Abstract
Mutations in presenilin-1 and presenilin-2 (PS1 and PS2) are the most common cause of familial Alzheimer disease. PS1 and PS2 are the presumptive catalytic components of the multisubunit γ-secretase complex, which proteolyzes a number of type I transmembrane proteins, including the amyloid precursor protein (APP) and Notch. APP processing by γ-secretase produces β-amyloid peptides (Aβ40 and Aβ42) that accumulate in the Alzheimer disease brain. Here we identify a pathogenic L435F mutation in PS1 in two affected siblings with early-onset familial Alzheimer disease characterized by deposition of cerebral cotton wool plaques. The L435F mutation resides in a conserved C-terminal PAL sequence implicated in active site conformation and catalytic activity. The impact of PS1 mutations in and around the PAL motif on γ-secretase activity was assessed by expression of mutant PS1 in mouse embryo fibroblasts lacking endogenous PS1 and PS2. Surprisingly, the L435F mutation caused a nearly complete loss of γ-secretase activity, including >90% reductions in the generation of Aβ40, Aβ42, and the APP and Notch intracellular domains. Two nonpathogenic PS1 mutations, P433L and L435R, caused essentially complete loss of γ-secretase activity, whereas two previously identified pathogenic PS1 mutations, P436Q and P436S, caused partial loss of function with substantial reductions in production of Aβ40, Aβ42, and the APP and Notch intracellular domains. These results argue against overproduction of Aβ42 as an essential property of presenilin proteins bearing pathogenic mutations. Rather, our findings provide support for the hypothesis that pathogenic mutations cause a general loss of presenilin function.
Keywords: Alzheimer Disease, Amyloid, Neurodegeneration, Neurological Diseases, Notch Pathway, Presenilin, Secretases, Cotton Wool Plaques, Early Onset Familial Alzheimer Disease, γ-Secretase
Introduction
Alzheimer disease (AD)3 is the most common cause of both dementia and neurodegeneration. Occurrence of AD is largely sporadic, typically affecting individuals over age 65, but a small minority of cases (∼5%) displays familial inheritance with early onset. Dominantly inherited missense mutations in presenilin-1 (PS1), presenilin-2 (PS2) or the amyloid precursor protein (APP) cause familial AD with complete penetrance (1–3). PS1 mutations account for the majority of cases of familial AD, and more than 170 pathogenic mutations have been identified thus far throughout the PS1 coding sequence.
Presenilins (PS) are essential components of the multiprotein γ-secretase complex, which catalyzes the intramembranous proteolysis of a number of type I transmembrane proteins, including APP and Notch (for review, see Refs. 3–5). Before processing of APP and Notch by γ-secretase, proteolytic removal of an N-terminal ectodomain by distinct proteases (β-secretase and tumor necrosis factor-α converting enzyme, respectively) is required to activate the transmembrane domain as a γ-secretase substrate. Intramembranous proteolysis of APP by γ-secretase acting at the “γ-sites” produces a heterogeneous population of Aβ peptides, whereas more distal cleavage at the “ϵ site” releases an intracellular C-terminal domain termed AICD. Analogous cleavage of Notch by γ-secretase acting near the cytoplasmic face of the plasma membrane (the “S3 site”) releases an intracellular domain termed NICD, which translocates to the nucleus and activates transcription of Notch effector genes.
Considerable evidence suggests that γ-secretase is an aspartic protease with the presenilin subunit contributing two critical active-site aspartate residues (3–5). PS are conserved throughout metazoan evolution and share limited homology with another family of intramembranous aspartic proteases, the signal peptide peptidases. PS and signal peptide peptidases display similar topology with nine transmembrane domains, but conservation of primary sequence between PS and signal peptide peptidases is limited to the YD and GXGD motifs containing the putative catalytic aspartate residues as well as a C-terminal PAL motif of uncertain function (PS1 sequence 433PAL435) (6). Recent evidence suggests that these PAL residues are essential for catalytic activity and active site conformation (7–12).
The clinical and neuropathological features of familial AD (FAD) closely resemble those of sporadic AD. AD is characterized neuropathologically by neuronal degeneration and deposition of amyloid plaques and neurofibrillary tangles in affected brain regions, most notably the cerebral cortex. Neurofibrillary tangles are intraneuronal fibrillary aggregates composed of hyperphosphorylated forms of the microtubule-binding protein Tau. Amyloid plaques are extracellular aggregates composed of 40–42-residue β-amyloid peptides (designated Aβ40 and Aβ42) derived from sequential proteolytic processing of APP by β-secretase and γ-secretase. The “neuritic” amyloid plaques characteristic of AD contain a dense congophilic amyloid core and dystrophic neuronal processes. Interestingly, a small number of PS1 mutations cause an unusual variant of familial AD characterized by the deposition of large, diffuse “cotton wool” amyloid plaques (CWPs) lacking a dense core and associated neuritic changes (13, 14). In such cases, dementia is often but not always accompanied by spastic paraparesis. CWPs typically display strong immunoreactivity for Aβ42 but weak immunoreactivity for Aβ40, leading to the suggestion that CWP formation reflects overproduction of Aβ42.
The pathogenesis of familial AD caused by PS and APP mutations has been attributed to toxic effects associated with the overproduction and aggregation of Aβ peptides, particularly the more hydrophobic Aβ42 (15). This hypothesis was originally based on the premise that pathogenic mutations increase the production of Aβ42. However, it has become clear that many pathogenic PS mutations impair γ-secretase activity, bringing about an overall reduction in Aβ production, and some PS mutations have been found to decrease Aβ40 production without affecting Aβ42 production (for review, see Refs. 16). In an attempt to address these inconsistencies, the focus of the amyloid hypothesis has shifted to emphasize the potential pathogenicity of an elevated Aβ42/Aβ40 ratio (4, 5). It has been suggested that partial loss of function conferred by PS mutations shifts the cleavage specificity of the mutant enzyme to favor Aβ42 at the expense of Aβ40, resulting in a “toxic gain of function” caused by preferential production of Aβ42 (4, 5).
An alternative explanation for familial AD pathogenesis is provided by the presenilin hypothesis, which proposes that loss of essential PS functions may be the primary trigger of neurodegeneration and associated synaptic and cognitive dysfunction (16). This hypothesis helps to explain the observations that inactivation of presenilin function in the adult mouse brain is sufficient to cause widespread neurodegeneration, whereas overproduction of Aβ42 in the adult mouse brain produces extensive cerebral amyloidosis but fails to cause significant neurodegeneration (17–19). This hypothesis incorporates growing evidence that PS mutations in FAD cause a loss of PS function and that active site-directed inhibitors of γ-secretase can mimic the effects of pathogenic mutations, elevating the production of Aβ42 while suppressing production of Aβ40. Given the importance ascribed to overproduction of Aβ42 in the amyloid hypothesis, an important unresolved question is whether preservation of Aβ42 production is an essential property of PS bearing pathogenic mutations, and indeed whether mutant PS invariably retains sufficient catalytic activity to bring about a putative toxic gain of function caused by preferential production of Aβ42. If the primary effect of pathogenic mutations is to cause an intrinsic loss of PS function, one might expect to find mutations that cause a general impairment of the mutant enzyme's catalytic activity, including the capacity to generate Aβ42.
Here we describe a PS1 mutation altering the conserved PAL motif in two siblings with an unusual variant of familial AD. Genetic and neuropathological analysis indicated that an L435F missense mutation in PS1 causes AD associated with deposition of CWPs in this pedigree. Detailed analysis of the intrinsic effects of this mutation on PS1 function revealed a nearly complete loss of γ-secretase activity, including virtual elimination of APP processing and the generation of both Aβ40 and Aβ42. Our results are inconsistent with the view that pathogenic mutations in PS invariably enhance the relative or absolute production of Aβ42 by the mutant protein. These findings provide support for the hypothesis that loss of PS function plays a key role in FAD pathogenesis.
