Abstract
Consensus design is an appealing strategy for the stabilization of proteins. It exploits amino acid conservation in sets of homologous proteins to identify likely beneficial mutations. Nevertheless, its success depends on the phylogenetic diversity of the sequence set available. Here we show that randomization of a single protein represents a reliable alternative source of sequence diversity essentially free of phylogenetic bias. A small number of functional protein sequences selected from binary-patterned libraries suffices as input for consensus design of active enzymes that are easier to produce and substantially more stable than individual members of the starting data set. Although catalytic activity correlates less consistently with sequence conservation in these extensively randomized proteins, less extreme mutagenesis strategies might be adopted in practice to augment stability while maintaining function.
Keywords: protein stabilization, multiple sequence alignments, consensus mutation, binary patterning, chorismate mutase
Utilization of proteins outside of their normal biological context – for example in diagnostic, medical, or industrial applications or for the creation of novel receptors and catalysts1; 2 – often requires optimization of biophysical properties like stability.3; 4 For this purpose, engineering methods that exploit statistical amino acid frequencies from multiple sequence alignments (MSAs) are widely used.5; 6 Data-driven consensus design is based on the simple assumption that the frequency of a given residue in an MSA of homologous proteins correlates with that amino acid’s contribution to protein stability.7 An artificial protein possessing the most frequent residue at each position should accordingly show maximum stability. Given the difficulty of predicting how individual residues contribute to overall stability,8 this approach to protein stabilization is often preferable to classical rational design, particularly as it does not depend on the availability of structural information.
If an MSA were composed of fully independent protein sequences all selected for stable folding to the same structure, and if individual residues contributed additively to stability, then the stability contribution of a particular amino acid at a given position should be a roughly logarithmic function of its frequency in the MSA.9 Indeed, approaches based on this idea have proven broadly successful at creating more stable proteins.5;6;10 However, because the sequences of natural proteins generally derive from a common ancestor, they tend to be heavily biased by evolutionary relationships. This lack of statistical independence among different protein sequences violates one of the key assumptions underlying the idea of the logarithmic relationship between an amino acid’s stability contribution and its frequency in an MSA. As a consequence, the purely statistical approach of simply replacing all non-consensus residues in conserved positions of a sequence or sequence motif with their consensus counterparts may often fail to yield a more stable protein6 unless large numbers of functionally and structurally similar proteins are available to minimize phylogenetic bias11;12 or structural factors are considered in the design process.13;14 Various mathematical algorithms have been employed to correct for such bias, ranging from simply reducing the weighting of highly similar sequences10 to complex likelihood-based methods that fully account for phylogeny.15 Labor-intensive mutational analysis of conserved residues is another common strategy for determining individual contributions to stability,16;17;18;19 although it does not necessarily guarantee success.20
Combinatorial methods have also been employed to identify stabilizing consensus mutations in natural homologs. For example, shuffling experiments with conserved residues in β-lactamases21 or larger fragments of cytochrome P450 enzymes22 have shown that in vitro-generated sequence diversity can be a viable alternative to MSAs of natural proteins as input for consensus design. Extrapolating from these results, libraries created from a single protein via sequence randomization and selection should be an attractive source of fully unbiased sequence diversity. Since the resulting variants would be phylogenetically unrelated a priori, the frequency of a given residue in the randomized stretch should directly reflect its contribution to stability according to a standard Boltzmann-like relationship. To test this hypothesis, we have generated consensus enzymes based on functional clones isolated from libraries of individual, extensively randomized proteins.
