Abstract
Interaction and cross-talk between microtubules and actin microfilaments are important for numerous processes during plant growth and development, including the control of cell elongation and tissue expansion, but little is known about the molecular components of this interaction. Plant kinesins with the calponin-homology domain (KCH) were recently identified and associated with a putative role in microtubule-microfilament cross-linking. The putative biological role of the rice KCH member OsKCH1 is addressed here using a combined approach with Tos17 kch1 knock-out mutants on the one hand, and a KCH1 overexpression line generated in tobacco BY-2 cells. It is shown that OsKCH1 is expressed in a development and tissue-specific manner in rice and antagonistic cell elongation and division phenotypes as a result of knock-down and overexpression are reported. Further, the dynamic repartitioning of OsKCH1 during the cell cycle is described and it is demonstrated that KCH overexpression delays nuclear positioning and mitosis in BY-2 cells. These findings are discussed with respect to a putative role of KCHs as linkers between actin filaments and microtubules during nuclear positioning.
Keywords: KCH, nuclear positioning, plant kinesins, rice
Introduction
Many processes in plant cell growth and development rely on cross-talk and co-ordination between the two highly dynamic arrays of microtubules and actin microfilaments (Wasteneys and Galway, 2003; Collings, 2008). Microscopic studies in fixed and living cells of a variety of different plant species have reported co-alignment and cross-bridging of microfilaments to cortical microtubules (Petrasek and Schwarzerova, 2009; Deeks et al., 2010). Interactions between both types of filaments are suggested to contribute to the control of cell elongation and tissue expansion. As shown by pharmacological manipulation, microfilament and microtubule disrupting agents reduced cell- and overall-elongation in Arabidopsis roots (Collings et al., 2006), maize roots (Blancaflor, 2000) and cotton fibres (Seagull, 1990), and additionally disturb growth anisotropy in Arabidopsis (Baskin and Bivens, 1995) and rice roots and coleoptiles (Giani et al., 1998; Wang and Nick, 1998).
Despite the extensive studies on the microtubule-association of microfilaments, relatively little research has been conducted on the proteins that might mediate them. In animals and fungi a number of proteins have been identified that mediate such interactions, either in a bifunctional way through binding to both cytoskeletal elements or as complexes from two or more monofunctional proteins binding either microtubules or actin (Goode et al., 2000; Rodriguez et al., 2003). In plants, however, the situation has remained unclear. Recently, kinesins with a calponin-homology domain (KCH) were identified as a subset of the kinesin-14 family in plants and have been associated with a putative role for microtubule–microfilament interaction (Tamura et al., 1999; Preuss et al., 2004; Frey et al., 2009; Xu et al., 2009). In addition to the characteristic microtubule-binding kinesin motor domain, KCH-proteins possess a conserved calponin-homology (CH) domain, well known as actin-binding motif from a variety of actin-associated proteins such as α-actinin, spectrin, and fimbrin. Thus, KCHs are strong candidates for a bifunctional mediation between both cytoskeletal elements, and several studies confirmed that they can bind in fact to both elements of the cytoskeleton. Preuss et al. (2004) have described a KCH member from cotton, GhKCH1 and have shown that this specific type of kinesin interacts with actin filaments in the developing cotton-fibre using a combination of immunofluorescence and binding studies that employ recombinant fragments of KCH. Most recently, an additional member of KCH from cotton, GhKCH2, has been identified (Xu et al., 2009). GhKCH2 binds actin filaments with high affinity and is able to bundle and to cross-link them with microtubules in vitro. Recently, the first monocotyledon member of this plant-specific type of kinesin could be characterized in rice (Frey et al., 2009). OsKCH1 is associated with cortical microtubules and actin microfilaments in vivo, binds to microtubules and actin microfilaments in vitro in a domain-dependent way, and has been shown to oligomerize both in vivo and in vitro. This provides evidence that the microtubule-microfilament binding ability observed in cotton fibres is not limited to a highly specialized cell type but rather represents a general and conserved function of KCHs, found in both elongating and cycling cells (Frey et al., 2009).
Although members of KCH are highly conserved in higher plants and the moss Physcomitrella patens (Richardson et al., 2006; Frey et al., 2009), biological functions of KCH have so far remained elusive. Xu et al. (2009) have suggested that GhKCH2 might play a role in cotton fibre elongation through cross-linking of microtubules and microfilaments, but no further evidence has been provided. In this study, therefore, the putative biological role of the rice KCH member OsKCH1 is addressed using both rice Tos17 kch1 knock-out mutants and a KCH overexpression line generated in tobacco BY-2 cells. It is shown that OsKCH1 is expressed in a development and tissue-specific manner in rice and antagonistic cell elongation and division phenotypes as the result of knock-out and overexpression are reported. Further, it is described that OsKCH1 is repartitioned during the cell cycle and it is demonstrated that the overexpression of KCH delays nuclear positioning and mitosis in BY-2.
Materials and methods
Phylogenetic analysis of KCHs
Candidate proteins from Arabidopsis thaliana, Gossypium hirsutum, Oryza sativa, Physcomitrella patens, Populus trichocarpa, Ricinus communis, and Vitis vinifera, putatively belonging to the KCH subgroup, were identified in Swiss-Prot/TrEMBL via a BLAST search using AtKatD (Tamura et al., 1999) as a template. The protein sequences were aligned with the ClustalW2 tool at EBI (http://www.ebi.ac.uk/Tools/clustalw2/index.html), and the alignment transferred to PhyML (Guindon et al., 2005) for further phylogenetic analysis using a maximum-likelihood approach. The phylogenetic tree was obtained by a heuristic search method with a random stepwise addition of sequences. Bootstrap-support values were obtained from 100 replicates. The resulting tree file was visualized using iTOL (Letunic and Bork, 2007) and rooted arbitrarily using the kinesin-14 family member AtKatA (Mitsui et al., 1993; Liu et al., 1996) as the outgroup.
Protein sequence data used in the phylogenetic analysis can be accessed in Swiss-Prot/TrEMBL under the following accession numbers: OsKCH1 (Q0IMS9), Q10MN5, A3CDK9, Q5JKW1, B9FFM0, B9FL70, A5BH78, A7Q9E6, A5C0F0, A5BG13, A5APK4, GhKCH1 (Q5MNW6), GhKCH2 (A4GU96), AtKatD (O81635), AtKP1 (Q8W1Y3), O80491, Q9FHD2, Q9SS42, Q84W97, O22260, A9SJ46, A9SDI6, A9SPL2, B9I798, B9GH20, B9GSE0, B9H9K3, B9I2M3, B9GWJ1, B9T1P9, B9T5B8, B9RCC3, B9RFF9, B9SAW6, AtKatA (Q07970).