MATERIALS AND METHODS
Generation of cDNA Constructs
Point mutations (D257A, P433L, L435F, L435R, P436Q, P436S) were introduced by site-directed mutagenesis of wild-type human PS1 cDNA cloned into pCI expression vector using the GeneTailor system (Invitrogen). Incorporation of mutations was verified by bidirectional sequencing of cDNAs. APP C99-myc and NotchΔE-myc constructs were kindly provided by A. Goate (Washington University School of Medicine) and have been previously described (9, 58).
Cell Culture
PS-null murine embryonic fibroblasts (MEFs) were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Hyclone), penicillin/streptomycin, and l-glutamine (20, 21). Cells were transiently cotransfected with expression vectors encoding substrate and either PS1 variant or vector alone using Lipofectamine 2000 (Invitrogen). Cells were washed in phosphate-buffered saline and lysed in ice-cold lysis buffer 24 h post-transfection (50 mm Tris, pH 8.0, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 1 mm dithiothreitol, and CompleteMini protease inhibitor mixture (Roche Applied Science)). Insoluble material was removed by centrifugation (16,000 × g, 10 min, 4 °C). Protein concentration of lysates was measured using the Bio-Rad protein assay.
Gel Electrophoresis and Western Analysis
Cell lysates were separated by SDS-polyacrylamide gel electrophoresis, and proteins were transferred to nitrocellulose membranes. After blocking in Tris-buffered saline 0.1% Tween-20, 5% Blotto (Santa Cruz Biotechnology), membranes were incubated overnight with primary antibodies and developed the following day with horseradish peroxidase-linked anti-mouse or anti-rabbit secondary antibodies and LumiGLO chemiluminescent reagent (Cell Signaling Technology). To control for equal loading, membranes were stripped and incubated with anti-α-tubulin antibody and developed as described. Band intensity was quantified using ImageJ software (National Institutes of Health), and results were normalized to β-tubulin levels. Antibodies used included mouse anti-PS1-CTF (Chemicon, clone 5232), mouse anti-Myc (Sigma, clone 9E10), mouse anti-N-cadherin (BD Biosciences, clone 32), rabbit anti-cleaved Notch (Val1744) (Cell Signaling Technology), and mouse anti-β-tubulin (Sigma, clone B512).
Enzyme-linked Immunosorbent Assay (ELISA)
PS-null MEFs were transfected with expression vectors encoding PS1 variants and C99 as described above. Medium was replaced with Dulbecco's modified Eagle's medium, 10% fetal bovine serum supplemented with 10 μm phosphoramidon (Sigma) 4 h post-transfection, and cells were returned to 37 °C. Conditioned medium was collected 24 h later, centrifuged to remove insoluble material (10,000 × g, 10 min, 4 °C), frozen on dry ice, and stored at −80 °C until analysis. Sandwich Aβ ELISA was performed as previously described (22). Monoclonal antibodies directed against the C terminus of Aβ40 or Aβ42 were used for specific capture of Aβ species. A biotinylated monoclonal secondary antibody (4G8) recognizing Aβ residues 17–24 was used for detection of both Aβ40 and Aβ42 with a reporter system of streptavidin-conjugated alkaline phosphatase (Promega) and AttoPhos reagent (Amersham Biosciences). AttoPhos fluorescence was measured with excitation at 444 nm and emission at 555 nm. Aβ40 and Aβ42 synthetic peptide standards diluted in cell culture medium were included in each analysis and were used to quantify Aβ levels, expressed as the concentration in pm of Aβ40 or Aβ42.
Cell Surface Protein Isolation
PS-null MEFs were transiently transfected with expression vectors encoding wild-type or L435F mutant PS1 with an N-terminal 3×FLAG epitope tag. 20 h post-transfection, cells were incubated with membrane-impermeable Sulfo-NHS-SS-Biotin to label cell surface proteins. Cell lysis and affinity precipitation of biotinylated proteins was performed with Neutravidin resin as described by the manufacturer (Pierce/Thermo Scientific). Proteins were analyzed by gel electrophoresis and Western analysis with mouse anti-FLAG (M2) or rabbit anti-Nicastrin (1660) antibodies (Sigma).
Statistical Analysis
Statistical significance of the data was assessed by Student's t test (two-tailed, unequal variance). Data are presented as the means ± S.E. Group differences with p < 0.05 were considered statistically significant.
RESULTS
Identification of a Novel Mutation in PS1 Linked to Early- Onset Familial Alzheimer Disease
Genetic and neuropathological analysis was performed on two affected siblings from a pedigree with early-onset familial Alzheimer disease (Fig. 1A). The mean age of onset among the three siblings in the second generation was 47 years, and the mean age at death was 56 years. Clinical features in all cases included early and progressive memory deficits and aphasia, with subsequent development of functional impairment and parkinsonism. Sequencing of PSEN1 exons from genomic DNA revealed an identical heterozygous C to T transversion in exon 12 in both siblings examined, resulting in the substitution of phenylalanine for leucine at residue 435 (c.1303C>T, p.L435F). This L435F missense mutation alters the conserved PAL motif to the sequence PAF. Interestingly, the same nucleotide and amino acid substitutions in PSEN1 were previously reported in a single individual with AD in a large screening study for PSEN1 mutations (23). However, no information regarding family history or the clinical or neuropathological features in this affected individual was reported.
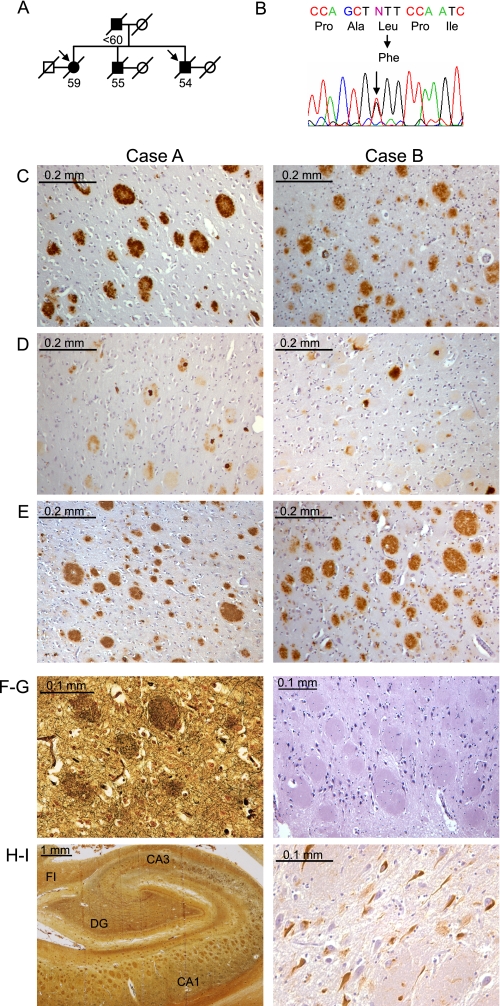
FIGURE 1.
