Abstract
Backgrounds
Admixture is of great relevance to the clinical application of pharmacogenetics and personalized medicine, but unfortunately these studies have been scarce in Puerto Ricans. Besides, allele frequencies for clinically relevant genetic markers in warfarin response (i.e., CYP2C9 and VKORC1) have not yet been fully characterized in this population. Accordingly, this study is aimed at investigating whether a correlation between overall genetic similarity and CYP2C9 and/or VKORC1 genotypes could be established.
Methods
98 DNA samples from Puerto Ricans were genotyped for major CYP2C9 and VKORC1 polymorphisms and tested on a physiogenomic (PG)-array to infer population structure and admixture pattern.
Results
Analysis affirmed that Puerto Ricans are broadly admixed. A genetic distance dendrogram was constructed by clustering those subjects with similar genetic profiles. Individual VKORC1 and CYP2C9 genotypes were visually overlaid atop the three dendrogram sectors. Sector-1, representing Amerindian ancestry, showed higher VKORC1-1639G>A variant frequency than the rest of the population (p=0.051). Although CYP2C9*3 allele frequencies matched the expected HapMap values, admixture may explain deviations from published findings regarding VKORC1-1639G>A and CYP2C9*2 allele frequencies in sector-3.
Conclusions
Results suggest that the observed inter-individual variations in ancestral contributions have significant implications for the way each Puerto Rican responds to warfarin therapy. Our findings provide valuable evidence on the importance of controlling for admixture in pharmacogenetic studies of Puerto Rican Hispanics.
Keywords: Admixture-matching, Puerto Ricans, Pharmacogenetics, VKORC1, CYP2C9 genotypes
Introduction
In January 2010, the U.S. Food and Drug Administration (FDA) revised the warfarin label to include pharmacogenomic dosing recommendations based on CYP2C9 and VKORC1 gene polymorphisms. Accordingly, there is a need to translate this guidance into public health impact based on the prevalence of combinatorial genotypes predicting dose adjustments in substantial segments of the population with cardiovascular morbidity. To date, few studies have ventured to determine whether a need exists for admixture-matching to improve the accuracy and quality of clinical pharmacogenetic in heterogeneous populations, such as Puerto Ricans.
Several single nucleotide polymorphisms (SNPs) in the gene of the major protein Vitamin K epoxide reductase complex subunit 1 (VKORC1) have been associated with a deficiency in vitamin-K dependent clotting factors, resulting in either increased sensitivity to warfarin or warfarin resistance [1–3]. Five haplotypes have been found to commonly occur in the general population [3]. Two of these haplotypes (H1 and H2) are typically grouped together and associated with lower warfarin dose requirements (haplotype A, sensitivity), whereas another 3 haplotypes (H7, H8 and H9) have been associated with higher warfarin dose requirements (haplotype B, resistance) [3,4]. According to most studies, VKORC1 haplotypes account for approximately 25% of the warfarin dose variability [5].
Among genes affecting warfarin response, the first to be identified was CYP2C9 [6]. CYP2C9 encodes the cytochrome P450, subfamily IIC, polypeptide 9, an enzyme responsible for metabolizing the S-enantiomer of warfarin [7]. At least 6 SNPs of CYP2C9 have been found that may influence enzyme activity and subsequently account for slower metabolism [7]. Studies show that the CYP2C9*2 (Arg144Cys, C430T SNP at exon 3) and CYP2C9*3 (Ile359Leu, A1075C SNP at exon 7) variants have 12–70% and 5% of the enzyme activity of the wild-type allele, respectively [8]. These alleles can cause alterations in initial warfarin dose sensitivities, delays in achieving a stable maintenance dose, and increased risk of serious or life-threatening bleeding complications [5,9]. The warfarin dose required to maintain a stable INR decreases in the presence of one or more of the low-dose warfarin alleles [10–11]. In univariate analyses, CYP2C9 genotype alone appears to account for approximately 15–20% of the overall variability in warfarin dose [5,12–14].
The prevalence of CYP2C9 polymorphisms varies across different ethnic groups [13–16]. VKORC1 variants have also been widely studied in a variety of ethnic populations [3–5,17–19]. Clinical data and observations reveal that Caucasian patients require warfarin doses 31–40% greater than those required by Asian patients [5]. Since deficient CYP2C9 genotypes are not as common in Asians as they are in Caucasians, VKORC1 variations likely explain the observed inter-ethnic differences in dosing requirements. Some evidence indicates that Africans, African-Americans and in all probability the admixed populations of Hispanics in the American continent, have a high incidence of haplotypes that have not yet been described and have unknown functions [5,19].