Plant material and growth conditions
The rice (Oryza sativa L. japonica) cultivar Nipponbare represented the background cultivar for the kch1 Tos17 mutants and was therefore used as the wild-type control in most experiments. The mutant lines kch1-1 (NF9840) and kch1-2 (NG1558) were obtained from the Tos17 mutant database (http://tos.nias.affrc.go.jp/) (Miyao et al., 2003) and plants were grown in the greenhouse under long-day conditions in the summer time. Plants were genotyped using the two gene-specific primer sets kch1-1fw/kch1-1rv and kch1-2fw/kch1-2rv (kch1-1fw: TTTGTGCAGAGTGGACCGAAAGTT; kch1-1rv: CATCTAGTTCCGGGCTTACGTG; kch1-2fw: TACAATGAGCAAGTGAGGGATCTC; kch1-2rv: CATCTAGTTCCGGGCTTACGTG) and the insert specific primer Tos17RB (Tos17RB CTGTATATTGGCCCATGTCCAG) together with either kch1-1fw or kch1-2fw, respectively. For seed amplification purposes, plants were propagated in growth chambers under controlled conditions. Rice seedlings for gene expression studies and physiological experiments were raised at 25 °C for 7 d in photobiological darkness (using black boxes, black cloth, and isolated dark chambers) either on floating meshes as described by Nick and Furuya (1993) or on 0.6% water agar in agripots under sterile conditions. Usually seed germination was higher than 95% and seedling length among the population varied by less than 5%.
Tobacco Bright Yellow 2 wild-type (BY-2 Wt) cells were maintained in the dark in liquid MS medium at 25 °C under constant shaking and were subcultured weekly. Preparation and microscopic characterization of BY-2 cells stably expressing a nearly full-length version (aa 1-800) of OsKCH1 (BY-2 KCH1) under the control of the constitutive CaMV 35S promoter has been described by Frey et al. (2009). Cell suspension cultures of BY-2 KCH1 were maintained in liquid MS medium supplemented with 100 μg ml−1 kanamycin and subcultured weekly. BY-2 KCH1 cells used for phenotyping experiments were cultivated in the absence of the antibiotic.
Phenotyping experiments with rice and tobacco BY-2
Coleoptile length was determined in etiolated rice seedlings 7 d after germination. Macroscopic images of the seedlings were recorded and coleoptiles measured using the periphery tool of ImageJ (NIH, Bethesda, USA). At least 50 seedlings, collected cumulatively over a minimum of three independent experimental series, were investigated and mean values and standard errors (SE) calculated. Differences between mutants and wild types were tested for significance using Students t test for unpaired data sets at a confidence level of 95%. For cell length determination, cross walls per cell file (see Fig. 1E for an illustration) were counted and used to calculate mean cell length and SE. A total of 150 analysed cell files, collected cumulatively from 15 coleoptiles, were scored and the observed differences were tested for significance by t tests as described above. The number of cells per coleoptile was determined by dividing the total length of the coleoptile by the respective mean cell length. Again, mean values and SE were determined from a total of 150 analysed cell files, collected cumulatively from 15 coleoptiles and all observed differences were tested for significance by Student's t tests.
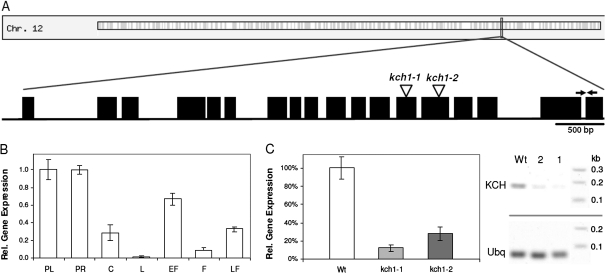
Fig. 1.
Expression pattern of OsKCH1 in rice and Tos17 insertion mutants. (A) Schematic representation of the OsKCH1 gene structure. Introns are shown as lines, exons as boxes. Positions of Tos17 insertion sites in kch1-1 and kch1-2 are indicated by arrow heads. Positions of the primers used for gene expression analysis are indicated by arrows. (B) Expression of OsKCH1 in rice seedlings and adult plants of the wild type measured by real-time PCR. PL, primary leaf; PR, primary root; C, coleoptile; L, adult leaf; EF, young flower bud; F, fully developed fower; LF, flower after pollination. (C) Expression of OsKCH1 in Tos17 insertion mutants quantified by real-time PCR in RNA extracts from complete seedlings of the mutant lines kch1-1 and kch1-2 as compared to the wild-type control. In addition, a representative RT-PCR shows the abundance of OsKCH transcripts in seedlings of the wild type as compared to the mutants kch1-1 (1) and kch1-2 (2). Ubiquitin (Ubq) was used as internal standard in all samples.
Cell length and width in BY-2 cells were determined using the length measurement function of the AxioVision software (Zeiss, Jena, Germany). DIC images were recorded at day 3 after subcultivation from three independent experimental series comprising 300 cells each, mean values and SE were calculated, and the results were tested for significance by Student's t test for unpaired data at a 95% confidence level. Mitotic indices (MI) were determined microscopically in tobacco BY-2 cell suspensions after fixation and Hoechst staining as described below. MI were calculated as the number of cells in mitosis divided by the total number of counted cells. For each time point 450 cells obtained from three independent experimental series were scored and the results tested for significance by Student's t test as described above. Nuclear positions (NP) in tobacco BY-2 were assessed at different time points after subcultivation (0–6 d) from DIC images of central sections of the cells using the length measurement function of the AxioVision software (Zeiss, Jena, Germany). NP were determined as relative values by division of the shortest distance between the middle of the nucleus and the total cell diameter (see Fig. 7D for an illustration) Typical values for mean NP ranged between 0.15 and 0.5 and were clustered into three main intervals indexed by the attributes lateral (NP=0.15–0.28), intermediate (NP=0.3–0.4), and central (NP=0.4–0.5). For each time point and cell line, three times 500 cells were measured, mean values, SE, and occurrence distributions calculated, and the results tested for significance by Student's t test.
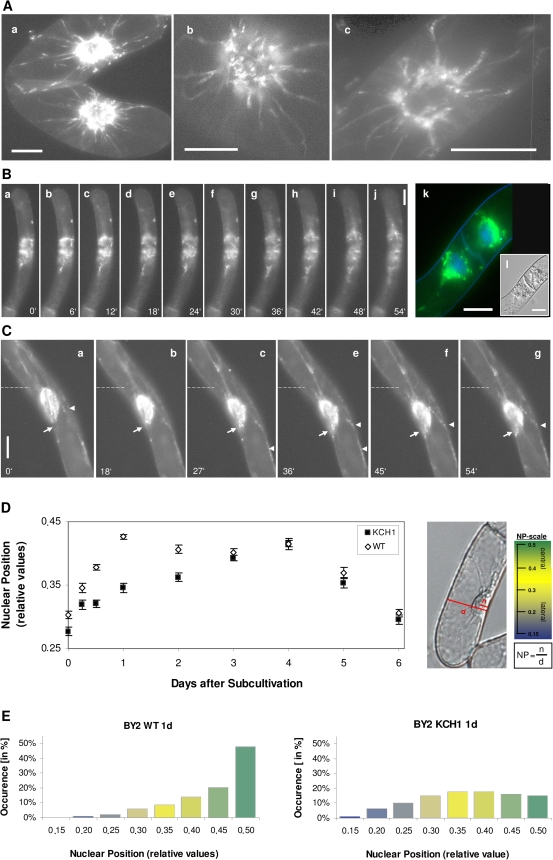
Fig. 7.
