Abstract
Proteins of the 14-3-3 family regulate a divergent set of signalling pathways in all eukaryotic organisms. In this study, several cDNAs encoding 14-3-3 proteins were isolated from a cotton fibre cDNA library. The Gh14-3-3 genes share high sequence homology at the nucleotide level in the coding region and at the amino acid level. Real-time quantitative RT-PCR analysis indicated that the expression of these Gh14-3-3 genes is developmentally regulated in fibres, and reached their peak at the stage of rapid cell elongation of fibre development. Furthermore, overexpression of Gh14-3-3a, Gh14-3-3e, and Gh14-3-3L in fission yeast promoted atypical longitudinal growth of the host cells. Yeast two-hybrid analysis revealed that the interaction between cotton 14-3-3 proteins is isoform selective. Through yeast two-hybrid screening, 38 novel interaction partners of the six 14-3-3 proteins (Gh14-3-3a, Gh14-3-3e, Gh14-3-3f, Gh14-3-3g, Gh14-3-3h, and Gh14-3-3L), which are involved in plant development, metabolism, signalling transduction, and other cellular processes, were identified in cotton fibres. Taking these data together, it is proposed that the Gh14-3-3 proteins may participate in regulation of fibre cell elongation. Thus, the results of this study provide novel insights into the 14-3-3 signalling related to fibre development of cotton.
Keywords: 14-3-3, cotton (Gossypium hirsutum), fibre development, gene expression, protein–protein interaction
Introduction
Proteins of the 14-3-3 family were originally identified by Moore and Perez (Moore and Perez, 1967). The name 14-3-3 was given to the proteins in brain tissue of mammals on the basis of their migration pattern on starch gel electrophoresis. 14-3-3 proteins are small (27–32 kDa), acidic regulatory proteins that are highly conserved in all eukaryotes (Chaudhri et al., 2003). Highly functional conservation of 14-3-3 proteins was demonstrated by the successful functional complementation of four Arabidopsis homologues in yeast mutants (van Heusden et al., 1996). 14-3-3 isoforms are usually made up of homodimers and heterodimers that form a clamp-shaped structure to regulate their target proteins in cells (Yaffe et al., 1997). They participate in various signal transduction and regulatory processes by interacting with their targets in a sequence-specific and phosphorylation-dependent manner. A 14-3-3 dimer can interact with one protein at two different sites or interact with two target proteins at the same time by binding to phosphoserine/phosphothreonine motifs of the type RSXpSXP and RXY/FXpSXP (Braselmann and McCormick, 1995; Muslin et al., 1996). Through these binding reactions, 14-3-3 proteins regulate their target proteins by altering their localization, stability, phosphorylation state, activity, and molecular interactions (Bachman et al., 1996; Muslin and Xing, 2000).
Crystal structure data indicated that 14-3-3 proteins bind to multiple phosphoserine-containing targets through a conserved amphipathic groove (Liu et al., 1995; Muslin et al., 1996). The structural conservation of 14-3-3 isoforms does not extend to the N- and C-terminal regions, although their sequences are highly conserved (Liu et al., 1995; Petosa et al., 1998). A critical residue Lys49 (K49), which appears to be part of a binding interface with general importance for ligand binding, is identified in the amphipathic groove of mammalian 14-3-3ζ. The K49E mutation disrupts the interaction between 14-3-3ζ and Raf-1 kinase (Zhang et al., 1997). Previous studies indicated that C-terminal differences affected the interactions between 14-3-3s and their targets, and may be related to the isoform-specific function of 14-3-3 proteins (Shen et al., 2003; Visconti et al., 2008). Removal of the last 22 amino acid residues of GF14-6 increases binding to H+-ATPase and stimulation of its activity (Visconti et al., 2008). Another divergent region, the N-terminal region, is responsible for the dimerization of 14-3-3 proteins. An earlier study revealed that dimerization occurs through two N-terminal fragments, but not an N-terminus–C-terminus interaction of two 14-3-3 proteins (Jones et al., 1995).
In the years since their discovery, 14-3-3 proteins have risen to occupy a very important position in cell biology. It was demonstrated that 14-3-3 proteins play crucial roles in the control of many cellular functions (such as metabolism, signal transduction, cell cycle control, apoptosis, protein trafficking, transcription, stress responses, and malignant tumour transformation) (van Hemert et al., 2001). Likewise, plant 14-3-3 proteins participate in various cellular processes through binding to a wide range of transcription factors and signalling proteins. Previous studies revealed that 14-3-3 proteins are involved in the response to biotic and abiotic stress by directly interacting with signalling proteins (reviewed by Roberts et al., 2002). GF14c acts as a negative regulator of flowering in rice by interacting with Hd3a (Purwestri et al., 2009). Barley 14-3-3 proteins specifically interact with the catalytic A-subunit of V-ATPase, and are involved in both blue light perception and the subsequent activation of P-type ATPase (Klychnikov et al., 2007). A previous study on the interaction between H+-ATPase and 14-3-3 protein revealed that the binding of 14-3-3 protein to the autoinhibitory domain released the autoinhibitory action of H+-ATPase (Maudoux et al., 2000). The activation of H+-ATPase by the binding of 14-3-3 proteins in tobacco cells is negatively controlled by phosphorylation of two residues in the H+-ATPase 14-3-3 protein-binding site (Duby et al., 2009). Through interacting with AREB/ABF/ABI5 proteins which act as master signal mediators in the abscisic acid (ABA) signal transduction pathway, 14-3-3 proteins play important roles in controlling ABA signalling during barley seed germination (Schoonheim et al., 2007a). Tobacco 14-3-3 proteins negatively modulated RSG by sequestering it in the cytoplasm to respond to GA (gibberellic acid) signalling. In contrast, the mutant of RSG could not bind to 14-3-3 proteins and, consequently, it exhibited a higher activity than did wild-type RSG (Iqarashi et al., 2001; Ishida et al., 2004). It was believed that 14-3-3 proteins regulate the intracellular localization of the target proteins in plant cells. Previous studies revealed that 14-3-3 proteins mediate brassinosteroid (BR) signal transduction by regulating the nuclear export of a transcriptional factor BZR1 in Arabidopsis (Gampala et al., 2007; Ryu et al., 2007). Similarly, rice 14-3-3 proteins directly regulate OsBZR1 function in BR signalling by reducing its nuclear localization. Mutation of a 14-3-3-binding site in the OsBZR1 protein abolished its interaction with 14-3-3 proteins, resulting in an increased nuclear distribution of the protein (Bai et al., 2007).
Cotton (Gossypium hirsutum) is a widely grown textile crop which is cultivated mainly for its fibres. Cotton fibres, which are seed hairs, are single-cell trichomes that arise from the epidermal layer of the seed coat. Approximately 30% of the ovule epidermal cells elongate and develop into single-cell fibres at anthesis. Fibre development is a highly complicated and programmed process that can be divided into four distinct but overlapping phases: initiation [from 2 d before anthesis to 5 d post-anthesis (DPA)], cell elongation (1–20 DPA), secondary wall synthesis (16–40 DPA), and maturation (40–50 DPA) (Basra and Malik, 1984), associated with many functional proteins that are involved in signal transduction and transcriptional regulation. In recent years, many genes that are expressed in fibres have been identified in cotton. Up to now, however, the roles of only a few genes in fibre development have been investigated in detail. It was reported that the genes related to the actin and tubulin cytoskeletons are essential for fibre elongation (Li et al., 2005, 2007). The cotton sucrose synthase gene (Sus) plays an important role in fibre initiation and elongation. Delayed or insufficient Sus expression leads to delayed initiation and distinctly reduced fibre elongation (Ruan et al., 2005). GhMYB109, identified as an important transcription factor in cotton, participates in fibre development. Suppression of its expression results in a substantial reduction in fibre length (Pu et al., 2008). Although the 14-3-3 proteins participating in various signalling pathways have been well characterized in a few plants (such as Arabidopsis, rice, and barley), little is known so far about how the cotton 14-3-3 proteins regulate fibre development through interacting with target proteins. Our previous study reported that Gh14-3-3L is predominantly expressed in cotton fibre (Shi et al., 2007), but its role, especially how it interacts with other proteins, in fibre development still remains unclear. Here, isolation and characterization of six cotton 14-3-3s and their possible target proteins during fibre development are reported.
Materials and methods
Collection of plant materials
Cotton (G. hirsutum cv. Xuzhou142 and Coker312) seeds were surface sterilized with 70% (v/v) ethanol for 60 s and 10% (v/v) H2O2 for 1–2 h, followed by washing with sterile water. The sterilized seeds were germinated on half-strength Murashige and Skoog (MS) medium (pH 5.8) containing 0.8% agar under a 16 h light/8 h dark cycle at 28 °C for 5–6 d. Roots, cotyledons, and hypocotyls were collected from a portion of sterile seedlings. The remaining seedlings were transplanted into soil for further growth to maturation in the field, and the other tissues (such as leaves, stems, petals, anthers, ovules, and fibres) were derived from these cotton plants for RNA isolation.