Due to its remarkable heterogeneity and trichotomous ancestral genetic admixture, the Puerto Rican population may significantly differ from other earlier pharmacogenetically characterized populations with respect to the frequency, distribution and combination of allelic variants in genes associated with drug response and diseases [20]. Recently, we have published a physiogenomic analysis to infer structure and ancestry in the Puerto Rican population [21]. The Puerto Rican sample was found to be broadly heterogeneous, with three main clusters reflecting the historical admixture from Amerindian, African, and European ancestors. Our results matched previously published estimations of Puerto Rican admixture that were ascertained using more traditional ancestral genetic markers [22–24]. The study provided a set of 384 physiologically informative SNPs from 222 cardio-metabolic and neuro-endocrine genes that can be used to facilitate the translation of genome diversity into personalized medicine and control for admixture in Puerto Ricans [21]. The observed large variance in admixture proportions suggested that this population is ideal for admixture matching studies.
Admixture is of great relevance to the clinical application of pharmacogenetics and personalized medicine, but unfortunately these studies have been scarce. As physiogenomic-guided multi-gene models are developed to predict drug response, the range of possible allelic combinations in the Puerto Rican population is certain to exceed that in populations without admixture. In addition, the allele frequencies for both the CYP2C9 and VKORC1 gene markers in this population have not been fully characterized. Accordingly, in an effort to investigate whether a correlation between overall genetic similarity and CYP2C9 and/or VKORC1 genotypes could be established, 98 DNA samples were sent from Puerto Rico to the Laboratory of Personalized Health (LPH) at Genomas, Inc (Hartford, CT) to be genotyped. This work provides valuable evidence on the importance of controlling for admixture in pharmacogenetic studies of Puerto Rican Hispanics.
Methods
Human genomic DNA samples (40–60 ng/μl) were extracted and purified from existing dried blood spots on Guthrie cards supplied by the Puerto Rico Newborn Screening Program (PRNSP), where >95% of Puerto Rican newborns are screened for common hereditary diseases. Accordingly, this survey (protocol #A4070107) was exempt from IRB review under FDA and OHRP guidelines based on category 4, 45CFR46.118. A controlled stratified-by-region random sampling protocol was followed, taking into consideration the percentage of birth at each region around the Puerto Rican Island based on the 2004 national register of total births.
The extracted genomic DNA samples were first genotyped using Tag-It Mut detection technology on Luminex® 100-xMAP™ platform, targeting 12 combined polymorphisms in 2 genes known to have a major effect on warfarin variability [25], namely VKORC1 and CYP2C9. Table 1 depicts relevant aspects of these 12 SNPs genotyped: 5 alleles in CYP2C9 and 7 alleles in VKORC1, apart from the corresponding wild types. Once the samples were genotyped for SNPs on these two warfarin-related genes, samples were genotyped on a physiogenomic (PG) array detecting 384 SNPs from 222 cardio-metabolic and neuro-endocrine genes spanning their entire genome [21,26,27]. Careful manual analysis was performed on the alignments underlying the genotype calls using GenCall 6.1.3.24 and 50 SNPs with even a slight degree of uncertainty about calling accuracy were not included in the analysis, leaving 332 SNPs from 196 genes. Genotyping was accomplished using Illumina® BeadArray™ technology [28]. Analysis of the results using the Structure v2.2 software package was used to cluster those subjects with similar genetic profiles [29–31]. A detailed explanation of the analytics underlying the clustering of the samples as well as a full list of the genes and SNPs in the PG array is provided in our physiogenomic analysis study [21].
Table 1.
CYP2C9 and VKORC1 variants detected with the HILOMet Warfarin system on Luminex® 100 xMap™ technology. Effects on enzymatic activity are also depicted.
| CYP2C9 | VKORC1 | ||||||
|---|---|---|---|---|---|---|---|
| Alleles | DNA variant | Change to Protein | Activity | Alleles | DNA variant | Change to Protein | Activity |
| *1 | Wild Type# | Reference | Normal | WT | Wild Type# | Reference | Normal |
| *2 | 430C→T | Arg144Cys | Decreased | −1639 | G→A | Promoter | Deficient |
| *3 | 1075A→C | Ile359Leu | ~Null | +85 | G→T | Val29Leu | ~Null |
| *4 | 1076T→C | Ile359Tyr | Decreased | +121 | G→T | Ala41Ser | ~Null |
| *5 | 1080C→G | Asp360Glu | Decreased | +134 | T→C | Val45Ala | ~Null |
| *6 | 818delA | Frameshift | ~Null | +172 | A→G | Arg58Gly | ~Null |
| +1331 | G→A | Val66Met | ~Null | ||||
| +3487 | T→G | Leu128Arg | ~Null | ||||
Wild-types are assigned as a result of the absence of other SNPs
Test for deviations from Hardy-Weinberg equilibrium (HWE) were performed. Departure from HWE were estimated under the null hypothesis of the predictable segregation ratio of specific matching genotypes (p>0.05) by use of χ2 goodness-of-fit test with one degree of freedom.