Identification of an L435F mutation in PS1 in familial Alzheimer disease with cotton wool plaques. A, the pedigree of a family with early-onset AD is shown. Arrows indicate probands, filled symbols indicate affected individuals, and numbers below symbols indicate age at death. Circles, female; squares, male; diagonal slash, deceased. B, sequence analysis of PS1 exons revealed heterozygosity for a C to T transition resulting in substitution of phenylalanine for leucine at amino acid 435. C–E, detection of Aβ in frontal cortex of both probands. C, detection of Aβ42 with C-terminal specific antibody revealed numerous CWPs that were strongly immunoreactive for Aβ42. D, diffuse staining of CWPs with Aβ40-specific antibody is shown. Occasional plaques displayed a dense core of Aβ40. E, detection of total Aβ revealed numerous CWPs, with a staining pattern similar to that for Aβ42. F–H, analysis of the hippocampus (Case A) revealed numerous large CWPs throughout CA1-CA3 regions, visible by silver staining (F and H) and hematoxylin-eosin (G). I, phospho-Tau-positive neurofibrillary tangles were abundant in CA1-CA3 and dentate gyrus (DG) regions. FI, fimbria; CA, cornu ammonis.
Neuropathological analysis of the two siblings examined was remarkable for numerous and widespread CWPs deposited throughout the neocortex, hippocampus, and deep cerebral nuclei. As previously described, these CWPs were large and eosinophilic with little associated neuritic dystrophy and apparent displacement of surrounding tissue (Fig. 1, F–H) (13, 14). CWPs in both cases displayed strong immunoreactivity for Aβ42 and weak or absent immunoreactivity for Aβ40 (Fig. 1, C–E). Occasional plaques displayed central regions of dense Aβ40-positive staining, but dense cores were absent in the vast majority of plaques. Neurofibrillary tangles were relatively scarce in the cerebral cortex, but more numerous neurofibrillary tangles were observed in the entorhinal cortex and hippocampus (Fig. 1, H–I). Cerebral atrophy and neuronal loss were moderately severe in both cases, particularly in hippocampal area CA1, where CWPs completely replaced the pyramidal cell layer. Both cases also displayed mild cerebral amyloid angiopathy. In contrast to some prior reports of CWP neuropathology caused by PS1 mutations, there was no clinical evidence of spastic paraparesis in either case, but mild gliosis was observed in the brainstem corticospinal tracts in one sibling. Corresponding to the clinical finding of parkinsonism, both cases displayed depigmentation, neuronal loss, and gliosis in the substantia nigra pars compacta. Collectively, the results of clinicopathologic and genetic analysis suggest that a pathogenic PS1 mutation in the conserved PAL motif causes early-onset familial AD with CWP pathology in this pedigree.
The PS1 L435F Mutation Results in Loss of γ-Secretase-dependent PS1 Processing
To evaluate the effects of the L435F mutation on γ-secretase activity, we employed transient transfection in PS-null MEFs lacking endogenous γ-secretase activity (20, 21). Previous studies have shown that overexpression can compensate for the functional impairments caused by PS1 mutations (24). To obtain reliable estimates of the intrinsic effects of PS1 mutations on γ-secretase activity, we expressed limiting quantities of PS1 that resulted in a linear dose-response relationship for PS1 and γ-secretase product.4 For comparison with the pathogenic PS1 L435F mutation identified above, we analyzed the effects of a series of mutations in neighboring residues. The nonpathogenic P433L mutation was analyzed because prior studies have shown that proline to leucine substitution in the PAL motif (altering PAL to LAL) causes a loss of function in Caenorhabditis elegans, Drosophila, and cultured MEFs (7, 9, 25, 26). Similarly, the nonpathogenic L435R substitution has been shown to cause a loss of γ-secretase activity in cultured MEFs (9). We also analyzed two pathogenic mutations, P436S and P436Q, affecting the proline residue immediately C-terminal to the PAL motif (433PALP436), which is conserved in all known presenilins but not in signal peptide peptidase. Interestingly, the P436Q mutation has also been associated with CWP neuropathology (27). As a negative control for γ-secretase activity, we employed a nonpathogenic D257A mutation eliminating one of the transmembrane aspartate residues required for catalytic activity.
An important step in maturation of the γ-secretase complex is endoproteolysis of the presenilin holoprotein to produce N-terminal and C-terminal fragments (NTF and CTF). This cleavage event, which occurs within the cytoplasmic loop domain between TM6 and TM7, appears to be autocatalytic and is normally required for γ-secretase activity. For example, γ-secretase inhibitors interfere with presenilin cleavage, and PS1 mutants defective in presenilin endoproteolysis are generally also deficient in γ-secretase activity (28). However, a mutant PS1 protein lacking the cytoplasmic loop domain containing the cleavage site (ΔE9, in-frame deletion of exon 9) retains proteolytic activity, suggesting that presenilin cleavage may function to relieve steric inhibition imposed by the cytoplasmic loop in the presenilin holoprotein (29).
To assess the impact of the L435F mutation and neighboring mutations in the PAL motif on presenilin function, we first evaluated presenilin endoproteolysis. The L435F mutation resulted in nearly complete loss of PS1 holoprotein cleavage, with a reduction in the level of PS1 CTF to ∼5% of the wild-type level (Fig. 2, A and B). Similarly, the P433L, L435R, and D257A mutations showed essentially complete loss of presenilin endoproteolysis, whereas the pathogenic mutations at P436 showed a more modest reduction in formation of PS1 CTF.
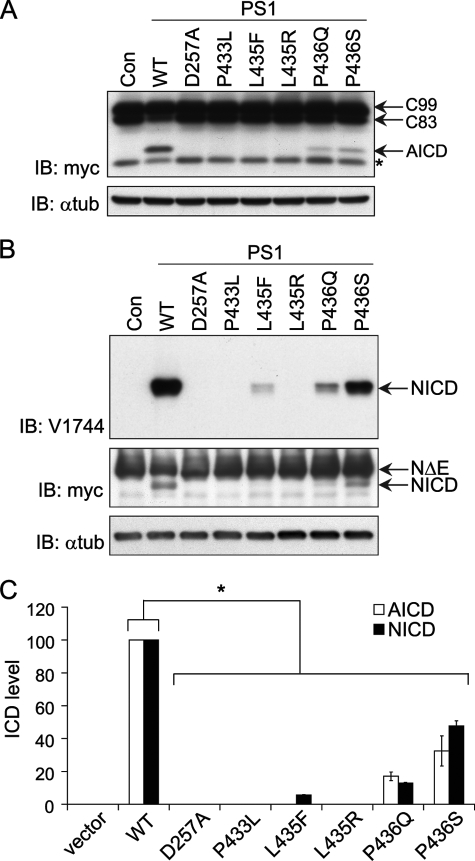
FIGURE 2.
The PS1 L435F mutant displays deficient endoproteolysis but normal cell surface expression. Wild-type (WT) and mutant PS1 were transiently expressed in PS-null MEFs to assess endoproteolytic activity and cell surface localization. Con (A, C) and vector (B) indicate transfection with empty rather than PS1-expressing vector. A, detection of full-length PS1 and PS1-CTF by Western analysis is shown. Proteins were detected with antibodies recognizing the C terminus of PS1 (top panel) and α-tubulin (α-tub, lower panel). Arrows indicate PS1 holoprotein (holo) and CTF. IB, immunoblot. B, quantification of PS1-CTF production is shown. PS1-CTF levels were normalized to α-tubulin and expressed as % of WT levels. Data are the means of three independent experiments (*, p < 0.05 compared with WT PS1-CTF level; n = 3). C, PS1-L435F is delivered to the cell surface and rescues cell-surface localization of mature Nicastrin. PS1-WT and PS1-L435F bearing N-terminal FLAG epitope tags were transiently expressed in PS-null MEFs, and cell surface proteins were isolated by biotinylation and affinity precipitation. PS1and Nicastrin (Nct) were detected by Western analysis of total cell lysates and biotinylated cell surface fractions. The cytosolic protein α-tubulin served as a control for non-cell surface protein. Arrows indicate PS1 holoprotein (holo), PS1 N-terminal fragment (NTF), and immature (imm) and mature (m) forms of Nicastrin.
