Summary
In mammalian and Drosophila cells, heat stress strongly reduces general protein translation while activating cap-independent translation mechanisms to promote the expression of stress-response proteins. In contrast, in Saccharomyces cerevisiae general translation is only mildly and transiently reduced by heat stress and cap independent translation mechanisms have not been correlated with the heat stress response. Recently we have identified direct target genes of the heat shock transcription factor, HSF, including genes encoding proteins thought to be important for general translation. One gene activated by HSF during heat stress encodes the enhancer of decapping protein, Edc2, previously shown to enhance mRNA decapping under conditions when the decapping machinery is limited. In this report we show that strains lacking Edc2, as well as the paralogous protein Edc1, are compromised for growth under persistent heat stress. This growth deficiency can be rescued by expression of a mutant Edc1 protein deficient in mRNA decapping indicative of a decapping independent function during heat stress. Yeast strains lacking Edc1 and Edc2 are also sensitive to the pharmacological inhibitor of translation paromomycin and exposure to heat stress and paromomycin functions synergistically to reduces yeast viability, suggesting that in the absence of Edc1 and Edc2 translation is compromised under heat stress conditions. Strains lacking Edc1 and Edc2 have significantly reduced rates of protein translation during growth under heat stress conditions, but not under normal growth conditions. We propose that Edc1 and the stress responsive isoform Edc2 play important roles in protein translation during stress.
Introduction
All organisms encounter environmental, chemical or physiological stresses in various forms. In response to these stresses eukaryotic cells promote changes in macromolecular synthesis, trafficking and degradation until more favorable conditions are met (Lindquist, 1992). The exposure of eukaryotic cells to elevated temperatures, or heat stress, results in the inhibition of general transcription, a reduction in mRNA splicing, the retention of specific mRNAs in the nucleus, the inhibition of overall protein translation and the induced expression of stress-response genes (Lindquist, 1986, Morano et al., 1998). These changes promote a remodeling of the proteome to increase the concentration of proteins important for stress survival while reducing the levels of proteins not essential for stress survival (Lindquist, 1986).
Previous studies in cultured mammalian and Drosophila cells show that a stress-dependent reduction of proteins that do not specifically contribute to stress-survival is achieved through the inhibition of cap-dependent protein translation (McCormick & Penman, 1969, Lindquist, 1981) by mechanisms that include the inactivation of the translation initiation factors eIF2α and eIF4E as well as the activation of the translation repressive 4E binding protein (4E-BP) (Sierra & Zapata, 1994, Kleijn et al., 1998). In conjunction with the repression of cap-dependent translation, the preferential translation of proteins required for heat stress survival is promoted through the activation cap-independent translation mechanisms (Sierra & Zapata, 1994).
In Saccharomyces cerevisiae cap-dependent translation is repressed in response to a variety of stresses including osmotic shock and nutrient starvation (Ashe et al., 2000, Uesono & Toh, 2002). However, overall translation in yeast is only modestly reduced during heat stress (McAlister & Finkelstein, 1980, Uesono & Toh, 2002). Rather, only transient reductions in the levels of some proteins are observed shortly after exposure to elevated temperatures (Miller et al., 1982). In general, these reductions are promoted through reductions in transcription and mRNA stability (McAlister & Finkelstein, 1980, Miller et al., 1982, Herrick et al., 1990, Gasch et al., 2000). Consistent with this notion, a cap independent-translation mechanism has not been described in yeast in response to heat stress.
An important aspect of the response to heat stress is the activation of genes encoding proteins required for heat stress survival. This activation is mediated in part by heat shock transcription factors (HSF), whose fundamental architecture and heat shock element (HSE) binding site are conserved from yeast to humans, (Wu, 1995, Littlefield & Nelson, 1999, Akerfelt et al., 2007). Through chromatin immunoprecipitation in conjunction with microarray analysis we have recently identified 165 direct HSF target genes in S. cerevisiae (Hahn et al., 2004). While many of the identified HSF target genes encode protein chaperones, newly identified targets suggest an important role for HSF in many of the molecular events that occur in response to heat stress including protein folding and degradation, ion transport, signal transduction, energy generation, carbohydrate metabolism, vesicular transport and cytoskeleton formation (Hahn et al., 2004). Consistent with the finding that general translation is not repressed during heat stress in yeast, HSF was shown to bind the promoters of several genes encoding proteins linked to general translation, including SOL1, encoding a protein involved in tRNA export from the nucleus (Shen et al., 1996), TMA10 encoding a protein that associates with poly-ribosomes (Fleischer et al., 2006), ZPR1 encoding a protein that interacts with eEF1α and is important for rRNA biogenesis in S. pombe (Galcheva-Gargova et al., 1998, Gangwani et al., 1998) and YGR250c, encoding a protein homologue of the poly-A binding protein Pab1 (Buchan et al., 2008).
While much is known about gene transcription in response to heat stress, relatively little is known about post-transcriptional events. Recently we identified EDC2 as a direct target of yeast HSF (Hahn et al., 2004, Hahn & Thiele, 2004). EDC2 encodes an RNA-binding protein previously identified in conjunction with the homologous protein Edc1 as multi-copy suppressors of strains containing temperature sensitive alleles of the mRNA decapping complex (Dunckley et al., 2001). Subsequent studies showed that Edc1 and Edc2 interact with the mRNA decapping machinery in an mRNA-dependent manner and enhance the rate of mRNA decapping through as of yet unknown mechanisms (Schwartz et al., 2003, Steiger et al., 2003). Interestingly, Edc1 and Edc2 are only required for mRNA decapping and decay when the activity of the decapping complex is limited (Dunckley et al., 2001, Schwartz et al., 2003). In addition, localization of Edc1 and Edc2 to P-bodies, the predominant extranuclear sites of mRNA decapping and decay and translation inhibition, has not been described.