Isolation of 14-3-3 cDNAs and their corresponding genes
To identify the genes that are functionally expressed in cotton fibres, >10 000 cDNA clones were randomly selected from a cotton fibre cDNA library constructed by Li et al. (2002) for sequencing. Some cotton 14-3-3 cDNAs with partial or full-length sequences were identified from these clones. Subsequently, a 400 bp fragment of each Gh14-3-3 partial sequence (including the partial coding region and 3'-untranslated region) was labelled with [α-32P]dCTP and used as probe to screen the cotton fibre cDNA library according to standard procedures. A total of 2×105 cDNA clones were screened, and >50 clones were identified. Among them, the full-length clones were sequenced and analysed. In total, six unique clones with complete Gh14-3-3 cDNA sequences were obtained.
The corresponding Gh14-3-3 genes were amplified from the genomic DNA of cotton by PCR, using Pfu DNA polymerase and gene-specific primers that were designed according to the sequence of each Gh14-3-3 cDNA (Supplementary Table S1 available at JXB online). In total, four Gh14-3-3 genes were obtained.
RNA isolation and real-time quantitative RT-PCR analysis
Total RNA was isolated from roots, hypocotyls, cotyledons, leaves, petals, anthers, 10 DPA ovules, and developing fibres (3–20 DPA) of cotton by the method described previously (Li et al., 2002). A 2–4 g aliquot of each cotton tissue was randomly collected from 3–10 plants (see ‘Collection of plant materials’) for RNA isolation. The concentration and purity of total RNA were identified by NanoDrop spectrophotometry and agarose gel electrophoresis. RNA samples were stored at – 80 °C until use.
Expression profiling of the Gh14-3-3 genes in different cotton tissues (such as roots, hypocotyls, cotyledons, leaves, petals, anthers, 10 DPA ovules, and 10 DPA fibres) and during different stages of fibre development (including 3, 5, 6, 9, 10, 12, 15, 18, and 20 DPA fibres) was carried out by real-time quantitative RT-PCR using the fluorescent intercalating dye SYBR-Green in the detection system (Option 2, MJ Research, Waltham, MA, USA). A cotton polyubiquitin gene (GhUBI1, GenBank accession no. EU604080) was used as a standard control in the RT-PCRs. A two-step RT-PCR procedure was performed in all experiments using a previously described method (Li et al., 2005). In brief, total RNA was reverse transcribed into cDNA and used as a template in real-time PCRs with gene-specific primers (Table 1). The real-time PCR was performed using SYBR-Green Real-time PCR Master Mix according to the manufacturer's instructions (Toyoba Co. Ltd, Osaka, Japan). The Ct (cycle threshold), defined as the PCR cycle at which a statistically significant increase of reporter fluorescence is first detected, is used as a measure of the starting copy number of the target gene. The relative quantity of the target Gh14-3-3 expression level was determined using the comparative Ct method. The relative value for the expression level of each Gh14-3-3 gene was calculated by the equation Y=10ΔCt/3.75×100% (ΔCt is the differences in Ct between the control GhUBI1 products and the target Gh14-3-3 products, i.e. ΔCt=CtGhUBI1−CtGh14-3-3, and 3.75 is a parameter for the cycle number which represents a 10-fold expression difference between the target gene and control gene). To achieve optimal amplification, PCR conditions for every primer combination were optimized for annealing temperature and Mg2+ concentration. PCR products were confirmed on an agarose gel. The efficiency of each primer pair was detected by using Gh14-3-3 cDNA as standard templates, and the RT-PCR data were normalized with the relative efficiency of each primer pair.
Table 1.
Primer pairs used in gene-specific RT-PCR of the six Gh14-3-3 genes
| Gene no. | Primers |
| Gh14-3-3L | 5′-GCAGATGAACCTCAGGCTGAGAG-3′ |
| 5′-GTCATAAAGACTATAAAGTGTAAC-3′ | |
| Gh14-3-3a | 5′-GGATTTGTGACCTATGGCTTG-3′ |
| 5′-CCACACTCTAGGGAGCTGCAT-3′ | |
| Gh14-3-3e | 5′-CAACTTCTCCGGGACAACCTG-3′ |
| 5′-CGCACTATTTTAGTACTAGCATG-3′ | |
| Gh14-3-3f | 5′-GCTTGACACGCTGGGAGAAGAG-3′ |
| 5′-TAGAGTATAGCATCATGCCATG-3′ | |
| Gh14-3-3g | 5′-ATTGGGTGAGGAATCCTACAAG-3′ |
| 5′-ACCCAAATTCATGTCCGAATGG-3′ | |
| Gh14-3-3h | 5′-GCTTTTAAGGGACAACCTTAC-3′ |
| 5′-TTGATCACAGAGGAAGGATg-3′ | |
| GhUBI1 | 5′-CTGAATCTTCGCTTTCACGTTATC-3′ |
| 5′-GGGATGCAAATCTTCGTGAAAAC-3′ |
DNA sequencing and protein analysis
The sequences of the isolated cotton 14-3-3 genes (cDNAs) and their deduced proteins were analysed using DNAstar software (DNAstar Inc., Madison, WI, USA), and protein sequence homology analysis was performed with ClustalW (http://www.ebi.ac.uk/clustalw/). Phylogenetic analysis was employed to investigate the evolutionary relationships among the Gh14-3-3 proteins and Arabidopsis 14-3-3 proteins. A minimum evolution tree was generated in MEGA3.1 (Kumar et al., 2004). A bootstrap analysis with 1000 replicates was performed to assess the statistical reliability of the tree topology.
Overexpression of Gh14-3-3 genes in fission yeast
The open reading frame (ORF) of each Gh14-3-3 gene was amplified by PCR using Pfu DNA polymerase with gene-specific primers (Supplementary Table S2 at JXB online), and was cloned into the yeast vector pREP5N (Alfa et al., 1993) with XhoI/BamHI sites. Then the construct was transferred into yeast (Schizosaccharomyces pombe) cells by electroporation (Bio-Rad MicroPulser Electroporation Apparatus, USA) according to the manufacturer's instructions. Overexpression of Gh14-3-3 genes in the transformed yeast cells was induced by the method described previously (Li et al., 2007). The transformants were selected on plates containing minimal medium (MM) with 2 μM thiamine at 30 °C. Then 10 colonies for each construct were picked out to incubate in liquid MM with 2 μM thiamine, which represses the nmt-1 promoter activity, until mid-log phase in a shaker (220 rpm, 30 °C). Subsequently, the yeast cells were harvested and washed three times with MM without thiamine to de-repress the promoter, and then incubated in the same thiamine-free MM for 20 h (220 rpm, 30 °C). The yeast cells were observed and photographed under a Nikon microscope (Japan), and the lengths of 50 cells per transformant (single colony) were measured for statistical evaluation of the cell elongation, using empty pREP-5N transformants as controls.
Yeast two-hybrid analysis
For directed yeast two-hybrid assays of protein–protein interaction between cotton 14-3-3s, the coding sequences of Gh14-3-3 genes amplified by PCR using Pfu DNA polymerase and gene-specific primers (Supplementary Table S3 at JXB online) were cloned into the NdeI/BamHI restriction sites of yeast two-hybrid vectors pGBKT7 (bait vector) and pGADT7 (prey vector), creating fusions to the binding domain and activation domain of the yeast transcriptional activator GAL4, respectively. Each pGBKT7-Gh14-3-3 construct was introduced singly into the yeast strain Y187 using the high-efficiency lithium acetate transformation procedure (Gietz et al., 1992), and each pGADT7-Gh14-3-3 construct was transferred into the yeast strain AH109. Mating reactions were performed between the two haploid strains containing pGBKT7-Gh14-3-3 and pGADT7-Gh14-3-3 constructs, respectively, and plated on double dropout medium (DDO medium, SD/–Leu/–Trp) (BD Biosciences Clontech, Palo Alto, CA, USA). The transformants were further streaked on quadruple dropout medium (QDO medium, SD/–Trp/–Leu/–His/–Ade) and confirmed with colour change on a β-galactosidase filter paper using the flash-freezing filter assay (James et al., 1996).
A yeast two-hybrid library of cotton fibre cDNAs from mRNAs of 10 DPA fibres was constructed, using the Clontech BD Matchmaker Library Construction and Screening Kits according to the manufacturer's instruction (BD Biosciences Clontech). The first-strand cDNAs were reversely synthesized by reverse transcriptase and amplified into double-stranded cDNAs using long-distance PCR. pGADT7-Rec vector (prey vector) and cotton fibre cDNAs were co-transferred into yeast strain AH109, and then the yeast cells were cultivated on SD/−Leu medium at 30 °C for 5 d for selection of transformants. Finally, the transformed cells as the prey library were suspended in fresh SD/–Leu medium and stored as glycerol stock at −80 °C.
