Summary
Dicer is a multidomain ribonuclease III enzyme involved in the biogenesis of microRNAs (miRNAs) in the vast majority of eukaryotes. In human, Dicer has been shown to interact with cellular proteins via its N-terminal domain. Here, we demonstrate the ability of Dicer C-terminus to interact with 5-lipoxygenase (5LO), an enzyme involved in the biosynthesis of inflammatory mediators, in vitro and in cultured human cells. Yeast two-hybrid and GST binding assays delineated the smallest 5-lipoxygenase binding domain (5LObd) of Dicer to its C-terminal 140 amino acids comprising the double-stranded RNA (dsRNA) binding domain (dsRBD). The Dicer 5LObd-5LO association was disrupted upon Ala substitution of Trp residues 13, 75 and 102 in 5LO, suggesting that the Dicer 5LObd may recognize 5LO via its N-terminal C2-like domain. Whereas a catalytically active 5LObd-containing Dicer fragment was found to enhance 5LO enzymatic activity in vitro, human 5LO modified the miRNA precursor processing activity of Dicer. In addition to revealing the dual RNA and protein binding properties of Dicer C-terminus, our results may provide a link between miRNA-mediated regulation of gene expression and inflammation.
Keywords: 5-Lipoxygenase, Dicer, yeast two-hybrid system, protein interactions, enzyme activity
Introduction
As a member of the ribonuclease (RNase) III family of enzymes, Dicer is a large multidomain protein that plays an essential role in the biogenesis of microRNAs (miRNAs) in the vast majority of eukaryotes, including humans [1–4]. Human Dicer is a protein of 1912 amino acid residues composed of several domains: an N-terminal putative ATPase/helicase domain containing a DECH box, a domain of unknown function (DUF283), a central PIWI/Ago/Zwille (PAZ) domain, and a C-terminal RNase III domain, composed of tandem RNase III motifs and a C-terminal double-stranded RNA (dsRNA) binding domain (dsRBD) (reviewed in [5]). Human Dicer has been proposed to recognize the terminal 3′ overhangs of miRNA precursors (pre-miRNAs) through its PAZ domain [6] and to process its substrates through intramolecular dimerization of its two RNase III motifs, assisted by the flanking RNA binding domains PAZ and dsRBD [4].
Although the RNase III processing activity and enzyme properties of human Dicer have been well characterized [2–4], the assessment and validation of its anticipated multiple functionalities remains incomplete. While the structure of its C-terminal RNase III domain (residues 1660–1852) has been reported [7], characterization of the several predicted domains of the 217-kDa human Dicer remains challenging and is currently being extrapolated mainly from studies performed on RNase III-related enzymes [8], ancestral, minimal or reengineered forms of Dicer from distantly related lower eukaryotes [6], such as Giardia intestinalis [9, 10]. Highly conserved through evolution, Dicer exhibits striking differences at various levels. For example, the human genome harbors, like that of S. pombe and C. elegans, a single Dicer form, in contrast to two and four Dicer isoforms in Drosophila melanogaster and Arabidopsis thaliana, respectively [5]. These enzymes also differ in terms of sequences, domain composition and functionalities. For instance, the function of the dsRBD present in Dicer enzymes remains unclear in view of the observations that (i) the C-terminal dsRBD is dispensable for processing activity in E. coli RNase III [11], (ii) Giardia Dicer lacks a dsRBD domains [9], and (iii) an RNase III naturally lacking the dsRBD functions in B. subtilis [12].
Concerning the molecular context in which Dicer operates in cells, three mammalian Dicer-interacting proteins have been identified so far: Argonaute 2 (Ago2), transactivating response RNA-binding protein (TRBP) and PACT. Ago2 was found in immunoprecipitates prepared from Drosophila S2 cells expressing an epitope-tagged version of Dicer-1 (Dcr-1) [13]. A study by Tahbaz et al. [14] extended these findings to mammalian cells and determined that Dicer•Ago2 complex formation may involve a direct interaction between a subregion of the PIWI domain of Ago2 and the RNase III domain of Dicer. Using coimmunoprecipitation strategies in cultured human cells, two research groups reported the identification of TRBP as a Dicer-interacting protein [15, 16]. TRBP was shown to facilitate Dicer-mediated cleavage of pre-miRNAs in vitro and to be required for optimal RNA silencing in vivo [15, 16]. A similar role was proposed for PACT [17]. In fact, both TRBP and PACT were found to interact with the N-terminal region of Dicer that contains the putative ATPase/helicase domain.
Human Dicer partial cDNA clones were initially isolated from a yeast two-hybrid screen using 5-lipoxygenase (5LO) as bait [18]. In humans, 5LO is expressed mainly in differentiated inflammatory cells, such as granulocytes, monocytes/macrophages, mast cells, dendritic cells, and B lymphocytes, as reviewed in [19]. This lipoxygenase catalyzes the first two steps in the biosynthesis of leukotrienes, which are potent mediators of inflammation [20, 21]. The 5LO enzyme activity depends on prosthetic iron in the C-terminal catalytic domain (residues 121–673), whereas its C2-like N-terminal β-sandwich (residues 1–114) binds Ca2+, leading to Ca2+ stimulation of enzyme activity [22]. Previously shown to bind phosphatidylcholine (PC) [36], the 5LO β-sandwich was also found to mediate interaction with proteins, such as Coactosin-like Protein (CLP) [23].
In this study, we sought to validate and characterize the interaction between human Dicer and 5LO, and identified a 140-amino acid (a.a.), dsRBD-containing, C-terminal domain of Dicer (hereafter referred to as 5-lipoxygenase binding domain, or 5LObd) as a protein interacting module recognized by the N-terminal C2-like domain of 5LO. Modulating each others enzymatic activity, the functional implications of the interaction between human Dicer and 5LO may provide a link between miRNA-mediated regulation of gene expression and inflammatory processes.
Materials and Methods
Plasmid DNA constructs
Various deletion mutants of human Dicer were amplified by polymerase chain reaction (PCR) and cloned in frame into the BamHI/SalI or SalI sites of pACT2 (Clontech). The pGBT9-5LO 62-673 vector was prepared by cloning the PCR-amplified inserts in frame into the EcoRI/SalI sites of pGBT9 (Clontech). The presence and orientation of the insert was verified by restriction analysis and at least two bacterial clones were tested.
The open reading frames of human platelet-type 12LO (acc. no. M58704), rat brain 12LO (acc. no. L06040) and human 15LO type I (acc. no. M23892) were directionally cloned into pGBT9 and sequenced.
A cDNA fragment encoding human Dicer C-terminal domain (C-term; composed of a.a. 1238-1912) was amplified by PCR, digested and ligated into the BamHI/XhoI restriction sites of the pcDNA3.1-5′Flag vector, as described previously [3]. A pcDNA3.1-5LO-HA expression construct was created by inserting the human 5LO open reading frame into a pcDNA3.1 vector containing a C-terminal HA epitope inserted into the XhoI/ApaI restriction sites. The constructs were verified by DNA sequencing.
Yeast two-hybrid system
Yeast two-hybrid strain (PJ69-4A), vectors (pGBT9-5LO, pGBT9-SNF1, pACT2-SNF4) and reporter gene assays were described previously [18, 24, 25]. The known two-hybrid interactors SNF1 and SNF4 were used as a positive control [18].
