Abstract
The protein kinase C (PKC) signaling pathway is a major regulator of cellular functions and is implicated in pathologies involving extracellular matrix remodeling. Inflammatory joint disease is characterized by excessive extracellular matrix catabolism, and here we assess the role of PKC in the induction of the collagenases, matrix metalloproteinase (MMP)-1 and MMP-13, in human chondrocytes by the potent cytokine stimulus interleukin-1 (IL-1) in combination with oncostatin M (OSM). IL-1 + OSM-stimulated collagenolysis and gelatinase activity were ameliorated by pharmacological PKC inhibition in bovine cartilage, as was collagenase gene induction in human chondrocytes. Small interfering RNA-mediated silencing of PKC gene expression showed that both novel (nPKCδ, nPKCη) and atypical (aPKCζ, aPKCι) isoforms were involved in collagenase induction by IL-1. However, MMP1 and MMP13 induction by IL-1 + OSM was inhibited only by aPKC silencing, suggesting that only atypical isoforms play a significant role in complex inflammatory milieus. Silencing of either aPKC led to diminished IL-1 + OSM-dependent extracellular signal-regulated kinase (ERK) and signal transducer and activator of transcription (STAT) 3 phosphorylation, and c-fos expression. STAT3 gene silencing or ERK pathway inhibition also resulted in loss of IL-1 + OSM-stimulated c-fos and collagenase expression. Silencing of c-fos and c-jun expression was sufficient to abrogate IL-1 + OSM-stimulated collagenase gene induction, and overexpression of both c-fos and c-jun was sufficient to drive transcription from the MMP1 promoter in the absence of a stimulus. Our data identify atypical PKC isozymes as STAT and ERK activators that mediate c-fos and collagenase expression during IL-1 + OSM synergy in human chondrocytes. aPKCs may constitute potential therapeutic targets for inflammatory joint diseases involving increased collagenase expression.
Keywords: Collagen, Cytokine Action, Extracellular Matrix, Fos, Gene Expression, Gene Silencing, MAP Kinases (MAPKs), Matrix Metalloproteinase, Protein Kinase C (PKC), STAT Transcription Factor
Introduction
Inflammation is a major characteristic of joint diseases such as rheumatoid arthritis and osteoarthritis (OA),3 during which inflammatory mediators released by infiltrating immune cells as well as resident joint cells induce alterations in gene expression that can lead to ECM degradation (1, 2). Cytokines such as interleukin (IL-)1, IL-17, and tumor necrosis factor α (TNFα) are key mediators thought to be involved in promoting inflammatory responses in such destructive joint diseases (3, 4). We have shown that the IL-6 family cytokines oncostatin M (OSM) and IL-6 markedly exacerbate the catabolic potential of these mediators, synergistically promoting cartilage ECM catabolism both in vitro and in vivo (5–9).
Chondrocytes are the only resident cell type in normal articular cartilage and function to preserve homeostasis. This is achieved by balancing the expression of ECM components with catabolic factors such as the matrix metalloproteinases (MMPs), which collectively can degrade all the ECM macromolecules. During inflammatory joint diseases, chondrocytes are stimulated to secrete elevated levels of MMPs that, once activated, mediate the proteolysis of tendon, bone, and cartilage (10, 11). MMP-1 and MMP-13 are collagenolytic MMPs that have been most strongly associated with cartilage collagenolysis, a key proteolytic event in inflammatory joint diseases because it is essentially irreversible (12). A marked synergistic induction of collagenase gene expression occurs in human chondrocytes following IL-1 + OSM stimulation (8, 13, 14), and we proposed this to be via interplay of signal transduction pathways whereby signal transducers and activators of transcription (STAT), c-fos, and alterations in activator protein-1 (AP-1) composition are important (15). MMP gene expression is certainly reliant on transcription factors such as erythroblastosis 26 and nuclear factor-κB (NFκB) as well as AP-1 (16), and we have previously demonstrated the importance of the phosphatidylinositol 3′-kinase (PI3K) pathway in this process (17). However, relatively little is still known about the signaling mechanisms upstream of transcriptional activation, and in particular about points of cross-talk that occur during cytokine synergy.
Protein kinase C (PKC) comprises a family of serine/threonine protein kinases with key roles in fundamental cellular processes such as cell survival and apoptosis, proliferation and migration, as well as malignant transformation (18). The PKC family is divided into groups according to their domain structure, which determines their requirement for allosteric activators. Conventional PKC (cPKC) isoforms, designated α, βI, βII, and γ, require Ca2+ for activation, as well as diacylglycerol (DAG) and the lipid phosphatidylserine. Novel PKC (nPKC) isoforms (δ, ϵ, η, θ) lack a Ca2+ binding domain, but still require DAG and phosphatidylserine. Finally, atypical PKC (aPKC) isoforms (ζ, ι) lack the ability to bind DAG but still require a lipid cofactor. PKC isozymes are subject to “priming” phosphorylation by phosphoinositide-dependent kinase-1, stabilizing them and facilitating subsequent allosteric activation (18), such that PKC is influenced by the PI3K pathway.
The PKC pathway is a major drug target for cancer therapy (19, 20) because some isoforms are important in survival/proapoptotic pathways. Altered PKC expression is observed in many tumors, and there is a large body of evidence linking PKC to invasion involving MMPs including MMP-9 (21, 22). PKC activity, in particular that of nPKCθ, is of great interest with regard to T cell activation and consequently inflammatory diseases such as asthma, inflammatory bowel disease, and multiple sclerosis (23). PKC inhibitors are being trialled (24) as drugs for psoriasis and lymphoma (25–28).
Some associations between PKC and arthritis have been described. The δ, ϵ, η, and θ isoforms of PKC are all activated in Lyme disease (29). PKC activity is necessary for neutrophil activation in gouty arthritis (30), and linked to OA for over a decade (31–33). However, only recently has isoform-specific information, essential for informing future therapeutic design, begun to emerge. aPKCζ appears to regulate both NFκB and Janus kinase (JAK)/STAT pathways in inflammation (34). aPKCζ is reported to be required for NFκB activation in chondrocytes, is up-regulated in OA (35), and its inhibition reduces aggrecanase-1 expression (36). nPKCδ has been implicated in cross-talk with mitogen-activated protein kinases (MAPKs) for MMP13 expression (37). In other systems, PKC enhances extracellular signal-regulated kinase (ERK) signaling via phosphorylation of the Raf kinase inhibitor protein (RKIP) (38, 39). nPKCδ stimulates transcriptional activity of STAT1, another target for cancer therapy (40) via Ser-727 phosphorylation (41). Therefore, PKC can influence multiple signaling pathways that are important in disease processes involving dysregulated MMP expression. However, little is still known regarding the role of specific PKC isoforms in collagenase gene expression during cartilage ECM destruction in rheumatoid arthritis or OA.
In this study we assess the role of PKC activity in cartilage degradation stimulated by IL-1 + OSM, and investigate the role of individual PKC isoforms in collagenase expression in human chondrocytes. We show that aPKC isoforms play a major role in this process, via STAT3 and ERK activation, and subsequent expression of c-fos.
EXPERIMENTAL PROCEDURES
Materials
All chemicals were obtained from Sigma unless otherwise stated and of the highest purity available. All cytokines were recombinant human. IL-1α was a generous gift from Dr. Keith Ray (GlaxoSmithKline, Stevenage, UK). OSM was prepared in-house using expression vectors kindly provided by Prof. J. Heath (University of Birmingham, UK) and methods described in Ref. 42. Gö6976, Gö6983, and calphostin C were from Merck Chemicals (Nottingham, UK). Kinase inhibitors and small interfering RNA (siRNA) reagents were screened for toxicity using the Toxilight assay of adenylate kinase release (Lonza, Wokingham, UK), and always used at concentrations that did not affect cell viability over the assay period. Expression vectors for c-fos (43) and c-jun were derived from pCMV2 and were generously provided by Dr. I. Verma (Salk Institute for Biological Studies, San Diego, CA) and Prof. Paul Dobner (University of Massachusetts Medical School, Worcester, MA), respectively.
Chondrocytes
Human chondrocytes were obtained by enzymatic digestion of macroscopically normal articular cartilage from OA patients undergoing joint replacement surgery as described (44). All subjects gave informed consent and the study was approved by the Newcastle and North Tyneside Joint Ethics Committee. Bovine cartilage was dissected from nasal septi obtained from a local abattoir as described (45). T/C28a4 immortalized human chondrocytes (46) were used in some experiments as indicated. Chondrocytes were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 IU penicillin, 100 μg/ml of streptomycin, 40 units/ml of nystatin.
Cartilage Degradation Assay
Bovine nasal septum cartilage discs were incubated in serum-free medium for 14 days in the presence of IL-1 ± OSM (± inhibitors), changing medium after 7 days as previously described (45). The cartilage remaining at day 14 was digested with papain (45). Lack of toxicity for treatments including inhibitors was confirmed by the Toxilight assay.
Collagen, Collagenase, and Gelatinase Assays
Hydroxyproline measurements (47) were used as an estimate of cartilage collagen, and the cumulative release was calculated and expressed as a percentage of the total for each well (48). Collagenolytic activity present in the culture medium from cartilage explants was determined using a diffuse fibril assay with 3H-acetylated collagen (49). One unit of collagenase activity degrades 1 μg of collagen per min at 37 °C. Gelatin zymography was performed as described previously (50).
