Abstract
Mannosylphospho dolichol synthase (DPMS) plays a critical role in Glc3Man9GlcNAc2-PP-Dol (lipid-linked oligosaccharide, LLO) biosynthesis, an essential intermediate in asparagine-linked (N-linked) protein glycosylation. We have observed earlier that phosphorylation of DPMS increases the catalytic activity of the enzyme by increasing the Vmax as well as the enzyme turnover (kcat) without significantly changing the Km for GDP-mannose. As a result, LLO biosynthesis, turnover and protein N-glycosylation are increased. This is manifested in increased proliferation of capillary endothelial cells, i.e., angiogenesis. We have then asked if the phosphorylation event or the up-regulation of the DPMS due to over production of the enzyme is a key factor in up-regulating angiogenesis? This question has been answered by isolating a stable capillary endothelial cell clone overexpressing the DPMS gene. Our results indicate that the DPMS overexpressing clone has a high level DPMS mRNA judged by QRT-PCR. The clone also expresses nearly four-times higher DPMS protein over the clone transfected with pEGFP-N1 vector only (i.e., control) as analyzed by western blotting. Most importantly, the overexpressing DPMS clone has ~108% higher DPMS activity than that of the vector control. Immunofluorescence microscopy with Texas-Red conjugated WGA indicates a high level expression of GlcNAc-β-(1,4)-GlcNAc)1-4-β-GlcNAc-NeuAc glycans on the external surface of the capillary endothelial cells overexpressing DPMS. Increased cellular proliferation and accelerated healing of the wound induced by a mechanical stress of the DPMS overexpressing clone unequivocally supports DPMS for angiogenesis.
Keywords: Mannosylphospho dolichol synthase, angiogenesis, capillary endothelial cells, breast cancer, wound healing, gene expression
Introduction
Mannosylphospho dolichol synthase (DPMS; EC 2.4.1.83), a 31kDa phosphoprotein, catalyzes the transfer reaction GDP-mannose + Dol-P < == > Dol-P-Man + GDP in the rough endoplasmic reticulum (ER; 1-7). The synthesis occurs at the cytoplasmic face of the ER membrane (8–10). Dol-P-Man then flips into the lumen of the endoplasmic reticulum to serve as a ‘key’ mannosyl donor in the assembly of the precursor oligosaccharide-lipid Glc3Man9GlcNAc2-PP-Dol (LLO) in N-glycosylation of proteins. Precisely, Dol-P-Man participates in the conversion of Man5GlcNAc2-PP-Dol to Man9GlcNAc2-PP-Dol. In addition, Dol-P-Man serves as a mannosyl donor (i) in the synthesis of glycosylphosphatidylinositol (GPI) anchors, (ii) in O-glycosylation of proteins in yeast, and (iii) in C-mannosylation of Trp-7 in human ribonuclease 2 (RNase 2).
DPMS is a structural gene in Saccharomyces cerevisiae and it is essential for their viability because disruption of the Dpm1 gene is lethal and does not allow the organism to grow at non-permissive temperature (11). Similarly, a partial deficiency of DPMS has been found to be associated with some variants of congenital disorder of glycosylation (CDG) (12,13). These patients exhibit developmental delay, seizures, hypotonia, and dysmorphic function (14). Mutation of DPMS has also been reported in human lymphoma Thy-1- class E cells and are defective in Dol-P-Man biosynthesis (15). DPMS activity has also been found associated with the development in mammalian system. For example, the Km for GDP-mannose in rat parotid acinar cells in young (3-6 months) and aged (22-24 months) did not change but the Vmax was reduced nearly 50% in aged animals (16). On the other hand, the Km for GDP-mannose was reduced by ~33% without changing the Vmax when the microenvironment of the capillary endothelial cells growth potential was changed (17).
Our interest has been in the regulation of N-linked protein glycosylation and its relationship to angiogenesis. Angiogenesis is the formation of new blood vessels, an essential physiological event during growth and development, and also for wound healing. It is equally important for tumor progression and metastasis. We had used earlier a β-receptor agonist, isoproterenol to enhance the intracellular cAMP level to dissect the molecular details of the angiogenic process. cAMP turned out to be an activator of angiogenesis and a positive regulator of N-linked glycosylation, especially that of the DPMS (18) β-Adrenoreceptor is a trimeric G-protein coupled receptor and when activated the rate of LLO biosynthesis and its turnover are increased. As a result, the N-glycosylation of capillary endothelial cells Factor VIIIc (eFVIIIc) is enhanced (19). cDNA cloning of capillary endothelial cell DPMS and its deduced amino acid sequence indicated the presence of a consensus sequence for phosphorylation by cAMP-dependent protein kinase (20; GenBank #GQ367549).