Previous studies have shown that PS1 bearing PAL motif mutations display normal stability, high molecular weight complex formation, and subcellular distribution (8, 9). To confirm that PS1 L435F undergoes normal trafficking and γ-secretase complex assembly, we performed cell surface biotinylation of PS-null MEFs transiently expressing PS1 wild-type and L435F and then analyzed cell surface proteins for levels of PS1 and Nicastrin. Nicastrin is an essential component of the γ-secretase complex, and PS1 is required for Nicastrin maturation in the Golgi compartment and its subsequent trafficking to the cell surface (30). As shown in Fig. 2C, PS1 wild-type and PS1 L435F were expressed in similar levels at the cell surface. Consistent with the results in Fig. 2A, wild-type PS1 was detected at the cell surface as a mixture of holoprotein and N-terminal fragment, whereas PS1 L435F was detected almost exclusively as holoprotein. Moreover, PS1 wild type and PS1 L435F were similarly effective in inducing Nicastrin maturation and localization to the cell surface. These results indicate that the L435F mutation does not compromise the delivery of PS1 to the plasma membrane or its interaction with mature Nicastrin.
The PS1 L435F Mutation Causes a Nearly Complete Loss of γ-Secretase-dependent Cleavage of APP and Notch
To determine the effect of the L435F mutation and neighboring PALP mutations on cleavage of type I transmembrane proteins by γ-secretase, we evaluated the ability of mutants to rescue γ- secretase-dependent processing of APP and Notch in PS-null MEFs. To focus our analysis specifically on γ-secretase activity, we employed N-terminal-truncated forms of APP or Notch lacking the ectodomains as direct substrates for γ-secretase cleavage (APP C99 and NotchΔE, respectively), and we monitored the ability of PS1 variants to rescue production of AICD and NICD in PS-null MEFs. Fusion of a myc epitope tag to the C terminus of both substrates enabled specific detection by Western analysis of the intracellular domains released by γ-secretase cleavage.
Transfection of PS-null MEFs with expression vector encoding wild-type PS1 reconstituted γ-secretase activity, as evidenced by detection of AICD released from APP C99 (Fig. 3A). In contrast, we were unable to detect production of AICD in PS-null MEFs expressing PS1 L435F, P433L, or L435R. The P436Q and P436S mutations partially rescued AICD production to ∼20 and 30% of wild-type levels, respectively. As expected, no AICD production was detected with the inactive D257A mutant PS1 nor was AICD detected in the absence of exogenous PS1.
FIGURE 3.
PS1 L435F and neighboring mutations markedly impair γ- secretase-dependent proteolytic cleavage of APP and Notch. To assess cleavage activity, WT and mutant PS1 were co-expressed with APP C99-myc or NotchΔE-myc by transient transfection in PS-null MEFs, and full-length and cleaved substrates were detected by Western analysis. Con (A, B) and vector (C) indicate transfection with empty rather than PS1-expressing vector. Representative expression of PS1 for the same series of experiments is shown in Fig. 2A. A, APP C99 and AICD were detected with antibody recognizing the myc epitope tag (*, nonspecific background band). IB, immunoblot; α-tub, α-tubulin. B, γ-secretase-dependent Notch cleavage was detected by Western analysis with antibody recognizing cleaved Notch ICD (top panel). Full-length NotchΔE and NICD were detected with anti-myc antibody (middle panel). C, quantification of AICD and NICD production by PS1 mutants is shown. AICD and NICD levels were normalized to α-tubulin and expressed as % of WT levels. Data are the means of three independent experiments (*, p < 0.05 versus AICD or NICD level in cells expressing WT PS1; n = 3). ICD, intracellular domain.
We then assessed Notch processing as an independent measure of γ-secretase activity using NotchΔE as a substrate (Fig. 3B). The L435F mutation nearly abolished γ-secretase-mediated Notch processing, reducing NICD production to ∼5% of the wild-type level. Notch processing was completely inactivated by the P433L, L435R, and D257A mutations with no detectable NICD produced. The P436Q and P436S mutations caused a partial impairment of Notch cleavage that was comparable to the reduction observed for APP C99 processing.
The PS1 L435F Mutation Fails to Rescue γ-Secretase-dependent Cleavage of Endogenous N-cadherin in PS-null MEFs
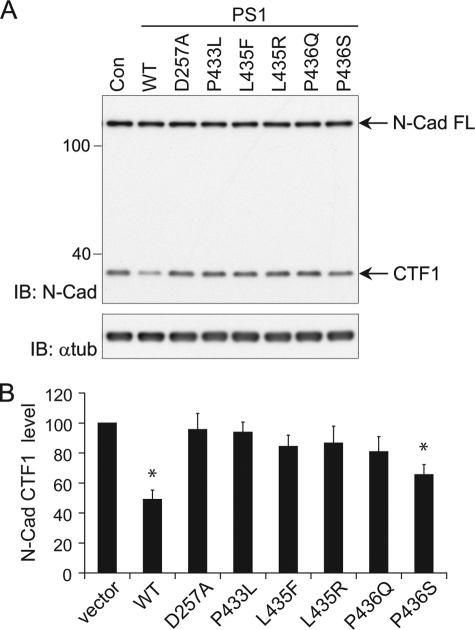
To confirm the impairments observed in the above assays, which relied on expression of exogenous substrates, we next examined γ-secretase-dependent processing of endogenous N-cadherin in PS-null MEFs transiently transfected with PS1 mutant constructs. Intramembranous cleavage of N-cadherin by γ-secretase depends on prior proteolytic removal of its ectodomain by the ADAM10 metalloprotease (31). The intracellular C-terminal fragment released by γ-secretase cleavage (CTF2) is rapidly degraded, but disappearance of the ADAM10-cleaved product (CTF1) provides a measure of the efficiency of processing by γ-secretase. N-cadherin was detected by Western analysis with antibody recognizing both full-length N-cadherin and CTF1. Expression of wild-type PS1 in PS-null MEFs restored γ-secretase-dependent processing of N-cadherin, as indicated by an ∼50% reduction in CTF1 compared with the level in PS-null MEFs transfected with empty vector (Fig. 4, A and B). The residual CTF1 may be attributable to cells that do not express PS1 because of the transfection efficiency of PS-null MEFs. In contrast to the restoration of CTF1 processing by wild-type PS1, the L435F mutant did not significantly reduce the level of CTF1 below that in PS-deficient MEFs, indicating a loss of γ-secretase activity toward this endogenous substrate. The D257A, P433L, L435R, and P436Q mutants were similarly deficient in γ-secretase-mediated CTF1 processing. Only the P436S mutant displayed an ability to process CTF1, albeit at a reduced efficiency relative to wild-type PS1 (Fig. 4, A and B).
FIGURE 4.
PS1 L435F and neighboring mutations inhibit γ-secretase-dependent cleavage of endogenous N-cadherin. WT and mutant PS1 were transiently expressed in PS-null MEFs. Disappearance of N-cadherin (N-Cad) CTF1 was monitored as a measure of γ-secretase activity toward endogenous substrate. Con (A) and vector (B) indicate transfection with empty rather than PS1-expressing vector. A, endogenous levels of full-length (FL) N-Cad and CTF1 were detected with antibody recognizing a sequence in the intracellular domain. IB, immunoblot; α-tub, α-tubulin. Con, control. B, quantification of N-Cad CTF1 levels is shown. CTF1 levels were normalized to α-tubulin and expressed as % of control (empty vector) levels. Data are the means of three independent experiments (*, p < 0.05 compared with transfection with empty vector; n = 3).
