 days. For RNA interference (RNAi) design and construction of plasmids for shRNA synthesis, 19 base-pair gene-specific regions were designed based on the algorithm of Reynolds et al23. Oligonucleotides were cloned into pSuper.puro (Oligoengine). All sequences were analyzed by BLAST to ensure specificity. Tbx3 RNAi sequence: GAGCCAACGATATCCTGAA; Control RNAi sequence: GATGAAATGGGTAAGTACA.
Cell fusion
For PEG-mediated fusions, cells of each type (~1 × 106) were mixed in serum-free DMEM, pelleted, and the supernatant removed. The pellet was resuspended in 300 μl of 50% w/v PEG1500, and left for 3 min with occasional tapping to mix. Then, 2 ml of medium was added and the cells were spun down, and supernatant discarded. The pellet was resuspended in ESC medium and plated on 10 cm2 dishes. Puromycin (1 μg ml-1) and neomycin (300 mg ml-1) were added after 24 h.
Gene expression microarray and analysis
Cells were rinsed twice in ice-cold PBS. Total RNA was extracted using Trizol (Invitrogen) and column-purified with RNeasy kits (Qiagen). Expression profiling of coding genes was carried out using Illumina MouseRef-8v1.1 BeadArrays as per manufacturer's instructions. Array data is deposited for public access with GEO repository accession number: updating in progress. All data were subtracted from background intensities and were normalized across chips using the cross-correlation method24. The normalized data were first log2 transformed and then subtracted from the mean of the median intensities of the two groups (i.e. Nanog OE with control vector, Tcf3 RNAi with control RNAi). Prior to clustering, the data were further sorted based on direct targets of Nanog or Tcf3, and subsequently arranged in the descending order of the fold change. The clustering tree was generated using hierarchical clustering with average linkage.
ChIP sequencing analysis
ChIP assay was carried out as described previously25. Briefly, the cells were cross-linked with 1% (w/v) formaldehyde for 10 min at room temperature, and formaldehyde was then inactivated by the addition of 125 mM glycine. Chromatin extracts containing DNA fragments with an average size of 500 bp were immunoprecipitated using anti-Tbx3 antibody (sc-31657; Santa Cruz Biotechnology). The ChIP-enriched DNA was then decrosslinked and analyzed by realtime PCR using the ABI PRISM 7900 sequence detection system and SYBR green master mix. For ChIP-seq assay, 10 ng of ChIP DNA was end polished with T4 DNA polymerase and kinase. An A base was added to the polished DNA fragments followed by the Qiaquick column clean up. Solexa adaptors were ligated to the ChIP DNA fragments and enriched by 15 cycles of PCR amplification. 150-300 bp size fractions were selectively cut out from the gel and eluted by Qiagen gel extraction kit. The extracted DNA was quantified by picogreen assay and subjected to Solexa sequencing according to the manufacturer's instruction. Tbx3 binding sites peak calling was done by MACS program using specific chromatin IP and control input sequencing data from the same cells as describe in the Supplement. Colocalization of ChIP-seq TFs binding sites in mouse genome was done as described previously18. The ChIP-seq data are deposited in GEO under accession number: updating in progress.
Mouse molecular genetics
Oct4-GFP transgenic mice (Jackson's Lab, stock no. 004654) were used for MEF isolation at E13.5. Albino embryos were isolated at 2-cell stage, matured to 4-8-cell stage and microinjected with iPS cells using the Piezo Micro Manipulator (PMAS-CT150, PMM) under the fluorescent microscope (Olympus) to generate chimeras. Injected embryos were cultured in KSOM media (Specialty Media) to the blastocyst stage and then transferred to the uterine horns of E2.5 pseudopregnant F1 (CBA × C57BL/6J) females. Chimeric embryos were harvested at E13.5 for analysis of GFP expression in the gonads.