An all-helical, homodimeric chorismate mutase from Escherichia coli, EcCM,23 served as our starting point (Fig 1). In a previous study, its N-terminal H1 helix, which is 42 amino acids long and forms a dimer-spanning coiled coil, was replaced with a module of randomized sequence.24 The library maintained the polar/apolar binary pattern25 of the original H1 helix but replaced hydrophobic residues with mixtures of leucine (Leu), isoleucine (Ile), methionine (Met), and phenylalanine (Phe) and hydrophilic residues with mixtures of lysine (Lys), glutamate (Glu), aspartate (Asp), and asparagine (Asn). Only three highly conserved active site residues in the helix and two amino acids needed for library construction purposes were held constant. Functional clones were isolated from the library by genetic complementation of chorismate mutase-deficient bacteria. Here, we subjected the sequences of 26 catalytically active variants (Supplementary Table 1 and Supplementary Fig. 1a) to statistical analysis, calculating the conservation energy E for an amino acid at position i from its frequency (f) in the sequence alignment according to equation 1:
| (1) |
The consensus protein H1-E-cons (Fig. 2a) was then designed by choosing the amino acid that appeared most frequently in the selected proteins at each position of the H1 helix. In cases where no consensus could be determined due to equal occurrence of two or more residues, the residue encoded by the least frequent codon in the design set was chosen. Summing the individual contributions of all the consensus residues over the length of the helix affords the conservation energy for the entire binary patterned module, Ec (equation 2), which serves as a measure of sequence conservation:
| (2) |
The gene for the consensus design was expressed in E. coli and the resulting protein was biochemically characterized. For comparison, representative library members possessing low (H1-E-low), medium (H1-E-med), and high (H1-E-high) conservation energies were also produced (Fig. 2a).
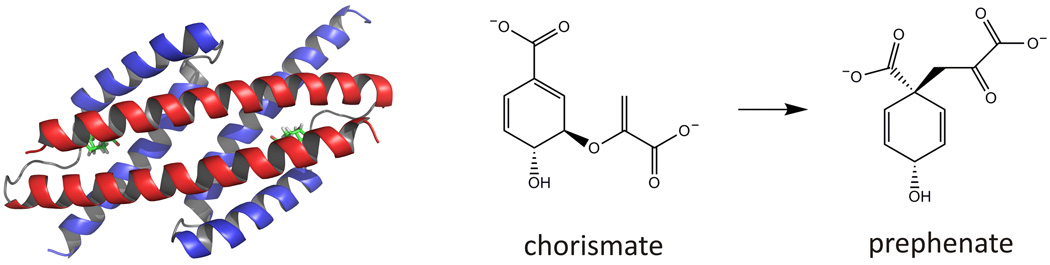
Figure 1.
Structural model of the homodimeric, helical bundle chorismate mutases (left) and the reaction they catalyze (right). The H1 and H2/3 helices of the protein are shown in red and blue, respectively. A transition state analog inhibitor bound at the active sites is highlighted in green. The protein graphic was made with PyMol (DeLano Scientific LLC) based on the crystal structure of the E. coli chorismate mutase EcCM (PDB ID: 1ECM).
