Abstract
Differentially expressed nucleolar transforming growth factor-β1 target (DENTT), also known as testis-specific protein Y-encoded-like (TSPYL-2) and cell division autoantigen-1, is a member of the testis-specific protein Y-encoded (TSPY)/TSPY-L/SET/nucleosome assembly protein-1 superfamily. DENTT is expressed in various tissues including normal human lung. Here, we investigate the involvement of DENTT in cancer promotion and progression. DENTT messenger RNA (mRNA) and protein levels were shown to be markedly downregulated in human and mouse primary tumors and in human tumor cell lines. Overexpression of DENTT in human lung (A549-DENTT) and breast (MCF-7-DENTT) cancer cells resulted in diminished growth potential in anchorage-dependent growth assays and reduced capacity to form colonies under anchorage-independent culture conditions. The migratory potential of A549-DENTT and MCF-7-DENTT cells was reduced when compared with empty vector control cells. Treating human lung cell lines with demethylating agents increased DENTT expression significantly. DENTT expression pattern paralleled that of transforming growth factor-β1 (TGF-β1) in normal and malignant tissue and ectopic expression or treatment with TGF-β1 in lung cancer cells was followed by increased DENTT mRNA and protein levels. Collectively, our results suggest a role for DENTT as a suppressor of the tumorigenic phenotype.
Introduction
We identified differentially expressed nucleolar transforming growth factor-β1 target (DENTT) in a transforming growth factor-β1 (TGF-β1)-responsive epithelial lung cancer cell line by differential messenger RNA (mRNA) analysis (1). DENTT was first described as se20-4 (2) and is also known as cell division autoantigen-1 and CASK-interacting nucleosome assembly protein (3,4). DENTT belongs to the testis-specific protein Y-encoded (TSPY)/testis-specific protein Y-encoded-like (TSPY-L)/SET/nucleosome assembly protein-1 (NAP-1) superfamily whose members, such as TSPY, NAP-1 and SET oncoprotein, have been implicated in cancer cell cycle regulation and chromatin remodeling. TSPY was shown to potentiate cell proliferation and expedite the transition of cells through the G2–M phase (5), whereas both SET and NAP-1 were shown to regulate the G2–M transition by binding to B-type cyclins and modulating cyclin-dependent kinase 1 activity (6,7). A functional role for TSPY in tumorigenesis was proposed based on its loss of expression during tumor progression in melanoma (8). More recent reports have also shown loss of the TSPY gene in 44.4% of primary prostate tumors (9) and hypermethylation of its promoter in prostate cancer cell lines (10). DENTT has been shown to arrest cell growth, to cause a decline of DNA synthesis measured by a bromodeoxyuridine incorporation assay and to inhibit DNA synthesis in S phase in HeLa cells (3). This effect is mediated through upregulation of p21 by activation of p53 and MEK/ERK1/2 MAPK pathways (11). DENTT was also recently found to be downregulated in primary glioblastoma tumors when compared with normal brain tissue (12). Interestingly, treatment of a glioblastoma cell line (T98G) with demethylating agents caused a marked increase in DENTT expression, suggesting that hypermethylation of DENTT promoter is related to its suppression in tumor cells (12).
All the earlier findings suggest possible roles for members of the TSPY/TSPY-L/SET/NAP-1 superfamily in tumorigenesis. Here, we investigate the expression of DENTT in normal versus malignant lung tissue and cell lines and study the effect of its ectopic expression on growth, colony formation and migration potential of lung and breast cancer cells. We also investigate the association between DENTT and TGF-β1 in tumors and the possible role of methylation in DENTT's expression in tumor cells.
Materials and methods
Cell culture
Lung cancer cell lines comprised non-small cell lung cancer cells H23, H2087, A549, H676, H125, H1299, H1155, H1395, H460, H157 and H727 and small cell lung cancer cells N417, H345, H209, H446, H510, H187, H82, H720 and H69 and breast cancer cell line MCF-7 were obtained from the American Type Culture Collection (ATCC) (Manassas, VA) and cultured in RPMI supplemented with 10% fetal bovine serum (FBS). The normal human bronchial epithelial cell line BEAS2B ATCC was maintained in LHC-9 serum-free medium (Biosource International, Camarillo, CA) supplemented with BEGM SingleQuots (Clonetics, San Diego, CA). Human tracheal epithelial cell lines, CEPAC and CFTE, and human bronchial epithelial cell line, HBE135, a kind gift from Dr Catherine Jozwik, were cultured in Dulbecco's modified Eagle's medium and Eagle minimum essential medium, respectively, supplemented with 10% FBS. Human bronchial epithelial cells, IB3, were obtained from ATCC and cultured in LHC-8 serum-free medium (Biofluids, Rockville, MD) with 5% FBS. All cells were supplemented with 1% streptomycin–ampicillin (Invitrogen, Carlsbad, CA). Recombinant TGF-β1 was obtained from R&D Systems (Minneapolis, MN).