Results
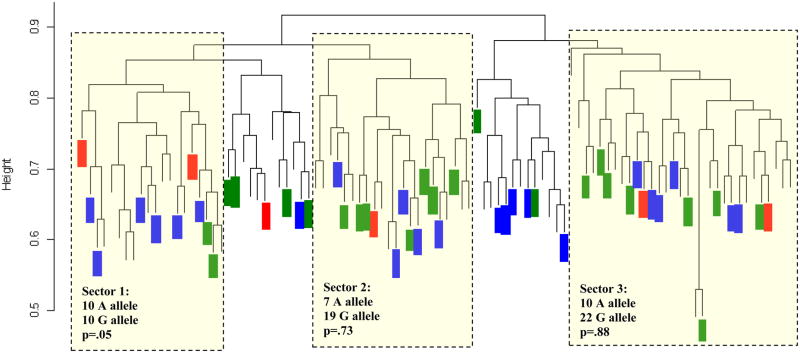
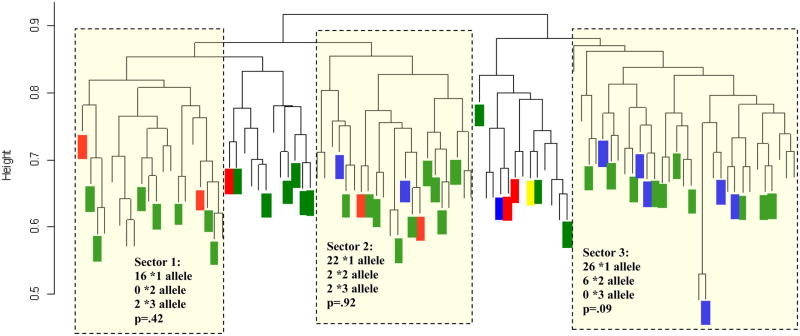
Results are represented graphically with a genetic distance dendrogram (Fig. 1), similar to a phylogenetic tree. Those individuals who had many similar polymorphisms were grouped together in one “sector” while those with whom they had very few similarities lie on the other side of the “tree’; greater distances denoting lesser degrees of common ancestry. To determine if there are relationships between genetic clusters defined by Strucutre analysis [21] and warfarin-related genotypes, we superimposed the individual warfarin genotypes for CYP2C9 and VKORC1 on the dendrogram. Figures 1-Panel A and B represent less than 98 patients due to incomplete combinatorial genotyping data in some patients during the initial physiogenomic array (PG Array) analysis which precluded definitive assignment to any of the 3 Structure clusters. Furthermore, only those samples with strong associations with any one of the three clusters were included in the superimposition analysis seen in Figure 1, yielding 52 subjects.
Figure 1.
Panel A: Individual VKORC1(1639 G→A) genotypes, overlaid on the genetic distance dendrogram for the samples from the Puerto Rican population (dendrogram taken from previously published physiogenomic population analysis[21]). Green color represents G/G genotype; whereas, blue and red colors are for the G/A and A/A genotypes, respectively. P-values were calculated by a chi-squared test comparing observed allele frequencies with expected frequencies given the overall allelic ratios. The VKORC1 SNP 1639 G→A is in high linkage disequilibrium with haplotype A[3], which has been associated with a significant decrease in the warfarin dose per allele.
Panel B: Individual CYP2C9 genotypes overlaid on the genetic distance dendrogram for the samples from the Puerto Rican population (dendrogram taken from previously published physiogenomic population analysis [21]). Green color represents wild-type *1/*1 genotype; blue color denotes *1/*2 and red colors indicates the *1/*3 genotype. The yellow rectangle highlights the one *1/*6 genotype observed. P-values were calculated by a chi-squared test comparing observed allele frequencies within the sector with expected frequencies given the overall allelic ratios.
