Abstract
Protein scaffolds play an important role in signal transduction, regulating the localization of signaling components and mediating key protein interactions. Here, we report that the major binding partners of the Connector Enhancer of KSR 1 (CNK1) scaffold are members of the cytohesin family of Arf guanine nucleotide exchange factors, and that the CNK1/cytohesin interaction is critical for activation of the PI3K/AKT cascade downstream from insulin and insulin-like growth factor 1 (IGF-1) receptors. We identified a domain located in the C-terminal region of CNK1 that interacts constitutively with the coiled-coil domain of the cytohesins, and found that CNK1 facilitates the membrane recruitment of cytohesin-2 following insulin stimulation. Moreover, through protein depletion and rescue experiments, we found that the CNK1/cytohesin interaction promotes signaling from plasma membrane-bound Arf GTPases to the phosphatidylinositol 4-phosphate 5-kinases (PIP5Ks) to generate a PIP2-rich microenvironment that is critical for the membrane recruitment of insulin receptor substrate 1 (IRS1) and signal transmission to the PI3K/AKT cascade. These findings identify CNK1 as a new positive regulator of insulin signaling.
Keywords: Protein scaffold, signal transduction, insulin pathway
The Connector Enhancer of KSR (CNK) protein family was initially discovered through genetic studies performed in Drosophila (Therrien et al. 1998). One CNK protein is present in Drosophila and Caenorhabditis elegans, whereas three distinct CNK homologs are found in mammals (CNK1, CNK2, and CNK3) (Therrien et al. 1998; Yao et al. 1999; Douziech et al. 2003). Members of the CNK family possess multiple protein interaction domains (Fig. 1A), and, as a result, they have been proposed to function as protein scaffolds. All of the CNK family members contain a sterile α motif (SAM), a conserved region in CNK (CRIC) domain, and a PSD-95/DLG-1/ZO-1 (PDZ) domain, and, with the exception of mammalian CNK3, they all have a pleckstrin homology (PH) domain (Therrien et al. 1998; Douziech et al. 2003). In addition, the mammalian CNK2 splice variant CNK2A has a PDZ domain-binding motif at its C terminus (Yao et al. 1999), and Drosophila CNK (D-CNK) uniquely possesses a domain known as the Raf-interacting region (RIR) that mediates binding of the Drosophila Raf kinase (Douziech et al. 2003). In Drosophila, where there is only one CNK protein, D-CNK has been shown to be essential for Ras signaling, and to contribute to the Raf activation process by facilitating both the membrane recruitment of Raf and the interaction of Raf with the KSR scaffold (Therrien et al. 1998; Anselmo et al. 2002; Douziech et al. 2006). Experiments in flies further revealed that D-CNK may function in more than one signaling pathway, in that overexpression of D-CNK was found to cooperatively enhance the action of RasV12G37, a Ras effector loop mutant that selectively activates the Ral pathway (Therrien et al. 1999).
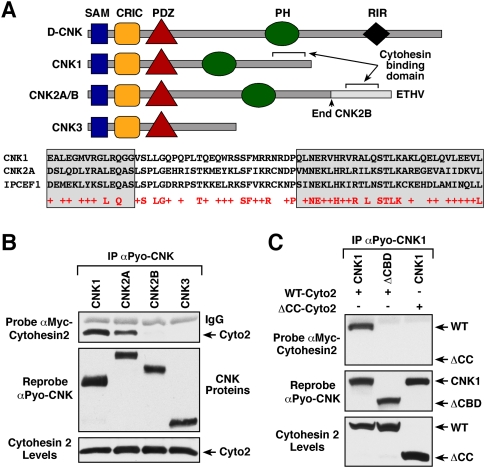
Figure 1.
Identification of a cytohesin-binding domain in CNK1 and CNK2A. (A) The domain structure of D-CNK and the mammalian CNK proteins is depicted. The CNK2A splice variant contains a C-terminal PDZ domain-binding domain motif (ETHV), and D-CNK uniquely possesses a RIR. The sequence alignment of the cytohesin-binding region of IPCEF1 and sequences in the C-terminal region of CNK1 (568–637) and CNK2A (907–976) is also shown. Gray-shaded boxes indicate predicted CC regions. (B) Myc-cytohesin-2 (Cyto-2) was coexpressed with the indicated Pyo-CNK constructs in 293T cells. After cell lysis, the Pyo-CNK proteins were immunoprecipitated and probed for the presence of Myc-cytohesin-2 by immunoblot analysis. (C) Pyo-CNK proteins were immunoprecipitated from 293T cells coexpressing the indicated CNK1 and cytohesin-2 proteins, and were probed for the presence of Myc-cytohesin-2. ΔCBD-CNK1 lacks amino acid residues 564–713 of CNK1. CC-Cyto-2 is a deletion mutant lacking the N-terminal CC domain of cytohesin-2.
Of the mammalian CNK proteins, CNK1 is expressed primarily in epithelial tissues (Jaffe et al. 2004; Rabizadeh et al. 2004), whereas CNK2A (also known as MAGUIN-1) and the alternatively spliced CNK2B (also known as MAGUIN-2) are found almost exclusively in neuronal tissues (Yao et al. 1999; Lanigan et al. 2003). Although there are no reports characterizing CNK3, EST databases suggest that CNK3 is more broadly expressed. To date, studies examining the mammalian CNKs indicate that, as with D-CNK, these proteins may function in a multiple signaling cascade. For example, CNK1 has been found to interact with various Ral and Rho pathway components and to influence both Rho-mediated gene transcription and activation of the JNK cascade (Jaffe et al. 2004, 2005), as well as to impact Src-mediated Raf-1 activation (Ziogas et al. 2005). The CNK2 proteins also associate with components of the Ral pathway (Lanigan et al. 2003), and have been reported to contribute an essential function to PC12 differentiation that is distinct from their effects on ERK cascade activation (Bumeister et al. 2004). In addition, the CNK2 splice variant CNK2A has been implicated in post-synaptic density (PSD) signaling through its interactions with Densin-180 and members of the membrane-associated guanylate kinase (MAGUK) protein family (Yao et al. 1999; Ohtakara et al. 2002).
Because regulated protein–protein interactions play an important role in the transmission of cellular signals, scaffolding molecules have emerged as key modulators of the signaling process. Therefore, to gain further insight as to the role of the CNK proteins in signal transduction, we took a proteomics approach to determine the binding partners present in mammalian CNK scaffold complexes. Strikingly, we found that members of the cytohesin family of Arf guanine nucleotide exchange factors (GEFs) are the major binding partners of CNK1, and that CNK1 is an critical regulator of cytohesin function during insulin signaling.