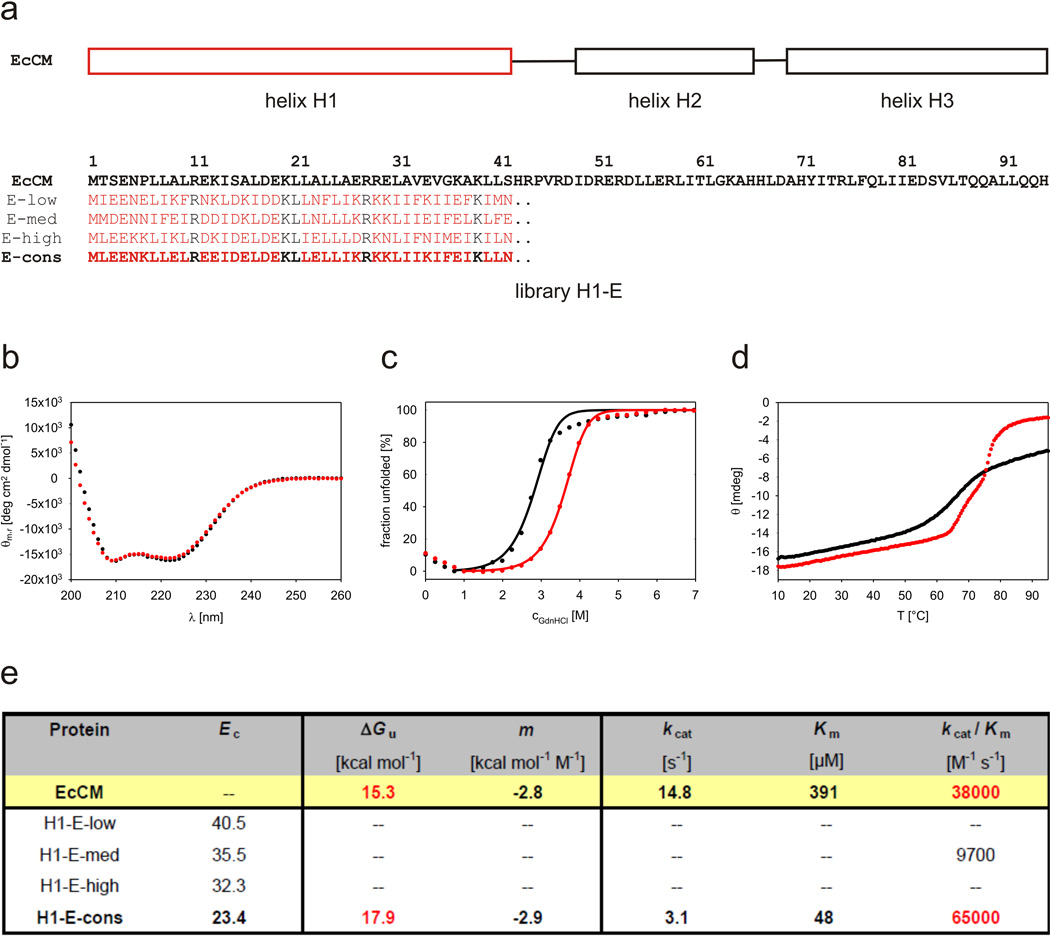
Figure 2.
Consensus design of a stable EcCM variant. (a) Protein sequences of representative H1-E library members with low (H1-E-low), medium (H1-E-med), and high (H1-E-high) conservation energies as well as the consensus protein (H1-E-cons). Amino acid positions randomized and held constant are shown in red and black, respectively. The H1-E-low, H1-E-med, and H1-E-high variants could not be produced as soluble dimeric proteins at concentrations necessary for complete biochemical characterization. CD-spectra (b) and chemical (c) and thermal (d) denaturation curves were thus only recorded for wild-type EcCM (black) and H1-E-cons (red) at 5 µM protein concentration in PBS at pH 7.5. In contrast to EcCM, thermal melting of H1-E-cons is irreversible due to precipitation of the denatured protein, which may explain its quantitative denaturation above 90°C. (e) Stability parameters (free energy of unfolding ΔGu and cooperativity of unfolding m) as well as steady state parameters for the EcCM-based proteins. Experimental details about gene construction and molecular cloning, protein production and purification, as well as biophysical characterization are described in the supporting information.
While none of the characterized proteins from the original library of patterned EcCM variants was sufficiently stable for detailed biophysical studies, H1-E-cons was readily produced as a helical dimer as judged by CD spectroscopy (Fig. 2b) and size-exclusion chromatography. Moreover, even though only eight different amino acids were used for randomization of the H1 helix, chemical denaturation experiments with guanidinium chloride revealed H1-E-cons to be 2.6 kcal/mol more stable than the parent EcCM protein (Fig. 2c/e). The thermal melting profile of the consensus protein exhibits two transitions (Fig. 2d), the first matching that of wild-type EcCM at 67°C and a second at 76°C, suggesting that the newly designed H1/H1’ coiled-coil unfolds at a significantly higher temperature than the rest of the protein. Notably, the improvement in stability was not achieved at the cost of catalytic efficiency. An 8-fold decrease in Km compensates for a 5-fold lower kcat, so the consensus design exhibits an apparent bimolecular rate constant kcat/Km that is two times larger than that of EcCM itself (Fig. 2e and Supplementary Fig. 5a).