Human and murine tissue
Mouse lung tissue was dissected, snap frozen and kept at −80°C. Mice included C57BL/6NCr mice that have heterozygous (HT) deletion of a TGF-β1 allele (TGF-β1±) (TGF-β1 HT) (13,14), C57BL/6/129/sv mice with a latent-activatable (LA) K-ras mutation (K-ras LA) (15) and wild-type mice. Mouse lung tumors from TGF-β1 HT mice were available from an earlier study in which mice were injected with ethyl carbamate to generate lung tumors (16, 17). Mice were treated in accordance with the guidelines for animal care and use established by the National Institutes of Health. Normal and tumor human lung matched sets (11 each) were a kind gift from Dr Fred Hirsch.
RNA extraction, reverse transcription and real-time quantitative polymerase chain reaction
RNA from tissues and cell lines was isolated using an RNeasy mini kit (Qiagen, Valencia, CA) and was reverse-transcribed using the SuperScript First-Strand Synthesis system (Invitrogen). Real-time quantitative polymerase chain reaction (RT-Q-PCR) was performed in an Opticon 2 cycler (MJ Research, Waltham, MA) using Sybr Green PCR Master Mix (Applied Biosystems, Foster City, CA). Thermocycling was performed in a final volume of 25 μl containing 2 μl of complementary DNA (1:10 dilution) and 400 nmol/l of primers. The 18S ribosomal RNA was run for every sample as a normalizing housekeeping gene. Samples were run in triplicates using the following cycle scheme: after initial denaturation at 95°C for 2 min, samples were run for 46 cycles of 95°C for 30 s, 60°C for 30 s and 72°C for 45 s. Fluorescence was measured in every cycle and a melting curve was performed after the PCR by increasing temperature from 50 to 96°C (0.5°C increments). A defined single peak was obtained for all amplicons confirming specificity of amplification.
Cloning of human DENTT
The DENTT open reading frame was cloned and sequenced to confirm the previously published sequence (3). The coding region of DENTT gene was PCR amplified in a final volume of 50 μl containing 1 μl complementary DNA (see section above for protocol), 400 nM of primers (forward: 5′-ATGAAACTGGTTCACGTAGCG-3′ and reverse: 5′-TCAAACTCCGAGGACGGGGGC-3′) and 1 μl of Pfu Turbo Hotstart DNA polymerase (Stratagene, La Jolla, CA). The amplified product was then gel purified and cloned into the pcDNA 3.1/V5-His TOPO TA vector (Invitrogen) and transformed into One Shot TOP10 Chemically Competent Escherichia coli strain (Invitrogen). Positive clones were identified by restriction analysis with BamHI and BglII. Sequencing of the vector containing DENTT complementary DNA was performed (MWG Biotech, High Point, NC) and the final construct is referred to as pDENTT.
Transient and stable transfections
A549 and MCF-7 cell lines were transfected with both pDENTT and empty vector (EV) pcDNA 3.1 using FuGENE6 (Roche Diagnostics, Nutley, NJ). They are referred to as A549-DENTT, MCF-7-DENTT, A549-EV and MCF-7-EV. After 72 h, cells for stable transfection were exposed to appropriate media containing 800 μg/ml of geneticin (Invitrogen). A549 were also transiently transfected with human TGF-β1 (pTGF-β1) and human K-ras (pK-ras) expression vectors along with control EV (pCMV6) from OriGene Technologies, (Rockville, MD). Transfectants are referred to as A549-TGF-β1, A549-K-ras and A549-CMV6.
In vitro transcription–translation
One microgram of pcDNA 3.1 and pDENTT vectors was added to a 50 μl translation reaction containing rabbit reticulocyte lysate (Promega, Madison, WI) for 90 min at 30°C and followed by electrophoretic separation on a 4–12% sodium dodecyl sulfate–polyacrylamide gel of a 10 μl aliquot to confirm the presence of translation product. Translated product signal was detected by 35S-methionine autoradiography (Life Science, Amersham, Boston, MA) or western blot analysis.
Antibody production and characterization and western blotting
The DENTT antibody was generated in chicken against the synthetic peptide CZSDDDDRDIEYYEK for representing amino acid position 596–608 (Z is aminocaproic acid). The antibody was affinity purified with the same antigen employed to immunize the chicken.
Cells were seeded onto six-well plates (Corning Costar, Cambridge, MA) and harvested in lysis buffer. Equal amounts of protein were separated by electrophoresis on 4–12% Nupage Bis–Tris gels (Invitrogen) and transferred to nitrocellulose membranes (Life Science, Amersham). For signal generation, the membrane was incubated at room temperature first for 1 h with 5% non-fat-dried milk in Tris-buffer with 0.05% Tween 20 (Sigma, St Louis, MO) and then overnight at 4°C with DENTT antibody diluted 1:1000 in Tris-buffer with 0.05% Tween 20. The membrane was washed and incubated for 1 h at room temperature with horseradish peroxidase-conjugated anti-chicken IgG goat antibody (Life Science) diluted 1:1000 in Tris-buffer with 0.05% Tween 20. Antibody binding was visualized using the ECL Plus chemiluminescence kit (Amersham Pharmacia, Piscataway, NJ) or the horseradish peroxidase color development reagent,4-chloro-1-naphthol (Bio-Rad, Hercules, CA).
Enzyme-linked immunosorbent assay
The levels of immunoreactive TGF-β1 were measured using the TGF-β1 E max ImmunoAssay System (Promega) (sensitivity, 32 pg/ml for TGF-β1). Samples taken from the supernatant were acid treated according to the manufacturer's instructions.