By previous analyses, the dendrogram sectors 1, 2 and 3 in Figure 1 correspond to Amerindian, Caucasian, and West African heritage, respectively [21]. For the 52 cases with warfarin genotypes, the VKORC1-1639 G allele frequency was 69/104 or 66%, whereas the A allele frequency was 35/104 or 34%. Statistical analyses to compare each sector to the overall allelic ratios, revealed that sector 1 (left-most portion of the genetic distance dendrogram in Fig. 1, Panel A), showed a lower frequency of VKORC1 -1639 G, than the rest of the population (50% compared to an expected 70%). (p=0.051). Sectors 2 and 3 in Figure 1, Panel A, show -1639 G allele frequencies of 73% and 68%, respectively. There is a low frequency of 8–22% for VKORC1 -1639 G allele in Asian populations [4,15]. The low G allele frequency of 50% in sector 1 is not as low as expected for a population of purely Asian ethno-geographic origin, no surprise given the heterogeneity of the Puerto Rican population.
Sector 3 is associated with African heritage [16,21]. The HapMap VKORC1 -1639 G frequencies for African people are 98% for the Yoruba people of Nigeria (YRI) and 89% for African Americans in Southwest US (ASW), the latter a population with greater chance for admixture. The present study reports a VKORC1 -1639 G frequency of 68% in sector 3, consistent with the possible interpretation that the Puerto Rican population reflects an even greater admixture with Caucasian and Asian populations than ASW. Similarly, the slightly inflated G allele frequency of the primarily Caucasian sector 2 may be explained by increased admixture with the African population.
With respect to the CYP2C9 gene (Fig. 1-Panel B), the *2 allele frequencies were 0%, 7.7% and 18.8% in sectors 1, 2 and 3, respectively. The results are complex when comparing CYP2C9 allele frequencies of the 3 dendrogram sectors (1 – Amerindian; 2- Caucasian; 3 – West African) with the HapMap and other published values. In sector 1, there were no instances of the *2 allele which is consistent with the HapMap report of a 0% CYP2C9 allele frequency for the Han Chinese of Beijing [16]. Furthermore, the CYP2C9 *2 allele frequency in sector 2 was 7.7% compared to 10% for Caucasian residents of Utah documented in the HapMap database [16]. However, in sector 3 the CYP2C9 *2 allele frequency was 18.8%, considerably higher than the HapMap CYP2C9 *2 allele frequency of 0% for the Nigerian YRI population and the 2.2% rate reported by Limdi et al. [32] for African-Americans.
Greater admixture and heterogeneity in the Puerto Rican population as compared to the African-American population or certainly the Yoruba population allow for the presence of the six *2 alleles observed in the 16 individuals in sector 3 (Fig. 1-Panel B) to have come from partial European ancestry, as no individual in that sector was of purely West-African descent [21]. The observed trend may also be an artifact of chance given the small sample size. The reported CYP2C9*3 allele frequencies of 11.1% for sector 1, 7.6% for sector 2 and 0% for sector 3 compare to the expected HapMap values of 5%, 6% and 0%, respectively. No statistically significant deviations from HWE were found with respect to the distribution frequencies of both VKORC1 and CYP2C9 polymorphisms.
Discussion
Since no departures from HWE were observed and considering that our study cohort is island-wide, chosen by a controlled, stratified-by-region, representative sampling from the Puerto Rican population, we can expect the observed frequencies of the VKORC1 and CYP2C9 polymorphisms to be representative for the rest of this population. The Puerto Rican population is a three-way admixed population that experienced migration, which might lead us to expect increased heterozygosity. However, such deviations from HWE quickly subside within a single generation under the assumption of random mating. The size and geography of Puerto Rico do not favor isolated subpopulations, and social stratification is likewise not excessive. Considering that the primary admixing events (Spanish settlement for Caucasian influx and the slave trade for African admixture) are many generations past, observing no overt deviations from HWE stands to reason.
We previously demonstrated that the left-most portion of the dendrogram (sector 1) is associated with a cluster representing proportionally greater Amerindian ancestry (Han Chinese in origin based on HapMap database) [21]. The link between the Amerindians and the Asians originates with the “Bering Strait” theory [33]. It is known that the “A” allele is the most common in Asians, thus, the increased prevalence of this allele in sector 1 of the dendrogram is consistent with our assignment of sector 1 as primarily of Amerindian descent. Further investigation into the geographic and ethnic origin of the samples in this cluster must be conducted as part of a validation study; should a significant association be observed future studies will incorporate admixture-matching in order to probe variations in warfarin response across different genotypes (i.e., stratification of the Puerto Rican population). The uniqueness of the frequency distribution of the VKORC1 haplotypes in the Amerindians as compared to Asian, African, and European populations has previously been postulated by Perini et al. [34]. However, given the vast diversity of Amerindians across different Latino-American populations, available data should not be interpreted as representative of other groups. They found that two Brazilian Amerindian populations (i.e., Kaingang and Guaranies) comprise high proportions of individuals carrying VKORC1 haplotypes requiring reduced warfarin doses [34].
