Abstract
Estrogen-related receptor γ (ERRγ) regulates the perinatal switch to oxidative metabolism in the myocardium. We wanted to understand the significance of induction of ERRγ expression in skeletal muscle by exercise. Muscle-specific VP16ERRγ transgenic mice demonstrated an increase in exercise capacity, mitochondrial enzyme activity, and enlarged mitochondria despite lower muscle weights. Furthermore, peak oxidative capacity was higher in the transgenics as compared with control littermates. In contrast, mice lacking one copy of ERRγ exhibited decreased exercise capacity and muscle mitochondrial function. Interestingly, we observed that increased ERRγ in muscle generates a gene expression profile that closely overlays that of red oxidative fiber-type muscle. We further demonstrated that a small molecule agonist of ERRβ/γ can increase mitochondrial function in mouse myotubes. Our data indicate that ERRγ plays an important role in causing a shift toward slow twitch muscle type and, concomitantly, a greater capacity for endurance exercise. Thus, the activation of this nuclear receptor provides a potential node for therapeutic intervention for diseases such as obesity, which is associated with reduced oxidative metabolism and a lower type I fiber content in skeletal muscle.
Keywords: Gene Regulation, Metabolic Regulation, Mitochondrial Metabolism, Nuclear Receptors, Skeletal Muscle Metabolism, Exercise Regulation
Introduction
The estrogen-related receptors (ERRs)2 are orphan nuclear receptors that were identified through their homology to the estrogen family of receptors (1). ERRs are expressed in tissues with high metabolic demand, with ERRα being more ubiquitously expressed, whereas ERRβ and ERRγ show a more restricted pattern of expression. The levels of ERR activity are dramatically enhanced by the addition of peroxisome proliferator-activated receptor γ-coactivator 1α (PGC-1α) (2), such that the PGC-1 proteins are sometimes referred to as protein ligands for this class of nuclear receptors.
ERRα is an important mediator of adaptive mitochondrial biogenesis under situations of increased physiological stress as evidenced by the inability of ERRα knock-out mice in regulating body temperature upon cold challenge (3). The ERRα null mice are also more susceptible to cardiac occlusion by banding (4). Phenotypic analyses of the perinatal lethal ERRγ knock-out mouse model implicates ERRγ as essential for the transition from glycolytic to fatty acid utilization in cardiac muscle during early postnatal life (5). Microarray and ChIP-on-chip analyses in cardiomyocytes indicate a significant overlap in genes regulated by ERRα and ERRγ (6). Recently, gene expression profiling in ERRγ null mice described a role in the regulation of potassium homeostasis in the heart, kidney, and stomach (7). Although several studies describe the functional role for ERRs in energy metabolism in cardiac muscle and adipose tissue (1), the significance in skeletal muscle is not clearly elucidated.
Structural studies provide evidence that ERRs are constitutively active. ERRγ assumes an activated conformation in the absence of added ligand (8). Despite this constitutive activity, it is feasible to further transcriptionally activate ERRγ using a small molecule ligand (9). Although the existence of an endogenous ligand for ERRγ is unknown, the antiestrogens 4-hydroxytamoxifen and diethylstilbestrol bind as antagonists and abrogate transcriptional activity (10). The endocrine disruptor bisphenol A was shown to bind ERRγ with a high affinity, with the IC50 for this interaction in the low nanomolar range (11). In lieu of recent epidemiological data linking bisphenol A levels in humans with type 2 diabetes and metabolic dysfunction (12), the understanding of the biological role of ERRγ in key metabolic tissues and its contribution to the effects elicited by bisphenol A merits study from a public health perspective.
A significant number of studies link mitochondrial dysfunction in skeletal muscle with type 2 diabetes and obesity (13, 14). Exercise interventions in humans increase mitochondrial number and function and can improve insulin sensitivity (15). Studies in humans have shown that mitochondrial number and function in muscle diminish with age, implicating mitochondrial dysfunction as a key pathological process in sarcopenia (16). The study of transgenic mice expressing higher levels of PGC-1α in skeletal muscle has added significantly to our understanding of its role in regulating exercise capacity and fuel uptake and metabolism in muscle (17, 18).
The lack of in vivo models for the ERRs is currently limiting to the understanding of the physiological role of these proteins. A recent publication demonstrated the treatment of C2C12 myotubes with an ERRβ/γ agonist, GSK4716, increases the expression of PGC-1α, suggesting that the activation of ERRγ may play a role in the induction of oxidative metabolism in skeletal muscle (19). We provide evidence that increased ERRγ expression in skeletal muscle is sufficient and necessary to increase treadmill endurance and mitochondrial function as well as induce expression of a coordinated network of fuel uptake and oxidation and mitochondrial and muscle structural genes.
EXPERIMENTAL PROCEDURES
Animal Studies
Mice were maintained on 12-h light/dark cycle and cared for in accordance with the Animal Care and Use Committee protocol in the Novartis Institutes for BioMedical Research animal facility. Body composition was determined using EchoMRI (Echo Medical Systems, Houston, TX). ERRγ heterozygous null mice were obtained from Deltagen (San Mateo, CA) and backcrossed to C57Bl/6 for four generations (7). ERRγ transgenic mice were generated on a C57Bl/6 background using standard procedures. The transgenic construct consists of the 4.8-kb mouse muscle creatine kinase promoter driving a hemagglutinin-tagged VP16-ERRγ fusion or a hemagglutinin-tagged ERRγ gene.
Primary Mouse Myotube Culture, Adenoviral Transduction, and Compound Treatment
Mouse myoblasts were isolated as described previously (20). Human ERRγ-IRES green fluorescent protein adenovirus was generated by Welgen (Worcester, MA). Following a 48-h differentiation period, myotubes were transduced with green fluorescent protein or ERRγ adenovirus for 48 h. The titers to achieve low and high levels of ERRγ expression were 1 × 108 and 5 × 108 virus particles/ml, respectively. GSK4716 was synthesized as described previously (9) and dissolved in DMSO for cell culture experiments. Cells treated with a corresponding volume of DMSO were used as controls.
Analysis of mRNA Expression and mtDNA Content
For both tissues as well as cell lysates, total RNA was isolated using TRIzol reagent (Invitrogen). For muscle tissue, tissues were pulverized, and the RNeasy® Fibrous Tissue Minikit (Qiagen, Valencia, CA) was utilized for isolation of RNA. Quantitative real time PCR was performed using Assay-on-Demand® primer probes (Applied Biosystems, Foster City, CA) (supplemental Table S1). Mitochondrial DNA levels were assessed as described previously (17).
Protein Analysis
Tissues or cell lysates were prepared in RIPA buffer and separated using standard techniques. Normalization for loading was done using Ponceau staining. The polyclonal anti-human ERRγ antibody was generated using a 229-amino acid peptide corresponding to the ligand-binding domain (Covance Research Products Inc, Denver, PA). A donkey anti-rabbit IgG horseradish peroxidase-linked secondary antibody (GE Healthcare) and SuperSignal West Dura extended duration ECL substrate (Pierce) were used for detection. Citrate synthase activity was determined using 2 μg of total protein as prepared above, using the citrate synthase assay kit (Sigma).