Influence of OsKCH1 on nuclear positioning. (A–C) Tobacco BY-2 cells stably expressing a GFP fusion of OsKCH1-800. (A) Optical sections from the cell midplane of interphasic cells. (B) Time-lapse series of cells after mitosis (a–j) (see Supplementary Video S3 at JXB online). In addition, a merge image is given in (k) showing GFP OsKCH1-800 fluorescence and DNA signals stained by Hoechst. The corresponding DIC image is given in (l). (C) Time-lapse series of nuclear movement in an interphasic cell (see Supplementary Video S4 at JXB online). The original position of the nucleus in each image is indicated by a dotted line. Arrows highlight filaments that span to the periphery while arrowheads point to OSKCH1 accumulation on cortical anchor points. (D) Time-course of nuclear positions (NP) in BY-2 KCH1 and BY-2 Wt through the culture cycle at different time points after subcultivation. The nuclear position is determined as a relative value by division of the shortest distance between the middle of the nucleus n and the total cell diameter d as displayed in the DIC image. The adjacent scale shows the typical values for mean NP, ranging between 0.15 and 0.5 together with a colour code. Three main intervals for the position of the nucleus in a cell of interest can be defined, indicating either a central location of the nucleus (NP=0.4–0.5; shaded in green), a lateral position (NP=0.15–0.28, shaded in blue), or an intermediate state (NP=0.3–0.4, shaded in bright yellow). For each time point and cell line, three times 500 cells were measured and mean values were plotted as a function of time together with the respective SE. Differences between mean NP values were positively tested for significance in unpaired t-tests and showed P <0.01 at days 0.5, 1, and 2. (E) Comparison between the NP value distributions in BY-2 Wt and BY-2 KCH1 at day 1. The histograms displays the NP values in ranges of 0.5, colour-coded as described in (D), together with the respective occurrence in a percentage of cells. Note that in BY-2 Wt at 1 d in about 70% of cells the nuclei are centrally located (NP values higher than 0.4) while at the same time point in BY-2 KCH1 this is only the case in about 28% of cells.
Fixation and staining of tobacco BY-2 cells
Microtubules were visualized by indirect immunofluorescence, basically as described by Nick et al. (2000) with minor modifications. After fixation for 60 min with 1.75% (w/v) paraformaldehyde in microtubule stabilizing buffer (MSB: 50 mM PIPES, 2 mM EGTA, 2 mM MgSO4, 0.1% Triton X-100, pH 6.9), cells were washed in PBS three times for at least 5 min to remove excess paraformaldehyde. Subsequently, the cell wall was digested using 1% (w/v) Macerozyme (Duchefa, Haarlem, The Netherlands) and 0.2% (w/v) Pectolyase (Fluka, Taufkirchen, Germany) in MSB for 7 min. After washing three times for 10 min with MSB, unspecific binding sites were blocked for 30 min with 0.5% (w/v) BSA diluted in PBS. Samples were subsequently directly transferred into a 1:1000 PBS-dilution of the primary antibodies DM1A and ATT (Breitling and Little, 1986), purchased from Sigma (Taufkirchen, Germany). The primary antibody was allowed to bind overnight at 4 °C. After removing unbound primary antibody by washing the cells three times for 10 min in PBS, the samples were incubated with a secondary TRITC-conjugated antibody (Sigma, Taufkirchen, Germany; 1:200 in PBS), targeted against mouse IgG, for 1 h at 37 °C. Again, unbound secondary antibody was removed by washing with PBS. If required, the DNA was also Hoechst stained as described below.
For the determination of MI, tobacco BY-2 cell suspensions were typically fixed in Carnoy fixative [3:1 (v/v) 96% (v/v) ethanol:glacial acetic acid, plus 0.25% Triton X-100]. The DNA was generally stained with 2'-(4-hydroxyphenyl)-5-(4-methyl-1-piperazinyl)-2,5'-bi(1Hbenzimidazole)-trihydrochloride (Hoechst 33258; Sigma, Taufkirchen, Germany). For this purpose, a final concentration of 1 μg ml−1 Hoechst, prepared as a 0.5 mg ml−1 filter-sterilized stock solution in distilled water, was added to either Carnoy-fixed or immunolabelled BY-2 cells and the samples were immediately investigated under the microscope. For staining of living cells, the samples were incubated with 1 μg ml−1 Hoechst 33258 under constant shaking for 12 h and subsequently investigated under the microscope.
Actin filaments were visualized by tetramethylrhodamine isothiocyanate (TRITC)-phalloidin (Sigma-Aldrich) staining of fixed BY- cells as previously published (Maisch et al., 2009)
Microscopy and image analysis
Cells were examined under an AxioImager Z.1 microscope (Zeiss, Jena, Germany) equipped with an ApoTome microscope slider for optical sectioning and a cooled digital CCD camera (AxioCam MRm) using either a ×63 plan apochromat oil-immersion objective, a ×40 or a ×20 objective. TRITC and GFP fluorescence were observed through the filter set 43 HE (excitation at 550 nm, beamsplitter at 570 nm, and emission at 605 nm) and 38 HE (excitation at 470 nm, beamsplitter at 495 nm, and emission at 525 nm), respectively. Hoechst staining was recorded using the filter set 49 DAPI (excitation at 365 nm, beamsplitter at 395 nm, and emission at 445 nm). For phenotyping, cells were examined with differential interference contrast (DIC) illumination. Images were processed for publication using the AxioVision Software (Zeiss, Jena, Germany).
RNA extraction and real-time PCR
Plant material was harvested, immediately frozen in liquid nitrogen, and ground in a TissueLyser (Qiagen, Hilden, Germany). Total RNA was extracted using an RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) including the on-column digest of genomic DNA with RNase-free DNAse I (Quiagen, Hilden, Germany) according to the protocol of the producer. All RNA preparations were checked for purity and integrity on 1% agarose gels. A Dynamo cDNA Synthesis Kit (Finnzymes, Espoo, Finland) was used for Reverse Transcription of RNA samples of 1 μg each.
Real-time PCR was set up with an iQ SYBR Green Supermix (Bio-Rad, München, Germany) according to the manufacturer. The PCR was run in a DNA Engine Opticon 2 cycler (Bio-Rad, München, Germany) following the recommended conditions [3 min 95 °C, 40× (10 s, 95 °C; 1 min, 60 °C)]. OsKCH1fw (5′-ATTACAGCAACGAAGATTCCA-3′) and OsKCH1rv (5′-TCTCACCTTGGCATAGTGG-3′) were used as primers to amplify OsKCH1 cDNA from rice. Ubiquitin was used as an endogenous control gene for normalization and was detected with the primers Rubg237F and Rubg304R (Riemann et al., 2008). Real-time PCR results were further analysed and quantified using the softwares MJ Opticon Monitor 3.1 (Bio-Rad, München, Germany) and Microsoft Office Excel 2003 (Microsoft, Unterschleißheim, Germany).
