Abstract
Pituitary tumor transforming gene (PTTG1, securin) is involved in cell-cycle control through inhibition of sister-chromatid separation. Elevated levels of PTTG1 were found to be associated with many different tumor types that might be involved in late stage tumor progression. However, the role of PTTG1 in early stage of tumorigenesis is unclear. Here we utilized the adenovirus expression system to deliver PTTG1 into normal human fibroblasts to evaluate the role of PTTG1 in tumorigenesis. Expressing PTTG1 in normal human fibroblasts inhibited cell proliferation. Several senescence-associated (SA) phenotypes including increased SA-β-galactosidase activities, decreased bromodeoxyuridine incorporation, and increased SA-heterochromatin foci formation were also observed in PTTG1-expressing cells, indicating that PTTG1 overexpression induced a senescent phenotype in normal cells. Significantly, the PTTG1-induced senescence is p53-dependent and telomerase-independent, which is distinctively different from that of replicative senescence. The mechanism of PTTG1-induced senescence was also analyzed. Consistent with its role in regulating sister-chromatid separation, overexpression of PTTG1 inhibited the activation of separase. Consequently, the numbers of cells with abnormal nuclei morphologies and chromosome separations were increased, which resulted in activation of the DNA damage response. Thus, we concluded that PTTG1 overexpression in normal human fibroblasts caused chromosome instability, which subsequently induced p53-dependent senescence through activation of DNA-damage response pathway.
Keywords: Aging, Cell Cycle, Chromosomes, DNA Damage, p53, PTTG1, Securin, Senescence
Introduction
The pituitary tumor transforming gene (PTTG1 or human securin) was first identified from rat pituitary tumor cells (1). It was then found to be a securin that is involved in cell-cycle control through inhibiting sister-chromatid separation (2). PTTG1 mRNA can be detected in adult testis, thymus, placenta, colon, small intestine, spleen, pancreas, brain, lung, and fetal liver (3, 4). However, PTTG1 protein levels are very low or undetectable in most normal human tissues. Notably, the expression of PTTG1 is elevated in many tumor types such as pituitary adenomas, primary epithelial neoplasms, and hemopoietic malignancies (for review, see Ref. 5) and is associated with metastasis and a poor clinical outcome with several types of tumors (6). Moreover, transfection of PTTG1 into NIH 3T3 cells results in anchorage-independent transformation in vitro and tumor formation in athymic nude mice (1). Thus, PTTG1 has been implicated as a proto-oncogene that would seem to be involved in tumorigenesis.
Increased PTTG1 levels cause mis-segregation of chromosomes and facilitate genome instability and thereby lead to cancer formation (7). In addition to its function in mitosis, where the protein binds separase to control the onset of sister-chromatid separation, PTTG1 also affects tumorigenesis through various other mechanisms. First, PTTG1 protein is able to bind p53 to regulate apoptosis and its transcriptional activity in tumor cells (8). Second, it might also interact with Ku protein, a regulatory subunit of the DNA-dependent protein kinase, to modulate the double strand DNA damage response (9) because it has been shown that the PTTG1-Ku interaction is disrupted upon DNA damage (9). Third, PTTG1 has been demonstrated both in vitro and in vivo to induce angiogenesis possibly through the induction of basic fibroblast growth factor secretion (4, 10). Thus, PTTG1 seems to render its effects on tumorigenesis through a combination of diverse mechanisms. However, because most experiments have been conducted in cancer cells that already have significant levels of chromosome aberrations, this scenario does not fully explain the role of PTTG1 in tumorigenesis. Immortal cells tend to respond to DNA damage or oncogenes by undergoing apoptosis or neoplastic transformation. In contrast, normal human fibroblasts respond to DNA damage by adopting a phenotype that closely resembles replicative senescence (11). Thus, normal human cells differ remarkably from immortal cells in their response to potentially oncogenic stimuli. Although higher expression of PTTG1 is frequently observed in malignant tumors, the role of PTTG1 in human cancers has yet to be clearly defined. The question of how PTTG1 overexpression affects normal cells has not been addressed.
To evaluate the role of PTTG1 in normal cells, we overexpressed PTTG1 in normal human fibroblasts and analyzed the cellular effects. Our results indicated that forced expression of PTTG1 inhibited the proliferation of normal human fibroblasts. This was accompanied by the induction of several senescence markers including senescence-associated acidic β-galactosidase (SA-β-gal)4 activity and the appearance of senescence-associated heterochromatin foci (SAHF) in these cells. This PTTG1-induced senescence is p53-dependent and telomerase-independent and is, thus, distinctively different from that of replicative senescence. It is similar to that of oncogene-induced senescence in that it serves as a failsafe mechanism to protect normal cells from transformation at the early stages of tumorigenesis. Thus, our results provide the first indication for the involvement of PTTG1 in the early stages of tumor formation.
EXPERIMENTAL PROCEDURES
Cell Culture
Human primary normal lung fibroblast IMR90 and foreskin fibroblast BJ-1 were maintained in minimum essential medium (Invitrogen) with 10% fetal bovine serum. Ad293 cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen) growth medium supplemented with 10% fetal bovine serum. BJ-hTERT cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) with Medium 199 (Invitrogen) at a 4:1 ratio with 10% fetal bovine serum.
Adenoviral-mediated Gene Transfer
The AdEasyTM XL Adenoviral Vector System (Stratagene) was used to generate recombinant adenoviruses. Approximately 50–70% confluence Ad293 cells were plated in 6-well plates for 24 h before transfection. PacI-digested recombinant adenoviral vector DNA (4 μg) was transfected into Ad293 cells using LipofectamineTM 2000 (Invitrogen). Viruses were collected 7–9 days after transfection. The viral lysates were freeze-thawed three times and then used to infect new Ad293 cells. After three rounds of infection, the viral titers were often high enough for later amplification. To generate higher titer viral stocks, viruses at a multiplicity of infection (m.o.i.) equal to 10 were used to infect 5 × 108 packaging cells. After 30–40 h the resultant viruses were then purified using the CsCl banding technique. The titer of the adenoviruses ranged between 106 and 107 plaque-forming units/μl.
Lentiviral Constructs, Virus Production, and Normal Cell Infection
All four lentiviral plasmids containing shRNA sequences against p53 were obtained from National RNAi core facility in Taiwan. The retroviral knockdown system was based on the RNAi Consortium (TRC) library pLKO.1 hairpin plasmid, the pCMV-ΔR8.91 packaging plasmid, and the pMD.G envelope plasmid (12). Lentiviral transductions were performed in 293T cells following the standard protocol.