Results
Mass spectrometry analysis of CNK protein complexes identifies the cytohesin proteins as the major binding partners of CNK1
To identify functionally important binding partners of the mammalian CNK1 proteins, Pyo-tagged CNK1, CNK2A, CNK2B, and CNK3 protein complexes were isolated from cycling 293T cells and analyzed using mass spectrometry (Supplemental Table 1). As verification of this approach, peptides from several of the known CNK-interacting proteins were detected in the CNK complexes, and the selective binding of certain proteins to specific CNKs was confirmed. For example, only complexes containing CNK2A were found to contain peptides derived from Densin-180 and SAP97, two PDZ domain-containing proteins that have been shown previously to interact with the PDZ-binding motif found exclusively in CNK2A (Yao et al. 1999; Ohtakara et al. 2002). Consistent with reports that CNK1 interacts with certain RhoGEFs (Jaffe et al. 2005) and the Src tyrosine kinase (Ziogas et al. 2005), peptides from the Src family kinase Fyn and the RhoGEFs RhoGEF11, RhoGEF12, and RhoGEF16 were found in CNK1 complexes. RhoGEF peptides were also found in the other CNK complexes, and all of the complexes contained peptides derived from the mammalian counterpart of the small SAM domain-containing protein Ave/Hyp. Ave/Hyp was discovered in Drosophila as a modifier of EGF signaling (Douziech et al. 2006; Roignant et al. 2006), and subsequent biochemical and structural studies have revealed that the SAM domains of Hyp/Ave and D-CNK can dimerize (Rajakulendran et al. 2008). In addition, all CNK complexes contained peptides from three subunits of the heterotrimeric serine/threonine phosphatase PP2A, implicating PP2A as a potential regulator of CNK phosphorylation. Strikingly, the most abundant binding partners detected in any of the CNK complexes were members of the cytohesin family. The cytohesins functions as GEFs for the Arf GTPases (Kolanus 2007), and peptides from cytohesin-1, ARNO/cytohesin-2, and GRP1/cytohesin-3 were detected in CNK1 and CNK2A complexes, but were completely absent from CNK2B or CNK3 complexes. It should be noted that the fourth cytohesin family member, cytohesin-4, is expressed primarily in peripheral blood leukocytes, and was not detected in any of the CNK complexes. As shown in Figure 1B, the selective interaction of the cytohesins with CNK1 and CNK2A was confirmed in coimmunoprecipitation assays using 293T cells coexpressing the various CNK scaffolds with a Myc-tagged cytohesin-2 protein.
Because alternative mRNA splicing generates the CNK2A and CNK2B proteins, selective binding of cytohesin to CNK2A indicates that the interaction is likely mediated by residues in the C-terminal region of CNK2A that are not present in CNK2B. When the sequence of this region was compared with the sequence of CNK1 and other previously identified cytohesin-binding proteins, an 83-amino-acid domain was identified in CNK2A and CNK1 that showed significant homology with the region in Interaction Protein for Cytohesin Exchange Factor 1 (IPCEF1) known to mediate cytohesin binding (Fig. 1A; Venkateswarlu 2003). As shown in Figure 1C, a CNK1 mutant that lacked these sequences (ΔCBD-CNK1) failed to interact with myc-cytohesin-2, identifying this region as a cytohesin-binding domain (CBD).
In addition to IPCEF1, the cytohesins have been found to interact with various other scaffold proteins, including GRASP/tamalin (Nevrivy et al. 2000; Kitano et al. 2002), CASP/Cybr/CYTIP (Mansour et al. 2002; Tang et al. 2002), and GRSP1 (Klarlund et al. 2001). Although there is no primary sequence in common among these scaffolds, binding is mediated by the N-terminal coiled-coil (CC) domain present in the cytohesins and regions in each scaffold that also possess CC structure. Based on structure prediction programs, the CBD of CNK1 likely adopts a CC structure; therefore, we examined whether the CNK1/cytohesin interaction requires the cytohesin CC domain. As shown in Figure 1C, a cytohesin-2 mutant lacking the CC domain (ΔCC-Cyto2) failed to interact with CNK1 in coimmunoprecipitation assays, further demonstrating the importance of the CC domain in localizing the cytohesins to signaling scaffolds.
Insulin induces the membrane recruitment of the CNK1 scaffold complex
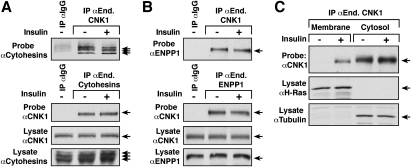
Another major constituent in the CNK1 complexes that bound selectively to CNK1 was ENPP1, an inhibitor of insulin receptor (IR) autophosphorylation (Belfiore et al. 1996; Maddux and Goldfine 2000). Interestingly, dysregulation of both ENPP1 and the cytohesins has been associated with parameters related to insulin resistance (Hafner et al. 2006; Goldfine et al. 2008). Thus, we examined the effect of insulin signaling on CNK1 protein interactions and localization. As shown in Figure 2A, endogenous CNK1 was found to interact constitutively with endogenous cytohesin-1, cytohesin-2, and cytohesin-3 in HepG2 cells, and the interaction was not altered by insulin treatment. Likewise, binding of endogenous CNK1 to endogenous ENPP1 was observed in both serum-starved and insulin-treated HepG2 cells (Fig. 2B). In cell fractionation experiments, endogenous CNK1 was found exclusively in the cytoplasmic fraction of serum-starved cells; however, in insulin-treated cells, a portion of endogenous CNK1 was detected in the membrane fraction (Fig. 2C).
Figure 2.
Effect of insulin treatment on endogenous CNK1 protein interactions and localization. (A,B) Serum-starved HepG2 cells were stimulated with insulin prior to lysis. Endogenous CNK1, cytohesin-1, cytohesin-2, or cytohesin-3, or ENPP1 proteins were immunoprecipitated and examined for the presence of the indicated proteins by immunoblot analysis. As a control, lysates were also incubated with a control rabbit IgG. Total lysates were probed for the presence of the endogenous CNK1, cytohesin-1, cytohesin-2, cytohesin-3, or ENPP1 proteins. (C) Membrane and cytoplasmic fractions were prepared from serum-starved and insulin-treated cells, following which endogenous CNK1 proteins were detected by immunoprecipitation and immunoblot analysis. As controls, the membrane and cytoplasmic fractions were probed for tubulin (cytoplasmic control) and H-Ras (membrane control).
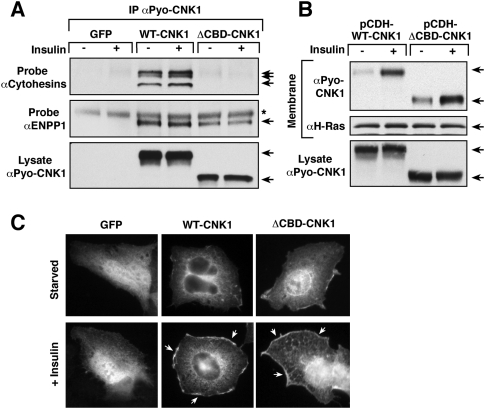
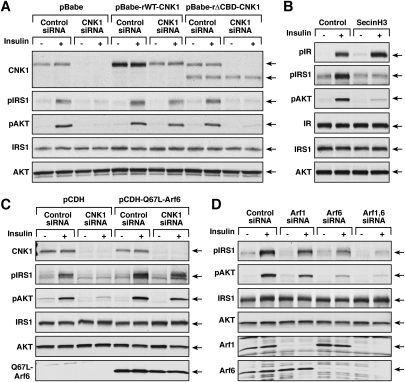
Using HepG2 cells that stably express either WT-CNK1 or ΔCBD-CNK1 at close to physiological levels, the CNK1/cytohesin interaction was not found to be required for either binding of ENPP1 to CNK1 or the membrane localization of CNK1. As shown in Figure 3A, although ΔCBD-CNK1 failed to bind the endogenous cytohesin proteins, it was fully competent to interact with ENPP1 (Fig. 3A). Moreover, as was observed for endogenous CNK1, a portion of both WT-CNK1 and ΔCBD-CNK1 was detected in the membrane fraction of insulin-treated cells, with some constitutive membrane localization observed for ΔCBD-CNK1 (Fig. 3B). Cell staining experiments further revealed the localization of both WT-CNK1 and ΔCBD-CNK1 at the plasma membrane in insulin-treated cells (Fig. 3C), indicating that insulin treatment recruits the CNK1 scaffold to the cell surface in a manner that does not require cytohesin binding.