These findings show that phylogenetically unbiased consensus design can lead to substantial stabilization of secondary structural motifs in mesostable proteins. Analogous experiments with a chorismate mutase from Methanococcus jannaschii (MjCM),26 an EcCM homolog, suggest that this strategy is general. Statistical analysis of sequences of 20 functional clones that were analogously selected from a binary patterned library of the MjCM H1 helix (Supplementary Table 2 and Supplementary Fig. 1b)24 afforded the consensus protein H1-M-cons, which was again produced, characterized, and compared to representative library proteins that varied in conservation energy (H1-M-low/med/high) (Fig. 3). In contrast to proteins from the EcCM library, all variants derived from the thermostable MjCM template yielded correctly folded homodimers (Supplementary Fig. 2b), enabling direct comparison of consensus and reference proteins. Chemical denaturation experiments showed that the consensus protein is more stable than the three characterized library variants by 2.8 to 3.7 kcal/mol (ΔΔGu) (Fig. 3b and Supplementary Fig. 3a). Consistent with this result, H1-M-low/med/high begin to denature at lower temperatures than H1-M-cons, although meaningful Tm values could not be determined from the melting profiles because only partial unfolding was observed below 95°C (Supplementary Fig. 4a). While H1-M-cons is not quite as robust as wild-type MjCM, its stability is comparable to that of H1-E-cons. H1-M-cons is also more active than the proteins selected directly from the patterned libraries, and in some cases considerably so. Relative to wild-type MjCM, the catalytic efficiency of library members of low and medium conservation is reduced 2,000 to 10,000-fold, whereas the consensus protein is only 30-fold less active (Fig. 3b and Supplementary Fig. 5b). The lower activity of H1-M-cons compared to the wild type may reflect a decreased tolerance of the thermostable scaffold to this particular simplified alphabet.
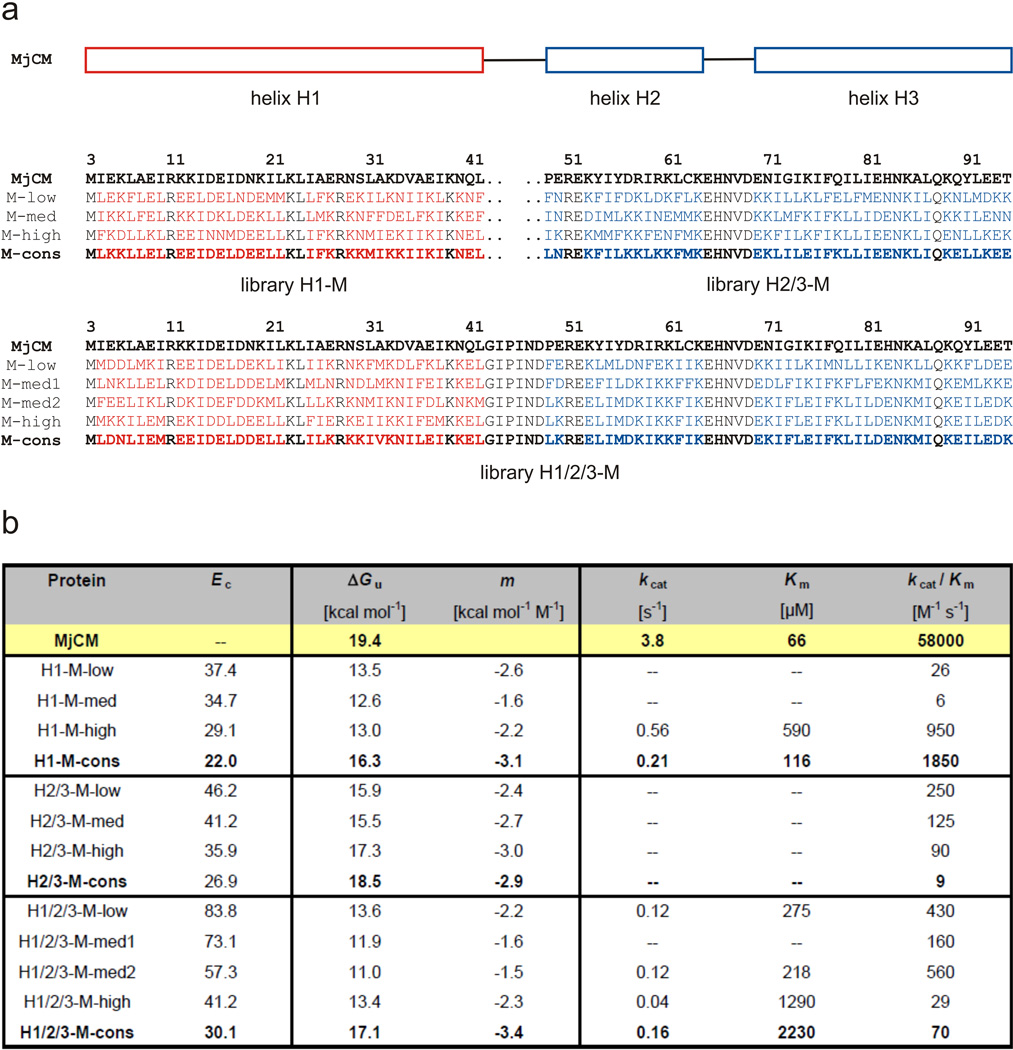
Figure 3.