SA-β-galactosidase Staining
The cells were washed with PBS, fixed for 3–5 min in 3% formaldehyde, and then incubated with fresh SA-β-gal stain solution containing 1 mg/ml X-gal, 40 mm citric acid, sodium phosphate, pH 6.0, 5 mm potassium ferrocyanide, 5 mm potassium ferricyanide, 150 mm NaCl, and 2 mm MgCl2 at 37 °C for 12–16 h. At least 250 cells were counted in randomly chosen fields for each sample.
Bromodeoxyuridine (BrdUrd) Incorporation Assay
About 104 cells were plated in 12-well plates using glass slides and labeling with 100 μm BrdUrd for 4 h. The cells were then fixed with 4% paraformaldehyde for 10 min followed by three washes using PBS. DNA within the cells were denatured on ice with 1 m HCl for 10 min, 2 m HCl at 25 °C for 10 min, and 2 m HCl at 37 °C for 20 min and then washed with PBS. Neutralization of the DNA was conducted using Tris borate-EDTA buffer, pH 8.4. The cells were then blocked in Tris-buffered saline solution containing 1% bovine serum albumin and 0.1% Triton X-100 for 1 h. Monoclonal antibody against BrdUrd (BU-33, Sigma) was then added and incubated at 4 °C overnight. Cells were washed and incubated with horseradish peroxidase-conjugated rabbit anti-mouse secondary antibody (Jackson ImmunoResearch) for 30 min. A solution containing 3,3′-diaminobenzidine tetraanhydrochloride (0.16 mg/ml) and 0.3% H2O2 was then added and incubated for 30 min in the dark. Cells were also stained with hematoxylin solution.
Expression and Purification of His6-tagged PTTG1 for Antibody Production
Plasmid pET6H-PTTG1, which was used to purify the recombinant PTTG1, was constructed by inserting the SmaI-SpeI fragment of pUC18-PTTG1 into SmaI-SpeI-digested pET6H (a gift from C.-H. Hu, National Marine University, Taipei, Taiwan). The resulting plasmid was used to express His6-tagged PTTG1 under the control of the T7 promoter. To purify His6-tagged PTTG1, a 1-liter culture of Escherichia coli harboring pET6H-PTTG1 was grown at 25 °C to an A600 of 0.5 and induced with the addition of 1 mm isopropyl-1-thio-β-d-galactopyranoside. The cells were grown at 25 °C for overnight before harvesting by centrifugation. Cells were resuspended in 10 ml of sonication buffer (50 mm NaH2PO4, pH 7.8, 300 mm NaCl, 1 mm β-mercaptoethanol, 1× protease inhibitors (Calbiochem)) and sonicated to release the cell contents. The sonicated cells were centrifuged at 13,000 × g for 15 min at 4 °C to obtain total cell free extracts. Nickel-nitrilotriacetic acid-agarose (Qiagen) (0.5 ml) was added to the total cell-free extracts and incubated at 4 °C for 1 h. The resin was washed and eluted with 2 ml of buffer containing 50 mm NaH2PO4, pH 8.0, 300 mm NaCl, and 20 mm imidazole. Purified protein was separated into aliquots and frozen by a dry ice-ethanol bath. The yield of PTTG1 was 20 mg from 1 liter of E. coli culture. Polyclonal antibodies against PTTG1 were generated using a rabbit.
Immunoblotting Analysis
Cells were washed with PBS and lysed in Nonidet P-40 lysis buffer (150 mm NaCl, 1.0% Nonidet P-40, 50 mm Tris-HCl, pH 8.0, 1 mm phenylmethylsulfonyl fluoride), 1 mm EDTA) containing a standard mixture of protease inhibitors. After 30 min on ice, the lysates were cleared by centrifugation. Samples corresponding to 50–100 μg of protein (Bio-Rad protein assay) were separated on 8 or 15% SDS-PAGE gels and transferred to nitrocellulose membranes. Western blot analysis was carried out according to standard procedures using ECL detection (PerkinElmer Life Sciences). The membranes were hybridized with the following antibodies: anti-p53 (Ab2, Oncogene), anti-Rb (Ab5, Oncogene), anti-p21waf1 (Ab3, Calbiochem), anti-p16 (Ab1, Calbiochem), anti-phosphohistone H2AX (Ser-139, Upstate Biotechnology), anti-separase (ab3762, Abcam), and anti-phospho-Rb (phosphoserine 780, Sigma). Equivalent loading of lanes was verified by hybridization with anti-GAPDH and anti-actin antibodies (Calbiochem). Horseradish peroxidase-conjugated donkey anti-rabbit or sheep anti-mouse antibodies (Amersham Biosciences) were used as the secondary antibodies.
Growth Curves
The proliferative capacity of cells was monitored by seeding of 2 × 105 into 30-mm dish containing 10% fetal bovine serum. Adenovirus carrying GFP or PTTG1 was added the next day as described above. Cells were then digested with trypsin, stained with 0.2% trypan blue, and counted using a hemocytometer.
Cell Cycle Analysis
Cell cycle analysis was performed by monitoring the DNA content at various time points by propidium iodide staining. Cells were fixed in 70% cold ethanol followed by washing with PBS. The cells were then incubated with 1 mg/ml RNase A and 2 μg/ml propidium iodide at 4 °C for 30 min and then analyzed using a FACScan flow cytometer (BD Biosciences). About 106 cells were acquired for analysis using Cell Quest software.
Immunofluorescent Staining
Cells were grew on slides, rinsed with PBS, fixed with 4% paraformaldehyde for 10 min, and permeabilized with blocking buffer (Tris-buffered saline with 1% bovine serum albumin and 0.1% Triton X-100). Primary antibody, anti-phosphohistone H2AX (Ser-139, Upstate Biotechnology), or anti-trimethylhistone H3-Lys-9 (Upstate Biotechnology) was added and incubated at 4 °C overnight. The cells were then washed and incubated with Rhodamine Red-X (RRX)-labeled goat anti-mouse IgG (Jackson ImmunoResearch) for 1 h. The cells were also stained with Hoechst 33258 (Sigma, 1 μg/ml) for 5 min. The cells were visualized using fluorescence or confocal microscopy. Cells expressing GFP were fixed and stained with Hoechst 33258 and visualized directly. The authors acknowledge the technical services (confocal microscopy) provided by Imaging Core Facility of Nanotechnology of the University System of Taiwan-National Yang-Ming University (UST-YMU).