Protein determination and immunoblot analysis
The protein concentrations were determined by the method of Bradford [26] using the Bio-Rad dye reagent, with bovine serum albumin as standard.
Yeast protein extracts or protein suspensions were fractionated by SDS-polyacrylamide gel electrophoresis (PAGE) using the Mini Protean system (Bio-Rad), transferred to nitrocellulose membranes (Hybond-C, Amersham Biosciences), immunoblotted, and the immunoreactive proteins visualized as described previously [18, 24, 25, 27].
Confocal immunofluorescence microscopy
Confocal immunofluorescence microscopy was performed essentially as described previously [3]. Briefly, HeLa cells were grown on sterile glass coverslips coated with poly-L-lysine in Dulbecco’s minimal essential medium supplemented with 10% (v/v) fetal bovine serum, 1 mM sodium pyruvate, 100 units/ml penicillin and 100 μg/ml streptomycin in a humidified incubator under 5% CO2 at 37 °C. Cells were transfected with plasmid constructs encoding epitope-tagged 5LO and Dicer C-term proteins using Lipofectamine 2000 (Invitrogen) and harvested 20 h post-transfection. Cells were then washed in phosphate buffered saline (PBS), fixed in 4% (w/v) paraformaldehyde, permeabilized with 0.1% Triton X-100 and incubated in blocking buffer (PBS containing 10% FBS). Cells were subsequently incubated with mouse monoclonal anti-HA clone 12CA5 (dilution 1/200; Roche) and rabbit polyclonal anti-Flag (dilution 1/200; Santa Cruz Biotechnologies, Inc.) antibodies, respectively. After extensive washing in PBS, the cells were incubated with Alexa Fluor 488 (green)-conjugated goat anti-mouse or Alexa Fluor 546 (red)-conjugated goat anti-rabbit secondary antibodies (dilutions 1/500; Molecular Probes). After extensive washing in PBS, the coverslips were mounted on slides with Prolong Gold antifade reagent (Molecular Probes). Labeling of HeLa cells was visualized with an inverted Olympus IX70 microscope (90X magnification), and images were prepared with Image J 1.38x software.
Immunoprecipitation experiments
HEK 293 cells were transiently transfected with plasmid constructs encoding epitope-tagged 5LO, 5LO W13/75/102A and/or Dicer C-term proteins by the calcium phosphate method and harvested 48 h later. Cells were washed twice with ice-cold PBS and solubilised with 1 ml of lysis buffer composed of 50 mM Tris•HCl, 137 mM NaCl, 1% Triton X-100, 1 mM PMSF, pH 8.0, supplemented with complete protease inhibitor cocktail (Roche). The lysate was kept on ice for 15 min, centrifuged at 13,000 g for 1 min, and the supernatant preserved. An aliquot of the supernatant was kept for protein determination by Bradford and analysis of 5LO and Dicer C-term protein expression by immunoblotting. Cell lysates (2 mg proteins) were incubated with 1 μg of anti-Flag M2 antibody (Sigma) for 1 h at 4 °C under continuous rotation. Ten (10) μl of pre-washed Protein G agarose beads (Roche) were added and the incubations continued for an additional 3 h. For anti-HA immunoprecipitations, 15 μl of anti-HA affinity matrix (rat anti-HA 3F10 linked to agarose beads) (Roche) was used. The beads were washed three times with lysis buffer and the immune complexes eluted by boiling in loading buffer for 5 min. The immunoprecipitated proteins were electrophoresed on a 7% SDS-PAGE gel, transferred on a PVDF membrane and immunoblotted with an anti-Flag or anti-5LO [24] antibody.
GST binding assays
Expression of recombinant GST-Dicer 1772-1912 (5LObd) fusion protein [3], GST alone [3], 5LO [28] and 5LO W13/75/102A [29] proteins have been described previously. For 5LO interaction studies, 5 μg of the GST-Dicer 5LObd fusion protein, or GST alone, coupled to glutathione (GSH)-Sepharose 4B beads (10 μl), was incubated with 0.5–10.0 μg 5LO or 0.5 μg 5LO W13/75/102A in buffer A (20 mM Tris•HCl, 100 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol, 1 mM ATP, 0.5 mg/ml bovine serum albumin, 0.01% NP-40, pH 7.5), and the binding assay performed as described previously [24, 25]. Bound 5LO was detected by IB analysis. In some experiments, incubations were performed in the absence or presence of 500-bp dsRNA (0.1–25.0 μg), which was prepared using the coactosin-like protein (CLP) (nt 150 to 649; acc. no. L54057) sequence, as in [3].
5LO activity assays
For 5LO activity assays, the reaction mixture (100 μl) contained 100 μM arachidonic acid, 10 μM 13-hydroperoxy-9,11-octadecadienoic acid, 1.3 mM CaCl2, 5 mM MgCl2, 1.2 mM EDTA, 1 mM ATP, 77 mM Tris (pH 7.5), 1.2 mM DTT and 1.5 pmol of recombinant human 5LO protein [22]. Formation of 5LO products was measured in the absence or presence of Dicer (1650-1912), or PC. After incubation for 10 min at room temperature, the reaction was stopped by addition of 300 μl of acetonitrile/water/acetic acid (60:40:0.1) containing 1 nmol internal standard 17(S)-hydroxy-7(Z),10(Z),13(Z),15(E)-docosatetraenoic acid (17-OH-22:4) (kindly provided by Mats Hamberg, Karolinska Institutet). After centrifugation of the samples at 13,000 g for 5 min at 4°C, 100 μl of the supernatant was applied onto a Nova-Pak C18-column (Waters) and eluted with acetonitrile/water/acetic acid (60:40:0.1) at 1 ml/min. Formation of 5(S)-hydroperoxy-6-trans-8,11,14-cis-eicosatetraenoic acid (5-HPETE) and 5(S)-hydroxy-6-trans-8,11,14-cis-eicosatetraenoic acid (5-HETE) was detected by monitoring at 234 nm and quantified using 17-OH-22:4 as internal standard. For analysis of leukotriene A4 hydrolysis products, 0.4 nmol of prostaglandin B2 internal standard was added to 200 μl of supernatant. An aliquot was applied to the same high pressure liquid chromatography (HPLC) column, eluted with acetonitril/metanol/water/acetic acid (40:30:30:0.1), and the products were detected at 270 nm.
Dicer activity assays
Recombinant human Dicer full-length or deletion mutant 1650–1912 proteins were incubated in the absence or presence of proteinase K for 10 min or human 5LO for 30 min prior to addition of a randomly, 32P-labeled microRNA precursor substrate (pre-let-7a-3) in a reaction buffer containing 20 mM Tris•HCl pH 7.5, 5 mM MgCl2, 1 mM dithiothreitol (DTT), 1 mM ATP, 5% Superase•In (Ambion), at 37°C for 1 h. The resulting RNA products were analyzed by denaturing PAGE and autoradiography, as described previously [30, 31].