Cell Fractionation and Immunoblotting
Chondrocyte lysates were prepared as described previously (17). In some experiments chondrocytes were subjected to subcellular fractionation using the NE-PER Nuclear and Cytoplasmic Protein Extraction Kit or Subcellular Protein Fractionation Kit (both from ThermoFisher Scientific, Loughborough, UK). Lysates or fractions were resolved by SDS-PAGE, transferred to polyvinylidene difluoride membranes, and subsequently probed using the following antibodies: PKCζ, PKCι, PKCζ/ι*T410/403, ERK1/2, ERK1/2*T202/Y204, phospho(Ser)-PKC substrate, STAT1, STAT1*Y701, STAT3, STAT3*Y705, STAT3*S727, PKD*S916, lamin A/C, caveolin-1, and Histone H3 were from Cell Signaling Technology (Danvers, MA); RKIP*S153, β-tubulin, PKCδ, and PKCδ*T507 were from Epitomics (Insight Biotechnology Ltd., Middlesex, UK); Fos and Jun were from Santa Cruz Biotechnology (Santa Cruz, CA); MARCKS and MARCKS*S158 were from GenScript USA Inc. (Piscataway, NJ), PKCη*T655 was from Millipore (Watford, UK).
Immunoassays
Primary human articular chondrocytes were stimulated with IL-1 ± OSM (± inhibitors preincubated with the cells for 1 h) for 24 h in serum-free conditions (9). The medium was removed and assayed by specific immunoassay for MMP-1 (51) and MMP-13 (52). The MMP assays detected both the pro- and active forms.
siRNA-mediated Gene Silencing
Primary human chondrocytes were prepared and cultured as above, and transfected as described previously (17). As an initial screen for modulation of MMP expression, siRNA reagents (3 duplexes per gene, pooled) from the MISSIONTM siRNA Human Kinase Panel (Sigma) were used. Dharmacon ON-TARGET plusTM SMARTpools® (ThermoFisher Scientific) of 4 specific siRNA duplexes (total of 100 nm siRNA) were used for follow-up studies where appropriate. Supplemental Table S1 details the target or siRNA sequences used. After transfection, cells were serum-starved for 24 h then stimulated for 45 min to measure c-fos and c-jun, or 24 h for MMP mRNAs. Depletion of gene-specific mRNA levels was calculated by comparison of expression levels with cells transfected with 100 nm siCONTROL (non-targeting siRNA 2, catalog number 001210-02; Dharmacon).
Real Time PCR of Relative mRNA Levels
RNA was stabilized in cell lysates in a 96-well format, cDNA synthesized and real time PCR assays were conducted using conditions described previously (17). Primer and probe sequences used are detailed in supplemental Table S2.
Transient Transfection and Reporter Gene Assay
The methods for transient transfection of T/C28a4 chondrocytes were modified from those described previously (53, 54) as detailed in Ref. 15. Transfected cells were stimulated overnight prior to determination of luciferase activity (Promega) using a 1450 Microbeta TriLux counter (Wallac, PerkinElmer, Milan, Italy). Luminescence values were normalized to the amount of plasmid per well, as determined by the Hirt assay (55). In co-transfection experiments, equal amounts of plasmid were used with the appropriate control vector.
Statistical Analyses
Statistical differences between sample groups were assessed using one-way analysis of variance with a post hoc Bonferroni's multiple comparison test or Student's two-tailed unpaired t test (1 sample t test for immunoblot quantification), where ***, p < 0.001; **, p < 0.01; *, p < 0.05. For clarity, only selected comparisons are presented in some figures.
RESULTS
Inhibition of PKC Activity Ameliorates IL-1 + OSM-stimulated Cartilage Degradation, Collagenase Activity, and Gelatinase Activity
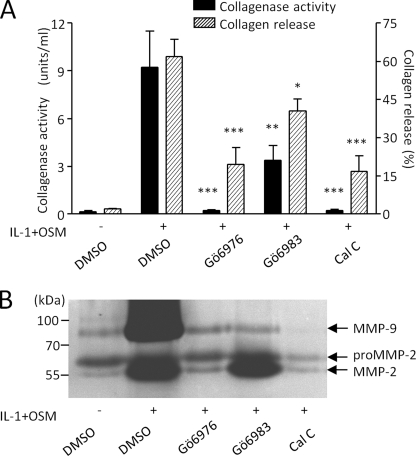
Because PKC has previously been reported to play a role in MMP expression in chondrocytes, we sought to assess whether this was the case for bovine cartilage stimulated using the potent synergistic cytokine combination, IL-1 + OSM. Many kinase inhibitors, including PKC inhibitors, have been shown to lack specificity (56). For this reason, we used inhibitors with relatively few reported selectivity issues, and with distinct mechanisms. Gö6976 and Gö6983 are related ATP-competitive indolylmaleimides. Calphostin C (Cal C) was also used, which acts via a DAG-competitive mechanism (57). However, it cannot be excluded that Cal C may act to inhibit DAG effectors other than PKC.
Fig. 1A shows that Gö6976 (10 μm), Gö6983 (20 μm), and Cal C (2 μm) all significantly inhibited IL-1 + OSM-stimulated collagen release from bovine cartilage. Furthermore, the levels of collagenase activity assayed in medium from these cultures were markedly inhibited (Fig. 1A). The much greater effect of PKC inhibition against collagenase activity compared with collagen release reflects our previous findings that residual collagenase activity exceeding 1 unit/ml is sufficient to catalyze efficient cartilage collagenolysis (58).
FIGURE 1.
PKC inhibition ameliorates IL-1 + OSM-stimulated cartilage degradation. Bovine nasal cartilage explant cultures were stimulated with IL-1 (1 ng/ml) and OSM (10 ng/ml) for 14 days, with fresh medium and cytokines at day 7, in the presence of dimethyl sulfoxide (DMSO) vehicle control or PKC inhibitors Gö6976 (10 μm), Gö6983 (20 μm), or calphostin C (2 μm). Medium was assayed for (A) collagen release and collagenase activity, or (B) gelatinase activity. Cumulative collagen released by day 14 was expressed as a percentage of the total for each treatment, and active collagenolytic activity in the day 14 culture supernatants was determined by bioassay (mean ± S.E.; ***, p < 0.001; **, p < 0.01; *, p < 0.05 compared with IL-1+OSM control). Data were pooled from 3 chondrocyte populations, each conducted in quadruplicate (A), or representative of two separate experiments (B).
We next used gelatin zymography to examine the effect of PKC inhibition on gelatinase activity, which may be viewed as the subsequent step to collagenase activity in the breakdown of cartilage collagen. Fig. 1B shows that PKC inhibition results in marked reduction of IL-1 + OSM-stimulated gelatinase activity in cartilage-conditioned medium, particularly that of the inducible gelatinase, MMP-9.
Effect of PKC Activity on IL-1 + OSM-dependent Collagenase Expression in Human Articular Chondrocytes
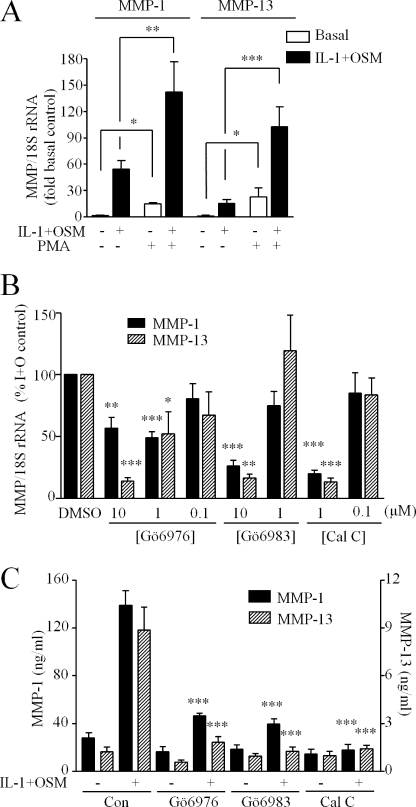
We next determined whether our findings could be extrapolated to human chondrocytes. First, we confirmed that the classical PKC activator phorbol 12-myristate 13-acetate markedly enhanced both basal and IL-1 + OSM-stimulated expression of both MMP1 and MMP13 genes in human chondrocytes (Fig. 2A). Preincubation of cells with each of the PKC inhibitors abrogated IL-1 + OSM-stimulated expression of both MMP1 and MMP13 in a concentration-dependent manner (Fig. 2B). Collagenase gene expression in response to a stimulus of IL-1 alone was similarly sensitive to the presence of PKC inhibitors (data not shown). The less selective compounds bisindolyl maleimide I, Ro31-8220, and chelerythrine had similar effects (not shown). The DAG kinase inhibitor, R59-022, had no effect on cytokine-stimulated expression of either MMP1 or MMP13 (data not shown), excluding a role for this major class of alternative, non-PKC DAG effectors.
FIGURE 2.