Consensus design of stable MjCM variants. (a) Protein sequences of representative H1-M, H2/3-M, and H1/2/3-M library members with low (M-low), medium (M-med), and high (M-high) conservation energies together with the derived consensus proteins (M-cons). Amino acid positions randomized in helices H1 and H2/3 are highlighted in red and blue, respectively. Residues held constant are shown in black. EcCM residue numbering was used for all MjCM-based proteins. The H1/2/3-M library was generated by shuffling the binary-patterned motifs of catalytically active selectants from libraries H1-M and H2/3-M, followed by a second round of genetic selection.27 Because a single H2/3 fragment was highly abundant in the set of 25 H1/2/3-M selectants, two sequences of medium conservation, one with (H1/2/3-M-med1) and one without (H1/2/3-M-med2) the conserved motif, were chosen for characterization. The original H2/3-M consensus protein was toxic in E. coli. To reduce its unusually high net charge from +12 to +1, six consensus lysines were mutated to Glu (five) or Asn (one), which were nearly as equally abundant in the sequence alignment. The redesigned H2/3-M-con could be produced in vivo as correctly folded protein in high yield. The H1/2/3-M library contained an unprogrammed valine at position 32 in 14 of the 25 sequences, which was consequently incorporated into the consensus protein. However, only reference proteins without this mutation were chosen for characterization to allow for unbiased frequency analysis and comparison of conservation energies. The protein originally chosen as H2/3-M-high could not be produced in E. coli and was therefore replaced by another variant from the same library that had a similar conservation energy. (b) Biochemical characterization of the MjCM-based proteins. Denaturation experiments were performed at 5 µM protein concentration in PBS at pH 7.5. Experimental details about gene construction and molecular cloning, protein production and purification, as well as biophysical characterization are described in the supporting information.
Because analogous selection experiments have also been reported for the H2/3 helices of MjCM alone and in combination with H1,27 it was possible to extend the consensus design approach to the entire MjCM scaffold. Proteins H2/3-M-cons and H1/2/3-M-cons as well as library members possessing low, medium, and high conservation energies were generated by the procedures described above (the input sequences are summarized in Supplementary Tables 3 and 4, and Supplementary Fig. 1c–e; CD-spectra of the proteins are shown in Supplementary Fig. 2c/d). Like the H1-M variants, both consensus proteins were obtained as soluble homodimers in good yield. Indeed, in contrast to wild-type MjCM, most of the binary patterned library proteins in this series exhibited favorable folding in vivo, suggesting that this property is actively selected for in the complementation experiments. While H2/3-M-cons and its progenitors show high thermostabilities comparable to that of wild-type MjCM (Tm > 90 °C), H1/2/3-M-cons begins to denature at a higher temperature than the input proteins (Supplementary Fig. 4b/c). Chemical denaturation experiments showed that both H2/3-M-cons and H1/2/3-M-cons have significantly higher ΔGu values than the original library variants, indicating considerable gains in stability (Fig. 3b and Supplementary Fig. 3b/c). Simultaneous optimization of all three helices gave a ΔΔGu of 4.6 ± 1.2 kcal/mol, which reflects roughly additive contributions of the H1-M (3.3 ± 0.5 kcal/mol) and H2/3-M (2.3 ± 0.9 kcal/mol) consensus designs. Additivity is also manifest in the stronger unfolding cooperativity (m) of H1/H2/H3-M-cons (−1.5 kcal mol−1 M−1) relative to H1-M-cons (−1.0 kcal mol−1 M−1) and H2/3-M-cons (−0.2 kcal mol−1 M−1).
