Abstract
The signaling molecule 15-deoxy-Δ12,14-prostaglandin J2 (15d-PGJ2) has been described as the “anti-inflammatory prostaglandin.” Here we show that substrates of the nuclear export receptor CRM1 accumulate in the nucleus in the presence of 15d-PGJ2, identifying this prostaglandin as a regulator of CRM1-dependent nuclear protein export that can be produced endogenously. Like leptomycin B (LMB), an established fungal CRM1-inhibitor, 15d-PGJ2 reacts with a conserved cysteine residue in the CRM1 sequence. This covalent modification prevents the formation of nuclear export complexes. Cells that are transfected with mutant CRM1 (C528S) are resistant to the inhibitory effects of LMB and 15d-PGJ2, demonstrating that the same single amino acid is targeted by the two compounds. Inhibition of the CRM1 pathway by endogenously produced prostaglandin and/or exogenously applied 15d-PGJ2 may contribute to its anti-inflammatory, anti-proliferative, and anti-viral effects.
Keywords: Cyclooxygenase (COX) Pathway, NFAT Transcription Factor, Nuclear Pore, Nuclear Transport, Prostaglandins, 15d-PGJ2, CRM1, LMB, Leptomycin B
Introduction
Prostaglandins are signaling molecules with a very diverse range of biological activities (1, 2). They play a role in inflammation, hemostasis, gastrointestinal secretion, thrombosis, and other cellular functions. Synthesis of prostaglandins is initiated by the release of arachidonic acid from membrane phospholipids by phospholipase A2. In a subsequent step, prostaglandin H2 (PGH2)4 is formed by the cyclooxygenase and peroxidase activities of prostaglandin peroxide H synthases (PGHS-1 and -2; also referred to as cyclooxygenases COX-1 and COX-2). PGH2 is the precursor for the synthesis of many other prostaglandins. The major signaling pathway of prostaglandins involves binding to G-protein-coupled receptors at the plasma membrane and to nuclear orphan receptors (2). Members of the cyclopentenone subclass of prostaglandins may have multiple additional targets. A prime example is 15-deoxy-Δ12,14-prostaglandin J2 (15d-PGJ2), which is generated from prostaglandin PGD2 by spontaneous or induced dehydration (3–5) and has unequivocally been shown to be synthesized in settings of inflammation (6). 15d-PGJ2 is described as having anti-inflammatory effects (3). It has a high affinity for PPARγ (7, 8). Upon binding of 15d-PGJ2, this transcription factor translocates from the cytoplasm to the nucleus, where it initiates expression of a large number of genes.
Many cellular effects of 15d-PGJ2 however, appear to be PPARγ-independent. Strikingly, 15d-PGJ2 can covalently modify several proteins, affecting their cellular function. One target of 15d-PGJ2 is IκB kinase, which phosphorylates IκB, the inhibitor of the transcription factor NF-κB (nuclear factor-κB). 15d-PGJ2 inhibits IκB kinase by binding to a specific free cysteine in its sequence. As a result, the pro-inflammatory effects of IκB kinase-activating cytokines are reduced, partially explaining the anti-inflammatory effects of 15d-PGJ2 (9, 10). Similarly, 15d-PGJ2 targets cysteine 184 in the GTP-binding protein H-Ras, leading to its activation (11). Furthermore, modification of cysteine 264 of the translation factor eIF4A by 15d-PGJ2 was shown to inhibit translation (12). A crucial feature of the prostaglandin for these modifications is the cyclopentenone ring with its α,β-unsaturated carbonyl group. This electrophilic center allows a Michael addition of the prostaglandin to nucleophiles, such as the sulfhydryl group of cysteine residues. Analogues of 15d-PGJ2, lacking the double bond in the ring (see Fig. 1A), also lack many of the biological effects of the prostaglandin. Besides its function as an endogenous signaling molecule, 15d-PGJ2 has anti-viral, anti-proliferative, and anti-inflammatory effects when added exogenously to cells (3). It has been shown to induce apoptosis, e.g. in neuronal cells and can induce oxidant stress, potentially through depletion of free glutathione (13).
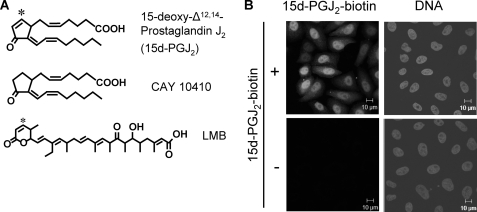
FIGURE 1.
15d-PGJ2 accumulates in nuclei. A, chemical structures of 15d-PGJ2, CAY 10410 (9,10-dihydro-15-deoxy-Δ12,14-prostaglandin J2) and LMB. The asterisks indicate the reactive C-atoms in 15d-PGJ2 and LMB. B, HeLa-cells were treated with 10 μm 15d-PGJ2-biotin for 1 h, fixed, and stained with Alexa488-streptavidin (left) and Hoechst (DNA, right).
Transport of proteins across the nuclear pore complex is mediated by importins and exportins, members of the importin β-superfamily (14). Importin β is the prototypic nuclear import receptor, which, together with the adaptor protein importin α, mediates nuclear import of proteins carrying a classical nuclear localization signal (cNLS). An alternative import receptor that does not require an adaptor protein is transportin, which recognizes the so-called M9-NLS (15). The major exportin is CRM1, a transport receptor that recognizes nuclear export signals (NESs). NESs, typically short hydrophobic stretches that are rich in leucines, were first identified in the viral protein HIV-1 Rev, (16), and in the inhibitor of cAMP-dependent protein kinase (PKI) (17). Together with the small GTP-binding protein Ran and the export substrate, CRM1 forms a trimeric complex that translocates across the nuclear envelope (18–21). On the cytoplasmic side of the nuclear pore complex (NPC), GTP-hydrolysis on Ran leads to dissociation of the export complex (22). Besides mediating nuclear export of a large number of cellular proteins, CRM1 also functions as a major export factor for certain RNAs, including rRNAs and certain viral and cellular mRNAs (23). Inhibition of the CRM1 pathway has been suggested as a therapeutic target, e.g. in certain myelomas (24).
The elucidation of the molecular mechanisms of nuclear export was strongly promoted by a fungal metabolite, leptomycin B (LMB). LMB, an unsaturated fatty acid with a terminal δ-lactone ring (Fig. 1A), induces nuclear accumulation of a number of proteins (25). The only known cellular target for LMB is CRM1, which is modified by the drug at a conserved cysteine residue (Cys-528 in humans) (26). The δ-lactone ring of LMB is required for its conjugation to Cys-528 of CRM1 by a Michael addition. LMB-modified CRM1 is unable to form trimeric export complexes, explaining the inhibitory effect of the drug on nuclear export. Similar to LMB, ratjadones, metabolites isolated from myxobacteria, modify CRM1, leading to inhibition of nuclear export (27, 28). Cys-528 in the CRM1 sequence is highly conserved, although it is not required for the export function of CRM1. These observations raise the question of whether this amino acid might also be targeted by endogenous, cellular compounds, allowing a rapid down-regulation of CRM1-mediated nuclear export.