Results and Discussion
5LO interacts with the C-terminal domain of Dicer in the yeast two-hybrid system
In our aim at identifying 5LO-interacting proteins, we opted for the two-hybrid system using yeast strain PJ69-4A [32]. This strain has the advantage of combining three reporter genes under the control of three different Gal4-responsive promoters (GAL1-HIS3, GAL2-ADE2, and GAL7-lacZ), thereby reducing the number of false positives [32] and providing an increased stringency to our protein interaction assays.
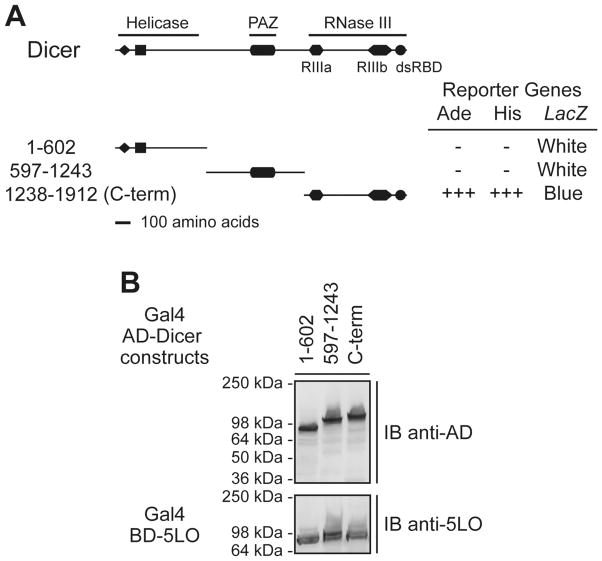
A yeast two-hybrid screen of a human lung cDNA library, using 5LO as a bait, led to the identification of partial cDNAs encoding human Dicer [18]. When tested in 5LO-interaction assays in yeast, only cells co-expressing 5LO and the C-terminal domain (C-term) of Dicer (a.a. 1238-1912), comprising the RNase III domains and dsRBD, showed robust growth on medium lacking adenine (Ade) or histidine (His) (in the presence of 3-amino-1,2,4-triazole; 3-AT), and turned blue when incubated on 5-bromo-4-chloro-3-indolyl β-D-galactoside-containing plates (see Figure 1A). Neither the N-terminus, bearing motifs of the putative helicase domain, nor the Piwi/Argonaute/Zwille (PAZ)-containing central portion of Dicer activated the reporter genes in combination with 5LO. Expression of the Gal4-AD and Gal4-BD fusion proteins was confirmed by immunoblotting (see Figure 1B). Hence, a lack of interaction could not be attributed to defective protein expression. These data suggest the presence of a 5LO-interacting module within the C-term domain of Dicer.
Figure 1.
5LO interacts with Dicer C-terminal (C-term) domain in the yeast two-hybrid system. (A–B) Yeast strain PJ69-4A was cotransformed with plasmids expressing the Gal4 BD-5LO and Gal4 AD-human Dicer deletion mutants. (A) Transformants were tested for the Ade, His and LacZ reporter genes. +++, denotes yeast growth visible at 2 days; −, denotes absence of yeast growth at 7 days. Color of the colonies was noted at 7 days. RIII, RNase III motifs; dsRBD, dsRNA binding domain. (B) Expression of the Gal4 AD-human Dicer deletion mutant (upper panel) and the Gal4 BD-5LO (lower panel) proteins was confirmed by immunoblot (IB) analysis.
5LO colocalizes with Dicer C-terminal domain in cultured human cells
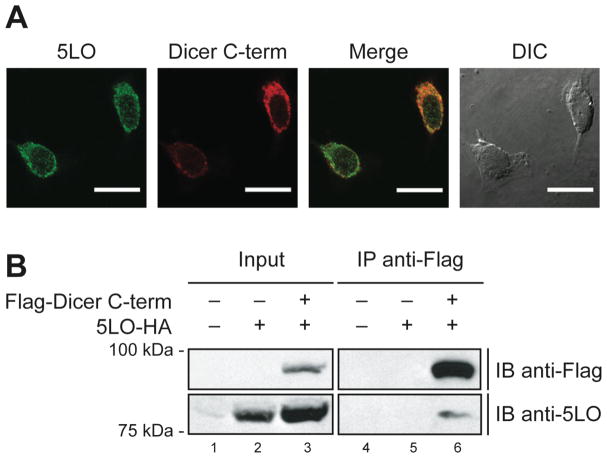
Yeast two-hybrid data should be interpreted with caution, since the expressed hybrid proteins carry nuclear localization sequences to be targeted to the yeast nucleus, where they can interact and activate transcription of the reporter genes. To investigate whether human 5LO interacts with the C-term of Dicer in cultured human cells, we initially performed colocalization studies using confocal immunofluorescence microscopy. HeLa cells were transiently transfected with mammalian expression constructs encoding epitope-tagged 5LO and Dicer C-term proteins. Expression of the proteins was confirmed by Western blotting (P. Provost and G. Pépin, unpublished data). In cells harvested 20 h after transfection, human 5LO showed a cytoplasmic staining more intense at the perinuclear region (see Figure 2A, left panel). This pattern closely resembles that of Dicer C-term protein (see Figure 2A, center-left panel), whose localization is similar to that previously reported for human Dicer [3]. Coenriched at the perinuclear region in HeLa cells, as observed in the merged image (see Figure 2A, right panel), human 5LO and Dicer C-term proteins may localize to the same subcellular compartments in vivo.
Figure 2.
Dicer C-terminal domain colocalizes and interacts with 5LO in transiently transfected human cells. (A) Confocal immunofluorescence microscopy of HeLa cells expressing epitope-tagged 5LO and Dicer C-terminal (C-term) proteins. The merged image demonstrates enrichment and colocalization of 5LO and Dicer C-term proteins at the perinuclear region of HeLa cells. Scale bar = 20 μm. (B) Immunoprecipitation experiments using HEK 293 cells expressing epitope-tagged 5LO and Dicer C-term proteins. Proteins present in the cell lysates (5% input) (lanes 1 to 3) and immunoprecipitates (lanes 4 to 6) were detected by immunoblot analysis (IB) using anti-Flag (upper panels) and anti-5LO (lower panels) antibodies.
5LO associates with Dicer C-terminal domain in cultured human cells
To determine if 5LO interacts with Dicer C-term protein in cultured human cells, we performed coimmunoprecipitation experiments using HEK 293 cells transiently expressing epitope-tagged versions of both proteins. IB analysis using anti-Flag and anti-5LO antibodies confirmed expression of both epitope-tagged Dicer C-term and 5LO proteins in HEK 293 cells upon transfection with the related expression vectors (see Figure 2B, left panels). Flag-Dicer C-term protein was immunoprecipitated from cell lysates by using anti-Flag antibody together with Protein G agarose beads (see Figure 2B, upper panel, lane 6). IB analysis of the anti-Flag immunoprecipitates with anti-5LO antibody revealed the presence of 5LO (see Figure 2B, lower panel, lane 6). No immunoreactivity to anti-Flag or anti-5LO antibody was observed in anti-Flag immunoprecipitates derived from HEK 293 cells expressing only 5LO (see Figure 2B, lane 5) or transfected with empty vectors (see Figure 2B, lane 4). These results indicate that the Dicer C-term is able to recognize 5LO in human cells, although we cannot exclude the possibility that epitope tagging of the proteins might have negatively altered their protein interacting abilities.