Growth assay
Proliferation of cells stably transfected with full-length DENTT or EV was assessed using a Quick Cell Proliferation Assay Kit (BioVision, Mountain View, CA). Cells were seeded onto 96-well polyvinylchloride plates at a concentration of 100 000 cells/ml (A549) and 50 000 cells/ml (MCF-7). The assay was performed in transferrin, insulin, selenium medium (18) with cells grown at 37°C, 5% CO2 in a humid incubator. The (4-[3-(4-Iodophenyl)-2-(4-nitrophenyl)-2H-5-tetrazolio]-1,3-benzene disulfonate) /Electro Coupling Solution reagent was added every day for five consecutive days. The Spectra Rainbow (Tecan, Raleigh, NC) plate reader and software were used to determine changes in the number of viable cells from dye reduction measured by absorbance at 480 nm.
Soft agar clonogenic assay
The anchorage-independent growth of A549-EV and A549-DENTT cells was examined by soft agar clonogenic assay. Cells were trypsinized in their log phase and 5000 cells of each cell line were resuspended in 1.5 ml of RPMI, 10% FBS and 0.35% agarose. The mix was plated in six-well plates (Corning Costar) containing 1.5 ml of presolidified culture media in 0.5% agar containing 10% FBS. Plates were incubated at 37°C in 5% CO2 atmosphere for 2–3 weeks and colonies >0.1 mm in diameter were counted.
Migration assay
Migration assay was performed in 8 μm pore 96-well ChemoTx plates (Neuroprobe, Cabin John, MD). A total of 100 000 cells for A549-EV and A549-DENTT and 50 000 of MCF-7-EV and MCF-7-DENTT were placed in the upper chambers after coating these membranes with 10 μg/ml fibronectin on both sides. The lower wells were filled with RPMI supplemented with 10% FBS. Plates were incubated at 37°C for 5 h, Non-migrating cells were wiped off the top surface and membranes were fixed and stained with Hema 3 (Biochemical Sciences, Swedesboro, NJ). Cells that migrated through the pores of the membranes were counted.
Immunohistochemical analysis
Deparaffinized sections of mouse lung tissue (5 μm in thickness) and A549 cells grown on glass slides were treated with 3% H2O2 and normal goat serum to lower non-specific binding. They were exposed to the chicken anti-DENTT antibody or anti-TGF-β1 antibody diluted 1:500 in phosphate-buffered saline overnight at 4°C. The following day, slides were processed using a Histostain-Plus Kit (Zymed Laboratories, Carlsbad, CA), counterstained with hematoxylin and photographed on an Olympus BX50 microscope (Olympus Optical Co., Melville, NY) equipped with a PDMC-2 camera (Polaroid, Waltham, MA). Negative controls included omission of the primary antibody and preabsorption in liquid phase with the antigen employed to generate the antibody.
5-Aza-2′-deoxycytidine and trichostatin A treatment and analysis of gene expression
Tumor cell lines were treated with demethylating agent AZA (Sigma) for 5 days at a concentration of 5 μM, histone deacetylases-inhibiting agent trichostatin A at a final concentration of 250 nM for the last 24 h or a combination of both. Total RNA was isolated from treated and untreated cells and mRNA was measured by RT-Q-PCR.
Statistical analysis
Results were presented as mean ± SD unless otherwise indicated. Statistical analysis was performed using the non-paired Student's t-test and statistical significance was at *P < 0.05, **P < 0.01 and ***P < 0.001.
Results
DENTT's expression is downregulated in lung tumors
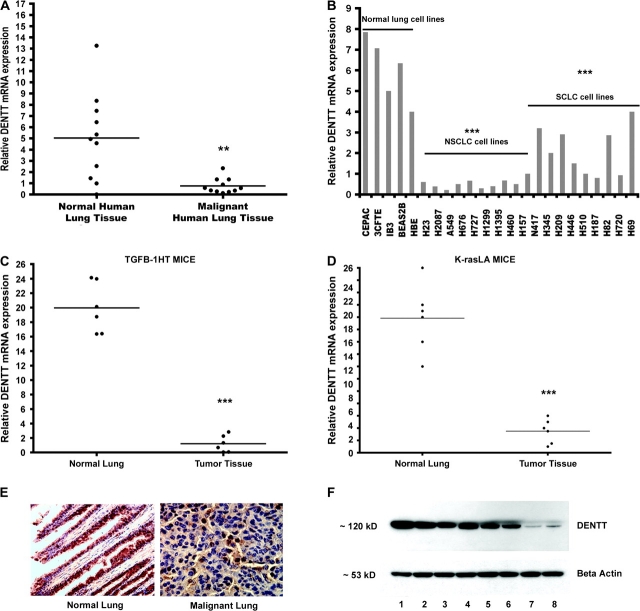
We have previously shown DENTT expression in normal human and mouse lung cell lines and tissue by northern blot and immunohistochemical staining (1,19). Here, we extended our investigation by comparing levels of DENTT in normal versus tumor specimens and we showed consistent downregulation of DENTT mRNA in tumor samples (Figure 1). First, we investigated DENTT's mRNA expression in human lung primary tumors (Figure 1A). All tumor samples showed significant downregulation of DENTT mRNA when compared with their normal matching counterpart (∼6-fold decrease). Levels of DENTT mRNA were also assessed in a panel of 24 human lung cell lines comprised non-small cell lung cancer, small cell lung cancer and normal immortalized human lung cell lines. An average 13- and 3-fold downregulation in non-small cell lung cancer and small cell lung cancer, respectively, compared with normal lung cells was observed (Figure 1B). Consistent downregulation of DENTT in human tumors encouraged us to investigate DENTT expression in mouse tumor models. In agreement with our human mRNA expression data, DENTT mRNA levels were found to be significantly reduced in primary murine lung tumors excised from different mouse tumor models, including TGF-β1 HT and K-ras LA (see Materials and Methods) (Figure 1C and D). Protein DENTT levels were also assessed using a newly generated DENTT antibody (see supplementary Figure S1, available at Carcinogenesis Online). Consistently, while normal lung tissue showed strong immunostaining for DENTT, its expression was markedly weaker in lung tumor cells (Figure 1E). Protein levels of selected tumor cell lines were also significantly decreased when compared with normal lung cell lines as shown by western blot analysis (Figure 1F).