Here we identify prostaglandin 15d-PGJ2 as an inhibitor of the CRM1-dependent export pathway that can be produced in mammalian cells. Very recently, a similar function has been described for S-nitrosylation of CRM1 (29). Like LMB, 15d-PGJ2 targets Cys-528 of CRM1. As a result, trimeric export complexes cannot form and CRM1-mediated nuclear export is inhibited. Our results may explain some of the anti-inflammatory, anti-proliferative, and anti-viral effects of 15d-PGJ2.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfections
HeLa-P4 cells (30) were grown in Dulbecco's modified Eagle's medium (Gibco) containing 4500 mg/liter glucose, 10% fetal calf serum, 2 mm glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. The cell line expressing GFP-NFAT has been described previously (31). For expression of reporter proteins and HA-CRM1, HeLa-P4 cells were transiently transfected using Polyfect (Qiagen), according to the instructions of the manufacturer. For treatment with prostaglandins and control substances, cells were washed with PBS and incubated with medium in the absence of FCS.
Prostaglandins
Initially, biotinylated 15d-PGJ2 was synthesized with help from Dr. Marc DeVocelle (Organic Synthesis Facility, Royal College of Surgeons, Ireland). Later, biotin-tagged 15d-PGJ2 and other prostaglandins were purchased from Cayman Chemical (Ann Arbor, MI).
Mass Spectrometry
HeLa cells were solubilized in lysis buffer, (0.2% Nonidet P-40, 150 mm NaCl, 50 mm Tris-HCl, pH 7.2). 300–600 μg of protein was used for each experiment. Neutravidin beads (50 μl) were washed three times in cold phosphate-buffered saline (PBS) at 4 °C for 10 min and then blocked overnight in 1% BSA in PBS at 4 °C, followed by washing with PBS and lysis buffer. Samples were precleared with 50 μl of neutravidin beads for 1.5 h at 4 °C. 10 μm of biotinylated 15d-PGJ2 or vehicle control was added to each lysate for 40 min at room temperature. Neutravidin beads were added to the sample and incubated overnight at 4 °C. The beads were washed with lysis buffer and proteins were eluted with SDS sample buffer and analyzed by SDS-PAGE, followed by Coomassie Blue staining. Gel spots were destained with NH4HCO3 and cysteines were reduced/alkylated with 10 mm DTT, 50 mm iodoacetamide, and digested with trypsin overnight at 37 °C. Samples were run on a Finnigan LTQ mass spectrometer connected to a Surveyor chromatography system. MS scans were performed to select the 5 most intense ions prior to MS/MS analysis. For peptide data analysis, MS/MS data were searched using BioWorks 3.2 software against the Uniprot-Swissprot Homo sapiens data base, and the peptides were filtered according to the following parameters: Xcorr above 1.90 (+1), 2.50 (+2), and 3.20 (+3). Protein identification was performed using Proline, an in-house proteomic data analysis platform. SEQUEST was used in combination with Peptide-Protein Prophet for additional validation. Proteins with a minimum of 2 peptides and Sequest/protein prophet scores of 0.8 or greater were deemed identified, each peptide with a minimum peptide probability of 0.8, minimum length of 4 residues, >25% matched ions, >5 ions, and a spectrum intensity >200. Single peptide hits were validated manually.
The synthetic peptide DLLGLCEQK (amino acids sequence 523–531 of CRM1) was solubilized in water (pH 7.4) at a concentration of 100 μm and 50 μm of 15d-PGJ2 was subsequently added for 1 h at room temperature. Samples were diluted 10-fold using H2O:CH3CN:formic acid (1:1:0.1 v/v) and the analysis was performed using a Finnigan LTQ equipped with a nanoelectrospray ionization (nanoESI) source by direct infusion through non-coated silica tips with internal diameters of 20 μm (New Objective, Inc. Woburn, MA), at a flow rate of 1 μl/min. The mass spectrometer was operated in positive ion mode with a capillary temperature of 200 °C, a capillary voltage of 46 V, a tube lens voltage of 140 V, and with a potential of 1800 V applied to the frit. Mass spectra were acquired from m/z 450 to 1500.
Cell Lysates for Western Blotting
5 × 106 HeLa cells were suspended in HB (20 mm Hepes-KOH, pH 7.3, 110 mm KOAc, 2 mm Mg(OAc)2, 1 mm EGTA, 2 mm DTT, 1 μg/ml each of aprotinin, leupeptin, and pepstatin) and permeabilized with 0.01% digitonin. After the addition of 10 μm biotinylated 15d-PGJ2, cells were incubated at 20 °C for 30 min, washed with HB and lysed in 1 ml of radioimmune precipitation assay buffer (50 mm Tris, pH 7.2, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS). The lysate was cleared by centrifugation (100,000 × g for 30 min), and biotinylated proteins were captured on neutravidin beads and analyzed by SDS-PAGE followed by Western blotting. The ECL system (Pierce) was used for visualization of proteins.
Plasmids
The coding sequence for human CRM1 was PCR-amplified and cloned into a pcDNA 3.1 vector containing a sequence coding for an HA tag. The C528S mutation was introduced by site-directed mutagenesis. The plasmid coding for GFP-NES was generated using annealed oligonucleotides corresponding to the NES of HIV-1 Rev (amino acids 68–90). Plasmid NES-GFP2-M9core, containing the Rev-NES and the M9-NLS was described before (32). Plasmids coding for GFP-TFIIAα and RFP-NC2β were kindly provided by Dr. Jörg Kahle and Dr. Detlef Doenecke.
Protein Purification
CRM1 (33), RanGAP, (34), and Ran (35) were expressed as described. All proteins were dialyzed against HB, frozen in liquid nitrogen, and stored at −80 °C.