Semi-quantitative RT-PCR
For semi-quantitative RT-PCR a two step protocol was applied. Reverse Transcription was performed as described above and was followed by a PCR step with standard Taq polymerase (NEB, Frankfurt, Germany) according to the manufacturer's instructions using the following PCR conditions [2 min 94 °C, 32× (30 s, 94 °C; 1 min, 60 °C; 30 s 72 °C), 5 min 72 °C]. The cycle numbers for PCR were chosen such that the amplification of templates for all primers were still in an exponential range (data not shown) and the products clearly visible on agarose gels, making quantification possible and reproducible. OsKCH1 cDNA from rice was amplified with the primers OsKCH1fw and OsKCH1rv (see above). For the detection of KCH cDNA levels in tobacco BY-2, a set of degenerated primers was designed (KCHfw: CACGTAAGCCCGGAACTAGATGCT; KCHrv: GCCTTGAGGGTAGCAATCTGT). For this purpose an alignment of KCH sequences from rice, Arabidopsis, cotton, Vitis, and Populus was used as template and primers were set to highly conserved sequence parts outside the CH and the motor domain. The degenerated primers were tested on rice, tobacco, and tobacco BY-2 cDNA for their functionality and products were verified by sequencing prior to application of the primers in semi-quantitative RT-PCR. Ubiquitin was generally used as the endogenous control gene and was detected with the primers Rubg237F and Rubg304R (Riemann et al., 2008) in both rice and tobacco BY-2. PCR products were separated by 2% agarose gels, stained with SYBR Safe (Invitrogen, Karlsruhe, Germany) and images were taken. Bands were quantified by grey value analysis in ImageJ (NIH, Bethesda, USA) and the ratios between sample RNA and Ubiquitin calculated to normalize for initial variations in sample concentration, and as a control for reaction efficiency. Mean values and SE were calculated from at least three independent experiments.
Results
Expression of OsKCH1 is tissue and development specific
In order to get an insight into the potential physiological roles of OsKCH1 (SwissProt/TrEMBL accession Q0IMS9) (Frey et al., 2009), the expression of OsKCH1 was quantified in rice in different tissues and during different developmental stages by real-time PCR. Clear differences in gene expression patterns were detected, as shown in Fig. 1B. The highest abundance of transcripts was found in tissues with meristematic activity such as primary leaf, primary root, and the developing flower. In addition, transcripts were present in the coleoptile, a rapidly expanding organ exclusively growing via cell elongation.
For further characterization of OsKCH1 a loss-of-function approach was chosen, and the rice Tos17 database (http://tos.nias.affrc.go.jp/) (Miyao et al., 2003) screened for putative mutants. Two independent mutant lines NF9840 (kch1-1) and NG1558 (kch1-2) were identified with retrotransposon insertion sites in the 13th and 14th exons of the OsKCH1 locus, respectively (Fig. 1A). Heterozygous seeds were obtained from the National Institute of Agrobiological Science (NIAS, Tsukuba, Japan) and segregating mutants and wild types were identified by genotyping. Seedlings of both mutant and wild type were analysed for expression of OsKCH1. As shown in Fig. 1C, transcript abundance in entire rice seedlings was clearly reduced to about 10–25% of wild-type expression in both Tos17 lines. Whether the residual transcripts will yield functional or partially functional proteins, is not known.
kch1 insertion mutants show impaired cell expansion and increased division
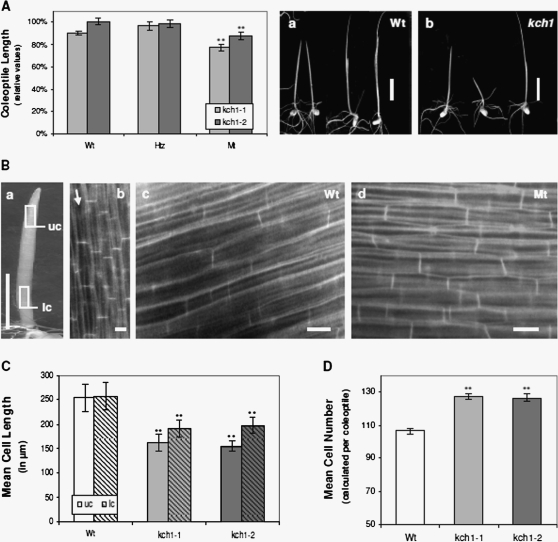
Next, etiolated coleoptiles of kch1-1 and kch1-2 were analysed for morphological phenotypes. Rice coleoptiles represent a system highly suitable for phenotyping studies as they grow rapidly and homogenously and are physiologically characterized in great detail (Holweg et al., 2004). The cells that constitute the coleoptile are all formed during late embyogenesis, and, following germination, no further division occurs. Thus, rice coleoptile growth relies exclusively on cell elongation and therefore records sensitively even small differences in cell growth rates. As shown in Fig. 2A, coleoptiles were about 10–15% shorter in kch1-1 and kch1-2 mutants, compared with the segregating wild types, to heterozygous plants, or to non-transformed Oryza sativa L. japonica cv. Nipponbare wild-type seedlings. This observed phenotype might either result from a diminished or delayed elongation growth, producing shorter cells in the mutants, or from a reduced number of cells. In order to discriminate between the two alternatives, average cell length was measured in tangential sections from either the apical or the basal third of the coleoptile (Fig. 2Ba, b). As visible in Fig. 2C, the average cell length was clearly shorter in both mutant lines with respect to wild-type coleoptiles. This difference was most prominent in the apical region, where mutant cells reached only about 60% of the length observed in the wild types. The observed strong reduction in cell length in the kch1 insertion mutants was, however, almost compensated by a concomitant increase in the average cell number per coleoptile (Fig. 2D).
Fig. 2.