Microarray Analysis
RNA isolated from myotubes transduced with adenovirus or intact gastrocnemius muscle were used for microarray analysis. RNA was hybridized to mouse 430_2 Affymetrix chip (Affymetrix, Santa Clara, CA) in triplicate.
Histology, Immunohistochemical Staining, and Morphometrics
Muscle samples were rapidly frozen by submerging for 20 s in a chilled isopentane bath wrapped in pre-cooled plastic wrap and placed in a −80 °C freezer for storage until cryo-sectioning. Samples were sectioned at 6 μm in a cryo-microtome and mounted on glass slides immediately prior to staining. Succinate dehydrogenase activity was measured histochemically according to published protocols (21), and muscle fiber size was measured by immunohistochemical staining using a polyclonal rabbit anti-laminin antibody (Sigma) at room temperature for 1 h at a 1:800 dilution. Detection was performed using a rabbit-on-rodent APL polymer (Biocare, Concord, CA) and Vulcan Fast Red chromogen (Biocare).
Stained slides were digitized using an Aperio ScanScope XT slide scanner and analyzed using algorithms from Aperio (color deconvolution for succinate dehydrogenase staining intensity and positive pixel count for total tissue area) as well as a custom watershed algorithm (ASTORIA) developed to segment the muscle fibers from the laminin-stained sections. Area of positive staining was reported as the percent tissue area that stained positive for succinate dehydrogenase, and staining intensity was reported as the total optical density of all positive pixels divided by the total tissue area.
Transmission Electron Microscopy
Transmission electron microscopy was performed using standard techniques. Gastrocnemius muscles were dissected unilaterally and placed in a modified Karnovski's fixative. Samples were routinely processed and were embedded in EMbed (Epon) 812 such to maintain a longitudinal orientation of the resulting sections. Thick sections were stained with toluidine blue to identify the best longitudinal block for each animal. Ultrathin sections cut for transmission electron microscopy survey were double-stained with uranyl acetate and lead citrate and examined using a FEI Tecnai G2 BioTwin electron microscope. Photomicrographs were captured using an Olympus-SIS Morada digital camera.
Exercise Protocols
C57Bl/6 mice (age 15 weeks) were trained for a period of 8 days by running on a treadmill (10 m/min, zero degree incline) for 2 h each day. Tissues were isolated immediately after the last bout of exercise training. To determine exercise capacity, 4-month old male mice were placed on a 6-lane treadmill (Columbus Instruments, Columbus, OH) and run with a fixed upward slope of 10°. The speed during the 1st h was 10 m/min and increased 2 m/min every 15 min thereafter. Work, peak VO2, and respiratory exchange ratios were assessed as described previously (17).
Data Analysis
Statistical analysis was performed using a two-tailed Student's t test or analysis of variance as appropriate and was considered statistically significant if p < 0.05. For analysis of microarray data, the Probe Logarithmic Intensity Error (22) method was used to normalize the raw data and generate the summarized probe-set level gene expression values. A moderated t test (23) was applied to identify the significantly altered genes using the “limma” package from Bioconductor. p values were adjusted using the BH (24) method to control for false discovery rate in multiple testing. Functional enrichment analyses were performed at DAVID (Data base for Annotation, Visualization and Integrated Discovery) (25).
RESULTS
Expression of ERRγ in Oxidative Skeletal Muscle
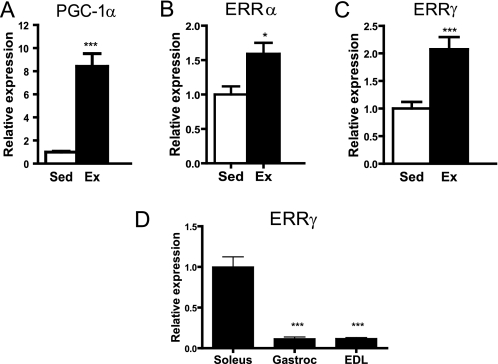
Exercise is a physiological intervention that results in the dynamic induction of mitochondrial biogenesis in skeletal muscle. We showed that both PGC-1α and ERRα were elevated with our exercise protocol (Fig. 1, A and B), as reported previously (26–28). Interestingly, ERRγ expression is induced by exercise (Fig. 1C). The expression of ERRγ transcript was found to correlate with the oxidative potential of skeletal muscle; it is expressed at high levels in the soleus (type I, oxidative muscle) and at low levels in the gastrocnemius (type I and II mixed fiber muscle) and extensor digitorum longus (type II, glycolytic muscle) (Fig. 1D).
FIGURE 1.
ERRγ expression in skeletal muscle is induced by exercise. Expression of PGC-1α (A), ERRα (B), and ERRγ (C) in mouse gastrocnemius muscle in sedentary (Sed) (n = 8) and exercised (Ex) (n = 8) mice. Mice were run on a treadmill for 2 h/day at 12–16 m/h for 8 days. D, expression of ERRγ is higher in soleus muscle as compared with gastrocnemius (Gastroc) or extensor digitorum longus (EDL) muscle in C57Bl/6 mice (n = 3 tissues per group). Data are presented as mean ± S.E. Statistical significance is indicated as follows: *, p < 0.05; ***, p < 0.005.
Expression of ERRγ in Primary Myotubes Results in an Up-regulation in Mitochondrial Function
Primary mouse myotubes were transduced with green fluorescent protein or GFP-IRES-ERRγ using two concentrations of adenovirus per treatment. The levels of ERRγ protein obtained are shown in supplemental Fig. 1A. Quantitative real time PCR demonstrated that induction of ERRγ in myotubes resulted in a down-regulation of ERRα and PGC-1α (supplemental Fig. 1B), suggesting the presence of a negative feedback mechanism to regulate levels of mitochondrial biogenesis. Additionally, genes of fatty acid metabolism, including carnitine palmitoyltransferase 1b (Cpt1b) and fatty acid-binding protein 3 (Fabp3), were also increased, whereas those of oxidative phosphorylation were mildly up-regulated (supplemental Fig. 1B). Interestingly, one of the most highly up-regulated genes is Epas1 (encoding endothelial PAS domain protein 1). Citrate synthase activity was increased with ERRγ expression (supplemental Fig. 1C). These data indicate that the expression of ERRγ results in a coordinate increase in mitochondrial function in primary mouse myotubes. Gene set enrichment analysis of microarray data of the ERRγ-transduced cells indicated a coordinated increase in genes of the mitochondrial electron transport chain, tricarboxylic acid cycle, and fatty acid oxidation (supplemental Fig. 1D) as reported previously in cardiac muscle (6). In addition, genes encoding pathways for muscle calcium handling and contractile proteins for slow muscle were also found to be up-regulated by ERRγ.