Figure 3.
Cytohesin binding is not required for CNK1 to localize to the plasma membrane. (A) Serum-starved HepG2 cells stably expressing GFP, WT-CNK1, or ΔCBD-CNK1 were stimulated with insulin prior to lysis. Pyo-CNK1 proteins were immunoprecipitated and examined for the presence of endogenous cytohesins or endogenous ENPP1 by immunoblot analysis. (B) Serum-starved cells stably expressing WT-CNK1 or ΔCBD-CNK1 were treated or not with insulin. Membrane fractions were prepared and probed for the presence of the CNK1 proteins and H-Ras by immunoblot analysis. CNK1 protein levels in total cell lysates are also shown. (C) Localization of GFP, WT-CNK1, and ΔCBD-CNK1 proteins in serum-starved and insulin-treated HepG2 cells was determined by immunofluorescent staining. Insulin-induced plasma membrane staining was observed for both WT-CNK1 (83 out of 100 cells) and ΔCBD-CNK1 (97 out of 100 cells).
CNK1 depletion inhibits insulin-mediated IR substrate 1 (IRS1)/PI3K/AKT signaling
To investigate whether CNK1 has a function in insulin signaling, the effect of CNK1 depletion on the activation of various signaling components in insulin-treated cells was examined. Using either transfected siRNAs or a lentivirus vector-based shRNA directed against human CNK1, we were able to achieve ≥90% knockdown of CNK1 protein levels in several epithelial cell lines, including HepG2, MCF7, and HeLa cells (Fig. 4A; Supplemental Fig. S1). When cells were treated with insulin or insulin-like growth factor-1 (IGF-1), activation and tyrosine phosphorylation of neither the IR nor the IGF-1 receptor was altered, yet phosphorylation of both IRS1 and AKT was reduced significantly in all CNK1-depleted cells (Fig. 4A; Supplemental Figs. S1, S2). CNK1 depletion did not simply alter the kinetics of IRS1 and AKT phosphorylation, but rather inhibited their phosphorylation at all time points. Signaling downstream from AKT through the TORC1 complex was also affected, in that phosphorylation of S6K and S6 was reduced (Fig. 4A). In contrast, the only change observed in ERK activation was that basal phosphoERK levels were elevated approximately twofold in CNK1-depleted cells (Fig. 4A, Supplemental Figs. S1, S2). The elevated basal levels of ERK activation were not found to contribute to the reduction in IRS1/PI3K/AKT pathway signaling observed in the CNK1-depleted cells, in that defects in AKT phosphorylation were still detected when ERK activation was blocked by pretreating cells with the MEK inhibitor U0126 (Supplemental Fig. S2B).
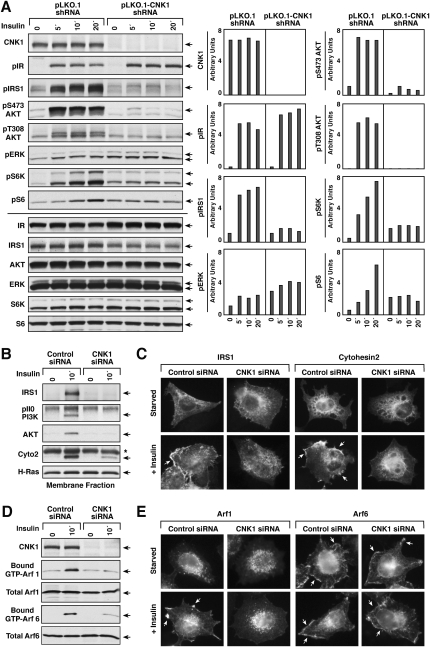
Figure 4.
CNK1 depletion alters insulin-mediated signaling through the IRS1/PI3K/AKT pathway. (A) Lentivirus-infected HeLa cells expressing either the pLKO.1 vector or pLKO.1-CNK1 shRNA were serum-starved and then treated with insulin for the indicated times prior to lysis. Lysates were then examined for CNK1, pIR, pIRS1, pS473AKT, pT308AKT, and pERK levels, as well as total IR, IRS1, AKT, and ERK levels by immunoblot analysis. A representative experiment is shown, with quantification of the blots (relative phosphorylation levels normalized to total protein levels) presented on the right. (B) Membrane fractions were prepared from serum-starved and insulin-treated HeLa cells that had been transfected with control or CNK1-siRNAs. Endogenous IRS1, p110-PI3K, AKT, or cytohesin-2 proteins were immunoprecipitated from the membrane fractions and then detected by immunblot analysis. (C) HeLa cells transfected with control or CNK1-siRNAs were treated with insulin, and the localization of IRS1 and cytohesin-2 was determined by immunofluorescent staining. CNK1 depletion inhibited the membrane localization of IRS1 in 85 out of 100 cells and cytohesin-2 in 73 out of 100 cells. (D) HeLa cells transfected with control or CNK1-siRNAs were treated with insulin prior to lysis. Lysates were incubated with glutathione beads containing GST-GGA, and binding of endogenous GTP-bound Arf1 or Arf6 to GST-GGA was determined by immunoblot analysis. Total Arf1 and Arf6 levels are also shown. (E) HeLa cells expressing HA-Arf1 or Arf6 were transfected with control or CNK1-siRNAs. Cells were treated as indicated, and the localization of the HA-Arf proteins was determined by immunofluorescent staining. Insulin-induced plasma membrane staining of Arf1 was observed in 68 out of 100 cells, and CNK1 depletion inhibited the membrane localization of Arf1 in 100 out of 100 cells.
IRS1 is recruited to the plasma membrane in response to insulin stimulation, and phosphorylation of IRS1 by the IR generates docking sites that can bind the PI3K p85 subunit, thus recruiting PI3K to the cell surface and initiating signaling through the PI3K/AKT pathway. As shown by immunofluorescent staining and cell fractionation, the insulin-mediated membrane recruitment of IRS1 was dramatically impaired in CNK1-depleted cells (Fig. 4B,C). In addition, little to no PI3K p110 subunit or AKT was detected in membrane fractions of insulin-treated cells lacking CNK1 (Fig. 4B).
