Ste24 is a membrane-integral CaaX metalloprotease of the ER and its only known substrate in yeast is the mating factor a. We describe a novel role for Ste24 in modulating chitin levels at the bud neck by delocalization of Chs3.
Abstract
Ste24 is a membrane-integral CaaX metalloprotease residing in the endoplasmic reticulum (ER). In yeast, the only known substrate of Ste24 is the mating factor a precursor. A global screening for protein–protein interactions indicated that Ste24 interacts with chitin synthesis deficient (Chs)3, an enzyme required for chitin synthesis. We confirmed this interaction by yeast two-hybrid analyses and mapped the interacting cytoplasmic domains. Next, we investigated the influence of Ste24 on chitin synthesis. In sterile (ste)24Δ mutants, we observed resistance to calcofluor white (CFW), which was also apparent when the cells expressed a catalytically inactive version of Ste24. In addition, ste24Δ cells showed a decrease in chitin levels and Chs3-green fluorescent protein localized less frequently at the bud neck. Overexpression of STE24 resulted in hypersensitivity to CFW and a slight increase in chitin levels. The CFW phenotype of ste24Δ cells could be rescued by its human and insect orthologues. Although Chs3 binds to Ste24, it seems not to be a substrate for this protease. Instead, our data suggest that Chs3 and Ste24 form a complex in the ER that facilitates protease action on prenylated Chs4, a known activator of Chs3 with a C-terminal CaaX motif, leading to a more efficient localization of Chs3 at the plasma membrane.
INTRODUCTION
Although chitin is only a minor constituent of the yeast cell wall (∼2% dry weight), its synthesis is essential, because it has vital functions during cell division and sporulation. During cell division chitin is first concentrated in a ring-like structure at the site where the bud emerges, and then it is deposited as a disk, forming the primary septum (Lesage and Bussey, 2006). Chitin is also found in the ascospore wall, where it is modified by spore-specific chitin deacetylases and mediates spore wall resistance (Briza et al., 1988; Christodoulidou et al., 1999). Chitin synthesis in yeast is catalyzed by three chitin synthases encoded by the genes chitin synthesis deficient (CHS)1, CHS2, and CHS3 (Cabib et al., 1993). Chs1 acts as a repair enzyme that is involved in remodeling of the cell wall during cell division, and Chs2 forms the chitin in the central disk of the primary septum. Chs3 accounts for ∼90% of the chitin produced in vivo (Shaw et al., 1991) and is required for chitin synthesis at the lateral cell wall and for the formation of the chitin ring at the bud neck (Cabib et al., 1996). Chs3 is a membrane-integral protein with its catalytic domain facing the cytosol, where it catalyzes the transfer of the sugar moiety of UDP-N-acetylglucosamine (UDP-GlcNAc) to the nonreducing end of the growing chitin chain. The transmembrane domains may be involved in the translocation of nascent chitin across the plasma membrane (Merzendorfer, 2006).
The precise number and topology of transmembrane helices and soluble domains of Chs3 are uncertain. Fluorescence microscopic studies revealed that Chs3 is localized in smaller amounts at the plasma membrane, in higher amounts in the membranes of post-Golgi vesicles, and during cell division in a ring-like structure at the bud neck of small-budded cells (Cabib et al., 1993). Chs3 is synthesized by ribosomes and inserted into the lipid bilayer of the endoplasmic reticulum (ER), where it eventually attains its native conformation. This process requires the presence of the ER chaperone Chs7, which seems to prevent Chs3 aggregation (Trilla et al., 1999). After processing of Chs3 in the ER and Golgi apparatus, including glycosylation and palmitoylation (Santos and Snyder, 1997; Lam et al., 2006), it is transported from the trans-Golgi network to the cell surface, a process that requires Chs5 and Chs6, which are part of an exomer coat-complex (Wang et al., 2006). At the bud neck, Chs3 is linked to septins via Chs4 and Bni4 (DeMarini et al., 1997). Bni4 also recruits the catalytic subunit of protein phosphatase 1 (Glc7) to the bud neck in a temporal and spatial restricted manner, a process that assists in recruiting Chs3 by a yet unidentified substrate (Larson et al., 2008). Chs3 is not degraded in vacuoles but accumulates in chitosomes, which are specific secretory vesicles for chitin synthase transport to the plasma membrane (Ruiz-Herrera et al., 1977). They seem to act also as a trans-Golgi reservoir that is replenished by the endocytotic turnover of the enzyme (Ziman et al., 1996). Endocytotic turnover seems to be impaired by Chs4, which binds to Chs3 at the plasma membrane and regulates chitin synthase activity (Trilla et al., 1997; Reyes et al., 2007). Chs4 is not only required for Chs3 activity but also for the interaction between Chs3 and Bni4, which promotes chitin synthesis and proper chitin localization (DeMarini et al., 1997). Chs4 is transported to the plasma membrane independently of the Chs3 trafficking rout (Reyes et al., 2007). Two groups have reported that Chs4 is a prenylated protein. However, the role of this prenylation is controversial. Although Grabinska et al., (2007) suggest that Chs4 prenylation is required for Chs3 activity and chitin biosynthesis but not for membrane association, Reyes et al., (2007) concluded that prenylation is required for its membrane association and lateral diffusion but not for its biological function in chitin biosynthesis. The latter conclusions were mainly drawn by analyzing yeast mutants expressing Chs4 with an altered C-terminal CaaX motive. It is the cysteine of the CaaX motif that becomes prenylated by the farnesyltransferase. Prenylation is a prerequisite for the subsequent steps of CaaX processing, which include prenylation-dependent cleavage of the CaaX motif by a CaaX protease and carboxy-methylation by a methyl transferase (Wright and Philips, 2006).
To gain more insight into the role of post-prenylation steps of CaaX processing of Chs4, we investigated the CaaX protease Ste24, which we showed to bind to Chs3 and hence seems related to chitin synthesis. Ste24 is a metalloprotease that resides in the membranes of the ER, and it is, in addition to Rce1, the only CaaX protease identified in yeast (Huyer et al., 2006). The only substrate of Ste24 in yeast so far known is the precursor of the mating factor a (MFa). Ste24 cleaves the MFa precursor at the N terminus, and at the C terminus in a prenylation-dependent manner (Tam et al., 2001). CaaX processing of the MFa precursor (prenylation by Ram1/Ram2, cleavage by Rce1/Ste24 and carboxy-methylation by Ste14) is essential for the nonconventional export of MFa by an ATP-binding cassette transporter Ste6 (Chen et al., 1997). In this study, we provide evidence for a new function of Ste24 in modulating chitin synthesis by delocalization of Chs3, possibly involving CaaX cleavage of Chs4.
MATERIALS AND METHODS
General
All chemical reagents were of analytical grade and purchased from local distributors. Primers (Supplemental Table S1) were synthesized by Eurofins MWG Operon (Ebersberg, Germany). Calcofluor white (CFW) was purchased from Sigma-Aldrich (St. Louis, MO). YPD medium was prepared with deionized water and contained 1% (wt/vol) yeast extract, 2% (wt/vol) peptone, and 2% (wt/vol) glucose. YPG was prepared as YPD medium adding 2% (wt/vol) galactose instead of glucose. Sequencing was performed by Sequence Laboratories (Göttingen, Germany). Prediction of transmembrane helices, domain topology, and palmitoylation sites was done with different programs (DAS, HMMTOP, Predict Protein, SOSUI, TMAP, TMHMM, TMPred, TOPpred, and CSS-Palm2.0) linked at http://www.expasy.org/tools.