The success of library-based consensus design is notable given that the binary-patterned input proteins do not exhibit a consistent correlation between the calculated conservation energy Ec and the experimentally determined stability ΔGu (Fig. 3b). This is presumably due to the fact that the contributions of individual sites to stability are not identical. As a consequence, a library protein with a low Ec can be less stable than a protein with a higher Ec if a few key residues that contribute disproportionately to stability are lacking in the former but are present in the latter. In contrast, the consensus sequence, which has the lowest Ec by definition, always contains the optimal (most frequent) residue at each variable position and can thus be expected to lead to a protein with enhanced stability. In the H2/3-M series Ec is also a poor predictor of catalytic activity. In contrast to H1-E and H1-M, the kcat/Km values for the H2/3-M enzymes show a weak inverse correlation with sequence conservation (i.e. with -Ec), and this trend carries over to the fully remodeled H1/2/3-M proteins (Fig. 3b and Supplementary Fig. 5c/d). Given the severely restricted set of amino acids used to construct the binary patterned libraries and the large fraction of the protein that was mutagenized, this reduction in catalytic activity is not terribly surprising. Some of the stabilizing mutations may subtly alter the placement of key catalytic residues or impair substrate access to the completely buried active site. Such explanations would be consistent with the common observation that activity is a local property, often resulting from highly synergistic interactions of a few residues,28 whereas protein stability arises from small but additive effects distributed over the entire molecule.8; 29
Overall, our results show that consensus design based on unbiased input sequences derived from binary patterned libraries can reliably predict stabilized proteins. Any phylogenetic bias was precluded a priori by randomization of just a single parental sequence followed by functional selection. The four libraries we tested were based on two different scaffolds and involved partial or complete diversification of secondary structural elements in the protein; each directly afforded consensus designs that were substantially more stable than any of the input proteins. Because side chain diversity was greatly restricted through the use of a simplified alphabet, these effects could be achieved by statistical analysis of a relatively small number of binary-patterned sequences (≤ 30).
The fact that the consensus approach, which traditionally relies on many sequences of naturally evolved proteins, works efficiently with a small set of artificially randomized sequences generated by "synthetic evolution" experiments is notable. It indicates that simple stochastic sampling of residues selected from unbiased libraries composed of a restricted alphabet is a valid alternative to the consensus analysis of evolutionarily related proteins. Successful protein stabilization by a purely statistical method – independent of any specific mechanism operating during natural evolution – represents an intriguing, potentially general finding.
Because library-based consensus design does not depend on the availability of structurally and functionally related natural homologs, it represents a potentially powerful strategy for stabilizing proteins of industrial or therapeutic interest. For practical applications, the extreme mutagenesis strategy adopted in our proof-of-principle study is unlikely to be necessary. Instead, as suggested by our experiments with EcCM, the design of relatively small focused libraries should suffice to achieve significant increases in stability without loss of biological function. Optimizing the amino acid alphabet used for randomization with respect to structural propensities and functional diversity, or taking covariation into account in the design process,30 are additional strategies that might be exploited to maximize stabilization while preserving activity.
Supplementary Material
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Christian Jäckel, Email: jaeckel@org.chem.ethz.ch.
Jesse D Bloom, Email: jesse.bloom@gmail.com.
Peter Kast, Email: kast@org.chem.ethz.ch.
Frances H Arnold, Email: frances@cheme.caltech.edu.
Donald Hilvert, Email: hilvert@org.chem.ethz.ch.
