Abstract
Despite general observations of non-random genomic distribution of new genes, it is unclear whether or not new genes preferentially occur in certain genomic regions driven by related molecular mechanisms. Using 1.5 Mb of genomic sequences from short arms of chromosome 3 of Oryza glaberrima and O. punctata, we conducted a comparative genomic analysis with the reference O. sativa ssp. japonica genome. We identified a 60-kb segment located in the middle of the subtelomeric region of chromosome 3, which is unique to the species O. sativa. The region contained gene duplicates that occurred in Asian cultivated rice species that diverged from the ancestor of Asian and African cultivated rice one million years ago (MYA). For the 12 genes and one complete retrotransposon identified in this segment in O. sativa ssp. japonica, we searched for their parental genes. The high similarity between duplicated paralogs further supports the recent origination of these genes. We found that this segment was recently generated through multiple independent gene recombination and transposon insertion events. Among the 12 genes, we found that five had chimeric gene structures derived from multiple parental genes. Nine out of the 12 new genes seem to be functional, as suggested by Ka/Ks analysis and the presence of cDNA and/or MPSS data. Furthermore, for the eight transcribed genes, at least two genes could be classified as defense or stress response-related genes. Given these findings, and the fact that subtelomeres are associated with high rates of recombination and transcription, it is likely that subtelomeres may facilitate gene recombination and transposon insertions and serve as hot spots for new gene origination in rice genomes.
Keywords: comparative genomics, gene duplication, Oryza sativa, subtelomere, new genes
INTRODUCTION
Previous studies have provided evidence of the significant role that novel genetic elements have played in organismal diversification and speciation (Ohno, 1970; Long et al., 2003). Various mechanisms, such as retroposition, exon shuffling, tandem gene duplication, and transposon-mediated gene duplication, have been proposed for the creation of novel genetic elements in numerous organisms (see reviews by Long et al., 2003; Fan et al., 2007a). However, detailed analysis of gene duplication and novel gene evolution in plants is still lacking.
Comparative genomics is a powerful tool to search for gene duplication events across entire genomes, and has been applied in the analysis of several organisms (e.g. Betran et al., 2002; Marques et al., 2005; Zhang et al., 2005; Wang et al., 2006). Comparing closely related species is particularly powerful for the detection of recent gene duplications. For example, the recently released 12 wild Drosophila genomic sequences have provided excellent opportunities to decipher gene and genome duplication at the phylogenetic level within a single genus Drosophila (Drosophila 12 Genomes Consortium, 2007). However, the few plant species for which genome sequences are available are distantly related, and thus a search for new genes has been hindered by insufficient resolution for short evolutionary time intervals.
The genus Oryza, which contains the world's most important food crop—rice (O. sativa)—is an ideal plant model system to study detailed gene and genome evolution, due to small genome size and the availability of genome sequences from both subspecies of cultivated rice, japonica and indica (IRGSP, 2005; Yu et al., 2002). Findings from rice research can also inform studies on other cereal crops, such as corn and wheat, which, despite sharing a common ancestor 50 MYA, have genome sizes of six to 38 times larger than rice, respectively. Thus, rice provides the central comparative genomics core for monocot research (Bennetzen, 2007; Paterson et al., 2005; Wing et al., 2005).
The genus Oryza is composed of 23 species that are classified into 10 distinct genome types (six diploid and four allotetraploid) (Ge et al., 1999), and the phylogenetic relationships among these genome types are well resolved (Ge et al., 1999; Zhu and Ge, 2005) and cover an approximate 17 million year time span. Such broad diversification over such a relatively short period of time indicates the potential for new gene creation is relatively high.
In 2004, we were funded to create a genus level comparative genomics system for the genus Oryza composed of 11 bacterial artificial chromosome (BAC)-based fingerprint/end sequence physical maps, representative of the 10 genome types, aligned to the rice RefSeq (Wing et al., 2005; Ammiraju et al., 2006; Kim et al., 2008). The Oryza Map Alignment Project (OMAP) system now provides immediate access to virtually any region of the collective Oryza genomes for detailed comparative investigation. As part of an international effort to functionally characterize all rice genes, we are focusing on a detailed analysis of the short arm of chromosome 3 and have used the BAC-based physical maps to select minimum tiling paths of BAC clones across the chromosome 3 short arms of O. glaberrima [AA], O. punctata [BB], O. officinalis [CC], and O. minuta [BBCC] for sequencing. As part of an initial pilot project to sequence these short arms, we sequenced and finished ∼1.5-Mb BAC tiles from O. glaberrima and O. punctata and compared these sequences with the O. sativa ssp. japonica RefSeq. With such a genomic scale comparison, we were able to identify a recently evolved 60-kb DNA segment in O. sativa that contained a number of young genes that originated within the last one million years (Myr).