Fig. 1.
DENTT is downregulated in lung tumor specimens. DENTT mRNA levels in matched human tumor–normal lung tissue sets (A), in a panel of 24 lung cell lines (B), in primary mouse lung tumors and adjacent normal mouse lung tissues from TGF-β1 HT mice (C) and K-ras LA mice (D). DENTT protein expression is shown by immunohistochemical staining in normal mouse and tumor lung tissue (×20 and ×40, respectively) (E) and in selected human cell lines by western blot analysis (F). DENTT protein levels are shown to be higher in lanes 1, 2, 3 and 4 representing normal cell lines: CEPAC, 3CFTE, IB3 and HBE, respectively, compared with lanes 5, 6, 7 and 8, which correspond to tumor lung cells lines: H720, H446, H23 and H2087, respectively.
Overexpression of DENTT mitigates growth potential, substrate-independent colony formation and cell motility in tumor cells
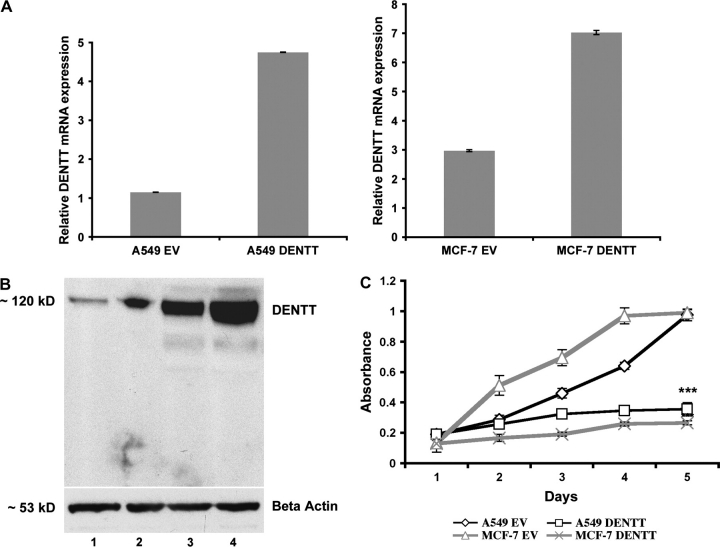
The marked downregulation of DENTT in tumor cells led us to investigate its function in two different tumor cell types. Overexpression of DENTT in lung cancer A549 (A549-DENTT) and breast cancer MCF-7 (MCF-7-DENTT) cells was confirmed by RT-Q-PCR (Figure 2A) and western blot analysis (Figure 2B).
Fig. 2.
(A) DENTT mRNA and protein expression in A549- and MCF-7-transfected cells. A549-DENTT and MCF-7-DENTT cell lines show a 4- and 7-fold increase, respectively, in DENTT mRNA level when compared with their EV counterpart. (B) Western blot showing an increase in DENTT protein level in A549-DENTT (lane 2) and MCF-7-DENTT (lane 4) compared with A549 (lane 1) and MCF-7 (lane 3) EV cells. (C) Overexpression of DENTT decreases the growth rate of A549 and MCF-7 cells (six repeats per measurement).