The PS1 L435F Mutation Severely Impairs Generation of Both Aβ40 and Aβ42
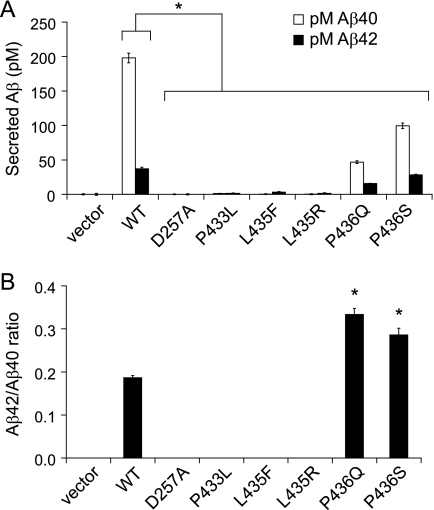
Because we were unable to detect AICD production by PS1 L435F, suggesting inactivation of ϵ cleavage activity, we were interested to determine whether this mutant displayed evidence of γ cleavage activity and particularly whether Aβ42 production was affected. The levels of Aβ40 and Aβ42 secreted by PS-null MEFs cotransfected with APP C99 and PS1 mutant constructs were measured by ELISA performed on conditioned medium (Fig. 5A). Surprisingly, Aβ42 production was essentially inactivated by the PS1 L435F mutation, with Aβ42 levels falling at the limit of detection (∼1 pm). Similarly, PS1 L435F did not produce detectable levels of Aβ40, consistent with a general loss of γ-secretase activity. Production of both Aβ40 and Aβ42 was also eliminated by the P433L, L435R, and D257A mutations. Based on the Aβ detection limits relative to the levels produced by wild-type PS1, each of these mutations reduced generation of Aβ40 by >99% and Aβ42 by >94%. The P436Q and P436S mutations caused partial but substantial reductions in the production of both Aβ40 and Aβ42, and the magnitude of reduction in total Aβ was comparable to the reduction in AICD for both mutations (cf. Fig. 2). Because of the relatively greater reduction in Aβ40 than in Aβ42, the Aβ42/Aβ40 ratio was elevated for the P436Q and P436S mutations.
FIGURE 5.
PS1 L435F and neighboring mutations cause marked reductions in the generation of both Aβ40 and Aβ42. WT and mutant PS1 were co-expressed with APP C99-myc by transient transfection in PS-null MEFs, and the levels of secreted Aβ40 and Aβ42 were quantified by ELISA. Vector indicates transfection with empty rather than PS1-expressing vector. Representative expression of PS1 and APP C99 for the same series of experiments is shown in Figs. 2A and 3A. A, Aβ levels (pm) in conditioned medium were measured by sandwich ELISA with antibodies specific for Aβ40 and Aβ42. The levels of Aβ40 and Aβ42 were at or below the threshold of detection for D257A, P433L, L435F, and L435R mutations. B, the concentration ratio of Aβ42/Aβ40 was calculated for mutations producing detectable levels of both cleavage products. Although total amounts of Aβ were significantly reduced for all mutations, the P436Q and P436S mutations resulted in an elevated Aβ42/Aβ40 ratio because of a greater reduction in Aβ40 than in Aβ42. Data shown are means of three separately transfected replicates and are a representative example from three independent experiments (A, *, p < 0.05 versus Aβ40 or Aβ42 levels, respectively, in cells transfected with WT PS1; B, *, p < 0.05 versus Aβ42/40 ratio for WT PS1; n = 3).
DISCUSSION
We have identified a PS1 L435F missense mutation in an early-onset FAD pedigree with cerebral deposition of CWPs. The pathogenicity of this mutation is supported by a number of observations. Two affected siblings from this pedigree displayed an identical heterozygous nucleotide substitution in the PSEN1 gene accompanied by highly similar clinical and neuropathological features. The L435F mutation is a nonconservative substitution that perturbs the evolutionarily conserved PAL motif previously shown to be important for normal PS function (7–12). An L435F mutation due to the same nucleotide substitution (c.1303C>T) was previously identified in a single individual with early onset AD in a large referral-based screen, and this mutation was not observed as a normal variant or polymorphism in control individuals (23). However, the role of this mutation as a cause of familial AD has been somewhat uncertain because no information was available regarding the clinical or neuropathological features or possible familial inheritance of AD in this affected individual. Our findings demonstrate a clear association of the L435F mutation with early-onset familial AD and further show that this mutation can lead to AD with variant neuropathology characterized by cotton wool plaques.
Our functional analysis revealed that the PS1 L435F mutation caused nearly complete loss of γ-secretase cleavage of multiple substrates, including APP, Notch, and endogenous N-cadherin. The severity of the loss of γ-secretase activity caused by this mutation was comparable to the effects of an inactivating mutation (D257A) in one of the putative catalytic aspartate residues. Although PS1 bearing the L435F mutation retained a low level (∼5%) of Notch processing activity, APP processing was effectively abolished. Surprisingly, we found that the L435F mutation essentially inactivates Aβ generation, reducing both Aβ40 and Aβ42 to undetectable levels. Although FAD-linked PS mutations have been shown to cause variable impairments of γ-secretase activity, inactivation of Aβ42 production has not been previously reported. Rather, investigations of clinical FAD mutations have typically documented two different patterns of altered Aβ production; that is, increased Aβ42 with unchanged or decreased Aβ40 or unchanged Aβ42 with decreased Aβ40 (32–40).
The L435F mutation, thus, represents the first demonstration of a FAD-causing mutation that effectively abolishes the ability of the mutant protein to produce Aβ42. Importantly, our analysis was designed to evaluate the intrinsic effects of PS1 mutations on γ-secretase activity while avoiding technical issues that could have led to overestimation of the activity of mutant PS in prior studies such as the presence of endogenous PS, viral overexpression of PS, and the use of mutant forms of APP that exert independent effects on APP processing. These technical differences may account in part for the remarkable loss of Aβ production that we observed for the L435F mutation. The loss of function conferred by the L435F mutation is unlikely to be due to a defect in trafficking or γ-secretase complex assembly, as the mutation did not compromise the cell surface delivery of PS1 or its ability to associate with mature Nicastrin. Collectively, our data suggest that the L435F mutation causes a particularly severe loss of intrinsic catalytic activity, likely reflecting a crucial role for the PAL motif in γ-secretase function.