RESULTS
Comparative Analysis of a 1.5-Mb Region from the Short Arm of Chromosome 3 in O. sativa ssp. japonica, O. glaberrima, and O. punctata
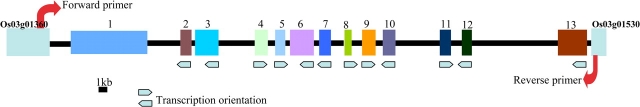
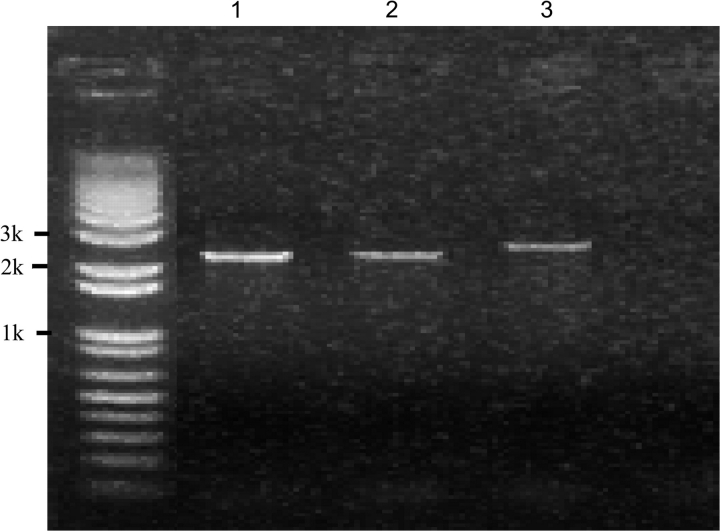
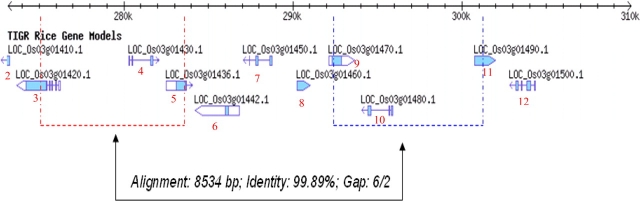
A unique and contiguous 60-kb region from the subtelomeric region of the short arm of chromosome 3 of the Asian cultivated rice, O. sativa ssp. japonica, was identified by comparing with 1.5-Mb orthologous regions sequenced from the AA genome of African cultivated rice O. glaberrima and the BB genome of O. punctata. This unique region in O. sativa was found to contain 12 candidate genes and one complete 11-kb long TyGypsy LTR retrotransposon that was annotated as three independent retrotransposons in The Institute for Genomic Research (TIGR) gene ontology database (Figure 1 and Table 1). To determine if the 60-kb region was unique to japonica rice or could be found in its putative progenitor species, O. nivara and O. rufipogon, we searched BAC end sequence (BES) datasets for these species (Kim et al., 2008) for sequences similar to the 60-kb sequence using BLAST. This analysis identified orthologous BESs to Os03g01410 in both O. nivara and O. rufipogon, and Os03g01420 in O. nivara (Table 2), thus providing evidence that at least part of the unique 60-kb sequence could be found in these two wild species. This finding was further supported by analysis of the complete and partial orthologous sequences from O. nivara and O. rufipogon, respectively, which revealed the presence of both genes 2 and 5 (Yu et al., unpublished data). Since BES datasets and sequence data were not available for two additional AA genome species, O. barthii (the wild progenitor of O. glaberrima) and O. glumaepatula (a wild species from South America), we designed a pair of diagnostic PCR primers to detect the presence or absence of the O. sativa ssp. japonica unique 60-kb region at this location on chromosome 3 (Figure 1). The expected PCR amplification band size between primers Os03g01360F and Os03g01530R for O. sativa, as shown in Figure 1, is 65 Kb, which is too big for the regular PCR amplification. If this segment is missing, the expected size of the PCR band would be 2.5–3 kb, which is exactly what we detected using genomic DNA isolated from the O. glaberrima control and O. barthii and O. glumaepatula (Figure 2). We further constructed a synteny map using the chromosome 3 sequences to check whether this 60-kb region was located outside the syntenic chain based on the Chain and Net pipeline of UCSC. As expected, whether using the O. glaberrima or O. punctata genome as the reference sequence (data not shown), almost the entire region fell in the syntenic gap, which supports the hypothesis that most of the genes in this 60-kb region in O. sativa should be very young.
Figure 1.
Schematic Sketch of 60-kb Segment in O. sativa ssp. japonica.
The primer pair was used to amplify this segment for other AA species. See Table 1 for gene order.
Table 1.
The Detailed Information and Ka/Ks Analysis for 13 Annotated Genes.
| Gene order | Gene ID (TIGR) | Paralogs (identity and alignment length) | Annotation | Expression (no. of EST) | Ka | Ks | Ka/Ks (LRT p-value) |
| 1 | Os03g01380 | Os01g17360 (97.2%/2328) | TyGypsy retrotransposon | No (0) | NA | NA | NA |
| Os03g01390 | Os07g35630 (95.8%/988) | ||||||
| Os03g01400 | Os11g34420 (97.3%/1031) | ||||||
| 2 | Os03g01410 | Os01g72700 + de nova (93.8%/1190) | STK kinase | No (0) | 0.0250 | 0.1761 | 0.142/0.011 |
| 3 | Os03g01420# | Os03g01490 (100%/267) | Expression protein | FL-cDNA and EST transcript (65) | 0 | 0 | ∞ |
| 4 | Os03g01430 | Os03g01480 (100%/303) | Hypothetical protein | No (0) | 0 | 0 | ∞ |
| 5 | Os03g01436 | Os03g01470 (99.7%/1173) | Conserved hypothetical protein | FL-cDNA and EST transcript (1) (leaf of seedling 4 min gamma-irradiation, after 6 h) | NA | NA | NA |
| 6 | Os03g01442# | Os01g69904# (97.6%/2647) | Expression protein | FL-cDNA and EST transcript (12) | 0.0277 | 0.0003 | 99/0.13 |
| 7 | Os03g01450* | Os04g32150 (93.6%/643) + Os06g11900 (92.1%/478) | Hypothetical protein | EST transcript (3) (leaf of seedling 4 min gamma-irradiation, after 6 h) | NA | NA | NA |
| 8 | Os03g01460 | No close homology | Hypothetical protein | No (0) | NA | NA | NA |
| 9 | Os03g01470 | Os03g01436 | Expression protein | FL-cDNA and EST transcript (6) (shoot and callus) | NA | NA | NA |
| 10 | Os03g01480 | Os03g01430 | Hypothetical protein | No (0) | 0 | 0 | ∞ |
| 11 | Os03g01490*# | Os03g01420+ | Expression protein | FL-cDNA and EST transcript (46) | 0 | 0 | ∞ |
| 12 | Os03g01500* | Os12g24870 (94.0%/1050) | Hypothetical protein | EST transcript (2) | NA | NA | NA |
| 13 | Os03g01520*# | No close homology | Hypothetical protein | EST transcript (3)(2/2 callus) | NA | NA | NA |
Notes: * Denotes mis-annotated gene models described in the TIGR rice annotation database. # Denotes genes with multiple alternative splicing isoforms. The cDNA from each parental gene and young gene were aligned with BLASTN using a low complexity filter off. Identity and alignment length were calculated using the chained SearchIO module of BioPerl. NA stands for non-feasible substitution analysis due to retrotransposon, no candidate parental gene, or distinct gene structure.
Table 2.
BLAST Analysis Results Using the Unique O. sativa ssp. japonica 60-kb Segment as a Query against the O. rufipogon and O. nivara BES Datasets.
| Species of BAC | BAC ID (location of BAC) | Identity (%) | Length of alignment (bp) | O. sativa orthologues |
| O. nivara | OR_BBa0096N24 | 99.21 | 629 | Os03g01420 |
| O. nivara | OR_BBa0036B16 | 99.83 | 590 | Os03g01420 |
| O. nivara | OR_BBa0122N16 | 97.79 | 768 | Os03g01420 |
| O. nivara | OR_BBa0059M20 | 98.94 | 661 | Os03g01410 |
| O. rufipogon | OR_CBa0020A22 | 98.97 | 677 | Os03g01410 |
Figure 2.