For screening target proteins of cotton 14-3-3s, each Gh14-3-3 bait clone was mated with the prey library by incubation for 24 h at 30 °C, and positive clones were selected on QDO medium (SD/–Trp/–Leu/–His/–Ade) at 3 °C. After further cultivation for 7 d, the independent positive clones were restreaked on QDO 2–3 times and LacZ activity was further tested by the flash-freezing filter assay (James et al., 1996).
To analyse interactions of each Gh14-3-3 and its targets, the selected cotton cDNA clones that showed activation of both reporter genes were then inoculated in DDO liquid medium for plasmid DNA isolation by using the Yeast Plasmid Isolation kit (Tiangen Biotech Co. Ltd, Beijing, China). The isolated plasmid DNAs (pGADT7 vector containing cotton cDNA fragments) were then transferred into bacteria (Escherichia coli) strain DH5α on LB medium with 100 mg l−1 ampicillin. The cDNA clones were sequenced by Sunny Biotech. Co. Ltd (Shanghai, China). Similarities of cDNA sequences were analysed by blasting against the NCBI public databases using the BLASTX program and the DFCI Cotton Gene Index (http://compbio.dfci.harvard.edu/cgi-bin/tgi/gimain.pl?gudb=cotton) using the blastn program. Expressed sequence tags (ESTs) of each gene were collected and classified.
Results
Isolation and characterization of Gh14-3-3 cDNAs
To isolate the genes that might be involved in the regulation of fibre development, >10 000 cDNA clones from the cotton fibre cDNA library constructed by Li et al. (2002) were randomly sequenced. More than 30 14-3-3 cDNA clones were identified from these clones. Among them, three clones contain full-length 14-3-3 cDNA sequences, and the remainder have partial 14-3-3 sequences. Subsequently, another three complete sequences were obtained by screening the fibre cDNA library using 400 bp fragments (including the partial coding region and 3'-untranslated region) of each Gh14-3-3 cDNA as probes (see Materials and methods). In total, six unique full-length cDNAs encoding putative 14-3-3 proteins were identified, including one cDNA (Gh14-3-3L) that was previously reported (Shi et al., 2007) and the remainder designated as Gh14-3-3a, Gh14-3-3e, Gh14-3-3f, Gh14-3-3g, and Gh14-3-3h (GenBank accession nos: EU189220, EU189224, GU451708, GU451709, and GU451710). As shown in Fig. 1, sequence analysis predicted that two Gh14-3-3 genes (Gh14-3-3e and Gh14-3-3g) encode a 262 amino acid polypeptide, whereas the other four (Gh14-3-3L, Gh14-3-3a, Gh14-3-3f, and Gh14-3-3h) encode polypeptides consisting of 253, 251, 252, and 258 amino acids, respectively. The Gh14-3-3 genes share high sequence homology at the nucleotide level (62−91% identity) in the coding region and at the amino acid level (62−96% identity). Sequence alignment revealed that all of the predicted proteins encoded by these genes contain nine highly conserved antiparallel α-helices (Fig. 1). In contrast, both the N- and the C-terminal region of Gh14-3-3 proteins display relatively high variability (Fig. 1).
Fig. 1.
Sequence alignment among the six Gh14-3-3 proteins. Sequences of Gh14-3-3s were aligned. The amino acid residues identical among the sequences are indicated in black, while similar residues are shown in grey, and the regions of the nine conserved antiparallel α-helices (α1–α9) are underlined.
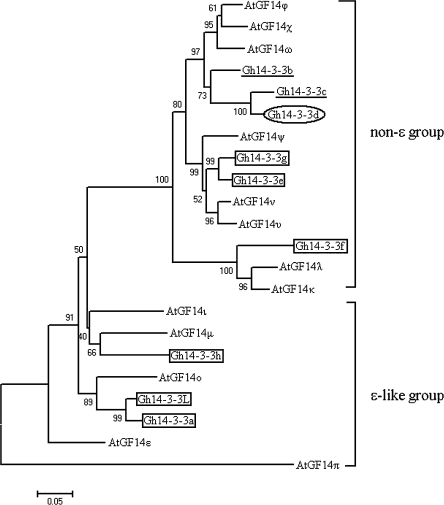
Phylogenetic relationship of Gh14-3-3 proteins
To investigate the evolutionary relationships of the cotton 14-3-3 proteins with other 14-3-3 proteins, all of the known Arabidopsis 14-3-3s and cotton 14-3-3s were selected from GenBank for phylogenetic analysis. Plant 14-3-3 proteins can be divided into the ε-like group and the non-ε group (Wu et al., 1997; Sehnke et al., 2002a, b). As shown in Fig. 2, these proteins obviously split into two subgroups. Gh14-3-3L, Gh14-3-3a, and Gh14-3-3h together with the Arabidopsis ε-like isoforms form one subgroup, while Gh14-3-3b, Gh14-3-3c, Gh14-3-3d, Gh14-3-3e, Gh14-3-3f, and Gh14-3-3g are located in the non-ε branch of the tree. These results suggest that cotton contains both ε-like and non-ε 14-3-3 proteins, and the divergence in the ε-like and non-ε group could have occurred before the differentiation of the two species. One protein pair (Gh14-3-3e and Gh14-3-3g) share high sequence homology, resulting in an independent branch which is basal to the AtGF14ν/μ clade, indicating that the divergence between Gh14-3-3e and Gh14-3-3g occurred relatively late during evolution. Likewise, Gh14-3-3a and Gh14-3-3L form a distinct clade which is basal to the AtGF14° clade, suggesting that the two isoforms have a close evolutionary relationship. On the other hand, Gh14-3-3f occupies a distinct branch that is basal to the AtGFλ/κ clade, implying that 14-3-3f diverged earlier from the other cotton 14-3-3s. In addition, cotton 14-3-3b, 14-3-3c (Wei et al., 2009), and 14-3-3d (unpublished data), which are preferentially expressed in roots, form another independent branch basal to Arabidopsis GF14ω, GF14ϕ, and GF14χ in the non-ε group.
Fig. 2.
Phylogenetic relationship of cotton 14-3-3 proteins to Arabidopsis 14-3-3 proteins. The minimum evolution tree was constructed in MEGA3.1 from 1000 bootstrap replicates. The accession numbers of cotton and Arabidopsis 14-3-3 proteins in GenBank are: Gh14-3-3a (EU189220), Gh14-3-3b (EU189221), Gh14-3-3c (EU189222), Gh14-3-3d (EU189223), Gh14-3-3e (EU189224), Gh14-3-3f (GU451708), Gh14-3-3g (GU451709), Gh14-3-3h (GU451710), Gh14-3-3L (DQ402076), AtGF14κ (AAD51783), AtGF14λ (AAD51781), AtGF14χ (AAA96254), AtGF14ϕ (AAB62224), AtGF14ω (AAA96253), AtGF14ν (AAD51782), AtGF14υ (AAB62225), AtGF14ψ (AAA96252), AtGF14μ (AAD51784), AtGF14ι (AAK11271), AtGF14° (AAG47840), AtGF14ε (AAD51785), and AtGF14π (NP-565174).
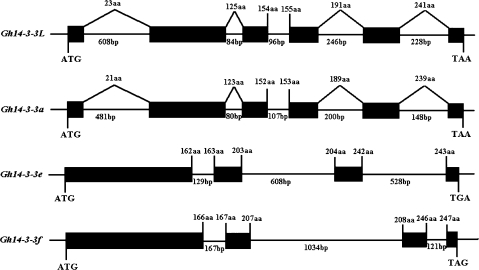
Isolation and characterization of Gh14-3-3 genes
Four Gh14-3-3 genes (14-3-3L/a/e/f) were amplified from the cotton (G. hirsutum) genome by PCR, using gene-specific primers. Sequence comparison between the cDNAs and their corresponding genes revealed that ε-like and non-ε 14-3-3 genes share highly conserved structures (as shown in Fig. 3). Gh14-3-3a and Gh14-3-3L which belong to the ε-like group contain five introns splitting their ORF into six exons. The intron positions are identical (within glutamate, within methionine, between glutamate and alanine, within arginine, and within glycine, respectively) in the two genes, although the length of each intron is slightly variable in both genes. In contrast, Gh14-3-3e and Gh14-3-3f belonging to the non-ε group contain three introns splitting their ORF into four exons. The positions of all three introns are conserved. The first intron is located between codons encoding glutamine and aspartate, and the second intron is positioned between codons encoding glutamine and alanine in the two genes. However, the third intron is inserted between codons encoding theonine and aspartate in Gh14-3-3e, but between codons encoding glutamine and aspartate in Gh14-3-3f.
Fig. 3.