Our findings are compatible with prior evidence suggesting that the PAL motif is essential for the normal conformation and activity of the γ-secretase catalytic site. First, the only regions displaying absolute sequence conservation among all known PS homologs and signal peptide peptidase family members are the PAL motif and the putative catalytic aspartates and flanking residues, suggesting that the PAL sequence may play an important role in catalytic activity. Second, cysteine substitution and cross-linking analysis has located the PAL residues in close proximity to the presumptive active site of PS1 and particularly the critical Asp-257 residue required for catalytic activity (11, 12). The maximum distance between Pro-433 and either Leu-250 or Asp-257 was estimated to be ≤5.2 Å, whereas Ala-434 and Leu-435 were both estimated to reside ∼2 Å from Asp-257 in the folded protein structure (11, 12). Third, mutations in the PAL motif have been shown to interfere with binding of transition-state analog inhibitors to PS1. P433L, A434D, and L435R mutations all blocked the binding of PS1 to a transition-state analog inhibitor that targets the enzyme active site (8). Conversely, binding of a transition-state analog inhibitor to PS1 blocked the accessibility of Cys-substituted residues within and adjacent to the PS1 PAL motif (e.g. A434C, L435C) to cross-linking reagents (11). Fourth, nonconservative mutations in the PAL motif can markedly impair γ-secretase activity. Mutation of the proline in the PAL motif to leucine in both Drosophila and C. elegans PS homologs (Dps P507L and SPE-4 P440L, respectively) caused phenotypes indistinguishable from those of null alleles (25, 26). The analogous mutations in human PS1 and PS2 (P433L and P414L, respectively) abolished endoproteolysis and production of AICD and NICD in PS-null MEFs (7, 9, 10). Similarly, mutagenesis of the PS1 PAL sequence has revealed a general requirement for small side chains at each position, and nonconservative mutations typically result in reduced catalytic function (8). Mutations in the PAL motif do not appear to interfere with the assembly, stability, or subcellular distribution of the γ-secretase complex (8, 9). Rather, these observations collectively suggest that PAL mutations impair catalytic function directly by altering the conformation of the active site.
Consistent with the importance of the PAL motif for catalytic function, we also found that the nonpathogenic P433L and L435R mutations cause a severe general loss of proteolytic activity comparable to the effect of the D257A mutation. These results confirm and extend prior studies showing that nonconservative PAL mutations typically compromise γ-secretase activity. Specifically, our results agree with previous reports that mutant PS1 bearing the P433L, L435R, or L435F mutations is inactive for PS1 endoproteolysis and production of AICD and NICD when tested in PS-null MEFs (7–10). The reported effects of PAL mutations on Aβ production have been more variable, possibly due to differences in experimental approach that could influence the observed levels of Aβ40 and/or Aβ42 production, such as those outlined above (7–12). Our finding that the PAL mutations examined in this study essentially abolish production of Aβ40 and Aβ42 is consistent with their inactivation of AICD and NICD production, suggesting a general loss of proteolytic activity due to conformational disruption of the active site.
We also investigated the impact of FAD-associated PS1 mutations affecting the proline residue immediately C-terminal to the PAL motif (Pro436) on γ-secretase activity. This proline residue is conserved throughout PS homologs but not among signal peptide peptidase family members, suggesting that it plays an important role in PS function but may not be as critical as the adjacent PAL residues. Indeed, the P436Q and P436S mutations caused substantial but incomplete loss of γ-secretase activity, with P436Q exerting a relatively greater inhibitory effect. The deficit in PS1 endoproteolysis caused by the P436S mutation was surprisingly mild relative to its inhibition of APP and Notch processing, suggesting that loss of endoproteolysis may not account entirely for the deleterious effects of some mutations on γ-secretase activity. Presenilinase cleavage may also be somewhat more permissive or at least more tolerant of amino acid substitutions than other substrate cleavage events.
PS mutations have been presumed to cause FAD by enhancing production of Aβ42 by the mutant protein, thereby conferring a toxic gain of function (15). However, a number of recent studies have shown that clinical PS mutations can impair Aβ40 production without affecting Aβ42 production, leading to the revised view that pathogenic PS mutations invariably shift the cleavage specificity of the mutant protein to favor production of Aβ42 at the expense of Aβ40 (4, 41). Our findings, however, are incompatible with the view that absolute or relative overproduction of Aβ42 is an essential property of PS bearing pathogenic mutations. Rather, our results suggest that at least some FAD-associated PS mutations can cause a nearly complete loss of the mutant protein's ability to support γ-secretase activity. Whether the L435F mutation represents a relatively unique example due to the importance of the PAL motif for catalytic function or whether similar properties may be ascribed to a broader spectrum of PS mutations is presently unclear.
Juxtaposition of the findings from our clinicopathological and molecular investigations raises an interesting paradox; the L435F mutation virtually abolishes the ability of the mutant protein to produce Aβ42, and yet affected individuals with the L435F mutation develop widespread CWPs. In reconciling these apparently conflicting observations, the effects of the heterozygous mutation must be considered in the context of wild-type PS expressed from the remaining PS1 and PS2 alleles. Even if the L435F mutant PS1 produced trace amounts of Aβ40 and/or Aβ42 below the detection limits of our assays, such trace amounts would not appreciably alter the levels of Aβ40 and Aβ42 produced by wild-type PS expressed from the remaining PS alleles or the resulting Aβ42/Aβ40 ratio. Thus, the Aβ42 deposited in cerebral CWPs in individuals with the L435F mutation is presumably the product of wild-type PS. Simple deletion of a single PS1 allele in mice does not alter the Aβ42/Aβ40 ratio or promote amyloid plaque formation; on the contrary, PS1 deficiency ameliorates amyloid deposition in the brains of transgenic mice expressing mutant human APP (42–45). Therefore, the intrinsic or intramolecular loss of PS1 function caused by the L435F mutation is unlikely by itself to account for the observed Aβ deposition. The most straightforward resolution of this apparent paradox is that mutant PS1 influences wild-type PS1/PS2 in a manner that alters Aβ production and promotes CWP formation. It is also possible that the L435F mutation may act in some way to inhibit Aβ clearance and promote plaque formation; for example, loss of γ-secretase activity has been shown to impair expression and activity of the Aβ-degrading enzyme neprilysin, and neprilysin deficiency promotes Aβ accumulation (46–49).
The ability of γ-secretase inhibitors to enhance Aβ42 production offers a precedent for the notion that proteolytically inactive γ-secretase can modulate Aβ production by wild-type γ-secretase (16). Numerous studies have shown that low to moderate concentrations of active site-directed inhibitors of γ-secretase increase Aβ42 production while decreasing both Aβ40 and total Aβ production (50–54). Furthermore, the Aβ42/Aβ40 ratio is typically increased across the entire range of inhibitor concentration, even at higher concentrations at which production of both Aβ42 and Aβ40 is diminished. Because bound inhibitor presumably occupies the enzyme active site and precludes proteolytic activity, these observations suggest that inactive γ-secretase complexes with bound inhibitor stimulate Aβ42 production by the remaining uninhibited γ-secretase complexes. At the same time, Aβ40 production is reduced as inhibitor-bound γ-secretase is inactivated. Thus, the enhancement of Aβ42 production induced by γ-secretase inhibitors is likely to represent an intermolecular rather than intramolecular effect. This phenomenon is suggestive of an allosteric effect of inhibitor on a multimeric enzyme; indeed, several lines of evidence suggest that PS physically interacts to form a dimeric catalytic core within the γ-secretase complex (55–57).
The opposing effects of γ-secretase inhibitors on Aβ42 and Aβ40 imply that elevation of the Aβ42/Aβ40 ratio, which is thought to promote amyloid plaque formation, is a manifestation of impaired γ-secretase activity (16). Recent evidence obtained from analysis of PS1 knock-in mice suggests that amyloid plaque pathology may similarly be a manifestation of impaired PS function. Introduction of a heterozygous PS1 M146V knock-in mutation elevated the Aβ42/Aβ40 ratio and enhanced amyloid plaque formation in the brains of APP transgenic mice, and deletion of the remaining wild-type PS1 allele produced a further increase in the Aβ42/Aβ40 ratio and markedly exacerbated the amyloid plaque pathology (42). Taken together, these observations suggest that partial loss of PS function may be primarily responsible for elevation of the Aβ42/Aβ40 ratio and consequent amyloid deposition in the FAD brain.