The PCR Amplification Using Primers Spanning in the Flanking Region of the 60-kb O. sativa Specific Segment in Three Oryza AA Genome Species.
(1) O. glaberrima; (2) O. barthii; (3) O. glumaepatula.
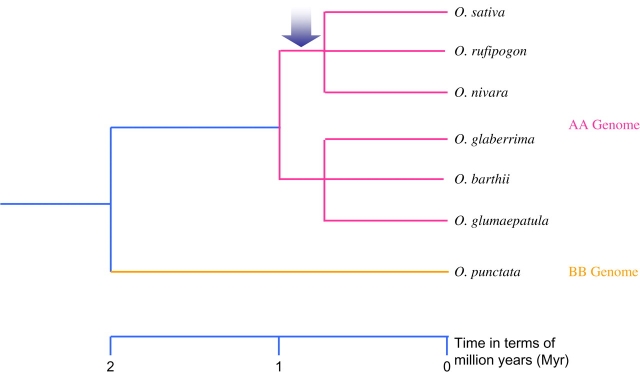
Thus, all data indicated that the origin of this region predates the divergence of the O. sativa–O. rufipogon–O. nivara clade but occurred after the divergence of this clade from the ancestral O. glaberrima, O. barthii, and O. glumaepatula clade (Figure 3). The phylogenetic distribution of this region suggests that the genes encoded in the segment originated around 1 MYA or later.
Figure 3.
The Phylogenetic Relationship of AA Genome Oryza Species Rooted by BB Genome Species O. punctata.
The blue arrow indicates the origination point of this 60-kb segment.
Gene Structure and Origination of Annotated Genes
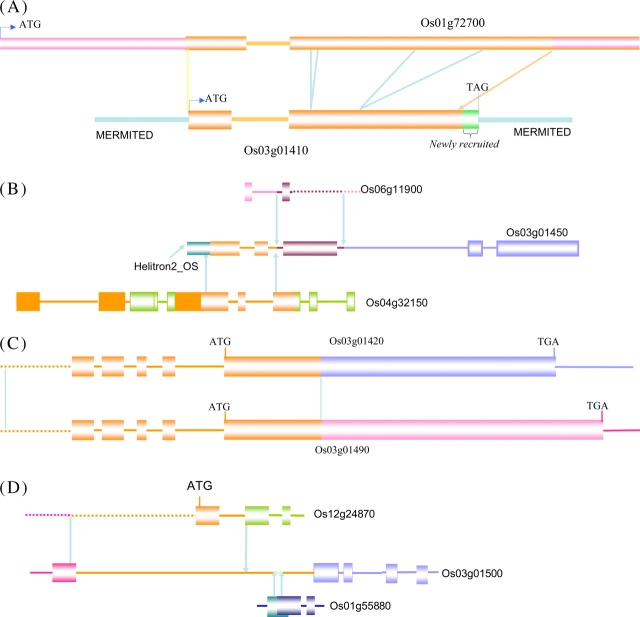
Twelve candidate genes, two embedded in PackMULEs (Os03g01410 and Os03g01520) and one embedded in a Helitron (Os03g01442), and one complete TyGypsy retrotransposon were annotated in the 60-kb segment in O. sativa (Figure 1 and Table 1). Among the genes, 11 were classified as hypothetical or expressed genes. Only Os03g01410, contained within a PackMULE, could be assigned a putative function, based on TIGR ontology assignments, and was proposed to encode a ‘STK kinase’ (Table 1). In order to detect the origin of these 12 genes, we searched the O. sativa ssp. japonica genome and were able to identify candidate parental genes for 10 genes, and all the paralogs had high overall DNA sequence identity (greater than 93%), which is suggestive of being derived from recent duplication and transpositional events (Table 1). The two genes (Os03g01460 and Os03g01520) without paralogs elsewhere in the O. sativa genome may have originated de novo, or the parental genes from which they were derived may have subsequently been deleted, rearranged, or degraded.
By thoroughly comparing the gene structures of the 10 pairs of paralogs in O. sativa ssp. japonica, two distinct classes were revealed. First, five genes (Os03g01410, Os03g01420, Os03g01450, Os03g01490, and Os03g01500) had chimeric gene structures in which part of the parental gene was combined with additional sequences. Specifically, Os03g01410 was a chimera composed of part of the Os01g72700 gene and a 60-bp newly recruited sequence at the 3’ end (Figure 4A and Supplemental Figure 1). Os01g72700 was annotated as an ATP binding protein and its corresponding mRNA has been identified in several different rice tissues. Os03g01420 and Os03g01490 shared 450 bp of partial homologous coding sequence and appeared to have been created by an inverse tandem duplication event (Figures 4B and 5, and Supplemental Figure 2). However, we could not locate the homologous sequence for the remaining coding sequences for these two genes. Gene Os03g01450 was found to have originated from three separate sequences. Os04g32150 and Os06g11900 recombined to form the first three exon of Os03g01450, and then combined with the flanking sequence to generate the entire gene structure (Figure 4C). The first intron of Os03g01500 was very large (>10 kb) and appears to have originated from three independent sequences composed of: (1) the first exon (270 bp) and almost all of the first intron (780) of gene Os12g24870, (2) part of the first exon of Os01g55880 (110 bp), and (3) 9 kb of flanking sequence. However, the remaining sequence of Os03g01500 had no paralogous sequence identified (Figure 4D).
Figure 4.
The Schematic Sketch of Five Chimeric Gene Origination Patterns.
(A) Os03g01410.
(B) Os03g01450.
(C) Os03g01490 and Os03g01420.
(D) Os03g01500.
The homologous regions are shown in the same color. Solid boxes stand for exon, and solid lines indicate introns. The genes showing both start and stop codons have complete gene structure, otherwise only partial genes were drawn.
Figure 5.
A Region Showing the Recent Tandem Inverse Duplication (in Dash Line Boxes).