Strains
All strains used in this study are listed in Table 1. BY4741 (mating type [MAT]a his3Δ1 leu2Δ0 met15Δ0 ura3Δ0); its isogenic knockout strains ste24Δ, chs4Δ, chs3Δ, rce1Δ, and CEN.PK2–1D (MATα ura3-52 trp1-289 leu2-3_112 his3Δ MAL2-8C SUC2), with its knockout strain ste24Δ, were obtained from Euroscarf (Frankfurt, Germany). Green fluorescent protein (GFP)-fusion protein strains (Chs3GFP and Ste24GFP) were obtained from Invitrogen (Carlsbad, CA). The genomic point mutation chs4C693S was generated by homologous recombination using the primers chs4C693S-F and chs4C693S-R (Supplemental Table S1) and the vector pFA6a-GFP(S65T)-KanMX4 as a template (McElver and Weber, 1992). After transformation of BY4741 cells and selection on appropriate growth media, the point mutation was verified by sequencing of a polymerase chain reaction (PCR)-generated fragment. The double mutants rce1Δ ste24Δ, Chs3GFP ste24Δ, Chs3GFP chs4Δ, and chs4C693G ste24Δ were constructed by mating of the strains CEN.EN13-3C ste24Δ, BY4741 rce1Δ, Chs3GFP, BY4742 ste24Δ, BY4742 chs4Δ, and BY4741chs4C693S. Resulting diploid strains were sporulated, and segregants carrying double mutations were selected on SD-His + G418 plates. The double mutations and mating types were confirmed by direct PCR (Huxley et al., 1990).
Table 1.
Strains
| Yeast strain | Genotype/description | Source or reference |
|---|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Euroscarf (Winzeler et al., 1999) |
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Euroscarf (Winzeler et al., 1999) |
| BY4741 chs3Δ | As for BY4741, chs3Δ::kanMX4 | Euroscarf (Winzeler et al., 1999) |
| BY4741 ste24Δ | As for BY4741, ste24Δ::kanMX4 | Euroscarf (Winzeler et al., 1999) |
| BY4742 ste24Δ | As for BY4742, ste24Δ::kanMX4 | Euroscarf (Winzeler et al., 1999) |
| BY4741 chs4Δ | As for BY4741, chs4Δ::kanMX4 | Euroscarf (Winzeler et al., 1999) |
| BY4742 chs4Δ | As for BY4742, chs4Δ::kanMX4 | Euroscarf (Winzeler et al., 1999) |
| BY4741 rce1Δ | As for BY4741, rce1 Δ::kanMX4 | Euroscarf (Winzeler et al., 1999) |
| BY4742 sst2Δ | As for BY4742, sst2 Δ::kanMX4 | Euroscarf (Winzeler et al., 1999) |
| CEN.PK2-1D | MATα ura3-52 trp1-289 leu2-3_112 his3Δ1 MAL2-8CSUC2 | Euroscarf (Entian et al., 1999) |
| CEN.EN13-3C ste24Δ | As for CEN.PK2, YJR117w::HIS3 | Euroscarf (Entian et al., 1999) |
| Ste24TAP | MATa ade2 arg4 leu2-3_112 trp1-289 ura3-52 | CellZone AG (Heidelberg, Germany) |
| Chs3GFP | As for BY4741, CHS3-GFP::HIS3 | Invitrogen (Huh et al., 2003) |
| Ste24GFP | As for BY4741, STE24-GFP::HIS3 | Invitrogen (Huh et al., 2003) |
| Chs3GFP ste24Δ | As for BY4741, CHS3-GFP::HIS3 ste24Δ::kanMX4 | This study |
| Chs3GFP chs4Δ | As for BY4741, CHS3-GFP::HIS3 chs4Δ::kanMX4 | This study |
| rce1Δ ste24Δ | As for BY4741, rceIΔ::kanMX4, ste24Δ::HIS3 | This study |
| AH109 | MATa trp1-901 leu2-3_112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1UAS-GAL1TATA-HIS3 GAL2UAS-GAL2TATA-ADE2 URA3:: MEL1UAS-MEL1TATA-lacZ MEL1 | Clontech (James et al., 1996) |
| BY4741 CHS4C693S | As for BY4741, CHS4C693G chs4- terminator:kanMX4 | This study |
Plasmids
All plasmids used in this study are listed in Supplemental Table S2. The open reading frames encoding Chs3, Chs4, Ste24, zinc metalloprotease Ste24 homologue (ZmpSte24), and TcSte24 were amplified from yeast genomic DNA, or human and Tribolium cDNAs by PCR using specific primers (Supplemental Table S1) and cloned into centromeric vectors pAG503, pRS415, pGREG576, or the 2μ vector pJJH71 under the control of the PKF2 promoter (Raben et al., 1995). The nucleotide sequences of the GAL1 promoter or the 13MYC epitope tag were amplified by PCR using specific primers (Supplemental Table S1) and cloned into the respective vectors. Yeast strains were transformed via electroporation (1 pulse, 1.5 kV, 25 μF, 200 Ω) with 1 μg of plasmid DNA per transformation performed in 50 μl of 1 M sorbitol. Plasmids were assayed for complementation of chs3Δ, chs4Δ, or ste24Δ on the basis of the CFW resistance phenotype or by CFW staining. The pRS415ste24E269G plasmid was created by site directed mutagenesis using the QuikChange kit (Stratagene, Cedar Creek, TX) and pRS415ste24 as a template. The point mutation was confirmed by nucleotide sequencing. For generation of the STE24↑ strain overexpressing STE24, the plasmid pAG503 GAL-STE24↑ placing STE24 under the control of the GAL1 promoter was used to transform BY4741 cells.
CFW Serial Drop Dilution Assay
Each 5 μl of a yeast suspension was spotted at different concentration (103-107cells/ml) onto solid rich medium (YPD or YPG) plates containing 50 μg/ml CFW. After incubation at 30°C for 3 d, colony growth was documented using a Versa Doc Imaging System (Bio-Rad Laboratories, Hercules, CA) and the Quantity One, version 4.6 (Bio-Rad Laboratories).
Fluorescent Calcofluor White Assay
To estimate chitin levels, a CFW fluorescence assay was used according to a modified assay published by Lam et al. (2006). In brief, each 5 μl of a yeast suspension (concentrated to 109cells/ml) was spotted onto solid rich medium (YPD or YPG) plates containing 50 μg/ml CFW. After incubation at 30°C for 3 d, the fluorescence was quantified densitometrically using the Versa Doc imaging system (λex = 356 nm, 520LP filter; Bio-Rad Laboratories) and Quantity One, version 4.6 (Bio-Rad Laboratories). The optical densities from constant areas within the spots were averaged over 15–40 independent experiments and corrected for the local background. The mean optical density averaged over 40 spots of wild-type cells was set to 100%, whereas the mean optical density averaged over 40 spots of chs3Δ cells was set to 0%. Relative chitin amounts (RCA; ± SE) from various mutants were calculated according to RCA = x-chs3Δ/WT-chs3Δ, with x is the mean optical density averaged over 20 spots of the respective yeast mutant.
Measurement of the Chitin Content
The chitin content of different yeast cell strains was determined by the Morgan–Elson method as described in Bulik et al. (2003), with some minor modifications. KOH-treated cell pellets were incubated for 48 h with 5 μl (20 mg/μl) of Streptomyces griseus chitinase (Sigma-Aldrich). Colorimetric determination of GlcNAc was performed in microtiter plates, of which each slot was loaded with 150 μl of the samples treated with Ehrlich's reagent.
Microscopy
Cells were grown in YPD or SD media until the early logarithmic phase was reached. When gene expression was controlled by a GAL1 promoter, cells were grown in glucose-free medium containing 1% (wt/vol) raffinose as a carbon source and analyzed in early logarithmic phase 3 h after induction of gene expression by adding 2% (wt/vol) galactose to the medium. For CFW staining, yeast cells were incubated in 0.02% (wt/vol) CFW solution for 30 min at room temperature and washed three times with deionized water. Microscopy was performed with a 100× oil-immersion objective (numerical aperture 1.36) and an IX70 fluorescence microscope (Olympus, Hamburg, Germany). Fluorescence was excited with a U-RFL-burner (Olympus), and appropriate filter cubes were used to set excitation and emission wavelengths. Images were captured with a CoolSNAP HQ2 digital camera (Roper Scientific, Tucson, AZ) using MetaMorph 6.2 software (Molecular Devices, Toronto, ON, Canada). Z-stack series of 12 optical layers were taken for each analyzed cell. Protein distribution was analyzed from Z-stack series comprising between 150 and 250 single cells. Measurement of local fluorescence intensities was performed with the Quantity One, version 4.6 (Bio-Rad Laboratories).