We report here isolation of a stable capillary endothelial cell line overexpressing DPMS by a genetic manipulation. Increased DPMS activity is associated with increased expression of DPMS protein and mRNA. The level of cell surface glycoproteins is also increased. Analysis of cellular proliferation and migration indicated increased angiogenesis.
Materials and Methods
Minimal essential medium with Earle's salt (EMEM), glutamine, trypsin-versine, anti-mycotic antibody mixture (penicillin-streptomycin-fungizone) and phosphate-buffer saline (PBS), pH 7.2 and 7.4 were from BioSource (Camarillo, CA). Fetal bovine serum (defined) was from HyClone, Logan, UT. Dimethylsulfoxide, nystatin, and ethidium bromide were obtained from Sigma Aldrich, St. Louis, MO. Texas-Red conjugated WGA was purchased from EY Laboratory, San Mateo, CA. iScript™ cDNA synthesis kit, Tween-20, biotinylated protein molecular weight markers, DNA markers and all electrophoresis reagents were obtained from BioRad Laboratories, Hercules, CA. Lipofectamine™ Reagent, Plus™ Reagent, G418 and TRIzol were from Invitrogen, Life Technologies, Carlsbad, CA. XhoI, BamHI, HindIII, T4 DNA ligase were from New England BioLab, Ipswich, MA. dNTPs and Taq polymerase were obtained from Promega, Madison, WI. Rabbit polyclonal anti-DPMS antibody was developed in-house. Horseradish peroxidase conjugated (HRP) goat anti-rabbit IgG, ECL chemiluminescence detection kit and GDP-[U-14C]-mannose was from GE Healthcare, Piscataway, NJ. Mouse monoclonal antibody to actin was obtained from BD Biosciences, San Jose, CA. PCR grade water and DNA decontamination kit were from Applied Biosystems, Austin,TX. All other chemicals and solvents were of the highest purity grade and obtained from commercial suppliers.
Culturing of endothelial cells and stable transfectants
Wild type cells were maintained in EMEM containing 10% fetal bovine serum (heat-inactivated), glutamine (2mM), penicillin (50 units/ml), streptomycin (50μg/ml), nystatin (1,000 units/ml) (21). Overexpressing DPMS stable cell clone and the cell clone carrying the vector alone were cultured in the same media but containing G418 (0.55μg/ml) at 37°C in an humidified incubator (5% CO2-95% air).
Construction of DPMS overexpression plasmid
Cellular RNA was isolated with TRIzol reagent and treated with DNase to remove any possible genomic DNA contamination. Total RNA was quantified in a NanoDrop Bioanalyzer. First strand cDNA was synthesized using the iScript™ cDNA synthesis kit according to the manufacturer's instructions. The primers for amplifying full DPMS gene were: 5′-ccgctcgagATGGCTGCCGAGGAAGCAAGTC-3′ (forward) and 5′-ccgggatccCTCCAGAAAACAGACTTGCCTTACCT-3′ (reverse), which provided with the XhoI and BamHI site. PCR reactions were carried out as follows: initial denaturation at 94°C for 4 min, 30 cycles at 94°C for 50s, 64°C for 50s, 72°C for 50s, and a final extension for 10 min at 72°C in 50μl containing 2μl each cDNA, 0.2μM each primer, 0.2mM dNTP, and 2 units of Taq DNA polymerase. 5 μl each reaction mixture was used for product identification and detected by 1% agarose gel electrophoresis followed by ethidium bromide staining. PCR products were digested with XhoI and BamHI, purified, and cloned into XhoI and BamHI sites of pEGFP-N1 vector to obtain pEGFP-N1-DPMS overexpression plasmid. The plasmids were confirmed by XhoI/BamHI double digestion and DNA sequencing.
Sequencing of DPMS overexpression plasmids
For sequencing, plasmids were isolated and 20μl sequencing reaction was designed according to the BigDye terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) containing 3.2 pmol primer [(DPMS overexpression plasmid sequencing primers: forward Primer: 5′-TTTCCAAAATGTCGTAACAACTCCG-3′; reverse Primer: 5′-TGAACTTGTGGCCGTTTACGT-3′)] and 300 ng plasmid. The reaction was carried out as follows: initial denaturation at 96°C for 1 min, 25 cycles at 96°C for 10s, 50°C for 50s, 60°C for 4 min, and rapid cooling to 4°C. This was followed using BigDye XTerminator™ Purification Kit (Applied Biosystems), and then electrophoresis on the ABI PRISM 310 Genetic Analyzer.