Import Ligand
RanGAP Assays
CRM1 at a concentration of 1.1 μm was preincubated for 30 min at 20 °C with a 50-fold excess of 15d-PGJ2 in the presence of 1.6% ethanol or ethanol alone as a solvent control. After the reaction, BSA was added to a final concentration of 3 mg/ml. Labeling of Ran with [γ-32P]GTP and RanGAP assays were essentially performed as described (37, 38). [γ-32P]RanGTP, CRM1, and 60 μm of the NES-peptide from the minute virus of mice NS2 protein (CVDEMTKKFGTLTIHDTEK) were preincubated for 10 min at 20 °C in a volume of 40 μl. The reaction was started by the addition of 10 μl of RanGAP to a final concentration of 20 nm and stopped after 10 min with stop solution (7% charcoal, 10% ethanol, 0.1 m HCl, 10 mm NaH2PO4). After centrifugation, the released [32P]phosphate in the supernatant was measured by scintillation counting. Background counts from a reaction without RanGAP were subtracted, and GTP hydrolysis was expressed as the percentage of the maximal value of recovered radioactivity.
Nuclear Transport Assays
For a quantitative analysis of nuclear transport in vivo, cells were transfected with plasmids coding for different reporter proteins. After treatment with prostaglandins or control substances, cells were analyzed for the subcellular distribution of the reporter protein and scored into the following categories: N>C (more protein in the nucleus), n = C (equal distribution of the reporter protein between nucleus and cytoplasm) and N<C (more reporter protein in the cytoplasm). When cells were co-transfected with HA-tagged CRM1, only HA-positive cells were included in the analysis.
For the analysis of nuclear transport in vitro, a stable cell line expressing GFP-NFAT was used (31). Briefly, cells expressing nuclear GFP-NFAT were permeabilized with digitonin and washed with HB buffer lacking DTT. ∼200.000 cells per reaction were preincubated with or without 30 μm of prostaglandins for 20 min at 20 °C. BSA was added to a concentration of 1 mg/ml, cells were washed with HB buffer (with DTT), and subjected to transport reactions in the presence of an ATP-regenerating system, ∼2 mg/ml of HeLa cytosol, and ∼2.5 μg/ml Cy5-BSA-NLS. After 30 min at 4 or 30 °C, cells were washed again, and the average nuclear fluorescence emitted from GFP-NFAT or Cy5-BSA-NLS in ∼10,000 cells was measured with a FACSCalibur flow cytometer (Becton and Dickinson). Reactions were standardized by arbitrarily assigning a value of 100 to the nuclear fluorescence of GFP-NFAT in a 4 °C control reaction and to the maximal nuclear fluorescence of Cy5-BSA-NLS after incubation at 30 °C.
Antibodies
For the detection of HA-tagged CRM1, a monoclonal mouse anti-HA antibody (clone 12CA5) or rabbit-anti-HA (Sigma) was used. For immunofluorescence, donkey-anti-mouse Alexa 594 (1:2000; Molecular Probes) was used as secondary antibody. For immunoblotting, horseradish peroxidase-coupled donkey anti-rabbit IgG (Dianova) was used as secondary antibody.
Immunofluorescence
Immunofluorescence staining was essentially performed as described (39). Cells were analyzed by fluorescence microscopy using a Zeiss Axioskop2 microscope and AxioVision software or a confocal microscope (510-Meta, Carl Zeiss, Jena, Germany) equipped with an argon laser and a 63x Plan-Neofluar 1.3NA water-corrected objective. Pictures were processed using Adobe Photoshop.
Live Cell Imaging
24 h after transfection with the NES-GFP2-M9 construct, HeLa cells grown on LabTec-chambers (Nunc) were treated with either 15 μm 15d-PGJ2 or ethanol as a solvent control for 3 h at 37 °C, and transferred to CO2-independent medium (Invitrogen) lacking the prostaglandin. FLIP experiments were carried out at 37 °C in a temperature controlled chamber attached to a confocal microscope (510-Meta, Carl Zeiss, Jena, Germany) equipped as above. For analysis of nuclear export, a cytoplasmic region was subjected to 180 bleach intervals (651 ms each) with the argon laser set to 100%. After each interval, the fluorescence emission was collected during a period of 154 ms at 2% laser intensity. Signal intensities were analyzed from nuclear regions of a bleached (Fbn) and a neighboring unbleached cell as a reference (Frn), and a background region (Fbg). Nuclear fluorescence intensities for each time point were expressed as F(t) = (Fbn − Fbg)/(Frn − Fbg), normalizing the pre-bleach fluorescence to 1. 10–20 cells were analyzed per condition. For analysis of nuclear import, the same settings were used bleaching a nuclear region and recording the reduction of fluorescence intensities from a cytoplasmic region.
Statistical Analysis
p values were obtained by performing a two-tailed, heteroscedastic t test. p < 0.05 was considered biologically significant.
RESULTS
15d-PGJ2 Accumulates in Nuclei of Living Cells
15d-PGJ2 has been reported to bind to intracellular proteins. We therefore examined the subcellular localization of biotinylated 15d-PGJ2, where the biotin is linked via the carboxylic acid, leaving the cyclopentenone ring free. Upon incubation of intact cells, the prostaglandin accumulated in the nucleus (Fig. 1B), similar to other cyclopentenones (40) and suggesting a predominant interaction with nuclear proteins.
CRM1 Is a Target for 15d-PGJ2
The α,β-unsaturated carbonyl group of cyclopentenones is a rather weak electrophile. Hence, only selected and/or very exposed nucleophilic groups in cellular macromolecules should be modified by 15d-PGJ2. The reduced sulfhydryl groups in glutathione, which occurs at millimolar concentrations in cells (41), can also be conjugated to cyclopentenone prostaglandins (42), thus competing with other reactive groups. This suggests that only a subset of cellular proteins with exposed cysteine residues should be targeted by 15d-PGJ2, e.g. predominantly nuclear proteins as seen in Fig. 1B. To identify potential conjugation partners of 15d-PGJ2, we incubated a cell lysate with the biotinylated prostaglandin. Modified proteins were bound to neutravidin beads and analyzed by SDS-PAGE, followed by LC-MS/MS. One of the proteins that could unequivocally be identified by several peptides (Fig. 2A) was the nuclear export receptor CRM1 (see also supplemental Fig. S1). We have previously shown that inhibition of the CRM1 pathway by LMB reduces the expression of pro-inflammatory genes in activated T-cells (43). Hence, inhibition of CRM1 could have anti-inflammatory effects under certain conditions, like 15d-PGJ2. In light of the structural similarities and the identical reaction mechanism of the anti-inflammatory prostaglandin 15d-PGJ2 and the well-described fungal drug LMB, we decided to focus our analysis on the LMB/15d-PGJ2-target CRM1. Other targets of 15d-PGJ2 will be described elsewhere. First, we confirmed the modification of CRM1 by 15d-PGJ2 by Western blotting. A cell lysate was incubated with biotinylated 15d-PGJ2, and biotinylated proteins were captured on neutravidin beads. Indeed, CRM1 bound to the neutravidin beads, suggesting its modification by the prostaglandin (Fig. 2B).