Herein we show that expression of EDC2 is activated in response to heat stress at both the mRNA and protein level, while expression of EDC1 is transiently reduced. Interestingly, we further show that despite only EDC2 expression being up-regulated by heat shock, both Edc1 and Edc2 are required for optimal growth during heat stress. In addition, we present evidence for potentially novel functions for Edc1 and Edc2 as proteins important for protein translation under heat stress conditions.
Results
EDC1 and EDC2 encode small, basic RNA binding proteins (Schwartz et al., 2003) (Fig. 1A) that have been reported to bind RNA non-specifically through an as of yet uncharacterized RNA binding domain. Our previous work has identified EDC2 as a direct binding target of HSF, expressed through a putative promoter HSE in response to heat stress (Hahn et al., 2004). No HSEs have been identified in the EDC1 promoter, consistent with the notion that EDC1 is not a target of HSF. As shown in Figure 1A, Edc1 and Edc2 share two distinct regions of homology. Previous experiments have shown that Edc1 truncation mutants, lacking the carboxyl-terminal domain (domain 2), retain the ability to bind RNA, yet are defective in their ability to enhance mRNA decapping (Schwartz et al., 2003). In addition, through primary sequence analysis of Edc1 and Edc2 we have identified a putative bipartite nuclear localization signal (NLS) in Edc2 that is lacking in Edc1 (Fig. 1A).
Fig. 1. Edc1 and Edc2 are small, basic RNA binding proteins that evolved as a result of whole genome duplication in an ancestor of S. cerevisiae.
A. Regions of homology between Edc1 and Edc2 are shown in grey. A putative bipartite nuclear localization signal (NLS) in Edc2 with the sequence, KKKSCKYKKKK, is shown in black. B. Summary of events in the evolution of the EDC1 and EDC2 loci. The phylogenetic tree on the left is not drawn to scale and shows the evolutionary relationships between various yeast species and is based on data reviewed by Wolfe, 2006. The line break represents the whole genome duplication event. In the table on the right, + indicates the presence of one, ++ indicates the presence of two and − indicates the absence of a locus, heat shock element (HSE) or NLS.
EDC1 and EDC2 were recently identified as ohnologs, or paralogous genes that were formed by a whole genome duplication event that occurred in an ancestor of S. cerevisiae (Kellis et al., 2004, Byrne & Wolfe, 2005). Sequence comparison with fungi that diverged from S. cerevisiae before the genome duplication (Wolfe, 2006), including S. kluyveri, K. waltii, K. lactis and A. gossypii, suggests that EDC1 is more closely related to the ancestral EDC gene (Fig 1B). This suggests that EDC2 arose from EDC1 as a result of genome duplication and was maintained, presumably because it conferred an adaptive advantage to the ancestral organism. In support of this hypothesis, several fungal species that diverged from S. cerevisiae after the genome duplication express paralogs of both EDC1 and EDC2. A notable exception to this is C. glabrata, which, despite diverging from S. cerevisiae after genome duplication (Wolfe, 2006), expresses only an EDC1 paralog (Fig 1B). The introduction of an HSE into the EDC2 promoter, as well as a putative NLS into the EDC2 ORF, likely conferred an adaptive advantage to an ancestral organism since both are absent from EDC1, but were retained in all EDC2 paralogs. Interestingly, the genome of S. castelli, which diverged from S. cerevisiae after the genome duplication (Wolfe, 2006), contains two EDC1-like genes neither of which has an HSE in the promoter or an NLS in the ORF. This suggests that the introduction of both the HSE and the NLS occurred after S. castelli diverged from S. cerevisiae.
EDC2 but not EDC1 is activated in response to heat stress
Previous experiments have shown that in response to heat shock, EDC2 is bound by HSF and transcriptionally activated (Hahn et al., 2004, Hahn & Thiele, 2004). To ascertain if EDC2 and the related gene EDC1 are co-regulated in response to heat shock, we analyzed the expression of both genes by RNA blotting after exposing a wild type strain to a 41°C heat shock for increasing times. As shown in Figure 2, EDC2 mRNA levels are transiently elevated in response to heat shock peaking within 5 min after exposure to elevated temperatures and then returning to slightly above basal levels. We detect the EDC2 mRNA as two distinctly migrating mRNA species that appear to be similarly regulated by heat stress. Consistent with our findings that EDC1 is not a target of HSF (Hahn et al., 2004), EDC1 expression was not upregulated in heat shocked yeast cells (Fig. 2). Interestingly, we also detect the EDC1 mRNA as two distinctly migrating species though unlike the EDC2 mRNA, the two EDC1 mRNA species appear to be transiently reduced upon exposure to heat shock (Fig. 2). While at this time the nature of the two mRNA species observed for the EDC1 and EDC2 remains unknown, it is possible that for EDC1 they represent the adenylated and de-adenylated state of the EDC1 mRNA which occur as a function of an extended poly-U tract within the 3’ UTR of EDC1 (Muhlrad & Parker, 2005).
Fig. 2. Gene expression of EDC1 and EDC2 is modulated by heat stress.
A. A wild type (WT), BY4741 strain, was heat shocked at 41°C for the indicated times. The expression of EDC1 and EDC2 was assayed by RNA blotting. ENO1 serves as a loading control. EDC1 and EDC2 mRNA species with differential mobility are indicated as Species 1(S1) and Species 2 (S2). B. EDC1 and EDC2 transcript levels were normalized to ENO1, from panel A.