Transfection of capillary endothelial cells with constructed plasmids
Transfection of capillary endothelial cells with pEGFP-N1-DPMS overexpression plasmid, and pEGFP-N1 vector were performed using Lipofectamine™ Reagent and Plus™ Reagent according to the manufacturer's instruction. After transfection, the cells were cultured in regular EMEM containing 0.55 mg/ml of G418 for 4 weeks, and cells overexpressing DPMS and the vector pEGFP-N1 alone (carrying a non-transcribable gene; control) were collected.
Mannosylphospho dolichol synthase assay
Enzymatic transfer of mannose from GDP-mannose was assayed using a procedure described earlier (22) with the following modification. Total cell lysates from overexpressing DPMS clone and the clone carrying vector only were incubated at 37°C for 5 min in 50 mM Tris-HCl (pH 7.0), 0.125M sucrose, 0.5mM EDTA, 2.5mM 5′-AMP, 10mM MnCl2, 10μg Dol-P and 2.5μM GDP–[U-14C]mannose in a total volume of 150μl. The reaction was stopped with 3ml of chloroform/methanol (2:1, v/v). Chloroform/methanol extracts containing Dol-P-Man were washed with 0.5 volume of 0.9% sodium chloride and twice with chloroform/methanol/water (3:47:48, v/v/v) to remove free GDP-[U-14C]mannose. A lower organic phase containing the Dol-P-Man was quantified in a liquid scintillation spectrometer.
Quantitative RT-PCR of DPMS
Total cellular RNA was extracted and the cDNA was synthesized using the iScript™ cDNA synthesis kit as mentioned above. Primers used for QRT-PCR were: DPMS gene forward primer 5′-GCTGAGCAGTTGGAGAAG-3′, reverse primer 5′-TGGATGGTGTGAGAGGTC-3′ (PCR product size: 153 bp); GAPDH gene forward primer 5′-TGACCCCTTCATTGACCTTC -3′, reverse primer 5′- GATCTCGCTCCTGGAAGATG -3′ (PCR product size: 143 bp) (XXIDT integrated DNA Technologies). QRT-PCR was performed in an iCycler (BioRad Laboratories) using iQ™ SYBR® Green Supermix (BioRad Laboratories) as a fluorescence dye for the double-stranded DNA. After optimizing the PCR conditions, reactions with SYBR® Green PCR master mix, 10μM forward/reverse primer and 100ng cDNA were performed following the instruction of manufacturer.
Quantitative PCR was accomplished under the following conditions: 95°C for 3 min, and 40 cycles of 95°C for 10s and 50°C for 60s. The relative gene expression for each sample was determined using the formula 2ΔΔ = 2ΔCt (GAPDH – target), which reflected the target gene expression normalized to GAPDH levels. Each sample was run three times. After amplification, 5 μl each reaction mixture was subjected to 1.5% agarose gel electrophoresis, and the bands were visualized by ethidium bromide staining. GAPDH expression was detected as the internal control.
Western blotting
To prepare whole cell extracts, stable cells were washed in PBS and resuspended in cell lysis buffer (10% SDS, 1M Tris.HCl, pH7.5, 100mM Na3VO4). After sonication and heating for 5 min, cell lysates were clarified by centrifugation at 9000 ×g for 10 min, and the supernatants were collected. Protein concentration was determined with BioRad DC Protein Assay using bovine serum albumin as a standard. Total protein (50μg) from the whole cell lysate was separated by 10% SDS-PAGE in a mini-gel. Proteins separated in the gel were transferred electrophoretically onto nitrocellulose membranes. After blocking with TTBS [50 mM Tris-HCl (pH 7.5), 0.15M NaCl, 0.1% Tween-20] containing 5% fat-free dry milk for 1 h, the membrane was incubated overnight with a specific polyclonal antibody directed against DPMS (6:1000 dilution) at 4°C. After washing with TTBS three times, 5 min/each, the membrane was incubated with horseradish peroxidase (HRP)-conjugated anti-mouse or anti-rabbit antibody (1:1000) for 1 h at 4°C. The membrane was developed with an enhanced chemiluminescence (ECL) detection system. The same blot was stripped and reprobed with anti-actin antibody (BD Biosciences) to confirm the equal loading (23).
Immunofluorescence microscopy
The stable clones were subcultured on cover glasses (15mm diameter, Warner Instruments, Hamden, CT) for 24 h under standard conditions. The cells were washed three times with PBS, pH 7.4 and fixed for 5 min in ice-cold methanol. The cells were washed three times with PBS, pH 7.4 and incubated with anti-DPMS antibody (rabbit polyclonal 1:50 dilution) for 1 h at room temperature. After washing three times with PBS, pH 7.4, the cells were incubated with rhodamine-conjugated goat anti-rabbit IgG (1:300 dilution) and Hoechst 33342 (1:10,000 dilution) for 30 min at room temperature. After washing three times with PBS, pH 7.4, the cover glasses were mounted and examined in a Zeiss AxiocamMRc (Carl Zeiss, Germany) fluorescent microscope at 40X magnification (24).