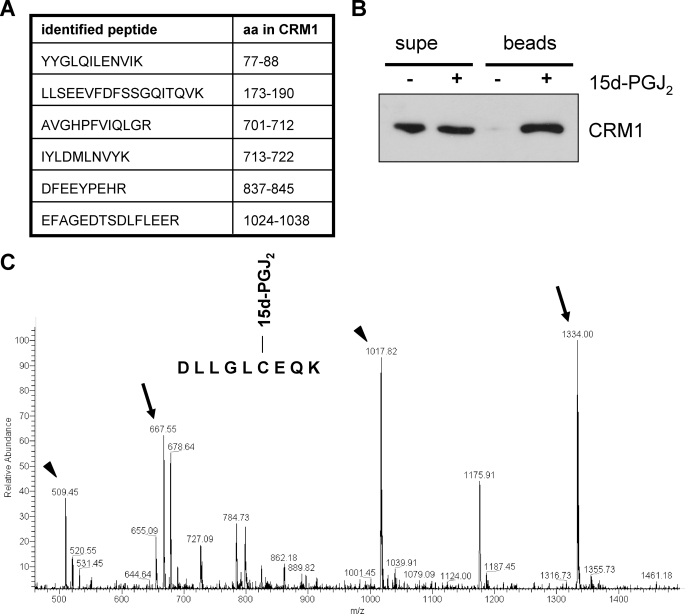
FIGURE 2.
CRM1 is a target of 15d-PGJ2. A, identified CRM1-peptides. Biotin-15d-PGJ2-modified proteins from a HeLa-cell lysate were subjected to LC-ESI-MS/MS. The identity of peptide LLSEEVFDFSSGQITQVK was confirmed by sequencing (compare supplemental Fig. S1). The amino acids correspond to the human CRM1-sequence. B, a cell lysate was treated with (+) or without (−) biotinylated 15d-PGJ2 and neutravidin-binding proteins (beads) were analyzed by Western blotting using an anti-CRM1 antibody. 1% of the supernatant (supe) was loaded. C, full mass spectrum of a CRM1 peptide modified by 15d-PGJ2. The peptide DLLGLCEQK (amino acids sequence 523–531 of CRM1) was treated with 15d-PGJ2, and the samples were analyzed by mass spectrometry. The arrows show the peaks 667 and 1334, corresponding to the modified peptide doubly and singly charged, respectively. The arrowheads show the peaks corresponding to the unmodified peptides.
To further characterize this modification, we performed a mass spectrometry analysis of a synthetic CRM1 peptide (amino acids 523–531; this peptide contains Cys-528, which is also the target for LMB; see below) that had been treated with 15d-PGJ2. The full mass spectrum of the peptide DLLGECEQK modified by 15d-PGJ2 (Fig. 2C) shows the peaks m/z 1017 and m/z 1334, respectively, corresponding to the unmodified and the modified peptide with a single positive charge. Also seen are the peaks m/z 509 and m/z 667, respectively, corresponding to the unmodified and the modified peptide each with a double charge. The mass shift of 316 between the peaks corresponds to the MW of 15d-PGJ2, demonstrating that the peptide is modified by the prostaglandin. Further evidence for the modification of the CRM1 peptide by 15d-PGJ2 was obtained by fragmentation of the precursor ions observed at m/z 667 and m/z 1334 (data not shown). Together, our results demonstrate that CRM1 can be covalently modified by 15d-PGJ2.
15d-PGJ2 Inhibits the Formation of CRM1-containing Export Complexes
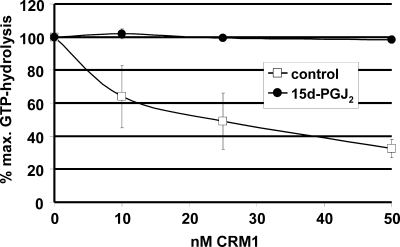
Modification of CRM1 by LMB inhibits the formation of trimeric export complexes containing the export receptor, an NES-substrate and RanGTP (18). To test whether 15d-PGJ2 has similar effects, we used RanGAP assays to measure the formation of export complexes. This assay is based on the observation that RanGTP in a complex with importin β-like transport receptors is resistant to GTP-hydrolysis, as induced by the GTPase-activating protein RanGAP (44). In the case of CRM1, it is the trimeric export complex that is resistant to RanGAP. As shown in Fig. 3, GTP hydrolysis on Ran was reduced in the presence of increasing concentrations of unmodified CRM1, indicating the formation of trimeric, RanGAP-resistant complexes. In reactions where CRM1 had been preincubated with 15d-PGJ2, GTP hydrolysis remained at a high level, even at the highest CRM1 concentration. This result shows that 15d-PGJ2 indeed inhibits the formation of trimeric export complexes, similar to LMB (39).
FIGURE 3.
15d-PGJ2 inhibits the formation of trimeric export complexes. RanGAP assays were performed in the presence of an NES-peptide and increasing concentrations of CRM1 that had been preincubated with 15d-PGJ2 (closed circles) or ethanol as a control (open squares). Trimeric export complexes are insensitive to GTPase activation on Ran by RanGAP. High levels of GTP hydrolysis indicate the presence of free RanGTP in the reaction. Bars indicate the variation from the mean of two independent experiments.
15d-PGJ2 Inhibits Nuclear Export in Vivo and in Vitro
The previous results suggested that 15d-PGJ2 may inhibit CRM1-dependent nuclear export. We therefore analyzed the subcellular localization of CRM1 substrates in control cells and in cells treated with the prostaglandin. RanBP1 is a Ran-binding protein that contains an NES and is actively exported from the nucleus to the cytoplasm (45). Incubation of cells with 15d-PGJ2 resulted in a clear shift of endogenous RanBP1 from a cytoplasmic localization, as observed in control cells, to an equal distribution between the nucleus and the cytoplasm (Fig. 4A). CAY 10410, which lacks the α,β-unsaturated carbonyl group of the reactive cyclopentenone ring of 15d-PGJ2 (Fig. 1A), did not lead to a redistribution of RanBP1 (Fig. 4A), suggesting that the mode of action of 15d-PGJ2 involves a Michael-addition to a cellular component. 15d-PGJ2 is known to lead to oxidative stress under certain conditions (46). Incubation of cells with H2O2, however, did not change the localization of RanBP1. Hence, oxidative stress does not affect nuclear export in our experimental system.
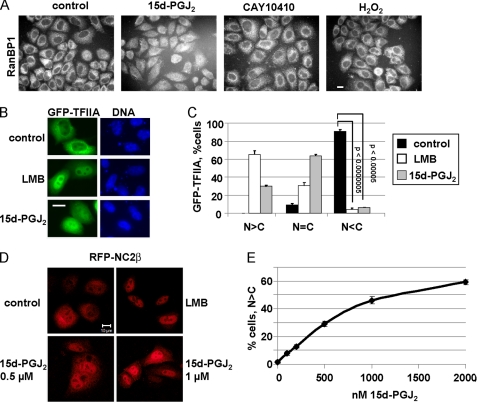
FIGURE 4.