RESULTS
PTTG1 Expression Inhibits Normal Cell Proliferation
To evaluate the role of PTTG1 in normal cells, we used an adenovirus delivery system to facilitate the expression of PTTG1 gene into normal fibroblasts. The percentages of adenoviral-transduced cells ranged from 60 to 90%, as estimated by parallel infection using adenovirus expressing the GFP gene. We have also expressed PTTG1 in E. coli and isolated recombinant PTTG1 protein to near homogeneity (supplemental Fig. S1A). The recombinant PTTG1 was then used to generate polyclonal antibodies from rabbit. Under our assay conditions, the generated antibodies successfully detect PTTG1 with high specificity in lung cancer H1299 cells (supplemental Fig. S1B). We were unable to detect PTTG1 in IMR90 and the two other normal human fibroblasts BJ1 and WI38 before transfection (supplemental Fig. S1C and data not shown). Using the adenovirus delivery system, we effectively introduced PTTG1 into IMR90 cells. The expression level of PTTG1 protein was increased in a dose-dependent manner (supplemental Fig. S1C). An m.o.i. value equivalent to 20 was chosen for all subsequent experiments as this m.o.i. efficiently expressed PTTG1 or GFP without causing virus infective stress. At an m.o.i. equivalent to 20, the expression level of PTTG1 peaked at ∼3–4 days after infection and then quickly decreased in the IMR90 cells (supplemental Fig. S1C). Thus, our established delivery systems enabled efficient introduction of PTTG1 into normal fibroblasts.
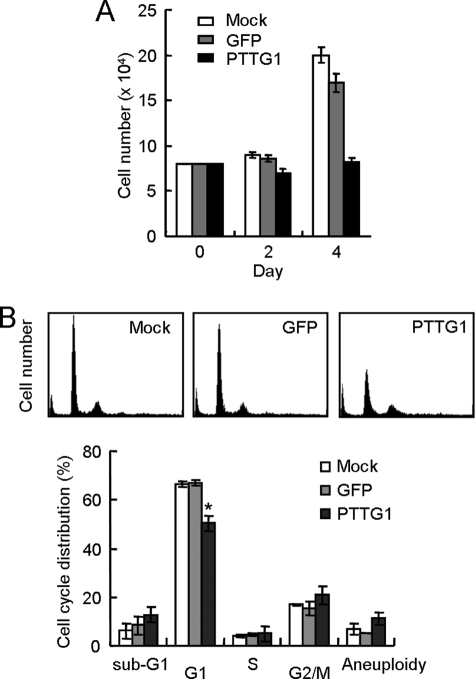
The cellular effects of forced PTTG1 expression were then evaluated in these infected normal fibroblasts. The proliferation potential of the normal cells upon PTTG1 expression was first examined. As shown in Fig. 1A, the cell number of PTTG1-expressing cells did not increase after infection, indicating that PTTG1 inhibited proliferation of normal cells. Flow cytometry analysis revealed that the proportion of G1 cells was reduced in PTTG1-expressing cells (Fig. 1B). The G1 fraction was decreased from ∼67% in the control sample to ∼50% in PTTG1-expressed cells. PTTG1 expression also slightly increased the number of aneuploid cells. Intriguingly, although PTTG1 is involved in mitosis progression, the proportion of aneuploid cells was modestly but reproducibly (albeit statistically insignificant) increased upon expression of this protein. The cell cycle profile was similar to that exhibited by senescence induced by overexpressing Raf-1 oncogene or DNA-damaging agents that the cell cycle defects were not limited to G1 phase (13, 14). It is also apparent that the number of sub-G1 cells was also not increased upon PTTG1 expression. This suggested that PTTG1 did not induce apoptosis in these normal cells. The distribution of cell cycle stages of PTTG1-expressing cells was also distinctly different from cells entering replicative senescence that arrests in G1 phase of the cell cycle. Together our results indicated that expression of PTTG1 induced a cessation of cell proliferation without causing apparent G2/M arrest or apoptosis in normal fibroblasts.
FIGURE 1.
PTTG1 overexpression inhibits normal cell proliferation. A, ∼1 × 105 IMR90 cells were infected with adenovirus carrying the GFP or PTTG1 gene at an m.o.i. equivalent to 20. The cell numbers were then counted at the times indicated. Values show the average of three experiments. B, the IMR90 cells were infected by adenoviruses carrying GFP or PTTG1 for 5 days and then analyzed by flow cytometry. The histograms of the propidium iodide-stained cells are presented (upper panel). Quantification of the cells at different cell cycle stages was then conducted. The average number for three independent experiments was determined and plotted (lower panel). An asterisk indicates p < 0.05 (p = 0.0098).