ERRγ Transgenic Mice Exhibit Skeletal Muscle Fiber-type Conversion and Enhance Exercise Capacity
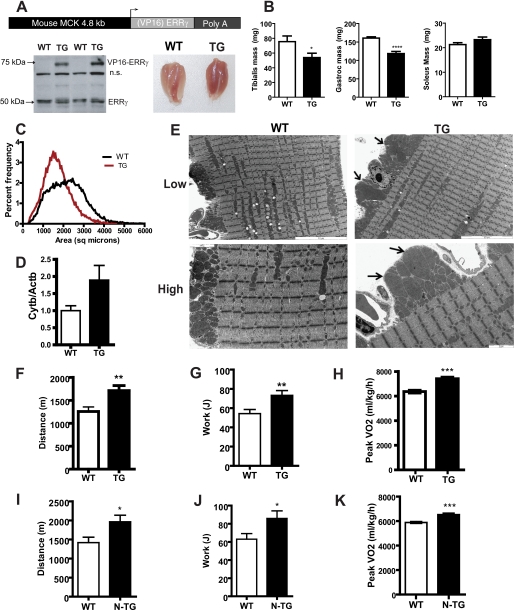
The muscle creatine kinase promoter was used to generate muscle-specific ERRγ transgenic mice (Fig. 2A, top panel). We analyzed two independent founder lines, one where the ERRγ was expressed as a fusion with the VP16 viral transactivator and the other expressing the “native” version of the protein. Expression of transcriptional factors as VP16 fusions is a strategy often utilized in vivo to ascertain effects elicited by increasing intrinsic activity of the factor beyond that achieved by enhanced protein levels alone (29, 30). Fig. 2A, lower left panel, demonstrates expression of the VP16ERRγ transgenic protein in founder line 431. The transgene was expressed at equal levels in gastrocnemius, soleus, and tibialis anterior muscle but was not expressed in the heart (supplemental Fig. 2A). We observed that VP1ERRγ transgenic muscles were noticeably redder and all hind limb muscles exhibited the same color as the soleus muscle (Fig. 2A, lower right panel). The animals had a similar body weight and body composition as compared with the control littermates (supplemental Fig. 2, B and C). The tibialis anterior (glycolytic type II fiber type muscle) and gastrocnemius (mixed fiber type muscle) muscles were smaller in the transgenics (Fig. 2B), although the mass of an oxidative type I fiber muscle such as soleus was not decreased. The transgenic line 339, expressing increased native ERRγ protein in muscle (supplemental Fig. 3A), exhibited a similar decrease in muscle weight of glycolytic and mixed fiber muscles but no change in soleus muscle, in the absence of any body weight changes (supplemental Fig. 3, B–E).
FIGURE 2.
Mice expressing ERRγ in skeletal muscle have “red” muscle, larger mitochondria, and improved oxidative capacity. A, schematic representing the structure of the transgene used to generate the ERRγ transgenic mice. The mouse muscle creatine kinase was used to drive the expression of native ERRγ or VP16ERRγ fusion protein. As shown in the lower left panel, in the VP16ERRγ transgenics, the protein was expressed in the skeletal muscle as observed by Western blot using ERRγ antibody. The endogenous ERRγ band at 50 kDa is indicated. The lower right panel shows a picture of the gastrocnemius-soleus muscle from the wild type (WT) and VP16ERRγ transgenic (TG) mice. n.s., not significant. B, individual muscle weights from the VP16ERRγ (TG) and WT mice (n = 12 mice per group). C, plot showing the size distribution of individual muscle fibers in VP16ERRγ transgenic (red) versus control littermate muscle (WT, black) (n = 5–6 mice per group). D, mitochondrial DNA content from gastrocnemius muscle from WT and VP16ERRγ TG mice (n = 8 mice per group). E, electron micrographs for gastrocnemius muscle from WT (left panels) and VP16ERRγ TG (right panels) mice. The upper panels represent a lower magnification (×5310), and the bottom panels are images taken at a higher magnification (×12,600). The scale for each image is embedded within the image. In the TG images, arrows point to enlarged mitochondria. Images shown are representative from n = 4 per genotype. Exercise capacity determined on a treadmill for the both VP16ERRγ (designated as TG, F–H) and ERRγ (designated as N-TG, I–K) transgenic mice along with their corresponding control WT mice. Data presented are distance run (F and I) and work performed (G and J) (n = 12 mice per group for VP16ERRγ strain and n = 10 for ERRγ strain). Peak oxidative capacity was assessed in WT and TG mice (H) and WT and N-TG mice (K), on a treadmill (n = 10 mice per group for both strains) under a peak VO2 protocol. Statistical significance is indicated as follows: *, p < 0.05; **, p < 0.01; ***, p < 0.005.
Individual fiber size was determined in the entire muscle cross-section by laminin staining. The fiber size distribution for the VP16ERRγ transgenic muscle was left-shifted, indicating a smaller muscle fiber size as evidenced by the histogram in Fig. 2C. Analysis of mitochondrial DNA levels in gastrocnemius muscle demonstrated a trend toward an increased content in the transgenic mice (Fig. 2D, p = 0.069). Electron microscopy of gastrocnemius muscle demonstrated an increase in numbers of large mitochondria, characterized by an enlargement of mitochondrial surface area in the transgenics (Fig. 2E). These large mitochondria were more pronounced in the subsarcolemmal region as opposed to the intramyofibrillar space (Fig. 2E, High, lower panels).
The VP16ERRγ transgenics did not show any alterations in energy expenditure, activity, or respiratory exchange ratios in the basal state (data not shown). When challenged with an endurance treadmill test, the VP16ERRγ transgenic mice demonstrated a significant increase in the work performed and distance traveled compared with the wild type controls (Fig. 2, F and G). In a peak VO2, the VP16ERRγ transgenic mice surpassed their control littermates (Fig. 2H). Interestingly, the respiratory exchange ratio values in the transgenic mice were lower during this exercise challenge, indicating a greater preference for fat oxidation (supplemental Fig. 2D). The respiratory exchange ratio values for the transgenics do not increase beyond a value of 1, implying that the transgenic mice are protected from the lactic acid-induced hyperventilation seen under maximal exercise. The non-VP16-tagged ERRγ transgenic mice also demonstrated an increase in distance run during an exercise capacity challenge and decreased peak oxidative capacity (Fig. 2, I–K).