15d-PGJ2 inhibits nuclear export in vivo. A, 15d-PGJ2 inhibits nuclear export of RanBP1. Cells were treated with 30 μm of 15d-PGJ2 or CAY10410 or 10 mm H2O2 for 1.5 h as indicated and analyzed for localization of endogenous RanBP1. B and C, cells were transfected with a plasmid coding for GFP-TFIIA and treated with ethanol as a control or 5 nm LMB or 30 μm 15d-PGJ2, as indicated. C, quantification of the subcellular localization of GFP-TFIIA. Whereas ∼95% of all cells showed a more cytoplasmic localization (N<C) of GFP-TFIIA under control conditions, this percentage significantly decreased in the presence of either LMB or 15d-PGJ2. D, HeLa cells were transfected with a plasmid coding for RFP-NC2β and treated for 2 h with ethanol (control), 5 nm LMB, or 0.5 or 1 μm 15d-PGJ2 as indicated. E, quantification of cells showing a clear nuclear localization of RFP-NC2β (N>C) in the presence of increasing concentrations of 15d-PGJ2. Error bars in C and E (sometimes too small to be seen) indicate the standard deviation from the mean of three independent experiments, counting ∼100 cells per condition. p values in E (in relation to 0 nm) were < 0.01 for concentrations of 15d-PGJ2 up to 500 nm and <0.001 for higher concentrations. Bars, 10 μm (A, B, D).
We also analyzed the subcellular localization of various transfected reporter proteins in the absence or presence of 15d-PGJ2. As a GFP fusion protein, the transcription factor TFIIAα is predominantly cytoplasmic (Fig. 4B, control). LMB lead to rapid nuclear accumulation of TFIIAα, implicating CRM1 as the export receptor that constantly transports TFIIAα out of the nucleus. Similar to LMB, 15d-PGJ2 induced nuclear accumulation of TFIIAα (Fig. 4, B and C). We also tested an artificial reporter protein containing GFP and an NES, mediating CRM1-dependent export (GFP-NES). In control cells, it localized either to the cytoplasm or showed an equal distribution between the nucleus and the cytoplasm (supplemental Fig. S2, A and B). Upon incubation of cells with 15d-PGJ2, almost all cells showed an equal distribution between the two compartments, suggesting that CRM1-dependent export was inhibited.
For the in vivo assays presented so far, we used 15d-PGJ2 concentrations in the range of 10–30 μm. These concentrations were based on effects of the prostaglandin in various cellular assays as described in the literature (9–12). We next searched for nucleocytoplasmic shuttling proteins whose subcellular localization might be affected by much lower concentrations of 15d-PGJ2. As shown in Fig. 4, D and E, the transcriptional regulator NC2β (negative cofactor 2β), a protein that can be exported from the nucleus in a CRM1-dependent manner (47), mostly exhibited an equal distribution between the nucleus and the cytoplasm in control cells. Incubation of transfected cells in the presence of LMB resulted in a strong nuclear accumulation of NC2β, indicating that CRM1 functions as the major if not the only export receptor for this protein. Similarly, NC2β localized predominantly to the nucleus (N>C) in many cells in the presence of 0.5 or 1.0 μm 15d-PGJ2 (Fig. 4D). A careful titration revealed that in the presence of 100–200 nm 15d-PGJ2 NC2β could be detected in the nucleus in a significant number of cells (Fig. 4E). Hence, comparatively low concentrations of 15d-PGJ2 (e.g. in the high nanomolar range) can affect the localization of certain proteins.
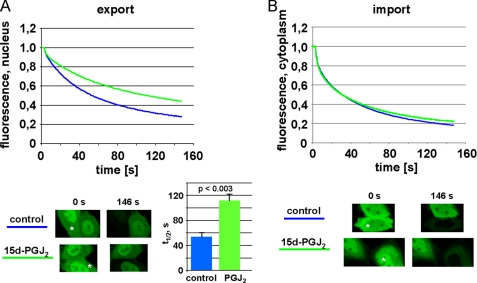
We then characterized the effects of 15d-PGJ2 on nucleocytoplasmic transport by live cell imaging. Our reporter protein for this assay, NES-GFP2-M9, uses transportin as an import receptor and CRM1 as an export receptor. As shown by FLIP- (fluorescence loss in photobleaching) analysis, 15d-PGJ2 had an inhibitory effect on the kinetics of nuclear export in living cells (Fig. 5A; see also supplemental Fig. S3). In control cells, it took ∼54 s until 50% of the nuclear fluorescence was lost upon bleaching a cytoplasmic region of the cell. In the presence of 15d-PGJ2, this t½ was doubled to ∼111 s. LMB, the established inhibitor of CRM1-dependent nuclear export, was only slightly more effective than 15d-PGJ2 in this assay (data not shown). Transportin-dependent nuclear import, on the other hand, was not affected by the prostaglandin (Fig. 5B).
FIGURE 5.
FLIP analysis of export inhibition by 15d-PGJ2. Cells were transfected with a plasmid coding for NES-GFP2-M9 and subjected to FLIP-analysis in the presence of 15 μm 15d-PGJ2 or ethanol as a control. The graphs show the mean of three (A, export) or two (B, import) independent experiments, analyzing 10–20 cells each. Error bars are omitted for clarity (see supplemental Fig. S3 for the original data set of a single experiment). A, a region in the cytoplasm was bleached to analyze loss of fluorescence from the nucleus, i.e. nuclear export. The bar graph shows t½, i.e. the time it takes to loose 50% of the nuclear fluorescence. The bars indicate the standard deviation from the mean of three independent experiments. B, a region in the nucleus was bleached to analyze loss of fluorescence from the cytoplasm, i.e. nuclear import. Single cells before the first (t = 0 s) and after the last (t = 146 s) bleach interval are depicted. Asterisks indicate the bleached compartments. Note that in the ethanol control (A) the bleached cell looses the vast majority of its fluorescence, indicating efficient export. The 15d-PGJ2-treated cell, by contrast, retains a significant level of fluorescence in the nucleus, indicating inhibited export.