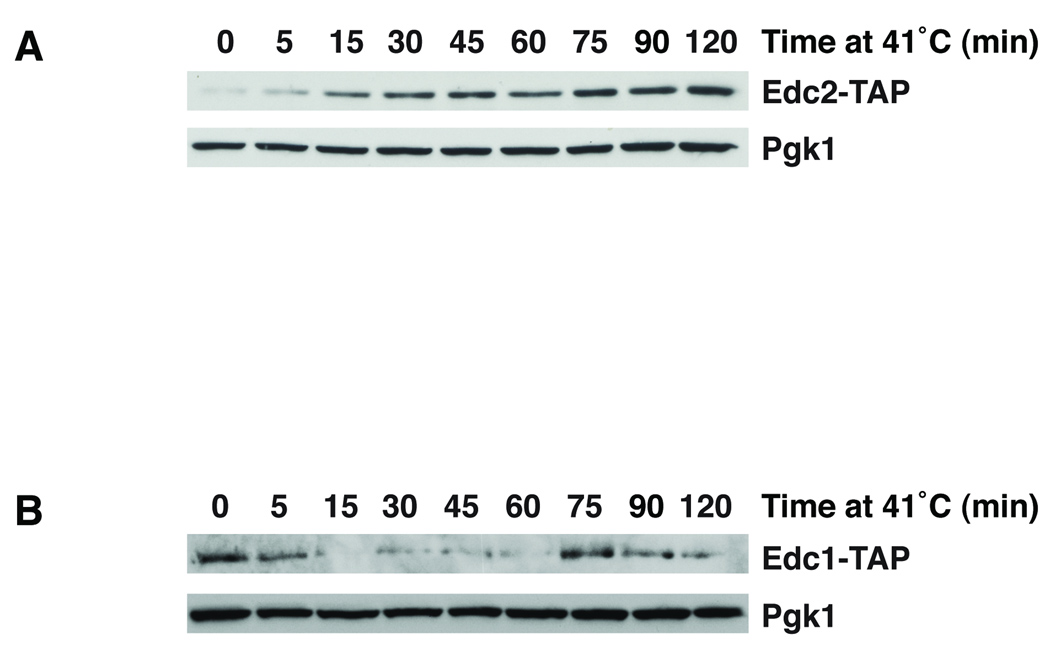
To determine if Edc1 and Edc2 protein levels change in response to heat shock, we assayed the expression of genomically TAP-tagged EDC1 and EDC2 alleles in response to heat stress by immunoblotting. As shown in Figure 3, Edc2 protein levels increased within 15 min after exposure to heat shock and continued to increase up to 75 min after heat shock exposure. In contrast, Edc1 protein levels, like the EDC1 mRNA, were transiently reduced in response to heat shock (Fig. 3B).
Fig. 3. Edc1 and Edc2 proteins are modulated by heat shock.
A. The EDC2-TAP strain was heat shocked at 41°C for the indicated times. The expression of Edc2-TAP was assayed by immunoblotting using an anti-TAP antibody. Pgk1 serves as a loading control. B. Expression of Edc1-TAP was analyzed as in A.
Edc1 and Edc2 are important for growth under stress conditions
To ascertain the functional significance of Edc1 and Edc2 during heat stress, yeast strains deleted for either EDC1 and/or EDC2 were generated and assayed for their ability to grow under normal conditions and heat stress conditions. Interestingly, despite only EDC2 being transcriptionally activated in response to heat stress, both the edc1Δ and the edc2Δ strain exhibited a reduction in growth under heat stress conditions as compared to the isogenic wild type parental strain (Fig. 4A). As shown in Figure 4A, the edc1Δ edc2Δ double mutant exhibited further growth reductions at 41°C than either of the single mutant strains. The significant growth reductions seen in edc1Δ strain suggest that an up-regulation of gene expression, as observed for EDC2, is not absolutely required for an EDC protein to have an important contribution in long-term growth under stress conditions.
Fig. 4. Edc1 and Edc2 are required for optimal growth under heat stress conditions.
A. Strains BY4741 (WT), DNY86 (edc1Δ), DNY25 (edc2Δ) and DNY63 (edc1Δ edc2Δ) were grown at 30°C to O.D.600 = 0.2 and 5-fold dilution of cells were spotted on SC medium and grown at either 30°C or 41°C. B. Strain BY4741 (WT) transformed with pRS416, and strain DNY63 (edc1Δ edc2Δ) transformed with either pRS416, pRS416-GPD-EDC1, or pRS416-GPD-EDC2 were grown and spotted as in A. on SC-URA medium and grown at either 30°C or 41°C. C. Strain BY4741 (WT) transformed with pRS425, and strain DNY63 (edc1Δ edc2Δ) transformed with either pRS425, pRP984 (2 μ EDC1) or pRP1056 (2 μ edc1Δ162–175) were grown and spotted as in A. on SC-LEU medium and grown at either 30°C or 41°C.
To test if Edc1 and Edc2 have similar functions during stress we assayed the ability of Edc1 or Edc2 overexpression to rescue the growth of the edc1Δ edc2Δ strain under heat stress conditions. As shown in Figure 4B, overexpression of either Edc1 or Edc2 from the constitutive GPD promoter could rescue the growth of the edc1Δ edc2Δ under thermal stress conditions, suggesting that Edc1 and Edc2 have, at least in part, redundant functions. The growth deficiency of the edc1Δ edc2Δ strain during heat stress was also rescued by overexpression of a mutant Edc1 protein that lacks domain 2 (edc1Δ162–175), known to be deficient in the enhancement of mRNA decapping (Schwartz et al., 2003) (Fig. 4C). These data suggest that Edc1 and Edc2 might have functions independent of mRNA decapping during heat stress. In support of this hypothesis, Edc1 was recently identified as a multicopy suppressor of a psk1Δ psk2Δ strain which lacks a functional PAS kinase, a positive modulator of translation initiation (Rutter et al., 2002), suggesting that Edc1 and, potentially Edc2, are involved in events underlying translation during heat stress.