Tetraploid complementation
Tetraploid (4n) embryos were generated using 2-cell fusion by Electro cell manipulator (ECM 2001, BTX Harvard Apparatus) and incubated in KSOM till 4-8 cell stage prior to aggregation with iPS cells. The zona pellucida of the tetraploid 4-8 cell stage embryos were removed by brief exposure to acid Tyrode's solution. Three tetraploid embryos and ~40 iPS cells were aggregated in a single well, and incubated in KSOM medium for 24 h to form a morula or blastocyst16,17. Approximately 10-14 embryos were transferred into the uterus (for blastocysts) or oviducts (for morulas) of CBAB6 (CBA × C57BL/6J) F1 pseudopregnant mice.
Supplementary Material
Acknowledgements
This work is supported by the Agency for Science, Technology and Research (Singapore) and the Singapore Stem Cell Consortium grant (SSCC-06-003). The work is also partially supported by National Institutes of Health (NIH) grants to B.L. (DK047636 and AI54973). W.L.T. is supported by the A*STAR Post-doctoral Fellowship. We acknowledge the Genome Technology and Biology group for technical support. We are grateful to Drs. Tara Huber, Paul Robson and Larry Stanton for critical comments, Dr Valere Cacheux-Rataboul for karyotype analyses, Yin Loon Lee, Phil Gaughwin and colleagues from the Stem Cell and Developmental Biology group for technical assistance. We also thank Dr Teru Wakayama from Riken, Kobe and Dr Ning Li from State Key Laboratory for Agrobiotechnology, Beijing, for imparting mouse embryo manipulation techniques.
Reference
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Huangfu D, et al. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795–7. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi Y, et al. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell. 2008;3:568–74. doi: 10.1016/j.stem.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Silva J, et al. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 2008;6:e253. doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–53. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 6.Daley GQ, et al. Broader implications of defining standards for the pluripotency of iPSCs. Cell Stem Cell. 2009;4:200–1. doi: 10.1016/j.stem.2009.02.009. author reply 202. [DOI] [PubMed] [Google Scholar]
- 7.Feng B, et al. Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nat Cell Biol. 2009 doi: 10.1038/ncb1827. [DOI] [PubMed] [Google Scholar]
- 8.Marson A, et al. Wnt signaling promotes reprogramming of somatic cells to pluripotency. Cell Stem Cell. 2008;3:132–5. doi: 10.1016/j.stem.2008.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wernig M, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–24. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 10.Chin MH, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–23. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitsui K, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–42. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 12.Silva J, Chambers I, Pollard S, Smith A. Nanog promotes transfer of pluripotency after cell fusion. Nature. 2006;441:997–1001. doi: 10.1038/nature04914. [DOI] [PubMed] [Google Scholar]
- 13.Tam WL, et al. T-cell factor 3 regulates embryonic stem cell pluripotency and self-renewal by the transcriptional control of multiple lineage pathways. Stem Cells. 2008;26:2019–31. doi: 10.1634/stemcells.2007-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivanova N, et al. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–8. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- 15.Hotta A, Ellis J. Retroviral vector silencing during iPS cell induction: an epigenetic beacon that signals distinct pluripotent states. J Cell Biochem. 2008;105:940–8. doi: 10.1002/jcb.21912. [DOI] [PubMed] [Google Scholar]
- 16.Eakin GS, Hadjantonakis AK. Production of chimeras by aggregation of embryonic stem cells with diploid or tetraploid mouse embryos. Nat Protoc. 2006;1:1145–53. doi: 10.1038/nprot.2006.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohta H, Sakaide Y, Yamagata K, Wakayama T. Increasing the cell number of host tetraploid embryos can improve the production of mice derived from embryonic stem cells. Biol Reprod. 2008;79:486–92. doi: 10.1095/biolreprod.107.067116. [DOI] [PubMed] [Google Scholar]
- 18.Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–17. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 19.Wernig M, et al. A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nat Biotechnol. 2008;26:916–24. doi: 10.1038/nbt1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 21.Loh YH, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–40. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J, et al. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat Cell Biol. 2006;8:1114–23. doi: 10.1038/ncb1481. [DOI] [PubMed] [Google Scholar]
- 23.Reynolds A, et al. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22:326–30. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 24.Chua SW, et al. A novel normalization method for effective removal of systematic variation in microarray data. Nucleic Acids Res. 2006;34:e38. doi: 10.1093/nar/gkl024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang J, et al. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol. 2008;10:353–60. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.