Modulation of PKC activity regulates collagenase expression in human chondrocytes. A, primary human articular chondrocytes were stimulated with IL-1 (0.02 ng/ml) + OSM (10 ng/ml) for 24 h ± 1 h preincubation with phorbol 12-myristate 13-acetate (100 nm). Cell lysates were subjected to reverse transcriptase-PCR (“Experimental Procedures”) and relative expression levels of MMP1 and MMP13 mRNA, normalized to the 18 S rRNA housekeeping gene, were determined. B, chondrocytes were preincubated (1 h) with concentrations of PKC inhibitors indicated prior to stimulation with IL-1 (0.02 ng/ml) + OSM (10 ng/ml) for 24 h and determination of MMP1 and MMP13 relative expression levels. C, chondrocytes were stimulated with IL-1 (0.02 ng/ml) ± OSM (10 ng/ml) for 24 h ± preincubation with PKC inhibitors at the concentrations indicated. Culture medium was assessed for MMP-1 and MMP-13 using specific immunoassays. Data (mean ± S.E.; ***, p < 0.001; **, p < 0.01; *, p < 0.05) were compared with dimethyl sulfoxide (DMSO) vehicle control (A), or control stimulated values (B and C). Data were pooled from three separate chondrocyte populations, each conducted in hextuplicate.
We then assessed whether the observed abrogation of collagenase gene induction by PKC inhibition in human chondrocytes was reflected in the amount of collagenase secreted into the culture medium by the cells. Measurement of MMP-1 and MMP-13 by enzyme-linked immunosorbent assay confirmed that PKC inhibition caused a marked reduction in IL-1 + OSM-stimulated collagenase production and secretion from chondrocytes (Fig. 2C).
IL-1 + OSM Enhances PKC Priming and Substrate Phosphorylation
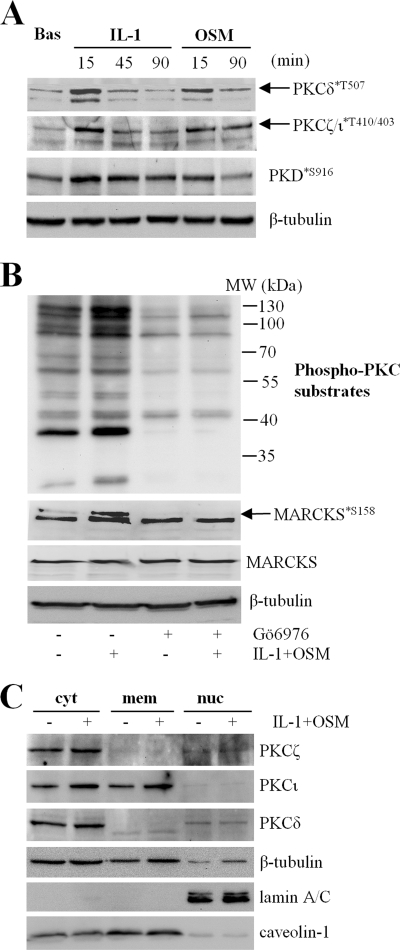
To assess whether PKC was actively stimulated by IL-1 + OSM, we next examined if stimulation of chondrocytes with these proinflammatory cytokines could enhance priming phosphorylation of PKC isoforms, which allows activation by allosteric factors and prevents ubiquitin-mediated degradation (59). nPKCδ and aPKC isoforms were analyzed because these have previously been investigated in relation to MMP expression (35–37). Fig. 3A shows that stimulation of chondrocytes with either IL-1 or OSM produced an increase in phosphorylation of PKCδ at Ser-507, and on atypical PKC isoforms (not distinguished) on residues Thr-410 (ζ) and −403 (ι), suggesting that both cytokines may play a role in PKC regulation.
FIGURE 3.
IL-1 + OSM stimulation enhances PKC priming and substrate phosphorylation. A and B, primary human articular chondrocytes were stimulated with IL-1 (0.2 ng/ml) or OSM (10 ng/ml) for the times indicated (A) or with IL-1 + OSM for 20 min ± preincubation with Gö6976 (B). Cell lysates were prepared and subjected to SDS-PAGE and immunoblotting (“Experimental Procedures”) using the antibodies indicated. Ser(P)-158 MARCKS is illustrated by the upper arrowed band. C, chondrocytes were stimulated with IL-1 (0.2 ng/ml) or OSM (10 ng/ml) for 20 min, harvested, and subjected to subcellular fractionation (“Experimental Procedures”). Fractions were resolved by SDS-PAGE and analyzed by immunoblotting using the antibodies indicated. Immunoblots shown are representative of three separate experiments each using chondrocyte cultures from different donors. Bas, basal; cyt, cytoplasm; mem, membrane; nuc, nucleus.
An alternative assessment of PKC activity is to examine phosphorylation of known PKC substrates. IL-1 in particular, and to a lesser extent OSM, increased protein kinase D (PKD) phosphorylation on Ser-916 (Fig. 3A). We also utilized an antibody that recognizes PKC-specific substrate motifs only containing phosphoserine. Multiple protein bands were enhanced following IL-1 + OSM stimulation of human chondrocytes, and were reduced in intensity in lysates from Gö6976-treated cells (Fig. 3B). Similarly, phosphorylation of myristoylated alanine-rich protein kinase C substrate (MARCKS), a commonly used surrogate marker of PKC activity, was enhanced by IL-1 + OSM in a Gö6976-sensitive manner (Fig. 3B, upper arrowed band). We also investigated the subcellular distribution of PKC isoforms, and observed enrichment of PKCι in the membrane fraction, and modest enrichment of each PKC isoform in the cytosol in most but not all cultures after IL-1 + OSM stimulation (Fig. 3C).
Effect of PKC Isoform Gene Silencing on IL-1- and IL-1 + OSM-stimulated Collagenase Induction
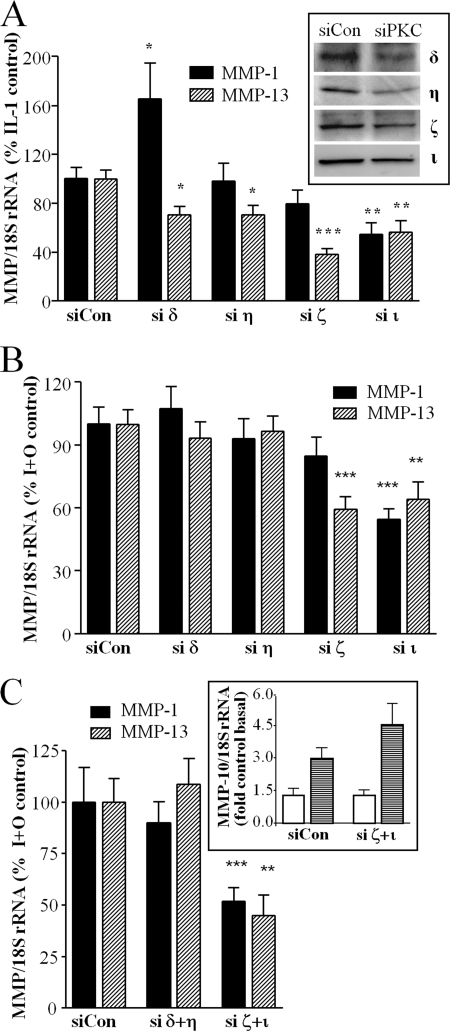
The data presented suggest strongly that IL-1 + OSM-stimulated collagenase expression is dependent on PKC activity. However, there are no reliable isoform-specific PKC inhibitors widely available (e.g. use of rottlerin as a PKCδ-specific inhibitor is now regarded as highly questionable (60)). To elucidate a role for specific PKC isoforms in collagenase gene expression, we used siRNAs to systematically assess all PKC isoforms for their ability to contribute to IL-1 + OSM-dependent gene expression. In the first instance we utilized siRNA pools from the MISSIONTM siRNA Human Kinase Panel and, based on these preliminary data, four PKC isoforms (δ, η, ζ, ι) were selected for further study. siRNA specific to other isoforms had no significant effect on collagenase expression (not shown).
Fig. 4A (inset) illustrates immunoblots to confirm siRNA for each PKC isoform was effective in reducing the abundance of the appropriate kinase in cell lysates, although the protein depletion achieved was incomplete. In addition, we confirmed each reagent effectively and specifically (compared with levels of other PKC isoform mRNA) silenced only the target PKC gene (not shown). After gene silencing, cell cultures were stimulated with IL-1, either alone or in combination with OSM. For IL-1 alone, MMP1 induction was significantly enhanced by silencing of nPKCδ, inhibited by silencing of aPKCι, whereas nPKCη and aPKCζ had no effect. In contrast, MMP13 induction was significantly inhibited by nPKCδ, nPKCη, aPKCζ, and aPKCι, with the aPKC isoforms having a more potent inhibitory effect (Fig. 4A). In contrast, when the combined stimulus (IL-1 + OSM) was used to synergistically induce collagenase expression, a different pattern emerged. In this case, silencing of neither nPKC isoform resulted in any significant effect. However, aPKCι silencing significantly decreased MMP1 induction, whereas silencing of either aPKCζ or aPKCι reduced MMP13 induction (Fig. 4B).
FIGURE 4.
PKC isoform silencing regulates collagenase expression in human chondrocytes. A and B, following transfection with siRNA specific to PKC isoforms nPKCδ, nPKCη, aPKCζ, aPKCι, or non-targeting siControl (100 nm), chondrocytes were stimulated with (A) IL-1 (0.02 ng/ml) or (B) IL-1 and OSM (10 ng/ml) for 24 h. Cells were lysed and real time reverse transcription-PCR was performed for MMP1 and MMP13, 72 h after transfection. Protein depletion of individual, specific PKC isoforms was confirmed by SDS-PAGE and immunoblotting (A, inset). In some experiments (C), chondrocytes were transfected with siRNA combined (100 nm total) targeting either nPKC (δ+η) or aPKC (ζ+ι) isoforms, prior to stimulation with IL-1 + OSM, and expression of MMP10 mRNA was measured (inset, open bars, basal; striped bars, stimulated) as well as MMP1 and MMP13. Data are pooled from at least three separate chondrocyte populations (each assayed in hextuplicate) presented as a percentage of the cytokine-induced expression (specific siPKC transfected versus siCon-transfected; mean ± S.E.; ***, p < 0.001; **, p < 0.01; *, p < 0.05).