Assessment of Mitochondrial Enzymatic Function and Gene Expression
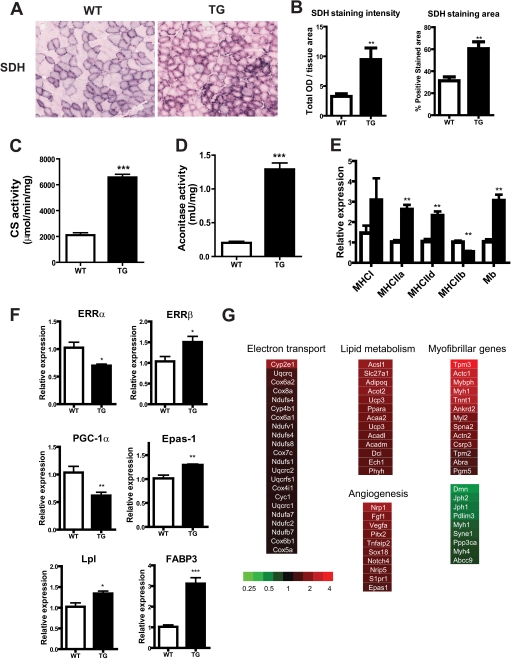
Analysis of succinate dehydrogenase enzyme activity in the muscle cross-sections demonstrated a robust increase in the succinate dehydrogenase positive area as well as intensity in the transgenic muscle (Fig. 3, A and B). Succinate dehydrogenase is situated in the mitochondrial inner membrane and is a component of the electron transport chain as well as the tricarboxylic acid cycle. Aconitase, like citrate synthase, is a mitochondrial matrix enzyme that is a component of the tricarboxylic acid cycle. The activities of both these enzymes were robustly elevated in the transgenic gastrocnemius muscle (Fig. 3, C and D). These data indicate that muscle mitochondrial activity in the ERRγ transgenic mice is strongly up-regulated.
FIGURE 3.
Transgenic VP16ERRγ mice have increased muscle mitochondrial function. A, representative images of succinate dehydrogenase (SDH) staining in tibialis muscle in littermate control (WT) (left panel) and VP16ERRγ transgenic (TG) (right panel) mice; B, staining intensity (left panel) and total staining area (right panel) of the muscle cross-sections (n = 5–6 mice per group). Enzymatic activities for citrate synthase (CS) (C) and aconitase (D) were determined in gastrocnemius muscle (n = 6–8 mice per group). Gene expression for MHCs (E) and metabolic and mitochondrial genes (F) (n = 6–8 mice per group) are shown. Data are presented as mean ± S.E. Statistical significance is indicated as follows: *, p < 0.05; **, p < 0.01; ***, p < 0.005. G, heat maps denoting individual genes and pathways that were altered in gastrocnemius muscle of TG mice relative to littermate control mice. The color scheme representing the fold change is shown.
Gastrocnemius muscle isolated from ERRγ transgenic mice exhibited decreased expression of MHC IIb and a concomitant increase in expression of MHC IId and MHC IIa and a trend toward an increase in slow MHC I, which did not reach statistical significance (Fig. 3E). This is indicative of a muscle fiber-type conversion from fast type II toward slow type I muscle fibers. Furthermore, we observed increased expression of myoglobin, a gene essential for the facilitated oxygen transport required by oxidative skeletal muscle (Fig. 3E). Candidate gene analysis shows decreases in Ppargc1a and Esrra but an increase in ERRβ, fatty acid oxidation genes (Lpl and Fabp3), and Epas1 (Fig. 3F). The down-regulation of Ppargc1a and Esrra suggests that there exist feedback regulatory loops for the control of mitochondrial function. Global gene expression was assessed in an unbiased fashion using microarrays, followed by gene set enrichment analysis. The top regulated pathways by their GO categories in the transgenic mice are described in Tables 1 and 2. Genes representing pathways of lipid metabolism, angiogenesis, and muscle calcium handling and contractility were robustly up-regulated in the transgenic muscle, whereas genes of the electron transport chain showed a smaller magnitude of up-regulation. Among the gene pathways that were down-regulated in transgenic muscle were genes representing fast twitch myosins and calcineurin Aα (Fig. 3G). Gene expression analyses using microarray in gastrocnemius muscle of the native ERRγ transgenics demonstrated similar patterns of changes in this muscle as in the VP16ERRγ muscle (supplemental Fig. 3F).
TABLE 1.
Over-represented GO terms for genes significantly up-regulated in VP16-ERRγ mice
Fold enrichment indicates the number of genes actually seen versus expected in the list given the distribution in the mouse genome of a specified GO term. p value indicates EASE score (p value of modified Fisher's exact test).
| GO category | Term | Fold enrichment | p value |
|---|---|---|---|
| Biological process | Angiogenesis | 3.47 | 3.98E-13 |
| Fatty acid metabolic process | 3.26 | 1.19E-12 | |
| Electron transport | 2.04 | 1.28E-09 | |
| Cell migration | 2.3 | 3.56E-09 | |
| Cellular compartments | Mitochondrion | 2.67 | 1.13E-47 |
| Actin cytoskeleton | 2.73 | 4.43E-11 | |
| Adherens junction | 4.02 | 7.11E-09 | |
| Basolateral plasma membrane | 3.71 | 2.01E-08 | |
| Contractile fiber | 3.61 | 3.48E-08 | |
| Myofibril | 3.6 | 7.31E-08 | |
| Mitochondrial lumen | 3.86 | 1.70E-07 | |
| Molecular function | Oxidoreductase activity | 1.98 | 7.52E-18 |
| Actin binding | 2.71 | 5.31E-13 | |
| FAD binding | 3.86 | 8.03E-8 | |
| Electron carrier activity | 2.32 | 3.78E-07 |
TABLE 2.
Over-represented GO terms for genes significantly down-regulated in VP16-ERRγ mice
Fold enrichment indicates the number of genes actually seen versus expected in the list given the distribution in the mouse genome of a specified GO term. p value indicates EASE score (p value of modified Fisher's exact test).