We recently proposed as part of the “presenilin hypothesis” that pathogenic PS mutations exert both intramolecular and intermolecular effects, bringing about an intrinsic loss of the mutant protein's function, which leads in turn to dominant-negative inhibition of wild-type PS and an overall impairment of γ-secretase activity (16). Dominant-negative inhibition by mutant γ-secretase was further proposed to alter the APP cleavage specificity of wild-type γ-secretase, resulting in heightened production of Aβ42 relative to Aβ40. Our findings demonstrating that the L435F mutation imposes a nearly complete loss of mutant PS1 activity while also engendering the deposition of amyloid plaques provide support for this proposal. The unusual composition of CWPs with abundant Aβ42 but scant Aβ40 suggests that the L435F mutation and perhaps other PS mutations associated with CWP neuropathology cause an especially pronounced loss of γ-secretase activity. It will be of interest in future studies to examine the hypothesis that PS1 bearing pathogenic mutations can inhibit the activity of wild-type PS1 and γ-secretase.
Acknowledgments
We thank Drs. Jun Wang and Alison Goate for helpful technical advice and the APP C99-myc construct, Dr. Raphael Kopan for the NotchΔE-myc construct, Dr. Bart De Strooper for the PS-null MEFs, Karlotta Fitch for advice on brain histochemistry, and members of the Kelleher and Shen laboratories for discussions.
This work was supported, in whole or in part, by grants from the Alzheimer's Association (to J. S. and R. J. K.) and by National Institutes of Health Grant R01 NS41783 (NINDS; to J. S.).
E. Heilig, J. Shen, and R. J. Kelleher, manuscript in preparation.
- AD
- Alzheimer disease
- FAD
- familial Alzheimer disease
- CWP
- cotton wool plaque
- PS
- presenilin
- PAL
- Pro-Ala-Leu (PS1 residues Pro-433—Ala-434—Leu-435)
- CTF
- C-terminal fragment
- WT
- wild type
- APP
- amyloid precursor protein
- Aβ
- β-amyloid
- AICD
- APP intracellular domain
- NICD
- Notch intracellular domain
- ELISA
- enzyme-linked immunosorbent assay
- MEF
- murine embryonic fibroblast.
REFERENCES
- 1.Tandon A., Rogaeva E., Mullan M., St George-Hyslop P. H. (2000) Curr. Opin. Neurol. 13, 377–384 [DOI] [PubMed] [Google Scholar]
- 2.Bertram L., Tanzi R. E. (2008) Nat. Rev. Neurosci. 9, 768–778 [DOI] [PubMed] [Google Scholar]
- 3.Brunkan A. L., Goate A. M. (2005) J. Neurochem. 93, 769–792 [DOI] [PubMed] [Google Scholar]
- 4.Selkoe D. J., Wolfe M. S. (2007) Cell 131, 215–221 [DOI] [PubMed] [Google Scholar]
- 5.Steiner H., Fluhrer R., Haass C. (2008) J. Biol. Chem. 283, 29627–29631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martoglio B., Golde T. E. (2003) Hum. Mol. Genet. 12, R201–R206 [DOI] [PubMed] [Google Scholar]
- 7.Tomita T., Watabiki T., Takikawa R., Morohashi Y., Takasugi N., Kopan R., De Strooper B., Iwatsubo T. (2001) J. Biol. Chem. 276, 33273–33281 [DOI] [PubMed] [Google Scholar]
- 8.Wang J., Beher D., Nyborg A. C., Shearman M. S., Golde T. E., Goate A. (2006) J. Neurochem. 96, 218–227 [DOI] [PubMed] [Google Scholar]
- 9.Wang J., Brunkan A. L., Hecimovic S., Walker E., Goate A. (2004) Neurobiol. Dis. 15, 654–666 [DOI] [PubMed] [Google Scholar]
- 10.Nakaya Y., Yamane T., Shiraishi H., Wang H. Q., Matsubara E., Sato T., Dolios G., Wang R., De Strooper B., Shoji M., Komano H., Yanagisawa K., Ihara Y., Fraser P., St George-Hyslop P., Nishimura M. (2005) J. Biol. Chem. 280, 19070–19077 [DOI] [PubMed] [Google Scholar]
- 11.Sato C., Takagi S., Tomita T., Iwatsubo T. (2008) J. Neurosci. 28, 6264–6271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tolia A., Horré K., De Strooper B. (2008) J. Biol. Chem. 283, 19793–19803 [DOI] [PubMed] [Google Scholar]
- 13.Karlstrom H., Brooks W. S., Kwok J. B., Broe G. A., Kril J. J., McCann H., Halliday G. M., Schofield P. R. (2008) J. Neurochem. 104, 573–583 [DOI] [PubMed] [Google Scholar]
- 14.Crook R., Verkkoniemi A., Perez-Tur J., Mehta N., Baker M., Houlden H., Farrer M., Hutton M., Lincoln S., Hardy J., Gwinn K., Somer M., Paetau A., Kalimo H., Ylikoski R., Pöyhönen M., Kucera S., Haltia M. (1998) Nat. Med. 4, 452–455 [DOI] [PubMed] [Google Scholar]
- 15.Hardy J., Selkoe D. J. (2002) Science 297, 353–356 [DOI] [PubMed] [Google Scholar]
- 16.Shen J., Kelleher R. J., 3rd (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 403–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsiao K., Chapman P., Nilsen S., Eckman C., Harigaya Y., Younkin S., Yang F., Cole G. (1996) Science 274, 99–102 [DOI] [PubMed] [Google Scholar]
- 18.Saura C. A., Choi S. Y., Beglopoulos V., Malkani S., Zhang D., Shankaranarayana, Rao B. S., Chattarji S., Kelleher R. J., 3rd, Kandel E. R., Duff K., Kirkwood A., Shen J. (2004) Neuron 42, 23–36 [DOI] [PubMed] [Google Scholar]
- 19.Irizarry M. C., Soriano F., McNamara M., Page K. J., Schenk D., Games D., Hyman B. T. (1997) J. Neurosci. 17, 7053–7059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herreman A., Hartmann D., Annaert W., Saftig P., Craessaerts K., Serneels L., Umans L., Schrijvers V., Checler F., Vanderstichele H., Baekelandt V., Dressel R., Cupers P., Huylebroeck D., Zwijsen A., Van Leuven F., De Strooper B. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 11872–11877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herreman A., Van Gassen G., Bentahir M., Nyabi O., Craessaerts K., Mueller U., Annaert W., De Strooper B. (2003) J. Cell Sci. 116, 1127–1136 [DOI] [PubMed] [Google Scholar]
- 22.Sun X., Beglopoulos V., Mattson M. P., Shen J. (2005) Neurodegener. Dis. 2, 6–15 [DOI] [PubMed] [Google Scholar]
- 23.Rogaeva E. A., Fafel K. C., Song Y. Q., Medeiros H., Sato C., Liang Y., Richard E., Rogaev E. I., Frommelt P., Sadovnick A. D., Meschino W., Rockwood K., Boss M. A., Mayeux R., St. George-Hyslop P. (2001) Neurology 57, 621–625 [DOI] [PubMed] [Google Scholar]
- 24.Levitan D., Doyle T. G., Brousseau D., Lee M. K., Thinakaran G., Slunt H. H., Sisodia S. S., Greenwald I. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 14940–14944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arduengo P. M., Appleberry O. K., Chuang P., L'Hernault S. W. (1998) J. Cell Sci. 111, 3645–3654 [DOI] [PubMed] [Google Scholar]