We then further characterized the PKC subclass dependence of IL-1 + OSM-stimulated collagenase induction by silencing both nPKC isoforms and both aPKC isoforms in combination (Fig. 4C). Silencing of both nPKCδ and nPKCη together still had no effect on induction of either MMP1 or MMP13 by IL-1 + OSM, whereas combination silencing of both aPKCζ and aPKCι appeared to have an increased inhibitory effect on collagenase expression compared with silencing either aPKC alone (Fig. 4C). Silencing of both aPKC isoforms did not significantly affect synergistic induction of MMP10 (stromelysin-2) by IL-1 + OSM (13) (Fig. 4C, inset), indicating that aPKC activity is not a pre-requisite for MMP expression.
Effect of aPKC Isoform Knockdown on IL-1 + OSM-stimulated Signaling Events
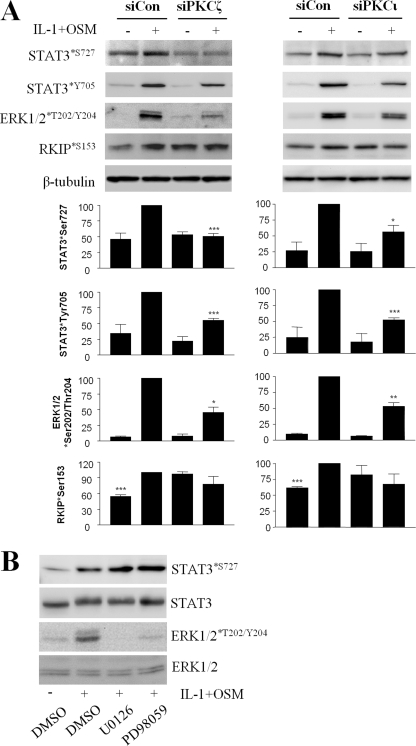
We next sought to elucidate the mechanism(s) by which loss of aPKCζ or aPKCι function might abrogate collagenase gene induction in chondrocytes. After transfection with the appropriate siRNA, we prepared cell lysates to examine IL-1 + OSM-stimulated ERK and STAT3 phosphorylation because these activities have been associated with both PKC activity (34, 37) and MMP gene regulation (15, 37). We found that silencing of either aPKCζ or aPKCι resulted in diminution of ERK1/2 phosphorylation. Silencing of expression of either aPKC also resulted in loss of STAT3 phosphorylation on both tyrosine and serine regulatory sites (Fig. 5A). STAT1 phosphorylation was also abrogated by aPKC silencing (not shown). Activation of the NFκB pathway was analyzed by IκBα degradation, because this is associated with PKC activity and MMP expression (36), but was not inhibited by silencing of either aPKC (data not shown).
FIGURE 5.
Atypical PKC gene silencing curtails IL-1 + OSM-stimulated STAT3 and ERK phosphorylation. A, primary human articular chondrocytes were transfected with siRNA (100 nm) specific to aPKCζ, aPKCι, or siCon non-targeting control, prior to stimulation with IL-1 (0.2 ng/ml) and OSM (10 ng/ml) for 20 min. Cells were lysed and proteins resolved by SDS-PAGE. Immunoblots were probed using the antibodies indicated. Blots shown are representative of three to six experiments using chondrocytes from different donors, and histograms show quantification of bands normalized to β-tubulin levels presented as a percentage of the cytokine-induced expression (siCon-transfected) (mean ± S.E.; ***, p < 0.001, **, p < 0.01; *, p < 0.05). Comparisons shown are stimulated (siCon versus siPKC) or siCon-stimulated versus all conditions for RKIP*S153. B, chondrocytes were incubated with MEK inhibitors UO126 or PD98059 (both 10 μm), or DMSO vehicle for 1 h prior to stimulation with IL-1 (0.02 ng/ml) and OSM (10 ng/ml). Cell lysates were prepared and proteins resolved by SDS-PAGE, and immunoblots were probed using the indicated antibodies. Blots shown are representative of three experiments using chondrocytes from different donors.
Because serine phosphorylation of STATs has been associated with PKC (41, 61), but also ERK activity (62), we assessed whether STAT3 phosphorylation might be dependent on ERK via PKC activity. This could occur via phosphorylation of RKIP by aPKC, therefore impinging on ERK activity at the MAPK kinase kinase level (39). Indeed, phosphorylation of RKIP on Ser-153 was increased by IL-1 + OSM stimulation of chondrocytes. When either aPKC gene was silenced, however, no stimulation was observed, suggesting that this may be the mechanism by which aPKC enhanced ERK activation (Fig. 5A), despite elevated basal phosphorylation of RKIP. We next assessed IL-1 + OSM-dependent STAT3 phosphorylation after incubation of cells with the MAPK kinase (MEK) inhibitors U0126 and PD98059. Inhibition of the MEK-ERK pathway resulted not in decreased but rather in increased STAT3 phosphorylation on Ser-727 (Fig. 5B), suggesting that aPKC-dependent serine phosphorylation of STAT3 was not mediated by ERK1/2. PKC inhibition using the inhibitor Gö6976, as expected, strongly inhibited STAT3 phosphorylation (not shown).
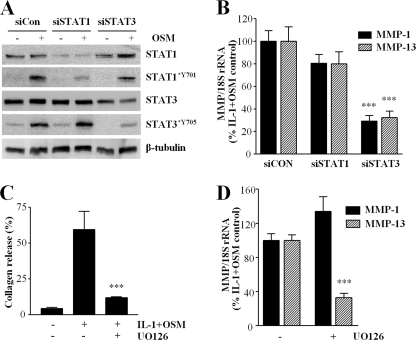
Effect of STAT Gene Silencing and ERK Pathway Inhibition on Collagenase Gene Expression and Cartilage Degradation
To ascertain whether the effects of aPKC depletion on STAT and ERK activation could explain their inhibition of IL-1 + OSM-stimulated collagenase gene expression, we silenced STAT1 and STAT3 genes in human chondrocytes before measuring cytokine-stimulated MMP1 and MMP13 levels. Fig. 6A shows that specific siRNA reagents were effective in selectively reducing the abundance of each STAT and consequently their OSM-dependent tyrosine phosphorylation. Fig. 6B illustrates that the consequence of STAT3 depletion was a marked impairment of IL-1 + OSM-stimulated MMP1 and MMP13, whereas STAT1 silencing had no significant effect. Use of the MEK inhibitor UO126 to prevent ERK activation resulted in reduced IL-1 + OSM-stimulated cartilage degradation (Fig. 6C; similar data were observed using the alternative MEK inhibitor PD98059, not shown), which was reflected in the ability of UO126 to inhibit IL-1 + OSM-stimulated expression of MMP13, but not MMP1, in human chondrocytes (Fig. 6D).
FIGURE 6.
STAT3 gene silencing and ERK pathway inhibition abrogate collagenase induction. A, primary human articular chondrocytes were transfected with siRNA (100 nm) specific to STAT1, STAT3, or siCon non-targeting control, prior to stimulation with OSM (10 ng/ml) for 20 min. Cells were lysed and proteins resolved by SDS-PAGE. Immunoblots were probed using the antibodies indicated. B, chondrocytes were transfected as above, but subsequently stimulated with IL-1 (0.02 ng/ml) and OSM (10 ng/ml) for 24 h. Real time reverse transcription-PCR from the isolated RNA was performed for MMP1 and MMP13, 72 h after transfection. C, bovine cartilage explant cultures were stimulated with IL-1 (1 ng/ml) and OSM (10 ng/ml) for 14 days, with fresh medium and cytokines at day 7, in the presence of DMSO vehicle control or MEK inhibitor UO126 (10 μm). Medium assayed for cumulative collagen release by day 14 were expressed as a percentage of the total for each treatment. D, chondrocytes were incubated with UO126 (3 μm) or DMSO vehicle for 1 h prior to stimulation with IL-1 (0.02 ng/ml) and OSM (10 ng/ml) for 24 h. Real time reverse transcription-PCR of the isolated RNA was performed for MMP1 and MMP13. Data are representative of (A) or pooled from (B-D) three separate chondrocyte populations (each assayed in at least quadruplicate) compared with stimulated control (mean ± S.E.; ***, p < 0.001).