Strains lacking Edc1 and Edc2 are sensitive to paromomycin and resistant to cycloheximide
To test if Edc1 and Edc2 may be involved in translation, we ascertained the ability of the edc1Δ edc2Δ mutant strain to grow in the presence of pharmacological inhibitors of translation. When the edc1Δ edc2Δ mutant was grown on medium containing 10 mg/ml paromomycin, an aminoglycoside antibiotic that interacts with the A site of the 16S ribosomal RNA and disrupts translation (Vicens & Westhof, 2001), a severe growth defect was observed (Fig. 5A). Because Edc1 and Edc2 are required for optimal growth under heat stress conditions, but not in the absence of stress, we reasoned that the sensitivity to translational inhibitors might be increased if the yeast are exposed to mild temperature increases. To test if heat stress could enhance the sensitivity to paromomycin, we assayed the edc1Δ edc2Δ strain on medium containing 2 mg/ml paromomycin under normal growth conditions (30°C) and under a mild heat stress (37°C). As shown in Figure 5B, when the edc1Δ edc2Δ strain was grown on low levels of paromomycin or under a mild heat stress, no growth reductions were observed. However, when the mild heat shock and low levels of paromomycin where combined, a pronounced growth deficiency of the edc1Δ edc2Δ strain was observed. In contrast to paromomycin, when the edc1Δ edc2Δ strain was grown on plates containing 500 ng/ml cycloheximide, an inhibitor of translational elongation (McKeehan & Hardesty, 1969), we observed a distinct growth advantage of the mutant strain in comparison to a wild type strain (Fig. 5C). Conversely, when either Edc1 or Edc2 were overexpressed in a wild type strain, the cells became strongly sensitized to cycloheximide (Fig. 5D). While the mechanistic basis for these phenotypes remains unclear, mutations in other proteins required for translation, for example the ribosome biogenesis factor Nmd3, result in similar cycloheximide resistance and paromomycin sensitivity (Ho & Johnson, 1999). Together these data suggest that Edc1 and Edc2 might have functions important for protein translation that are exacerbated during times of cells stress.
Fig. 5. edc1Δ edc2Δ strain is sensitive to growth on paromomycin but resistant to cycloheximide.
Strains BY4741 (WT), and DNY160 (edc1Δ edc2Δ) were grown at 30°C to O.D.600 = 0.2 and 5-fold dilutions of cells were spotted on (A) SC medium or SC medium supplemented with 10 mg/ml paromomycin (PM) and grown at 30°C, (B) SC medium or SC medium supplemented with 2 mg/ml paromomycin and grown at either 30°C or 37°C, or (C) SC medium or SC medium supplemented with 500 ng/ml cycloheximide (CHX) and grown at 30°C. D. Strain BY4741 transformed with pRS416-GPD-EDC2 was grown and diluted as in A. and spotted on SC-URA medium supplemented with 500 ng/ml cycloheximide and grown at 30°C.
Edc1 and Edc2 are required for efficient translation during stress
To test the importance of Edc1 and Edc2 in translation we assayed the incorporation of [35S]-methionine into trichloroacetic acid (TCA) precipitable proteins under normal growth and heat stress conditions. In the absence of thermal stress no defect in [35S]-methionine incorporation was observed (Fig. 6A), consistent with the notion that Edc1 and Edc2 have non-essential functions in global translation in the absence of stress. After exposure to a 2 hr heat shock, the rate of [35S]-methionine incorporation was reduced by approximately 2-fold in strains lacking either Edc1 or Edc2 and was reduced by approximately 4-fold in the edc1Δ edc2Δ strain (Fig. 6A). The rate of [35S]-methionine incorporation in the edc1Δ edc2Δ strain during heat stress could be rescued by overexpression of Edc1 and Edc2 but not the structurally distinct decapping enhancer protein Edc3 (Fig. 6B). Together these data support the hypothesis that Edc1 and Edc2 play important roles in effective protein translation during heat stress conditions.
Fig. 6. Edc1 and Edc2 are required for optimal translation under heat stress conditions.
A. Strains BY4742 (WT), DNY191 (edc1Δ), DNY192 (edc2Δ), DNY170 (edc1Δ edc2Δ) were grown to mid-log phase at 30°C and exposed to either 30°C or 41°C for 2 hr and [35S]-methionine incorporation was assayed. Data shown for all curves are averages of three experiments with associated standard deviations. B. Strain DNY170 (edc1Δ edc2Δ) transformed with pRS416, pRS416-GPD-EDC1 or pRS416-GPD-EDC2 was assayed for [35S]-methionine incorporation as in A. The rate of [35S]-methionine incorporation was determined by calculating the slope from 0 to 40 min.