Induction of Senescence by PTTG1 Overexpression in Normal Cells
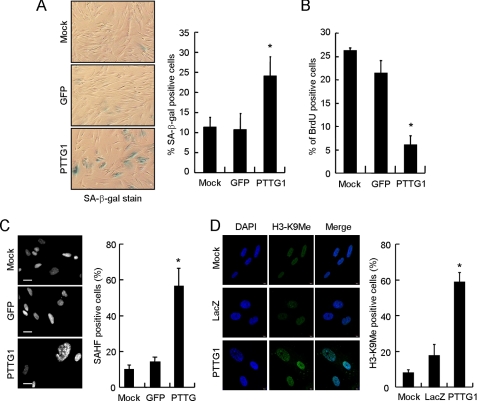
The morphology of the PTTG1-expressing normal fibroblasts was also observed. Four days after infection the PTTG1-expressed cells appeared to be flattened and enlarged, a phenotype similar to that of senescent cells (Fig. 2A and data not shown). The morphological changes were not limited to IMR90 as two other normal human fibroblast cell lines, BJ1 and WI38, also showed similar morphological changes (data not shown). These results suggested that PTTG1 overexpression induced senescent-like phenotype in normal fibroblasts. To confirm this observation, we assessed other senescence-associated features in the PTTG1-expressing fibroblasts. SA-β-gal activity (15) was assessed in PTTG1- and GFP-expressing IMR90 cells. Our results clearly indicated that IMR90 cells expressing PTTG1 showed an induction of SA-β-gal activity (Fig. 2A). The percentage of SA-β-gal-positive cells was similar for the GFP-expressing (∼11%) and the mock control cells (∼11%). In contrast, almost 25% of the PTTG1-expressing cells stained positive for SA-β-gal activities (Fig. 2A). Consistent with this, the proportion of BrdUrd-incorporating cells (16) was also reduced from ∼27 and ∼22% for normal and GFP-expressing cells to ∼7% for PTTG1-expressing cells (Fig. 2B). We further analyzed the presence of SAHF, a distinct DNA-dense heterochromatic structure that accumulates in senescent human fibroblasts, in these cells. Typically, one large nucleolus and punctuate DNA foci can be visualized by DAPI staining in senescent cells (17, 18). As shown in Fig. 2C, IMR90 cells overexpressing PTTG1 displayed an abnormal nuclear morphology with punctuate DNA foci. In comparison, GFP-expressing cells and mock control cells displayed a more uniform nuclear staining pattern (Fig. 2C). Quantitative analysis showed that there was a 4-fold increase in cells harboring SAHF foci in PTTG1-expressing cells compared with the control cells. To further confirm that the observed DNA foci were indeed SAHF, we conducted confocal fluorescence microscopy on PTTG1 senescent cells using modification-specific antibodies against histone H3-K9Me (17). The LacZ-expressing and mock control cells expressed H3-K9Me distributed throughout the nucleoplasm (Fig. 2D). In contrast, PTTG1-expressing cells showed a more distinctive localization of K9M-H3 signals, consistent with their preference for heterochromatic regions. Thus, the results demonstrated that the senescent program was initiated in response to PTTG1 overexpression in normal fibroblasts.
FIGURE 2.
Senescent phenotype observed in PTTG1-expressing normal human fibroblasts. IMR90 cells were transduced with adenoviruses carrying PTTG1 or GFP and then cultured at 37 °C for 4 days. A, the transduced cells were analyzed for SA-β-gal activity staining. Photographs of the X-gal-stained cells are shown. Quantification of the SA-β-gal positive-stained cells was conducted (right). Results were obtained from the average of three independent experiments. An asterisk indicates p < 0.05 (p = 0.0172). B, the virus-transduced cells were cultured in medium containing BrdUrd for 4 h. After labeling, the cells were fixed and stained with anti-BrdUrd antibody. The percentage of BrdUrd-positive cells is presented. Results were obtained from the average of three independent experiments. An asterisk indicates p < 0.05 (p = 0.000081). C, the virus-transduced cells were stained with DAPI and then visualized under a fluorescence microscope (left). The heterochromatin foci were quantified from three independent experiments (right). An asterisk indicates p < 0.05 (p = 0.003). D, shown are confocal images of indirect immunofluorescence of histone H3 methylation on lysine 9 (H3-K9Me) in mock control or LacZ- or PTTG1-expressed IMR90 cells. The DNA was also stained by DAPI. Quantification of the H3-K9Me foci-positive cells was conducted (right). An asterisk indicates p < 0.05 (p = 0.0003).
PTTG1 Induced Senescence in Normal Cells Depends on p53
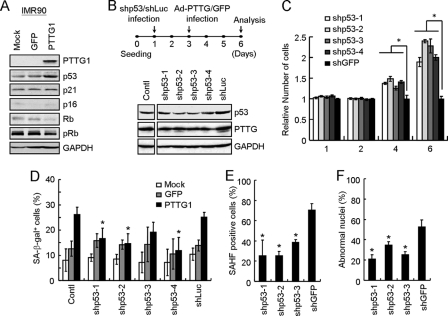
To gain insight into the molecular basis of PTTG1-induced growth arrest and senescence in normal fibroblasts, we examined the expression pattern of several key cell cycle regulators that are involved in senescence using immunoblotting analysis. In IMR90 cells, PTTG1 overexpression consistently induced p53 expression and inhibited Rb protein expression and phosphorylation (Fig. 3A). These results suggested that activation of p53 and reduction of Rb protein phosphorylation are involved in PTTG1-induced senescence. Interestingly, although p21 and p16 have been implicated as key mediators in senescence, the levels of these two proteins were not significantly altered in IMR90 cells. Nevertheless, our results suggested that p53/Rb senescent pathway was activated in response to PTTG1 overexpression in these normal cells.
FIGURE 3.
p53 is required of PTTG1-induced senescence in normal cells. A, IMR-90 cells were infected with adenoviruses carrying GFP or PTTG1. Cell extracts were prepared 4 days after infection and then analyzed by immunoblotting assays using antibodies against PTTG1, p53, p21, p16, Rb, phospho-Rb, or GAPDH. Bound antibodies were visualized by chemiluminescence using an ECL kit (Amersham Biosciences). B, IMR90 cells were infected with four different shRNAs against p53 (53sh1–4) and then cultured at 37 °C for 2 days. The cells were then infected with adenoviruses carrying PTTG1 or GFP and cultured at 37 °C for another 3 days. Cell extracts were prepared and analyzed by immunoblotting assays using antibodies against p53 or GAPDH. C, the PTTG1-expressing cells harboring shRNAs against p53 (shp53–1 to 4) or shGFP were prepared as describe above. The relative cell numbers were determined at the indicated times. An asterisk indicates p < 0.05. D, in a parallel experiment, the SA-β-gal activities were examined and quantified. The results from the average of three independent experiments are presented (lower panel). An asterisk indicates p < 0.05 (p values equal to 0.031, 0.016, and 0.015 for shp53–1, shp53–2, and shp53–4, respectively). E, the PTTG1-expressing cells harboring shRNAs against p53 (shp53 1–3) or shGFP were stained with DAPI and then visualized under a fluorescence microscope. The heterochromatin foci were quantified from more than 20 fields from three independent experiments. An asterisk indicates p < 0.05 (p values equal to 0.0090, 0.00052, and 0.0011 for shp53–1, shp53–2, and shp53–3, respectively). F, the PTTG1-expressing cells harboring shRNAs against p53 (shp53–1-3) or shGFP were stained with DAPI and observed under a fluorescence microscopy. Quantification of the nuclear morphology was also carried out to determine the percentage of abnormality. An asterisk indicates p < 0.05 (p values equal to 0.0025, 0.016, and 0.0033 for shp53–1, shp53–2, and shp53–3, respectively).