Phenotype of Tos17 insertion mutants. (A) Coleoptile length in etiolated seedlings 7 d after germination. For both mutant lines, kch1-1 (light grey), and kch1-2 (dark grey), in addition to homozygous mutant plants (Mt), heterozygotes (Htz), and the segregating wild types (Wt) were analysed. Coleoptile length plotted relative to the cultivar Oryza sativa L. japonica cv. Nipponbare that was the background for the mutants. Mean values ±SE of a total of at least 50 measurements are given, collected cumulatively over several independent experimental series. Asterisks indicate significant differences in both mutant lines between Mt and Wt/Htz as assessed by a t test for unpaired data; **P <0.01. Representative images of both Wt (a) and Mt (b) seedlings. Scale bar: 10 mm. (B, C) Cell length measurements in coleoptiles of etiolated seedlings 7 d after germination for kch1-1 (light grey), kch1-2 (dark grey) and wild-type (Wt, white) plants. Cell length were determined in the upper part (uc, open bars) and in the lower part (lc, hatched bars) of the coleoptile as marked in (a) by counting cross walls per cell file in the field of vision at the microscope as indicated in (b). Data represent mean values ±SE of a total of at least 150 analysed cell files, collected from 15 coleoptiles. Asterisks (**) indicate significant differences (P <0.01) between Mt and Wt as assessed by a t test for unpaired data. Representative fluorescence images of coleoptile cells are shown in (c) for the wild type and in (d) for the mutant. Scale bars:10 mm (a); 20 μm (b, c). (D) Mean cell numbers in coleoptiles of etiolated seedlings 7 d after germination for kch1-1 (light grey), kch1-2 (dark grey) and wild-type (Wt, white) plants. Mean values ±SE were estimated from the cell lengths determined in a total of at least 150 analysed cell files as described in (B). Asterisks (**) indicate significant differences (P <0.01) between Mt and Wt in t tests for unpaired data.
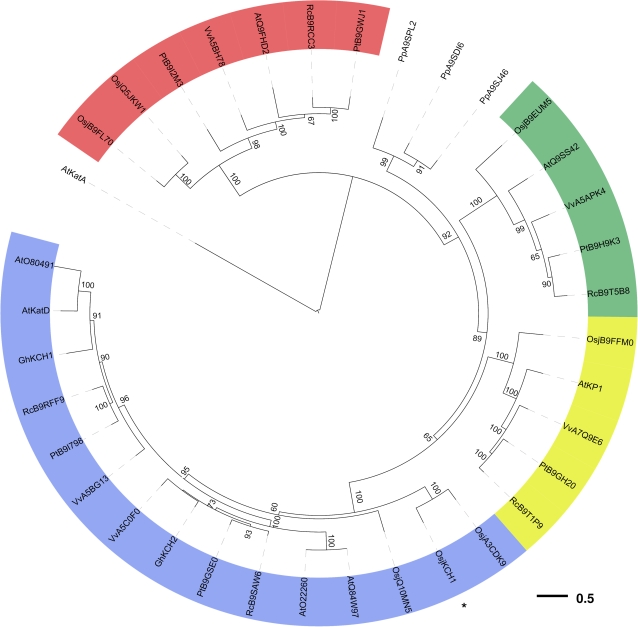
During subsequent development, no additional prominent phenotypes were identified. Both mutant and wild-type plants showed normal growth, flowering, and seed production under constant long-day conditions. To test whether the lack of a phenotype in adult plants might be due to functional redundancy, the molecular phylogeny of known KCHs was analysed. Figure 3 shows a phylogenetic tree displaying the relationship between KCHs from the grass model plant Oryza sativa, the Rosids Arabidopsis thaliana, Populus trichocarpa, Vitis vinifera, Ricinus communis, and the moss Physcomitrella patens. KCH from higher plants cluster into four different clades that each contains at least one KCH member from all species. OsKCH1 clusters into the most expanded branch, together with two additional KCH family members from rice, indicating a close relation and possibly conserved functions among these proteins. Interestingly, the KCH members from Physcomitrella were not resolved into the four prominent clades but formed a solitary branch, indicating evolutionary divergence of these proteins.
Fig. 3.
Phylogenetic analysis of KCH family members. Phylogenetic tree for the KCH family from several higher plants and from the moss Physcomitrella patens obtained by a maximum likelihood approach with random stepwise addition of protein sequences and arbitrary rooting using the Kin-14A family member AtKatA as outgroup. Bootstrap support values were obtained from 100 replicates and only values greater than 50 are shown in the tree. KCH members from higher plants cluster into four clades, all shaded in different colours. OsKCH1 (marked by an asterisk) resolves into the most expanded branch, together with two additional KCH family members from rice. Abbreviations: Osj, Oryza sativa japonica; At, Arabidopsis thaliana; Gh, Gossypium hirsutum; Vv, Vitis vinifera; Pp, Physcomitrella patens; Pt, Populus trichocarpa; Rc, Ricinus communis. All protein sequence data were obtained from Swiss-Prot/TrEMBL.
KCH1 overexpressors show stimulated cell elongation
The expression pattern of OsKCH1 in rice, along with the observed coleoptile phenotype of rice kch1 mutants, would allow two interpretations of the function of OsKCH1. The protein could either be involved in cell division during late embryogenesis when the coleoptile is laid down, or during post-germinative growth when the coleoptile cells expand. In order to discriminate further between these two possibilities, the effect of an overexpression of OsKCH1 on cell division or cell elongation should be investigated. Since rice coleoptiles grow exclusively via cell elongation, they do not represent an adequate model system for this question. We therefore turned towards the well-characterized and reliable tobacco BY-2 system as a widely studied model for cycling cells (Nagata et al., 1992; Nagata and Kumagai, 1999). A tobacco BY-2 cell line (BY-2 KCH1) was therefore generated that stably expressed a fusion of a GFP reporter with OsKCH1 driven by the constitutive CaMV 35S promoter. To verify the expression of the exogenous protein OsKCH1 in BY-2, the intracellular localization pattern of this GFP fusion was investigated microscopically. The protein clearly localized to both microtubules and actin filaments (see Supplementary Fig. S1 at JXB online), and reacted as sensitive to treatments with oryzalin and latrunculin B, underlining its functionality in the exogenous system (data not shown).
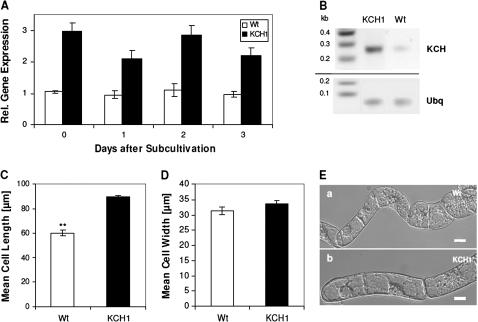
Since sequence information for tobacco homologues of OsKCH1 are not available yet, the expression of KCH genes in BY-2 had to be assessed using degenerated primers. The degenerated primers were designed using an alignment of KCH sequences from rice, Arabidopsis, cotton, Vitis, and Populus as template. The primers were tested for functionality on BY-2 Wt and rice cDNA and the obtained amplificates were verified by sequencing (data not shown). The abundance of KCH transcripts was then measured by semi-quantitative RT-PCR in both BY-2 Wt and BY-2 KCH1 cells at different stages of the cell cycle. In BY-2 KCH1, the KCH expression level was consistently elevated about 2.5–3-fold as compared to the non-transformed Wt cells (Fig. 4A, B), confirming overexpression of the transgene.
Fig. 4.