CNK1 depletion alters cytohesin localization and activity in insulin-treated cells
To begin to address whether the CNK1/cytohesin interaction contributes to the function of CNK1 in insulin signaling, we examined the effect of CNK1 depletion on cytohesin localization and activity. As shown in Figure 4, B and C, the insulin-dependent membrane recruitment of endogenous cytohesin-2 was reduced significantly in CNK1-depleted cells. Given that the cytohesins are known to function as exchange factors for Arf1 and Arf6, we examined the GTP loading of endogenous Arf1 and Arf6 as a measure of cytohesin activity. Using a GST-GGA protein to detect the activated Arf proteins (Santy and Casanova 2001), we found that, although insulin stimulated the GTP loading of both Arf1 and Arf6, an ∼87% reduction in Arf6 activation and an ∼97% reduction in Arf1 activation were observed in CNK1-depleted cells (Fig. 4D). Interestingly, Arf1 was detected in our proteomics analysis as a component of the CNK1 complexes (Supplemental Table 1), and, although Arf1 is known primarily for its functions at the Golgi, recent studies have shown that a fraction of Arf1 can localize to the plasma membrane in response to signaling events, and that Arf1 may be recruited to the plasma membrane by cytohesin-2 (Mitchell et al. 2003; Robertson et al. 2003; Cohen et al. 2007; Boulay et al. 2008). Consistent with these studies, we found that a portion of Arf1 was observed at the cell surface in insulin-treated cells, and that this membrane localization was reduced significantly when CNK1 was depleted (Fig. 4E). In contrast, Arf6, which localizes constitutively at the plasma membrane, was unaffected by knockdown of CNK1 (Fig. 4E), and membrane ruffling induced by insulin treatment was unaltered (Supplemental Fig. S3).
WT-CNK1 and constitutively active Q67L-Arf6 restore insulin-mediated PI3K/AKT signaling in CNK1-depleted cells
To confirm that the defects in insulin-mediated IRS1/PI3K/AKT signaling were due to the specific loss of CNK1, protein add-back experiments were performed using stable cell lines that express either WT-CNK1 or ΔCBD-CNK1 proteins resistant to the CNK1-siRNA. The cells were then depleted of endogenous CNK1 and examined for IRS1 and AKT phosphorylation after insulin treatment. As shown in Figure 5A, phosphorylation of IRS1 and AKT was restored in cells expressing WT-CNK1 but not in those expressing ΔCBD-CNK1. Moreover, consistent with the finding that the ability of CNK1 to interact with and modulate the function of the cytohesins is critical for insulin signaling through the IRS1/PI3K/AKT pathway, treatment of cells with SecinH3, an inhibitor that specifically inhibits the exchange function of the cytohesin proteins but not other ArfGEFs (Hafner et al. 2006), was found to block insulin-mediated IRS1 and AKT phosphorylation, but have no effect on IR activation (Fig. 5B).
Figure 5.
WT-CNK1 and Q67L-Arf6 restore insulin-mediated signaling to the PI3K/AKT pathway in CNK1-depleted cells. (A) HeLa cells stably expressing either vector control or siRNA-resistant WT-CNK1 or ΔCBD-CNK1 were transfected with control or CNK1-siRNAs prior to serum starvation and insulin treatment. Lysates were prepared and examined by immunoblot analysis as indicated. (B) Serum-straved HeLa cells were left untreated or were treated overnight with the cytohesin inhibitor SecinH3 (20 μM) prior to insulin stimulation. Lysates were prepared and examined by immunoblot analysis. (C) HeLa cells stably expressing vector control or Q67L-Arf6 were transfected with control or CNK1-siRNAs prior to serum starvation and insulin treatment. Lysates were prepared and examined by immunoblot analysis. (D) HeLa cells transfected with control, Arf1, Arf6, or Arf1, and Arf6-siRNAs were serum-starved and then treated or not with insulin prior to lysis. Lysates were examined by immunoblot analysis.
If CNK1 acts to facilitate the localization of the cytohesins to the plasma membrane where they can function as exchange factors for the Arfs, then it might be expected that expression of a constitutively active Arf6 protein would rescue the defects observed in CNK1-depleted cells. To address this possibility, a HeLa cell line was generated that stably expressed Q67L-Arf6 at low levels (note that high overexpression of Q67L-Arf6 was found to be toxic), following which the cells were depleted of CNK1. As shown in Figure 5C, insulin-mediated IRS1 and AKT phosphorylation were no longer reduced by CNK1 depletion in cells expressing Q67L-Arf6. Moreover, when cells were depleted of Arf1, Arf6, or both Arf1 and Arf6, reduced insulin-mediated IRS1 and AKT phosphorylation was observed, with Arf6 depletion and Arf1/Arf6 codepletion having the greatest effect (pIRS1 and pAKT levels were reduced ∼81% in Arf6-depleted cells and ∼92% in cells lacking both Arf1 and Arf6) (Fig. 5D).
CNK1 depletion alters insulin-induced changes in plasma membrane phosphoinositide levels
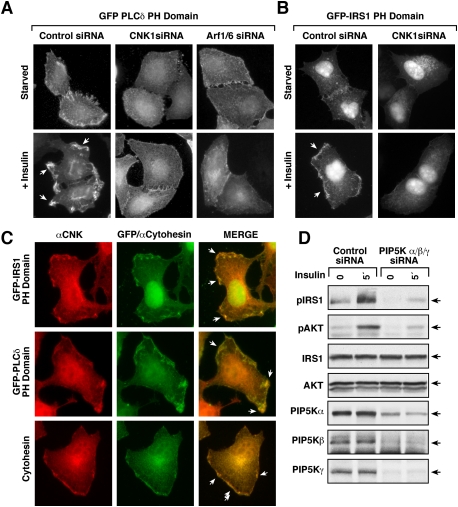
The above findings indicate that the CNK1/cytohesin interaction acts to promote Arf signaling, and further suggest that Arf activation is needed for insulin to efficiently recruit IRS1 to the cell surface. Activated Arf6 and Arf1 are known to function as allosteric activators of phosphatidylinositol 4-phosphate 5-kinases (PIP5Ks), which generate PIP2, a lipid second messenger that can function in the membrane recruitment of signaling proteins and is the substrate used by PI3K to generate PIP3. Therefore, we next examined the effect of CNK1 depletion on insulin-induced PIP2 generation using a GFP fusion protein containing the PH domain of PLCδ as a sensor for PIP2 levels. As shown in Figure 6A, insulin treatment strikingly increased the localization of the GFP-PLCδ PH domain to the plasma membrane, where strong punctate fluorescence was detected. Although some cell surface localization was seen in CNK1-depleted cells, little to no accumulation in discreet puncta was observed. As expected, depletion of Arf6/Arf1 also resulted in little accumulation of the PIP2 sensor in discreet puncta (Fig. 6A).
Figure 6.