Recent investigation into the genetic structure of the African continent has revealed a high resolution map of genetic diversity within Africa [35]. This study shows clear genetic differences in varying regions of the continent, a finding that may be responsible for the observed difference in the variant CYP2C9 allele frequency in the Afro-Caribbean population as compared to the African-American and Yoruba populations. An online database that documents 34,940 trans-Atlantic slave ship voyages from 1514 to 1866 reveals that there is indeed a difference in principal port of departure between ships that disembarked in the Greater Antilles and those that arrived in the United States [36]. Table 2 shows the principal locations of departure for the 25,569 voyages departing from Africa and arriving in the Americas.
Table 2.
Ports of departure and arrival for 25,569 trans-Atlantic slave voyages arriving in the United States, the Greater Antilles and the rest of the Americas between 1514 and 1866 according to the Voyages database [36]
| Port of Departure | Disembark in: Greater Antilles | % Greater Antilles | USA mainland | % USA | Other | % Other |
|---|---|---|---|---|---|---|
| Bight of Benin | 847 | 14.99% | 27 | 2.01% | 3611 | 19.44% |
| Gold Coast | 920 | 16.28% | 180 | 13.43% | 1561 | 8.40% |
| Other Africa | 364 | 6.44% | 63 | 4.70% | 669 | 3.60% |
| Sierra Leone | 292 | 5.17% | 183 | 13.66% | 652 | 3.51% |
| West Central Africa and St. Helena | 1396 | 24.71% | 233 | 17.39% | 7102 | 38.23% |
| Bight of Biafra and Gulf of Guinea islands | 1147 | 20.30% | 188 | 14.03% | 2229 | 12.00% |
| Senegambia and offshore Atlantic | 352 | 6.23% | 358 | 26.72% | 1517 | 8.17% |
| Southeast Africa and Indian Ocean islands | 142 | 2.51% | 24 | 1.79% | 683 | 3.68% |
| Windward Coast | 190 | 3.36% | 84 | 6.27% | 555 | 2.99% |
| Total | 5650 | 1340 | 18579 |
It is well-known that population-stratifying factors are potential confounders and might be a serious concern in pharmacogenetic association studies of candidate gene to complex traits, including drug response. Perini et al. [37] demonstrated that due to the highly admixed nature of Brazilians, self-reported race was not a reliable predictor of calculated warfarin dose. What proved useful, however, was a precise knowledge of individual admixture so as to place the patient on a continuum between “black” and “white”. The benefits of the continuum model were verified in a separate Brazilian study illustrating the impact of population structure on the GNB3 825C>T polymorphism [38]. Finally, Perini et al. [37] found that warfarin dosing algorithms derived from European, North-American, African-American and Japanese population studies performed poorly when applied to the Brazilian population, suggesting either a need for separate dosing algorithms for each admixed and heterogeneous population, or, admixture–matching of individuals so as to elucidate their proportional ancestries. Our research team is conducting a study of pharmacogenetic warfarin dosing algorithms in Puerto Rican patients on the island and the mainland US. It will be interesting to learn how well dosing algorithms based on CYP2C9 and VKORC1 variants perform in the Puerto Rican population.
These findings further substantiate the argument for admixture as a critical covariant in dosing algorithms for heterogeneous populations. Due to a lack of sufficient data in this population, we believe that the clinical benefit of knowing an individual’s genotype before initiating warfarin treatment in admixed Puerto Ricans is still an open question. A recent paper by the International Warfarin Pharmacogenetics Consortium developed a multi-ethnic warfarin dosing algorithm, but only included 24 Hispanics [39].
These results and further investigations involving warfarin-related clinical outcomes in Puerto Ricans build upon earlier findings published by Suarez-Kurtz et al. [20], who predicted a significant impact of genetic admixture on pharmacogenomics in the American continent. These authors concluded that a variable mosaic genome paradigm, which envisages the genome of any particular individual as a unique mosaic of variable haplotype blocks, has considerably higher explanation and predictive power for the populations of the Americas. Therefore, any extrapolation of pharmacogenomic data from well-defined ethnic groups such as Caucasians and Asians might be plagued with uncertainty due to the interethnic admixture in the Puerto Rican population. Admixed populations such as Puerto Ricans pose a greater challenge because it might be difficult to find a matching control for an individual with diverse ethnic origins; therefore, we will be forced to rely on multivariate adjustment models. That is, rather than allocate the subject to a single stratum in the analysis, it is desirable to construct a covariate for each stratum, giving the corresponding ancestral proportion derived from clustering analysis and then include these covariates as adjustment variables in a multiple logistic regression model [40].