Yeast Two-Hybrid Analysis
Mapping of the domains mediating the interaction of Chs3 and Ste24 was performed with the Matchmaker two-hybrid system according to the manufacturer's protocol and the Yeast Protocol Handbook (Clontech, St-Germain-en-Laye, France). Regions corresponding to the hydrophilic Chs3 domains C1, C3, C4, and C7 (amino acid positions 1–165, 226–452, 476–1000, and 1109–1165, respectively), and cytoplasmic Ste24 domains S2, S6, and S8 (amino acid positions 36–95, 221–304, and 384–453, respectively) were amplified with specific primers from yeast genomic DNA (Supplemental Table S1) and inserted into yeast two-hybrid vectors pGADT7 and pGBKT7 (see Figure 1A). Yeast cells of the strain AH109 were cotransformed with prey and bait plasmids (Supplemental Table S2).
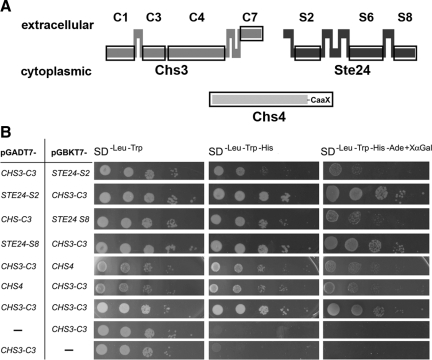
Figure 1.
Putative domain architectures of Chs3, Chs4, and Ste24 and yeast two-hybrid analysis to identify interacting domains. (A) Horizontal bars at the top represent extracellular domains, horizontal bars at the bottom intracellular domains, and vertical bars transmembrane helices. Soluble domains tested in the yeast two-hybrid analysis are marked with black squares. (B) AH109 cells were cotransformed with bait and pray vectors. Cells grown overnight in liquid SD−Leu −Trp medium were diluted with water to a final concentration of 1 × 107 cells/ml. Five microliters of each suspension and three subsequent 10-fold serial dilutions were individually spotted onto SD−Leu −Trp, SD−Leu −Trp −His and SD−Leu −Trp −His −Ade +X α-Gal plates for selection. Cells were incubated at 30°C for 2 d.
Halo Assays
The production of mature a-factor from various MATa strains was monitored by pheromone diffusion (halo) assay, modified after Trueblood et al., (2000). In brief, MATa wild-type cells were dropped at high concentrations onto agar plates with low concentrated MATα cells. Mating proficiency is indicated by growth inhibition of the MATα cells leading to the formation of a halo surrounding the MATa drops. To prepare a homogenous cell suspension, ∼5 × 106 cells of the MATα sst2 strain were mixed with 3 ml of 42°C YPD media containing 0.7% agarose (molecular grade; Bioline, Taunton, MA) and 0.04% Triton X-100 and spread onto solid rich medium (YPD) plates containing 0.04% Triton X-100.
Yeast Protein Extraction
Cells were grown in complete (YPD and YPG) or synthetic defined (SD) media until the early logarithmic phase was reached. Cells were centrifuged for 3 min at 3000 × g and 4°C. The pellet was resuspended in 50 mM Tris-HCl buffer, pH 7.0, containing 20 mM NaCl including Complete Protease Inhibitor Cocktail (Roche, Basel, Switzerland). After adding the same volume of glass beads, cells were vortexed for 5 min at 4°C. Subsequently, the suspension was centrifuged for 3 min at 3000 × g and 4°C, and the resulting pellet was resuspended in Laemmli buffer and boiled for 1 min (Laemmli, 1970).
Other Methods
Protein concentrations were determined by the Amido Black method (Wieczorek et al., 1990), and SDS-polyacrylamide gel electrophoresis was performed according to Laemmli (1970). Semidry electroblotting onto nitrocellulose membranes (Millipore, Schwalbach, Germany) was carried out essentially as described by Kyhse-Andersen (1984), with the modification that the buffers were supplemented with 20% (vol/vol) methanol. Blot membranes were stained with 0.02% (vol/vol) Ponceau S (Sigma-Aldrich, Taufkirchen, Germany). Immunoblots were performed as described previously (Zimoch and Merzendorfer, 2002). Primary antibodies used were polyclonal anti-Myc antibodies (1:100; AbD Serotec, Duesseldorf, Germany), and secondary antibodies were anti-mouse antibodies conjugated to alkaline phosphatase (1:30,000; Sigma-Aldrich).
RESULTS
Ste24 Interacts with Chs3 through Cytoplasmic Domains
Several studies have suggested that insect and fungal chitin synthases including yeast Chs3 are produced as zymogens requiring proteolytic cleavage for activation (reviewed in Merzendorfer (2006)). However, until now, no protease has been identified that cleaves the zymogenic form. Therefore, we screened different databases (BioGRID, BOND, BioPIXIE, DIP, and Yeast RC two-hybrid) for proteases of any kind that might interact with yeast Chs3. Among 95 proteins that potentially interact with Chs3, we detected only one likely protease, Ste24. Ste24 is a membrane-integral metalloprotease of the ER, which was identified to interact with Chs3 in a large-scale split ubiquitin screen (Miller et al., 2005). To confirm this interaction with an independent method and to map the interacting regions, we performed a yeast two-hybrid analysis testing different soluble domains of Chs3 and Ste24. As a positive control, we included Chs4, a known activator of Chs3, which was reported previously to interact with Chs3 in two independent studies (DeMarini et al., 1997; Ono et al., 2000).
We amplified the cDNA sequences encoding four soluble Chs3 domains, three soluble Ste24 domains, and the complete coding sequence of Chs4 (Figure 1A), and we ligated each of them into pGADT7-AD and pGBKT7-BD of the Matchmaker yeast two-hybrid system, to allow switching of bait and prey inserts. After negatively testing all constructs for endogenous activation of reporter gene activity (Figure 1B, examples shown in the two bottom rows), we tested the C1, C3, C4, and C7 domains of Chs3 for their ability to bind Chs4. In doing so, we could confirm the previously reported interaction between Chs3 and Chs4 and identify the C3 domain (amino acid positions 226–452) as the region binding to Chs4 (Figure 1B). No other Chs3 domains or Ste24 domains interacted with Chs4 (data not shown). Subsequently, we tested the C1, C3, C4, and C7 domains of Chs3 for their ability to interact with the cytosolic S2, S5, and S8 domains of Ste24 (Tam et al., 2001). We found that the C3 domain of Chs3 interacts with an interface of Ste24 formed at the cytoplasmic side by the S2 and S8 domains (Figure 1B). The observation that the C3 domain of Chs3 interacts with regions of proteins known to be exposed to the cytoplasm strongly supports a topology model placing this domain at the cytoplasmic site of the membrane (Figure 1A).
Ste24 Affects Chitin Levels but Not Its Cellular Distribution
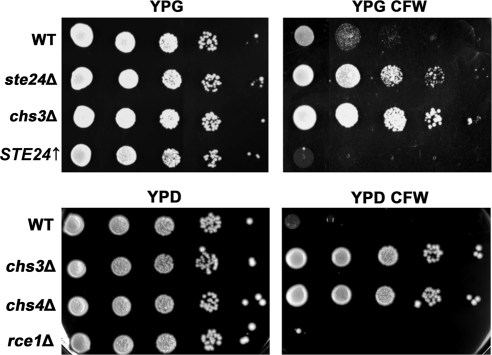
CFW is a fluorescent dye that binds primarily to chitin (Roncero and Duran, 1985). Because this compound is toxic to yeast cells, mutants with reduced chitin levels exhibit a CFW resistance phenotype. In vivo, the majority of chitin deposited in the yeast cell wall is synthesized by Chs3 (Shaw et al., 1991). Correspondingly, chs3Δ cells that produce significantly less chitin are CFW resistant. Therefore, changes in CFW resistance can be correlated with changes in Chs3 levels. To test whether Ste24 affects chitin levels, we analyzed wild-type and different mutant strains for their sensitivity to CFW. As expected, wild-type cells were highly sensitive toward CFW, whereas chs3Δ or chs4Δ cells exhibited increased resistance to CFW (Figures 2 and 4). We observed a clear, but moderate resistance to CFW, when we analyzed CFW sensitivity in ste24Δ cells. If Ste24 promotes chitin synthesis, overexpression of Ste24 should lead to hypersensitivity toward CFW. When we tested a corresponding yeast strain, we indeed observed hypersensitivity toward CFW (Figure 2). To test whether the observed effects on chitin levels are specific for the Ste24 protease, we examined rce1Δ cells for CFW sensitivity, a mutant lacking the other known yeast CaaX-protease. In contrast to ste24Δ cells, rce1Δ cells were highly sensitive toward CFW, comparable to the sensitivity of wild-type cells (Figure 2). These observations indicate that Ste24 affects chitin levels.