BY-2 cells overexpressing OsKCH1 are more elongated than wild-type cells. (A) Expression of KCH in non-transformed BY-2 wild-type cells (Wt, white) and BY-2 cells stably overexpressing OsKCH1 (KCH1, black). (B) Representative RT-PCR detection of KCH expression levels in BY-2 Wt and BY-2 KCH1. Ubiquitin (Ubq) was used as internal standard in all samples. (C) Cell length in BY-2 Wt and BY-2 KCH1. Mean values ±SE of a total of 300 analysed cells per sample, collected cumulatively in three independent experimental series are given. Asterisks (**) indicate P <0.01 as evaluated by a t test for unpaired data. (D) Cell width in BY-2 Wt and BY-2 KCH1. For details refer to (C). (E) Representative DIC images of (a) BY-2 Wt and (b) BY-2 KCH1 cells to show the differences in cell length. Scale bars: 20 μm.
The BY-2 KCH1 line grew homogeneously at a normal growth rate and generated the pluricellular files typical for BY-2. However, a detailed phenotypic investigation showed that BY-2 KCH1 cells were about 1.5-fold longer than the non-transformed BY-2 Wt cells (Fig. 4C, E). By contrast, cell width, was not significantly altered (Fig. 4D, E). The observed morphology in BY-2 KCH1 was underlined by investigation of two additional cell lines, BY-2 KCH1-2 and BY-2 KCH1-3, which had been independently transformed with the same reporter construct, and yielded similar phenotypes (data not shown).
To test, whether this altered morphology was caused by overexpression of a transgene per se, all phenotypical traits were assessed and quantified in a cell line stably expressing free-GFP under the control of a CaMV 35S promoter. However, this line did not show any differences to the non-transformed BY-2 Wt (data not shown), suggesting that the elevated cell elongation in BY-2 KCH1 cells is specific.
Mitosis in KCH1 overexpressor cells is delayed but morphologically normal
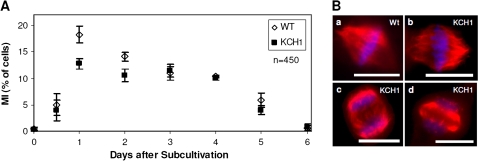
In order to investigate whether the observed increase in cell length might be caused by alterations in mitosis, the mitotic index (MI) was followed through the culture cycle in both BY-2 KCH1 and BY-2 Wt. As shown in Fig. 5A, the MI in BY-2 KCH1 was diminished at days 1 and 2 as compared with the non-transformed wild type. From day 3 on, however, the MI reached the wild-type level and remained similar to the wild-type levels for the rest of the investigation period. This indicated that the onset of the growth phase was delayed in BY-2 KCH1, but subsequently proceeded normally. The functionality of mitosis was further confirmed by microscopic investigation and comparison of mitotic spindle structures in BY-2 KCH1 and BY-2 Wt. For this purpose, microtubules were stained by immunofluorescence in both cell lines and chromosomes visualized by Hoechst. As visible in Fig. 5B, the spindle microtubules in BY-2 KCH1 appeared completely normal, and the spindle apparatus showed no structural alterations from the non-transformed Wt. Similarly, the shape of phragmoplasts appeared to be normal. Thus, the process of mitosis in general did not seem to be disturbed by overexpression of OsKCH1, its onset, however, was delayed.
Fig. 5.
Mitosis in BY-2 KCH1 cells is delayed but morphologically normal. (A) Time-course of mitotic indices (MI) in BY-2 Wt and BY-2 KCH1. Mean ±SE in per cent of cells of a total of 450 analysed cells per time point and sample, collected cumulatively from three independent experimental series with 150 cells per series are given. (B) Microscopic analysis of mitotic spindles in BY-2 Wt (a) and BY-2 KCH1 (b), as well as phragmoplasts (c, d). Microtubules were stained by immunofluorescence, the DNA visualized by Hoechst staining. Scale bars: 20 μm. (This figure is available in colour at JXB online.)
OsKCH1 is dynamically repartitioned during the cell cycle
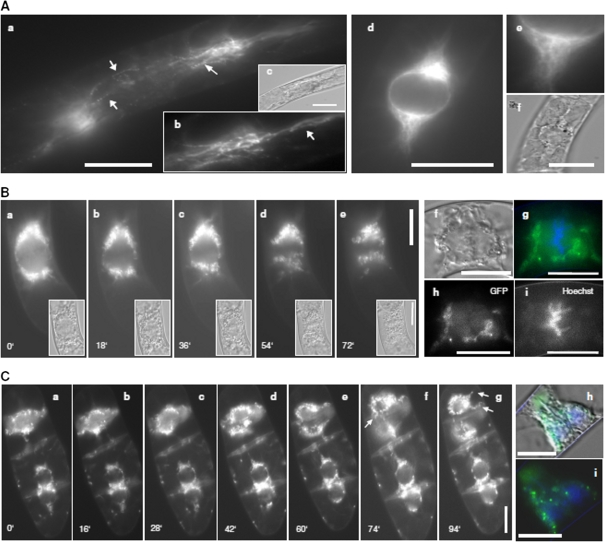
In order to assess a potential role of KCH1 in the cell cycle further, the localization pattern of GFP-OsKCH1 was followed through the cell cycle in tobacco BY-2 cells (Figs 6A–C, 7A–C). As visible in Fig. 6A, in premitotic cells OsKCH1 was clearly aligned as a punctate pattern along filamentous, mesh-like structures on both sides of the nucleus and on perinuclear filaments spanning over and surrounding the nucleus. At the onset of mitosis (Fig. 6B, C), OsKCH1 retracted and was mainly found at both sides of the nucleus. It localized neither to preprophase bands, nor the spindle apparatus, nor the division plate. During late telophase and the beginning of cytokinesis, OsKCH1 signals were repartitioned and subsequently were mainly found surrounding the newly forming nuclei and on filaments that tethered these nuclei to the periphery and the new cell wall. Similar signals were also found during later steps of cytokinesis, on filaments that span longitudinally from the nuclei towards the cell poles, as shown in Fig. 7B. Interestingly, the fluorescent signals were accumulating in the interspaces between these filaments and the leading edge of the migrating nuclei, as if the nucleus was guided. In interphase cells, OsKCH1 typically localized to fine filaments, tethering the nucleus to the periphery (Fig. 7A). Punctate signals accumulated at the nucleus, often located at the sites where the filaments reached the nuclear envelope or the sites where the filament attachment connected to the cell cortex (Fig. 7A).
Fig. 6.