In the second class, an internal duplication within the 60-kb unique region appears to have created a second copy of three genes. The respective gene pairs are Os03g01420–Os03g01490, Os03g01430–Os03g01480 and Os03g01436–Os03g01470, and a high level of sequence identity (99.89%) for this 8.5-kb region suggests this was a very recent duplication event, as shown in Figure 5. However, even though the tandem duplication was recent, two paralogous genes showed noteworthy divergence regarding gene structure. Specifically, the Os03g01420–Os03g01490 gene pair had similar alternative isoforms (an alternative second exon), but contained distinct 3’ terminal ends, of which Os03g01490 was 36 bp longer (Figure 4B). Finally, the Os03g01436 and Os03g01470 gene pair was found to have used the sense and anti-sense strands of one nearly identical DNA segment, respectively, encoding two totally distinct transcribed genes with open reading frames (ORFs) of more than 150 amino acid residues each.
Substitution Test between Paralogs
We investigated the functionality of six of the 12 O. sativa candidate genes that had clearly identifiable paralogues by calculating the Ka and Ks values (Table 1). For three of the remaining six candidate genes (Os03g01460, Os03g01500, and Os03g01520), we were not able to identify their corresponding parental genes (Os03g01460 and Os03g01520) or coding sequence (Os03g01500) as described above. Ka and Ks values for the final three genes (Os03g01436, Os03g01470, and Os03g01450) could not be calculated because they had totally different ORFs. That is, paralogous genes Os03g01436 and Os03g01470 used sense and antisense strands as ORFs, respectively, and Os03g01450 shared some sequences with Os04g32150 and Os06g11900, but their gene structures were different compared to either of them with non-alignable ORFs.
Ks values for two of the candidate genes, Os03g01410 and Os03g01442, were relatively small, with a value of 0.176 and 0.0003, respectively, and zero for paralogous gene pairs (Os03g01480 vs Os03g01430 and Os03g01420 vs Os03g01490) due to their nearly identical sequences. Using a synonymous substitution rate of 0.65/100 Myr/synonymous site, as estimated for the Adh loci of grasses (Gaut et al., 1996), we estimated that the gene duplication events for all five paralogous duplicate genes ranged from 14 Myr (Os03g01410) to 0.02 Myr (Os03g01442) (Table 1).
Os03g01410 showed a low Ka/Ks value (0.14) that significantly deviated from neutrality (= 0.5) with p ∼ 0.011 under the LRT test, indicating this gene is functionally constrained under purifying selection (Table 1). By contrast, Os03g01442 had an excess number of non-synonymous substitutions, with four replacement substitutions and zero synonymous substitutions. The LRT test gave a marginally significant p-value of 0.13, given an omega (Ka/Ks) = 1 under neutrality. Such an excess of non-synonymous substitutions suggests the possibility of adaptive evolution of gene Og03g01442 after gene duplication.
Expression Analysis
Eight of 12 O. sativa candidate genes appeared to be transcribed, as evidenced by the presence of either EST and/or FL-cDNA sequence in Genbank (Table 1 and Supplemental Figure 3). In all eight cases, at least two transcript sequences could be found in Genbank. All but two (Os03g01436 and Os03g01470) of these eight genes have multiple exons with conventional splice junctions. Finally, mRNA accumulation of three genes (Os03g01420, Os03g01442 and Os03g01490) appeared to be fairly high in vivo, as revealed by the presence of 65, 12, and 46 independent EST sequences in Genbank, respectively, as well as Massively Parallel Signature Sequencing (MPSS) expression signatures (Nakano et al., 2006).
The presence of abundant ESTs allowed us to analyze tissue-specific profiles of mRNA accumulation. As shown by the UniGene Profile Viewer (Wheeler et al., 2008), both Os03g01420 (with Chi square p ∼ 2 × 10−9) and Os03g01490 (p ∼ 8 × 10−6) were highly associated with ESTs derived from callus tissue (Supplemental Figure 4A and 4B). Os03g01442 mRNA was also found in callus tissue, but only a few copies were detected, which could not ensure a robust statistical test (Supplemental Figure 4C). However, MPSS data derived from mRNA isolated from various rice tissues, 6 h post inoculation with Xathomonas oryzae pv. oryzae, revealed that mRNA from the Os03g01442 gene was present in leaf tissue only with a normalized tag count of 34. Interestingly, although the numbers of ESTs were quite low, mRNA accumulation for genes Os03g01470 and Os03g01520 were only detected in callus tissue and mRNA accumulation for genes Os03g01436 and Os03g01450 were only detected in leaf tissue subjected to gamma radiation. In total, except for Os03g01500, whose ESTs data lack specific tissue information, all other seven genes appear to be involved in manipulated tissue (callus), general defense, resistance responses, or various other responses that may have caused the cell death. In contrast, all paralogs showed no or less mRNA accumulation from defense response-derived tissues (see Supplemental Figure 4D–4G).
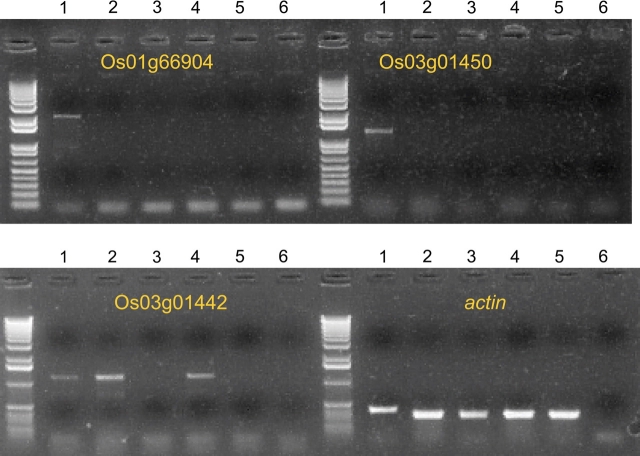
We further conducted an experimental profiling for two of the genes (Os03g01442 and Os03g01450), for which we also generated polymorphism data. The mRNA accumulation profile of Os03g01442 was found to be quite different when compared to its parental gene. RT–PCR results show that Os03g01442 mRNA could be detected at moderate levels in leaf and root tissues but not in stems or flowers (Figure 6). In contrast, we did not detect any mRNA in these tissues for Os01g69904, the paralogous counterpart of Os03g01442 (Figure 6). No mRNA was detected for Os03g01450 under normal growth conditions (Figure 6), which is consistent with MPSS data showing that Os03g01450 tags could be detected in leaf tissue subjected to gamma radiation.
Figure 6.
Expression Analysis of Os3g01442, Os03g01450, Os01g69904, and Transcript Internal Control actin (with 35 Cycles of PCR).
(1) Nipponbare genomic DNA; (2) Nipponbare leaf cDNA; (3) Nipponbare stem cDNA; (4) Nipponbare root cDNA; (5) Nipponbare flower cDNA; (6) Negative control.