Dicer C-terminal 140 amino acids are sufficient for interaction with 5LO
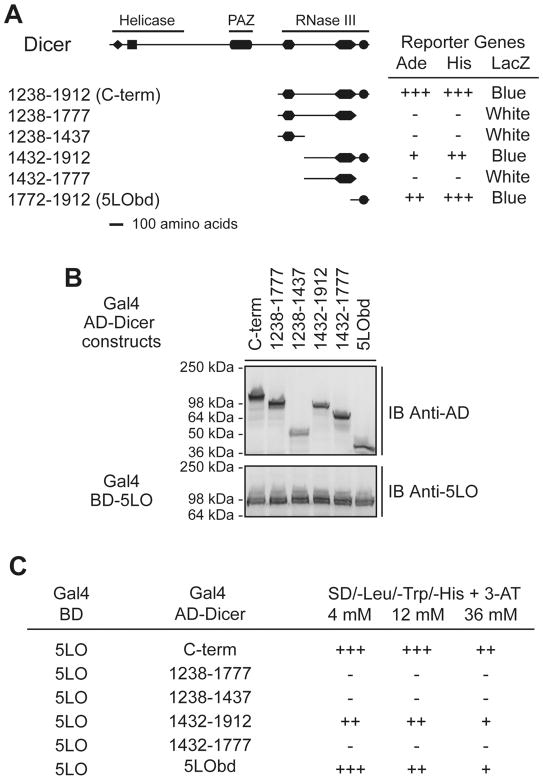
Next, we made use of deletional mutagenesis with the yeast two-hybrid system in order to delineate the minimal region of Dicer mediating 5LO binding. Accordingly, the Dicer C-term was subdivided further into fragments containing various combinations of the RNase IIIa, RNase IIIb and/or dsRBD. Only the fragments containing the C-terminal 140 a.a. (a.a. 1772-1912) comprising the dsRBD of Dicer interacted with 5LO, whereas those lacking it did not (see Figure 3A). Expression of the various interacting as well as non-interacting hybrid proteins was confirmed by immunoblotting (see Figure 3B). No activation of the reporter genes was observed when the various Dicer constructs were cotransformed with SNF1 or the empty Gal4-BD vector (P. Provost, unpublished data). Further shortening of the 1772-1912 fragment from either the N- (a.a. 1828-1912) or C-terminus (a.a. 1772-1833) led to a loss in 5LO interaction (P. Provost, unpublished data).
Figure 3.
Delineation of the smallest 5LO-interacting domain of Dicer to its C-terminal 140 amino acids comprising the dsRBD in the yeast two-hybrid system. (A–C) Yeast strain PJ69-4A was cotransformed with plasmids expressing the Gal4 BD-5LO and Gal4 AD-Dicer deletion mutants. (A) Transformants were tested for the Ade, His and LacZ reporter genes. +++, denotes yeast growth visible at 2 days; −, denotes absence of yeast growth at 7 days. Color of the colonies was noted at 7 days. (B) Expression of the Gal4 AD-Dicer deletion mutant (upper panel) and the Gal4 BD-5LO (lower panel) proteins was confirmed by immunoblot (IB) analysis. (C) Transformants were tested for the His reporter gene in the presence of increasing concentrations of 3-AT. Growth of the colonies was scored when visible at 2 (+++), 4 (++) or 7 (+) days. −, denotes absence of yeast growth at 7 days.
We next evaluated the relative affinity between 5LO and the various Dicer C-terminal deletion mutants in a 3-AT sensitivity assay, in which an increased resistance of the yeast transformants to the 3-AT competitor correlates with the degree of His3 reporter activation. The data shown in Figure 3C support the C-term of Dicer as the strongest 5LO interactor, a property well retained by the smallest 5LO-interacting fragment identified (a.a. 1772-1912). Together, these findings map the minimal human 5LO binding region to Dicer C-terminal 140 a.a., which was defined as the 5-lipoxygenase binding domain (5LObd).
Dicer C-terminal 5LObd does not interact with other LOs
LOs form a family of enzymes that possess a catalytic domain containing a single atom of non-heme iron and catalyze oxygenation of arachidonic acid at various positions, hence their name (eg, 5LO introduces molecular oxygen at carbon-5, 12LO at carbon-12 and 15LO at carbon-15). We examined whether other LOs could interact with the Dicer 5LObd, by testing human platelet-type 12LO, rat brain 12LO or human 15LO type I. Yeast two-hybrid assays revealed that neither 12LOs nor 15LO interacted with the Dicer 5LObd (see Table 1). This may be reasonable, since the amino acid sequence of 5LO is the most divergent of the LOs examined (see Figure S1). These findings support the relative specificity of the Dicer 5LObd for binding 5LO.
Table 1.
Human Dicer 5LObd does not interact with the main LO types other than 5LO in the yeast two-hybrid system.
| Gal4 BD vector | Gal4 AD vector | Reporter genes | ||
|---|---|---|---|---|
| Ade | His | LacZ | ||
| SNF1 | SNF4 | +++ | +++ | Blue |
| Empty | H.s. Dicer 5LObd | - | - | White |
| SNF1 | H.s. Dicer 5LObd | - | - | White |
| 5LO | H.s. Dicer 5LObd | ++ | +++ | Blue |
| H.s. platelet-type 12LO | H.s. Dicer 5LObd | - | - | White |
| R.n. brain 12LO | H.s. Dicer 5LObd | - | - | White |
| H.s. 15LO type I | H.s. Dicer 5LObd | - | - | White |
Yeast growth visible at 2 (+++) or 4 (++) days; -, denotes absence of yeast growth at 7 days. Color of the colonies was noted at 7 days. H.s., Homo sapiens; R.n., Rattus norvegicus.
5LO interacts directly with Dicer 5LObd in vitro
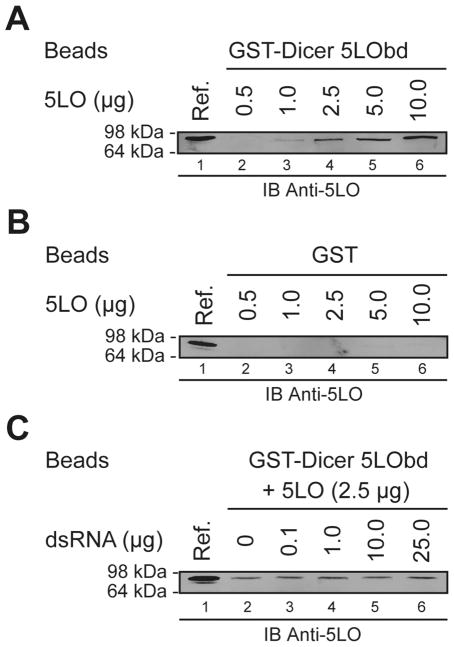
Considering the interaction between 5LO and Dicer 5LObd in the yeast two-hybrid system, the caveat has to be taken into account that other proteins and cellular components may contribute to favor or stabilize the interaction. We thus examined whether 5LO and Dicer 5LObd could interact directly in GST binding assays in vitro. As shown in Figure 4A (lanes 2 to 6), 5LO dose-dependently bound to beads bearing the GST-Dicer 5LObd fusion protein. In experiments performed in parallel, no binding of 5LO to GST beads was observed (see Figure 4B, lanes 2 to 6). These data indicate that 5LO interacts directly with the dsRBD-containing Dicer 5LObd moiety in vitro. Similar properties have been reported for the dsRBD of protein kinase R, which has been shown to play a role in protein dimerization [33] and heterotypic protein-protein interaction [34].