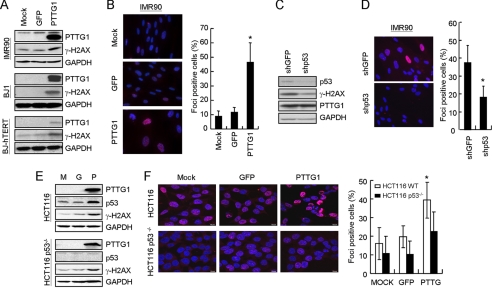
To further investigate the requirement for p53 as part of PTTG1-induced senescence in normal fibroblasts, we utilized a sequence-specific RNA interference approach to knock down the p53 levels in normal cells using a lentiviral delivery system (12). The IMR90 cells were first transduced with shRNAs targeting p53 or luciferase and then infected with adenovirus-carrying PTTG1. As shown in Fig. 3B, the level of PTTG1-activated p53 was decreased upon shRNA treatments. Significantly, the growth of PTTG1-expressing cells was increased upon knocking down p53 expression (Fig. 3C). The results indicated that reduced p53 function could restore PTTG1-induced growth inhibition in normal cells. The SA-β-gal activity was then checked to evaluate the effect of a lower p53 level on PTTG1-induced senescence. In the p53 knockdown cells, the number of SA-β-gal-positive cells was also significantly lower than that of the control cells (Fig. 3D). We next evaluated the SAHF and abnormal nuclei morphologies in the p53 knockdown cells. As shown in Fig. 3, E and F, the numbers of cells with SAHF and abnormal nuclei were also decreased in the p53 knockdown cells. Thus, our results indicated that p53 is required for PTTG1-induced senescence in normal cells.
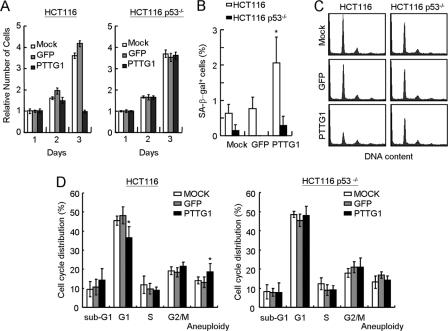
In an independent experiment, we also applied two isogenic colorectal cancer cell lines, HCT116 and its p53 knock-outs, in our studies (19). PTTG1 or GFP was introduced into these cells with the adenovirus delivery system, and the relative survivals were then examined. Upon PTTG1 overexpression, the HCT116 cells ceased to proliferate upon PTTG1 expression (Fig. 4A). The proliferation was recovered in HCT116 cells carrying p53−/− mutations. The senescent phenotype was also analyzed in PTTG1-overexpressing HCT116 cells. Although the HCT116 cells showed <1% of senescent cells, the population was consistently increased to ∼2% upon PTTG1 overexpression (Fig. 4B). The elevated senescent cells were not observed in HCT116 cells carrying p53−/− mutations, indicating that PTTG1-induced senescence is p53-dependent. It is also interesting to note that the level of PTTG1-induced senescent cells in HCT116 cells was relatively low compared with that in IMR90 cells. Although the reason is still unclear to us, HCT116 cells might have an additional mechanism to prevent from entering senescence. Flow cytometry analyses were also conducted to determine the cell cycle effects of HCT116 cells upon PTTG1 overexpression. Consistent with the results of normal cells, the results indicated that the proportion of G1 cells was decreased in PTTG1-expressing HCT116 cells and was recovered in p53−/− cells (Fig. 4, C and D). Thus, the inhibition of proliferation was partly due to the induction of senescence by PTTG1 overexpression. Together, our results indicated that p53 is required for PTTG1-induced growth inhibition in both normal and cancer cells.
FIGURE 4.
p53 is required of PTTG1-induced growth arrest in cancer cells. A, ∼1 × 105 HCT116 cells carrying wild-type or p53−/− were transduced with adenovirus carrying PTTG1 or GFP at an m.o.i. value equal to 2. The relative survivals were determined using MTT assays at the indicated times. B, HCT116 cells were transduced with adenoviruses carrying PTTG1 or GFP and then cultured at 37 °C for 4 days. The transduced cells were analyzed for SA-β-gal activity staining. Quantification of the SA-β-gal-positive-stained cells was conducted. An asterisk indicates p < 0.05 (p = 0.0317). C, HCT116 cells were infected by adenoviruses carrying GFP or PTTG1 for 3 days and then analyzed by flow cytometry. The histograms of the propidium iodide-stained cells are presented. D, quantification of the cells at different cell cycle stages was then conducted. The average number for three independent experiments was determined and plotted. An asterisk indicates p < 0.05 (p = 0.0073 for G1 and p = 0.04 for aneuploid cells, respectively).