The structures of four Gh14-3-3 genes. Exons are denoted by black boxes. Introns, 5′-flanking regions, and 3′-UTRs are denoted by lines. The length of the intron in base pairs is indicated. The position of substitution is denoted by a diagonal line.
Expression of Gh14-3-3 genes is developmentally regulated in fibres
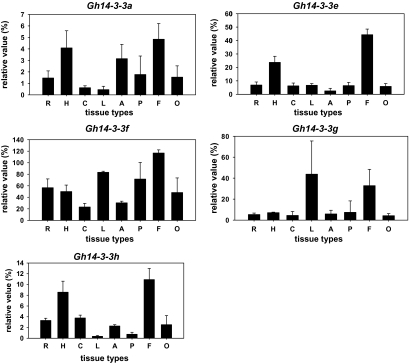
To analyse the expression patterns of the isolated Gh14-3-3 genes, mRNA levels of these genes in cotton tissues were quantified by real-time quantitative SYBR-Green RT-PCR using gene-specific primers (Table 1). As shown in Fig. 4, four genes (Gh14-3-3a, Gh14-3-3e, Gh14-3-3f, and Gh14-3-3h) showed fibre-preferential expression patterns, while the other one (Gh14-3-3g) exhibited its relatively high expression level in fibres. Gh14-3-3a was preferentially expressed in both fibres and hypocotyls, and showed relatively high expression levels in anthers, whereas its expression activity was relatively lower in other tissues. Strong expression of the Gh14-3-3e gene was detected in fibres, and moderate expression was found in hypocotyls, but relatively weak signals were detected in other tissues such as roots, cotyledons, leaves, anthers, petals, and ovules. Gh14-3-3h also displayed a similar expression pattern to Gh14-3-3e, but its peak value (10.91) in fibres is much lower than that (44.35) of Gh14-3-3e. Gh14-3-3f was expressed at the highest level in fibres, and at relatively high levels in leaves and petals, but moderate to weak expression was detected in other tissues. On the other hand, Gh14-3-3g was preferentially expressed in leaves, and showed a relatively high expression level in fibres, but its activity was very low in other tissues. In addition, a previous study revealed that the Gh14-3-3L gene was expressed at high levels in fibres, ovules, and petals, but at low levels in the other tissues (Shi et al., 2007).
Fig. 4.
Real-time quantitative RT-PCR analysis of expression of Gh14-3-3 genes in cotton tissues. Relative values of Gh14-3-3 gene expression in cotton tissues are shown as a percentage of GhUBI1 expression activity. R, roots; H, hypocotyls; C, cotyledons; L, leaves; A, anthers; P, petals; F, 10 DPA (days post-anthesis) fibres; O, 10 DPA ovules.
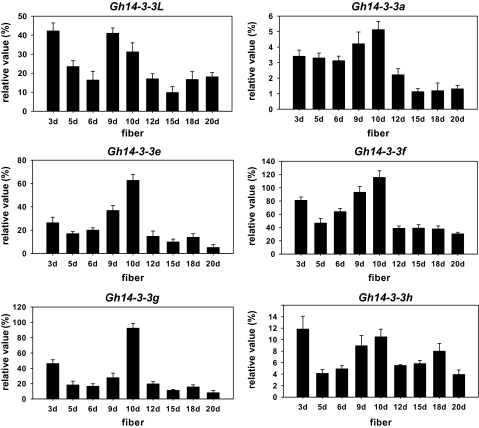
To investigate further the expression profiling of these 14-3-3 genes in fibre development, mRNA accumulation of the Gh14-3-3 genes during fibre development was determined by quantitative RT-PCR (Fig. 5). The experimental results revealed that Gh14-3-3a, Gh14-3-3e, Gh14-3-3f, and Gh14-3-3g were expressed at relatively high levels in the early stage (3 DPA) of fibre development, and their transcripts reached the highest levels in 9–10 DPA fibre cells. As fibre developed further, the expression levels of these genes declined dramatically. Similarly, Gh14-3-3L and Gh14-3-3h genes were also expressed at high levels in early stages (3–10 DPA) of fibre development, but the expression activities of both genes gradually declined to relatively low levels with further development of fibre cells (12–20 DPA). The results suggested that the expression of these 14-3-3 genes was developmentally regulated in fibre cells of cotton.
Fig. 5.
Expression of Gh14-3-3 genes during fibre development. Relative values of Gh14-3-3 gene expression in developing fibres of cotton are shown as a percentage of GhUBI1 expression activity. 3d, 5d, 6d, 9d, 10d, 12d, 15d, 18d, and 20d refer to 3, 5, 6, 9, 10, 12, 15, 18, and 20 DPA fibres.
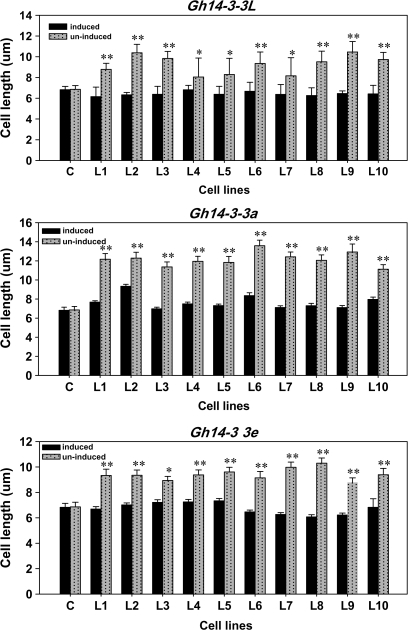
Overexpression of Gh14-3-3 genes in yeast promotes cell elongation
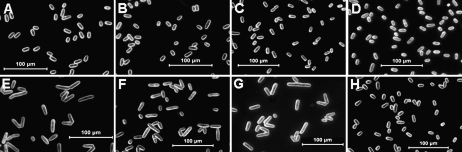
To investigate whether the Gh14-3-3 isoforms play a role in cell elongation, three cotton 14-3-3 genes were heterologously expressed in yeast (S. pombe) cells. Two genes (Gh14-3-3a and Gh14-3-3L) represent the ε-like group, while the other one (Gh14-3-3e) represents the non-ε group of the cotton 14-3-3 family. The coding sequences of the three Gh14-3-3 genes were cloned separately into the yeast vector pREP5N and transferred into yeast cells. As shown in Fig. 6, morphological studies indicated that there were significant differences in cell length between the transformed cell lines and controls under induction conditions. Transformed cells expressing Gh14-3-3 genes grown in induction medium (Fig. 6E–G) were remarkably longer than those harbouring the same vector but grown in uninduced conditions (Fig. 6A–C). In contrast, control cells with the empty pREP5N vector displayed normal length in both induced (Fig. 6H) and uninduced conditions (Fig. 6D). Measurement of cell length of the control and 10 randomly selected transformed yeast cell lines revealed that the length of the Gh14-3-3a-transformed cells was 1.6- to 2.0-fold greater than that of the control cells, while the length of Gh14-3-3e- and Gh14-3-3L-transformed cells was 1.2- to 1.5-fold longer than that of the control cells. Statistical analysis indicated that there were significant differences in cell length between the transformed lines and control (P-value <0.05 or <0.01 by t-test) (Fig. 7). On the other hand, statistical analysis demonstrated that there was no significant difference in the percentage of cell division between the transformed lines and controls (data not shown). The results suggested that overexpression of the cotton 14-3-3 genes stimulates the longitudinal growth of the host cells.
Fig. 6.
Overexpression of Gh14-3-3 genes stimulated the longitudinal growth of fission yeast cells. Micrographs were taken using dark-field microscopy. Yeast cells harbouring the constructed vectors pREP5N-Gh14-3-3L (A and E), pREP5N-Gh14-3-3a (B and F), and pREP5N-Gh14-3-3e (C and G), and empty plasmid (pREP5N; D and H) were cultured in non-induction medium (A–D) or induction medium (E–H). Bar=100 μm.
Fig. 7.
Statistical analysis of the length of various fission yeast cell lines. C, controls (yeast cells with empty vectors); L1–L10, transformed cell lines (yeast cells harbouring Gh14-3-3 genes). A single asterisk represents a significant difference (P-value <0.05), and double asterisks represent very significant differences (P-value <0.01) between the transformed cell lines and the control by t-test. The experiments were repeated three times; means represent average values of cell length (n=100 cells each line), and bars show standard errors.