| GO category | Term | Fold enrichment | p value |
|---|---|---|---|
| Biological process | Post-translational protein modification | 2.05 | 8.85E-27 |
| Ubiquitin cycle | 2.81 | 7.78E-23 | |
| Protein amino acid phosphorylation | 1.67 | 3.73E-06 | |
| Chromatin modification | 2.44 | 3.99E-06 | |
| Molecular functions | Transcription factor binding | 2.51 | 1.53E-08 |
| Magnesium ion binding | 1.93 | 4.71E-06 | |
| Transforming growth factor β receptor activity | 8.39 | 6.20E-05 | |
| Cytoskeletal protein binding | 1.77 | 6.47E-05 | |
| Ubiquitin thiol esterase activity | 2.95 | 4.16E-04 | |
| Cellular compartments | Contractile fiber part | 3.56 | 3.52E-06 |
| Myofibril | 3.33 | 9.76E-06 | |
| Nucleoplasm part | 1.63 | 5.92E-05 | |
| Endoplasmic reticulum | 1.52 | 1.05E-04 | |
| Histone deacetylase complex | 4.82 | 1.24E-04 |
Gene Expression Profile in ERRγ Transgenic Mice Resembles That Observed in Type I Muscle
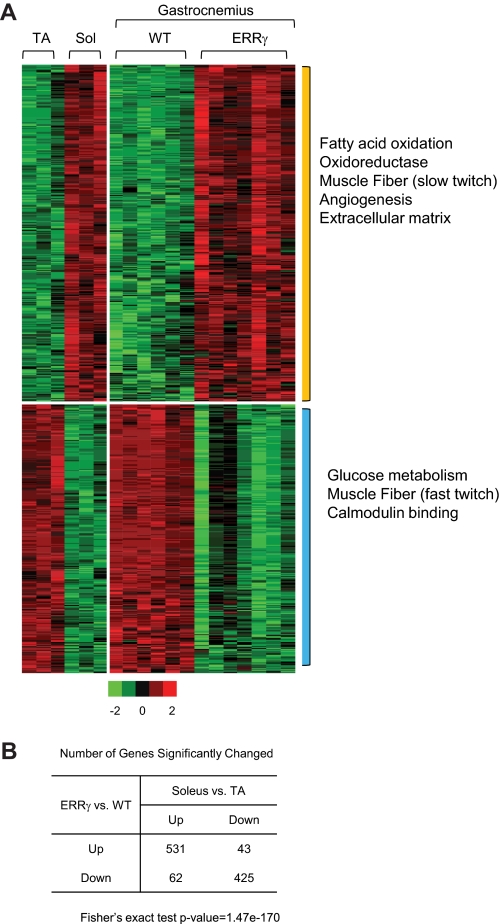
We analyzed our microarray data in context of the gene signatures observed in two distinct muscle fiber types (Fig. 4A, left panel) that were generated based on a published data set described for tibialis anterior muscle, a representative of type II fast twitch glycolytic muscle, and soleus muscle, which is a slow type I fiber oxidative muscle NCBI GEO accession number GSE10347) (31). Classically, the definition of the skeletal muscle fiber type is based on MHC profile distribution, as well as determination of ATPase activity (32). Interestingly, we observed that the majority of gene signature up-regulated in soleus muscle is also induced in the ERRγ transgenic mice; conversely, the gene pathways decreased in soleus but increased in the tibialis are decreased in the ERRγ transgenic muscle (Fig. 4A, right panel). Gene pathways over-represented in the soleus and transgenic muscle were those of fatty acid oxidation, oxidoreductases, muscle fiber type, angiogenesis, and extracellular matrix components. Gene pathways representing glucose metabolism, fast twitch muscle, and calmodulin binding were lower in soleus as compared with the tibialis muscle and were similarly lower in the transgenic muscle as compared with the wild type. Statistical analysis of this cross-comparison yielded a high a degree of significance (Fig. 4B). Thus, we can closely overlay the molecular genetic signature of ERRγ onto that of a pure oxidative muscle.
FIGURE 4.
Gene expression profile of ERRγ transgenic muscle is similar to that of type I oxidative muscle. A, heat map representing the coordinate gene expression pathways in tibialis anterior (TA) and soleus (Sol) (left panel) and wild type (WT) and VP16ERRγ transgenic (right panel) gastrocnemius muscle. Each individual column within the heat map represents a single mouse subject (n = 3 each for the soleus and tibialis anterior, n = 6 for WT and 7 for TG mice). The expression values are standardized within the data sets. Red color means up-regulation; green is down-regulation, and black is no change. The color scale is represented in Fig. 5A. The significantly over-represented GO terms by each group of genes is marked on the right. The p values used to determine significance for the individual genes are as follows: p < 0.01 for TG versus WT and p < 0.05 for soleus versus tibialis anterior. B, table represents the number of genes significantly changed in both datasets. The p value of Fisher's exact test is 1.47e-170 for the null hypothesis that the genes regulated by VP16ERRγ are unrelated to those differentially expressed between soleus and tibialis anterior (TA) muscle.
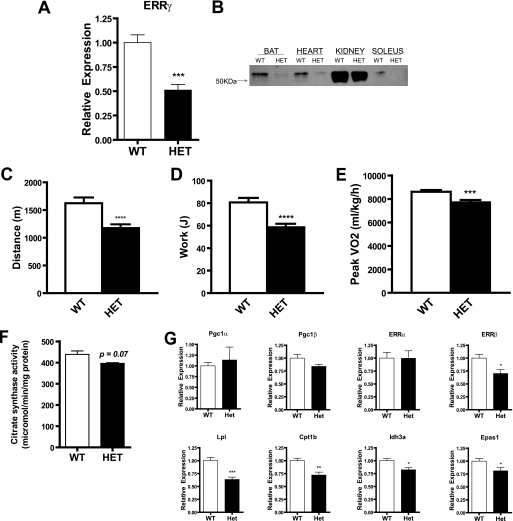
ERRγ Heterozygotes Exhibit Impairments in Muscle Function and Oxidative Capacity
Tissues from ERRγ heterozygous mice displayed a 50% reduction in ERRγ mRNA (Fig. 5A) and protein (Fig. 5B). No significant differences in muscle, heart, and total body weights were observed in these mice (data not shown). These mice did not show any alterations in energy expenditure, activity, or respiratory exchange ratios in the basal state (data not shown). The ERRγ heterozygotes ran a shorter distance and performed less work during an exercise capacity test (Fig. 5, C and D). These mice attained a reduced maximum speed (16.71 ± 0.24 versus 21.22 ± 0.29 m/min) compared with their wild type controls. Furthermore, ERRγ heterozygotes demonstrated a lower peak VO2 measurement (Fig. 5E). Citrate synthase levels trended to be lower, although the difference did not reach statistical significance (Fig. 5F). Gastrocnemius muscle demonstrated no changes in levels of Ppargc1a and Esrra to compensate for the partial loss of ERRγ in the heterozygous mice, although Esrrb levels were reduced. Additionally, fatty acid uptake and oxidation genes such as Lpl and Cpt1b were decreased in the muscle of these mice, indicating that these animals may have impairment in the utilization of fatty acids as fuel.
FIGURE 5.
ERRγ heterozygous mice have lower exercise capacity and impaired mitochondrial oxidative metabolism. A, ERRγ mRNAs are decreased in gastrocnemius muscle of ERRγ heterozygous (HET) mice as compared with controls (WT) (n = 4–5 mice per group). B, protein levels for ERRγ are decreased in brown adipose, heart, kidney, and muscle from ERRγ heterozygous mice and corresponding littermate control mice. Nuclear extract was used for protein analysis. Distance run (C) and work performed (D) by ERRγ heterozygous mice are compared with WT mice on a treadmill (n = 12 mice per group). E, peak oxidative capacity assessed in ERRγ heterozygous mice on a treadmill (n = 7–8 mice per group) under a peak VO2 protocol. F, citrate synthase activity in gastrocnemius muscle of ERRγ heterozygous mice. Mice were 12 weeks of age (n = 4–5 mice per group). G, mitochondrial gene expression in gastrocnemius muscle of ERRγ heterozygous mice. Mice utilized for these analyses were the same as in F above. Data are presented as mean ± S.E. Statistical significance is indicated as follows: *, p < 0.05; **, p < 0.01; ***, p < 0.005; ****, p < 0.001.