PTTG1 Overexpression Induced Chromosome Instability
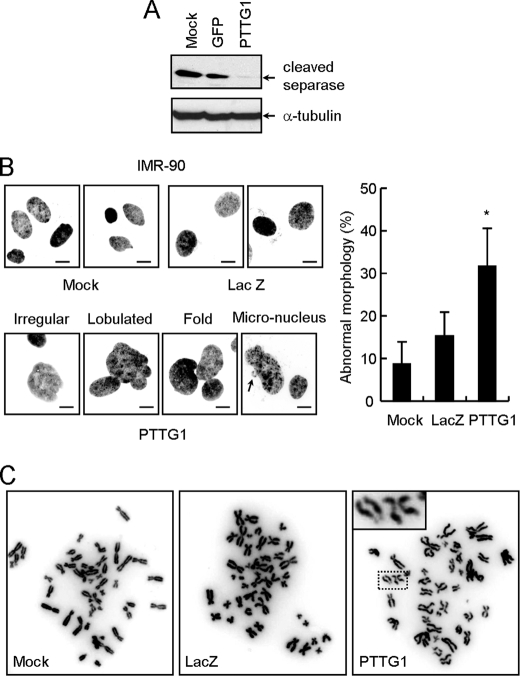
The mechanism of how PTTG1 expression induces senescence in normal fibroblasts was investigated. The PTTG1-induced senescence did not appear to be mediated through telomere as the telomerase catalytic subunit hTERT-immortalized cell line BJ-hTERT also showed similar senescent phenotype (data not shown). Moreover, telomerase activity was not affected by PTTG1 overexpression in BJ-hTERT cells (data not shown). We then focused on the normal cellular function of PTTG1. Upon anaphase initiation, PTTG1 protein was destroyed by APC (anaphase-promoting complex), an ubiquitin-ligase, to activate separase. The activated separase then promotes the separation of sister chromatids by degrading cohesion molecules. Because PTTG1 is involved in inhibiting separase until anaphase initiation, the level of active separase was analyzed in PTTG1-overexpressing cells. Here, the auto-cleaved product of separase was used as an indicator for separase activation (20). As shown in Fig. 5A, the active form of separase was greatly reduced upon PTTG1 overexpression. Thus, overexpression of PTTG1 might mimic a condition similar to separase depletion. Because both PTTG1 and separase are important regulators that control sister-chromatin separation and maintain genome stability, the effect of PTTG1 overexpression on the nuclear morphology of normal fibroblasts was next analyzed. Cells that expressed PTTG1 were stained with Hoechst 33258 to visualize the nuclear morphology under a fluorescence microscopy. Nuclear dysmorphism, such as irregular contour and multiple lobules and folds, was clearly observed in cells expressing PTTG1. The proportion of cells with enlarged nuclei or micronuclei was also increased in PTTG1-expressing cells (Fig. 5B). Upon scoring the number of abnormal nuclei, we found that a total of about 32% of the PTTG1-expressing cells showed abnormal nuclei, a 2-fold increase over the LacZ-expressing cells (Fig. 5B). These results suggested that PTTG1-expressing cells might undergo severe chromosome instability. We, thus, prepared metaphase chromosomes to examine chromosome integrity in the PTTG1-expressing cells. The structure of the chromosomes was observed and scored individually using a fluorescence microscopy. When the control and LacZ-expressed cells were examined, they showed a normal chromosomes spread (Fig. 5C). However, the chromosomes of the PTTG1-expressing cells showed a high level of abnormalities including an elevated level of aneuploidy, end-to-end fusions, and chromosome breaks (data not shown). The most striking feature observed in the PTTG1-expressing cells was partially paired sister chromatids with centromere separation, which is an indication of premature anaphase sister chromosome separation (Fig. 5C). By scoring the number of cells with abnormal chromosomes, it was found that ∼12.7% of the PTTG1-expressing cells showed abnormal chromosomes, which contrasted with 2.7 and 3.8% of the mock control and LacZ-expressing cells, respectively (data not shown). Thus, our results clearly indicated that PTTG1 overexpression in normal fibroblasts caused chromosome abnormalities.
FIGURE 5.
Decreasing separase activity and increasing abnormal chromosomes in PTTG1-expressing fibroblasts. A, IMR-90 cells were infected with adenoviruses carrying GFP or PTTG1. Cell extracts were prepared 4 days after infection and then analyzed by immunoblotting assays using antibodies against separase (ab3762, Abcam) or α-tubulin. Bound antibodies were visualized by chemiluminescence using an ECL kit (Amersham Biosciences). Interphase nuclei (B) and metaphase chromosomes of mock, LacZ- or PTTG1-expressing cells (C) were collected, stained with Hoechst 33258, and observed under a fluorescence microscopy. Quantification of the nuclear morphology was also carried out to determine the percentage of abnormality An asterisk indicates p < 0.05 (p = 0.001).
We next test if the abnormal chromosomes observed in PTTG1-expressing cells could induce DNA-damage responses that lead to senescence. The phosphorylation level of H2AX (γ-H2AX) was examined, as it is known to play a very early and important role in the cellular response to DNA double-strand breaks (21). As shown in Fig. 6A, a significant induction of γ-H2AX was observed in response to PTTG1 overexpression in the three different normal cell lines. Immunofluorescence microscopy analysis also revealed that up to ∼45% of the PTTG1-expressing cells were stained positive for γ-H2AX foci formation (Fig. 6B). To determine the role of p53 in PTTG1-induced DNA damage response, the level of γ-H2AX was also determined in PTTG1-expressing cells with reduced p53 expression using shRNA. As shown in Fig. 6C, reduction of p53 is accompanied by reduction of γ-H2AX. Immunostaining analysis also showed that the γ-H2AX foci-positive cells were decreased in PTTG1 expression cells that also expressed p53 shRNA (Fig. 6D). The results indicated that a p53-dependent DNA damage response pathway was induced in PTTG1-expressing normal fibroblasts.
FIGURE 6.
Detection of DNA damage response signals in PTTG1-expressing fibroblasts. A, cell extracts were prepared from IMR90, BJ1, and BJ-hTERT 4 days after PTTG1 expression. They were then assayed by immunoblotting analysis using antibodies against PTTG1, γ-H2AX, or GAPDH in the three fibroblast lines. B, in a parallel experiment, PTTG1-expressing IMR90 cells were stained with antibody against γ-H2AX. Staining of DNA by Hoechst 33258 was employed to locate the positions of the nuclei. The percentage of γ-H2AX-positive cells was quantified (right). An asterisk indicates p < 0.05 (p = 0.0256). C, IMR90 cells were infected with shRNA against p53 or GFP and then cultured at 37 °C for 2 days. The cells were then infected with adenoviruses carrying PTTG1 and cultured at 37 °C for another 3 days. Cell extracts were prepared and analyzed by immunoblotting assays using antibodies against p53, γ-H2AX, or GAPDH. D, procedures were as in C; merged images of shp53/PTTG1 and shGFP/PTTG1 cells are presented. The percentage of γ-H2AX-positive cells was quantified (right). An asterisk indicates p < 0.05 (p = 0.046). E, HCT116 cells (M) were infected with adenoviruses carrying GFP (G) or PTTG1 (P). Cell extracts were prepared 3 days after infection and then analyzed by immunoblotting assays using antibodies against PTTG1, p53, γ-H2AX, or GAPDH. Bound antibodies were visualized by chemiluminescence using an ECL kit (Amersham Biosciences). F, procedures were as in B; merged images of HCT116and HCT116 p53−/− cells are presented. The percentage of γ-H2AX-positive cells was quantified (right). An asterisk indicates p < 0.05 (p = 0.039).
Similar to that of normal fibroblasts, the p53 level was increased upon PTTG1 expression in HCT116 p53+/+ cells (Fig. 6E). The level of γ-H2AX in PTTG1-expressing cells was also increased in p53+/+ cells, suggesting that a DNA-damage response was activated in these cells. Similar to that in normal cells, immunofluorescence microscopy analysis also revealed that up to ∼40% of the PTTG1-expressing HCT116 p53+/+ cells were stained positive for γ-H2AX foci formation (Fig. 6F). Together these results indicated that the DNA-damage response was triggered when PTTG1 was overexpressed in normal fibroblasts.