Differential localization of Edc1 and Edc2
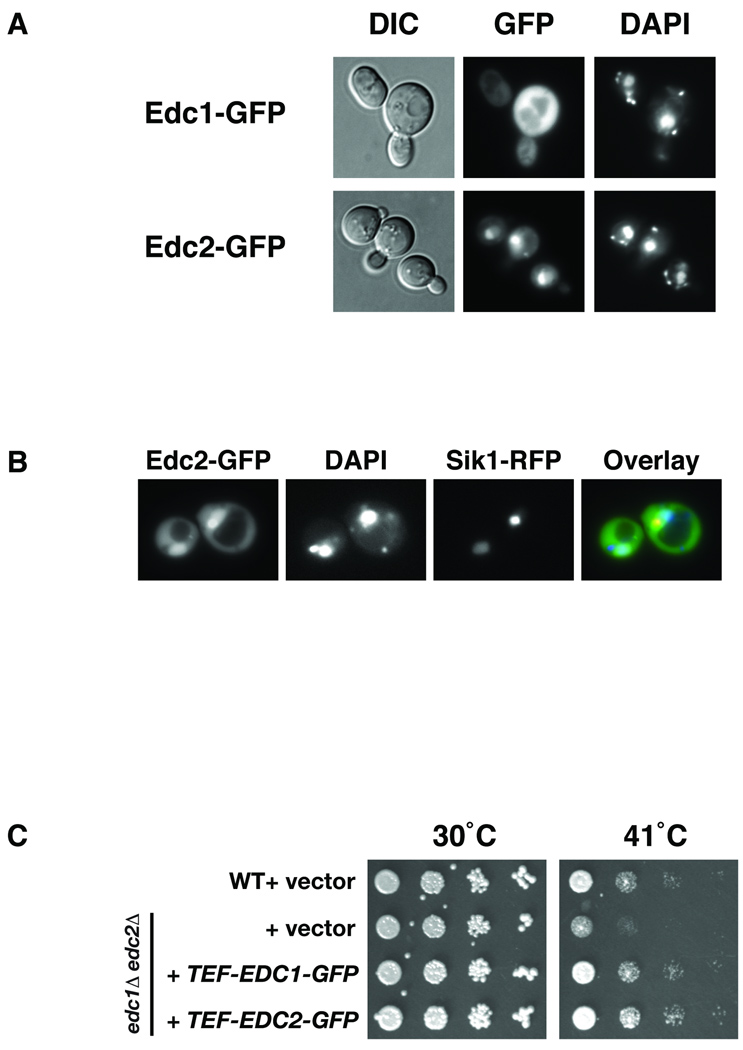
In order to better understand the potential roles of Edc1 and Edc2 in translation we examined the subcellular localization of the Edc1 and Edc2 proteins. For Edc1 and Edc2 localization analysis, proteins carboxyl-terminally fused to green fluorescent protein (GFP) were expressed from a plasmid under control of the constitutively active TEF promoter. This allowed us to examine Edc1 and Edc2 localization independent of stress-induced transcriptional activation or repression. Edc1-GFP was localized throughout the cell in the absence of stress, though a diminution of Edc1 in the nucleus was observable (Fig. 7A). The localization of Edc1-GFP did not change when the cells were exposed to heat shock (data not shown). In contrast to Edc1, Edc2-GFP appeared to be localized largely to the nucleus and a peri-nuclear compartment (Fig. 7A), indicative of nucleolar localization. Like Edc1-GFP, the localization of Edc2-GFP did not change in response to heat shock (data not shown). While expression of the Edc1-GFP and Edc2-GFP fusion proteins from the constitutive TEF-promoter was substantially higher than expression from the native promoter (data not shown), the localization of genomically tagged GFP fusion proteins was indistinguishable from those expressed from plasmids (data not shown). To independently test whether Edc2-GFP is localized to the nucleolus, we expressed Edc2-GFP in a strain co-expressing the nucleolar Sik1 protein fused to red fluorescence protein (RFP). As shown in Figure 7B, Edc2-GFP partially co-localized with Sik1-RFP, indicative of Edc2-GFP being partially localized to the nucleolus. These data correlate well with a recent finding suggesting that Edc2 is complexed with a variety of nucleolar proteins involved in ribosome biogenesis (Krogan et al., 2006). In addition, the presence of a putative NLS in Edc2 but not in Edc1 (Fig. 1A) fits well with our data demonstrating that Edc2-GFP but not Edc1-GFP, is localized to the nucleus and nucleolus. To confirm the functionality of the Edc1-GFP and Edc2-GFP fusion proteins, both proteins were expressed in the edc1Δ edc2Δ strain and assayed for their ability to promote growth during heat stress conditions. As shown, in Figure 7C, expression of both Edc1-GFP an Edc2-GFP rescued growth of the edc1Δ edc2Δ strain under heat stress conditions.
Fig. 7. Differential localization of Edc1 and Edc2.
A. Strain BY4741 transformed with either pRS416-TEF-EDC1-GFP, or pRS416-TEF-EDC2-GFP was grown to mid-log phase and the localization of Edc1-GFP and Edc2-GFP were assessed by fluorescence microscopy. B. A SIK1-RFP strain transformed with pRS416-TEF-EDC1-GFP was grown to mid-log phase and the localization of Edc2-GFP and Sik1-RFP was assessed by fluorescence microscopy. C. Strain BY4741 (WT) transformed with pRS416 and strain DNY63 (edc1Δ edc2Δ) transformed with either pRS416, pRS416-TEF-EDC1-GFP or pRS416-TEF-EDC2-GFP were grown at 30°C to O.D.600 = 0.2 and 5-fold dilution of cells were spotted on SC-URA medium and grown at either 30°C or 41°C
Edc1 and Edc2 localize to P-bodies in an xrn1Δ strain
Because many proteins involved in mRNA decapping localize to P-bodies (Sheth & Parker, 2003) we tested if Edc1 and Edc2 could also localize to P-bodies under specific circumstances, using Dcp2-RFP as a marker for P-body localization (Teixeira et al., 2005). In a wild-type strain grown under normal growth or heat stress-conditions, Edc1-GFP and Edc2-GFP did not co-localize with Dcp2-RFP (data not shown). However, when Edc1-GFP or Edc2-GFP expressing strains were starved for glucose (data not shown) or when Edc1-GFP or Edc2-GFP where expressed in an xrn1Δ strain (Fig. 8), P-body co-localization was readily observed. Interestingly, for Edc2-GFP P-body localization in the xrn1Δ strain did not occur at the expense of the nuclear localization suggesting that only a subset of Edc2-GFP is being localized to P-bodies. These data support a role for Edc1 and Edc2 in mRNA decapping and decay but suggests the notion that Edc1 and Edc2 are multifunctional proteins important for many aspects of mRNA biology.