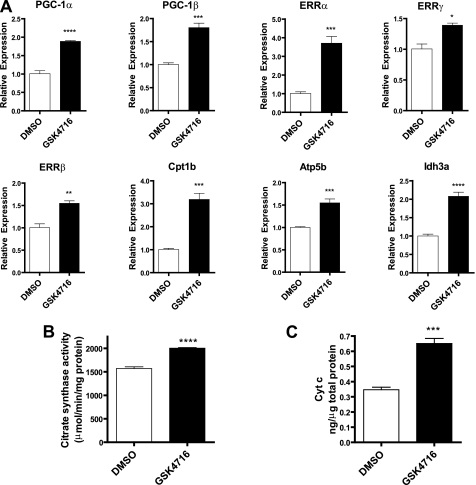
Effects of ERRβ/γ Agonist on Mitochondrial Gene Expression and Function in Primary Mouse Myotubes
Treatment of primary mouse myotubes with GSK4716, an ERRβ/γ agonist (9), resulted in a concerted increase in the expression levels of Ppargc1a, Ppargc1b, and the Esrr genes (Fig. 6A). Furthermore, Cpt1b, Atp5b, and Idh3, genes in key mitochondrial pathways, were also induced by GSK4716 (Fig. 6A). The concentration of the compound used in our study is similar to that observed for the activation of both ERRβ and ERRγ in the Gal4 transactivation assay (9). Additionally, GSK4716 increased citrate synthase activity (Fig. 6B) and cytochrome c protein levels (Fig. 6C). These results indicate that the activation of ERRβ/γ transcriptional activity with a small molecule agonist can produce a coordinate increase in mitochondrial gene expression and function.
FIGURE 6.
Primary mouse myotubes were treated with ERRβ/γ agonist, GSK4716 (10 μm), for 48 h. A, gene expression was determined using real time PCR. The levels of the test genes were expressed relative to the housekeeping gene, B2M, and for each sample normalized to the average signal for vehicle-treated cells. Citrate synthase activity (B) and cytochrome c (Cyt c) (C) protein levels were determined in cells treated with GSK4716. Data are expressed as mean ± S.E. (n = 3). Statistical significance is indicated as follows: *, p < 0.05; **, p < 0.01; ***, p < 0.005; ****, p < 0.001.
DISCUSSION
We demonstrate that ERRγ expression in skeletal muscle is sufficient and necessary to increase exercise capacity and activate mitochondrial function. In addition to its role in the regulation of mitochondrial and metabolic gene levels, ERRγ controls the expression of key genes controlling angiogenic, myofibrillar, and calcium handling pathways in skeletal muscle that are required for longer term adaptation to exercise. Thus, ERRγ emerges as a key regulator of the “slow, oxidative” muscle phenotype, serving as an integrator to ensure the concerted modulation of ultrastructural and metabolic transcriptional pathways that determine the identity of this muscle subtype.
Studies have shown impaired mitochondrial function via diminished ATP production in skeletal muscle of individuals with a family history of diabetes and in older insulin-resistant individuals (14). Additional studies have shown a decrease in mitochondrial number and function as a correlation of age (16), focusing the attention on improving mitochondrial function as a potential therapeutic modality for the treatment of age-related disease. Muscle from obese and type 2 diabetic subjects demonstrated a reduction in mitochondrial numbers as well as impairment in functional capacity, even after normalization for mitochondrial content (33). Exercise training results in a greater increase in mitochondrial activity in the subsarcolemmal mitochondrial fraction (34). PGC-1α levels are increased by exercise via up-regulation of AMP-activated protein kinase activity, which phosphorylates PGC-1α and increases its transcriptional activity (35). This mechanism allows for the modulation of coordinated gene networks through a feed forward loop in response to an increase in energy demand (36). We speculate that the increase in ERRγ levels by endurance-type exercise may occur through the induction of PGC-1α expression or via direct activation of ERRγ by AMP-activated protein kinase.
Epas1 (or hypoxia-inducible factor-2α (Hif-2α)), a transcription factor that is activated when oxygen demand is high under hypoxic conditions (37), is increased as an acute response to exercise, and is proposed to be a key mediator of exercise-induced gene expression (37). The Hif families of proteins are thought to be primarily regulated at the protein and not the transcriptional level (38). Levels of Wisp2, a Hif-2α-specific target gene (39), and pan-Hif-responsive genes are increased in transgenic muscle, although that of Hif-1α and its specific targets genes Pgk1 (phosphoglycerate kinase 1) and Ldha (lactate dehydrogenase A) (40) were unchanged (supplemental Fig. 3). Thus, it is predominantly the Hif-2α transcriptional program that is up-regulated by ERRγ. Hif-2α null mice have abnormal myofibrillar arrangement in skeletal muscle and demonstrate the presence of lipid droplets in muscle and heart, suggesting that the mitochondria may be dysfunctional in the absence of Hif-2α (41). Our data suggest that Hif-2α works as a downstream effector of the adaptive response in skeletal muscle during exercise-induced hypoxia. Interestingly, a recent publication described a supplementation of the Hif transcriptional response via a direct interaction of ERRs with Hifα/β heterodimers in human cancer cells (42).
PGC-1α regulates the expression of mitofusin-2 (Mfn2) in an ERRα-dependent manner (43), and the overexpression of PGC-1α in skeletal muscle causes an increase in the number of mitochondria (44). The levels of Mfn2 in skeletal muscle are decreased in obese individuals as compared with lean individuals (45). These data have led to the hypothesis that Mfn2 links mitochondrial dysfunction to the etiology of disease (43). Interestingly, through our unbiased gene expression profiling efforts, we discovered that the ERRγ transgenic muscle does not demonstrate an increase in Mfn2 and other genes of mitochondrial fusion/fission. This was further evidenced by the lack of an increase in the number of mitochondria observed by electron microscopy. Our data imply that the absence of increased mitochondrial fusion/fission does not negatively impact the energy production/functionality of the mitochondria at the organismal level. Furthermore, increased subsarcolemmal mitochondria as seen in the transgenics imply an increase in ATP supply for membrane activities, including substrate and ion transport and fatty acid oxidation (46).
Increased ERRγ expression in skeletal muscle results in a decrease in size of fast twitch muscle and a distinct shift toward slow fiber type and greater endurance. Unbiased gene expression profiling uncovered striking increases in pathways of slow type myosin heavy chain, tropomyosins, and cytoskeletal component proteins, which are activated by MEF2/histone deacetylase, key regulators of fiber type specification. Similar to our muscle creatine kinase-VP16ERRγ transgenic model, transgenic expression of activated MEF2 in muscle increases endurance capacity and increases expression of the slow contractile proteins (29). Nuclear factor of activated T-cells is a crucial partner of MEF2 in skeletal muscle, and its activity is controlled by calcineurin. Calcineurin signaling via nuclear factor of activated T-cells has been demonstrated to be essential for fiber-type switching from a fast/glycolytic phenotype to a slow/oxidative phenotype in skeletal muscle (47). Furthermore, the genetic ablation of the calcineurin inhibitor, calsarcin-2, in skeletal muscle results in a decrease in fast glycolytic muscle mass and an increase in endurance exercise in mice (48).
Given the role of ERRγ in the myocardial switch to oxidative metabolism, the effects on exercise capacity observed with ERRγ haploinsufficiency may be due to diminished cardiac function. We did not observe a decrease in the heart or muscle size in adult ERRγ heterozygous mice in contrast to what was observed in E18.5 embryos, indicating a postnatal compensation in these animals (5). Unlike the heart, we did not see a compensatory increase in ERRα or PGC-1α in skeletal muscle in the heterozygous mice. The study of tissue type-specific ERRγ null mouse models will contribute significantly toward the understanding of the role of ERRγ in cardiac versus skeletal muscle.