CNK1 depletion inhibits insulin-induced changes in plasma membrane lipid microenvironments. (A, B) HeLa cells transfected with control or CNK1-siRNAs were subsequently transfected with constructs encoding either a GFP-PLCδ PH domain (A) or GFP-IRS1 PH domain (B). A GFP-PLCδ PH domain was also expressed in cells transfected with Arf1/Arf6 siRNAs. Localization of the GFP fusion proteins was then visualized in serum-starved and insulin-treated cells by immunofluorescent microscopy. The phenotypes shown were observed in >90% of cells expressing equivalent levels of the GFP fusion proteins. (C) Localization of the CNK1 and GFP-IRS1 PH domain, GFP-PLCδ PH domain, or cytohesin-2 was determined in insulin-treated cells by immunofluorescent staining. The phenotypes shown were observed in >80% of cells expressing equivalent levels of the GFP fusion proteins. (D) HeLa cells transfected with control or PIP5Kα/β/γ siRNAs were serum-starved and then treated or not with insulin prior to lysis. Lysates were examined as indicated.
The IRS adaptor proteins possess PH domains that are critical for their membrane recruitment, and, because the PH domains of the IRS proteins have been found to bind both PIP2 and PIP3 (Razzini et al. 2000; Vainshtein et al. 2001), we next examined the effect of CNK1 depletion on the localization of a GFP fusion protein containing the PH domain of IRS1. As shown in Figure 6B, and as reported previously (Razzini et al. 2000), the GFP-IRS1 PH domain was found to accumulate in membrane ruffles of insulin-treated cells; however, little membrane localization was observed in cells lacking CNK1. Cell staining experiments further revealed that CNK1 colocalized with both the GFP-IRS1 and GFP-PLCδ PH domain fusion proteins as well as cytohesin-2 in insulin-treated cells (Fig. 6C), indicating the presence of the CNK1 scaffold in these lipid-rich membrane microenvironments. Moreover, as further support that the CNK1/cytohesin interaction functions to stimulate signaling from membrane-localized Arf6 and Arf1 to the PIP5Ks in order to generate a local lipid-rich environment, depletion of the human type I PIP5K enzymes (α, β, γ) was found to inhibit IRS1 and AKT phosphorylation (Fig. 6D; Supplemental Fig. S4) to a level similar to that observed in CNK1-depleted cells.
CNK1 is required for IR to IRS/PI3K/AKT signaling but not for PDGF to PI3K/AKT signaling
The above findings suggest a signaling cascade whereby CNK1 acts through its interaction with the cytohesins to promote Arf activation, and that both Arf activation and PIP5K activity are needed to efficiently recruit IRS1 to the cell surface and instigate signaling through the PI3K/AKT pathway. Therefore, constitutively targeting IRS1 to the plasma membrane might bypass the need for this signaling cascade and, as a result, be unaffected by CNK1 depletion. To investigate this possibility, HeLa cell lines were generated that stably express either Flag-tagged WT-IRS1 or an IRS1 protein that is constitutively targeted to the plasma membrane by an N-terminal myristoylation motif (Myr-IRS1) (Supplemental Fig. S5). Cells were then depleted of endogenous CNK1 and monitored for insulin-mediated IRS1 and AKT activation. As shown in Figure 7A, CNK1 depletion still inhibited IRS1 and AKT phosphorylation in cells expressing the WT-IRS1 protein, whereas it had no significant effect in cells expressing the membrane targeted Myr-IRS1.
Figure 7.
CNK is required for IR to IRS/PI3K/AKT signaling but not for PDGF to PI3K/AKT signaling. (A) HeLa cells stably expressing vector control, WT-IRS1, or Myr-IRS1 were transfected with control or CNK1-siRNAs prior to serum starvation and insulin treatment. Lysates were prepared and examined by immunoblot analysis. (B) HeLa cells stably expressing PDGFR were transfected with control or CNK1-siRNAs prior to serum starvation and treatment with either insulin or PDGF. Lysates or membrane fractions were prepared and examined by immunoblot analysis. (C) Model for CNK1 function in insulin signaling (see the text for details).
Activation of many receptor tyrosine kinases (RTKs) initiates signaling through the PI3K/AKT pathway. However, in contrast to IR and IGF-1R that engage the PI3K/AKT pathway through the IRS adaptor proteins, the PDGF receptor (PDGFR) can directly activate the PI3K/AKT pathway due to autophosphorylation sites on the PDGFR itself that bind the PI3K p85 subunit. Therefore, to compare the effects of CNK1 depletion on insulin- and PDGF-mediated AKT activation, a stable HeLa cell line expressing human PDGFRβ was generated. The cells were then transfected with either control or CNK1-siRNAs, following which they were serum-starved and treated with either insulin or PDGF. As shown in Figure 7B, CNK1 depletion had no effect on the activation and phosphorylation of either receptor, nor did it alter PDGF-induced ERK activation. Strikingly, AKT activation was not significantly affected when the CNK1-depleted cells were treated with PDGF. In addition, cell fractionation experiments revealed that the PI3K p110 subunit was recruited efficiently to the plasma membrane in PDGF-treated CNK1-depleted cells, and that AKT recruitment was only minimally reduced (Fig. 7B). These findings are consistent with the ability of the PDGFR to directly engage the PI3K/AKT pathway, and indicate that CNK1 depletion primarily inhibits insulin-mediated PI3K/AKT signaling by impacting the ability of the IR to signal to IRS1. Moreover, these finding indicate that the contribution of CNK1 to RTK-mediated PI3K/AKT signaling will likely vary depending on the type of RTK and how the receptor engages the PI3/AKT pathway.
Discussion
To gain a better understanding of how the mammalian CNK proteins function in cell signaling, we took a proteomics approach to determine the binding partners present in individual CNK scaffold complexes. Not surprisingly, given the similar domain structure of the CNK family members, this analysis identified several common CNK-interacting proteins; however, it also revealed key differences in the CNK complexes that suggest important functional diversity. Aided by this proteomics study, here we define a specific function for CNK1 in insulin signaling.
Characterization of the CNK scaffold complexes by mass spectrometry revealed that members of the cytohesin family were the most abundant binding partners detected in any of the CNK complexes. The cytohesins were not previously known to interact with CNK proteins, and they showed selective binding to CNK1 and CNK2A. The cytohesins function as GEFs for the Arf GTPases, and possess a common domain structure consisting of an N-terminal CC domain, a central Sec-7 domain, and a C-terminal PH domain (Kolanus 2007). Of the four mammalian family members, cytohesin-4 is expressed primarily in peripheral blood leukocytes (Ogasawara et al. 2000) and was not detected in the CNK proteomics analysis, whereas peptides derived from cytohesin-1, cytohesin-2, and cytohesin-3 were detected in both CNK1 and CNK2A complexes. Binding of the cytohesins was found to be constitutive, mediated by a predicted CC domain present only in the C-terminal region of CNK1 and CNK2A. The cytohesins are also known to interact with several other signaling scaffolds, including GRASP/tamalin, CASP/Cybr/CYTIP, and GRSP1, and binding of the cytohesins to all of these scaffolds, including the CNKs, requires the cytohesin CC domain. Thus, it appears that a critical function of the CC domain is to mediate binding of the cytohesins to scaffold proteins, which in turn can localize the cytohesins to specific signaling environments.