Fig. 8. Edc1 and Edc2 localize to P-bodies in an xrn1Δ strain.
A. Strain DNY253 (xrn1Δ) transformed with pRS416-TEF-EDC1-GFP and pRP1167 (DCP2-RFP) was grown to mid-log phase and the localization of Edc1- GFP and Dcp2-RFP were assessed by fluorescence microscopy. B. Strain DNY253 (xrn1Δ) transformed with pRS416-TEF-EDC2-GFP and pRS415-DCP2-RFP was analyzed as in A.
Discussion
In response to heat stress eukaryotic cells increase the concentration of proteins important for stress survival while reducing those proteins less essential for survival under stress conditions (Lindquist, 1986). In mammalian and Drosophila cells this is achieved primarily through repression of cap-dependent protein translation and the activation of cap-independent translation mechanisms (Sierra & Zapata, 1994). Unlike mammalian and Drosophila cells, S. cerevisiae cap-dependent protein translation is not interrupted by heat stress (McAlister & Finkelstein, 1980, Uesono & Toh, 2002), and HSF promotes the activation of genes that are thought to play an important role in general translation under heat stress (Hahn et al., 2004). In this report we have begun to characterize the role of two ohnologous RNA binding proteins, Edc1 and Edc2, whose function is important for effective protein translation and growth during heat stress but not under normal growth conditions.
While the mechanistic basis underlying the importance of Edc1 and Edc2 in translation during heat stress remains unclear, the nucleolar localization of Edc2 makes it tempting to speculate that Edc2 and potentially Edc1 are involved in ribosome biogenesis. This notion is supported by sensitivity and resistance of the edc1Δ edc2Δ mutant strain to paromomycin and cycloheximide respectively, phenotypes which are also observed in response to mutations in other ribosome biogenesis factors (Ho & Johnson, 1999). Also suggesting a role in ribosome biogenesis are recent findings that suggest Edc2 interacts with the ribosome biogenesis factors Mak5, Nop14 and Kri1 (Krogan et al., 2006). However, our experiments testing the export of Rps2-GFP and Rpl11b-GFP from the nucleolus to the cytoplasm (Stage-Zimmermann et al., 2000, Milkereit et al., 2003) have not detected reductions in ribosome biogenesis in the edc1Δ edc2Δ mutant strain under normal growth or heat stress conditions (data not shown). Nevertheless, at this time we cannot rule out the possibility that Edc1 and Edc2 affect ribosome function without affecting ribosome biogenesis. Consistent with a potential role in ribosome function, SDS-PAGE analysis of [35S]-methionine labeled proteins in the edc1Δ edc2Δ strain suggests that exposure to thermal stress results in a global reduction in protein translation (data not shown).
Previous studies have elegantly shown that Edc1 enhances the activity of the decapping enzyme and is required for the degradation of specific mRNA in response to changes in carbon sources (Schwartz et al., 2003). The ability of Edc1 to enhance the function of the decapping enzyme is strongly dependent on the last 14 amino acids which constitute domain 2. In striking contrast, our data show that this domain is not required for the ability of Edc1 to rescue the growth deficiency of the edc1Δ edc2Δ strain during heat stress thereby suggesting that under heat stress conditions Edc1 is functioning independent of mRNA decapping. Therefore, Edc1 as well as Edc2 are likely to have important functions in multiple aspects of RNA biology including mRNA decapping and translation. This idea is supported in part by the apparent non-specificity of Edc1 and Edc2 in RNA binding (Schwartz et al., 2003) suggesting that these proteins can act on many target RNAs. Our findings showing that Edc1 and Edc2 are important for translation only during cells stress in addition to previous findings that Edc1 and Edc2 are required for mRNA decapping only when the activity of the decapping complex is limited (Dunckley et al., 2001, Schwartz et al., 2003) evokes an interesting comparison to protein chaperones which in many instances act to stabilize misfolded proteins or promote the accurate folding of nascent polypeptides (Morimoto, 2008) and many of which only become essential for growth under stress conditions. Could Edc1 and Edc2 be acting as RNA chaperones and stabilizing RNA in specific confirmations? Indeed several instances of RNA chaperones have been described in prokaryotes, acting to stabilize RNA structure during non-optimal growth conditions (Rajkowitsch et al., 2007). Interestingly, like Edc1 and Edc2 many prokaryotic RNA chaperones are small basic proteins, with little or no RNA binding specificity (Rajkowitsch et al., 2007). Future experiments will test the RNA chaperone functions of Edc1 and Edc2 and will explore if Edc1 and Edc2 affect translation directly or indirectly, potentially through roles in ribosome biogenesis.
Experimental procedures
Yeast strains
The strains used in this study are listed in Table 1 and were derived from BY4741 and BY4742, which are considered wild type, except for strain DNY111 which is derived from W303. Gene deletions were generated by homologous recombination with either the loxP-spHIS5-loxP or loxP-kanMX-loxP cassettes derived by PCR from plasmid pUG27 and pUG6 respectively (Guldener et al., 1996). For strains, DNY160 and DNY170, the spHIS5 and kanMX genes were removed via recombination through expression of the Cre-recombinase from plasmid pSH47 (Guldener et al., 1996). Strain DNY100 was obtained from the TAP-tagged collection (Ghaemmaghami et al., 2003). To generated strain DNY111, DNA encoding the TAP-tag and selectable marker was generated by PCR from plasmid pBS1479 (Caspary et al., 1999) and inserted by homologous recombination. The Sik1-RFP strain was previously described (Huh et al., 2003).
Table 1.