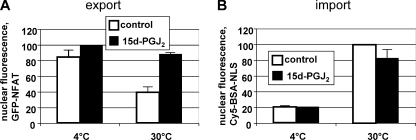
Next, we analyzed the effect of 15d-PGJ2 on nuclear transport in digitonin-permeabilized cells. To this end, we used our well-established in vitro nuclear transport assay that allows simultaneous quantification of nuclear import and export (31). It is based on HeLa cells expressing a GFP-tagged version of the transcription factor NFAT (nuclear factor of activated T cells). Export of NFAT is mediated by the export receptor CRM1 (31). Briefly, permeabilized cells with nuclear GFP-NFAT are incubated in the presence of a cytosolic extract and a fluorescently labeled, importin α/β-dependent nuclear import substrate. After the reaction, nuclear fluorescence of import and export cargoes is analyzed by flow cytometry. As shown in Fig. 6, 15d-PGJ2 strongly inhibited nuclear export of GFP-NFAT. Import of the cNLS-import substrate, by contrast, was only slightly inhibited by the prostaglandin. Together, these results suggest that 15d-PGJ2 exerts an inhibitor effect on nuclear export that involves a Michael addition to CRM1. The importin α/β- and the transportin-dependent nuclear import pathways, by contrast, are not compromised by 15d-PGJ2.
FIGURE 6.
15d-PGJ2 inhibits nuclear export, but not import, in vitro. After digitonin permeabilization and preincubation with 15d-PGJ2 (black bars) or ethanol as a control (gray bars), GFP-NFAT-expressing cells were subjected to nuclear transport reactions on ice or at 30 °C, as indicated. A, nuclear export was quantified by measuring the residual nuclear GFP-NFAT fluorescence. B, nuclear import was quantified by measuring nuclear Cy5-BSA-NLS fluorescence. Note that the same cells were analyzed in A and B. Error bars show the variation from the mean of two independent experiments.
15d-PGJ2 and LMB Target Cysteine 528 of CRM1
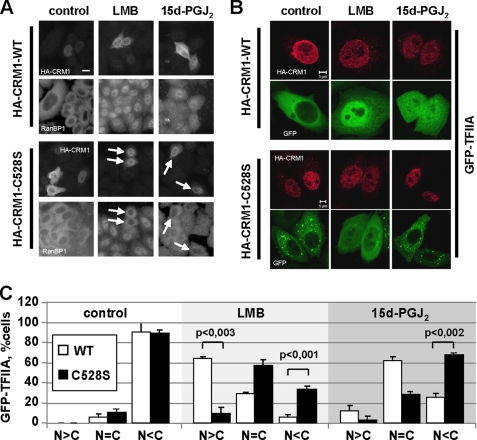
LMB is a very potent inhibitor of CRM1-dependent export. It very selectively binds to Cys-528 of human CRM1. Yeast cells carrying a mutation at the corresponding site are LMB-resistant (26). We therefore asked the question, whether the same amino acid in CRM1 is targeted by 15d-PGJ2. We expressed an HA-tagged version of CRM1, both as a wild-type protein and as a C528S mutant. The cysteine-serine mutation should render the protein LMB-resistant and, possibly, also insensitive to 15d-PGJ2. Cells expressing wild-type HA-CRM1 showed a cytoplasmic localization of the endogenous reporter protein RanBP1, demonstrating that exogenous CRM1 does not interfere with nuclear export (Fig. 7A). Upon addition of LMB or 15d-PGJ2 to the cells, RanBP1 localization was shifted toward the nucleus, as described above. Strikingly, cells expressing HA-CRM1-C528S were resistant to LMB and also to 15d-PGJ2 treatment, as RanBP1 localized predominantly to the cytoplasm in these cells (Fig. 7A).
FIGURE 7.
CRM1-C528S rescues nuclear export in the presence of LMB or 15d-PGJ2. A, cells were transfected with plasmids coding for HA-tagged wild-type CRM1 (HA-CRM1-WT) or the CRM1 mutant C528S (HA-CRM1-C528S) as indicated. After treatment with 5 nm LMB or 30 μm 15d-PGJ2 for 1.5 h, cells were fixed and stained with antibodies against RanBP1 and the HA tag. LMB/15d-PGJ2-resistant cells expressing CRM1C528S are indicated by arrows. B, cells were co-transfected with plasmids coding for CRM1 (mutant or wild-type, as indicated) and the reporter protein GFP-TFIIA. After treatment with 5 nm LMB or 15 μm 15d-PGJ2 for 1.5 h, cells were fixed and stained with antibodies against the HA tag. Bars, 10 μm (A) or 5 μm (B). C, quantitative analysis of the subcellular localization of GFP-TFIIA in B. Only cells expressing wild-type HA-CRM or mutant HA-CRM1-C528S were included in the analysis. Error bars indicate the standard deviation from the mean of three independent experiments, counting >100 cells per condition.
We also analyzed the subcellular localization of our reporter protein GFP-TFIIAα in cells treated with LMB or 15d-PGJ2 and co-expressing either wild-type or mutant CRM1. Again, CRM1-C528S partially rescued the export defect in the presence of LMB. Similarly, the majority of cells expressing HA-CRM1-C528S showed a cytoplasmic localization of the reporter protein in the presence of 15d-PGJ2 (Fig. 7, B and C), in contrast to cells expressing only endogenous CRM1 (compare Fig. 4, B and C) or transfected wild-type HA-CRM1 (Fig. 7, B and C). A similar rescue of nuclear export in the presence of LMB or 15d-PGJ2 was also observed in cells co-expressing HA-CRM1-C528S and GFP-NES (supplemental Fig. S4) or RFP-NC2β (data not shown). Together, our results show that the cellular prostaglandin 15d-PGJ2, like the fungal metabolite LMB, inhibits CRM1-mediated nuclear protein export by binding to a critical amino acid, cysteine 528.
DISCUSSION
In this study, we show that prostaglandin 15d-PGJ2 functions as an inhibitor of CRM1-dependent nuclear export. Interestingly, 15d-PGJ2 has recently been shown to induce nuclear accumulation of cytidylyltransferase, a key enzyme in the synthesis of phosphatidylcholine (48). The underlying molecular mechanisms, however, have not been analyzed. In contrast to nuclear export, the importin α/β- and the transportin-dependent nuclear protein import pathways are not affected by 15d-PGJ2, demonstrating the specificity of the effect.
Several CRM1 inhibitors have been described, besides LMB (27, 28, 49). 15d-PGJ2, however, is a CRM1 inhibitor that can be produced by higher eukaryotic cells and may therefore function as an endogenous regulator of CRM1-mediated nuclear protein export. Furthermore, 15d-PGJ2 and/or derivatives of this prostaglandin are potential therapeutic agents and the elucidation of their molecular functions will be key for further clinical applications.