For glycoprotein detection, cells were seeded on cover glasses as mentioned above, and washed three times with buffer (50mM Tris-HCl, pH, 0.15M NaCl, 4mM CaCl2). Texas-Red conjugated WGA (100ug/ml in Tris-CaCl2 buffer) was then added and the cells were incubated for 30 min at room temperature. After washing three times, the cover glasses were mounted and the images were captured in a Zeiss AxiocamMRc (Carl Zeiss, Germany) fluorescent microscope at 40X magnification.
Cell proliferation and migration assay
The vector (carrying no transcribable gene) and DPMS overexpressing clones were seeded in 24-well plates (2×104 cells/well) in 1 ml of normal medium containing 10% serum and 0.55 mg/ml G418. After 24 h the medium was removed, cells were washed three-times with PBS (pH 7.4) and incubated in a serum-free medium for 24 h. At the end of incubation, the medium was replaced with normal medium containing 10% serum and 0.55 mg/ml G418. The cell numbers were counted after every 24 h for seven days. The experiment was conducted in triplicate wells and averaged after counting the cells in four quadrants for each well (25).
For migration assay cells (1×106) were plated into 6-well cell culture plate in normal media containing 0.55 μg/ml G418. After 24h the media was removed, cells were washed with PBS, pH7.4 and changed to a serum-free medium. The cells were continued to incubate for another 24h when the media were removed and replaced with the normal media. After 48h, a 1-mm wide scratch (reminiscent of a wound) was made across the cell monolayer using a pipette tip. The monolayer was washed with PBS, pH 7.4; normal media were added and photographed. After incubating at 37°C for 6 h the plates were again photographed. Quantitative analyses of the scratches were performed by counting the cells migrated towards the scratch. Results are presented as mean ± S.D. averaged from three low power fields (magnification 10X).
Statistical analysis
The results were analyzed statistically using the student t-test.
Results
Cloning and Identification of DPMS Gene in a Capillary Endothelial Cell Clone Overexpressing DPMS
DPMS gene was cloned and confirmed by RT-PCR followed by agarose gel electrophoresis (Figure 1). pEGFP-N1 vector used to isolate the overexpressing DPMS clone has a green fluorescence protein (GFP) construct in-frame but downstream of the DPMS gene. In stable clone DPMS was not expressed as a fusion protein but the expression of GFP was indicative of DPMS expression (Figure 2).
Figure 1. Cloning and identification of over-expressing DPMS clones.
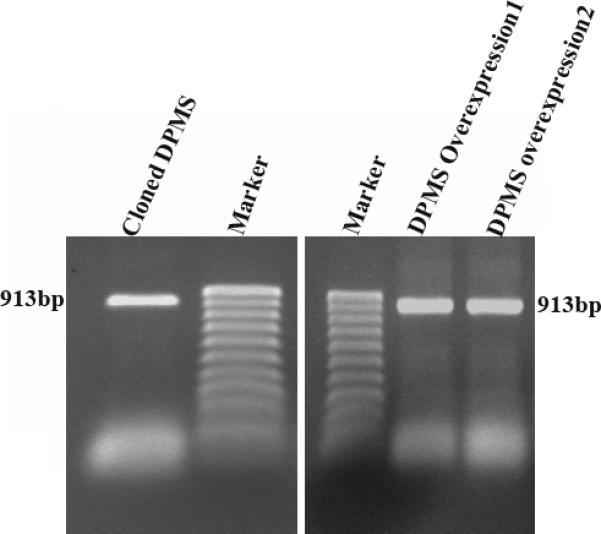
The DPMS gene was cloned and confirmed by RT-PCR followed by agarose gel electrophoresis and ethidium bromide staining. Left panel, 1.5% agarose gel profile of the RT-PCR product; Right panel, 1.5% agarose gel profile of the overexpression plasmid from DPMS overexpressing clone-1 and clone-2.
Figure 2. Verification of DPMS gene transfection by GFP expression in capillary endothelial cells.

To monitor the DPMS expression cells were cultured overnight on cover glasses, washed with PBS, pH 7.4, mounted and examined in a Zeiss AxiocamMRc (Carl Zeiss, Germany) fluorescent microscope at 10X magnification for GFP expression. Upper panel: Vector; and lower panel: overexpressing clone.