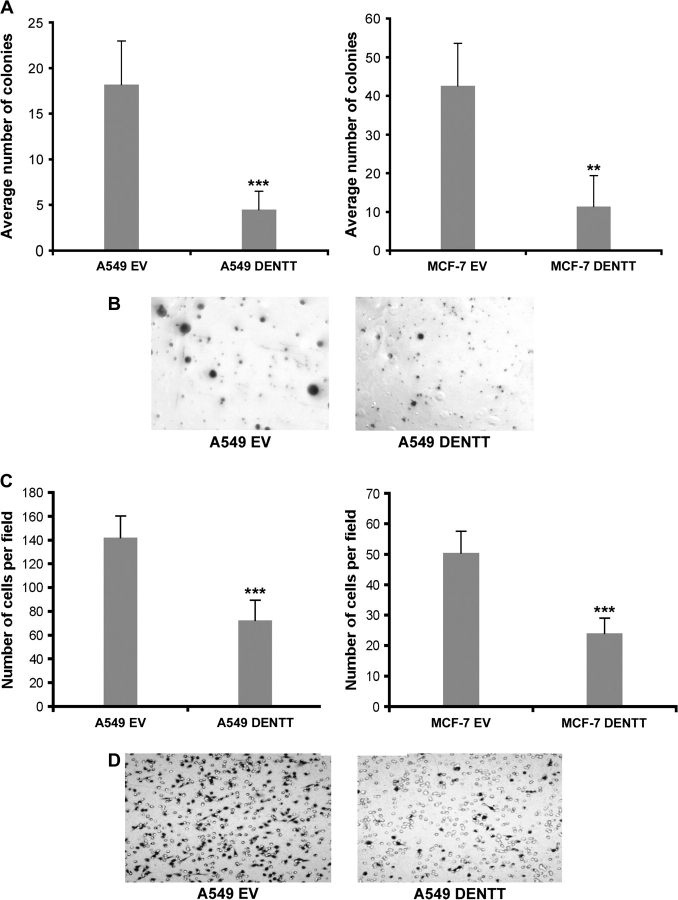
A549-DENTT and MCF-7-DENTT showed diminished cell growth potential as assessed by (4-[3-(4-Iodophenyl)-2-(4-nitrophenyl)-2H-5-tetrazolio]-1,3-benzene disulfonate) /Electro Coupling Solution proliferation assay. A significant difference of 4- and 5-fold in growth was observed between A549-DENTT and MCF-7-DENTT and their respective controls transfected with EV (Figure 2C). In a separate experiment, overexpression of DENTT reduced the ability of A549-DENTT and MCF-7-DENTT to form colonies in anchorage-independent clonogenic assays by 40 and 75%, respectively (Figure 3A). Colonies appeared to be larger in A549-EV control wells (Figure 3B) compared with A549-DENTT.
Fig. 3.
(A) Overexpression of DENTT decreases the potential of A549 and MCF-7 to form colonies in growth anchorage-independent clonogenic assays. (B) Representative assay for A549-EV (left) and A549-DENTT (right). (C) Overexpression of DENTT reduces migratory potential of tumor cells. Overexpression of DENTT in A549 (left panel) and MCF-7 (right panel) significantly reduces the number of migrated cells when compared with their EV control cells. Bars represent the mean and standard deviation of six independent measurements. Two representative images of the membranes were used (D).
Cell motility is important for metastasis of tumor cells. Migration assays comparing A549-DENTT, MCF-7-DENTT and EV controls demonstrated that overexpession of DENTT blocks cell motility (Figure 3C and D). Cells transfected with DENTT showed a 50% reduction in the number of cells migrating through a 8 μm pore membrane compared with EV control cells, suggesting that DENTT may also play a role in mitigating tumor cell migration.
DENTT is hypermethylated in lung tumors
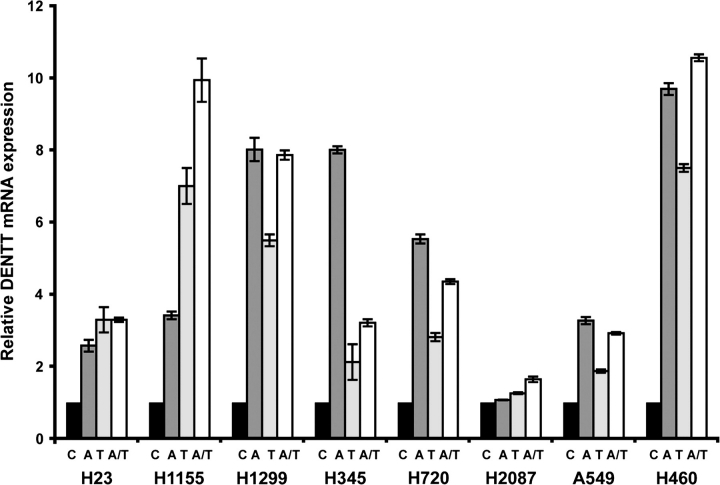
Downregulation of DENTT in tumor cells has a profound impact on their physiology. We, therefore, investigated the mechanisms underlying the regulation of its expression in tumor cells. Methylation has been proposed as a possible mechanism for DENTT suppression in glioblastoma cells (12). Here, we treated eight human lung cancer cell lines with demethylating agents, 5-aza-2′-deoxycytidine and/or trichostatin A, and found consistent increase in DENTT expression, suggesting methylation as a potential mechanism for DENTT downregulation in lung tumors (Figure 4).
Fig. 4.
DENTT expression is enhanced in human lung cancer cells after exposure to demethylating agents. Cells were treated with 5-aza-2′-deoxycytidine (A), histone deacetylases-inhibiting agent trichostatin A (T) or a combination of both (A/T).