Yeast strains used in this study.
| Strain | Genotype |
|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 |
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 |
| DNY25 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 edc2Δ::loxP-kanMX-loxP |
| DNY63 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 edc1Δ::loxP-HIS5-loxP edc2Δ::loxP-kanMX-loxP |
| DNY86 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 edc1Δ::loxP-HIS5-loxP |
| DNY100 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 EDC2-TAP::HIS3 |
| DNY111 | MATa leu2-3, 112 trp1-1 can1-100 ura3-1 ade2-1 his3-11, 15 EDC1-TAP::TRP1 |
| DNY160 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 edc2Δ::loxP |
| DNY170 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 edc1Δ::loxP edc2Δ::loxP |
| DNY191 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 edc1Δ::loxP |
| DNY192 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 edc2Δ::loxP |
| DNY253 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 xrn1Δ::KanMX4 |
| SIK1-RFP | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 SIK1-RFP::kanMX |
RNA blotting and immunoblotting analysis
Total RNA was extracted from 10 ml yeast cultures by glass bead lysis utilizing LETS buffer (0.1 M LiCl, 10 mM EDTA, 10 mM Tris (pH 8.0), 0.5% SDS) and phenol:chloroform. RNA concentrations were quantified by measuring A260 and 10 µg of total RNA was analyzed as described (Neef & Kladde, 2003). Protein extracts were generated from 10 ml of yeast culture using cell lysis buffer (25 mM Tris, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 1 mM EDTA) supplemented with protease inhibitors and glass beads. Protein concentrations were quantified using the BCA assay and 10 µg of total protein was resolved by SDS-PAGE, transferred to a nitrocellulose membrane and proteins of interest detected by immunoblot analysis following standard procedures using anti-TAP and anti-Pgk1 antibodies.
Fluorescence microscopy
Yeast strains were grown to mid log-phase, the DNA was stained with 4',6-diamidino-2-phenylindole (DAPI) and fluorescence was analyzed using a Zeiss Axio Imager fluorescence microscope and images deconvoluted with MetaMorph software.
In vivo [35S]-methionine incorporation
Strains were grown overnight in CSM-Met or CSM-MET-URA to an O.D.600 = 0.2 and incubated at either 30°C or 41°C for 2 hr. At time zero, [35S]-methionine was added to each culture at a final concentration of 1 µCi/ml and the cultures were further incubated at 30°C or 41°C. At the indicated times, aliquots were removed and assayed for O.D.600 and methionine incorporation as previously described (Carr-Schmid et al., 1999).
Acknowledgements
We thank Dr. Roy Parker for plasmids pRP984, pRP1056 and pRP1167 as well as helpful suggestions and members of the Thiele laboratory for critical reading of this manuscript. This work was supported in part by the National Institutes of Health NRSA Postdoctoral Fellowship GM076954 (to D.W.N) and grant RO1-GM059911 (to D.J.T.).
References
- Akerfelt M, Trouillet D, Mezger V, Sistonen L. Heat shock factors at a crossroad between stress and development. Ann N Y Acad Sci. 2007;1113:15–27. doi: 10.1196/annals.1391.005. [DOI] [PubMed] [Google Scholar]
- Ashe MP, De Long SK, Sachs AB. Glucose depletion rapidly inhibits translation initiation in yeast. Mol Biol Cell. 2000;11:833–848. doi: 10.1091/mbc.11.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Muhlrad D, Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J Cell Biol. 2008;183:441–455. doi: 10.1083/jcb.200807043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne KP, Wolfe KH. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 2005;15:1456–1461. doi: 10.1101/gr.3672305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr-Schmid A, Durko N, Cavallius J, Merrick WC, Kinzy TG. Mutations in a GTP-binding motif of eukaryotic elongation factor 1A reduce both translational fidelity and the requirement for nucleotide exchange. J Biol Chem. 1999;274:30297–30302. doi: 10.1074/jbc.274.42.30297. [DOI] [PubMed] [Google Scholar]
- Caspary F, Shevchenko A, Wilm M, Seraphin B. Partial purification of the yeast U2 snRNP reveals a novel yeast pre-mRNA splicing factor required for pre-spliceosome assembly. EMBO J. 1999;18:3463–3474. doi: 10.1093/emboj/18.12.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunckley T, Tucker M, Parker R. Two related proteins, Edc1p and Edc2p, stimulate mRNA decapping in Saccharomyces cerevisiae. Genetics. 2001;157:27–37. doi: 10.1093/genetics/157.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischer TC, Weaver CM, McAfee KJ, Jennings JL, Link AJ. Systematic identification and functional screens of uncharacterized proteins associated with eukaryotic ribosomal complexes. Genes Dev. 2006;20:1294–1307. doi: 10.1101/gad.1422006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galcheva-Gargova Z, Gangwani L, Konstantinov KN, Mikrut M, Theroux SJ, Enoch T, et al. The cytoplasmic zinc finger protein ZPR1 accumulates in the nucleolus of proliferating cells. Mol Biol Cell. 1998;9:2963–2971. doi: 10.1091/mbc.9.10.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangwani L, Mikrut M, Galcheva-Gargova Z, Davis RJ. Interaction of ZPR1 with translation elongation factor-1alpha in proliferating cells. J Cell Biol. 1998;143:1471–1484. doi: 10.1083/jcb.143.6.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Guldener U, Heck S, Fielder T, Beinhauer J, Hegemann JH. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn JS, Hu Z, Thiele DJ, Iyer VR. Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol Cell Biol. 2004;24:5249–5256. doi: 10.1128/MCB.24.12.5249-5256.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn JS, Thiele DJ. Activation of the Saccharomyces cerevisiae heat shock transcription factor under glucose starvation conditions by Snf1 protein kinase. J Biol Chem. 2004;279:5169–5176. doi: 10.1074/jbc.M311005200. [DOI] [PubMed] [Google Scholar]