DPMS Expression in Stable Clones. (a) By immunofluorescence microscopy
Capillary endothelial cells harboring the DPMS overexpressing plasmid are expected to express DPMS at a level higher than the vector. The expression of DPMS was monitored by indirect immunofluorescence microscopy. The results indicated that the overexpressing clone indeed expressed DPMS at a higher level (Figure 3). (b) By western blotting and QRT-PCR: When compared with that of the vector (pEGFP-N1) by western blotting the level of DPMS per unit protein in an overexpressing clone was higher than that of the corresponding vector (Figure 4A). Analysis of DPMS mRNA expression by QRT-PCR indicated that the overexpressing clone expressed 4-fold higher level of mRNA (Figure 4B).
Figure 3. Immunofluorescence microscopy of capillary endothelial cells over-expressing DPMS.
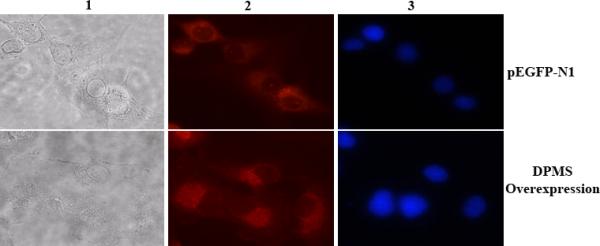
Column 1 = phase contrast microscopy; column 2 = DPMS staining; column 3 = Stained nucleus with Hoeschst dye. Magnification: 40X.
Figure 4. Monitoring DPMS expression in capillary endothelial cell clone by western blotting and QRT-PCR.
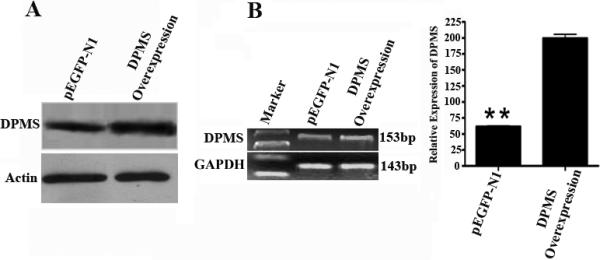
Western blotting: Blots were developed with enhanced chemiluminescence (ECL) detection system. Actin was used as an internal control. QRT-PCR: QRT-PCR products were identified by 1% agarose gel electrophoresis. The bands were visualized by ethidium bromide staining. GAPDH expression was used as an internal control. **: p <0.01.
Expression of Cell Surface Glycoproteins
The expression of cell surface glycoproteins was analyzed by immunofluoresence microscopy using Texas-Red conjugated WGA. WGA is specific for (GlcNAc-β-(1,4)-GlcNAc)1-4-β-GlcNAc-NeuAc glycan. Our results indicated that the DPMS overexpressing cells expressed a high level of complex glycans on the cell surface as compared to the vector (Figure 5).
Figure 5. Analysis of cell surface glycoproteins.
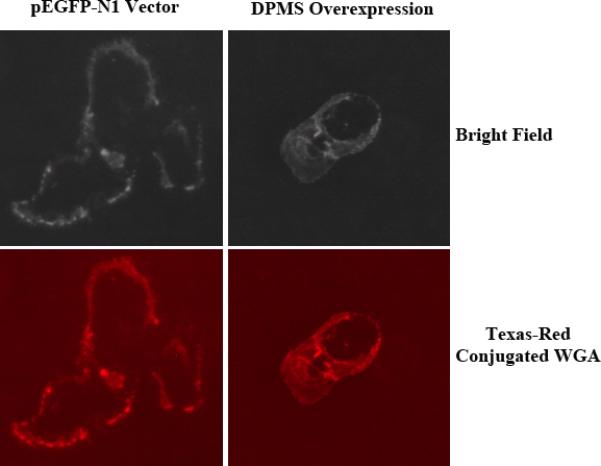
DPMS over-expressing and vector transfected cells were plated on cover glasses (15mm diameter). After 24 h, cells were washed and incubated with Texas-Red conjugated WGA for 30 min and monitored under a fluorescence microscope.
Mannosylphospho Dolichol Synthase Activity
Increased glycan expression as well as that of the protein and mRNA expression raised the question of the catalytic activity of the DPMS. DPMS activity was assayed in vitro in the presence of a fixed concentration of dolichyl monophosphate. The results (Table 1) indicated that the catalytic activity of DPMS was nearly 108% higher in the over-expressing capillary endothelial cells compared to the vector control.
Table 1. Dol-P-Man synthase activity in capillary endothelial cell clone overexpressing DPMS.