Localization of KCH1 during mitosis. (A) Transient expression of a YFP-fusion with OsKCH1-800 in tobacco BY-2 cells. Optical sections through the cell midplanes of premitotic BY-2 cells at different focus levels (a, b, d, e). The corresponding DIC image is given in (c, d). Arrows highlight punctate pattern on perinuclear filaments. (B, C) Preprophasic and prophasic tobacco BY-2 cells stably expressing a GFP-fusion with OsKCH1-800. The DNA was stained with Hoechst to determine the stage of mitosis. (B) A time-lapse series (see Supplementary Video S1 at JXB online) shows GFP OsKCH1 signals in tobacco BY2 cells during the early stages of mitosis, from prophase to early anaphase, together with the corresponding DIC images (a–e). In addition, a double-staining of GFP OsKCH1 (h) with Hoechst (i) during metaphase is shown (see merge in g). The corresponding DIC image is given in (f). (C) Time-lapse series (see Supplementary Video S2 at JXB online) of GFP OsKCH1 localization in tobacco BY-2 cells during late telophase and the beginning of cytokinesis. A double-staining of GFP OsKCH1 with Hoechst is shown in (i) together with a merge including the DIC image (h). Arrows highlight filaments that tether the nuclei to the periphery (g), and bridge the nucleus and the new cell wall (f). Scale bars: 20 μm. (This figure is available in colour at JXB online.)
Nuclear positioning is delayed in BY-2 KCH1
Phenotypic and microscopic analysis had shown that mitosis in general in BY-2 KCH1 was not severely altered. Mitotic spindles had morphologically normal appearances and at most of the time points investigated, mitosis occurred at a normal rate indicating that the cells, in principle, do not have any problems with the division process itself. Interestingly, however, the onset of mitosis was clearly delayed in BY-2 KCH1, resulting in a reduced MI at early time-points after subcultivation. In plant cells that prepare for division, typically a migration of the nucleus occurs towards the site where the future cell plate will form (for a review, see Nick, 2008). Especially for the first division, the nucleus has to cover a considerable distance from the cell periphery. During the subsequent divisions, however, it remains near the cell centre and premitotic migration is then, as a consequence, less prominent. Interestingly, detailed microscopic investigations of the localization of GFP OsKCH1 in tobacco BY-2 cells have revealed that OsKCH1 signals during nuclear migration in cytokinetic and premitotic cells were predominantly found at the leading edge of the migrating nucleus (Fig. 7C), pointing towards a possible connection to this process.
In order to study if premitotic migration might be impaired in BY-2 KCH1, mean nuclear positions (NP) were followed in BY-2 KCH1 and BY-2 Wt during the culture cycle as described by Katsuta et al. (1990) with minor modifications (Fig. 7D). Due to the addition of exogenous 2,4-D to the auxin-depleted stationary cells, division activity reinitiates from about 0.5–1 d after subcultivation with a high degree of homogeneity. At days 0.5, 1, and 2, after subcultivation, the mean NP in BY-2 Wt and BY-2 KCH1 were significantly different (Fig. 7D). While in the non-transformed Wt the nuclei had already shifted from their original lateral into a more central position at day 0.5, most BY-2 KCH1 nuclei were still located at the periphery, indicating a delay in premitotic nuclear movement in the OsKCH1 overexpressor. At days 1 and 2, in the majority of BY-2 Wt cells, nuclei had already reached the cell centre while, in BY-2 KCH1, this position was only reached at day 3 after subcultivation in the majority of cells. The histograms in Fig. 7E exhibit the percentage occurrence of the NP values in 1800 cells of both the BY-2 Wt and the BY-2 KCH1 cell line at day 1, respectively, and illustrate the differences in nuclear positions even more clearly. Thus, in Wt cells at 1 d, in about 70% of the cells the nuclei were centrally located, while at the same time point in KCH1 cells this was the case only for about 28% of the population.
Discussion
Plant kinesins with the calponin-homology domain (KCHs) have recently been identified as plant-specific family of kinesins (Tamura et al., 1999; Preuss et al., 2004) and have been associated with a role in microtubule–microfilament interaction (Preuss et al., 2004; Frey et al., 2009; Xu et al., 2009). Co-ordination and cross-talk between microtubules and microfilaments is necessary for a couple of processes in plant growth and development such as the control of cell elongation and tissue expansion (for a recent review see Collings, 2008). The cellular role of KCH proteins in microtubular–actin interaction is, however, still far from understood. In this study, the potential biological role of the rice KCH member OsKCH1 was therefore addressed. Tissue and developmental regulation of OsKCH1 gene expression, and the phenotypes of rice Tos17 kch1 knock-out mutants on the one hand, and a KCH1 overexpression line generated in tobacco BY-2 cells were analysed. It was observed that, in both systems, cell elongation and cell division were altered antagonistically as the result of knock-out and overexpression, respectively. In addition, dynamic repartitioning of OsKCH1 during the cell cycle is reported and it is demonstrated that KCH overexpression delays nuclear positioning and mitosis in BY-2.
Our gene expression studies show that the transcription of OsKCH1 is regulated in a tissue- and development-specific way with high transcript abundances in young and developing tissues, pointing towards a possible involvement of the protein in cell division and development. For other members of KCH, similar variations of protein expression in different developmental stages have been reported. Preuss et al. (2004), for example, show that the GhKCH1 protein in cotton fibres was predominantly abundant at 10–17 d post-anthesis (DPA), whereas the levels strongly decreased at time points later than 21 DPA. Furthermore, Tamura et al. (1999) could detect the Arabidopsis KatD protein only in total protein extracts of flowers and not in any other organ including of leaflets, stems, roots, or siliques.
A morphological investigation revealed that rice kch1 Tos17 insertion mutants show alterations with respect to cell elongation and cell division in coleoptiles. The average cell length in coleoptiles was clearly reduced in the mutants. In addition, in contrast to the wild type, the apical cells were still not fully expanded when the primary leaves emerged. This indicates reduced and delayed elongation growth, producing generally shorter cells. In the upper third of mutant coleoptiles, cell length reached only about 60% of the length observed in the wild types. This observed strong reduction in cell length in the kch1 insertion mutants was, however, almost compensated by a concomitant increase in the average cell number per coleoptile, indicating an increased division during late embryogenesis when the coleoptile is laid down.
During subsequent development, no additional prominent phenotypes were identified, similar to the situation in Arabidopsis, where no significant phenotypes of null mutations in AtKatD had been observed, as mentioned by Preuss et al. (2004). This indicates functional redundancy existing among KCHs. Richardson et al. (2006) already conducted a thorough phylogenetic analysis of kinesins in different photosynthetic organisms and detected seven copies of KCH in Arabidopsis thaliana and Oryza sativa that have presumably emerged through gene duplication during plant evolution. Our phylogenetic analysis, including KCHs from the grass model plant Oryza sativa, the Rosids Arabidopsis thaliana, Populus trichocarpa, Vitis vinifera, and Ricinus communis, and the moss Physcomitrella patens, shows that the proteins cluster into four different clades, each containing at least one copy of KCH from all species, with the exception of the KCHs from Physcomitrella that only form a branch, indicating a possible higher evolutionary divergence of these proteins. OsKCH1 clusters into the most expanded branch, together with two other closely related rice KCH family members, such that functional redundancy is to be expected.