Population Genetics Analysis
To determine the pattern of DNA variation in this 60-kb segment in O. sativa, we investigated the nucleotide polymorphism spectrum of annotated protein genes for Os03g01442 and Os03g01450 across 30 difference O. sativa ssp. japonica accessions that represented broad geographical distribution (Table 3). We observed a relatively low polymorphism rate in the two genes tested, consistent with previous studies (Caicedo et al., 2007). Furthermore, all neutrality tests (Tajima's D, Fu and Li's D, and Fay and Wu's H) with both negative values, and a coalescence simulation test revealed a biased frequency spectrum that deviated significantly from neutrality for Os03g01442. Os03g01450 showed the same trend with a negative Tajima's D, although the p-value was marginally significant, at 0.04 (Table 3).
Table 3.
Levels of Polymorphism of Two Genes in O. sativa ssp. japonica and Neutrality Tests on the Site Frequency Spectrum.
| Summary statistic | Os03g01450 | Os03g01442 |
| N | 30 | 30 |
| L | 820 | 1341 |
| S | 5 | 7 |
| Π | 0.00063 | 0.00035 |
| Θ | 0.00154 | 0.00132 |
| Tajima's D | –1.62, p = 0.04* | –2.17, p = 0.0.001** |
| Fu and Li's D* | –1.48, p = 0.15 | –3.60, p = 0.003** |
| Fay and Wu's H | 0.48, p = 0.52 | –7.27, p = 0.003** |
Note: The Fay and Wu's H was calculated using paralogous sequences as outgroup. N, population size; L, gene length (bp); S, the number of segregation sites. * The significance as P < 0.05; ** the significance as P < 0.01.
DISCUSSION
Chimeric genes can be generated through DNA-level recombination or retroposition-targeting mechanisms (Arguello et al., 2007). Chimeric genes derived from multiple parental loci, due to their potential to evolve novel functions, have provided a very useful system to study gene evolution and its role in species diversification. For DNA-level recombination, several molecular mechanisms (homologous and non-homologous) have been observed to recombine different genic and non-genic regions to create chimeric genes (Roth and Wilson, 1988; Stankiewicz and Lupski, 2002). In retroposition-based chimeric gene formation, retroposed copies can recruit target sequences to form novel gene structures. In a previous effort to systematically detect retroposed genes in the rice genome (O. sativa ssp. indica), Wang et al. (2006) detected extensive origination activities that led to the formation of a large number of chimeric retroposed genes. Subsequent studies confirmed that the majority of these retroposed genes are also present in the O. sativa ssp. japonica genome (Fan et al., unpublished). In this study, consistently with previous findings, we annotated 12 putative young genes and a single TyGypsy retrotransposon in a contiguous 60-kb sequence in O. sativa ssp. japonica, five of which appeared to be chimeric genes created by DNA-level recombination. We found two genes (Os03g01450 and Os03g01500) that were created by a combination of two genes, and one gene (Os03g01410) that was formed from a paralogous gene and a flanking sequence. The homologous gene pair, Os03g01420 and Os03g01490, appears to contain chimeric gene structures with shared partial homologous sequences. An excess of chimeric gene formation appears be a phenomenon of the grass species. This is in contrast to data from Arabidopsis and other dicot species, where no chimeric retroposed genes were identified (Fan et al., 2007b) and few chimeric gene structures, formed through DNA-level exon shuffling, have been reported (Drea et al., 2006; Domon and Steinmetz, 1994). The higher rate of chimeric gene formation through gene duplication and the generation of a larger number of functional genes in rice may demonstrate that the diversification of the grass species is a mirror of their broad ecological adaptation and morphological complexity.
Our analysis of the automated gene models found in the TIGR rice annotation database in combination with the EST/cDNA evidence suggests that at least eight of the gene models need to be manually re-annotated. Specifically, the gene model for Os03g01450 is in total conflict with the splicing structure revealed by EST sequences (Supplemental Figure 3E). Moreover, it constitutes a sense/antisense pair with Os03g01470 and shares at least 700 base pairs of sequence. Similarly, the gene structures of Os03g01500 and Os03g01520 also form a non-exonic overlapping gene pair (Supplemental Figure 3D). Lastly, four genes (Os03g01420, Os03g01442, Os03g01490, and Os03g01520) have alternative splice sites, thereby encoding two splicing isoforms each.
Although the species tree indicates the evolutionary age of this 60-kb region should be around 1 Myr, estimations of gene duplication events based on Ks ranged from 0.02 to 14 Myr. Such a conflict might be caused by the following factors. First, there may be a greater variation for substitution rates among genes than previously thought, so the rate we used derived from Adh may not be appropriate for other genes. An interesting alternative would be that, in some cases, such as with Os03g01410 embedded within a PackMULE, a gene might have actually emerged 14 MYA and then moved to its current location more recently. This latter hypothesis predicts that one would find many homologous sequences in non-syntenic regions in several of the wild relatives of rice. It will be possible to test this idea more systematically when complete genomic sequences from the other Oryza species become available.
Both comparative genomics and experimental analysis showed that this 60-kb segment in O. sativa ssp. japonica started evolving after the divergence of Asian (O. sativa, O. rufipogon, and O. nivara) and African species (O. glaberrima and O. barthii) about 1 MYA. Given this segment contained 12 relatively young putative genes in O. sativa, the most straightforward explanation for its origination is a single segmental duplication. This hypothesis seems highly unlikely for a number of reasons. First, as shown above, an inverse tandem duplication occurred, which contributed three pairs of genes. Second, candidate parental genes of the 12 genes were distributed across the entire genome (i.e. chromosomes 1, 3, 4, 6, and 12). Third, the overall identity between paralogous duplicates fluctuated from 93 to 100%, which points to different evolutionary ages, corresponding to 0.02 Myr (Ks = 0.0003) and 14 Myr (Ks = 0.17). Fourth, three of the candidate genes are embedded in transposable elements, PackMULEs and Helitrons, known to be associated with new gene formation and movement. Fifth, the region contains a complete TyGypsy LTR retrotransposon that was estimated to have inserted about 0.5 MYA. Finally, we found that the orthologous regions in O. nivara and O. rufipogon were smaller than in O. sativa and contained fewer genes (Yu et al., unpublished data), which suggests that the evolution of this segment is still ongoing through recurrent recombination and transposition via transposable elements.