Figure 4.
Dicer 5LObd interacts directly with 5LO in vitro. (A–C) GST binding assays (lanes 2 to 6) using GST-Dicer 5LObd (A and C), or GST (B) protein, coupled to GSH-Sepharose 4B beads and incubated in the presence of increasing amounts of recombinant 5LO proteins (A and B). (C) Beads bearing the GST-Dicer 5LObd fusion protein was incubated with 5LO protein in the absence (lane 2) or presence of dsRNA (0.1–25.0 μg) (lanes 3 to 6). Bound 5LO was visualized by immunoblot (IB) analysis using an anti-5LO antibody, and compared with a reference of 5LO (0.02 μg) (lane 1 of each panel).
We have previously documented the ability of the GST-Dicer 5LObd to bind dsRNA [3] in vitro, raising the possibility of competitive binding with 5LO. When this issue was examined in GST binding assays, we observed that addition of increasing amounts of soluble dsRNA, sufficient to displace bound dsRNA from the GST-Dicer 5LObd (P. Provost, unpublished data), had no effect on the 5LO binding properties of the Dicer 5LObd (see Figure 4C). Conversely, 5LO did not displace bound dsRNA from the GST-Dicer 5LObd fusion protein (P. Provost, unpublished data). These findings suggest that the Dicer dsRBD-containing, C-terminal 5LObd may have the ability to bind dsRNA and 5LO in an independent, noncompetitive manner. Our finding that the Dicer dsRBD contributes to protein interaction is of interest in relation to a model for human Dicer in which the dsRBD may only play an accessory role for RNase III activity [4]. In addition, the observations that Escherichia coli RNase III, amputated of its dsRBD retains its catalytic efficiency and specificity for dsRNA [11], and that an RNase III that naturally lacks the dsRBD functions in B. subtilis [12] are also supportive of this scenario.
5LO interacts with Dicer C-terminal domain via its N-terminal β-sandwich
To determine the region(s) or amino acid residue(s) of 5LO involved in binding Dicer 5LObd, we made use of deletional mutagenesis with the yeast two-hybrid system. N-terminal trimming of 5LO (composed of 673 a.a.), yielding 5LO 62-673 mutant, abrogated binding to Dicer C-term (see Table 2), suggesting the possible involvement of the C2-like N-terminal β-sandwich (residues 1–114) of 5LO in mediating binding to Dicer.
Table 2.
The amino terminus of 5LO is required for interaction with human Dicer C-terminal (C-term) domain in the yeast two-hybrid system.
| Gal4 BD vector | Gal4 AD vector | Reporter genes | ||
|---|---|---|---|---|
| Ade | His | LacZ | ||
| SNF1 | SNF4 | +++ | +++ | Blue |
| Empty | Dicer C-term | - | - | White |
| SNF1 | Dicer C-term | - | - | White |
| 5LO | Dicer C-term | +++ | +++ | Blue |
| 5LO 62-673 | SNF4 | - | - | White |
| 5LO 62-673 | Dicer C-term | - | - | White |
denotes yeast growth visible at 2 days;
denotes absence of yeast growth at 7 days. Color of the colonies was noted at 7 days.
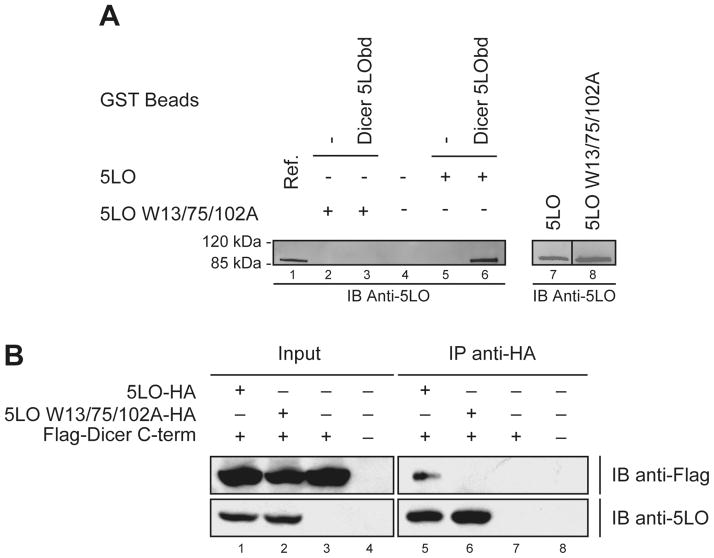
This N-terminal region of 5LO, which resembles the C2 domain of cytosolic phospholipase A2 (cPLA2), functions in the calcium regulation of enzyme activity [22]. Ca2+ has been shown to increase the hydrophobicity of 5LO as well as the affinity of the N-terminal β-sandwich for membrane phospholipids [28, 35, 36]. Three Trp ligands in the ligand binding loops of the 5LO N-terminal C2-like domain were shown to be important for binding of the isolated 5LO β-sandwich to PC [36] as well as for activation of 5LO by 1-oleoyl-2-acetylglycerol (OAG) [29]. Recently, the same three Trp residues (13, 75 and 102) were found to be required for binding of Coactosin-like Protein (CLP) to 5LO [23]. Likewise, we observed that the 5LO W13/75/102A mutant lost its ability to interact with Dicer 5LObd in GST binding assays in vitro (see Figure 5A, lane 3 vs lane 6). Trp residues 13, 75 and 102 are thus important for interactions of 5LO with both lipids and proteins.
Figure 5.
5LO interacts with Dicer 5LObd via its N-terminal non-catalytic domain. (A) GST binding assays were performed, as described in the legend of Figure panels 4A and 4B, using 0.5 μg 5LO (lanes 5 and 6) or 5LO W13/75/102A (lanes 2 and 3) recombinant protein. (A, left panel) Bound 5LO protein was analyzed by immunoblotting (IB), and compared with a reference (Ref.) of 5LO (0.05 μg) (lane 1). (A, right panel) 5LO and 5LO W13/75/102A mutant proteins (0.15 μg each), analyzed on the same gel by IB analysis, exhibited similar immunoreactivities (lanes 7 and 8). (B) Immunoprecipitation experiments using HEK 293 cells expressing epitope-tagged Dicer C-term and 5LO or 5LO W13/75/102A mutant proteins. Proteins present in the cell lysates (5% input) (lanes 1 to 4) and immunoprecipitates (lanes 5 to 8) were detected by IB using anti-Flag (upper panels) and anti-5LO (lower panels) antibodies.