Figure 2.
Calcofluor white resistance phenotypes of different yeast strains. Wild-type, chs3Δ, ste24Δ, rce1Δ, chs4Δ, and STE24↑cells were grown overnight in liquid YPD medium and diluted with water to a final concentration of 1 × 107 cells/ml. Five microliters of each suspension and three subsequent 10-fold serial dilutions were spotted onto YPD or YPD plates with or without 50 μg/ml CFW. Cells were incubated at 30°C for 3 d.
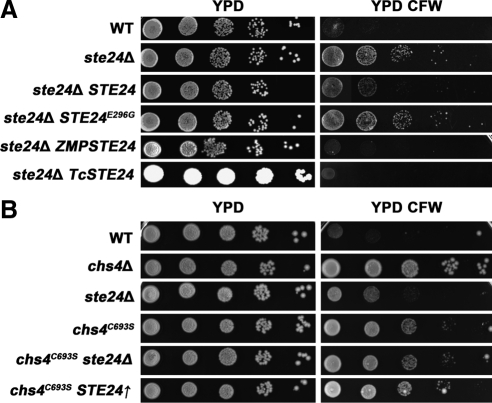
Figure 4.
Calcofluor white sensitivity in different yeast strains. (A) Functional tests of a catalytic inactive Ste24 mutant, and human and insect orthologues. Wild-type, ste24Δ, ste24Δ pRS415ste24, ste24Δ pRS415ste24E296G, ste24Δ pJJH71-ZMPSTE24, and ste24Δ pJJH71-TcSTE24 cells were grown overnight in liquid YPD medium and diluted to a final concentration of 1 × 107cell/ml in water. Five microliters of each suspension and three subsequent 10-fold serial dilutions were spotted onto YPD plates with or without 50 μg/ml CFW. Cells were incubated at 30°C for 3 d. (B) Calcofluor white sensitivities in mutants defective in Chs4 prenylation. Wild-type, chs4C693S, ste24Δ, ste24Δ chs4C693S, and chs4C693S pJJH71-STE24 cells were grown and diluted as described above.
Next, we performed a fluorescence assay based on the specific binding of CFW to estimate relative chitin levels in the cell wall (Lam et al., 2006; Figure 3B). When we measured CFW fluorescence in wild-type and chs3Δ cells, we observed a decrease in total CFW fluorescence of ∼70%. The difference in CFW fluorescence between wild-type and chs3Δ was set to 100% to represent chitin synthesis mediated by Chs3. Consistently with the results from our growth tests, ste24Δ cells showed significantly less CFW fluorescence and hence reduced chitin levels (78 ± 1.8%; n = 20). STE24 overexpressing cells showed a slightly higher CFW fluorescence and hence increased chitin levels (110 ± 2.6%; n = 15). Cells that were defective in CHS4 encoding an activator of Chs3, displayed a very low CFW fluorescence that was comparable with that of chs3Δ cells (2 ± 1.4%; n = 20). To evaluate the results from the CFW fluorescence assay, we measured chitin amounts independently by the method of Morgan–Elson (Figure 3B). Wild-type cells exhibited a chitin content of 5.2 nmol GlcNAc/mg cells (±0.2; n = 6), which was significantly reduced to 1.8 nmol GlcNAc/mg cells (±0.1; n = 6) in chs3Δ cells. Both values are in good agreement with previous measurements (Bulik et al., 2003). In line with the CFW fluorescence assay the chitin content in ste24Δ was reduced to 4.4 nmol GlcNAc/mg cells (±0.2; n = 6), which are ∼76% of the chitin produced by Chs3. As observed in the CFW fluorescence assay, STE24-overexpressing cells increased chitin levels of 6.4 GlcNAc/mg cells (±0.3; n = 6), which corresponds to 135% of the chitin produced by Chs3 and is somewhat higher than estimated from CFW fluorescence measurements. The chitin content of chs4Δ cells was with 2.0 nmol GlcNAc/mg cells (±0.3; n = 6) in the same range as that of chs3Δ cells. Overall, the chemical determination of chitin content was in good agreement with the estimate based on CFW fluorescence.
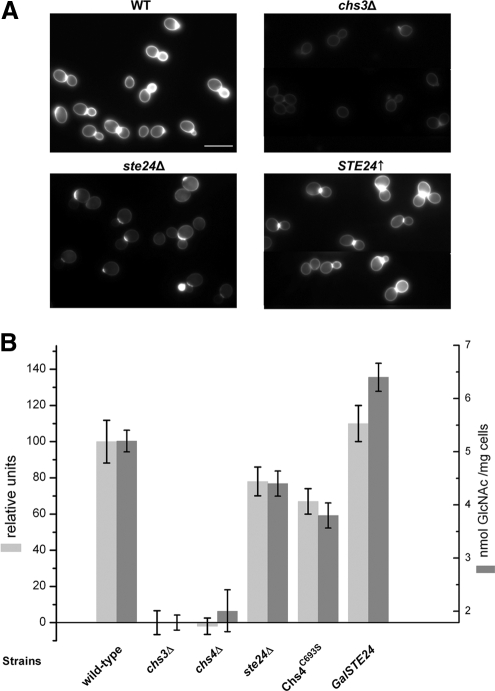
Figure 3.
Chitin deposition and amounts in yeast cells defective in different genes. (A) Wild-type, ste24Δ, chs3Δ, and STE24↑ cells were grown overnight in liquid YPD or YPG medium and stained with 0.02% CFW. Bar, 10 μm. (B) Quantification of chitin levels. Wild-type (n = 40), ste24Δ (n = 40), chs3Δ (n = 20), chs4Δ (n = 20), Chs4C693S (n = 20), and STE24↑ (n = 15) cells (109/ml) were grown for 2 d on YPD or YPG plates containing 50 μg/ml CFW. CFW fluorescence was excited by UV-light, and fluorescence was quantified densitometrically (light gray bars). Relative CFW fluorescence is given in percentage as mean values of deviations from wild-type cells (±SE). Chitin amounts were additionally quantified by the Morgan–Elson method (dark gray bars). Data represent averages (±SE) obtained from six independent experiments for each yeast strain. One-way ANOVA and Tukey's HSD test revealed significant differences for all variations from wild-type cells (HSD 0.05 = 7.57 and HSD 0.01 = 9.08 for the CFW assay; HSD 0.05 = 0.44 and HSD 0.01 = 0.54 for the Morgan–Elson assay).
To visualize chitin deposition in growing wild-type, ste24Δ, chs3Δ, and STE24↑ cells, we performed a CFW staining and monitored the cells under a fluorescence microscope. The results shown in Figure 3A more or less reflect the chitin levels as they were determined by the CFW fluorescence and Morgan–Elson assays. To test for possible differences in chitin distribution, we measured local fluorescence intensities at the bud neck and in the cell walls and calculated their ratios for wild-type and mutant cells. The fluorescence intensity ratios were 1.7 (±0.3; n = 10) in wild-type, 1.5 (±0.5; n = 8) in chs3Δ, 1.4 (±0.2; n = 9) in ste24Δ and 2.0 (±0.5; n = 10) in STE24-overexpressing cells. Thus, although chitin content in various mutant cells differed significantly from that of wild-type cells its distribution did not change significantly as indicated by analysis of variance (ANOVA) tests (honestly significant difference [HSD] 0.05 = 0.50; HSD 0.01 = 0.63) of the combined data (Figure 3A).