Effect of aPKC Isoform Knockdown on IL-1 + OSM-stimulated AP-1 Transcription Factors
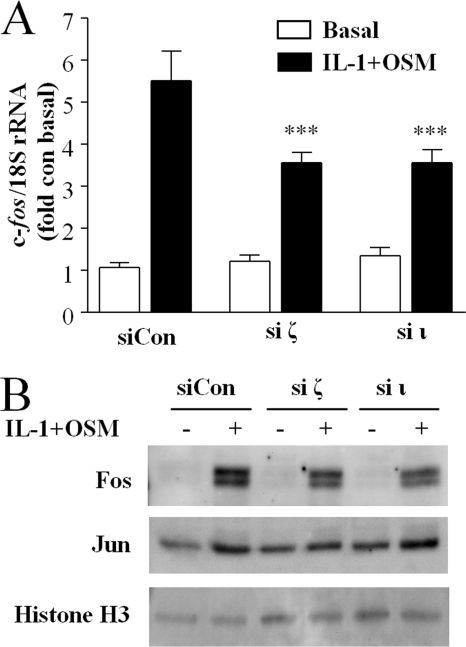
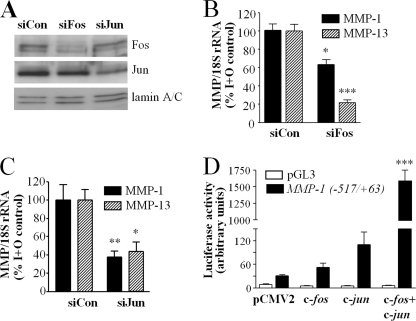
It has been reported widely that transcription factors of the AP-1 family are important for MMP gene regulation (63) and both ERK, through activation of the Elk-1 transcription factor (64), and STAT (65) activity have been shown to be important in c-fos gene expression. Therefore, we sought to determine the role of aPKC activity on IL-1 + OSM-stimulated c-fos expression and its canonical AP-1 partner c-jun in human chondrocytes. Silencing of either aPKCζ or aPKCι resulted in a substantial decrease in expression of c-fos in response to IL-1 + OSM at both the mRNA (Fig. 7A) and protein (Fig. 7B) levels. IL-1 + OSM only marginally induced c-jun mRNA expression in human chondrocytes (not shown), and neither basal nor cytokine-induced Jun abundance in chondrocyte nuclei was significantly affected by aPKC silencing (Fig. 7B). Therefore, we focused on the regulation of c-fos expression in further experiments.
FIGURE 7.
Atypical PKC gene silencing inhibits IL-1 + OSM-stimulated c-fos expression. A, following transfection with siRNA specific to aPKCζ, aPKCι, or non-targeting siControl (100 nm), chondrocytes were stimulated with IL-1 (0.02 ng/ml) and OSM (10 ng/ml) for 45 min. Real time reverse transcription-PCR of the isolated RNA was performed for c-fos. Data were pooled from two separate chondrocyte populations (each assayed in hextuplicate) presented as a percentage of the cytokine-induced expression (specific siPKC transfected versus siCon-transfected; mean ± S.E.; ***, p < 0.001). B, human chondrocytes were transfected with siRNA (100 nm) specific to aPKCζ, aPKCι, or siCon non-targeting control, prior to stimulation with IL-1 (0.2 ng/ml) and OSM (10 ng/ml) for 3 h. Nuclear extracts were isolated and proteins resolved by SDS-PAGE. Immunoblots were probed using the antibodies indicated. Blots shown are representative of three experiments using chondrocytes from different donors.
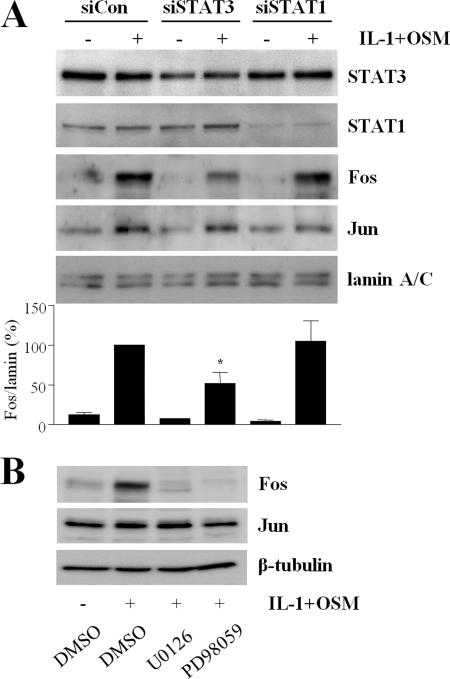
STAT3 Depletion and MEK-ERK Pathway Inhibition Curtails IL-1 + OSM-dependent c-fos Expression
We next sought to determine whether STAT3 and ERK activation, consequences of IL-1 + OSM-dependent aPKC activity, were necessary for c-fos expression in human chondrocytes. We have previously proposed a novel mechanism whereby STAT3 regulates c-fos expression in human chondrocytes by binding to the sis-inducible element of the c-fos promoter (15). Fig. 8A shows that siRNA-mediated depletion of STAT3, but not STAT1, resulted in a marked loss of nuclear Fos protein, whereas Jun abundance was unaffected. MEK-ERK signaling was inhibited by UO126 and PD98059, and these interventions resulted in total loss of IL-1 + OSM-dependent Fos expression (Fig. 8B). The data suggest that both STAT3 and ERK signaling are required for IL-1 + OSM-dependent expression of c-fos in human chondrocytes.
FIGURE 8.
STAT3 gene silencing and ERK pathway inhibition curtail Fos expression. A, primary human articular chondrocytes were transfected with siRNA (100 nm) specific to STAT1, STAT3, or siCON non-targeting control, prior to stimulation with IL-1 (0.2 ng/ml) and OSM (10 ng/ml) for 3 h. Nuclear extracts were isolated, and proteins were resolved by SDS-PAGE. Immunoblots were probed using the antibodies indicated. Blots shown are representative of four experiments using chondrocytes from different donors, and the histogram shows quantification of bands normalized to lamin A/C levels presented as a percentage of the cytokine-induced expression (siCon-transfected), mean ± S.E.; *, p < 0.05). Comparisons shown are stimulated (siCon versus siSTAT). B, chondrocytes were incubated with MEK inhibitors UO126 or PD98059 (both 10 μm), or DMSO vehicle for 1 h prior to stimulation with IL-1 (0. 2 ng/ml) and OSM (10 ng/ml) for 3 h. Nuclear extracts were isolated, and proteins were resolved by SDS-PAGE. Immunoblots were probed using the antibodies indicated. Blots are representative of three experiments using chondrocytes from different donors.
Effect of AP-1 Factor Expression Levels on MMP Gene Expression
We sought to confirm whether levels of Fos (subject to regulation by aPKC via STAT3 and ERK) and Jun could influence collagenase expression in human chondrocytes. Loss of nuclear Fos and Jun after gene silencing was confirmed by immunoblotting of IL-1 + OSM-stimulated nuclear proteins (Fig. 9A). Silencing of c-fos and consequent abrogation of Fos expression resulted in loss of IL-1 + OSM-stimulated MMP1 gene induction (Fig. 9B), complementary to our previous findings where c-fos overexpression enhanced IL-1 + OSM-dependent MMP1 promoter activity (15). Furthermore, we show here that c-fos silencing also curtailed MMP13 mRNA induction. Silencing of c-jun gene expression and consequent loss of Jun protein also inhibited induction of both MMP1 and MMP13 by IL-1 + OSM (Fig. 9C). We co-transfected T/C28a4 human chondrocytes with pCMV2 plasmid constructs for overexpression of c-fos, c-jun, or both c-fos and c-jun, together with a pGL3-derived construct harboring the proximal (−517/+63) region of the human MMP1 promoter controlling expression of a luciferase reporter gene (15). MMP1 promoter activity was greatly increased when both c-fos and c-jun were overexpressed, but not by overexpression of either alone, in the absence of any cytokine stimulus (Fig. 9D).
FIGURE 9.
Collagenase expression and MMP1 promoter activity are dependent upon canonical AP-1 components. A, primary human articular chondrocytes were transfected with siRNA (100 nm) specific to c-fos, c-jun, or siCon non-targeting control, prior to stimulation with IL-1 (0.2 ng/ml) and OSM (10 ng/ml) for 3 h. Nuclear extracts were isolated, and proteins were resolved by SDS-PAGE. Immunoblots were probed using the antibodies indicated. Blots shown are representative of two experiments using chondrocytes from different donors. B and C, following transfection with siRNA specific to c-fos, c-jun, or siCon (100 nm), chondrocytes were stimulated with IL-1 (0.02 ng/ml) and OSM (10 ng/ml) for 24 h and real time reverse transcription-PCR of the isolated RNA was performed for MMP1 and MMP13, 72 h after transfection. Data are pooled from at least three separate chondrocyte populations (each assayed in hextuplicate) presented as a percentage of the cytokine-induced expression (specific siPKC transfected versus siCon-transfected; mean ± S.E.; ***, p < 0.001; **, p < 0.01; *, p < 0.05). D, T/C28a4 human chondrocytes were transiently co-transfected with pCMV2 plasmid alone or constructs for overexpression of c-fos, c-jun, or both c-fos and c-jun, together with a pGL3-derived construct harboring the proximal (−517/+63) region of the human MMP1 promoter controlling luciferase expression or empty vector control. Luciferase activity was determined and data are representative of two experiments conducted in quadruplicate; mean ± S.E.; ***, p < 0.001 compared with pCMV alone.