The disruptive effects of the Trp mutations was confirmed in transiently transfected human cells, where epitope-tagged 5LO, but not the 5LO W13/75/102A mutant, coimmunoprecipitated with Flag-Dicer C-term protein (see Figure 5B). These data, together with those of Figure 2B, also illustrate our ability to perform 5LO/Dicer C-term coimmunoprecipitation in a reciprocal manner. Moreover, loss of the Dicer C-term interacting properties of 5LO upon Ala substitution of Trp residues cannot be attributed to gross protein misfolding or perturbation of the 5LO structure, since the 5LO W13/75/102A mutant exhibits an enzymatic activity similar to wild-type 5LO [23]. Among the mutated Trp residues, only W102 is conserved in other noninteracting LOs (see Figure S1), suggesting that residues W13 and W75 are the most important for binding Dicer 5LObd. In view of these data, we cannot exclude the possibility that binding to the C2 domain is required for subsequent Dicer interaction with the catalytic domain of 5LO.
A C-terminal, 5LObd-containing fragment of human Dicer enhances 5LO enzymatic activity
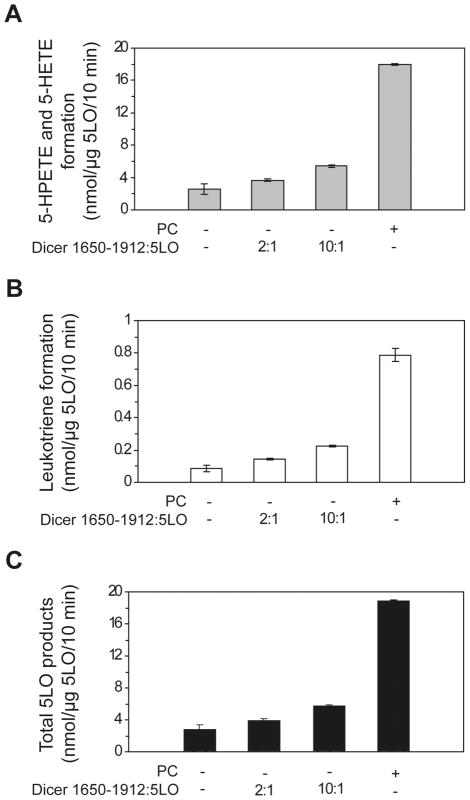
In order to explore the functional implications of this interaction, we first assessed whether the catalytically active, 5LObd-containing Dicer 1650-1912 fragment could influence the enzymatic activity of 5LO in in vitro assays using recombinant human 5LO protein. In assays performed in the absence of PC or Dicer 1650-1912 fragment, the amount of total 5LO products was low (see Figure 6). However, we observed a ~2-fold increase in the formation of 5LO products (5-HPETE, 5-HETE and leukotrienes) when 5LO was coincubation in the presence of the Dicer 1650-1912 fragment (see Figure 6). Whereas PC mimicking cellular membrane strongly enhanced 5LO enzyme activity (see Figure 6), as previously described [23], they had no additive effect (V. Dincbas-Renqvist and O. Rådmark, unpublished data). Indicating that the Dicer 1650-1912 fragment can partially substitute for PC, these results suggest that 5LO can be active in the absence of nuclear membrane association, i.e. when forming a complex with Dicer, which, in turn, may facilitate the enzymatic conversion of arachidonic acid by 5LO by acting as a scaffold protein for the N-terminal β-sandwich of 5LO in a way similar to, but not as efficient as, that reported for CLP [23].
Figure 6.
The smallest 5LObd-containing fragment of Dicer exerts stimulatory effects on 5LO activity. (A–C) 5LO enzyme activity assays. Recombinant human 5LO was incubated in the absence or presence of human Dicer 1650-1912 deletion mutant at a 2:1 or 10:1 ratio, or PC, prior to addition to the 5LO activity assays. After incubation for 10 minutes at room temperature, the reactions were stopped and the 5-HPETE and 5-HETE (A), leukotriene (B) formation and total 5LO products (C) were measured by HPLC by using 17-OH-22:4 (A) or prostaglandin B2 (B) as the internal standard.
5LO modifies the pre-miRNA processing activity of human Dicer
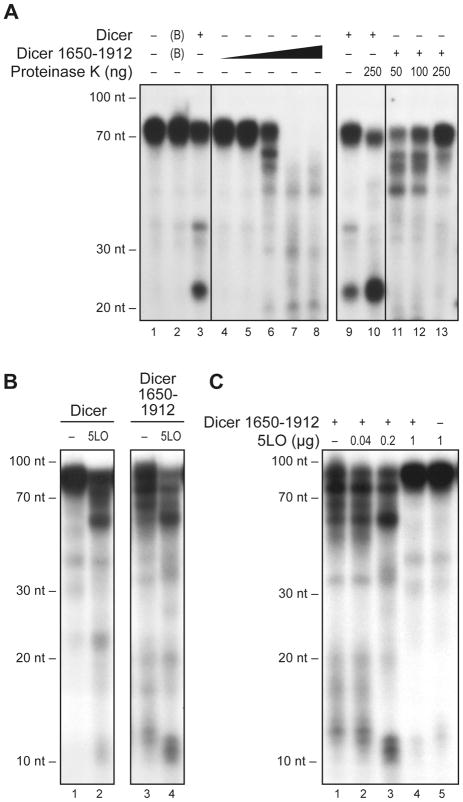
Finally, we asked if 5LO could influence the RNase III activity of human Dicer. For that purpose, we utilized the full-length Dicer protein as well as a 5LObd-containing fragment of Dicer (deletion mutant 1650-1912). As shown in Figure 7A (lanes 4 to 8), this fragment exhibited RNase III activity and processed the 32P-labeled hsa-let-7a-3 pre-miRNA substrate into smaller RNA species. In contrast to the situation observed with full-length Dicer protein (see Figure 7A, lanes 1 to 3), there was no preferential accumulation of RNAs of the size expected for mature miRNAs, i.e. ~22 to 23-nt, an observation that can be related to the absence, in the Dicer 1650-1912 fragment, of the central PAZ domain, which is known to be required for anchoring the base of the hsa-let-7a-3 pre-miRNA stem. The proteinaceous nature of the dsRNase activity exerted by Dicer fragment 1650-1912 was confirmed by its sensitivity to proteinase K treatment (see Figure 7A, lanes 11 to 13). Treatment of the full-length Dicer protein, however, considerably enhanced dsRNase activity (see Figure 7A, lanes 9 and 10), as initially reported [3]. This effect induced by proteolysis is likely due to the removal of the putative helicase domain located in the N-terminal region of human Dicer, which has been reported to exert auto-inhibitory effects on Dicer activity [37].
Figure 7.
5LO modifies the pre-miRNA processing activity of human Dicer. (A–C) Dicer RNase activity assays. Recombinant human Dicer full-length or 1650-1912 mutant protein was incubated in the absence or presence of human 5LO prior to addition of a 32P-labeled microRNA precursor substrate (pre-let-7a-3). (A) Recombinant human Dicer full-length (66 ng) or increasing amounts of the 1650-1912 mutant (lanes 4–8: 5, 20, 100, 500 or 2500 ng and lanes 11–13: 2.5 μg) proteins were incubated in the absence (lanes 1–9) or presence (lanes 10–13) of proteinase K for 10 min at 37°C, prior to addition of a 32P-labeled pre-miRNA substrate (pre-let-7a-3). BSA (1 μg) is annotated as (B) in the figure. (B–C) Recombinant human Dicer full-length (33 ng) or 1650-1912 mutant (100 ng) protein was incubated in the absence (5LO buffer only, denoted as −) or presence of 5LO (panel B, 200 ng) for 30 min at room temperature (B), or at 4°C (C), prior to addition of 32P-labeled pre-let-7a-3. After a 1-h incubation at 37°C, the samples were analyzed by denaturing PAGE and autoradiography. A 10-nt RNA size marker was used.