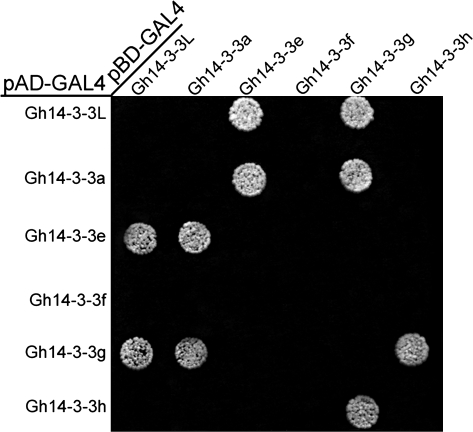
Gh14-3-3s display isoform selectivity in protein–protein interaction
To understand how cotton 14-3-3 proteins interact with each other, yeast two-hybrid technology was employed to analyse the interactions among these Gh14-3-3 proteins. The ORF of each Gh14-3-3 gene was cloned into pGBKT7 which contains the GAL4-binding domain, and into pGADT7 which contains the GAL4-activation domain. Empty vectors containing the activation domain or binding domain were used as negative controls. Gh14-3-3 proteins showed no self-activation of transcription. As shown in Fig. 8, Gh14-3-3a interacted with both Gh14-3-3e and Gh14-3-3g, and Gh14-3-3L also interacted with these two proteins, while Gh14-3-3g could interact with three proteins (Gh14-3-3a, Gh14-3-3h, and Gh14-3-3L). However, Gh14-3-3f did not interact with any of the other five Gh14-3-3 proteins. In addition, interaction of the same 14-3-3 proteins was not detected in the yeast two-hybrid assays (Fig. 8). These results suggest that each of the six cotton 14-3-3s may display isoform selectivity in formation of heterodimers, and may not combine into its homodimer in cells during fibre development of cotton.
Fig. 8.
Isoform-specific interactions among the Gh14-3-3 proteins. The coding sequences of the Gh14-3-3 genes were cloned into the yeast two-hybrid vectors pGADT7 and pGBKT7, and introduced into yeast cells (see Materials and methods). Interactions among the Gh14-3-3 proteins were analysed by yeast mating. Transformants were assayed for growth on QDO nutritional selection medium.
Interaction partners of Gh14-3-3 proteins identified in a yeast two-hybrid screen
To identify interaction partners of cotton 14-3-3 proteins, yeast two-hybrid analysis was performed using the six isolated Gh14-3-3 proteins as bait to screen the two-hybrid library of cotton fibre cDNAs constructed on the prey vector. Each of the six 14-3-3 proteins was used in a separate screen. As shown in Table 2, 38 unique proteins (the sequences are provided in Supplementary Table S4 at JXB online) were identified as positive clones in these six screens for both of the reporter genes ADE and LacZ. All positive clones were checked for the presence of a cDNA–AD fusion and further confirmed in the one-to-one interaction analysis.
Table 2.
Identified cDNA clones from the Gh14-3-3 yeast two-hybrid screen
| Spot no. | Gh14-3-3 proteins used as bait | Homologous protein and species | E-value | Homology | Score | Total ESTs | Percentage of ESTs expressed in fibre |
| 1 | L, e, h | Nucleic acid-binding protein (Ricinus communis) | 6e-52 | Id. 85%; Po. 94% | 207 | 57 | 80.7 |
| 2 | e | Putative R2R3-Myb transcription factor (Citrus sinensis) | 9e-38 | Id. 61%; Po. 69% | 160 | 98 | 53.1 |
| 3 | L, e, f, g, h | BRASSINAZOLE-RESISTANT 1 protein (Ricinus communis) | 2e-66 | Id. 83%; Po. 91% | 254 | 78 | 73.1 |
| 4 | h | Auxin-repressed protein (Gossypium barbadense) | 5e-40 | Id. 68%; Po. 81% | 167 | 218 | 59.6 |
| 5 | h | Abscisic acid responsive element-binding protein 2 (Populus suaveolens) | 2e-31 | Id. 69%; Po. 80% | 138 | 65 | 86.2 |
| 6 | g | 14-3-3h protein (Gossypium hirsutum) | 2e-113 | Id. 100%; Po. 100% | 412 | 139 | 79.1 |
| 7 | e | 14-3-3a protein (Gossypium hirsutum) | 3e-136 | Id. 99%; Po. 99% | 488 | 141 | 80.1 |
| 8 | a, f, h | 40S Ribosomal protein S20 (Ricinus communis) | 4e-25 | Id. 100%; Po. 100% | 117 | 76 | 82.9 |
| 9 | g | 40S Ribosomal protein S20 (Ricinus communis) | 1e-24 | Id. 100%; Po. 100% | 115 | 103 | 85.4 |
| 10 | L, a, e | 26S Proteasome regulatory subunit 7, psd7 (Ricinus communis) | 1e-99 | Id. 91%; Po. 95% | 272 | 103 | 28.2 |
| 11 | a, g | Ubiquitin (Gossypium hirsutum) | 1e-28 | Id. 96%; Po. 98% | 129 | 14 | 78.6 |
| 12 | h | Aspartic proteinase (Theobroma cacao) | 1e-58 | Id. 82%; Po. 95% | 278 | 321 | 50.2 |
| 13 | h | KEU (keule); protein transporter (Arabidopsis thaliana) | 2e-43 | Id. 73%; Po. 83% | 178 | 4 | 75 |
| 14 | f, h | Sulphite oxidase (Hibiscus cannabinus) | 2e-100 | Id. 92%; Po. 96% | 368 | 32 | 71.9 |
| 15 | f | Multicopper oxidase (Ricinus communis) | 1e-71 | Id. 84%; Po. 90% | 272 | 52 | 78.8 |
| 16 | f | Cytosolic phosphoglucomutase (Solanum tuberosum) | 1e-46 | Id. 93%; Po. 96% | 189 | 74 | 64.9 |
| 17 | f | Transferase (Ricinus communis) | 2e-05 | Id. 52%; Po. 67% | 52.0 | 44 | 75 |
| 18 | g, h | Carbonic anhydrase (Gossypium hirsutum) | 2e-42 | Id.45%; Po. 62% | 176 | 495 | 39.4 |
| 19 | h | Neutral/alkaline invertase (Coffea Arabica) | 2e-13 | Id. 94%; Po. 97% | 78.6 | 96 | 51 |
| 20 | L, e, f, h | Autoinhibited H+ ATPase (Populus trichocarpa) | 1e-11 | Id. 98%; Po. 98% | 73.2 | 78 | 48.7 |
| 21 | L, e, g, h | ATP-binding protein (Ricinus communis) | 3e-46 | Id. 88%; Po. 92% | 187 | 112 | 58.9 |
| 22 | h | Signal transducer (Ricinus communis) | 2e-47 | Id. 75%; Po. 83% | 191 | 41 | 63.4 |
| 23 | h | Signal transducer (Ricinus communis) | 3e-16 | Id. 37%; Po. 47% | 89.0 | 211 | 46.4 |
| 24 | h | Calmodulin-binding protein (Ricinus communis) | 2e-42 | Id. 56%; Po. 68% | 176 | 102 | 84.3 |
| 25 | h | Plant synaptotagmin (Populus trichocarpa) | 2e-39 | Id. 83%; Po. 93% | 164 | 31 | 74.2 |
| 26 | L, a, e, f | Root phototropism protein (Ricinus communis) | 7e-65 | Id. 74%; Po. 83% | 250 | 83 | 54.2 |
| 27 | h | Photosystem I P700 apoprotein A2 (Gossypium hirsutum) | 5e-64 | Id. 98%; Po. 98% | 246 | 21 | 61.9 |
| 28 | h | Harpin-induced 1 (Medicago truncatula) | 3e-23 | Id. 55%; Po. 73% | 111 | 64 | 76.6 |
| 29 | h | Clathrin coat assembly protein ap-1 (Ricinus communis) | 4e-63 | Id. 90%; Po. 96% | 243 | 33 | 66.7 |
| 30 | h | Hypothetical protein (Zea mays) | 2e-44 | Id. 87%; Po. 88% | 182 | 17 | 70.6 |
| 31 | h | Conserved hypothetical protein (Ricinus communis) | 2e-38 | Id. 62%; Po. 73% | 161 | 29 | 55.2 |
| 32 | g | Predicted protein (Populus trichocarpa) | 5e-86 | Id. 76%; Po. 85% | 322 | 21 | 76.2 |
| 33 | e | Predicted protein (Populus trichocarpa) | 4e-43 | Id. 60%; Po. 75% | 179 | 66 | 77.3 |
| 34 | L, a | Predicted protein (Populus trichocarpa) | 6e-25 | Id. 57%; Po. 66% | 117 | 38 | 84.2 |
| 35 | f, h | Predicted protein (Laccaria bicolor S238N-H82) | 8.3 | Id. 30%; Po. 43% | 33.5 | 63 | 74.6 |
| 36 | L | Hypothetical protein isoform 2 (Vitis vinifera) | 2e-22 | Id. 70%; Po. 81% | 108 | 8 | 87.5 |
| 37 | h | Unnamed protein product (Vitis vinifera) | 1e-35 | Id. 59%; Po. 72% | 152 | 21 | 71.4 |
| 38 | g | Hypothetical protein (Vitis vinifera) | 1e-25 | Id. 50%; Po. 58% | 119 | 62 | 58.1 |
The clones that showed activity of both two reporter genes were identified as positive clones, and DNA was isolated. Sequences were BLAST searched against the NCBI databases.