Thus, multiple independent duplications and TE-mediated transpositions are a plausible explanation for the origination of all these new genes in this region. Our hypothesis is supported by the location of these genes and retrotransposons. This region is located between 250 and 310 Kb at the subtelomeric region of chromosome 3 in O. sativa ssp. japonica. It has been reported that subtelomeric regions, usually around 500 Kb near the tip of each chromosome, have much higher recombination activity and tend to be gene-rich and highly transcriptionally active (Mizuno et al., 2006). Thus, frequent recombination renders it possible for such a short region to accumulate more duplicated sequences in a short time span, and the local environment makes these duplicates more likely to maintain or evolve a new transcriptional activity, like recruiting new exons or forming sense/antisense gene pairs.
What mechanism(s) is implicated in these numerous recombination and transposition events? A straightforward possibility is repeat element-mediated homologous recombination, given that subtelomeres and telomeres are known to be enriched with transposon elements, which have been reported to be important for the creation of new genes in the Drosophila genomes (Anderson et al., 2008) and plant genomes (Hudson et al., 2003; Wang et al., 2006; Hollister and Gaut, 2007; Gaut and Ross-Ibarra, 2008). One of the genes within this 60-kb region (Os03g01442) is embedded within a Helitron, an autonomous DNA transposon. Consistent with this, its parental gene, Os01g69904, is also located adjacent to one Helitron repeat. Helitrons are known to be able to shuffle pre-existing genes in plants (Lal et al., 2003; Kapitonov and Jurka, 2001, 2007; Hollister and Gaut, 2007). Furthermore, the highly conserved sequences found in the flanking regions of O. glaberrima, O. barthii, and O. glumaepatula (Supplemental Figure 5) and the presence of apparent transposon insertion sites in the flanking region of O. sativa (Supplemetal Figure 6) further support our conclusion that recombination and/or transposition via transposable elements were highly active mechanisms that led to the recent creation of young genes in O. sativa.
It has been reported that the cultivated rice genome encodes 898 functional retrogenes (Wang et al., 2006), which is in contrast to the prevailing view that plant genomes contain only a few retrogenes. Moreover, about 100 of the 898 retrogenes are very young, as suggested by their Ks values, which are smaller than 0.1 (less than 10 Myr). In our study, we identified 12 candidate young genes and one retrotransposon in O. sativa. Except for retrotransposons, whose transcriptional activity might be repressed by the host genome, nine of 12 genes showed evidence of functionality based on EST/cDNA/MPSS data and/or evolutionary analyses. Remarkably, some (e.g. 03g01436 and 03g01450) appear to be involved in defense responses. The emergence of new genes related to stress and defense could be rationalized by the fact that rice has broad ecological adaptation and is under large selective pressures to protect itself against natural disasters and predators. Here, we only analyzed about the first 10% of the short arm of chromosome 3, and found a number of functional genes that originated recently through independent recombination and transpositional events. If the subtelomeric region of chromosome 3 is representative of the remaining 23 chromosome arms, then it is highly likely that we will identify many more young genes supporting our hypothesis that subtelomere serves as a hot bed for gene origination and evolution by recruiting DNA-level duplicated genes in rice.
We performed further detailed analyses for two of the identified 12 genes in the segment. The polymorphism spectrum of Os03g01450 showed a slight deviation from neutrality as revealed by a marginally significant p-value (0.04) using the Tajima's D test (Table 3). By contrast, both expression and population genetic analyses implied that Os03g01442 may be subject to positive selection during its origination and fixation. A differential mRNA accumulation pattern was observed for Os0301442, with no mRNA detected in stems and flowers and moderate mRNA levels in leaves and roots. In contrast, no transcript was detected for Os03g01442’s parental copy Os01g69904 in the same tissues. Furthermore, the biased frequency spectrum with an excess of both rare alleles and high frequency polymorphisms in Os03g01442 also suggested the possibility that positive selection is acting on this gene. Although further analysis should be conducted in this region to rule out the possibility of demographic effects for the biased polymorphism spectrum (with a broader sample base, more young genes and flanking sequences), this case analysis provides an example that selection may be the acting force to drive the fixation of young genes in rice.
METHODS
Sequencing 1.5-Mb BAC Tiles from O. glaberrima and O. punctata
We selected two ∼1.5-Mb minimum tiling paths of overlapping BAC clones from O. glaberrima and O. punctata from the short arms of chromosome 3 utilizing previously described BAC libraries and BAC fingerprint/end-sequenced physical maps (Ammiraju et al., 2006; Kim et al., 2007, 2008). Each BAC clone was shotgun-sequenced, finished and sequence-validated using standard procedures as previously described (IRGSP, 2005), such that the final finished sequences had an error rate of less than one base in 10 000. Overlapping BAC sequences from each species were then manually assembled into ∼1.5-Mb pseudomolecules and used for further analysis.
Searching the O. sativa ssp. japonica Specific Sequence by Comparative Analysis
We performed genomic pairwise comparison between O. sativa ssp. japonica genome and 1.5-Mb O. glaberrima chromosome 3 short arm sequences. The annotation and coding sequences (CDS) of O. sativa ssp. japonica were downloaded from TIGR (www.tigr.org/tdb/e2k1/osa1/). A MegaBLAST of 1.5-Mb O. glaberrima sequence to the CDS of O. sativa ssp. japonica was conducted. We searched orthologous sequences between two species; meanwhile, we also paid attention to the unique sequence found only in O. sativa ssp. japonica, but absent in O. glaberrima. We further searched the O. sativa ssp. japonica sequence to sequence of O. punctata. In such effort, we found an O. sativa ssp. japonica unique segment bearing sequence in length of 60 kb.
Sequence Analysis of the O. sativa Specific 60-kb Segment
To determine if the 60-kb region identified in O. sativa ssp. japonica was unique to the japonica genome, we used two approaches. To probe the wild Asian species, we used BLAST to identify any BESs from the O. rufipogon and O. nivara BAC libraries that were similar to the 60-kb japonica sequence, then detected those BACs that locate in the suntelomeric region of the chromosome 3 short arm using the Finger Printed Contigs (FPC)-based physical map and SyMAP synteny browser (Kim et al., 2008; Soderlund et al., 2006). For the African and South American species, we performed PCR amplification on genomic DNA isolated from O. glaberrima, O. barthii, and O. glumeatulata, using a pair of primers specific to O. glaberrima and O. sativa spp. japonica located in the flanking regions of the 60-kb sequence (see Figure 1). Based on the TIGR rice annotation database (Release 5), we were able to identify 12 genes and one retrotransposon in this segment. In order to understand the evolution pattern and history of these genes, we implemented a robust strategy to search for their candidate parental genes (paralogs) in the O. sativa ssp. japonica genome. In brief, we searched each gene together with its 10K flanking sequence against the whole genome using BLASTN. We then mapped the 1.5-Mb chromosome 3 sequence of O. sativa ssp. japonica to the rest of the japonica RefSeq using the ChainSelf pipeline developed by UCSC. We manually checked the hits generated by both methodologies and retrieved the best hits to serve as the parental genes.