In RNase activity assays, human 5LO was found to enhance the conversion of the hsa-let-7a-3 pre-miRNA substrate by Dicer fragment 1650-1912 and favored the production of ~55-nt and ~10 to 12-nt RNA species (see Figure 7B, lanes 3 and 4). Similar results were obtained when using the full-length Dicer protein (see Figure 7B, lanes 1 and 2). The effect of 5LO on the RNase III activity of Dicer fragment 1650-1912 appears to be dose-dependent (see Figure 7C, lanes 1 to 3), up to a concentration at which the action of 5LO is rather inhibitory (see Figure 7C, lane 4). As for 5LO itself, it had no effect on the pre-miRNA substrate (see Figure 7C, lane 5). Our results indicate that Dicer interacts with 5LO via its 5LObd in a manner that affects each others enzymatic activities and suggest that the occupancy of the Dicer C-terminal 5LObd by 5LO may modify the RNase III properties of the adjacent dual RNase III domains that have been shown to function through intramolecular dimerization [4].
Human Dicer 5LObd may have evolved to acquire 5LO-interacting properties
Although Dicer is evolutionarily conserved from fission yeast to human, species-specific structural and functional differences in the protein may have arisen during the course of evolution. In fission yeast, for example, Dicer generates small RNAs through processing of long dsRNAs derived from centromeric repetitive sequences whereas, in human, Dicer preferentially converts ~60 to 70-nt pre-miRNAs into miRNAs. Absent from the fission yeast enzyme, the PAZ domain has been reported to act as an anchor for the 2-nt 3′overhang of duplex pre-miRNA substrates [6]. Human Dicer may thus have evolved and adapted a structure-specific recognition module in its PAZ domain that conferred to the protein the ability to process genome-encoded pre-miRNAs. Emergence of a LO-interacting module specific to 5LO within the C-term of human Dicer, which is defined as the 5-lipoxygenase binding domain, or 5LObd, is consistent with the expression of 5LO in human and the absence of 5LO or 5LO-related sequences in fission yeast and nematode genomes, and argues further for the specialization and acquisition of new functionalities by Dicer during evolution.
Conclusion
In addition to broadening the repertoire of mammalian Dicer-interacting proteins, including Argonaute 2, transactivating response RNA-binding protein (TRBP) and PACT, our results with 5LO suggest that the Dicer C-terminal 5LObd, which was previously shown to bind RNA [3], may also function as a protein interaction module that contributes to enhance 5LO enzyme activity. The nature and impact of the interaction of Dicer with 5LO may differ from that with TRBP and PACT which, in contrast to 5LO, are dsRNA binding proteins known to interact with Dicer N-terminal region containing the putative helicase domain [17]. Although it remains to be determined whether they interact in primary leukocytes, the relatively weak coimmunoprecipitation between 5LO and Dicer proteins suggests that this Dicer 5LObd-mediated interaction may be transient, conditional and/or require structural changes in Dicer conformation. Whether Dicer 5LObd becomes exposed to the surface (i) of an alternatively spliced variant form of Dicer, (ii) following protease-directed cleavage, as observed in the case of calpain-mediated release of Dicer fragments [38], (iii) in association with an enhanced enzyme activity induced by limited proteolysis [3], or (iv) upon covalent modification, such as phosphorylation, also remains to be determined.
In contrast to Dicer, which is expected to be present in every miRNA-expressing cell, 5LO expression is restricted in mammals to inflammatory cells, but also in various cancer cells such as those of prostate and pancreas origin [39]. In the context that chronic inflammation can preceed cancer, the relationship between 5LO and Dicer may provide an intriguing link between inflammation and miRNA-guided regulation of gene expression.
Supplementary Material
Acknowledgments
We wish to thank Philip James for providing yeast strain PJ69-4A, Lutz Fischer for the 5LO W13/75/102A mutant, and Agneta Nordberg for excellent technical assistance. P.P. is a New Investigator of the Canadian Institutes of Health Research (CIHR) and a Junior 2 Research Scholar of the Fonds de la Recherche en Santé du Québec. This work was supported by grants from the Institute of Infection and Immunity (III) of the CIHR (NIP-67463) (P.P.), the Natural Sciences and Engineering Research Council of Canada (262938-08) (P.P.), the Swedish Research Council (03X-217) (O.R., B.S.), and the European Union (LSHM-CT-2004-005033) (O.R.).
Abbreviations
- AD
activating domain
- Ade
adenine
- a.a
amino acids
- ATP
adenosine triphosphate
- BD
binding domain
- C-terminal domain
C-term
- ds
double-stranded
- dsRBD
dsRNA binding domain
- DTT
dithiothreitol
- GSH
glutathione
- GST
glutathione S-transferase
- His
histidine
- HPLC
high pressure liquid chromatography
- IB
immunoblot
- LO
lipoxygenase
- miRNA
microRNA
- nt
nucleotide
- OAG
1-oleoyl-2-acetylglycerol
- PAZ
Piwi/Argonaute/Zwille
- PC
phosphatidylcholine
- PCR
polymerase chain reaction
- RNAi
RNA interference
- RNase
ribonuclease
- 17-OH-22
4, 17(S)-hydroxy-7(Z),10(Z),13(Z),15(E)-docosatetraenoic acid
- 5LObd
5-lipoxygenase binding domain
- 5-HPETE
5(S)-hydroperoxy-6-trans-8,11,14-cis-eicosatetraenoic acid
- 5-HETE
5(S)-hydroxy-6-trans-8,11,14-cis-eicosatetraenoic acid
References
- 1.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 2.Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. Embo J. 2002;21:5875–5885. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Provost P, Dishart D, Doucet J, Frendewey D, Samuelsson B, Radmark O. Ribonuclease activity and RNA binding of recombinant human Dicer. Embo J. 2002;21:5864–5874. doi: 10.1093/emboj/cdf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 5.Ouellet DL, Perron MP, Gobeil LA, Plante P, Provost P. MicroRNAs in Gene Regulation: When the Smallest Governs It All. J Biomed Biotechnol. 2006;2006:69616. doi: 10.1155/JBB/2006/69616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacRae IJ, Zhou K, Doudna JA. Structural determinants of RNA recognition and cleavage by Dicer. Nat Struct Mol Biol. 2007;14:934–940. doi: 10.1038/nsmb1293. [DOI] [PubMed] [Google Scholar]
- 7.Takeshita D, Zenno S, Lee WC, Nagata K, Saigo K, Tanokura M. Homodimeric structure and double-stranded RNA cleavage activity of the C-terminal RNase III domain of human dicer. J Mol Biol. 2007;374:106–120. doi: 10.1016/j.jmb.2007.08.069. [DOI] [PubMed] [Google Scholar]