In the case of the CNK1/cytohesin interaction, our studies indicate that CNK1 and its ability to bind the cytohesins is critical for insulin signaling. We found that CNK1 strongly accumulates at the cell surface in response to insulin treatment, and that, when CNK1 is depleted from cells, numerous defects in insulin signaling are observed. Most strikingly, despite no alterations in IR activation, membrane recruitment and activation of the PI3K/AKT pathway is inhibited. The loss of PI3K/AKT signaling observed in CNK1-depleted cells was found to be due to a defect in the membrane recruitment of IRS1, and could be restored by constitutively localizing IRS1 to the cell surface by the addition of an N-terminal myristoylation motif. Moreover, in cells lacking CNK1, the defects in signal transmission from IR to IRS1 and the PI3K/AKT cascade could be restored by re-expression of a siRNA-resistant WT-CNK1 protein, but not one lacking the cytohesin-binding domain, demonstrating the biological importance of the CNK1/cytohesin interaction.
Interestingly, this is not the first time that cytohesin function has been implicated in the insulin pathway. Previous studies using the cytohesin-specific inhibitor SecinH3 have shown that cytohesin activity is needed for insulin-mediated AKT activation, and that inhibition of cytohesin activity can lead to insulin resistance in mice (Fuss et al. 2006; Hafner et al. 2006). These studies, however, did not elucidate the precise mechanism of cytohesin function in insulin signaling. Here, we found that the CNK1 scaffold helps localize the cytohesins to the cell surface, such that they can modulate Arf GTPase activity to generate changes in the plasma membrane lipid environment. We found that not only was the insulin-mediated membrane recruitment of cytohesin-2 reduced in CNK1-depleted cells, so was cytohesin activity as measured by the GTP loading of Arf1 and Arf6. We also found that inhibition of cytohesin activity as well as Arf6 or Arf1/Arf6 depletion significantly reduced IRS1/PI3K/AKT signaling, and the defects in IRS1/PI3K/AKT signaling observed in CNK1-depleted cells could be rescued by expressing constitutively active Q67L-Arf6. Both Arf1 and Arf6 are known to be allosteric activators of PIP5K and phospholipase D (PLD) (Myers and Casanova 2008), which generate the lipid second messengers PIP2, and phosphatidic acid, respectively. The Arf-dependent membrane recruitment of PIP5K activates PIP5K and results in enhanced local production of PIP2, which can serve as a binding lipid for certain PH domain-containing proteins and is the substrate used by PI3K to generate PIP3.
Based on the following findings, we conclude that alterations in the localized generation of PIP2 leads to the deficits in insulin signaling observed in the CNK1-depleted cells. First, using a GFP-PLCδ PH domain fusion protein to monitor PIP2 levels, we found that PIP2 levels are reduced in CNK1-depleted cells to a similar extent as is observed in Arf6/Arf1-depleted cells, and that little to no intense areas of PIP2 generation are observed in response to insulin treatment. Second, as with CNK1 depletion, knockdown of PIP5Kα, PIP5Kβ, and PIP5Kγ protein levels were found to inhibit insulin-mediated IRS1 and AKT phosphorylation. CNK1 was also found to colocalize with cytohesin-2 and the PIP2 sensor at discreet membrane areas in insulin-treated cells. Moreover, we found that CNK1 colocalizes with a GFP-IRS1 PH domain fusion protein, and that the insulin-mediated membrane recruitment of this protein was severely impaired in CNK1-depleted cells. Previous studies have shown that the IRS1 PH domain is critical for the membrane recruitment of IRS1, and that it can bind both PIP2 and PIP3. Thus, we propose that, in these PIP2-rich membrane regions, the PIP2 would first act as a binding lipid for the IRS1 PH domain, allowing IRS1 to interact with the IR and become phosphorylated on effector binding sites that would mediate the membrane localization and activation of PI3K. The PIP2-rich regions would then provide the substrate for PI3K-mediated PIP3 generation, which in turn could recruit additional IRS1 proteins and serve as the binding lipid for the PH domains of PDK and AKT, thus mediating the translocation of these kinases to the plasma membrane for activation.
The insulin pathway is vital for energy metabolism and growth, and its dysregulation is a major contributor to human disease. Our findings here provide important new mechanistic insight regarding insulin pathway regulation, and define a function for CNK1 as key regulator of cytohesin function during insulin signaling. We found that the CNK1 scaffold facilitates the insulin-induced recruitment of the cytohesins to the plasma membrane, where they modulate Arf signaling to PIP5Ks, ultimately resulting in the generation of a PIP2-rich membrane microenvironment that is critical for IRS1/PI3K/AKT signaling (Fig. 7C). One aspect of CNK1 function that is yet to be determined deals with the mechanism by which CNK1 is recruited to the cell surface. Our results indicate that the insulin-induced translocation of CNK1 does not require cytohesin binding, in that the CNK1 mutant unable to interact with the cytohesins still localized to the plasma membrane in insulin-treated cells. Moreover, we were unable to detect an interaction between CNK1 and the IR by either mass spectrometry analysis or coimmunprecipitation assays, suggesting that it is unlikely that the activated IR directly recruits the CNK1 scaffold to the cell surface. CNK1 does contain PH and PDZ domains that might contribute to its membrane localization, or it is possible that other CNK1-binding partners may play a role in this process. Further experimentation will be required to resolve this issue, and to determine whether CNK1, through its other binding interactions, may have additional functions that contribute to insulin signaling.
Materials and methods
Antibodies, reagents, and DNA constructs
Insulin receptor IRβ, pIRS1, ERK2, Arf6, PIP5K1β, CNK1, and c-myc (9E10) antibodies were from Santa Cruz Biotechnology; CNK1 and PI3K p110α antibodies were from BD Biosciences; pIR, pAKT S473, pAKT T308, pS6 Kinase, pS6, pERK, AKT, S6 Kinase, S6, pPDGF receptor β, PIP5K1α, and PIP5K1γ antibodies were from Cell Signaling; IRS1, pTyr (4G10), and PDGFRβ were from Millipore; Arf1 antibody was from Abcam; pan-cytohesin antibody and insulin were from Sigma-Aldrich; cytohesin-2 was from Dr. J. Cassanova; HA antibody was from Covance; and ENPP1 antibody was from IMGENEX. The antibody recognizing the Pyo epitope has been described previously (Therrien et al. 1995). SecinH3 was a generous gift from Dr. A. Schmitz. Human CNK1 cDNA was provided by Dr. B. Seed, and human CNK2 cDNA constructs were from Dr. K.-L. Guan. CNK3 sequences were isolated from mouse brain mRNA by RT–PCR. Two copies of the Pyo epitope tag were added to the N terminus of the CNKs by PCR amplification, and sequences encoding the Pyo-CNK proteins were inserted into pcDNA3. Plasmids encoding Myc-cytohesin-2 (from J. Wilson); HA-Arf1 and HA-Arf6 (from P. Randazzo); Flag- IRS1 (from R. Roth); PDGFR (from J. Cooper); GST-GGA3 (from J. Parent); GFP-IRS1 PH domain (from T. Maffucci); and GFP-PLCδ PH domain (from T. Balla) were generous gifts. Pyo-ΔCBD-CNK1, Myc-ΔCC-cytohesin-2, and Myr-IRS1-Flag were generated by PCR amplification, and siRNA-resistant CNK1 proteins were generated by site-directed mutagenesis.