Catalytically Inactive Ste24 Does Not Restore CFW Sensitivity in ste24Δ Cells
To examine whether the observed effects on chitin levels depend on the catalytic activity of Ste24, we generated a catalytically inactive mutant Ste24E298G and tested whether it can restore CFW sensitivity in ste24Δ cells. After cloning STE24 into a centromeric vector and placing it under the control of its endogenous promoter, we mutated the catalytic glutamate within the conserved HExxH motif and changed it to a glycine residue. MATa cells carrying mutations in this motif are unable to process the MFa precursor and hence are mating-deficient (Fujimura-Kamada et al., 1997). To assess whether a mutant expressing a catalytically inactive form of Ste24 had been successfully generated, we performed a halo assay described in Trueblood et al. (2000). In this assay, the growth of MATα cells is suppressed when active MFa is produced by cleavage of the MFa precursor by active Ste24 producing a halo. The halo will be absent if Ste24 is not active. Because rce1Δ and ste24Δ single mutants still exhibit a halo due to the activity of the remaining CaaX protease, they are almost indistinguishable from wild-type cells (Supplemental Figure S1). However, in rce1Δ ste24Δ double deletion mutants, no halos were visible. When we transformed the rce1Δ ste24Δ double deletion cells with a vector carrying STE24, we could restore the halo phenotype of wild-type cells. In contrast, transformation of ste24Δ cells with a vector carrying ste24E298G showed no halo, indicating that the catalytic activity of Ste24 was abolished (Supplemental Figure S1). Next, we tested CFW resistance in ste24Δ cells expressing either Ste24 or the catalytically inactive Ste24E298G. As shown in Figure 4A, transformation with the wild-type STE24 vector restored CFW sensitivity of ste24Δ cells, whereas transformation with the catalytically inactive ste24E298G vector did not. Thus, the observed effect on chitin synthesis in ste24 mutants depends on the catalytic activity of Ste24.
Human and Insect Homologues of STE24 Restore CFW Sensitivity in ste24Δ Cells
Ste24 is a conserved CaaX protease and homologues can be found ubiquitously in eukaryotes. Despite a rather low amino acid sequence identity of ∼36% between yeast Ste24 and its human homologue ZmpSte24, Leung et al. (2001) demonstrated that human ZmpSte24 can restore the halo phenotype in the rce1Δ ste24Δ cells. The insect Ste24 homologue from the red flour beetle, Tribolium castaneum (TcSte24) fully restores the halo phenotype in the rce1Δ ste24Δ cells (Supplemental Figure S1). To test whether the STE24 homologues could restore the CFW phenotype, we expressed ZmpSTE24 and TcSTE24 in ste24Δ cells and plated them on plates containing CFW. These growth tests indicated that ste24Δ cells expressing human or Tribolium STE24 exhibit the same sensitivity to CFW as wild-type cells (Figure 4A), suggesting that they are functional orthologues of yeast STE24.
Immunoblots Do Not Support That Chs3 Is a Substrate for Ste24
Ste24 has dual roles in protein processing. In addition to its function of cleaving the C-terminal CaaX motif, it is also known to cleave the N terminus of the MFa precursor (Trueblood et al., 2000). Because Chs3 lacks a C-terminal CaaX motif, Ste24 evidently does not cleave the C terminus of Chs3. Therefore, we addressed the question whether Ste24 cleaves Chs3 at all. For this purpose, we prepared total cellular extracts from wild-type and ste24Δ cells expressing Chs3-13Myc from centromeric plasmids and performed Western blot analysis. In both cell extracts, we could detect two protein bands of ∼100 and 160 kDa, both of which did not shift in response to deletion of STE24 (Supplemental Figure S2). This result suggests that Chs3 is not cleaved by Ste24 and further substantiates the findings of Cos et al. (1998), who also observed no differences in the migration behavior of Chs3 tagged with HA-epitopes either at the N or C terminus. However, we cannot totally exclude the possibility that a small peptide is cleaved off by Ste24 at the N or C terminus.
Prenylation of Chs4 Is Required for Ste24 Effects on Chitin Synthesis
The results described above imply that the proteolytic activity of Ste24 is required to maintain wild-type chitin levels. Although Chs3 binds to Ste24, direct processing of Chs3 by the CaaX protease turned out to be unlikely. However, Ste24 may process another protein required for chitin synthesis. An obvious candidate is Chs4, a known activator of Chs3 (DeMarini et al., 1997; Trilla et al., 1997). Unlike Chs3, Chs4 possesses a C-terminal CaaX motive (CVIM). Thus, Chs4 fulfils all sequence requirements to be cleaved by Ste24 (Trueblood et al., 2000). Furthermore, Chs4 is known to be prenylated, which is a prerequisite for removal of the C-terminal tripeptide by the CaaX protease. Direct biochemical proof corroborating that Chs4 is a substrate of Ste24 is difficult to obtain, because protein analysis is hampered by prenylation and the fact that only a tripeptide is removed yielding only small differences in molecular masses. Therefore, we used a genetic approach to address this issue based on the observation that mutants expressing a nonprenylatable version of Chs4, in which the cysteine of the CaaX motif is replaced by a serine (Chs4C693S), exhibit a CFW resistance phenotype (Grabinska et al., 2007). When we repeated this experiment we could confirm the CFW resistance phenotype. CFW resistance of chs4C693S cells was more pronounced than that of ste24Δ cells but less pronounced than that of chs4Δ cells (Figure 4B). The chitin content in chs4C693S cells was reduced by 33%, which was 11% more than the reduction of the chitin content in ste24Δ cells (Figure 3B). If Ste24-mediated cleavage of the CaaX motif of Chs4 is required for chitin synthesis, the CFW phenotype of cells expressing a nonprenylated form of Chs4 should be unchanged when STE24 is overexpressed or deleted. As shown in Figure 4B, Chs4C693S cells exhibit the same CFW phenotype as ste24Δ Chs4C693S or STE24 overexpressing Chs4C693S cells.
Membrane Association of Chs4 Requires Prenylation but Is Independent of Further CaaX Processing
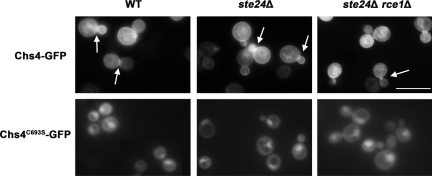
To further analyze the role of Chs4 prenylation and CaaX cleavage, we performed localization studies using GFP-tagged proteins. To investigate the contribution of prenylation and cleavage independently, we expressed Chs4-GFP and Chs4C693S-GFP in wild-type, ste24Δ and ste24Δ rce1Δ cells. As shown in Figure 5, no significant difference in Chs4-GFP localization was detectable in wild-type, ste24Δ, or ste24Δ rce1Δ suggesting that Ste24-mediated cleavage of the CaaX motif is not required for membrane association of Chs4. However, Chs4C693S-GFP showed distinct differences in localization compared with wild-type Chs4-GFP. The majority of Chs4C693S-GFP localized in the cytoplasm and no signal could be observed at the plasma membrane (Figure 5). The deletion of the CaaX proteases had, as expected, no influence on Chs4C693S-GFP localization. Hence, membrane association of Chs4 depends on prenylation but not on subsequent steps of CaaX processing.
Figure 5.
Intracellular localization of Chs4-GFP and Chs4C693S-GFP in wild-type, ste24Δ, and ste24Δ rce1Δ cells. Cells were grown overnight in liquid media containing 1% raffinose and were observed 3 h after adding 2% galactose to initiate expression of Chs4-GFP or Chs4C693S-GFP. Chs4-GFP and Chs4C693S-GFP were localized by fluorescence microscopy. Arrows point to increased fluorescence at the bud necks. Bar, 10 μm.