DISCUSSION
IL-1 + OSM is the most potent cytokine stimulus reported to effect cartilage ECM degradation (48). We have now shown that PKC activity is a key regulator of IL-1 + OSM-dependent cartilage destruction and collagenase induction in human chondrocytes. The PKC inhibitor Gö6983 was rather more potent in inhibiting MMP levels (Fig. 2C) than collagenase activity, probably due to the additional requirement for pro-MMP activation, or impaired penetration of this compound into cartilage. Rather than basal PKC activity being a permissive requirement for IL-1 + OSM-stimulated MMP-1 and MMP-13 expression, these cytokines appear to increase PKC activity as assessed by increases in PKC priming and substrate phosphorylation. The presence of nuclear PKC suggests possible downstream transcriptional effects (66). The presence of aPKCι at the membrane suggests interactions with other membrane-bound proteins, because aPKC isoforms cannot migrate to the membrane via DAG binding, although they may bind other lipid species such as phosphatidylinositol 3,4,5′-trisphosphate or ceramide (59). Indeed, aPKCι is well known to bind to the cell polarity scaffold protein PAR6, and the disruption of this interaction by anti-rheumatic gold compounds such as aurothiomalate has been proposed as a potential mechanism of action for these agents (67).
The use of pharmacological PKC inhibitors yields little useful information regarding the role of specific isoforms. Gö6976 acts with greater potency against cPKC isoforms, but is equally selective for PKD and is an effective inhibitor of n- and aPKC at higher concentrations (68). Rottlerin has been used widely in the past as a PKCδ-specific inhibitor, but in recent years has been shown to have marked off-target effects including the collapse of cellular ATP levels (60). Our gene silencing studies have shown that only aPKC isoforms are required for IL-1 + OSM-stimulated collagenase induction, unlike the situation for the IL-1 only stimulus, where nPKC isoforms contributed also. This suggests that when the combined IL-1 + OSM stimulus was used to induce collagenase expression, additional signaling pathways activated by OSM diminish the contribution of nPKC activity to MMP expression compared with a simple IL-1 stimulus. This has implications for the adoption of PKC isoforms as therapeutic targets for inhibiting MMP expression. Pathological inflammatory milieus in vivo will invariably comprise a complex array of cytokines, rather than a single stimulus. Therefore, it is likely to be more instructive to assess the contribution of particular signaling moieties to more complex stimuli.
Because we have identified aPKC as key in IL-1 + OSM-stimulated collagenase induction, it is curious that Cal C, a DAG-competitive inhibitor, was as effective as the bismaleimide compounds in inhibiting this process. Cal C would also target other DAG effectors including the PKC substrate PKD. Because PKD has itself very recently been implicated in MMP gene regulation for the first time (69), it is plausible that inhibition of PKD could explain the effects of Cal C on collagenase expression. Conversely, it is notable that Gö6983 was relatively ineffective at inhibiting MMP-2 activity compared with MMP-9 (Fig. 1B). This could possibly indicate a requirement for PKD activity for MMP-2 expression.
This is, to our knowledge, the first report of a role for aPKCι in collagenase gene expression. A role for aPKCι signaling in MMP-10 expression has been noted in lung cancer cells (70), although our data indicate that this relationship does not exist in chondrocytes. The role of aPKCζ in expression of aggrecanase-1 in human chondrocytes has been examined previously (36). In this report, however, aPKCι was not considered, likely because this group had previously reported aPKCι to be present in only very small quantities in OA chondrocytes compared with aPKCζ, thus unlikely to exert any significant effect (35). It is possible that some experimental variable explains this apparent discrepancy with the present work, for instance, our use of macroscopically normal versus diseased tissue. We suggest that the ratio of expression of each aPKC varies between subjects, because gene silencing of either aPKC had variable consequences for the level of PKCζ/ι*T410/403 (data not shown). We have not attempted to accurately quantify levels of each aPKC isoform, but we detected transcripts and protein for both aPKCζ and aPKCι quite readily, albeit using different reagents compared with these other workers. This group reported no “reproducible” effects on MMP-13 levels after aPKCζ silencing (36), which infers that this may have been observed in some experiments. These workers also reported a role for aPKCζ in nitric-oxide synthase expression, and aPKC-mediated STAT activation might provide a mechanism for this finding (71, 72).
Also of note is the effect of nPKCδ silencing, in enhancing MMP-1 expression, but inhibiting MMP-13 expression. Not only does this confirm that differential regulatory mechanisms exist in chondrocytes for these genes as previously reported (17, 73), but is consistent with the findings of a previous report describing a positive role for nPKCδ in basic fibroblast growth factor-stimulated MMP-13 expression (37). MMP-1 expression was not considered in this report. In any case, our findings indicate that nPKCδ does not play a major role in collagenase expression induced by a complex cytokine stimulus.
Our data show that both ERK and STAT activation are compromised when aPKC isoforms are down-regulated in chondrocytes. Atypical PKC activity has previously been associated with regulation of both STAT (34) and ERK (74). We have previously shown STAT3-dependent stimulation of c-fos induction by IL-1 + OSM in human chondrocytes, and that overexpression of c-fos results in increased MMP1 promoter activity (15). Here, we show that silencing of either aPKCζ or aPKCι inhibits c-fos induction, and that silencing of c-fos expression inhibits cytokine-induced expression of both MMP1 and MMP13 genes. Silencing of one aPKC gene did not affect expression of the other, therefore the impact of each appears to be independent, a view reinforced by their distinct subcellular localizations. STAT3 silencing reproduced the effects of aPKC silencing on c-fos and collagenase expression, suggesting that aPKC-mediated STAT3 activity may explain the effects of aPKC down-regulation. In this regard it is of interest to note that, in metastatic melanoma, defective activation of PKC and STAT3 are associated with OSM resistance (61).
Prevention of ERK activation using MEK inhibitors inhibited cartilage degradation and induction of MMP13, but not MMP1, from human chondrocytes. Interestingly, induction of both collagenase genes was inhibited by these agents in bovine chondrocytes (not shown). ERK pathway inhibition also inhibited c-fos expression, and this effect was more pronounced than seen after STAT3 gene silencing, probably due to the less absolute depletion of activity using the latter method. It seems likely that ERK activation via aPKC contributes to IL-1 + OSM-stimulated collagenase expression, but is not itself sufficient, requiring STAT3 activation in addition.
The effect of aPKC in activating both STAT3 and ERK-dependent induction of c-fos may in part explain the mechanisms of synergy between IL-1 and OSM in driving collagenase expression and ECM destruction. We have already shown OSM-dependent signaling mechanisms such as JAK/STAT (15), PI3K (17), and ERK4 are able to enhance chondrocyte collagenase induction in the presence of IL-1 signaling, which itself drives MMP expression through mechanisms such as ERK, Jun N-terminal kinase (JNK), and p38 MAPKs and NFκB activation (75). Also, whereas we have found that only OSM stimulates Tyr-705 phosphorylation of STAT3, both IL-1 and OSM promote phosphorylation of the Ser-727 site (not shown). Our data indicate that both IL-1 and OSM may regulate aPKC activity, placing these kinases in a key position to mediate cross-talk of signaling networks initiated by inflammatory cytokine combinations that include IL-1. We have also shown that in addition to IL-1, OSM (and IL-6) synergize with TNFα and IL-17 to similarly promote profound release of collagen from the cartilage ECM (6, 7), and it is most likely that aPKC signaling also contributes to the catabolism mediated by these cytokine combinations, making aPKC signaling a key regulator in many inflammatory settings. Atypical PKC activity has also been linked to NFκB activation (36, 67). We did not find that IκBα degradation was inhibited when aPKC isoforms were down-regulated, although this observation alone is not sufficient to exclude a downstream role for NFκB signaling.
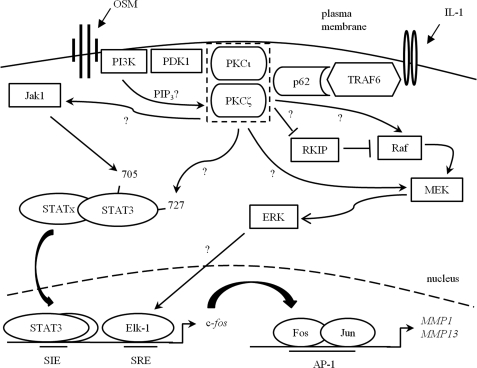
The exact mechanism(s) by which aPKC isoforms mediate collagenase induction are as yet unclear. Activation of PI3K activity by OSM (17) would explain increased priming phosphorylation by phosphoinositide-dependent kinase-1, which is particularly important for activity of aPKC, whereas phosphatidylinositol 3,4,5′-trisphosphate generation might also activate these isoforms directly (59). TNF receptor-associated factor-6 interacts with p62/sequestosome 1, providing a ready mechanism for aPKC activation by IL-1 (76). Because both aPKCζ and aPKCι appear to contribute to collagenase induction by IL-1 alone, there may be a functional overlap of aPKC isoforms, or alternatively one may act downstream of the other. We have not demonstrated direct substrate relationships here, but there is precedent for PKC phosphorylation of downstream effectors including STAT3 (77), and both Raf-1 (78) and RKIP (79) upstream of ERK. Furthermore, aPKC has been demonstrated to activate MEK5 directly (80), and MEK1 indirectly (74), whereas the mechanism for STAT tyrosine phosphorylation by aPKC must be indirect, and indeed aPKCζ has been shown to phosphorylate and activate Jak1 (81). Fig. 10 illustrates hypothetical mechanisms by which the activation of aPKC by IL-1 and OSM in chondrocytes may occur and lead to enhanced collagenase induction via c-fos expression.
FIGURE 10.