Since each individual genome in an admixed population is composed of ancestry mosaics, it provides an opportunity to unravel the genetic and environmental components of warfarin response. In the context of admixture ancestry matching, if a response allele is more common in one of the ancestral populations, then responders will share a greater level of ancestry from that population around the locus as compared with non-responders. In future studies, we expect to generate a detailed admixture map in the Puerto Rican population at very high resolution using all 1.2 million SNPs from a total genome (TG) array. Admixture studies at this resolution afford delineation of candidate genes for pharmacogenetic traits related not only to warfarin’s outcomes, but also to other cardiovascular conditions of high prevalence in Hispanics.
Fundamental information on human genome variation in the Hispanic populations is still lacking, potentially exacerbating healthcare disparities. In contrast, gathering such baseline information by conducting pharmacogenomic studies in admixed populations like Puerto Ricans could serve to advance DNA-guided personalized medicine in the Hispanic people of the US, a fast-growing and highly heterogeneous group where conventional ethno-geographic classification is imprecise. We hypothesize that genetic admixture studies in Puerto Ricans will result in a rich repertoire of combinatorial genotypes for key pharmacological pathways of warfarin response, rendering this population a better resource to develop DNA-guided dosing algorithms for the clinical management of warfarin. However, this long-term goal is unlikely to be attained if a population-based paradigm that clusters Hispanics into one racial entity emerges as the standard of “personalized medicine” as opposed to actual personalized therapy tailored to the individual’s genetic characteristics. Accordingly, it must be based on the recognition of inherent genetic individuality, rather than relying on inter-ethnic differences in the frequency of polymorphisms that affect the pharmacokinetics (e.g., CYP2C9) and targets (e.g., VKORC1) of drugs such as warfarin. This is particularly significant in admixed populations, in which the substructure created by inter-ethnic crosses further increases the fluidity of racial and/or ethnic labels [41].
Acknowledgments
This investigation was supported, in part, by a Research Centers in Minority Institution Award G12RR-03051 from the National Center for Research Resources, NIH and by the Puerto Rico Newborn Screening Program and Genomas internal research and development funds. The authors want to thank Dr. Pedro J Santiago-Borrero for kindly supply the samples for this work and Mrs. Yolanda Rodriguez for her support in collecting the samples.
Footnotes
Disclosure: Dr Ruaño is founder and President of Genomas, Inc. Dr Windemuth, Dr Seip, Mr. Kocherla, Mr. Villagra, Miss Gorowski and Miss Bogaard are full-time employees of Genomas. The rest of the authors have no potential conflicts of interest to disclose.
Statement: A glossary of genetic terminology is maintained by the National Human Genome Research Institute at www.genome.gov/glossary.cfm.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wadelius M, Chen LY, Downes K, et al. Common VKORC1 and GGCX polymorphisms associated with warfarin dose. Pharmacogenomics J. 2005;5(4):262–70. doi: 10.1038/sj.tpj.6500313. [DOI] [PubMed] [Google Scholar]
- 2.Rost S, Fregin A, Ivaskevicius V, et al. Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature. 2004;427(6974):537–41. doi: 10.1038/nature02214. [DOI] [PubMed] [Google Scholar]
- 3.Rieder MJ, Reiner AP, Gage BF, et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med. 2005;352(22):2285–93. doi: 10.1056/NEJMoa044503. [DOI] [PubMed] [Google Scholar]
- 4.Veenstra DL, You JH, Rieder MJ, et al. Association of Vitamin K epoxide reductase complex 1 (VKORC1) variants with warfarin dose in a Hong Kong Chinese patient population. Pharmacogenet Genomics. 2005;15(10):687–91. doi: 10.1097/01.fpc.0000174789.77614.68. [DOI] [PubMed] [Google Scholar]