The enzyme activity was assayed as described in Materials and Methods using GDP-[U-14C]mannose (Specific Activity 199cpm/pmol).
| Sample | Dol-P-Man (pmol/mg protein/5min) Mean ± S.D | p-value* |
|---|---|---|
| pEGFP-N1 vector | 44.06 ±1.2 | = 0.04 |
| DPMS Overexpression | 47.73 ±2.1 |
Analyzed by student t-test
Cellular Proliferation and Migration
The ability of endothelial cells to proliferate and differentiate into capillary-like structure is the “hall mark” of angiogenesis. It is an essential element in tumor growth and wound healing. After establishing that overexpression of catalytically active DPMS is intricately connected with increased glycan expression, we set out to investigate the angiogenic activity of these cells. Our results clearly demonstrated that the DPMS overexpressing clone has a faster growth rate compared to the vector control (Figure 6A). It was then tested for migration status to the wounded monolayer. Morphological studies showed that DPMS overexpressing cells moved faster towards the wounded surface (Figure 6B). To eliminate the possibility that closing of the wound was due to increased cell proliferation rather than cell migration, the cell numbers were counted in fixed areas of the wounded surface. The results indicated that the wound made on the DPMS over-expressing endothelial cell monolayer carries more cells than the control (i.e., vector with no transcribable gene) (Figure 6C). This was supported by the fact that (i) the doubling time of capillary endothelial cells is between 56h - 68h depending on the stimulus being present (25); and (ii) DPMS overexpressing endothelial cells exhibited a faster migratory rate (~ 2-fold) in a Matrigel™ invasion assay compared to the control, i.e., cells transfected with a vector carrying a non-transcribable gene (data not shown)
Figure 6. Proliferation of capillary endothelial cells overexpressing DPMS.
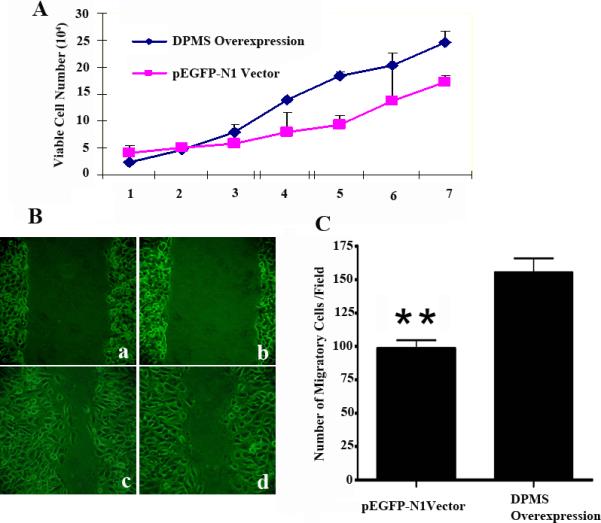
(A) Hemocytometer counts of synchronized cells over seven days. ■, overexpressing DPMS cell clone; ■, pEGFP-N1 Vector. (B) Quantitative scratch assays performed by counting migrated cells under low magnification. Results are presented as mean ± S.D. A 1-mm wide scratch was made across the cell layer using a pipette tip (B-a and B-b). Normal media was added after washing with PBS, pH 7.4. The plates were then photographed after incubating for 6 h. (B-c), DPMS overexpression clones; (B-d) pEGFP-N1 vector. **: p <0.01.
Discussion
Mannosylphospho dolichol synthase (DPMS), a glycosyltransferase of the protein N-glycosylation pathway is known to undergo regulation by PKA-mediated phosphorylation (23, 26). This post-translational modification alters the DPMS activity and impacts the cellular microenvironment. Phosphorylation of DPMS enhances the catalytic activity of the enzyme without appreciably altering the Km for GDP-mannose (23, 26). In addition, the phosphorylated DPMS exhibits a higher enzyme turnover (kcat) and enzyme efficiency (kcat/Km). Replacing serine-141 at the PKA phosphorylation site of the recombinant DPMS from Saccharomyces cerevisiae by alanine (i.e., S141A) by site-directed mutagenesis reduced the phosphorylation activation of the enzyme by ~50%. (26).
Protein kinase type I-deficient Chinese hamster ovary (CHO) cells have also exhibited reduced DPMS activity with a Km for GDP-mannose 160-400% higher than that of the wild type. The kcat for the DPMS reduced 2-4 fold in mutant cells, and exogenously added Dol-P failed to rescue the Km for GDP-mannose (27). This was reflected in down-regulation of LLO biosynthesis and turnover and impairment of protein glycosylation of many cellular glycoproteins (27). All of these, however, were corrected when the protein kinase deficiency was reversed (28). So far, there has been no study of the significance of DPMS on glycoprotein biosynthesis and regulation impacting angiogenesis after genetically modulating its level of expression. Our study presented here clearly demonstrates successful isolation of a stably transfected capillary endothelial cell clone over-expressing DPMS. There is no apparent change in cellular morphology between the over-expressing clone, one transfected with the vector carrying no transcribable gene and the wild type cells. To improve the selection process we selected a vector in which the GFP gene was in-frame with the DPMS gene. The GFP gene was down-stream of the DPMS gene but the DPMS was not expressed as a fusion protein. Both the vector and the DPMS over-expressing clones expressed GFP protein as indicated in Figure 2. The reason for a reduced green fluorescence in the over-expressing clone is not clear but we anticipate that over-expression of DPMS may cause a down-regulation of GFP expression. High expression of the DPMS protein as indicated by western blotting or its mRNA by QRT-PCR corroborated with the increased DPMS activity and the expression of increased cell surface N-glycans. This in turn correlated with increased cellular proliferation as well as increased migration during wound healing, supporting unequivocally a linear relationship between the DPMS-mediated protein N-glycosylation and angiogenesis. Further studies are needed to develop appropriate glycotherapeutics targeting DPMS for treating breast and other solid tumor growth.