Cell culture and generation of stable cell lines
HeLa and 293T cells were cultured in DMEM containing 10% FBS and 1% L-glutamine; HepG2 cells were cultured in EMEM containing 10% FBS, 1% L-glutamine, and 1% nonessential amino acids; and MCF7 cells were cultured in DMEM containing 10% FBS and 1% sodium pyruvate. Stable HeLa cell lines expressing siRNA-resistant CNK1 proteins were generated by retroviral infection, and stable lines expressing Q67L-Arf6, WT-IRS1, Myr-IRS1, or PDGFR were generated by lentiviral infection.
Mass spectrometry analysis of CNK scaffold complexes
Cycling 293T cells expressing Pyo-tagged CNK proteins were lysed in low-salt lysis buffer (30 mM Tris-HCl at pH 8.0, 75 mM NaCl, 10% glycerol, 1% TritonX-100, 0.15 U/mL aprotinin, 1 mM phenylmethylsulfonyl fluoride [PMSF], 20 μM leupeptin, 5 mM sodium orthovanadate). The CNK protein complexes were isolated from cell lysates using covalently coupled anti-Pyo sepharose beads, and were washed extensively in lysis buffer prior to examination by SDS-PAGE and Coomassie Brilliant Blue staining (Bio-Rad). Proteins were extracted from the gel matix by trypsin digestion, and peptides were desalted using C18 Zip Tips (Millipore) prior to mass spectrometry analysis as described in Zofall et al. (2009).
Coimmunoprecipiation and Arf activity assays
Cells were lysed in either low-salt lysis buffer or NP40 lysis buffer (30 mM Tris at pH 8.0, 137 mM NaCl, 10% glycerol, 1% NP-40, 0.15 U/mL aprotinin, 1 mM PMSF, 20 μM leupeptin, 5 mM sodium vanadate), and lysates were clarified by centrifugation. Lysates were incubated with the appropriate antibody plus protein G sepharose beads or with the appropriate affinity agarose resin for 3 h at 4°C. Immune complexes were washed and analyzed by immunoblotting. A GST fusion protein containing the N-terminal sequences of GGA (1–316 amino acids) was immobilized on glutathione resin as described in Giguere et al. (2006), and Arf activity assays were performed as described in Hafner et al. (2006).
Depletion of endogenous proteins by RNAi
For siRNA-mediated depletion, Invitrogen Stealth siRNA oligonucleotides designed against human CNK1 Arf1/6 or PIP5Kα/β/γ were transfected into cells using the reverse transfection method and Lipofectamine RNAiMAX transfection reagent (Invitrogen) according to the manufacturer's instructions. For shRNA-mediated depletion, a lentivirus-based pLKO.1 plasmid expressing human CNK1 shRNA (RMM3981-9590005, Open Biosource) was used, and viral particles were generated with the MISSION lentiviral packaging mix (Sigma).
Immunofluorescence and cell fractionation
Cell fractionation was performed as described previously (Stokoe and McCormick 1997). The purity of the membrane and cytoplasmic fractions were monitored by the presence of tubulin (cytoplasmic) and H-Ras (membrane). Protein localization was determined by immunoflurorescent staining or GFP visualization using a Leica Microsystems fluorescent microscope.
Acknowledgments
We thank Dan Ritt and Suzanne Specht for excellent technical assistance, and members of the Laboratory of Cell and Developmental Signaling for helpful discussions and comments. This project was supported with federal funds from the National Cancer Institute, National Institutes of Health, DHHS, in part under contract N01-CO-1240.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1904610.
Supplemental material is available at http://www.genesdev.org.
References
- Anselmo AN, Bumeister R, Thomas JM, White MA 2002. Critical contribution of linker proteins to raf kinase activation. J Biol Chem 277: 5940–5943 [DOI] [PubMed] [Google Scholar]
- Belfiore A, Costantino A, Frasca F, Pandini G, Mineo R, Vigneri P, Maddux B, Goldfine ID, Vigneri R 1996. Overexpression of membrane glycoprotein pc-1 in mda-mb231 breast cancer cells is associated with inhibition of insulin receptor tyrosine kinase activity. Mol Endocrinol 10: 1318–1326 [DOI] [PubMed] [Google Scholar]
- Boulay PL, Cotton M, Melancon P, Claing A 2008. Adp-ribosylation factor 1 controls the activation of the phosphatidylinositol 3-kinase pathway to regulate epidermal growth factor-dependent growth and migration of breast cancer cells. J Biol Chem 283: 36425–36434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumeister R, Rosse C, Anselmo A, Camonis J, White MA 2004. Cnk2 couples ngf signal propagation to multiple regulatory cascades driving cell differentiation. Curr Biol 14: 439–445 [DOI] [PubMed] [Google Scholar]
- Cohen LA, Honda A, Varnai P, Brown FD, Balla T, Donaldson JG 2007. Active arf6 recruits arno/cytohesin gefs to the pm by binding their ph domains. Mol Biol Cell 18: 2244–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douziech M, Roy F, Laberge G, Lefrancois M, Armengod AV, Therrien M 2003. Bimodal regulation of raf by cnk in drosophila. EMBO J 22: 5068–5078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douziech M, Sahmi M, Laberge G, Therrien M 2006. A ksr/cnk complex mediated by hyp, a novel sam domain-containing protein, regulates ras-dependent raf activation in drosophila. Genes Dev 20: 807–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuss B, Becker T, Zinke I, Hoch M 2006. The cytohesin steppke is essential for insulin signalling in drosophila. Nature 444: 945–948 [DOI] [PubMed] [Google Scholar]
- Giguere P, Rochdi MD, Laroche G, Dupre E, Whorton MR, Sunahara RK, Claing A, Dupuis G, Parent JL 2006. Arf6 activation by gαq signaling: Gαq forms molecular complexes with arno and arf6. Cell Signal 18: 1988–1994 [DOI] [PubMed] [Google Scholar]
- Goldfine ID, Maddux BA, Youngren JF, Reaven G, Accili D, Trischitta V, Vigneri R, Frittitta L 2008. The role of membrane glycoprotein plasma cell antigen 1/ectonucleotide pyrophosphatase phosphodiesterase 1 in the pathogenesis of insulin resistance and related abnormalities. Endocr Rev 29: 62–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Schmitz A, Grune I, Srivatsan SG, Paul B, Kolanus W, Quast T, Kremmer E, Bauer I, Famulok M 2006. Inhibition of cytohesins by secinh3 leads to hepatic insulin resistance. Nature 444: 941–944 [DOI] [PubMed] [Google Scholar]
- Jaffe AB, Aspenstrom P, Hall A 2004. Human cnk1 acts as a scaffold protein, linking rho and ras signal transduction pathways. Mol Cell Biol 24: 1736–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AB, Hall A, Schmidt A 2005. Association of cnk1 with rho guanine nucleotide exchange factors controls signaling specificity downstream of rho. Curr Biol 15: 405–412 [DOI] [PubMed] [Google Scholar]