- 5.Seip RL, Duconge J, Ruano G. Implementing genotype-guided antithrombotic therapy. Future Cardiol. 2010;6:409–424. doi: 10.2217/fca.10.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaminsky LS, Zhang ZY. Human P450 metabolism of Warfarin. Pharmacology and Therapeutics. 1997;73:67–74. doi: 10.1016/s0163-7258(96)00140-4. [DOI] [PubMed] [Google Scholar]
- 7.Rettie AE, Korzekwa KR, Kunze KL, et al. Hydroxylation of warfarin by human cDNA-expressed cytochrome P-450: a role for P-4502C9 in the etiology of (S)-warfarin-drug interactions. Chemical Research in Toxicology. 1992;5(1):54–59. doi: 10.1021/tx00025a009. [DOI] [PubMed] [Google Scholar]
- 8.Reynolds KK, Valdes R, Jr, Hartung BR, Linder MW. Individualizing Warfarin Therapy. Personalized Medicine. 2007;4(1):11–31. doi: 10.2217/17410541.4.1.11. [DOI] [PubMed] [Google Scholar]
- 9.Goldstein JA. Clinical relevance of genetic polymorphisms in the human CYP2C subfamily. Br J Clin Pharmacol. 2001;52:349–355. doi: 10.1046/j.0306-5251.2001.01499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furuya H, Fernandez-Salguero P, Gregory W, et al. Genetic polymorphism of CYP2C9 and its effect on warfarin maintenance dose requirement in patients undergoing anticoagulation therapy. Pharmacogenetics. 1995;5(6):389–392. doi: 10.1097/00008571-199512000-00008. [DOI] [PubMed] [Google Scholar]
- 11.Higashi MK, Veenstra DL, Kondo LM, et al. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. JAMA. 2002;287:1690–98. doi: 10.1001/jama.287.13.1690. [DOI] [PubMed] [Google Scholar]
- 12.Taube J, Halsall D, Baglin T. Influence of cytochrome P-450 CYP2C9 polymorphisms on warfarin sensitivity and risk of over-anticoagulation in patients on long-term treatment. Blood. 2000;96:1816–19. [PubMed] [Google Scholar]
- 13.Wu AHB. Use of genetic and non-genetic factors in warfarin dosing algorithms. Pharmacogenomics. 2007;8(7):851–861. doi: 10.2217/14622416.8.7.851. [DOI] [PubMed] [Google Scholar]
- 14.Hillman MA, Wilke RA, Caldwell MD, et al. Relative impact of covariates in prescribing warfarin according to CYP2C9 genotype. Pharmacogenetics. 2004;14:539–47. doi: 10.1097/01.fpc.0000114760.08559.dc. [DOI] [PubMed] [Google Scholar]
- 15.Wu AHB, Wang P, Haller C, Drake K, Linder M, Valdes R., Jr Dosing algorithm for warfarin using CYP2C9 and VKORC1 genotyping from a multi-ethnic population: comparison with other equations. Pharmacogenomics. 2008;9(2):169–178. doi: 10.2217/14622416.9.2.169. [DOI] [PubMed] [Google Scholar]
- 16.The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi H, Wilkinson GR, Nutescu EA, et al. Different contributions of polymorphisms in VKORC1 and CYP2C9 to intra- and inter-population differences in maintenance doses of warfarin in Japanese, Caucasians and African Americans. Pharmacogenet Genomics. 2006;16(2):101–10. doi: 10.1097/01.fpc.0000184955.08453.a8. [DOI] [PubMed] [Google Scholar]
- 18.Tham LS, Goh BC, Nafziger A, et al. A warfarin-dosing model in Asians that uses single-nucleotide polymorphism in vitamin K epoxide reductase complex and cytochrome P450 2C9. Clin Pharmacol Ther. 2006;80(4):346–55. doi: 10.1016/j.clpt.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 19.Marsh S, King CR, Porche-Sorbet RM, Scott-Horton TJ, Eby CS. Population variation in VKORC1 haplotype structure. J Thromb Haemost. 2006;4(2):473–4. doi: 10.1111/j.1538-7836.2006.01759.x. [DOI] [PubMed] [Google Scholar]
- 20.Suarez-Kurtz G, Pena SD. Pharmacogenomics in the Americas: the impact of genetic admixture. Curr Drug Targets. 2006;7(12):1649–58. doi: 10.2174/138945006779025392. [DOI] [PubMed] [Google Scholar]
- 21.Ruaño G, Duconge J, Windemuth A, et al. Physiogenomic Analysis of the Puerto Rican Population. Pharmacogenomics. 2009;10(4):565–577. doi: 10.2217/pgs.09.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bertoni B, Budowle B, Sans M, Barton SA, Chakraborty R. Admixture in Hispanics: distribution of ancestral population contributions in the Continental United States. Hum Biol. 2003;75:1–11. doi: 10.1353/hub.2003.0016. [DOI] [PubMed] [Google Scholar]