LMB affects nuclear export at nanomolar concentrations. For 15d-PGJ2, by contrast, higher concentrations are required to observe inhibitory effects, as reported for cytidyltransferase (48). Although we showed that exogenous 15d-PGJ2 can enter cells and localize in the nucleus, we do not know at present how efficiently it can passage the plasma membrane. Furthermore, it could also be re-exported from cells as glutathione S-conjugates (41), increasing the concentration that is required to see effects on nuclear export. In cell-free exudates, endogenous, unconjugated 15d-PGJ2 was detected at levels up to ∼16 nm (6). This, however, is probably an underestimation of initial, cellular 15d-PGJ2, which can be rapidly complexed to cellular components. When tissue samples were treated with Raney nickel to hydrogenate the lipid(s), increased amounts of 15d-PGJ2 could be detected (data not shown). Hence, it is conceivable that concentrations of endogenously generated 15d-PGJ2 can reach levels inside the producing cells that are sufficiently high to affect certain target proteins. Indeed, NC2β is a nucleocytoplasmic shuttling protein that accumulated in the nucleus at submicromolar concentrations of exogenously added 15d-PGJ2, i.e. 1–2 orders of magnitude below the 15d-PGJ2 concentration described in other experimental systems (9–12). The reason for the difference in sensitivity of transported proteins to 15d-PGJ2 is unclear. Apparently, NC2β is a member of a group of substrates with a rather low affinity for the export receptor CRM1, potentially explaining its sensitivity to CRM1 inhibition.
In the context of CRM1 inhibition, it is interesting to note that isoforms of PGHS, which form prostaglandin PGH2 from arachidonic acid, partially localize to the inner and the outer nuclear membrane (50). Similarly, cytosolic phospholipase A2, the arachidonic acid-releasing enzyme, translocates to nuclear membranes under certain conditions (51). Hence, the active concentration of reactive prostaglandin species could be sufficiently high in the vicinity of the nuclear envelope to modify CRM1, which can associate with components of the nuclear pore complex (22), leading to inhibition of the nuclear export pathway.
CRM1 function is required for the expression of a number of pro-inflammatory genes during a T-cell response (43). Inhibition of the CRM1-pathway by 15d-PGJ2 may contribute to the well-defined anti-inflammatory effects of the prostaglandin, together with its effects on the transcription factor PPARγ (3, 7) and the NFκB/IκB-signaling pathway (9, 52). One example for such a function of 15d-PGJ2 was demonstrated for the resolution of acute inflammations (6). Similarly, regulation of the CRM1 pathway by 15d-PGJ2 may be relevant in other cellular systems. One example are dendritic cells, where CRM1 activity is required for cell maturation (53). For CRM1-dependent genes, effects on transcriptional activation may contribute to their reduced expression under conditions of CRM1 inhibition. In these systems, subtle changes of CRM1 activity upon partial inhibition by endogenous or exogenous 15d-PGJ2 could have profound effects on gene expression and, hence, cellular functions.
Supplementary Material
Acknowledgments
We thank Dr. Frauke Melchior for general support of the project. We also thank Stephanie Roloff for cloning of HA-CRM1-C528S, Dr. Gerard Cagney for help with mass spectrometry, Sarah Wälde for help with FLIP-analysis, Dr. Marc DeVocelle for the initial synthesis of biotinylated 15d-PGJ2, the Mass Spectrometry Resource Conway Institute for informatics access, and Dr. Detlef Doenecke and Dr. Peter Rehling for comments on the manuscript.
This work was funded in part by a Research Program Grant, Faculty of Medicine, Georg-August-University, Göttingen (to R. H. K.) and by the Health Research Board of Ireland and the Cancer Research Ireland (to D. J. F.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- PGH2
- prostaglandin H2
- 15d-PGJ2
- 15-deoxy-Δ12,14prostaglandin J2
- FLIP
- fluorescence loss in photobleaching
- GFP
- green fluorescent protein
- LC-ESI-MS/MS
- liquid chromatography/ electrospray ionization tandem mass spectrometry
- NES
- nuclear export signal
- NFAT
- nuclear factor of activated T cells
- NLS
- nuclear localization signal
- PGHS
- prostaglandin peroxide H synthase
- DTT
- dithiothreitol
- HA
- hemagglutinin
- GFP
- green fluorescent protein
- BSA
- bovine serum albumin
- LMB
- leptomycin B.
REFERENCES
- 1.Smith W. L. (1989) Biochem. J. 259, 315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Funk C. D. (2001) Science 294, 1871–1875 [DOI] [PubMed] [Google Scholar]
- 3.Scher J. U., Pillinger M. H. (2005) Clin Immunol. 114, 100–109 [DOI] [PubMed] [Google Scholar]
- 4.Straus D. S., Glass C. K. (2001) Med. Res. Rev. 21, 185–210 [DOI] [PubMed] [Google Scholar]
- 5.Uchida K., Shibata T. (2008) Chem. Res. Toxicol. 21, 138–144 [DOI] [PubMed] [Google Scholar]
- 6.Rajakariar R., Hilliard M., Lawrence T., Trivedi S., Colville-Nash P., Bellingan G., Fitzgerald D., Yaqoob M. M., Gilroy D. W. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 20979–20984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forman B. M., Tontonoz P., Chen J., Brun R. P., Spiegelman B. M., Evans R. M. (1995) Cell 83, 803–812 [DOI] [PubMed] [Google Scholar]
- 8.Kliewer S. A., Lenhard J. M., Willson T. M., Patel I., Morris D. C., Lehmann J. M. (1995) Cell 83, 813–819 [DOI] [PubMed] [Google Scholar]
- 9.Rossi A., Kapahi P., Natoli G., Takahashi T., Chen Y., Karin M., Santoro M. G. (2000) Nature 403, 103–108 [DOI] [PubMed] [Google Scholar]
- 10.Vunta H., Davis F., Palempalli U. D., Bhat D., Arner R. J., Thompson J. T., Peterson D. G., Reddy C. C., Prabhu K. S. (2007) J. Biol. Chem. 282, 17964–17973 [DOI] [PubMed] [Google Scholar]