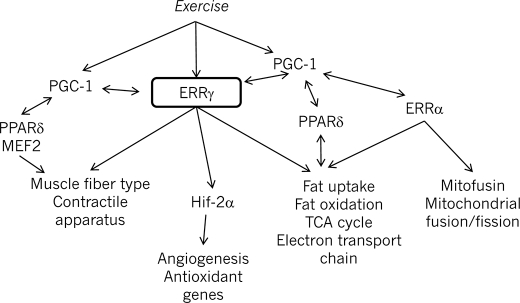
Because the phenotype of the transgenic ERRγ mice is strongly reminiscent of mouse models for peroxisome proliferator-activated receptor δ (30) or the MEF2/histone deacetylase/calcineurin pathway (29, 47), an important question that arises is whether these transcriptional factors have redundant roles in muscle. There are salient differences among these genetic models that point to a specific role for each protein in the control of muscle structure, function, and metabolism. Unlike the peroxisome proliferator-activated receptor δ transgenic models, we did not observe a resistance of our animals to a high fat diet, nor did they demonstrate an improvement in glucose tolerance.3 The MEF2 pathway controls expression of the slow muscle fiber genes but does not impact the mitochondrial or metabolic gene program. Our gene expression studies indicate that ERRγ plays a broader role in the control of biological processes such as angiogenesis and calcium handling in skeletal muscle. Thus, future studies will aim at comparing the gene expression patterns in these models directly to be able to tease apart the specific roles of these pathways in skeletal muscle. The schematic in Fig. 7 illustrates the complex transcriptional network regulating aspects of muscle gene expression.
FIGURE 7.
Schematic model displaying the central role for ERRγ in the transcriptional network regulating muscle fiber type and function.
Investigations into the molecular mechanism of ERRγ activation have shown that unlike other nuclear receptors ERRγ does not undergo rearrangement of its AF-2 helix upon agonist binding (49). Instead, it was observed that agonist binding increases thermal stability of the receptor, leading the authors to speculate that activation occurs via an increase in the cellular half-life of the protein. Thus, our studies with overexpression of the receptor provide a qualitative indication of the biological changes that would be elicited with an ERRγ agonist. It is noteworthy in this context that our results show that mice expressing the “activated” form of the receptor (VP16ERRγ) have a very similar phenotype compared with the native ERRγ-overexpressing strain. There may be a yet unknown endogenous agonist or antagonist ligand for ERRγ, in addition to common environmental contaminants that bind to the protein. Antagonistic agents would alter basal receptor activity and dampen downstream biological effects that may result in muscle or metabolic pathologies. From a therapeutic standpoint, we provide evidence that it is feasible to further activate ERRγ with a low molecular weight compound, despite its constitutive activity, and subsequently increase mitochondrial function in myocytes. Because of the lack of exposure in vivo, the use of GSK4716 for such investigations was not possible. The discovery of more potent and selective agonists with improved in vivo exposure will enable proof of concept studies linking ERRγ agonism to alleviation of disease symptoms. This will pave the path to the establishment of a novel mechanism in the arsenal against obesity and metabolic and muscle disease.
Supplementary Material
Acknowledgments
We acknowledge the contributions of Wilfried Frieauff for image analysis, Jolanta Dubauskaite and John Halupowski for generation and propagation of mouse strains, and Karen Killary and Beth Villarreal for electron microscopy. We thank Drs. Zhidan Wu, Chikwendu Ibebunjo, Sue Stevenson, and David Glass for discussions and comments on the manuscript.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4 and Table S1.
X. Wang and S. M. Rangwala, unpublished observations.
- ERR
- estrogen-related receptor
- Hif
- hypoxia-inducible factor
- MHC
- myosin heavy chain
- WT
- wild type
- TG
- transgenic.
REFERENCES
- 1.Giguère V. (2008) Endocr. Rev. 29, 677–696 [DOI] [PubMed] [Google Scholar]
- 2.Willy P. J., Murray I. R., Qian J., Busch B. B., Stevens W. C., Jr., Martin R., Mohan R., Zhou S., Ordentlich P., Wei P., Sapp D. W., Horlick R. A., Heyman R. A., Schulman I. G. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 8912–8917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Villena J. A., Hock M. B., Chang W. Y., Barcas J. E., Giguère V., Kralli A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 1418–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huss J. M., Imahashi K., Dufour C. R., Weinheimer C. J., Courtois M., Kovacs A., Giguère V., Murphy E., Kelly D. P. (2007) Cell Metab. 6, 25–37 [DOI] [PubMed] [Google Scholar]
- 5.Alaynick W. A., Kondo R. P., Xie W., He W., Dufour C. R., Downes M., Jonker J. W., Giles W., Naviaux R. K., Giguère V., Evans R. M. (2007) Cell Metab. 6, 13–24 [DOI] [PubMed] [Google Scholar]
- 6.Dufour C. R., Wilson B. J., Huss J. M., Kelly D. P., Alaynick W. A., Downes M., Evans R. M., Blanchette M., Giguère V. (2007) Cell Metab. 5, 345–356 [DOI] [PubMed] [Google Scholar]
- 7.Alaynick W. A., Way J. M., Wilson S. A., Benson W. G., Pei L., Downes M., Yu R., Jonker J. W., Holt J. A., Rajpal D. K., Li H., Stuart J., McPherson R., Remlinger K. S., Yi-Chang C., McDonnell D. P., Evans R. M., Billin A. N. (2010) Mol. Endocrinol. 24, 299–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greschik H., Wurtz J. M., Sanglier S., Bourguet W., van Dorsselaer A., Moras D., Renaud J. P. (2002) Mol. Cell 9, 303–313 [DOI] [PubMed] [Google Scholar]
- 9.Zuercher W. J., Gaillard S., Orband-Miller L. A., Chao E. Y., Shearer B. G., Jones D. G., Miller A. B., Collins J. L., McDonnell D. P., Willson T. M. (2005) J. Med. Chem. 48, 3107–3109 [DOI] [PubMed] [Google Scholar]
- 10.Coward P., Lee D., Hull M. V., Lehmann J. M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 8880–8884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takayanagi S., Tokunaga T., Liu X., Okada H., Matsushima A., Shimohigashi Y. (2006) Toxicol. Lett. 167, 95–105 [DOI] [PubMed] [Google Scholar]
- 12.Lang I. A., Galloway T. S., Scarlett A., Henley W. E., Depledge M., Wallace R. B., Melzer D. (2008) JAMA 300, 1303–1310 [DOI] [PubMed] [Google Scholar]