- Herrick D, Parker R, Jacobson A. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:2269–2284. doi: 10.1128/mcb.10.5.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho JH, Johnson AW. NMD3 encodes an essential cytoplasmic protein required for stable 60S ribosomal subunits in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:2389–2399. doi: 10.1128/mcb.19.3.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- Kleijn M, Scheper GC, Voorma HO, Thomas AA. Regulation of translation initiation factors by signal transduction. Eur J Biochem. 1998;253:531–544. doi: 10.1046/j.1432-1327.1998.2530531.x. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Lindquist S. Regulation of protein synthesis during heat shock. Nature. 1981;293:311–314. doi: 10.1038/293311a0. [DOI] [PubMed] [Google Scholar]
- Lindquist S. The heat-shock response. Annu Rev Biochem. 1986;55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- Lindquist S. Heat-shock proteins and stress tolerance in microorganisms. Curr Opin Genet Dev. 1992;2:748–755. doi: 10.1016/s0959-437x(05)80135-2. [DOI] [PubMed] [Google Scholar]
- Littlefield O, Nelson HC. A new use for the 'wing' of the 'winged' helix-turn-helix motif in the HSF-DNA cocrystal. Nat Struct Biol. 1999;6:464–470. doi: 10.1038/8269. [DOI] [PubMed] [Google Scholar]
- McAlister L, Finkelstein DB. Alterations in translatable ribonucleic acid after heat shock of Saccharomyces cerevisiae. J Bacteriol. 1980;143:603–612. doi: 10.1128/jb.143.2.603-612.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick W, Penman S. Regulation of protein synthesis in HeLa cells: translation at elevated temperatures. J Mol Biol. 1969;39:315–333. doi: 10.1016/0022-2836(69)90320-9. [DOI] [PubMed] [Google Scholar]
- McKeehan W, Hardesty B. The mechanism of cycloheximide inhibition of protein synthesis in rabbit reticulocytes. Biochem Biophys Res Commun. 1969;36:625–630. doi: 10.1016/0006-291x(69)90351-9. [DOI] [PubMed] [Google Scholar]
- Milkereit P, Strauss D, Bassler J, Gadal O, Kuhn H, Schutz S, et al. A Noc complex specifically involved in the formation and nuclear export of ribosomal 40 S subunits. J Biol Chem. 2003;278:4072–4081. doi: 10.1074/jbc.M208898200. [DOI] [PubMed] [Google Scholar]
- Miller MJ, Xuong NH, Geiduschek EP. Quantitative analysis of the heat shock response of Saccharomyces cerevisiae. J Bacteriol. 1982;151:311–327. doi: 10.1128/jb.151.1.311-327.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morano KA, Liu PC, Thiele DJ. Protein chaperones and the heat shock response in Saccharomyces cerevisiae. Curr Opin Microbiol. 1998;1:197–203. doi: 10.1016/s1369-5274(98)80011-8. [DOI] [PubMed] [Google Scholar]
- Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22:1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D, Parker R. The yeast EDC1 mRNA undergoes deadenylation-independent decapping stimulated by Not2p, Not4p, and Not5p. EMBO J. 2005;24:1033–1045. doi: 10.1038/sj.emboj.7600560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef DW, Kladde MP. Polyphosphate loss promotes SNF/SWI- and Gcn5-dependent mitotic induction of PHO5. Mol Cell Biol. 2003;23:3788–3797. doi: 10.1128/MCB.23.11.3788-3797.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkowitsch L, Chen D, Stampfl S, Semrad K, Waldsich C, Mayer O, et al. RNA chaperones, RNA annealers and RNA helicases. RNA Biol. 2007;4:118–130. doi: 10.4161/rna.4.3.5445. [DOI] [PubMed] [Google Scholar]
- Rutter J, Probst BL, McKnight SL. Coordinate regulation of sugar flux and translation by PAS kinase. Cell. 2002;111:17–28. doi: 10.1016/s0092-8674(02)00974-1. [DOI] [PubMed] [Google Scholar]
- Schwartz D, Decker CJ, Parker R. The enhancer of decapping proteins, Edc1p and Edc2p, bind RNA and stimulate the activity of the decapping enzyme. RNA. 2003;9:239–251. doi: 10.1261/rna.2171203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen WC, Stanford DR, Hopper AK. Los1p, involved in yeast pre-tRNA splicing, positively regulates members of the SOL gene family. Genetics. 1996;143:699–712. doi: 10.1093/genetics/143.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra JM, Zapata JM. Translational regulation of the heat shock response. Mol Biol Rep. 1994;19:211–220. doi: 10.1007/BF00986963. [DOI] [PubMed] [Google Scholar]
- Stage-Zimmermann T, Schmidt U, Silver PA. Factors affecting nuclear export of the 60S ribosomal subunit in vivo. Mol Biol Cell. 2000;11:3777–3789. doi: 10.1091/mbc.11.11.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger M, Carr-Schmid A, Schwartz DC, Kiledjian M, Parker R. Analysis of recombinant yeast decapping enzyme. RNA. 2003;9:231–238. doi: 10.1261/rna.2151403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uesono Y, Toh EA. Transient inhibition of translation initiation by osmotic stress. J Biol Chem. 2002;277:13848–13855. doi: 10.1074/jbc.M108848200. [DOI] [PubMed] [Google Scholar]
- Vicens Q, Westhof E. Crystal structure of paromomycin docked into the eubacterial ribosomal decoding A site. Structure. 2001;9:647–658. doi: 10.1016/s0969-2126(01)00629-3. [DOI] [PubMed] [Google Scholar]
- Wolfe KH. Comparative genomics and genome evolution in yeasts. Philos Trans R Soc Lond B Biol Sci. 2006;361:403–412. doi: 10.1098/rstb.2005.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. Heat shock transcription factors: structure and regulation. Annu Rev Cell Dev Biol. 1995;11:441–469. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]