- Kitano J, Kimura K, Yamazaki Y, Soda T, Shigemoto R, Nakajima Y, Nakanishi S 2002. Tamalin, a pdz domain-containing protein, links a protein complex formation of group 1 metabotropic glutamate receptors and the guanine nucleotide exchange factor cytohesins. J Neurosci 22: 1280–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klarlund JK, Holik J, Chawla A, Park JG, Buxton J, Czech MP 2001. Signaling complexes of the ferm domain-containing protein grsp1 bound to arf exchange factor grp1. J Biol Chem 276: 40065–40070 [DOI] [PubMed] [Google Scholar]
- Kolanus W 2007. Guanine nucleotide exchange factors of the cytohesin family and their roles in signal transduction. Immunol Rev 218: 102–113 [DOI] [PubMed] [Google Scholar]
- Lanigan TM, Liu A, Huang YZ, Mei L, Margolis B, Guan KL 2003. Human homologue of drosophila cnk interacts with ras effector proteins raf and rlf. FASEB J 17: 2048–2060 [DOI] [PubMed] [Google Scholar]
- Maddux BA, Goldfine ID 2000. Membrane glycoprotein pc-1 inhibition of insulin receptor function occurs via direct interaction with the receptor α-subunit. Diabetes 49: 13–19 [DOI] [PubMed] [Google Scholar]
- Mansour M, Lee SY, Pohajdak B 2002. The n-terminal coiled coil domain of the cytohesin/arno family of guanine nucleotide exchange factors interacts with the scaffolding protein casp. J Biol Chem 277: 32302–32309 [DOI] [PubMed] [Google Scholar]
- Mitchell R, Robertson DN, Holland PJ, Collins D, Lutz EM, Johnson MS 2003. Adp-ribosylation factor-dependent phospholipase d activation by the m3 muscarinic receptor. J Biol Chem 278: 33818–33830 [DOI] [PubMed] [Google Scholar]
- Myers KR, Casanova JE 2008. Regulation of actin cytoskeleton dynamics by arf-family gtpases. Trends Cell Biol 18: 184–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevrivy DJ, Peterson VJ, Avram D, Ishmael JE, Hansen SG, Dowell P, Hruby DE, Dawson MI, Leid M 2000. Interaction of grasp, a protein encoded by a novel retinoic acid-induced gene, with members of the cytohesin family of guanine nucleotide exchange factors. J Biol Chem 275: 16827–16836 [DOI] [PubMed] [Google Scholar]
- Ogasawara M, Kim SC, Adamik R, Togawa A, Ferrans VJ, Takeda K, Kirby M, Moss J, Vaughan M 2000. Similarities in function and gene structure of cytohesin-4 and cytohesin-1, guanine nucleotide-exchange proteins for adp-ribosylation factors. J Biol Chem 275: 3221–3230 [DOI] [PubMed] [Google Scholar]
- Ohtakara K, Nishizawa M, Izawa I, Hata Y, Matsushima S, Taki W, Inada H, Takai Y, Inagaki M 2002. Densin-180, a synaptic protein, links to psd-95 through its direct interaction with maguin-1. Genes Cells 7: 1149–1160 [DOI] [PubMed] [Google Scholar]
- Rabizadeh S, Xavier RJ, Ishiguro K, Bernabeortiz J, Lopez-Ilasaca M, Khokhlatchev A, Mollahan P, Pfeifer GP, Avruch J, Seed B 2004. The scaffold protein cnk1 interacts with the tumor suppressor rassf1a and augments rassf1a-induced cell death. J Biol Chem 279: 29247–29254 [DOI] [PubMed] [Google Scholar]
- Rajakulendran T, Sahmi M, Kurinov I, Tyers M, Therrien M, Sicheri F 2008. Cnk and hyp form a discrete dimer by their sam domains to mediate raf kinase signaling. Proc Natl Acad Sci 105: 2836–2841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razzini G, Ingrosso A, Brancaccio A, Sciacchitano S, Esposito DL, Falasca M 2000. Different subcellular localization and phosphoinositides binding of insulin receptor substrate protein pleckstrin homology domains. Mol Endocrinol 14: 823–836 [DOI] [PubMed] [Google Scholar]
- Robertson DN, Johnson MS, Moggach LO, Holland PJ, Lutz EM, Mitchell R 2003. Selective interaction of arf1 with the carboxy-terminal tail domain of the 5-ht2a receptor. Mol Pharmacol 64: 1239–1250 [DOI] [PubMed] [Google Scholar]
- Roignant JY, Hamel S, Janody F, Treisman JE 2006. The novel sam domain protein aveugle is required for raf activation in the drosophila egf receptor signaling pathway. Genes Dev 20: 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santy LC, Casanova JE 2001. Activation of arf6 by arno stimulates epithelial cell migration through downstream activation of both rac1 and phospholipase d. J Cell Biol 154: 599–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokoe D, McCormick F 1997. Activation of c-raf-1 by ras and src through different mechanisms: Activation in vivo and in vitro. EMBO J 16: 2384–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang P, Cheng TP, Agnello D, Wu CY, Hissong BD, Watford WT, Ahn HJ, Galon J, Moss J, Vaughan M, et al. 2002. Cybr, a cytokine-inducible protein that binds cytohesin-1 and regulates its activity. Proc Natl Acad Sci 99: 2625–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therrien M, Chang HC, Solomon NM, Karim FD, Wassarman DA, Rubin GM 1995. Ksr, a novel protein kinase required for ras signal transduction. Cell 83: 879–888 [DOI] [PubMed] [Google Scholar]
- Therrien M, Wong AM, Rubin GM 1998. Cnk, a raf-binding multidomain protein required for ras signaling. Cell 95: 343–353 [DOI] [PubMed] [Google Scholar]
- Therrien M, Wong AM, Kwan E, Rubin GM 1999. Functional analysis of cnk in ras signaling. Proc Natl Acad Sci 96: 13259–13263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainshtein I, Kovacina KS, Roth RA 2001. The insulin receptor substrate (irs)-1 pleckstrin homology domain functions in downstream signaling. J Biol Chem 276: 8073–8078 [DOI] [PubMed] [Google Scholar]
- Venkateswarlu K 2003. Interaction protein for cytohesin exchange factors 1 (ipcef1) binds cytohesin 2 and modifies its activity. J Biol Chem 278: 43460–43469 [DOI] [PubMed] [Google Scholar]
- Yao I, Hata Y, Ide N, Hirao K, Deguchi M, Nishioka H, Mizoguchi A, Takai Y 1999. Maguin, a novel neuronal membrane-associated guanylate kinase-interacting protein. J Biol Chem 274: 11889–11896 [DOI] [PubMed] [Google Scholar]
- Ziogas A, Moelling K, Radziwill G 2005. Cnk1 is a scaffold protein that regulates src-mediated raf-1 activation. J Biol Chem 280: 24205–24211 [DOI] [PubMed] [Google Scholar]
- Zofall M, Fischer T, Zhang K, Zhou M, Cui B, Veenstra TD, Grewal SI 2009. Histone h2a.Z cooperates with rnai and heterochromatin factors to suppress antisense rnas. Nature 461: 419–422 [DOI] [PMC free article] [PubMed] [Google Scholar]