DISCUSSION
Under normal culture conditions, normal diploid fibroblasts undergo a limited number of cell divisions and then cease proliferation. It is well documented that cellular senescence is caused by an attrition of the telomeres after repeated cycles of cell divisions (22). The shortened telomeres then induce a p53-dependent cell cycle arrest. Here we showed that forced expression of PTTG1 in normal human fibroblasts also induced senescent phenotype. However, because the senescent phenotype was also detected in PTTG1-expressing BJ-hTERT cells and telomerase was not affected upon PTTG1 expression, it is unlikely that telomere shortening is participated in PTTG1-induced senescence. Senescence induced by forced PTTG1 expression is different from that of replicative senescence. Here our results provide evidence that PTTG1-induced senescence is mediated by a p53-dependent DNA-damage response pathway. First, PTTG1 overexpression increased the formation of abnormal chromosomes, which is an indication of chromosome instability. Second, DNA damage-induced γH2AX foci were detected in PTTG1-expressing fibroblasts. Third, the PTTG1-induced senescence was associated with an elevation in p53 and a deceased in both Rb protein and phosphorylated Rb levels. Finally, RNA interference experiments demonstrated that down-regulation of p53 abolished the PTTG1-induced senescent phenotype and DNA damage response. Thus, the PTTG1-induced senescence closely resembles the senescent phenotype in response to oncogene expression or DNA damage stresses in normal human fibroblasts (11).
Forced expression of several oncogenes has been shown to cause senescence through a p53-dependent pathway. For example, oncogene ras has been shown to transform immortal rodent NIH 3T3 cells into a tumorigenic state; however, forced ras expression induces normal human or rodent cells to undergo senescence (23). It appears that these oncogenes have dual roles in tumorigenesis; they trigger the senescence pathway to protect normal cells from transformation at an early stage of tumorigenesis and then promote tumor progression at a late stage of tumorigenesis (11). Similarly, introduction of PTTG1 into NIH 3T3 cells results in tumor transformation both in vitro and in vivo (1), and we showed that overexpression of PTTG1-induced senescence in normal cells. Thus, it is likely that PTTG1 also has dual roles in tumorigenesis. In additional to its role in the late stage of promoting tumor progression, our results indicated that PTTG1 might also have a role in early tumorigenesis. To account for these results, it is anticipated that PTTG1 is overexpressed in the early stage of tumorigenesis. Indeed, it was shown that the rat PTTG was been found to be expressed coincidently with the early lactotrophic hyperplastic response, angiogenesis, and prolactinoma development (24). The senescence induced by PTTG1 overexpression might prevent the transformation of normal cells at early stage of tumorigenesis. Overcoming senescence during early tumor progression may be quite difficult, as it would require the inactivation of the DNA-damage checkpoint response pathways. However, chromosome abnormalities accumulated in PTTG1-expressing cells might provide a better chance for cells to escape senescence. Thus, PTTG1 overexpression might also promote further tumor progression. Alternatively or additionally, because it was been shown that senescent human fibroblasts stimulated hyperproliferation and the progression of pre-neoplastic epithelial cells as well as accelerating the tumorigenesis by neoplastic epithelial cells (25), the PTTG1-expressing senescent fibroblasts might themselves promote tumorigenesis of precancerous epithelial cells. Nevertheless, our results suggested that, in additional to its role in later stages of tumor progression, PTTG1 might also have a role in early stage of tumorigenesis.
Because PTTG1 has been shown to interact with different protein partners to mediate various functions, it is possible that the cellular effects observed in PTTG1-overexpressed normal cells are caused by these various interactions. Here we consider that the senescent phenotype observed in this study is primarily caused by its effects in mitotic cell cycle regulation. Separase is a cysteine protease responsible for cleaving the cohesion complex and is involved in chromatid separation during mitosis (26). Because a role of PTTG1 is to inhibit separase activity through protein-protein interactions, PTTG1 overexpression could induce cellular effects resembling separase defects. In budding yeast, failed sister-chromatid separation, gross chromosome mis-segregation, and decreased viability has been observed in cells with the separase esp1 mutation (27, 28). Similar observations were found in fission yeast, Drosophila, and zebrafish with the separase mutation (29–31). Here we showed that active separase level was indeed decreased upon PTTG1 overexpression. Moreover, we found that PTTG1 overexpression induced a cessation of cell proliferation and a senescent phenotype in normal fibroblasts. We also found that PTTG1 overexpression caused abnormal chromosome separation, consistent with the role of PTTG1 in regulating sister-chromatid separation. Thus, it is very likely that the cellular effects we observed in PTTG1-overexpressing cells are caused by inducing a situation similar to that of separase loss. Interestingly, unlike yeast cells, which lack separase, are not viable, and stop the cell cycle at M phase (32), the chromosome instability induced by PTTG1 overexpression causes normal human fibroblasts to enter a p53-dependent senescence. Because a role of PTTG1-p53 interaction is in the regulation of apoptosis in cancer cells (8), it is unclear whether the interaction might participate in determining senescence in normal cells. It is also interesting to note that although p53 appears to be a key factor in PTTG1-induced senescence, both p21 and p16 levels are not significantly altered upon PTTG1 overexpression in IMR90 cells. Thus, unlike p53, the roles of p21 and p16 in the PTTG1-induced cellular effects are less clear.
Supplementary Material
Acknowledgments
We thank members of the Institute of Biopharmaceutical Science for help and support. We thank Y. Su for the adenoviral system, T. H. Cheng for the HCT116 cells, and the National RNAi core facility for the shRNAs.
This work was supported by National Science Council Grants 97-2311-B-010-005 and 98-3112-B-010-006 (to J.-J. L.) and 96-2627-B002-010 (to P.-J. L.) and by the National Health Research Institute Grant NHRI-EX98-9625SI (to J.-J. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- SA-β-gal
- senescence-associated acidic β-galactosidase
- SAHF
- senescence-associated heterochromatin foci
- x-gal
- 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside
- m.o.i.
- multiplicity of infection
- shRNA
- short hairpin RNA
- BrdUrd
- bromodeoxyuridine
- PBS
- phosphate-buffered saline
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- GFP
- green fluorescent protein
- DAPI
- 4′,6-diamidino-2-phenylindole.