TGF-β1 and DENTT show parallel expression
As shown above, methylation seems to be an important mechanism for regulation of DENTT in tumor cells, although other mechanisms of regulation have been proposed. Previous studies have demonstrated a possible regulatory mechanism of DENTT by TGF-β1 (19,20). To extend our understanding of this relationship, levels of TGF-β1 mRNA were assessed by RT-Q-PCR in the same human and mouse lung tissue samples used to examine DENTT expression. TGF-β1 expression showed marked reduction in all human lung tumors compared with normal counterparts (Figure 5A). Human tumor lung cell lines also showed a statistically significant downregulation of TGF-β1 mRNA levels when compared with normal human lung cell lines (Figure 5B). Primary murine tumors excised from TGF-β1 HT and K-ras LA mice also showed a respective significant 25- and 12-fold downregulation of TGF-β1 mRNA levels (Figure 5C and D). Tissue sections taken from both tumor animal models also showed a markedly weaker staining for TGF-β1 protein in lung tumors (Figure 5E). Moreover, at 24 h, TGF-β1 levels detected in supernatants of normal human cell lines were significantly higher than in tumor cell lines as shown by enzyme-linked immunosorbent assay (Figure 5F).
Fig. 5.
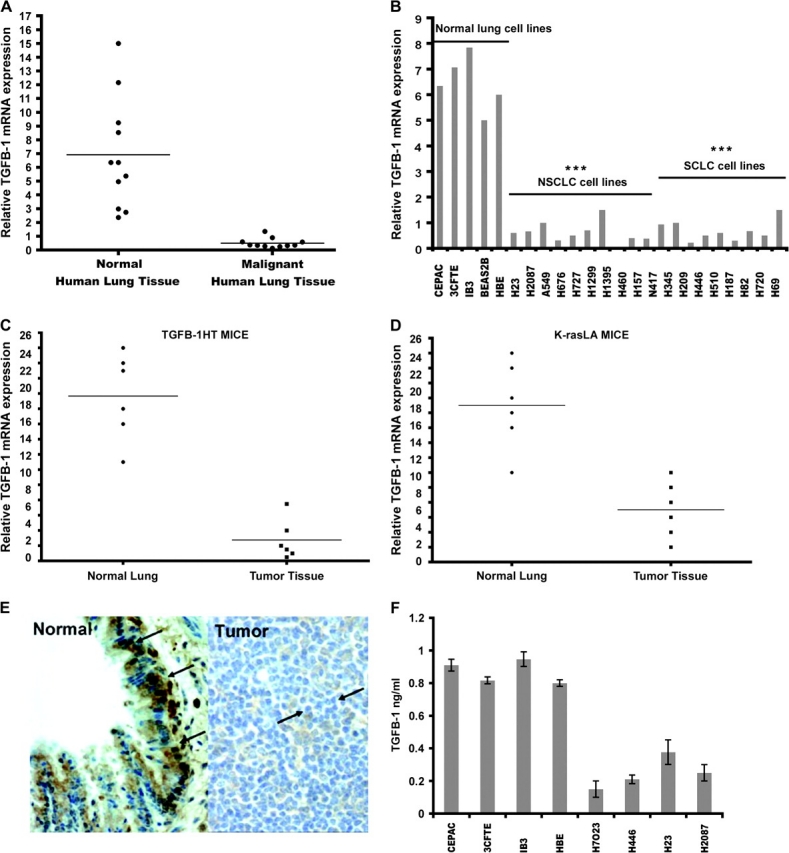
TGF-β1 parallels DENTT expression in lung tumors. TGF-β1 mRNA levels in matched human tumor–normal lung tissue (A) in a panel of 24 lung cell lines (B) and TGF-β1 HT (C) and K-ras LA (D) mouse lung tumor models. TGF-β1 protein expression is shown by immunohistochemical staining in normal (left panel) mouse and tumor (right panel) lung tissue (×40) (E). Arrows point to positive staining for TGF-β-1. Immunodetection of TGF-β1 in selected human cell lines show a significantly higher production in normal cell lines than in tumor cells (F).
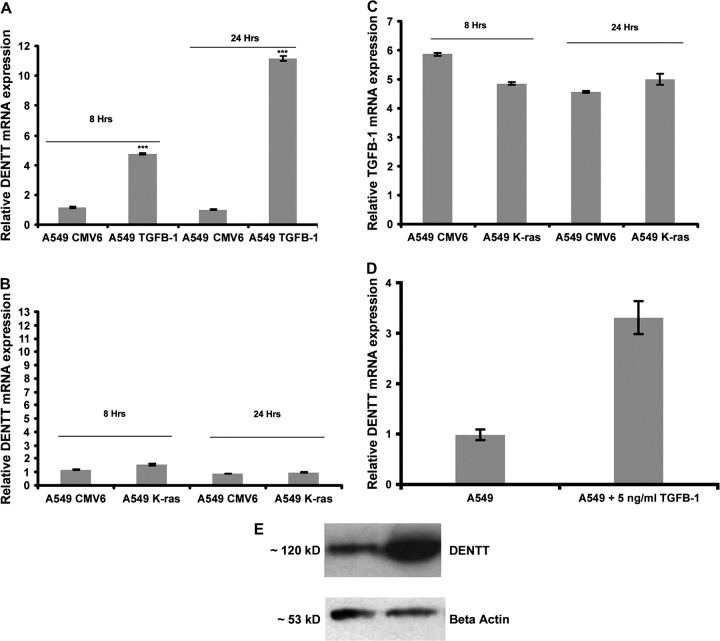
Our studies suggest a parallel pattern of expression of DENTT and TGF-β1 transcripts in tumor and normal lung tissue. Earlier reported data by our group demonstrated that exogenous addition of TGF-β1 to pituitary cells increased DENTT protein production (19). To study the effect of TGF-β1 on DENTT expression in an in vitro tumor model system, lung tumor cells were transiently transfected with TGF-β1 expression vector (A549-TGF-β1) and levels of DENTT mRNA were assessed by RT-Q-PCR. A549-TGF-β1 showed upregulation of DENTT mRNA after 8 and 24 h (Figure 6A). These data further suggest an association between DENTT and TGF-β1.