Deletion of ste24 Leads to Reduced Levels of Chs3 at the Bud Neck
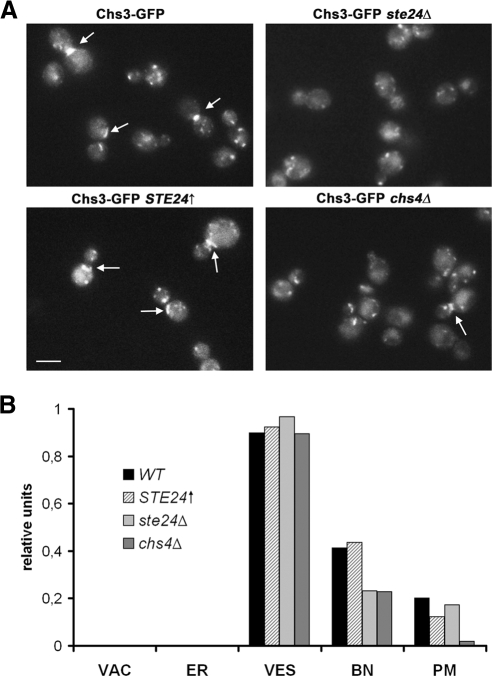
Finally, we addressed the question whether the different chitin levels in the cell walls of wild-type, ste24Δ, chs4Δ, and STE24↑ cells result from perturbations of the intracellular transport of Chs3. For this purpose, we examined the subcellular localization of Chs3-GFP in these strains, after we have shown that Chs3-GFP can restore the wild-type CFW phenotype in chs3Δ cells and thus is fully functional (data not shown). In all four strains, Chs3-GFP localizes mainly to vesicles (∼90% of examined cells; Figure 6), although in wild-type cells as well as in cells overexpressing STE24↑, ∼40% of all examined cells showed Chs3-GFP was located at the bud neck, but in ste24Δ cells, which produce significantly less chitin, only ∼20% of all examined cells showed Chs3-GFP at the bud neck (Figure 6). In chs4Δ cells, the Chs3-GFP signal was more diffuse than in the other examined strains and Chs3-GFP was less frequently at the bud neck as in the case of ste24Δ cells (∼20% of all examined cells). Thus our data indicate that the effects of ste24 on chitin synthesis maybe mediated by delocalization of Chs3.
Figure 6.
Intracellular localization of Chs3-GFP in WT, STE24↑, ste24Δ, and chs4Δ cells. (A) Fluorescence microscopy. Cells were observed in the early logarithmic growth phase. Chs3-GFP was visualized by fluorescence microscopy. Arrows point to increased fluorescence at the bud necks. Bar, 5 μm. (B) Quantitative analysis of changes in Chs3-GFP localization. The distribution of Chs3-GFP was analyzed in WT, ste24Δ, and ste24Δ rce1Δ cells (150–250 single cells each strain). VAC, vacuole; PM, plasma membrane; BN, bud neck; VES, vesicle.
DISCUSSION
Protein interaction between Chs3 and Chs4 had been reported previously by classical yeast two-hybrid analysis (DeMarini et al., 1997; Ono et al., 2000), whereas that between Chs3 and Ste24 was detected in a large-scale split ubiquitin screening (Miller et al., 2005). DeMarini et al. (1997) further demonstrated that a 700-amino acid domain making up the N-terminal region of Chs3 mediates binding to Chs4, and homodimerization of Chs3. In this study, we were able to confirm these interactions and to narrow down the binding site of Chs3 to a region between amino acid positions 226–452 (C3 domain). Computer-based prediction of Chs3 topology with different programs yielded inconclusive results with six to nine postulated transmembrane helices and a C3 domain that is exposed either to the cytoplasm or to the extraplasmic space. Our yeast two-hybrid results strongly suggest that the C3 domain is located intracellularly: 1) Chs4 is a cytoplasmic protein and therefore can only interact with the C3 domain facing the cytoplasm. 2) The C3 domain interacts with a cytoplasmic interface of Ste24 formed by the S2 and S8 domains (Tam et al., 2001). 3) Lam et al. (2006) demonstrated that Chs3 is palmitoylated by Pfa4, whose catalytic domain faces the cytoplasm, and that this posttranslational modification is one prerequisite for its exit from the ER. Screening for palmitoylation sites with CSS-Palm2.0 identified only two putative type III motifs in Chs3, both located in the C3 domain (amino acid positions 227–231, 446–451; Ren et al., 2008), additionally supporting its cytosolic location. PhoA fusion studies performed with the rhizobial chitin synthase NodC showed that the catalytic domain of this β-glycosyltransferase is exposed to the cytoplasm (Barny et al., 1996). Therefore, we propose a topology model in which the C3 domain precedes the catalytic domain, both facing the cytoplasm (Figure 1A). Further studies have to be carried out to determine the correct topology and function of the remaining Chs3 domains.
Chs3 is synthesized at the rough ER and transported via the Golgi network and trans-Golgi vesicles to the plasma membrane of the bud neck (Lesage and Bussey, 2006). Ste24 and Rce1 are only known CaaX protease in yeast. Ste24 carries a C-terminal dilysine motif (KKXX) and thus is retained in the ER (Tam et al., 2001). An interaction between Chs3 and Ste24 can only take place at the ER, because it is the only membrane compartment in which both proteins are at least transiently located. Ste24 is also found at the inner nuclear membrane (Barrowman et al., 2008). In vertebrates, Ste24 orthologues cleave the nuclear filament lamin A, and mutations in the Zmpste24 gene have been linked to progeroid syndromes (Bergo et al., 2002; Pendas et al., 2002; Fong et al., 2004). A nuclear substrate of Ste24, however, has not been reported yet in yeast. The only proven substrate of Ste24 in yeast is the MFa precursor, which is cleaved by Ste24 at the C-terminal CaaX motive in a prenylation-dependent manner and at an undefined sequence at the N terminus (Tam et al., 1998). The sequence specificities for CaaX cleavage by Ste24 and Rce1 have been determined by site-directed mutagenesis and both proteases have overlapping but distinct substrate specificities (Trueblood et al., 2000). Genome analysis identified 98 yeast proteins that are potentially cleaved by CaaX proteases at their C termini, 35 of which were postulated to be cleaved by Ste24 (Trueblood et al., 2000). Because Chs3 does not possess a CaaX motif it is obviously not a substrate for prenylation-dependent cleavage by Ste24. Moreover, Western blot analysis performed in this work and by Cos et al. (1998) yielded no evidence for cleavage of Chs3 at the N terminus. How then does Ste24 modulate chitin synthesis? A plausible answer is that another protein involved in chitin synthesis might be a substrate for Ste24. Among the 35 CaaX proteins that theoretically can be cleaved by Ste24 we could identify only one which is linked to chitin synthesis. This protein is Chs4, a known activator of Chs3 (Trilla et al., 1997), which possesses a C-terminal CVIM motif recognized by Ste24 (Trueblood et al., 2000). How precisely Chs4 activates Chs3 is still unclear; however, this process requires an interaction between both proteins (DeMarini et al., 1997; Ono et al., 2000). Chs4 is required for Chs3 activity and/or its recruitment to the bud neck (DeMarini et al., 1997; Trilla et al., 1997). DeMarini et al. (1997) further hypothesized that Bni4 functions as a linker protein between Chs4 and septins of the bud neck. However, further analysis of this process yielded a more complex picture involving also the catalytic subunit of a type 1 serine/threonine protein phosphatase (Glc7), which is necessary for Chs3 recruitment to the bud neck (Kozubowski et al., 2003; Lesage et al., 2005; Larson et al., 2008).