- 11.Oliva J. L., Pérez-Sala D., Castrillo A., Martínez N., Cañada F. J., Boscá L., Rojas J. M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 4772–4777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim W. J., Kim J. H., Jang S. K. (2007) EMBO J. 26, 5020–5032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kondo M., Shibata T., Kumagai T., Osawa T., Shibata N., Kobayashi M., Sasaki S., Iwata M., Noguchi N., Uchida K. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 7367–7372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fried H., Kutay U. (2003) Cell. Mol. Life Sci. 60, 1659–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pollard V. W., Michael W. M., Nakielny S., Siomi M. C., Wang F., Dreyfuss G. (1996) Cell 86, 985–994 [DOI] [PubMed] [Google Scholar]
- 16.Fischer U., Huber J., Boelens W. C., Mattaj I. W., Lührmann R. (1995) Cell 82, 475–483 [DOI] [PubMed] [Google Scholar]
- 17.Wen W., Meinkoth J. L., Tsien R. Y., Taylor S. S. (1995) Cell 82, 463–473 [DOI] [PubMed] [Google Scholar]
- 18.Fornerod M., Ohno M., Yoshida M., Mattaj I. W. (1997) Cell 90, 1051–1060 [DOI] [PubMed] [Google Scholar]
- 19.Fukuda M., Asano S., Nakamura T., Adachi M., Yoshida M., Yanagida M., Nishida E. (1997) Nature 390, 308–311 [DOI] [PubMed] [Google Scholar]
- 20.Ossareh-Nazari B., Bachelerie F., Dargemont C. (1997) Science 278, 141–144 [DOI] [PubMed] [Google Scholar]
- 21.Stade K., Ford C. S., Guthrie C., Weis K. (1997) Cell 90, 1041–1050 [DOI] [PubMed] [Google Scholar]
- 22.Kehlenbach R. H., Dickmanns A., Kehlenbach A., Guan T., Gerace L. (1999) J. Cell Biol. 145, 645–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hutten S., Kehlenbach R. H. (2007) Trends Cell Biol. 17, 193–201 [DOI] [PubMed] [Google Scholar]
- 24.Turner J. G., Marchion D. C., Dawson J. L., Emmons M. F., Hazlehurst L. A., Washausen P., Sullivan D. M. (2009) Cancer Res. 69, 6899–6905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolff B., Sanglier J. J., Wang Y. (1997) Chem. Biol. 4, 139–147 [DOI] [PubMed] [Google Scholar]
- 26.Kudo N., Matsumori N., Taoka H., Fujiwara D., Schreiner E. P., Wolff B., Yoshida M., Horinouchi S. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 9112–9117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Köster M., Lykke-Andersen S., Elnakady Y. A., Gerth K., Washausen P., Höfle G., Sasse F., Kjems J., Hauser H. (2003) Exp. Cell Res. 286, 321–331 [DOI] [PubMed] [Google Scholar]
- 28.Meissner T., Krause E., Vinkemeier U. (2004) FEBS Lett. 576, 27–30 [DOI] [PubMed] [Google Scholar]
- 29.Wang P., Liu G. H., Wu K., Qu J., Huang B., Zhang X., Zhou X., Gerace L., Chen C. (2009) J. Cell Sci. 122, 3772–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Charneau P., Mirambeau G., Roux P., Paulous S., Buc H., Clavel F. (1994) J. Mol. Biol. 241, 651–662 [DOI] [PubMed] [Google Scholar]
- 31.Kehlenbach R. H., Dickmanns A., Gerace L. (1998) J. Cell Biol. 141, 863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hutten S., Wälde S., Spillner C., Hauber J., Kehlenbach R. H. (2009) J. Cell Sci. 122, 1100–1110 [DOI] [PubMed] [Google Scholar]
- 33.Kehlenbach R. H., Gerace L. (2002) Methods Mol. Biol. 189, 231–245 [DOI] [PubMed] [Google Scholar]
- 34.Mahajan R., Delphin C., Guan T., Gerace L., Melchior F. (1997) Cell 88, 97–107 [DOI] [PubMed] [Google Scholar]
- 35.Melchior F., Sweet D. J., Gerace L. (1995) Methods Enzymol. 257, 279–291 [DOI] [PubMed] [Google Scholar]
- 36.Paschal B. M., Gerace L. (1995) J. Cell Biol. 129, 925–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Askjaer P., Bachi A., Wilm M., Bischoff F. R., Weeks D. L., Ogniewski V., Ohno M., Niehrs C., Kjems J., Mattaj I. W., Fornerod M. (1999) Mol. Cell. Biol. 19, 6276–6285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kehlenbach R. H., Assheuer R., Kehlenbach A., Becker J., Gerace L. (2001) J. Biol. Chem. 276, 14524–14531 [DOI] [PubMed] [Google Scholar]
- 39.Hutten S., Kehlenbach R. H. (2006) Mol. Cell. Biol. 26, 6772–6785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Narumiya S., Ohno K., Fukushima M., Fujiwara M. (1987) J. Pharmacol Exp. Ther. 242, 306–311 [PubMed] [Google Scholar]
- 41.Meister A., Anderson M. E. (1983) Annu. Rev. Biochem. 52, 711–760 [DOI] [PubMed] [Google Scholar]
- 42.Cox B., Murphey L. J., Zackert W. E., Chinery R., Graves-Deal R., Boutaud O., Oates J. A., Coffey R. J., Morrow J. D. (2002) Biochim. Biophys. Acta 1584, 37–45 [DOI] [PubMed] [Google Scholar]
- 43.Schütz S., Chemnitz J., Spillner C., Frohme M., Hauber J., Kehlenbach R. H. (2006) J. Mol. Biol. 358, 997–1009 [DOI] [PubMed] [Google Scholar]
- 44.Bischoff F. R., Görlich D. (1997) FEBS Lett. 419, 249–254 [DOI] [PubMed] [Google Scholar]
- 45.Richards S. A., Lounsbury K. M., Carey K. L., Macara I. G. (1996) J. Cell Biol. 134, 1157–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kondo M., Oya-Ito T., Kumagai T., Osawa T., Uchida K. (2001) J. Biol. Chem. 276, 12076–12083 [DOI] [PubMed] [Google Scholar]
- 47.Kahle J., Piaia E., Neimanis S., Meisterernst M., Doenecke D. (2009) J. Biol. Chem. 284, 9382–9393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ryan A. J., Chen B. B., Vennalaganti P. R., Henderson F. C., Tephly L. A., Carter A. B., Mallampalli R. K. (2008) J. Biol. Chem. 283, 24628–24640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Daelemans D., Afonina E., Nilsson J., Werner G., Kjems J., De Clercq E., Pavlakis G. N., Vandamme A. M. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 14440–14445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spencer A. G., Woods J. W., Arakawa T., Singer, Smith W. L. (1998) J. Biol. Chem. 273, 9886–9893 [DOI] [PubMed] [Google Scholar]
- 51.Perisic O., Paterson H. F., Mosedale G., Lara-González S., Williams R. L. (1999) J. Biol. Chem. 274, 14979–14987 [DOI] [PubMed] [Google Scholar]
- 52.Straus D. S., Pascual G., Li M., Welch J. S., Ricote M., Hsiang C. H., Sengchanthalangsy L. L., Ghosh G., Glass C. K. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 4844–4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chemnitz J., Turza N., Hauber I., Steinkasserer A., Hauber J. (2009) Immunobiology 20, 370–379 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.