Hypothetical mechanism for IL-1 + OSM-stimulated collagenase induction via aPKC activity in chondrocytes. PKCζ or PKCι become activated after IL-1 stimulation (via TNF receptor-associated factor 6 (TRAF6) and p62/sequestosome 1 activity) and/or OSM (via PI3K-dependent phosphatidylinositol 3,4,5′-trisphosphate increases or phosphoinositide-dependent kinase-1 (PDK-1) activity). The proximity of PKCζ and PKCι in the schematic does not denote association with each other, merely proximity to potential activators. Once activated, aPKC may activate STAT3, directly by phosphorylation on Ser-727 and/or indirectly via Jak1 activation, leading to phosphorylation on Tyr-705. aPKC may also lead to ERK activation, by inhibition of RKIP or activation of Raf or MEK. Activated STAT3 homo- or heterodimers and ERK then lead to induction of c-fos, by binding the Sis-inducible element (SIE) or activation of Elk-1 and subsequent serum response element (SRE) binding, respectively, at the c-fos promoter. Translated Fos then mediates collagenase promoter activation via AP-1 binding in partnership with c-Jun. The scheme is based on the current study and input from Refs. 15, 17, 59, 64, 74, and 76–81.
In conclusion, this study highlights a novel role for aPKC signaling in mediating, at least in part, the pathological cartilage ECM catabolism mediated by members of the IL-6-type cytokine family when in combination with other key pro-inflammatory cytokines (5–9, 14, 48). Therapeutic interventions targeted at this pathway may provide a means of treatment for inflammatory joint diseases as well as other pathologies associated with aPKC-dependent collagenase expression and ECM catabolism.
Supplementary Material
Acknowledgments
We thank all the collaborators mentioned for their generous provision of reagents.
This work was supported by Arthritis Research UK Grant 18726, the Nuffield Foundation (Oliver Bird Fund), the Dunhill Medical Trust, the John G. W. Patterson Foundation, and a UK National Institute of Health Research Biomedical Research Centre for Aging and Age-Related Disease award to the Newcastle upon Tyne Hospitals National Health Service Foundation Trust.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2.
M. Jefferson and A. D. Rowan, unpublished data.
- OA
- osteoarthritis
- AP-1
- activator protein-1
- aPKC
- atypical PKC
- ECM
- extracellular matrix
- Cal C
- calphostin C
- cPKC
- conventional PKC
- DAG
- diacylglycerol
- ERK
- extracellular signal-regulated kinase
- IL
- interleukin
- JAK
- Janus kinase
- JNK
- Jun N-terminal kinase
- MAPK
- mitogen-activated protein kinase
- MARCKS
- myristoylated alanine-rich C-kinase substrate
- MEK
- MAPK kinase
- MMP
- matrix metalloproteinase
- NFκB
- nuclear factor-κB
- nPKC
- novel PKC
- PKD
- protein kinase D
- OSM
- oncostatin M
- PI3K
- phosphatidylinositol 3′-kinase
- PKC
- protein kinase C
- RKIP
- raf kinase-inhibitory protein
- siRNA
- small interfering RNA
- STAT
- signal transducer and activator of transcription
- TNFα
- tumor necrosis factor-α
- DMSO
- dimethyl sulfoxide.
REFERENCES
- 1.Sakkas L. I., Platsoucas C. D. (2007) Arthritis Rheum. 56, 409–424 [DOI] [PubMed] [Google Scholar]
- 2.Scrivo R., Di Franco M., Spadaro A., Valesini G. (2007) Ann. N. Y. Acad. Sci. 1108, 312–322 [DOI] [PubMed] [Google Scholar]
- 3.Goldring M. B., Goldring S. R. (2007) J. Cell Physiol. 213, 626–634 [DOI] [PubMed] [Google Scholar]
- 4.Koenders M. I., Joosten L. A., van den Berg W. B. (2006) Ann. Rheum. Dis. 65, Suppl. 3, iii29–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hui W., Cawston T., Rowan A. D. (2003) Ann. Rheum. Dis. 62, 172–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hui W., Rowan A. D., Richards C. D., Cawston T. E. (2003) Arthritis Rheum. 48, 3404–3418 [DOI] [PubMed] [Google Scholar]
- 7.Koshy P. J., Henderson N., Logan C., Life P. F., Cawston T. E., Rowan A. D. (2002) Ann. Rheum. Dis. 61, 704–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rowan A. D., Hui W., Cawston T. E., Richards C. D. (2003) Am. J. Pathol. 162, 1975–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rowan A. D., Koshy P. J., Shingleton W. D., Degnan B. A., Heath J. K., Vernallis A. B., Spaull J. R., Life P. F., Hudson K., Cawston T. E. (2001) Arthritis Rheum. 44, 1620–1632 [DOI] [PubMed] [Google Scholar]
- 10.Burrage P. S., Mix K. S., Brinckerhoff C. E. (2006) Front. Biosci. 11, 529–543 [DOI] [PubMed] [Google Scholar]
- 11.Page-McCaw A., Ewald A. J., Werb Z. (2007) Nat. Rev. Mol. Cell Biol. 8, 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jubb R. W., Fell H. B. (1980) J. Pathol. 130, 159–167 [DOI] [PubMed] [Google Scholar]
- 13.Barksby H. E., Hui W., Wappler I., Peters H. H., Milner J. M., Richards C. D., Cawston T. E., Rowan A. D. (2006) Arthritis Rheum. 54, 540–550 [DOI] [PubMed] [Google Scholar]
- 14.Koshy P. J., Lundy C. J., Rowan A. D., Porter S., Edwards D. R., Hogan A., Clark I. M., Cawston T. E. (2002) Arthritis Rheum. 46, 961–967 [DOI] [PubMed] [Google Scholar]
- 15.Catterall J. B., Carrère S., Koshy P. J., Degnan B. A., Shingleton W. D., Brinckerhoff C. E., Rutter J., Cawston T. E., Rowan A. D. (2001) Arthritis Rheum. 44, 2296–2310 [DOI] [PubMed] [Google Scholar]
- 16.Vincenti M. P., Brinckerhoff C. E. (2002) Arthritis Res. 4, 157–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Litherland G. J., Dixon C., Lakey R. L., Robson T., Jones D., Young D. A., Cawston T. E., Rowan A. D. (2008) J. Biol. Chem. 283, 14221–14229 [DOI] [PubMed] [Google Scholar]
- 18.Griner E. M., Kazanietz M. G. (2007) Nat. Rev. Cancer 7, 281–294 [DOI] [PubMed] [Google Scholar]
- 19.Chu F., Chen L. H., O'Brian C. A. (2004) Carcinogenesis 25, 585–596 [DOI] [PubMed] [Google Scholar]
- 20.Hofmann J. (2004) Curr. Cancer Drug Targets 4, 125–146 [DOI] [PubMed] [Google Scholar]
- 21.Liu J. F., Crépin M., Liu J. M., Barritault D., Ledoux D. (2002) Biochem. Biophys. Res. Commun. 293, 1174–1182 [DOI] [PubMed] [Google Scholar]
- 22.Urtreger A. J., Grossoni V. C., Falbo K. B., Kazanietz M. G., Bal de Kier Joffé E. D. (2005) Mol. Carcinog. 42, 29–39 [DOI] [PubMed] [Google Scholar]
- 23.Boschelli D. H. (2009) Curr. Top. Med. Chem. 9, 640–654 [DOI] [PubMed] [Google Scholar]
- 24.Shen G. X. (2003) Curr. Drug Targets Cardiovasc. Haematol. Disord. 3, 301–307 [DOI] [PubMed] [Google Scholar]
- 25.Budhiraja S., Singh J. (2008) Fundam. Clin. Pharmacol. 22, 231–240 [DOI] [PubMed] [Google Scholar]
- 26.Clarke M., Dodson P. M. (2007) Best Pract. Res. Clin. Endocrinol. Metab. 21, 573–586 [DOI] [PubMed] [Google Scholar]
- 27.Robertson M. J., Kahl B. S., Vose J. M., de Vos S., Laughlin M., Flynn P. J., Rowland K., Cruz J. C., Goldberg S. L., Musib L., Darstein C., Enas N., Kutok J. L., Aster J. C., Neuberg D., Savage K. J., LaCasce A., Thornton D., Slapak C. A., Shipp M. A. (2007) J. Clin. Oncol. 25, 1741–1746 [DOI] [PubMed] [Google Scholar]
- 28.Skvara H., Dawid M., Kleyn E., Wolff B., Meingassner J. G., Knight H., Dumortier T., Kopp T., Fallahi N., Stary G., Burkhart C., Grenet O., Wagner J., Hijazi Y., Morris R. E., McGeown C., Rordorf C., Griffiths C. E., Stingl G., Jung T. (2008) J. Clin. Invest. 118, 3151–3159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shin O. S., Behera A. K., Bronson R. T., Hu L. T. (2007) Cell. Microbiol. 9, 1987–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Popa-Nita O., Proulx S., Paré G., Rollet-Labelle E., Naccache P. H. (2009) J. Immunol. 183, 2104–2114 [DOI] [PubMed] [Google Scholar]