Does Ste24 cleave Chs4 at its CaaX motif and why does Ste24 bind to Chs3? Because CaaX cleavage depends on prenylation, we generated a mutant in which the cysteine of the CaaX motif was replaced by serine and examined its CFW phenotype in ste24Δ or STE24 overexpressing cells. Although yeast cells expressing wild-type CHS4 showed CFW resistance or hypersensitivity in response to deletion or overexpression of STE24, we observed no effects in cells expressing nonprenylated Chs4C693S. Because we could not detect any interactions between Chs4 and Ste24 in our yeast two-hybrid analysis (data not shown), we propose that Chs3, which evidently interacts with Ste24, helps to recruit Chs4 and hence facilitates cleavage of its CaaX motif. Chs4 has been detected in the cytoplasm and at the plasma membrane accumulating at the bud neck during cytokinesis (Reyes et al., 2007). However, we could show that Chs4 is also found at the ER (Figure 5). Perhaps Chs3, Chs4, and Ste24 are part of transient complex formed at the ER that is required for Chs4 processing. This complex could also involve other ER proteins such as Chs7 and Ste14. The ER chaperone Chs7 binds to Chs3 and controls its export from the ER, and Ste14 is required for the last step of CaaX processing, i.e., carboxyl methylation (Romano et al., 1998; Trilla et al., 1999). Prenylation of Chs4 by the Ram1/Ram2 complex may be required also for ER tethering. Prenylation is difficult to analyze in yeast as deletion mutants of ram2 are inviable. The role of Chs4 prenylation has been analyzed previously with inconsistent results. Although Grabinska et al. (2007) showed that yeast cells expressing a nonprenylated mutant of Chs4 exhibit 30% less chitin in their cell walls than wild-type cells, Reyes et al. (2007) claimed that the expression of nonprenylated Chs4 has only minor effects on the chitin content. Reyes et al. (2007) also suggested that prenylation of Chs4 mediates membrane association. In contrast, Grabinska et al. (2007) stated that prenylation of Chs4 does not mediate plasma membrane association of this protein. These inconsistent results could be due to different genetics backgrounds and technical differences (Reyes et al., 2007). Our findings are in accordance with Grabinska et al., (2007), demonstrating that the chitin content in cells expressing nonprenylated Chs4C693S is decreased by ∼30% and that CFW resistance is significantly increased (Figure 4B). In contrast, we observed that prenylation of Chs4 seems to be necessary for membrane association, because nonprenylated Chs4 was clearly found in the cytosol as also was reported by Reyes et al. (2007) (Figure 5). As Chs4 is not delocalized in ste24Δ cells or in the ste24Δ rce1Δ double mutants (Figure 5), cleavage of prenylated Chs4 or subsequent steps of CaaX processing seem not to be involved in membrane association. Nevertheless, deletion of ste24 affects proper localization of Chs3-GFP at the bud neck and decreases chitin levels. Therefore, next to prenylation subsequent steps of CaaX processing of Chs4 may contribute to the correct localization and activation of Chs3. However, deletion of ste24 moderately affects chitin synthesis and does not result in a complete loss of Chs3-GFP at the bud neck. Therefore, yeast cells may have mechanisms that partially compensate for the deficiency in Ste24 mediated CaaX cleavage, which may furthermore process additional substrates involved in chitin synthesis.
Supplementary Material
ACKNOWLEDGMENTS
The technical help of Margret Düvel is gratefully acknowledged. We also are grateful to Jürgen Heinisch and Gunnar Broehan for critically reading the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft (SFB 431).
Abbreviations used:
- CFW
calcofluor white
- Chs
chitin synthesis deficient
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- MFa
mating factor a
- MAT
mating type
- ste
sterile
- Zmpste24
zinc metalloprotease Ste24 homologue.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-01-0080) on May 26, 2010.
REFERENCES
- Barny M. A., Schoonejans E., Economou A., Johnston A. W., Downie J. A. The C-terminal domain of the Rhizobium leguminosarum chitin synthase NodC is important for function and determines the orientation of the N-terminal region in the inner membrane. Mol. Microbiol. 1996;19:443–453. doi: 10.1046/j.1365-2958.1996.382911.x. [DOI] [PubMed] [Google Scholar]
- Barrowman J., Hamblet C., George C. M., Michaelis S. Analysis of prelamin A biogenesis reveals the nucleus to be a CaaX processing compartment. Mol. Biol. Cell. 2008;19:5398–5408. doi: 10.1091/mbc.E08-07-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergo M. O., Ambroziak P., Gregory C., George A., Otto J. C., Kim E., Nagase H., Casey P. J., Balmain A., Young S. G. Absence of the CAAX endoprotease Rce 1, effects on cell growth and transformation. Mol. Cell. Biol. 2002;22:171–181. doi: 10.1128/MCB.22.1.171-181.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briza P., Ellinger A., Winkler G., Breitenbach M. Chemical composition of the yeast ascospore wall. The second outer layer consists of chitosan. J. Biol. Chem. 1988;263:11569–11574. [PubMed] [Google Scholar]
- Bulik D. A., Olczak M., Lucero H. A., Osmond B. C., Robbins P. W., Specht C. A. Chitin synthesis in Saccharomyces cerevisiae in response to supplementation of growth medium with glucosamine and cell wall stress. Eukaryot. Cell. 2003;2:886–900. doi: 10.1128/EC.2.5.886-900.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabib E., Mol P. C., Shaw J. A., Choi W. J. Biosynthesis of cell wall and septum during yeast growth. Arch. Med. Res. 1993;24:301–303. [PubMed] [Google Scholar]
- Cabib E., Shaw J. A., Mol P. C., Bowers B., Choi W. J. Chitin biosynthesis and morphogenetic processes. In: Brambl R., Marzluf G. A., editors. The Mycota, Vol. III: Biochemistry and Molecular Biology. Berlin, Germany: Springer-Verlag; 1996. pp. 243–267. [Google Scholar]
- Chen P., Sapperstein S. K., Choi J. D., Michaelis S. Biogenesis of the Saccharomyces cerevisiae mating pheromone a-factor. J. Cell Biol. 1997;136:251–269. doi: 10.1083/jcb.136.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christodoulidou A., Briza P., Ellinger A., Bouriotis V. Yeast ascospore wall assembly requires two chitin deacetylase isozymes. FEBS Lett. 1999;460:275–279. doi: 10.1016/s0014-5793(99)01334-4. [DOI] [PubMed] [Google Scholar]
- Cos T., Ford R. A., Trilla J. A., Duran A., Cabib E., Roncero C. Molecular analysis of Chs3p participation in chitin synthase III activity. Eur. J. Biochem. 1998;256:419–426. doi: 10.1046/j.1432-1327.1998.2560419.x. [DOI] [PubMed] [Google Scholar]
- DeMarini D. J., Adams A. E., Fares H., De Virgilio C., Valle G., Chuang J. S., Pringle J. R. A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiae cell wall. J. Cell Biol. 1997;139:75–93. doi: 10.1083/jcb.139.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entian K. D., et al. Functional analysis of 150 deletion mutants in Saccharomyces cerevisiae by a systematic approach. Mol. Gen. Genet. 1999;262:683–702. doi: 10.1007/pl00013817. [DOI] [PubMed] [Google Scholar]
- Fong L. G., et al. Heterozygosity for Lmna deficiency eliminates the progeria-like phenotypes in Zmpste24-deficient mice. Proc. Natl. Acad. Sci. USA. 2004;101:18111–18116. doi: 10.1073/pnas.0408558102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura-Kamada K., Nouvet F. J., Michaelis S. A novel membrane-associated metalloprotease, Ste24p, is required for the first step of NH2-terminal processing of the yeast a-factor precursor. J. Cell Biol. 1997;136:271–285. doi: 10.1083/jcb.136.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabinska K. A., Magnelli P., Robbins P. W. Prenylation of S. cerevisiae Chs4p affects chitin synthase III activity and chitin chain length. Eukaryot. Cell. 2007;6:328–336. doi: 10.1128/EC.00203-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Huxley C., Green E. D., Dunham I. Rapid assessment of S. cerevisiae mating type by PCR. Trends Genet. 1990;6:236. doi: 10.1016/0168-9525(90)90190-h. [DOI] [PubMed] [Google Scholar]