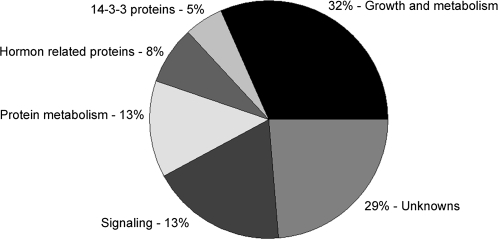
The targets of Gh14-3-3 proteins were related to various aspects of plant development, metabolism, and signal transduction. Some of the identified proteins, such as BRASSINAZOLE-RESISTANT protein (BZR, spot 3), H+-ATPase (spot 20), ABA responsive element-binding protein (spot 5), and 40S ribosomal protein (spots 8 and 9), were characterized as 14-3-3 targets in previous studies (Gampala et al., 2007; Ryu et al., 2007; Schoonheim et al., 2007b), but the majority were identified as novel interaction partners for cotton 14-3-3s. As shown in Fig. 9, the 38 identified proteins were divided into seven classes. Of the 38 identified targets, nine (spots 30–38) represented unknown proteins. Two 14-3-3 proteins (5%) were identified in the yeast two-hybrid screens as Gh14-3-3 interactors, of which one (spot 7) is Gh14-3-3a and another (spot 6) is Gh14-3-3h identified in this study. Growth and metabolism-related proteins (32%), such as sulphite oxidase (spot 14), carbonic anhydrase (spot 18), neutral/alkaline invertase (spots 19), and photosystem I P700 apoprotein A2 (spot 27), were found to be the major class of 14-3-3 protein interactors, and 13% of the 38 interactors were signalling-related proteins (spots 21–25). Furthermore, five proteins (spots 8–12) related to protein metabolism were identified, while three proteins (spots 3–5) involved in hormone response were detected in interaction with Gh14-3-3 proteins.
Fig. 9.
Venn diagram of classification of the isolated Gh14-3-3 interactors. The proteins identified in cotton fibres by yeast two-hybrid screening were classified into seven classes.
A systematic approach was employed to evaluate the publicly available cotton resources, and to help in the selection of specific subsets of the target genes. Using this approach, an expression summary of targets of each 14-3-3 protein was obtained from the cotton gene index (Table 2). The results indicated that there are a large number of ESTs for all 38 genes encoding the proteins targeted by Gh14-3-3s. Based on the percentage of ESTs detected in EST databases of cotton fibres, it was demonstrated that almost all targets are predominantly expressed in fibres, except 26S proteasome regulatory subunit 7 (28.2%) and carbonic anhydrase (39.4%).
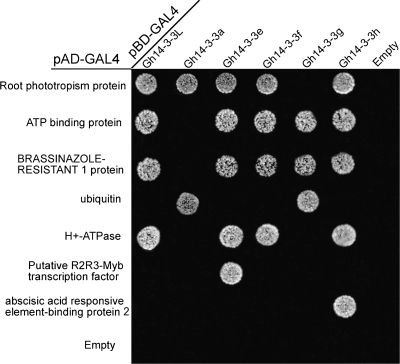
To understand whether the interactions between Gh14-3-3s and their target proteins are isoform selective, the direct yeast two-hybrid assay was employed to analyse the interactions between each Gh14-3-3 and its target proteins. Isoform-specific interactions were observed in this study. As shown in Fig. 10, some proteins (such as root phototropism protein, ubiquitin, ATP-binding protein, H+-ATPase, and BZR1) showed high affinity with more than one 14-3-3 protein, whereas the others (such as putative R2R3-Myb transcription factor and ABA responsive element-binding protein 2) interacted with only one 14-3-3 isoform.
Fig. 10.
Isoform-specific interactions between Gh14-3-3s and target proteins. Seven interactors were selected to analyse the interaction with each Gh14-3-3 protein by yeast mating. Transformants were assayed for growth on QDO nutritional selection medium.
Discussion
Plant 14-3-3 proteins belong to a large conserved family which can be divided into two groups (ε-like group and non-ε group) based on phylogenetic clustering (Sehnke et al., 2002a, b). In Arabidopsis, there are fifteen 14-3-3 genes, of which 13 are found to be functionally expressed, and the intron positions in the gene family are also conserved (Wu et al., 1997; Rosenqust et al., 2001). Similarly, the present data indicated that both ε-like and non-ε 14-3-3 proteins exist in cotton. Furthermore, the gene structure splicing patterns of the isolated Gh14-3-3 genes are consistent with phylogenetic analysis of the ε-like and non-ε clustering (Figs 2 and 3), like the Arabidopsis 14-3-3 family. Gh14-3-3a and Gh14-3-3L genes encoding ε-like 14-3-3 proteins contain three introns, whereas Gh14-3-3e and Gh14-3-3f genes which encode non-ε 14-3-3 isoforms have five introns. All the introns in cotton 14-3-3 genes are positioned in conserved codons, compared with the known plant 14-3-3 genes (Rosenqust et al., 2001). In addition, the data presented in Fig. 1 revealed that cotton 14-3-3 proteins contain the expected conserved structure, the nine antiparallel α-helical regions, but show great divergence in their N- and C-terminal regions, like all the other 14-3-3 proteins.
A previous study indicated that Arabidopsis 14-3-3 gene expression is restricted to specific tissues (Daugherty et al., 1996). Similarly, the present results showed that the mRNAs of all the six Gh14-3-3 genes are abundant in cotton fibres. The transcripts of these genes reached their peak values in fibres at 9–12 DPA during fibre development, but remarkably declined to lower levels as fibre cells developed further, indicating that the high expression levels of Gh14-3-3 genes are consistent with rapid elongation of cotton fibres. Furthermore, overexpression of Gh14-3-3L, Gh14-3-3a, and Gh14-3-3e in fission yeast significantly induced the longitudinal growth of the host cells, like the cotton fibre-specific TUA9 gene (Li et al., 2007). These data suggest that Gh14-3-3 proteins participate in regulation of cell polar growth and may play an important role in fibre development.
Dimerization is important for the function of 14-3-3 proteins. A recent study showed that two aldosterone-induced 14-3-3 isoforms, β and ε, interact with phosphor-Nedd4-2 as an obligatory heterodimer. Knockdown of either 14-3-3β or 14-3-3ε reduced the association of Nedd4-2 with the other isoform (Liang et al., 2008). In this study, interactions were found in five cotton 14-3-3 protein pairs, Gh14-3-3L–Gh14-3-3g, Gh14-3-3L–Gh14-3-3e, Gh14-3-3a–Gh14-3-3e, Gh14-3-3a–Gh14-3-3g, and Gh14-3-3g–Gh14-3-3h, but no homodimer was detected in these proteins. Each pair of the interacting proteins may form a heterodimer involved in the same signal transduction during fibre development of cotton.
From the yeast two-hybrid screening, 38 cotton 14-3-3-interacting proteins which were related to plant growth, development, metabolism, and signal transduction were identified in cotton fibres (Table 2). By searching in the cotton EST index, it was verified that these target proteins are very abundant in cotton fibres. Among them, some targets have been characterized as 14-3-3 target proteins. A previous study revealed that 40S ribosomal protein and 26S protease regulatory subunit 7 are 14-3-3 interactors in barley, and the interaction between ABF and 14-3-3 proteins was involved in the ABA signal transduction pathway in barley (Schoonheim et al., 2007a). The autoinhibitory action of H+-ATPase was released by binding of 14-3-3 protein (Maudoux et al., 2000). Likewise, the present data revealed that the above interactions in fibre cells may be important for fibre development of cotton.
Cotton 14-3-3 proteins may regulate fibre development by participating in phytohormome signal transduction. Fibre development of cotton is positively affected by a number of phytohormones, including auxin, GA, brassinosteroid (BR), and ethylene, but negatively impacted by ABA and cytokinin (Lee et al., 2007). SAR5 was first isolated as an auxin-repressed protein (ARP) by differential screening of auxin-deprived strawberry receptacles (Reddy and Poovaiah, 1990). It was reported that auxin levels precede fibre initiation, decline after anthesis (1–3 DPA), and rebound after 4 DPA during fibre development (Guinn and Brummett, 1988). Furthermore, knockdown of tobacco ABP1 expression causes deficient auxin-induced cell elongation (Chen et al., 2001). Previous studies revealed that BZR1 is a key transcription factor in BR signal transduction. Interaction between BZR1 and 14-3-3 protein affects the cytoplasmic translocation of BZR1 in Arabidopsis and rice (Bai et al., 2007; Ryu et al., 2007). Treatment of cotton floral buds with a BR inhibitor Brz resulted in the complete absence of fibre differentiation, whereas exogenous BL promoted fibre elongation, suggesting that BRs are essential for cotton fibre initiation and development (Sun et al. 2005). In this study, three proteins (ABA responsive element-binding protein, ARP, and BZR1 protein) involved in the response to phytohormone signalling were identified as Gh14-3-3 interactors. These data suggested that the interaction between Gh14-3-3s and the phytohormone-related proteins may be crucial for responding to phytohormone signalling or maintaining an appropriate phytohormone levels for rapid fibre cell elongation during fibre development of cotton.