We compared both CDS and genomic sequence for both paralogs to gain insight into gene structure and origin, and we further calculated the Ka/Ks ratio using maximum likelihood algorithm using the PAML package (Yang, 2007). The significance of Ka/Ks that deviated from neutrality (= 0.5) were tested using the LRT (Emerson et al., 2004). Specifically, we aligned protein sequences of the parental gene and the daughter gene with MUSCLE (Edgar, 2004) and converted the protein alignment into the codon-based nucleotide alignment with the Pal2nal script (Suyama et al., 2006). We then used codeml with fixed and free omega models to test whether any of the young genes detected were under selective constraint, namely Ka/Ks was significantly smaller than 0.5 (Yang, 2007).
We re-annotated four incorrect TIGR gene models using the UniGene rice EST/mRNA dataset. ESTs were mapped to the genome by BLAT (Kent, 2002) and all ambiguous or low-quality mappings were discarded (Zhang et al., 2006). Then, we re-mapped these hits to the genome using SIM4 (Florea et al., 1998) and GeneSeqer (Schlueter et al., 2003) with the rice scoring matrix in order to refine the entire splicing structure. Gene structures with the highest identity and the most standard splicing junction were retained.
DNA Extraction, PCR Amplification, and Sequencing
Total genomic DNA was extracted from fresh leaves of a single plant using the Qiagen DNeasy kit following the manufacturer's protocol. PCR reactions were performed using Invitrogen Taq polymerase, with annealing temperature adjusted based on the length of fragments with 1 kb min−1. Double-stranded PCR products were purified using either the Qiagen PCR purification or Qiagen miniprep Gel purification kits. Purified PCR products were sequenced using the ABI-3730XL 96-capillary automated DNA sequencer. Sequences were edited and assembled. Clustal X was used to align sequences for further analyses (Thompson et al., 1997). Manual adjustments were made where necessary.
Expression Analysis
As mentioned above, we mapped the latest UniGene rice EST/mRNA dataset to the complete genome, which consists of more than 1 000 000 entries. The expression profiles for two genes were further investigated using reverse transcription (RT)–PCR in different tissues grown under normal conditions. Total RNA was extracted from leaf, root, stem, and entire flower bud using a Qiagen total RNA extraction kit. cDNA were generated using the Invitrogene RT–PCR kit and full description of RT–PCR was described previously. The constitutively expressed gene actin was used as internal control to quantify the density of cDNA.
Population Genetics Analysis
We sampled the worldwide collection of O. sativa ssp. japonica accessions to generate a nucleotide frequency spectrum for population genetics analysis. Most O. sativa ssp. japonica accessions were chosen from a wide range of Asia, with a few samples from Africa. Basic population genetic analysis was implemented in DnaSP (Rozas et al., 2003). Sequence diversity was quantified as nucleotide diversity (π) (Nei, 1987) and Watterson's θ (1975). Tests of deviation from neutrality were conducted using Tajima's D (1989), Fu and Li's D (1993), and Fay and Wu's H (2000) tests. We further used coalescent simulation to assess the significance of the statistic for the all parameters generated. The neutral coalescent process was simulated using 2000 replicates, with the number of segregating sites set to that observed in the data.
SUPPLEMENTARY DATA
Supplementary Data are available at Molecular Plant Online.
FUNDING
This work was supported by National Science Foundation grant DBI-0321678 (to R.A.W. and S.R.), the Bud Antle Endowed Chair (to R.A.W.), and the National Science Foundation CAREER award (MCB0238168) and National Institute of Health R01 grants R01GM065429–01A1 and 1R01GM078070–01A1 (to M.L.).
Supplementary Material
Acknowledgments
No conflict of interest declared.
References
- Ammiraju JS, et al. The Oryza bacterial artificial chromosome library resource: construction and analysis of 12 deep-coverage large-insert BAC libraries that represent the 10 genome types of the genus Oryza. Genome Res. 2006;16:140–147. doi: 10.1101/gr.3766306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JA, Song YS, Langley CH. Molecular population genetics of Drosophila subtelomeric DNA. Genetics. 2008;178:477–487. doi: 10.1534/genetics.107.083196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arguello JR, Fan C, Wang W, Long M. Origination of chimeric genes through DNA-level recombination. Genome Dyn.: Protein and Gene Evolution. 2007;3:131–146. doi: 10.1159/000107608. [DOI] [PubMed] [Google Scholar]
- Bennetzen JL. Patterns in grass genome evolution. Curr. Opin. Plant Biol. 2007;10:176–81. doi: 10.1016/j.pbi.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Betran E, Thornton K, Long M. Retroposed new genes out of the X in Drosophila. Genome Res. 2002;12:1854–1859. doi: 10.1101/gr.604902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo AL, et al. Genome-wide patterns of nucleotide polymorphism in domesticated rice. PLoS Genet. 2007;3:1745–1756. doi: 10.1371/journal.pgen.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domon C, Steinmetz A. Exon shuffling in anther-specific genes from sunflower. Mol. Gen. Genet. 1994;244:312–317. doi: 10.1007/BF00285459. [DOI] [PubMed] [Google Scholar]
- Drea SC, Lao NT, Wolfe KH, Kavanagh TA. Gene duplication, exon gain and neofunctionalization of OEP16-related genes in land plants. Plant J. 2006;46:723–735. doi: 10.1111/j.1365-313X.2006.02741.x. [DOI] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson JJ, Kaessmann H, Betran E, Long M. Extensive gene traffic on the mammalian×chromosome. Science. 2004;303:537–540. doi: 10.1126/science.1090042. [DOI] [PubMed] [Google Scholar]
- Fan C, Emerson JJ, Long M. The origin of new gene. In: Pagel M, Pomiankowski A, editors. Evolutionary Genomics and Proteomics. Sunderland, Massachusetts, USA: Sinauer Associates, Inc.; 2007a. pp. 27–44. [Google Scholar]
- Fan C, Vibranovski M, Chen Y, Long M. A microarray-based genomic hybridization method for identification of new genes in plants: case analyses of Arabidopsis and Rice. J. Integ. Plant Biol. 2007b;49:915–926. [Google Scholar]