- 23.Martinez-Cruzado JC, Toro-Labrador G, Viera-Vera J, et al. Reconstructing the population history of Puerto Rico by means of mtDNA phylogeographic analysis. Am J Phys Anthropol. 2005;128:131–55. doi: 10.1002/ajpa.20108. [DOI] [PubMed] [Google Scholar]
- 24.Choudhry S, Coyle NE, Tang H, et al. Population stratification confounds genetic association studies among Latinos. Hum Genet. 2006;118:652–64. doi: 10.1007/s00439-005-0071-3. [DOI] [PubMed] [Google Scholar]
- 25.Gordon J, Merante F, Weiss S, Zastawny R. Pharmacogenetic P-450 Screening Using the Tag-It Universal Bead-Based Array Platform. In: Wong S, Linder M, Valdes R Jr, editors. Pharmacogenomics and Proteomics: Enabling the Practice of Personalized Medicine. Washington, DC: AAC Press; 2006. [Google Scholar]
- 26.Ruaño G, Windemuth A. Physiogenomic Method for Predicting Clinical Outcomes of Treatments in Patients. 20060278241. USA patent. 2006
- 27.Ruaño G, Windemuth A, Holford T. Physiogenomics: Integrating Systems Engineering and Nanotechnology for Personalized Medicine. In: Bronzino J, editor. The Biomedical Engineering Handbook. CRC Press; 2005. pp. 281–289. [Google Scholar]
- 28.Oliphant A, Barker DL, Stuelpnagel JR, Chee MS. BeadArray technology: enabling an accurate, cost-effective approach to high-throughput genotyping. Biotechniques. 2002;56–8:60–1. [PubMed] [Google Scholar]
- 29.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multi-locus genotype data. Genetics. 2000;155:945–59. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Falush D, Stephens M, Pritchard JK. Inference of population structure using multi-locus genotype data: dominant markers and null alleles. Mol Ecol Notes. 2007;7:574–8. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.The University of Chicago Department of Biological Sciences. Pritchard Lab; http://pritch.bsd.uchicago.edu. [Google Scholar]
- 32.Limdi NA, McGwin G, Goldstein JA, et al. Influence of CYP2C9 and VKORC1 1173C/T Genotype on the Risk of Hemorrhagic Complications in African-American and European-American Patients on Warfarin. Clin Pharmacol Ther. 2008;83:312–21. doi: 10.1038/sj.clpt.6100290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jennings J. Across an Arctic Bridge. In: Billard J, editor. The World of the American Indian. Washington, DC: National Geographic Society; 1979. [Google Scholar]
- 34.Perini JA, Petzl-Erler ML, Tsuneto LT, Suarez-Kurtz G. VKORC1 polymorphisms in Amerindian populations of Brazil. Pharmacogenomics. 2008;9:1623–9. doi: 10.2217/14622416.9.11.1623. [DOI] [PubMed] [Google Scholar]
- 35.Tishkoff SA, Reed FA, Friedlaender FR, et al. The Genetic Structure and History of Africans and African Americans. Science. 2009;324:1035–44. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.The Trans-Atlantic Slave Trade Voyages Database. The National Endowment for the Humanities. Emory University Digital Library Research Initiative; [Accessed on July 2009]. http://www.slavevoyages.org/tast/database/search.faces. [Google Scholar]
- 37.Perini JA, Struchiner CJ, Silva-Assuncao E, et al. Pharmacogenetics of warfarin: development of a dosing algorithm for Brazilian patients. Clin Pharmacol Ther. 2008;84:722–728. doi: 10.1038/clpt.2008.166. [DOI] [PubMed] [Google Scholar]
- 38.Vargens DD, Almendra L, Struchiner CJ, Suarez-Kurtz G. Distribution of the GNB3 825C>T polymorphism among Brazilians: impact of population structure. Eur J Clin Pharmacol. 2008;64:253–6. doi: 10.1007/s00228-007-0413-2. [DOI] [PubMed] [Google Scholar]
- 39.Klein TE, Altman RB, Eriksson N, et al. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360:753–764. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomas DC, Witte JS. Population stratification: A problem for case-control studies of candidate-gene associations? Cancer Epidemiol. Biomark Prev. 2002;11:505–512. [PubMed] [Google Scholar]
- 41.Suarez-Kurtz G. Ethnic differences in drug therapy: a pharmacogenomics perspective. Exp Rev Clin Pharmacol. 2008;1:337–339. doi: 10.1586/17512433.1.3.337. [DOI] [PubMed] [Google Scholar]