- 8.Blaszczyk J, Gan J, Tropea JE, Court DL, Waugh DS, Ji X. Noncatalytic assembly of ribonuclease III with double-stranded RNA. Structure (Camb) 2004;12:457–466. doi: 10.1016/j.str.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 9.Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 10.MacRae IJ, Doudna JA. Ribonuclease revisited: structural insights into ribonuclease III family enzymes. Curr Opin Struct Biol. 2007;17:138–145. doi: 10.1016/j.sbi.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 11.Sun W, Jun E, Nicholson AW. Intrinsic double-stranded-RNA processing activity of Escherichia coli ribonuclease III lacking the dsRNA-binding domain. Biochemistry. 2001;40:14976–14984. doi: 10.1021/bi011570u. [DOI] [PubMed] [Google Scholar]
- 12.Redko Y, Bechhofer DH, Condon C. Mini-III, an unusual member of the RNase III family of enzymes, catalyses 23S ribosomal RNA maturation in B. subtilis. Mol Microbiol. 2008;68:1096–1106. doi: 10.1111/j.1365-2958.2008.06207.x. [DOI] [PubMed] [Google Scholar]
- 13.Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 14.Tahbaz N, Kolb FA, Zhang H, Jaronczyk K, Filipowicz W, Hobman TC. Characterization of the interactions between mammalian PAZ PIWI domain proteins and Dicer. EMBO Rep. 2004;5:189–194. doi: 10.1038/sj.embor.7400070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. Embo J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Provost P, Samuelsson B, Radmark O. Interaction of 5-lipoxygenase with cellular proteins. Proc Natl Acad Sci U S A. 1999;96:1881–1885. doi: 10.1073/pnas.96.5.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radmark O, Werz O, Steinhilber D, Samuelsson B. 5-Lipoxygenase: regulation of expression and enzyme activity. Trends Biochem Sci. 2007;32:332–341. doi: 10.1016/j.tibs.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 20.Radmark O. Arachidonate 5-lipoxygenase. Prostaglandins Other Lipid Mediat. 2002;68-69:211–234. doi: 10.1016/s0090-6980(02)00032-1. [DOI] [PubMed] [Google Scholar]
- 21.Peters-Golden M, Henderson WR., Jr Leukotrienes. N Engl J Med. 2007;357:1841–1854. doi: 10.1056/NEJMra071371. [DOI] [PubMed] [Google Scholar]
- 22.Hammarberg T, Provost P, Persson B, Radmark O. The N-terminal domain of 5-lipoxygenase binds calcium and mediates calcium stimulation of enzyme activity. J Biol Chem. 2000;275:38787–38793. doi: 10.1074/jbc.M006136200. [DOI] [PubMed] [Google Scholar]
- 23.Rakonjac M, Fischer L, Provost P, Werz O, Steinhilber D, Samuelsson B, Radmark O. Coactosin-like protein supports 5-lipoxygenase enzyme activity and up-regulates leukotriene A4 production. Proc Natl Acad Sci U S A. 2006;103:13150–13155. doi: 10.1073/pnas.0605150103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Provost P, Doucet J, Hammarberg T, Gerisch G, Samuelsson B, Radmark O. 5-Lipoxygenase interacts with coactosin-like protein. J Biol Chem. 2001;276:16520–16527. doi: 10.1074/jbc.M011205200. [DOI] [PubMed] [Google Scholar]
- 25.Doucet J, Provost P, Samuelsson B, Radmark O. Molecular cloning and functional characterization of mouse coactosin-like protein. Biochem Biophys Res Commun. 2002;290:783–789. doi: 10.1006/bbrc.2001.6236. [DOI] [PubMed] [Google Scholar]
- 26.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 27.Provost P, Doucet J, Stock A, Gerisch G, Samuelsson B, Radmark O. Coactosin-like protein, a human F-actin-binding protein: critical role of lysine-75. Biochem J. 2001;359:255–263. doi: 10.1042/0264-6021:3590255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammarberg T, Radmark O. 5-lipoxygenase binds calcium. Biochemistry. 1999;38:4441–4447. doi: 10.1021/bi9824700. [DOI] [PubMed] [Google Scholar]
- 29.Hornig C, Albert D, Fischer L, Hornig M, Radmark O, Steinhilber D, Werz O. 1-Oleoyl-2-acetylglycerol stimulates 5-lipoxygenase activity via a putative (phospho)lipid binding site within the N-terminal C2-like domain. J Biol Chem. 2005;280:26913–26921. doi: 10.1074/jbc.M500068200. [DOI] [PubMed] [Google Scholar]
- 30.Plante I, Davidovic L, Ouellet DL, Gobeil LA, Tremblay S, Khandjian EW, Provost P. Dicer-Derived MicroRNAs Are Utilized by the Fragile X Mental Retardation Protein for Assembly on Target RNAs. J Biomed Biotechnol. 2006;2006:64347. doi: 10.1155/JBB/2006/64347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ouellet DL, Plante I, Landry P, Barat C, Janelle ME, Flamand L, Tremblay MJ, Provost P. Identification of functional microRNAs released through asymmetrical processing of HIV-1 TAR element. Nucleic Acids Res. 2008;36:2353–2365. doi: 10.1093/nar/gkn076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cosentino GP, Venkatesan S, Serluca FC, Green SR, Mathews MB, Sonenberg N. Double-stranded-RNA-dependent protein kinase and TAR RNA-binding protein form homo- and heterodimers in vivo. Proc Natl Acad Sci U S A. 1995;92:9445–9449. doi: 10.1073/pnas.92.21.9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Daher A, Longuet M, Dorin D, Bois F, Segeral E, Bannwarth S, Battisti PL, Purcell DF, Benarous R, Vaquero C, Meurs EF, Gatignol A. Two dimerization domains in the trans-activation response RNA-binding protein (TRBP) individually reverse the protein kinase R inhibition of HIV-1 long terminal repeat expression. J Biol Chem. 2001;276:33899–33905. doi: 10.1074/jbc.M103584200. [DOI] [PubMed] [Google Scholar]
- 35.Reddy KV, Hammarberg T, Radmark O. Mg2+ activates 5-lipoxygenase in vitro: dependency on concentrations of phosphatidylcholine and arachidonic acid. Biochemistry. 2000;39:1840–1848. doi: 10.1021/bi9919246. [DOI] [PubMed] [Google Scholar]
- 36.Kulkarni S, Das S, Funk CD, Murray D, Cho W. Molecular basis of the specific subcellular localization of the C2-like domain of 5-lipoxygenase. J Biol Chem. 2002;277:13167–13174. doi: 10.1074/jbc.M112393200. [DOI] [PubMed] [Google Scholar]
- 37.Ma E, MacRae IJ, Kirsch JF, Doudna JA. Autoinhibition of human dicer by its internal helicase domain. J Mol Biol. 2008;380:237–243. doi: 10.1016/j.jmb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lugli G, Larson J, Martone ME, Jones Y, Smalheiser NR. Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J Neurochem. 2005;94:896–905. doi: 10.1111/j.1471-4159.2005.03224.x. [DOI] [PubMed] [Google Scholar]
- 39.Furstenberger G, Krieg P, Muller-Decker K, Habenicht AJ. What are cyclooxygenases and lipoxygenases doing in the driver’s seat of carcinogenesis? Int J Cancer. 2006;119:2247–2254. doi: 10.1002/ijc.22153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.