- 31.Chu S. C., Yang S. F., Lue K. H., Hsieh Y. S., Wu C. L., Lu K. H. (2004) Connect. Tissue Res. 45, 142–150 [DOI] [PubMed] [Google Scholar]
- 32.Tanaka S., Hamanishi C., Kikuchi H., Fukuda K. (1998) Semin. Arthritis Rheum. 27, 392–399 [DOI] [PubMed] [Google Scholar]
- 33.Tardif G., Pelletier J. P., Dupuis M., Geng C., Cloutier J. M., Martel-Pelletier J. (1999) Arthritis Rheum. 42, 1147–1158 [DOI] [PubMed] [Google Scholar]
- 34.Moscat J., Rennert P., Diaz-Meco M. T. (2006) Cell Death Differ. 13, 702–711 [DOI] [PubMed] [Google Scholar]
- 35.LaVallie E. R., Chockalingam P. S., Collins-Racie L. A., Freeman B. A., Keohan C. C., Leitges M., Dorner A. J., Morris E. A., Majumdar M. K., Arai M. (2006) J. Biol. Chem. 281, 24124–24137 [DOI] [PubMed] [Google Scholar]
- 36.Chockalingam P. S., Varadarajan U., Sheldon R., Fortier E., LaVallie E. R., Morris E. A., Yaworsky P. J., Majumdar M. K. (2007) Arthritis Rheum. 56, 4074–4083 [DOI] [PubMed] [Google Scholar]
- 37.Im H. J., Muddasani P., Natarajan V., Schmid T. M., Block J. A., Davis F., van Wijnen A. J., Loeser R. F. (2007) J. Biol. Chem. 282, 11110–11121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caraglia M., Tassone P., Marra M., Budillon A., Venuta S., Tagliaferri P. (2006) Ann. Oncol. 17, Suppl. 7, vii124–127 [DOI] [PubMed] [Google Scholar]
- 39.Keller E. T., Fu Z., Brennan M. (2004) Biochem. Pharmacol. 68, 1049–1053 [DOI] [PubMed] [Google Scholar]
- 40.Quesnelle K. M., Boehm A. L., Grandis J. R. (2007) J. Cell Biochem. 102, 311–319 [DOI] [PubMed] [Google Scholar]
- 41.Uddin S., Sassano A., Deb D. K., Verma A., Majchrzak B., Rahman A., Malik A. B., Fish E. N., Platanias L. C. (2002) J. Biol. Chem. 277, 14408–14416 [DOI] [PubMed] [Google Scholar]
- 42.Staunton D., Hudson K. R., Heath J. K. (1998) Protein Eng. 11, 1093–1102 [DOI] [PubMed] [Google Scholar]
- 43.Carrozza M. L., Jacobs H., Acton D., Verma I., Berns A. (1997) Oncogene 14, 1083–1091 [DOI] [PubMed] [Google Scholar]
- 44.Cleaver C. S., Rowan A. D., Cawston T. E. (2001) Ann. Rheum. Dis. 60, 150–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shingleton W. D., Ellis A. J., Rowan A. D., Cawston T. E. (2000) J. Cell Biochem. 79, 519–531 [PubMed] [Google Scholar]
- 46.Goldring M. B., Birkhead J. R., Suen L. F., Yamin R., Mizuno S., Glowacki J., Arbiser J. L., Apperley J. F. (1994) J. Clin. Invest. 94, 2307–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bergman I., Loxley R. (1963) Anal. Chem. 35, 1961–1965 [DOI] [PubMed] [Google Scholar]
- 48.Cawston T. E., Curry V. A., Summers C. A., Clark I. M., Riley G. P., Life P. F., Spaull J. R., Goldring M. B., Koshy P. J., Rowan A. D., Shingleton W. D. (1998) Arthritis Rheum. 41, 1760–1771 [DOI] [PubMed] [Google Scholar]
- 49.Koshy P. J., Rowan A. D., Life P. F., Cawston T. E. (1999) Anal. Biochem. 275, 202–207 [DOI] [PubMed] [Google Scholar]
- 50.Bigg H. F., Rowan A. D., Barker M. D., Cawston T. E. (2007) FEBS J. 274, 1246–1255 [DOI] [PubMed] [Google Scholar]
- 51.Clark I. M., Powell L. K., Wright J. K., Cawston T. E., Hazleman B. L. (1992) Matrix 12, 475–480 [DOI] [PubMed] [Google Scholar]
- 52.Morgan T. G., Rowan A. D., Dickinson S. C., Jones D., Hollander A. P., Deehan D., Cawston T. E. (2006) Ann. Rheum. Dis. 65, 184–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Botelho F. M., Edwards D. R., Richards C. D. (1998) J. Biol. Chem. 273, 5211–5218 [DOI] [PubMed] [Google Scholar]
- 54.Korzus E., Nagase H., Rydell R., Travis J. (1997) J. Biol. Chem. 272, 1188–1196 [DOI] [PubMed] [Google Scholar]
- 55.Hirt B. (1967) J. Mol. Biol. 26, 365–369 [DOI] [PubMed] [Google Scholar]
- 56.Davies S. P., Reddy H., Caivano M., Cohen P. (2000) Biochem. J. 351, 95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kobayashi E., Nakano H., Morimoto M., Tamaoki T. (1989) Biochem. Biophys. Res. Commun. 159, 548–553 [DOI] [PubMed] [Google Scholar]
- 58.Shingleton W. D., Jones D., Xu X., Cawston T. E., Rowan A. D. (2006) Rheumatology 45, 958–965 [DOI] [PubMed] [Google Scholar]
- 59.Steinberg S. F. (2008) Physiol. Rev. 88, 1341–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Soltoff S. P. (2007) Trends Pharmacol. Sci. 28, 453–458 [DOI] [PubMed] [Google Scholar]
- 61.Lacreusette A., Barbieux I., Nguyen J. M., Pandolfino M. C., Dréno B., Jacques Y., Godard A., Blanchard F. (2009) J. Pathol. 217, 665–676 [DOI] [PubMed] [Google Scholar]
- 62.Wierenga A. T., Vogelzang I., Eggen B. J., Vellenga E. (2003) Exp. Hematol. 31, 398–405 [DOI] [PubMed] [Google Scholar]
- 63.Clark I. M., Swingler T. E., Sampieri C. L., Edwards D. R. (2008) Int. J. Biochem. Cell Biol. 40, 1362–1378 [DOI] [PubMed] [Google Scholar]
- 64.Babu G. J., Lalli M. J., Sussman M. A., Sadoshima J., Periasamy M. (2000) J. Mol. Cell. Cardiol. 32, 1447–1457 [DOI] [PubMed] [Google Scholar]
- 65.Leaman D. W., Pisharody S., Flickinger T. W., Commane M. A., Schlessinger J., Kerr I. M., Levy D. E., Stark G. R. (1996) Mol. Cell. Biol. 16, 369–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Martelli A. M., Evangelisti C., Nyakern M., Manzoli F. A. (2006) Biochim. Biophys. Acta 1761, 542–551 [DOI] [PubMed] [Google Scholar]
- 67.Fields A. P., Regala R. P. (2007) Pharmacol. Res. 55, 487–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gschwendt M., Dieterich S., Rennecke J., Kittstein W., Mueller H. J., Johannes F. J. (1996) FEBS Lett. 392, 77–80 [DOI] [PubMed] [Google Scholar]
- 69.Eiseler T., Döppler H., Yan I. K., Goodison S., Storz P. (2009) Breast Cancer Res. 11, R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Frederick L. A., Matthews J. A., Jamieson L., Justilien V., Thompson E. A., Radisky D. C., Fields A. P. (2008) Oncogene 27, 4841–4853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guo Z., Shao L., Du Q., Park K. S., Geller D. A. (2007) FASEB J. 21, 535–542 [DOI] [PubMed] [Google Scholar]
- 72.Pérez-Rodríguez R., Roncero C., Oliván A. M., González M. P., Oset-Gasque M. J. (2009) J. Neurochem. 108, 1083–1096 [DOI] [PubMed] [Google Scholar]
- 73.Mengshol J. A., Vincenti M. P., Coon C. I., Barchowsky A., Brinckerhoff C. E. (2000) Arthritis Rheum. 43, 801–811 [DOI] [PubMed] [Google Scholar]
- 74.Regala R. P., Weems C., Jamieson L., Copland J. A., Thompson E. A., Fields A. P. (2005) J. Biol. Chem. 280, 31109–31115 [DOI] [PubMed] [Google Scholar]
- 75.Vincenti M. P., Brinckerhoff C. E. (2007) J. Cell Physiol. 213, 355–364 [DOI] [PubMed] [Google Scholar]
- 76.Sanz L., Diaz-Meco M. T., Nakano H., Moscat J. (2000) EMBO J. 19, 1576–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jain N., Zhang T., Kee W. H., Li W., Cao X. (1999) J. Biol. Chem. 274, 24392–24400 [DOI] [PubMed] [Google Scholar]
- 78.Kolch W., Heidecker G., Kochs G., Hummel R., Vahidi H., Mischak H., Finkenzeller G., Marmé D., Rapp U. R. (1993) Nature 364, 249–252 [DOI] [PubMed] [Google Scholar]
- 79.Lorenz K., Lohse M. J., Quitterer U. (2003) Nature 426, 574–579 [DOI] [PubMed] [Google Scholar]
- 80.Diaz-Meco M. T., Moscat J. (2001) Mol. Cell. Biol. 21, 1218–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Martin P., Villares R., Rodriguez-Mascarenhas S., Zaballos A., Leitges M., Kovac J., Sizing I., Rennert P., Márquez G., Martínez-A. C., Diaz-Meco M. T., Moscat J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 9866–9871 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.