Acknowledgement
The work was partly supported by the funds from the NIH U54-CA096297and the Susan G. Komen for the Cure BCTR0600582 (DKB) and the NIH/NCRR/RCMI grant G12-RR03035 (KB). The technical assistance of Miss Neysha Sanchez and Jésus Santiago is greatly appreciated.
Footnotes
Part of this work was presented at the 8th International Carbohydrate Bioengineering Meeting in Ischia, Naples, Italy, May 10-13, 2009.
References
- 1.Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 2.Tannner W, Lehle L. Protein glycosylation in yeast. Biochim. Biophys. Acta. 1987;906:81–99. doi: 10.1016/0304-4157(87)90006-2. [DOI] [PubMed] [Google Scholar]
- 3.Orlean P. Dolichol phosphate mannose synthase is required in vivo for glycosyl phosphatidylinositol membrane anchoring, O mannosylation, and N glycosylation of protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 1990;10:5796–5806. doi: 10.1128/mcb.10.11.5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Menon AK, Mayor S, Schwarz RT. Biosynthesis of glycosylphosphatidylinositol lipids in Trypanosoma brucei: involvement of mannosylphosphoryldolichol as the mannose donor. EMBO J. 1990;9:4249–4258. doi: 10.1002/j.1460-2075.1990.tb07873.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Englund PT. The structure and biosynthesis of glycosyl phosphatidylinositol protein anchors. Annu. Rev. Biochem. 1993;62:121–138. doi: 10.1146/annurev.bi.62.070193.001005. [DOI] [PubMed] [Google Scholar]
- 6.Herscovics A, Orlean P. Glycoprotein biosynthesis in yeast. FASEB J. 1993;7:540–550. doi: 10.1096/fasebj.7.6.8472892. [DOI] [PubMed] [Google Scholar]
- 7.Doucey MA, Hess D, Cacan R, Hofsteenge J. Protein C-mannosylation is enzyme-catalysed and uses dolichyl-phosphate-mannose as a precursor. Mol. Biol. Cell. 1998;9:291–300. doi: 10.1091/mbc.9.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clarke BL, Naylor C, Lennarz WJ. Comparative studies on mannosylphosphoryl dolichol and glucosylphosphoryl dolichol synthases. Chem. Phys. Lipid. 1989;51:239–247. doi: 10.1016/0009-3084(89)90011-x. [DOI] [PubMed] [Google Scholar]
- 9.Hirschberg CB, Snider MD. Topography of glycosylation in the rough endoplasmic reticulum and Golgi apparatus. Annu. Rev. Biochem. 1987;56:63–87. doi: 10.1146/annurev.bi.56.070187.000431. [DOI] [PubMed] [Google Scholar]
- 10.Abeijon C, Hirschberg CB. Topography of glycosylation reactions in the endoplasmic reticulum. Trends Biol. Sci. 1992;17:32–36. doi: 10.1016/0968-0004(92)90424-8. [DOI] [PubMed] [Google Scholar]
- 11.Orlean P, Albright C, Robbins PW. Cloning and sequencing of the yeast gene for dolichol phosphate mannose synthase, an essential protein. J Biol Chem. 1988;263:17499–17507. [PubMed] [Google Scholar]
- 12.Marquardt T, Denecke J. Congenital disorders of glycosylation: review of their molecular bases, clinical presentations and specific therapies. Eur. J. Pediatr. 2003;162:359–379. doi: 10.1007/s00431-002-1136-0. [DOI] [PubMed] [Google Scholar]
- 13.Kim S, Westphal V, Srikrishna G, Mehta DP, Peterson S, Filiano J, Karnes PS, Patterson MC, Freeze HH. Dolichol phosphate mannose synthase (DPM1) mutations define congenital disorder of glycosylation Ie (CDG-Ie). J. Clin. Invest. 2000;105:191–198. doi: 10.1172/JCI7302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nozaki M, Ohishi K, Yamada N, Kinoshita T, Nagy A, Takeda J. Developmental abnormalities of glycosylphosphatidylinositol-anchor deficient embryos revealed by Cre/loxP system. Lab. Invest. 1999;79:293–299. [PubMed] [Google Scholar]