It was demonstrated that MYB proteins participate in the regulation of sets of cell signalling in plants. Recently, a study by Pu et al. (2008) indicated that GhMYB109 is required for fibre development of cotton. Suppression of GhMYB109 activity led to a substantial reduction in fibre length. The fact that cotton 14-3-3 proteins interacted with MYB transcription factors suggests that 14-3-3 proteins may participate in fibre elongation by regulating MYB activity, controlling its intracellular localization or some other ways.
Gh14-3-3 proteins may regulate fibre cell elongation by modulating the pH of the cell wall. The interaction between the plasma membrane (PM) H+-ATPase and 14-3-3 proteins has been extensively explored. The PM H+-ATPase is known to play a major role in the control of various cell processes. The enzyme pumps protons from the cytoplasm to the cell exterior by using ATP as energy source and creates an electrochemical gradient across the PM that constitutes the driving force for nutrient uptake, phloem loading, water movements, and stomatal closure and opening (Comparot et al., 2003). The ‘Acid Growth Theory’ states that cells excrete protons into the wall at an enhanced rate when exposed to auxin, resulting in a decrease in apoplastic pH which activates the wall-loosening processes and causes cell elongation (Rayle et al., 1970, 1992). A study indicated that H+-ATPase was activated in wheat embryo by auxin, resulting in apoplastic acidification. This process contributes to cell wall loosening and cell elongation (Rober-Kleber et al., 2003). The binding of 14-3-3 proteins to the autoinhibitory domain of the H+-ATPase leads to the formation of a complex that activates the pump (Maudoux et al., 2000). Likewise, through the interaction with its target, the Gh14-3-3 protein may activate PM H+-ATPase, resulting in the loosening of cell walls for fibre cell elongation. On the other hand, the roles of the other target proteins of 14-3-3s in fibre development still need to be investigated in details. Gh14-3-3 proteins may regulate these target proteins by modulating their activities, localizations, or molecular interactions with other proteins for fibre development.
In conclusion, the data presented in this study indicated that the isolated Gh14-3-3 genes are fibre preferential and developmentally regulated in cotton, and overexpression of these genes in yeast promoted the longitudinal growth of the cells, suggesting that 14-3-3 proteins may participate in regulation of fibre elongation. Furthermore, the study revealed that the interaction between cotton 14-3-3 proteins is isoform selective, and 38 novel interaction partners for cotton 14-3-3 proteins were identified in cotton fibres by yeast two-hybrid screening. Thus, the results of this work provide novel insights into the 14-3-3 signalling related to fibre development of cotton.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Primer pairs used in isolation of Gh14-3-3 genes.
Table S2. Primer pairs used in overexpression of Gh14-3-3 genes in yeast cells.
Table S3. Primer pairs used in yeast two-hybrid analysis.
Table S4. Sequences of the 38 identified cDNAs encoding target proteins which interact with cotton 14-3-3 proteins.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Sciences Foundation of China (grant no. 30871317) and a project from the Ministry of Agriculture of China for transgenic research (grant no. 2008ZX08009-003, 2009ZX08009-117B).
References
- Alfa C, Fantes P, Hyams J, McLeod M, Warbrick E. Experiments with fission yeast: a laboratory course manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- Bachmann M, Huber JL, Athwal GS, Wu K, Ferl RJ, Huber SC. 14-3-3 proteins associate with the regulatory phosphorylation site of spinach leaf nitrate reductase in an isoform-specific manner and reduce dephosphorylation of Ser-543 by endogenous protein phosphatases. FEBS Letters. 1996;398:26–30. doi: 10.1016/s0014-5793(96)01188-x. [DOI] [PubMed] [Google Scholar]
- Bai MY, Zhang LY, Gampala SS, Zhu SW, Song WY, Chong K, Wang ZY. Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proceedings of the National Academy of Sciences, USA. 2007;104:13839–13844. doi: 10.1073/pnas.0706386104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basra AS, Malik CP. Development of the cotton fiber. International Review of Cytology. 1984;89:65–113. [Google Scholar]
- Braselmann S, McCormick F. Bcr and Raf form a complex in vivo via 14-3-3 proteins. EMBO Journal. 1995;14:4839–4848. doi: 10.1002/j.1460-2075.1995.tb00165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhri M, Scarabel M, Aitken A. Mammalian and yeast 14-3-3 isoforms form distinct patterns of dimers in vivo. Biochemical and Biophysical Research Communications. 2003;3:679–685. doi: 10.1016/s0006-291x(02)02902-9. [DOI] [PubMed] [Google Scholar]
- Chen JG, Ullah H, Young JC, Sussman MR, Jones AM. ABP1 is required for organized cell elongation and division in Arabidopsis embryogenesis. Genes and Development. 2001;7:902–911. doi: 10.1101/gad.866201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comparot S, Lingiah G, Martin T. Function and specificity of 14-3-3 proteins in the regulation of carbohydrate and nitrogen metabolism. Journal of Experimental Botany. 2003;54:595–604. doi: 10.1093/jxb/erg057. [DOI] [PubMed] [Google Scholar]
- Daugherty CJ, Rooney MF, Miller PW, Ferl RJ. Molecular organization and tissue-specific expression of an Arabidopsis 14-3-3 gene. The Plant Cell. 1996;8:1239–1248. doi: 10.1105/tpc.8.8.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duby G, Poreba W, Piotrowiak D, Bobik K, Derua R, Waelkens E, Boutry M. Activation of plant plasma membrane H+-ATPase by 14-3-3 proteins is negatively controlled by two phosphorylation sites within the H+-ATPase C-terminal region. Journal of Biological Chemistry. 2009;284:4213–4221. doi: 10.1074/jbc.M807311200. [DOI] [PubMed] [Google Scholar]
- Gampala SS, Kim TW, He JX, et al. An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Developmental Cell. 2007;2:177–189. doi: 10.1016/j.devcel.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D, St Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Research. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinn G, Brummett DL. Changes in abscisic acid and indoleacetic acid before and after anthesis relative to changes in abscission rates of cotton fruiting forms. Plant Physiology. 1988;3:629–631. doi: 10.1104/pp.87.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqarashi D, Ishida S, Fukazawa J, Takahashi Y. 14-3-3 proteins regulate intracellular localization of the bZIP transcriptional activator RSG. The Plant Cell. 2001;13:2483–2497. doi: 10.1105/tpc.010188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida S, Fukazawa J, Yuasa T, Takahashi Y. Involvement of 14-3-3 signaling protein binding in the functional regulation of the transcriptional activator REPRESSION OF SHOOT GROWTH by gibberellins. The Plant Cell. 2004;16:2641–2651. doi: 10.1105/tpc.104.024604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DH, Martin H, Madrazo J, Robinson KA, Nielsen P, Roseboom PH, Patel Y, Howell SA, Aitken A. Expression and structural analysis of 14-3-3 proteins. Journal of Molecular Biology. 1995;245:375–384. doi: 10.1006/jmbi.1994.0031. [DOI] [PubMed] [Google Scholar]
- Klychnikov OI, Li KW, Lill H, de Boer AH. The V-ATPase from etiolated barley (Hordeum vulgare L.) shoots is activated by blue light and interacts with 14-3-3 proteins. Journal of Experimental Botany. 2007;58:1013–1023. doi: 10.1093/jxb/erl261. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinformatics. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Lee JJ, Woodward AW, Chen ZJ. Gene expression changes and early events in cotton fibre development. Annals of Botany. 2007;7:1391–401. doi: 10.1093/aob/mcm232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Wang XL, Huang GQ, Li XB. Molecular characterization of cotton GhTUA9 gene specifically expressed in fibre and involved in cell elongation. Journal of Experimental Botany. 2007;58:3227–3238. doi: 10.1093/jxb/erm167. [DOI] [PubMed] [Google Scholar]