Fig. 6.
TGF-β1 modulates DENTT expression in A549 cells. (A) DENTT mRNA levels were upregulated in A549 cells transfected with TGF-β1 expression vector versus EV for 8 and 24 h. (B) DENTT mRNA levels were not statistically different in A549 cells after transfection with a K-ras expression vector nor were TGF-β1 mRNA levels (C). Treatment of A549 with 5 ng/ml of recombinant TGF-β1 protein resulted in upregulation of DENTT mRNA (D) and protein levels (E). Error bars represent three experiments.
Both TGF-β1 and DENTT mRNA levels were also downregulated in lung tumors induced by a K-ras mutation in K-ras LA mice, although no statistical differences in DENTT mRNA levels between A549-K-ras and A549-CMV6 were observed (Figure 6B). No statistical differences were also observed in TGF-β1 mRNA levels between A549-K-ras and A549-CMV6 (Figure 6C). Treating the parental cell line A549 with 5 ng/ml of TGF-β1 for 24 h shows a significant increase in DENTT mRNA (Figure 6D) as assessed by RT-Q-PCR and DENTT protein levels as shown by western blot analysis (Figure 6E).
Discussion
In this study, we provide novel information about the expression and role of DENTT in cancer. First, we find consistent downregulation of DENTT mRNA and protein levels in human lung tumors, in human lung cancer cell lines and, finally, in lung tumors from mice tumor models. By overexpressing DENTT in two different tumor cell lines, A549 and MCF-7, major traits of the cell's tumorigenic phenotype (21) were diminished. Overexpression of DENTT resulted in a marked inhibitory effect on cell growth and on clonogenic potential of both lung and breast cell lines. For the first time, we also provide evidence for the involvement of DENTT in cell migratory potential.
Our data on DENTT expression in tumors are consistent with a recently published study, which reported downregulation of DENTT and several members of the TSPY-L family in glioblastoma tumors (12). In this same study, the authors showed upregulation of DENTT in four glioblastoma cell lines after treatment with demethylating agents. Also in agreement with these data, we found upregulation of DENTT mRNA levels in various tumor lung cell lines after treatment with demethylating agents, suggesting that silencing of DENTT in tumor cells is epigenetically regulated through methylation of its promoter region.
We propose that regulation of DENTT expression in lung tumors could also be modulated by TGF-β1. We originally identified DENTT by differential mRNA analysis after treatment of lung adenocarcinoma H727 cells with TGF-β1. Exogenous addition of TGF-β1 has been shown to upregulate DENTT (1) and a similar expression pattern for DENTT and TGF-β1 has been shown in both embryonic and adult mice (19). Here, we demonstrate that TGF-β1 and DENTT have a parallel pattern of expression in human and mouse lung tumors. Interestingly, treating A549 cells with recombinant TGF-β1 protein and overexpression of TGF-β1 in this in vitro tumor model system but not other pro-oncogenic genes such as K-ras resulted in upregulation of DENTT mRNA and protein. Collectively, these data support the idea that TGF-β1 regulates DENTT's expression. Interestingly, tumors in K-ras-mutant mouse lungs show parallel downregulation of DENTT and TGF-β1 mRNA levels. However, overexpressing K-ras mutant in the in vitro tumor model system has no effect on TGF-β1 mRNA levels nor does it influence DENTT levels, suggesting that regulation of DENTT expression is related to TGF-β1 status in the tumor independent of the etiology of the tumor.
This study provides clinical, genetic, molecular and functional data supporting a role for DENTT in lung and breast tumorigenesis. Our collective data provide novel evidence suggesting that DENTT plays a suppressor role in lung cancer development and makes this protein an attractive target for future studies and development of therapies utilizing its tumor-suppressive potential.
Supplementary material
Supplementary Figure S1 can be found at http://carcin.oxfordjournals.org/
Funding
Intramural Research Program of the National Institutes of Health; National Cancer Institute; Center for Cancer Research.