- 15.Chapman A, Trowbridge IS, Hyman R, Kornfeld S. Structure of the lipid-linked oligosaccharides that accumulate in class E Thy-1-negative mutant lymphomas. Cell. 1979;17:509–515. doi: 10.1016/0092-8674(79)90259-9. [DOI] [PubMed] [Google Scholar]
- 16.Kousvelari EE, Banerjee DK, Murty L, Baum BJ. N-linked protein glycosylation in the rat parotid gland during aging. Mech. Aging and Development. 1988;42:173–181. doi: 10.1016/0047-6374(88)90072-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banerjee DK. Microenvironment of endothelial cell growth and regulation of protein N-glycosylation. Indian J. Biochem. Biophys. 1988;25:8–13. [PubMed] [Google Scholar]
- 18.Baksi K, Tavarez-Pagan JJ, Martinez JA, Banerjee DK. Unique structural motif supports mannosylphospho dolicol synthase: An important angiogenesis regulator. Current Drug Targets. 2008;9:262–271. doi: 10.2174/138945008783954916. [DOI] [PubMed] [Google Scholar]
- 19.Banerjee DK, Oliveira CM, Tavárez JJ, Katiyar VN, Saha S, Martínez JA, Banerjee A, Sánchez A, Baksi K. Importance of a Factor VIIIc-like Glycoprotein Expressed in Capillary Endothelial Cells (eFactor VIIIc) in Angiogenesis. In: Wu A, editor. Molecular Immunology of Complex Carbohydrates III. Springer; New York: 2009. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baksi K, Zhang Z, Banerjee A, Banerjee DK. Cloning and Expression of Mannosylphospho Dolichol Synthase from Bovine Adrenal Medullary Capillary Endothelial Cells. Glycoconj. J. 2009;26:635–645. doi: 10.1007/s10719-008-9214-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banerjee DK, Ornberg RL, Youdim MB, Heldman E, Pollard HB. Endothelial cells from bovine adrenal medulla develop capillary-like growth patterns in culture. Proc Natl Acad Sci (USA) 1985;82:4702–4706. doi: 10.1073/pnas.82.14.4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Banerjee DK. Amphomycin inhibits mannosylphosphoryldolichol synthesis by forming a complex with dolichylmonophosphate. J. Biol. Chem. 1989;264:2024–2028. [PubMed] [Google Scholar]
- 23.Banerjee DK, Carrasquillo EA, Hughey P, Schutzbach JS, Martínez JA, Baksi K. In vitro phosphorylation by cAMP-dependent protein kinase up-regulates recombinant Saccharomyces cerevisiae mannosylphosphodolichol synthase. J. Biol. Chem. 2005;280:4174–4181. doi: 10.1074/jbc.M406962200. [DOI] [PubMed] [Google Scholar]
- 24.Banerjee DK, Martinez JA, Baksi K. Significance of protein N-glycosylation in breast tumor angiogenesis. In: Maragoudakis ME, Papadimitriou E, editors. Angiogenesis. Basic Science and Clinical Application. Transworld Research Network; Trivandrum, Kerala, India: 2007. pp. 281–302. [Google Scholar]
- 25.Martinez JA, Torres-Negron I, Amigo LA, Banerjee DK. Expression of Glc3Man9GlcNAc2-PP-Dol is a prerequisite for capillary endothelial cell proliferation. Cell. Mol. Biol. (Noisy-le-grand) 1999;45:137–152. [PubMed] [Google Scholar]
- 26.Banerjee DK, Kousvelari EE, Baum BJ. cAMP-mediated protein phosphorylation of microsomal membranes increases mannosylphosphodolichol synthase activity. Proc. Natl. Acad. Sci. USA. 1987;84:6389–6393. doi: 10.1073/pnas.84.18.6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Banerjee DK, Aponte E, DaSilva JJ. Low expression of lipid-linked oligosaccharide due to a functionally altered Dol-P-Man synthase reduces protein glycosylation in cAMP-dependent protein kinase deficient Chinese hamster ovary cells. Glycoconj J. 2004;21:479–486. doi: 10.1007/s10719-004-5538-2. [DOI] [PubMed] [Google Scholar]
- 28.Banerjee DK. Requirement of protein kinase type I for cAMP-mediated up-regulation of lipid-linked oligosaccharide for asparagine-linked protein glycosylation. Cell Mol Biol (Noisy-le-grand) 2007;53:55–63. [PubMed] [Google Scholar]