- 13.Kelley D. E., He J., Menshikova E. V., Ritov V. B. (2002) Diabetes 51, 2944–2950 [DOI] [PubMed] [Google Scholar]
- 14.Morino K., Petersen K. F., Shulman G. I. (2006) Diabetes 55, S9–S15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodpaster B. H., Katsiaras A., Kelley D. E. (2003) Diabetes 52, 2191–2197 [DOI] [PubMed] [Google Scholar]
- 16.Short K. R., Bigelow M. L., Kahl J., Singh R., Coenen-Schimke J., Raghavakaimal S., Nair K. S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 5618–5623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calvo J. A., Daniels T. G., Wang X., Paul A., Lin J., Spiegelman B. M., Stevenson S. C., Rangwala S. M. (2008) J. Appl. Physiol. 104, 1304–1312 [DOI] [PubMed] [Google Scholar]
- 18.Wende A. R., Schaeffer P. J., Parker G. J., Zechner C., Han D. H., Chen M. M., Hancock C. R., Lehman J. J., Huss J. M., McClain D. A., Holloszy J. O., Kelly D. P. (2007) J. Biol. Chem. 282, 36642–36651 [DOI] [PubMed] [Google Scholar]
- 19.Wang S. C., Myers S., Dooms C., Capon R., Muscat G. E. (2010) Mol. Cell. Endocrinol. 315, 146–152 [DOI] [PubMed] [Google Scholar]
- 20.Kleiner S., Nguyen-Tran V., Baré O., Huang X., Spiegelman B., Wu Z. (2009) J. Biol. Chem. 284, 18624–18633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiernan J. A. (2004) Histological & Histochemical Methods, pp. 319–320, 331–334, 3rd Ed., Pergamon Press Inc., Tarrytown, NY [Google Scholar]
- 22.Affymetrix (2008) PLIER Technote, http://www.affymetrix.com/support/technical/technotes/plier_technote.pdf
- 23.Smyth G. K. (2004) in Statistical Application in Genetics & Molecular Biology, pp. 1–25, Vol. 3, The Berkeley Electronic Press, Barkeley, CA [Google Scholar]
- 24.Benjamini Y., Hochberg Y. (1995) J. R. Stat. Soc. Ser. B 57, 289–300 [Google Scholar]
- 25.Dennis G., Jr., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. (2003) Genome Biol. 4, P3. [PubMed] [Google Scholar]
- 26.Baar K., Wende A. R., Jones T. E., Marison M., Nolte L. A., Chen M., Kelly D. P., Holloszy J. O. (2002) FASEB J. 16, 1879–1886 [DOI] [PubMed] [Google Scholar]
- 27.Cartoni R., Léger B., Hock M. B., Praz M., Crettenand A., Pich S., Ziltener J. L., Luthi F., Dériaz O., Zorzano A., Gobelet C., Kralli A., Russell A. P. (2005) J. Physiol. 567, 349–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pilegaard H., Saltin B., Neufer P. D. (2003) J. Physiol. 546, 851–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Potthoff M. J., Wu H., Arnold M. A., Shelton J. M., Backs J., McAnally J., Richardson J. A., Bassel-Duby R., Olson E. N. (2007) J. Clin. Invest. 117, 2459–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y. X., Zhang C. L., Yu R. T., Cho H. K., Nelson M. C., Bayuga-Ocampo C. R., Ham J., Kang H., Evans R. M. (2004) PLoS Biol. 2, e294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lavery G. G., Walker E. A., Turan N., Rogoff D., Ryder J. W., Shelton J. M., Richardson J. A., Falciani F., White P. C., Stewart P. M., Parker K. L., McMillan D. R. (2008) J. Biol. Chem. 283, 8453–8461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bassel-Duby R., Olson E. N. (2006) Annu. Rev. Biochem. 75, 19–37 [DOI] [PubMed] [Google Scholar]
- 33.Ritov V. B., Menshikova E. V., He J., Ferrell R. E., Goodpaster B. H., Kelley D. E. (2005) Diabetes 54, 8–14 [DOI] [PubMed] [Google Scholar]
- 34.Menshikova E. V., Ritov V. B., Fairfull L., Ferrell R. E., Kelley D. E., Goodpaster B. H. (2006) J. Gerontol. A Biol. Sci. Med. Sci. 61, 534–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jäger S., Handschin C., St-Pierre J., Spiegelman B. M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 12017–12022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cantó C., Auwerx J. (2009) Curr. Opin. Lipidol. 20, 98–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lundby C., Gassmann M., Pilegaard H. (2006) Eur. J. Appl. Physiol. 96, 363–369 [DOI] [PubMed] [Google Scholar]
- 38.Naranjo-Suárez S., Castellanos M. C., Alvarez-Tejado M., Vara A., Landázuri M. O., del Peso L. (2003) J. Biol. Chem. 278, 31895–31901 [DOI] [PubMed] [Google Scholar]
- 39.Aprelikova O., Wood M., Tackett S., Chandramouli G. V., Barrett J. C. (2006) Cancer Res. 66, 5641–5647 [DOI] [PubMed] [Google Scholar]
- 40.Hu C. J., Sataur A., Wang L., Chen H., Simon M. C. (2007) Mol. Biol. Cell 18, 4528–4542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scortegagna M., Ding K., Oktay Y., Gaur A., Thurmond F., Yan L. J., Marck B. T., Matsumoto A. M., Shelton J. M., Richardson J. A., Bennett M. J., Garcia J. A. (2003) Nat. Genet. 35, 331–340 [DOI] [PubMed] [Google Scholar]
- 42.Ao A., Wang H., Kamarajugadda S., Lu J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 7821–7826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soriano F. X., Liesa M., Bach D., Chan D. C., Palacín M., Zorzano A. (2006) Diabetes 55, 1783–1791 [DOI] [PubMed] [Google Scholar]
- 44.Wenz T., Diaz F., Spiegelman B. M., Moraes C. T. (2008) Cell Metab. 8, 249–256 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45.Bach D., Pich S., Soriano F. X., Vega N., Baumgartner B., Oriola J., Daugaard J. R., Lloberas J., Camps M., Zierath J. R., Rabasa-Lhoret R., Wallberg-Henriksson H., Laville M., Palacín M., Vidal H., Rivera F., Brand M., Zorzano A. (2003) J. Biol. Chem. 278, 17190–17197 [DOI] [PubMed] [Google Scholar]
- 46.Hood D. A. (2001) J. Appl. Physiol. 90, 1137–1157 [DOI] [PubMed] [Google Scholar]
- 47.Parsons S. A., Millay D. P., Wilkins B. J., Bueno O. F., Tsika G. L., Neilson J. R., Liberatore C. M., Yutzey K. E., Crabtree G. R., Tsika R. W., Molkentin J. D. (2004) J. Biol. Chem. 279, 26192–26200 [DOI] [PubMed] [Google Scholar]
- 48.Frey N., Frank D., Lippl S., Kuhn C., Kögler H., Barrientos T., Rohr C., Will R., Müller O. J., Weiler H., Bassel-Duby R., Katus H. A., Olson E. N. (2008) J. Clin. Invest. 118, 3598–3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang L., Zuercher W. J., Consler T. G., Lambert M. H., Miller A. B., Orband-Miller L. A., McKee D. D., Willson T. M., Nolte R. T. (2006) J. Biol. Chem. 281, 37773–37781 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.