- 26.Guo Y., Livne-Bar I., Zhou L., Boulianne G. L. (1999) J. Neurosci. 19, 8435–8442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Houlden H., Baker M., McGowan E., Lewis P., Hutton M., Crook R., Wood N. W., Kumar-Singh S., Geddes J., Swash M., Scaravilli F., Holton J. L., Lashley T., Tomita T., Hashimoto T., Verkkoniemi A., Kalimo H., Somer M., Paetau A., Martin J. J., Van Broeckhoven C., Golde T., Hardy J., Haltia M., Revesz T. (2000) Ann. Neurol. 48, 806–808 [PubMed] [Google Scholar]
- 28.Campbell W. A., Iskandar M. K., Reed M. L., Xia W. (2002) Biochemistry 41, 3372–3379 [DOI] [PubMed] [Google Scholar]
- 29.Steiner H., Romig H., Grim M. G., Philipp U., Pesold B., Citron M., Baumeister R., Haass C. (1999) J. Biol. Chem. 274, 7615–7618 [DOI] [PubMed] [Google Scholar]
- 30.Leem J. Y., Vijayan S., Han P., Cai D., Machura M., Lopes K. O., Veselits M. L., Xu H., Thinakaran G. (2002) J. Biol. Chem. 277, 19236–19240 [DOI] [PubMed] [Google Scholar]
- 31.Uemura K., Kihara T., Kuzuya A., Okawa K., Nishimoto T., Ninomiya H., Sugimoto H., Kinoshita A., Shimohama S. (2006) Neurosci. Lett. 402, 278–283 [DOI] [PubMed] [Google Scholar]
- 32.Ancolio K., Dumanchin C., Barelli H., Warter J. M., Brice A., Campion D., Frébourg T., Checler F. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 4119–4124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen F., Gu Y., Hasegawa H., Ruan X., Arawaka S., Fraser P., Westaway D., Mount H., St. George-Hyslop P. (2002) J. Biol. Chem. 277, 36521–36526 [DOI] [PubMed] [Google Scholar]
- 34.Siman R., Reaume A. G., Savage M. J., Trusko S., Lin Y. G., Scott R. W., Flood D. G. (2000) J. Neurosci. 20, 8717–8726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walker E. S., Martinez M., Brunkan A. L., Goate A. (2005) J. Neurochem. 92, 294–301 [DOI] [PubMed] [Google Scholar]
- 36.Bentahir M., Nyabi O., Verhamme J., Tolia A., Horré K., Wiltfang J., Esselmann H., De Strooper B. (2006) J. Neurochem. 96, 732–742 [DOI] [PubMed] [Google Scholar]
- 37.Kumar-Singh S., Theuns J., Van Broeck B., Pirici D., Vennekens K., Corsmit E., Cruts M., Dermaut B., Wang R., Van Broeckhoven C. (2006) Hum. Mutat. 27, 686–695 [DOI] [PubMed] [Google Scholar]
- 38.Moehlmann T., Winkler E., Xia X., Edbauer D., Murrell J., Capell A., Kaether C., Zheng H., Ghetti B., Haass C., Steiner H. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 8025–8030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shimojo M., Sahara N., Murayama M., Ichinose H., Takashima A. (2007) Neurosci. Res. 57, 446–453 [DOI] [PubMed] [Google Scholar]
- 40.Kwok J. B., Halliday G. M., Brooks W. S., Dolios G., Laudon H., Murayama O., Hallupp M., Badenhop R. F., Vickers J., Wang R., Naslund J., Takashima A., Gandy S. E., Schofield P. R. (2003) J. Biol. Chem. 278, 6748–6754 [DOI] [PubMed] [Google Scholar]
- 41.Wolfe M. S. (2007) EMBO Rep. 8, 136–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang R., Wang B., He W., Zheng H. (2006) J. Biol. Chem. 281, 15330–15336 [DOI] [PubMed] [Google Scholar]
- 43.Jankowsky J. L., Slunt H. H., Gonzales V., Jenkins N. A., Copeland N. G., Borchelt D. R. (2004) Neurobiol. Aging 25, 885–892 [DOI] [PubMed] [Google Scholar]
- 44.Saura C. A., Chen G., Malkani S., Choi S. Y., Takahashi R. H., Zhang D., Gouras G. K., Kirkwood A., Morris R. G., Shen J. (2005) J. Neurosci. 25, 6755–6764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dewachter I., Reversé D., Caluwaerts N., Ris L., Kuipéri C., Van den Haute C., Spittaels K., Umans L., Serneels L., Thiry E., Moechars D., Mercken M., Godaux E., Van Leuven F. (2002) J. Neurosci. 22, 3445–3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pardossi-Piquard R., Dunys J., Yu G., St George-Hyslop P., Alves da Costa C., Checler F. (2006) J. Neurochem. 97, 1052–1056 [DOI] [PubMed] [Google Scholar]
- 47.Pardossi-Piquard R., Petit A., Kawarai T., Sunyach C., Alves da Costa C., Vincent B., Ring S., D'Adamio L., Shen J., Müller U., St. George-Hyslop P., Checler F. (2005) Neuron 46, 541–554 [DOI] [PubMed] [Google Scholar]
- 48.Iwata N., Tsubuki S., Takaki Y., Shirotani K., Lu B., Gerard N. P., Gerard C., Hama E., Lee H. J., Saido T. C. (2001) Science 292, 1550–1552 [DOI] [PubMed] [Google Scholar]
- 49.Iwata N., Tsubuki S., Takaki Y., Watanabe K., Sekiguchi M., Hosoki E., Kawashima-Morishima M., Lee H. J., Hama E., Sekine-Aizawa Y., Saido T. C. (2000) Nat. Med. 6, 143–150 [DOI] [PubMed] [Google Scholar]
- 50.Citron M., Diehl T. S., Gordon G., Biere A. L., Seubert P., Selkoe D. J. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 13170–13175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Durkin J. T., Murthy S., Husten E. J., Trusko S. P., Savage M. J., Rotella D. P., Greenberg B. D., Siman R. (1999) J. Biol. Chem. 274, 20499–20504 [DOI] [PubMed] [Google Scholar]
- 52.Sato T., Dohmae N., Qi Y., Kakuda N., Misonou H., Mitsumori R., Maruyama H., Koo E. H., Haass C., Takio K., Morishima-Kawashima M., Ishiura S., Ihara Y. (2003) J. Biol. Chem. 278, 24294–24301 [DOI] [PubMed] [Google Scholar]
- 53.Zhang L., Song L., Terracina G., Liu Y., Pramanik B., Parker E. (2001) Biochemistry 40, 5049–5055 [DOI] [PubMed] [Google Scholar]
- 54.Wolfe M. S., Xia W., Moore C. L., Leatherwood D. D., Ostaszewski B., Rahmati T., Donkor I. O., Selkoe D. J. (1999) Biochemistry 38, 4720–4727 [DOI] [PubMed] [Google Scholar]
- 55.Cervantes S., Saura C. A., Pomares E., Gonzàlez-Duarte R., Marfany G. (2004) J. Biol. Chem. 279, 36519–36529 [DOI] [PubMed] [Google Scholar]
- 56.Schroeter E. H., Ilagan M. X., Brunkan A. L., Hecimovic S., Li Y. M., Xu M., Lewis H. D., Saxena M. T., De Strooper B., Coonrod A., Tomita T., Iwatsubo T., Moore C. L., Goate A., Wolfe M. S., Shearman M., Kopan R. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 13075–13080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clarke E. E., Churcher I., Ellis S., Wrigley J. D., Lewis H. D., Harrison T., Shearman M. S., Beher D. (2006) J. Biol. Chem. 281, 31279–31289 [DOI] [PubMed] [Google Scholar]
- 58.Kopan R., Schroeter E. H., Weintraub H., Nye J. S. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 1683–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]