- Fay J, Wu C-I. Hitchhiking under positive Darwinian selection. Genetics. 2000;155:1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florea L, Hartzell G, Zhang Z, Rubin GM, Miller W. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998;8:967. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Li W-H. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut BS, Ross-Ibarra J. Selection on major components of angiosperm genomes. Science. 2008;320:484–486. doi: 10.1126/science.1153586. [DOI] [PubMed] [Google Scholar]
- Gaut BS, Morton BR, McCaig BC, Clegg MT. Substitution rate comparisons between grasses and palms: Synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc. Natl Acad. Sci. U S A. 1996;93:10274–10279. doi: 10.1073/pnas.93.19.10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge S, Sang T, Lu BR, Hong DY. Phylogeny of rice genomes with emphasis on origins of allotetraploid species. Proc. Natl Acad. Sci. U S A. 1999;96:14400–14405. doi: 10.1073/pnas.96.25.14400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollister JD, Gaut BS. Population and evolutionary dynamics of Helitron transposable elements in Arabidopsis thaliana. Mol. Biol. Evol. 2007;24:2515–2524. doi: 10.1093/molbev/msm197. [DOI] [PubMed] [Google Scholar]
- Hudson ME, Lisch DR, Quail PH. The FHY3 and FAR1 genes encode transposase-related proteins involved in regulation of gene expression by the phytochrome A-signaling pathway. Plant J. 2003;34:453–471. doi: 10.1046/j.1365-313x.2003.01741.x. [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project. The map based sequencing of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- Kim H, et al. Construction, alignment and analysis of twelve framework physical maps that represent the ten genome types of the genus Oryza. Genome Biol. 2008;9:R45. doi: 10.1186/gb-2008-9-2-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, San Miguel P, Nelson W, Collura K, Wissotski M, Walling JG, Kim JP, Jackson SA, Soderlund C, Wing RA. Comparative physical mapping between O. sativa (AA genome type) and O. punctata (BB genome type) Genetics. 2007;176:379–390. doi: 10.1534/genetics.106.068783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. Rolling-circle transposons in eukaryotes. Proc. Natl Acad. Sci. U S A. 2001;98:8714–8719. doi: 10.1073/pnas.151269298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. Helitrons on a roll: eukaryotic rolling-circle transposons. Trends Genet. 2007;23:521–529. doi: 10.1016/j.tig.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal SK, Giroux MJ, Brendel V, Vallejos CE, Hannah LC. The maize genome contains a helitron insertion. Plant Cell. 2003;15:381–391. doi: 10.1105/tpc.008375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M, Betran E, Thornton K, Wang W. The origin of new genes: glimpses from the young and old. Nat. Rev. Genet. 2003;4:865–875. doi: 10.1038/nrg1204. [DOI] [PubMed] [Google Scholar]
- Marques AC, Dupanloup I, Vinckenbosch N, Reymond A, Kaessmann H. Emergence of young human genes after a burst of retroposition in primates. PLoS Biol. 2005;3:e357. doi: 10.1371/journal.pbio.0030357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno H, Wu J, Kanamori H, Fujisawa M, Namiki N, Saji S, Katagiri S, Katayose Y, Sasaki T, Matsumoto T. Sequencing and characterization of telomere and subtelomere regions on rice chromosomes 1S, 2S, 2L, 6L, 7S, 7L and 8S. Plant J. 2006;46:206–217. doi: 10.1111/j.1365-313X.2006.02684.x. [DOI] [PubMed] [Google Scholar]
- Nakano M, Nobuta K, Vemaraju K, Tej SS, Skogen JW, Meyers BC. Plant MPSS databases: signature-based transcriptional resources for analyses of mRNA and small RNA. Nucleic Acids Res. 2006;34:D731–D735. doi: 10.1093/nar/gkj077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Ohno S. Evolution by Gene Duplication. Berlin: Springer; 1970. [Google Scholar]
- Paterson AH, Freeling M, Sasaki T. Grains of knowledge: genomics of model cereals. Genome Res. 2005;15:1643–1650. doi: 10.1101/gr.3725905. [DOI] [PubMed] [Google Scholar]
- Roth D, Wilson J. Illegitimate recombination in mammalian cells. In: Kucherlapati R, Smith GR, editors. Genetic Recombination. Washington, DC, USA: American Society of Microbiology; 1988. pp. 621–653. [Google Scholar]
- Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Schlueter SD, Dong Q, Brendel V. GeneSeqer@PlantGDB: gene structure prediction in plant genomes. Nucleic Acids Res. 2003;31:3597–3600. doi: 10.1093/nar/gkg533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderlund C, Nelson W, Shoemaker A, Paterson A. SyMAP: a system for discovering and viewing syntenic regions of FPC maps. Genome Res. 2006;16:1159–1168. doi: 10.1101/gr.5396706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR. Molecular–evolutionary mechanisms for genomic disorders. Curr. Opin. Genet. Dev. 2002;12:312–319. doi: 10.1016/s0959-437x(02)00304-0. [DOI] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical methods for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, et al. High rate of chimeric gene origination by retroposition in plant genomes. Plant Cell. 2006;18:1791–1802. doi: 10.1105/tpc.106.041905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson G. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Wheeler DL, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008;36:D13–D21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing RA, et al. The Oryza Map Alignment Project: the golden path to unlocking the genetic potential of wild rice species. Plant Mol. Biol. 2005;59:53–62. doi: 10.1007/s11103-004-6237-x. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yu J, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu XS, Liu QR, Wei L. Genome-wide in silico identification and analysis of cis natural antisense transcripts (cis-NATs) in ten species. Nucleic Acids Res. 2006;34:3465–3475. doi: 10.1093/nar/gkl473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wu Y, Liu Y, Han B. Computational identification of 69 retroposons in Arabidopsis. Plant Physiol. 2005;138:935–948. doi: 10.1104/pp.105.060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Ge S. Phylogenetic relationships among A-genome species of the genus Oryza revealed by intron sequences of four nuclear genes. New Phytol. 2005;167:249–265. doi: 10.1111/j.1469-8137.2005.01406.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.