- Huyer G., Kistler A., Nouvet F. J., George C. M., Boyle M. L., Michaelis S. Saccharomyces cerevisiae a-factor mutants reveal residues critical for processing, activity, and export. Eukaryot. Cell. 2006;5:1560–1570. doi: 10.1128/EC.00161-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E. A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozubowski L., Panek H., Rosenthal A., Bloecher A., DeMarini D. J., Tatchell K. A Bni4-Glc7 phosphatase complex that recruits chitin synthase to the site of bud emergence. Mol. Biol. Cell. 2003;14:26–39. doi: 10.1091/mbc.E02-06-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyhse-Andersen J. Electroblotting of multiple gels: a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J. Biochem. Biophys. Methods. 1984;10:203–209. doi: 10.1016/0165-022x(84)90040-x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lam K. K., Davey M., Sun B., Roth A. F., Davis N. G., Conibear E. Palmitoylation by the DHHC protein Pfa4 regulates the ER exit of Chs3. J. Cell Biol. 2006;174:19–25. doi: 10.1083/jcb.200602049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson J. R., Bharucha J. P., Ceaser S., Salamon J., Richardson C. J., Rivera S. M., Tatchell K. Protein phosphatase type 1 directs chitin synthesis at the bud neck in Saccharomyces cerevisiae. Mol. Biol. Cell. 2008;19:3040–3051. doi: 10.1091/mbc.E08-02-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage G., Bussey H. Cell wall assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2006;70:317–343. doi: 10.1128/MMBR.00038-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage G., Shapiro J., Specht C. A., Sdicu A. M., Menard P., Hussein S., Tong A. H., Boone C., Bussey H. An interactional network of genes involved in chitin synthesis in Saccharomyces cerevisiae. BMC Genet. 2005;6:8. doi: 10.1186/1471-2156-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung G. K., Schmidt W. K., Bergo M. O., Gavino B., Wong D. H., Tam A., Ashby M. N., Michaelis S., Young S. G. Biochemical studies of Zmpste24-deficient mice. J. Biol. Chem. 2001;276:29051–29058. doi: 10.1074/jbc.M102908200. [DOI] [PubMed] [Google Scholar]
- McElver J., Weber S. C. Flag N-terminal epitope overexpression of bacterial alkaline phosphatase and Flag C-terminal epitope tagging by PCR one-step targeted integration. Yeast. 1992;8:S627. [Google Scholar]
- Merzendorfer H. Insect chitin synthases: a review. J. Comp. Physiol. B. 2006;176:1–15. doi: 10.1007/s00360-005-0005-3. [DOI] [PubMed] [Google Scholar]
- Miller J. P., Lo R. S., Ben-Hur A., Desmarais C., Stagljar I., Noble W. S., Fields S. Large-scale identification of yeast integral membrane protein interactions. Proc. Natl. Acad. Sci. USA. 2005;102:12123–12128. doi: 10.1073/pnas.0505482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono N., Yabe T., Sudoh M., Nakajima T., Yamada-Okabe T., Arisawa M., Yamada-Okabe H. The yeast Chs4 protein stimulates the trypsin-sensitive activity of chitin synthase 3 through an apparent protein-protein interaction. Microbiology. 2000;146:385–391. doi: 10.1099/00221287-146-2-385. [DOI] [PubMed] [Google Scholar]
- Pendas A. M., et al. Defective prelamin A processing and muscular and adipocyte alterations in Zmpste24 metalloproteinase-deficient mice. Nat. Genet. 2002;31:94–99. doi: 10.1038/ng871. [DOI] [PubMed] [Google Scholar]
- Raben N., Exelbert R., Spiegel R., Sherman J. B., Nakajima H., Plotz P., Heinisch J. Functional expression of human mutant phosphofructokinase in yeast: genetic defects in French Canadian and Swiss patients with phosphofructokinase deficiency. Am. J. Hum. Genet. 1995;56:131–141. [PMC free article] [PubMed] [Google Scholar]
- Ren J., Wen L., Gao X., Jin C., Xue Y., Yao X. CSS-Palm 2.0, an updated software for palmitoylation sites prediction. Protein Eng. Des. Sel. 2008;21:639–644. doi: 10.1093/protein/gzn039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A., Sanz M., Duran A., Roncero C. Chitin synthase III requires Chs4p-dependent translocation of Chs3p into the plasma membrane. J. Cell Sci. 2007;120:1998–2009. doi: 10.1242/jcs.005124. [DOI] [PubMed] [Google Scholar]
- Romano J. D., Schmidt W. K., Michaelis S. The Saccharomyces cerevisiae prenylcysteine carboxyl methyltransferase Ste14p is in the endoplasmic reticulum membrane. Mol. Biol. Cell. 1998;9:2231–2247. doi: 10.1091/mbc.9.8.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncero C., Duran A. Effect of Calcofluor white and Congo red on fungal cell wall morphogenesis: in vivo activation of chitin polymerization. J. Bacteriol. 1985;163:1180–1185. doi: 10.1128/jb.163.3.1180-1185.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Herrera J., Lopez-Romero E., Bartnicki-Garcia S. Properties of chitin synthetase in isolated chitosomes from yeast cells of Mucor rouxii. J. Biol. Chem. 1977;252:3338–3343. [PubMed] [Google Scholar]
- Santos B., Snyder M. Targeting of chitin synthase 3 to polarized growth sites in yeast requires Chs5p and Myo2p. J. Cell Biol. 1997;136:95–110. doi: 10.1083/jcb.136.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J. A., Mol P. C., Bowers B., Silverman S. J., Valdivieso M. H., Duran A., Cabib E. The function of chitin synthases 2 and 3 in the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 1991;114:111–123. doi: 10.1083/jcb.114.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam A., Nouvet F. J., Fujimura-Kamada K., Slunt H., Sisodia S. S., Michaelis S. Dual roles for Ste24p in yeast a-factor maturation: NH2-terminal proteolysis and COOH-terminal CAAX processing. J. Cell Biol. 1998;142:635–649. doi: 10.1083/jcb.142.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam A., Schmidt W. K., Michaelis S. The multispanning membrane protein Ste24p catalyzes CAAX proteolysis and NH2-terminal processing of the yeast a-factor precursor. J. Biol. Chem. 2001;276:46798–46806. doi: 10.1074/jbc.M106150200. [DOI] [PubMed] [Google Scholar]
- Trilla J. A., Cos T., Duran A., Roncero C. Characterization of CHS4 (CAL2), a gene of Saccharomyces cerevisiae involved in chitin biosynthesis and allelic to SKT5 and CSD4. Yeast. 1997;13:795–807. doi: 10.1002/(SICI)1097-0061(199707)13:9<795::AID-YEA139>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Trilla J. A., Duran A., Roncero C. Chs7p, a new protein involved in the control of protein export from the endoplasmic reticulum that is specifically engaged in the regulation of chitin synthesis in Saccharomyces cerevisiae. J. Cell Biol. 1999;145:1153–1163. doi: 10.1083/jcb.145.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueblood C. E., Boyartchuk V. L., Picologlou E. A., Rozema D., Poulter C. D., Rine J. The CaaX proteases, Afc1p and Rce1p, have overlapping but distinct substrate specificities. Mol. Cell Biol. 2000;20:4381–4392. doi: 10.1128/mcb.20.12.4381-4392.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. W., Hamamoto S., Orci L., Schekman R. Exomer: a coat complex for transport of select membrane proteins from the trans-Golgi network to the plasma membrane in yeast. J. Cell Biol. 2006;174:973–983. doi: 10.1083/jcb.200605106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek H., Cioffi M., Klein U., Harvey W. R., Schweikl H., Wolfersberger M. G. Isolation of goblet cell apical membrane from tobacco hornworm midgut and purification of its Vacuolar-type ATPase. Methods Enzymol. 1990:608–616. doi: 10.1016/0076-6879(90)92098-x. [DOI] [PubMed] [Google Scholar]
- Winzeler E. A., et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Wright L. P., Philips M. R. Thematic review series: lipid posttranslational modifications. CAAX modification and membrane targeting of Ras. J. Lipid Res. 2006;47:883–891. doi: 10.1194/jlr.R600004-JLR200. [DOI] [PubMed] [Google Scholar]
- Ziman M., Chuang J. S., Schekman R. W. Chs1p and Chs3p, two proteins involved in chitin synthesis, populate a compartment of the Saccharomyces cerevisiae endocytic pathway. Mol. Biol. Cell. 1996;7:1909–1919. doi: 10.1091/mbc.7.12.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimoch L., Merzendorfer H. Immunolocalization of chitin synthase in the tobacco hornworm. Cell Tissue Res. 2002;308:287–297. doi: 10.1007/s00441-002-0546-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.