Supplementary Material
Acknowledgments
We thank Dr Catherine Jozwik, Uniformed Services University of Health Sciences, Bethesda, MD, for providing bronchial epithelial cell lines, Dr Fred Hirsch, University of Colorado Cancer Center, Denver, CO, for tumor samples and Christy Falco, National Cancer Institute, for assistance with immunocytochemistry. We also thank Drs Lucy Anderson, Kathy Flanders and William Stetler-Stevenson (National Cancer Institute) for helpful comments during preparation of the manuscript.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- DENTT
differentially expressed nucleolar transforming growth factor-β1 target
- EV
empty vector
- FBS
fetal bovine serum
- HT
heterozygous
- LA
latent activatable
- mRNA
messenger RNA
- NAP-1
nucleosome assembly protein-1
- RT-Q-PCR
real-time quantitative polymerase chain reaction
- TGF-β1
transforming growth factor-β1
- TSPY
testis-specific protein Y-encoded
- TSPY-L
testis-specific protein Y-encoded-like
References
- 1.Ozbun LL, et al. Identification of differentially expressed nucleolar TGF-beta1 target (DENTT) in human lung cancer cells that is a new member of the TSPY/SET/NAP-1 superfamily. Genomics. 2001;73:179–193. doi: 10.1006/geno.2001.6505. [DOI] [PubMed] [Google Scholar]
- 2.Eichmuller S, et al. Serological detection of cutaneous T-cell lymphoma-associated antigens. Proc. Natl Acad. Sci., USA. 2001;98:629–634. doi: 10.1073/pnas.021386498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chai Z, et al. SET-related cell division autoantigen-1 (CDA1) arrests cell growth. J. Biol. Chem. 2001;276:33665–33674. doi: 10.1074/jbc.M007681200. [DOI] [PubMed] [Google Scholar]
- 4.Wang GS, et al. Transcriptional modification by a CASK-interacting nucleosome assembly protein. Neuron. 2004;42:113–128. doi: 10.1016/s0896-6273(04)00139-4. [DOI] [PubMed] [Google Scholar]
- 5.Oram SW, et al. TSPY potentiates cell proliferation and tumorigenesis by promoting cell cycle progression in HeLa and NIH3T3 cells. BMC Cancer. 2006;6:154. doi: 10.1186/1471-2407-6-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canela N, et al. The SET protein regulates G2/M transition by modulating cyclin B-cyclin-dependent kinase 1 activity. J. Biol. Chem. 2003;278:1158–1164. doi: 10.1074/jbc.M207497200. [DOI] [PubMed] [Google Scholar]
- 7.Kellogg DR, et al. Members of the NAP/SET family of proteins interact specifically with B-type cyclins. J. Cell Biol. 1995;130:661–673. doi: 10.1083/jcb.130.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallagher WM, et al. Multiple markers for melanoma progression regulated by DNA methylation: insights from transcriptomic studies. Carcinogenesis. 2005;26:1856–1867. doi: 10.1093/carcin/bgi152. [DOI] [PubMed] [Google Scholar]
- 9.Vijayakumar S, et al. Detection of recurrent copy number loss at Yp11.2 involving TSPY gene cluster in prostate cancer using array-based comparative genomic hybridization. Cancer Res. 2006;66:4055–4064. doi: 10.1158/0008-5472.CAN-05-3822. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, et al. Survey of differentially methylated promoters in prostate cancer cell lines. Neoplasia. 2005;7:748–760. doi: 10.1593/neo.05289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tu Y, et al. Antiproliferative autoantigen CDA1 transcriptionally up-regulates p21(Waf1/Cip1) by activating p53 and MEK/ERK1/2 MAPK pathways. J. Biol. Chem. 2007;282:11722–11731. doi: 10.1074/jbc.M609623200. [DOI] [PubMed] [Google Scholar]
- 12.Kim TY, et al. Epigenomic profiling reveals novel and frequent targets of aberrant DNA methylation-mediated silencing in malignant glioma. Cancer Res. 2006;66:7490–7501. doi: 10.1158/0008-5472.CAN-05-4552. [DOI] [PubMed] [Google Scholar]
- 13.Kulkarni AB, et al. Transforming growth factor beta 1 null mutation in mice causes excessive inflammatory response and early death. Proc. Natl Acad. Sci., USA. 1993;90:770–774. doi: 10.1073/pnas.90.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang B, et al. Transforming growth factor-beta1 is a new form of tumor suppressor with true haploid insufficiency. Nat. Med. 1998;4:802–807. doi: 10.1038/nm0798-802. [DOI] [PubMed] [Google Scholar]
- 15.Johnson L, et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- 16.Kang Y, et al. Transforming growth factor-beta 1 and its receptors in human lung cancer and mouse lung carcinogenesis. Exp Lung Res. 2000a;26:685–707. doi: 10.1080/01902140150216765. [DOI] [PubMed] [Google Scholar]
- 17.Kang Y. Enhanced tumorigenesis and reduced transforming growth factor-beta type II receptor in lung tumors from mice with reduced gene dosage of transforming growth factor-beta1. Mol Carcinog. 2000b;29:112–26. doi: 10.1002/1098-2744(200010)29:2<112::aid-mc8>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 18.Miller MJ, et al. Adrenomedullin expression in human tumor cell lines. Its potential role as an autocrine growth factor. J. Biol. Chem. 1996;271:23345–23351. doi: 10.1074/jbc.271.38.23345. [DOI] [PubMed] [Google Scholar]
- 19.Martinez A, et al. Expression of differentially expressed nucleolar transforming growth factor-beta1 target (DENTT) in adult mouse tissues. Dev. Dyn. 2002;224:186–199. doi: 10.1002/dvdy.10096. [DOI] [PubMed] [Google Scholar]
- 20.Ozbun LL, et al. Differentially expressed nucleolar TGF-beta1 target (DENTT) shows tissue-specific nuclear and cytoplasmic localization and increases TGF-beta1-responsive transcription in primates. Biochim. Biophys. Acta. 2005;1728:163–180. doi: 10.1016/j.bbaexp.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 21.Hanahan D, et al. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.