REFERENCES
- 1.Pei L., Melmed S. (1997) Mol. Endocrinol. 11, 433–441 [DOI] [PubMed] [Google Scholar]
- 2.Zou H., McGarry T. J., Bernal T., Kirschner M. W. (1999) Science 285, 418–422 [DOI] [PubMed] [Google Scholar]
- 3.Domínguez A., Ramos-Morales F., Romero F., Rios R. M., Dreyfus F., Tortolero M., Pintor-Toro J. A. (1998) Oncogene 17, 2187–2193 [DOI] [PubMed] [Google Scholar]
- 4.Zhang X., Horwitz G. A., Prezant T. R., Valentini A., Nakashima M., Bronstein M. D., Melmed S. (1999) Mol. Endocrinol. 13, 156–166 [DOI] [PubMed] [Google Scholar]
- 5.Vlotides G., Eigler T., Melmed S. (2007) Endocr. Rev. 28, 165–186 [DOI] [PubMed] [Google Scholar]
- 6.Ramaswamy S., Ross K. N., Lander E. S., Golub T. R. (2003) Nat. Genet. 33, 49–54 [DOI] [PubMed] [Google Scholar]
- 7.Yu R., Lu W., Chen J., McCabe C. J., Melmed S. (2003) Endocrinology 144, 4991–4998 [DOI] [PubMed] [Google Scholar]
- 8.Bernal J. A., Luna R., Espina A., Lázaro I., Ramos-Morales F., Romero F., Arias C., Silva A., Tortolero M., Pintor-Toro J. A. (2002) Nat. Genet. 32, 306–311 [DOI] [PubMed] [Google Scholar]
- 9.Romero F., Multon M. C., Ramos-Morales F., Domínguez A., Bernal J. A., Pintor-Toro J. A., Tortolero M. (2001) Nucleic Acids Res. 29, 1300–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishikawa H., Heaney A. P., Yu R., Horwitz G. A., Melmed S. (2001) J. Clin. Endocrinol. Metab. 86, 867–874 [DOI] [PubMed] [Google Scholar]
- 11.Collado M., Blasco M. A., Serrano M. (2007) Cell 130, 223–233 [DOI] [PubMed] [Google Scholar]
- 12.Escarpe P., Zayek N., Chin P., Borellini F., Zufferey R., Veres G., Kiermer V. (2003) Mol. Ther. 8, 332–341 [DOI] [PubMed] [Google Scholar]
- 13.Olsen C. L., Gardie B., Yaswen P., Stampfer M. R. (2002) Oncogene 21, 6328–6339 [DOI] [PubMed] [Google Scholar]
- 14.Chang B. D., Xuan Y., Broude E. V., Zhu H., Schott B., Fang J., Roninson I. B. (1999) Oncogene 18, 4808–4818 [DOI] [PubMed] [Google Scholar]
- 15.Dimri G. P., Lee X., Basile G., Acosta M., Scott G., Roskelley C., Medrano E. E., Linskens M., Rubelj I., Pereira-Smith O., Peacocke M., Campisi J. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 9363–9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei W., Sedivy J. M. (1999) Exp. Cell Res. 253, 519–522 [DOI] [PubMed] [Google Scholar]
- 17.Narita M., Nũnez S., Heard E., Narita M., Lin A. W., Hearn S. A., Spector D. L., Hannon G. J., Lowe S. W. (2003) Cell 113, 703–716 [DOI] [PubMed] [Google Scholar]
- 18.Zhang R., Poustovoitov M. V., Ye X., Santos H. A., Chen W., Daganzo S. M., Erzberger J. P., Serebriiskii I. G., Canutescu A. A., Dunbrack R. L., Pehrson J. R., Berger J. M., Kaufman P. D., Adams P. D. (2005) Dev. Cell 8, 19–30 [DOI] [PubMed] [Google Scholar]
- 19.Bunz F., Dutriaux A., Lengauer C., Waldman T., Zhou S., Brown J. P., Sedivy J. M., Kinzler K. W., Vogelstein B. (1998) Science 282, 1497–1501 [DOI] [PubMed] [Google Scholar]
- 20.Waizenegger I., Giménez-Abián J. F., Wernic D., Peters J. M. (2002) Curr. Biol. 12, 1368–1378 [DOI] [PubMed] [Google Scholar]
- 21.Rogakou E. P., Boon C., Redon C., Bonner W. M. (1999) J. Cell Biol. 146, 905–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campisi J., d'Adda di Fagagna F. (2007) Nat. Rev. Mol. Cell. Biol. 8, 729–740 [DOI] [PubMed] [Google Scholar]
- 23.Serrano M., Lin A. W., McCurrach M. E., Beach D., Lowe S. W. (1997) Cell 88, 593–602 [DOI] [PubMed] [Google Scholar]
- 24.Heaney A. P., Horwitz G. A., Wang Z., Singson R., Melmed S. (1999) Nat. Med. 5, 1317–1321 [DOI] [PubMed] [Google Scholar]
- 25.Krtolica A., Parrinello S., Lockett S., Desprez P. Y., Campisi J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 12072–12077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nasmyth K., Peters J. M., Uhlmann F. (2000) Science 288, 1379–1385 [DOI] [PubMed] [Google Scholar]
- 27.McGrew J. T., Goetsch L., Byers B., Baum P. (1992) Mol. Biol. Cell 3, 1443–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ciosk R., Zachariae W., Michaelis C., Shevchenko A., Mann M., Nasmyth K. (1998) Cell 93, 1067–1076 [DOI] [PubMed] [Google Scholar]
- 29.Funabiki H., Kumada K., Yanagida M. (1996) EMBO J. 15, 6617–6628 [PMC free article] [PubMed] [Google Scholar]
- 30.Herzig A., Lehner C. F., Heidmann S. (2002) Genes Dev. 16, 2443–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shepard J. L., Amatruda J. F., Finkelstein D., Ziai J., Finley K. R., Stern H. M., Chiang K., Hersey C., Barut B., Freeman J. L., Lee C., Glickman J. N., Kutok J. L., Aster J. C., Zon L. I. (2007) Genes Dev. 21, 55–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baum P., Yip C., Goetsch L., Byers B. (1988) Mol. Cell. Biol. 8, 5386–5397 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.