References
- 1.Jäckel C, Kast P, Hilvert D. Protein design by directed evolution. Annu. Rev. Biophys. 2008;37:153–173. doi: 10.1146/annurev.biophys.37.032807.125832. [DOI] [PubMed] [Google Scholar]
- 2.Toscano MD, Woycechowsky KJ, Hilvert D. Minimalist active-site redesign: Teaching old enzymes new tricks. Angew. Chem. Int. Ed. 2007;46:3212–3236. doi: 10.1002/anie.200604205. [DOI] [PubMed] [Google Scholar]
- 3.Schmid A, Dordick JS, Hauer B, Kiener A, Wubbolts M, Witholt B. Industrial biocatalysis today and tomorrow. Nature. 2001;409:258–268. doi: 10.1038/35051736. [DOI] [PubMed] [Google Scholar]
- 4.Fasan R, Chen MM, Crook NC, Arnold FH. Engineered alkane-hydroxylating cytochrome P450(BM3) exhibiting nativelike catalytic properties. Angew. Chem. Int. Ed. 2007;46:8414–8418. doi: 10.1002/anie.200702616. [DOI] [PubMed] [Google Scholar]
- 5.Polizzi KM, Bommarius AS, Broering JM, Chaparro-Riggers JF. Stability of biocatalysts. Curr. Opin. Chem. Biol. 2007;11:220–225. doi: 10.1016/j.cbpa.2007.01.685. [DOI] [PubMed] [Google Scholar]
- 6.Lehmann M, Wyss M. Engineering proteins for thermostability: the use of sequence alignments versus rational design and directed evolution. Curr. Opin. Biotechnol. 2001;12:371–375. doi: 10.1016/s0958-1669(00)00229-9. [DOI] [PubMed] [Google Scholar]
- 7.Steipe B, Schiller B, Plückthun A, Steinbacher S. Sequence statistics reliably predict stabilizing mutations in a protein domain. J. Mol. Biol. 1994;240:188–192. doi: 10.1006/jmbi.1994.1434. [DOI] [PubMed] [Google Scholar]
- 8.Wintrode PL, Arnold PH. Adv.Protein Chem. Vol. 55. San Diego: Academic Press Inc; 2001. Temperature adaptation of enzymes: Lessons from laboratory evolution; pp. 161–225. [DOI] [PubMed] [Google Scholar]
- 9.Ohage EC, Graml W, Walter MM, Steinbacher S, Steipe B. β-Turn propensities as paradigm for the analysis of structural motifs to engineer protein stability. Protein Sci. 1997;6:233–241. doi: 10.1002/pro.5560060125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lehmann M, Kostrewa D, Wyss M, Brugger R, D'Arcy A, Pasamontes L, van Loon APGM. From DNA sequences to improved functionality: using protein sequences to rapidly design a thermostable consensus phytase. Protein Eng. 2000;13:49–57. doi: 10.1093/protein/13.1.49. [DOI] [PubMed] [Google Scholar]
- 11.Mosavi LK, Minor DL, Peng ZY. Consensus-derived structural determinants of the ankyrin repeat motif. Proc. Natl. Acad. Sci. U.S.A. 2002;99:16029–16034. doi: 10.1073/pnas.252537899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Main ERG, Xiong Y, Cocco MJ, D'Andrea L, Regan L. Design of stable alpha-helical arrays from an idealized TPR motif. Structure. 2003;11:497–508. doi: 10.1016/s0969-2126(03)00076-5. [DOI] [PubMed] [Google Scholar]
- 13.Binz HK, Stumpp MT, Forrer P, Amstutz P, Pluckthun A. Designing repeat proteins: Well-expressed, soluble and stable proteins from combinatorial libraries of consensus ankyrin repeat proteins. J. Mol. Biol. 2003;332:489–503. doi: 10.1016/s0022-2836(03)00896-9. [DOI] [PubMed] [Google Scholar]
- 14.Vazquez-Figueroa E, Chaparro-Riggers J, Bommarius AS. Development of a thermostable glucose dehydrogenase by a structure-guided consensus concept. ChemBioChem. 2007;8:2295–2301. doi: 10.1002/cbic.200700500. [DOI] [PubMed] [Google Scholar]