- Li XB, Fan XP, Wang XL, Cai L, Yang WC. The cotton ACTIN1 gene is functionally expressed in fibers and participates in fiber elongation. The Plant Cell. 2005;17:859–875. doi: 10.1105/tpc.104.029629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Butterworth MB, Peters KW, Walker WH, Frizzell RA. An obligatory heterodimer of 14-3-3beta and 14-3-3epsilon is required for aldosterone regulation of the epithelial sodium channel. Journal of Biological Chemistry. 2008;283:27418–27425. doi: 10.1074/jbc.M803687200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Bienkowska J, Petosa C, Collier RJ, Fu H, Liddinqton R. Crystal structure of the zeta isoform of the 14-3-3 protein. Nature. 1995;376:191–194. doi: 10.1038/376191a0. [DOI] [PubMed] [Google Scholar]
- Maudoux O, Batoko H, Oecking C, Gevaert K, Vandekerckhove J, Boutry M, Morsomme P. A plant plasma membrane H+-ATPase expressed in yeast is activated by phosphorylation at its penultimate residue and binding of 14-3-3 regulatory proteins in the absence of fusicoccin. Journal of Biological Chemistry. 2000;23:17762–17770. doi: 10.1074/jbc.M909690199. [DOI] [PubMed] [Google Scholar]
- Moore BW, Perez VJ. Specific acidic proteins of the nervous system. In: Carlson F, editor. Physiological and biochemical aspects of nervous integration. Woods Hole, MA: Prentice Hall; 1967. pp. 343–359. [Google Scholar]
- Muslin AJ, Tanner JW, Allen PM, Shaw AS. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84:889–897. doi: 10.1016/s0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- Muslin AJ, Xing H. 14-3-3 proteins: regulation of subcellular localization by molecular interference. Cellular Signaling. 2000;12:703–709. doi: 10.1016/s0898-6568(00)00131-5. [DOI] [PubMed] [Google Scholar]
- Petosa C, Masters SC, Bankston LA, Pohl J, Wang B, Fu H, Liddington RC. 14-3-3zeta binds a phosphorylated Raf peptide and an unphosphorylated peptide via its conserved amphipathic groove. Journal of Biological Chemistry. 1998;273:16305–16310. doi: 10.1074/jbc.273.26.16305. [DOI] [PubMed] [Google Scholar]
- Pu L, Li Q, Fan X, Yang W, Xue Y. The R2R3 MYB transcription factor GhMYB109 is required for cotton fiber development. Genetics. 2008;180:811–820. doi: 10.1534/genetics.108.093070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purwestri YA, Oqaki Y, Tamaki S, Tsuji H, Shimamoto K. The 14-3-3 protein GF14c acts as a negative regulator of flowering in rice by interacting with the florigen Hd3a. Plant and Cell Physiology. 2009;50:429–438. doi: 10.1093/pcp/pcp012. [DOI] [PubMed] [Google Scholar]
- Rayle DL, Cleland R. Enhancement of wall loosening and elongation by acid solutions. Plant Physiology. 1970;46:250–253. doi: 10.1104/pp.46.2.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayle DL, Cleland RE. The acid growth theory of auxin-induced cell elongation is alive and well. Plant Physiology. 1992;99:1271–1274. doi: 10.1104/pp.99.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AS, Poovaiah BW. Molecular cloning and sequencing of a cDNA for an auxin-repressed mRNA: correlation between fruit growth and repression of the auxin-regulated gene. Plant Molecular Biology. 1990;2:127–136. doi: 10.1007/BF00018554. [DOI] [PubMed] [Google Scholar]
- Rober-Kleber N, Albrechtová JTP, Fleig S, Huck N, Michalke W, Wagner E, Speth V, Neuhaus G, Fischer-Iglesias C. Plasma membrane H+-ATPase is involved in auxin-mediated cell elongation during wheat embryo development. Plant Physiology. 2003;131:1302–1312. doi: 10.1104/pp.013466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts MR, Salinas J, Collinge DB. 14-3-3 proteins and the response to abiotic and biotic stress. Plant Molecular Biology. 2002;50:1031–1039. doi: 10.1023/a:1021261614491. [DOI] [PubMed] [Google Scholar]
- Rosenquist M, Alsterfjord M, Larsson C, Sommarin M. Data mining the Arabidopsis genome reveals fifteen 14-3-3 genes. Expression is demonstrated for two out of five novel genes. Plant Physiology. 2001;127:142–149. doi: 10.1104/pp.127.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan YL, Llewellyn DJ, Furbank RT, Chourey PS. The delayed initiation and slow elongation of fuzz-like short fibre cells in relation to altered patterns of sucrose synthase expression and plasmodesmata gating in a lintless mutant of cotton. Journal of Experimental Botany. 2005;56:977–984. doi: 10.1093/jxb/eri091. [DOI] [PubMed] [Google Scholar]
- Ryu H, Kim K, Cho H, Park J, Choe S, Hwang I. Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. The Plant Cell. 2007;19:2749–2762. doi: 10.1105/tpc.107.053728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoonheim PJ, Sinniqe MP, Casaretto JA, Veiqa H, Bunney TD, Quatrano RS, de Boer AH. 14-3-3 adaptor proteins are intermediates in ABA signal transduction during barley seed germination. The Plant Journal. 2007a;2:289–301. doi: 10.1111/j.1365-313X.2006.02955.x. [DOI] [PubMed] [Google Scholar]
- Schoonheim PJ, Veiqa H, Pereira Dda C, Friso G, van Wijk KJ, de Boer AH. A comprehensive analysis of the 14-3-3 interactome in barley leaves using a complementary proteomics and two-hybrid approach. Plant Physiology. 2007b;2:670–683. doi: 10.1104/pp.106.090159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehnke PC, DeLille JM, Ferl RJ. Consummating signal transduction: the role of 14-3-3 proteins in the completion of signal-induced transitions in protein activity. The Plant Cell. 2002a;14:S339–S354. doi: 10.1105/tpc.010430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehnke PC, Rosenquist M, Alsterfjord M, DeLille J, Sommarin M, Larsson C, Ferl RJ. Evolution and isoform specificity of plant 14-3-3 proteins. Plant Molecular Biology. 2002b;50:1011–1018. doi: 10.1023/a:1021289127519. [DOI] [PubMed] [Google Scholar]
- Shen W, Clark AC, Huber SC. The C-terminal tail of Arabidopsis 14-3-3omega functions as an autoinhibitor and may contain a tenth alpha-helix. The Plant Journal. 2003;34:473–484. doi: 10.1046/j.1365-313x.2003.01739.x. [DOI] [PubMed] [Google Scholar]
- Shi H, Wang X, Li D, Tang W, Wang H, Xu W, Li X. Molecular characterization of cotton 14-3-3L gene preferentially expressed during fiber elongation. Journal of Genetics and Genomics. 2007;34:151–159. doi: 10.1016/S1673-8527(07)60016-2. [DOI] [PubMed] [Google Scholar]
- Sun Y, Allen RD. Functional analysis of the BIN 2 genes of cotton. Molecular Genetics and Genomics. 2005;1:51–59. doi: 10.1007/s00438-005-1122-0. [DOI] [PubMed] [Google Scholar]
- Sun Y, Veerabomma S, Abdel-Mageed HA, Fokar M, Asami T, Yoshida S, Allen RD. Brassinosteroid regulates fiber development on cultured cotton ovules. Plant and Cell Physiology. 2005;46:1384–1391. doi: 10.1093/pcp/pci150. [DOI] [PubMed] [Google Scholar]
- van Hemert MJ, Steensma HY, van Heusden GP. 14-3-3 proteins: key regulators of cell division, signalling and apoptosis. Bioessays. 2001;23:936–946. doi: 10.1002/bies.1134. [DOI] [PubMed] [Google Scholar]
- van Heusden GP, van der Zanden AL, Ferl RJ, Steensma HY. Four Arabidopsis thaliana 14-3-3 protein isoforms can complement the lethal yeast bmh1 bmh2 double disruption. FEBS Letters. 1996;391:252–256. doi: 10.1016/0014-5793(96)00746-6. [DOI] [PubMed] [Google Scholar]
- Visconti S, Camoni L, Marra M, Aducci P. Role of the 14-3-3 C-terminal region in the interaction with the plasma membrane H+-ATPase. Plant and Cell Physiology. 2008;49:1887–1897. doi: 10.1093/pcp/pcn172. [DOI] [PubMed] [Google Scholar]
- Wei XZ, Zhang ZT, Li Y, Wang XL, Shao SQ, Chen L, Li XB. Expression analysis of two novel cotton 14-3-3 genes in root development and in response to salt stress. Progress in Natural Science. 2009;19:173–178. [Google Scholar]
- Wu K, Rooney MF, Ferl RJ. The Arabidopsis 14-3-3 multigene family. Plant Physiology. 1997;114:1421–1431. doi: 10.1104/pp.114.4.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MB, Rittinqer K, Volinia S, Caron PR, Aitken A, Leffers H, Gamblin SJ, Smerdon SJ, Cantley LC. The structural basis for 14-3-3: phosphopeptide binding specificity. Cell. 1997;91:961–971. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- Zhang L, Wang H, Liu D, Liddington R, Fu H. Raf-1 kinase and exoenzyme S interact with 14-3-3zeta through a common site involving lysine 49. Journal of Biological Chemistry. 1997;272:13717–13724. doi: 10.1074/jbc.272.21.13717. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.