- 15.Bloom JD, Glassman MJ. Inferring stabilizing mutations from protein phylogenies: Application to influenza hemagglutinin. PLoS Comput. Biol. 2009;5 doi: 10.1371/journal.pcbi.1000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nikolova PV, Henckel J, Lane DP, Fersht AR. Semirational design of active tumor suppressor p53 DNA binding domain with enhanced stability. Proc. Natl. Acad. Sci. U.S.A. 1998;95:14675–14680. doi: 10.1073/pnas.95.25.14675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rath A, Davidson AR. The design of a hyperstable mutant of the Abp1p SH3 domain by sequence alignment analysis. Protein Sci. 2000;9:2457–2469. doi: 10.1110/ps.9.12.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loening AM, Fenn TD, Wu AM, Gambhir SS. Consensus guided mutagenesis of Renilla luciferase yields enhanced stability and light output. Protein Eng. Des. Sel. 2006;19:391–400. doi: 10.1093/protein/gzl023. [DOI] [PubMed] [Google Scholar]
- 19.Lehmann M, Loch C, Middendorf A, Studer D, Lassen SF, Pasamontes L, van Loon APGM, Wyss M. The consensus concept for thermostability engineering of proteins: further proof of concept. Protein Eng. 2002;15:403–411. doi: 10.1093/protein/15.5.403. [DOI] [PubMed] [Google Scholar]
- 20.Ryan BJ, O'Connell MJ, O'Fagain C. Consensus mutagenesis reveals that non-helical regions influence thermal stability of horseradish peroxidase. Biochimie. 2008;90:1389–1396. doi: 10.1016/j.biochi.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 21.Amin N, Liu AD, Ramer S, Aehle W, Meijer D, Metin M, Wong S, Gualfetti P, Schellenberger V. Construction of stabilized proteins by combinatorial consensus mutagenesis. Protein Eng. Des. Sel. 2004;17:787–793. doi: 10.1093/protein/gzh091. [DOI] [PubMed] [Google Scholar]
- 22.Li YG, Drummond DA, Sawayama AM, Snow CD, Bloom JD, Arnold FH. A diverse family of thermostable cytochrome P450s created by recombination of stabilizing fragments (vol 25, pg 1051, 2007) Nat. Biotechnol. 2007;25:1488–1488. doi: 10.1038/nbt1333. [DOI] [PubMed] [Google Scholar]
- 23.Lee AY, Karplus PA, Ganem B, Clardy J. Atomic structure of the buried catalytic pocket of Escherichia coli chorismate mutase. J. Am. Chem. Soc. 1995;117:3627–3628. [Google Scholar]
- 24.Besenmatter W, Kast P, Hilvert D. Relative tolerance of mesostable and thermostable protein homologs to extensive mutation. Proteins. 2007;66:500–506. doi: 10.1002/prot.21227. [DOI] [PubMed] [Google Scholar]
- 25.Kamtekar S, Schiffer JM, Xiong H, Babik JM, Hecht MH. Protein design by binary patterning of polar and nonpolar amino acids. Science. 1993;262:1680–1685. doi: 10.1126/science.8259512. [DOI] [PubMed] [Google Scholar]
- 26.MacBeath G, Kast P, Hilvert D. A small, thermostable, and monofunctional chorismate mutase from the archeon Methanococcus jannaschii. Biochemistry. 1998;37:10062–10073. doi: 10.1021/bi980449t. [DOI] [PubMed] [Google Scholar]
- 27.Taylor SV, Walter KU, Kast P, Hilvert D. Searching sequence space for protein catalysts. Proc. Natl. Acad. Sci. U.S.A. 2001;98:10596–10601. doi: 10.1073/pnas.191159298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holliday GL, Mitchell JBO, Thornton JM. Understanding the functional roles of amino acid residues in enzyme catalysis. J. Mol. Biol. 2009;390:560–577. doi: 10.1016/j.jmb.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 29.Wells JA. Additivity of mutational effects in proteins. Biochemistry. 1990;29:8509–8517. doi: 10.1021/bi00489a001. [DOI] [PubMed] [Google Scholar]
- 30.Magliery TJ, Regan L. Beyond consensus: statistical free energies reveal hidden interactions in the design of a TPR motif. J. Mol. Biol. 2004;343:731–745. doi: 10.1016/j.jmb.2004.08.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.