Consistent with the observed coleoptile phenotype in rice kch1 mutants, BY-2 cells overexpressing OsKCH1 were clearly more elongated than the non-transformed BY-2 wild-type cells and a cell line overexpressing free GFP. For the cotton homologues of KCH1 a potential role has been proposed for fibre elongation (Kim and Triplett, 2001; Preuss et al., 2004), because GhKCH1 was expressed during rapid cell expansion of the fibre where a parallel array of cortical microtubules is essential for the elongation process (Kim and Triplett, 2001). Similarly, GhKCH2 was proposed to maintain fibre elongation through cross-linking of microtubules and microfilaments (Xu et al., 2009). Our observations of knock-out and overexpression phenotypes for OsKCH1 confirm that this protein influences cell elongation. Inhibitor studies have demonstrated that actin filaments and microtubules interact closely during cell growth and elongation for various plant and cell systems including cotton fibres (Seagull, 1990), Arabidopsis roots (Baskin and Bivens, 1995; Collings et al., 2006), and seedlings of maize and rice (Giani et al., 1998; Wang and Nick, 1998; Blancaflor, 2000). The KCH1 proteins interact with both cytoskeletal elements and are, therefore, good molecular candidates for this interaction. Our observations would be consistent with a model, where KCH1 is required for efficient cell elongation by stabilizing transverse arrays of microtubules. Loss-of-function should then result in reduced cell length (which was observed in the coleoptiles of the Tos17 mutants; Fig. 2), whereas elevated abundance of KCH1 should result in increased cell length (as observed in the BY-2 OsKCH1 overexpressor; Fig. 4).
However, the reality is more complex than this simple working hypothesis. The observed changes in cell length in the mutants and overexpressor were accompanied by antagonistic changes in cell number. In the kch1 insertion mutants, the cell length was strongly reduced, and this reduction was almost compensated by a concomitant increase in the cell number. Conversely, in the BY-2 OsKCH1 overexpressor, the increased cell length was accompanied by a reduced mitotic index during the first days of the culture cycle. Therefore, our observations are equally compatible with a model, where the primary target of KCH1 is not cell expansion, but cell division. At first glance, this antagonism between cell elongation and cell division might appear trivial: cells are obviously larger when they have undergone a lower number of divisions. However, this trivial model ignores the fact that the regulation of cell division and cell expansion clearly differs in both Arabidopsis seedlings (Chen et al., 2001b; Perfus-Barbeoch et al., 2004) and tobacco cells (Chen et al., 2001a; Campanoni and Nick, 2005). Whereas auxin stimulates cell expansion via the receptor ABP1 and independently of G-protein signalling, cell division is controlled by an auxin receptor that differs with respect to ligand specificity and triggers G-protein dependent signalling (Ullah et al., 2001; Campanoni and Nick, 2005). The highest expression of OsKCH1 was found in young roots, young leaves, young flowers, and flowers post-pollination, in other words, in tissues endowed with meristematic activity, whereas it was almost absent in adult leaves (despite the fact that these leaves still expand throughout their life time), indicating a close link with cell division. When interpreting the relatively high expression of OsKCH1 in coleoptiles (where no cell division is observed following germination), one has to bear in mind that the coleoptile is the embryonic organ with the highest mitotic activity continuing until the very onset of dormancy (Nick et al., 1994).
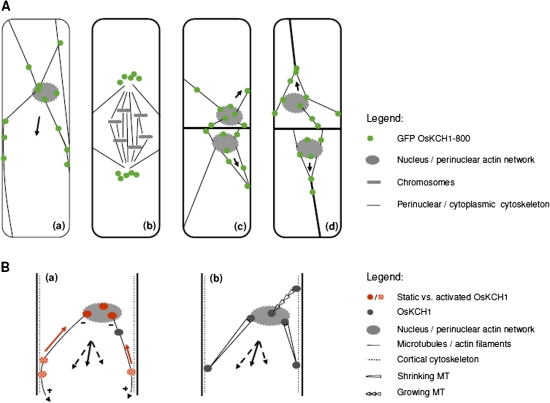
If cell division is a primary target for KCH1, it should be possible to observe directly that KCH1 is localized with mitotic structures or events, and that overexpression of KCH1 alters these structures or events. However, when mitosis was investigated in the BY-2 KCH1 overexpressor, spindle morphology was found to be identical to non-transformed wild-type cells, suggesting that mitosis in general was not structurally disturbed by overexpression of OsKCH1. However, the rate of mitosis during the first days of the culture cycle was clearly reduced in BY-2 KCH1, pointing towards an extended interphase and a delayed onset of mitosis. A microscopic analysis of GFP OsKCH1 localization in tobacco BY-2 revealed a dynamic repartitioning of the protein during the cell cycle. In interphasic and premitotic cells, a punctate OsKCH1 signal was clearly aligned with filaments that tethered the nucleus to the periphery. In addition, the signal accumulated at the sites, where these filaments emanate from the nuclear rim and where they merged into the cell cortex. Interestingly, at the onset of mitosis when the nuclear envelope breaks down, OsKCH1 retracted from the nucleus and the division plane such that it did not colocalize with mitotic microtubule structures. The signals of OsKCH1 during metaphase and anaphase, furthermore, did not appear as fine dots but rather accumulated in vesicular structures, indicating either an association of the kinesin with remnants of the nuclear envelope or even degradation of the protein following nuclear envelope breakdown. While OsKCH1 seems not to be involved in the mitotic processes that follow the nuclear envelope breakdown, a reorganization of the protein could be observed during cytokinesis. Thus, subsequent to the division, OsKCH1 reassembled around the newly formed daughter nulei and repopulated the filaments that joined the nuclei to the periphery and the newly formed cross wall (Fig. 8A). This dynamic relocalization pattern does not support a direct role of KCH1 in mitosis in the strict sense, but rather indicates a relationship with the position of the nucleus.
Fig. 8.
OsKCH1 localization throughout the cell cycle and working models for OsKCH1 function in nuclear positioning. (A) Dynamic redistribution of GFP OsKCH1 during the cell cycle. In interphasic and premitotic cells (a), OsKCH1 signals colocalized with a cytoskeletal network that tethers the nucleus to the periphery. Punctate signals accumulated at the sites, where the filaments reached the nuclear envelope or connected to the cell cortex. During mitosis (b), OsKCH1 retracted from nucleus and division plane and did not colocalize with mitotic microtubule structures. During cytokinesis, OsKCH1 signals were repartitioned and subsequently mainly found surrounding the newly formed nuclei, again along the cytoskeletal network that tethered these nuclei to the periphery or connected the nuclei with the cell poles. Arrows indicate the observed directions of nuclear movement. (B) Schematic models for the function of OsKCH1 in nuclear positioning. A ‘sliding model’ (a) is based on two different populations of OsKCH1. Static OsKCH1 proteins anchor the minus (–) end of radial microtubules at the nuclear envelope, possibly via interaction with the perinuclear actin network. The plus (+) ends of the microtubules are captured at the cortex by protein complexes involving activated OsKCH1 motors that move towards microtubule minus-ends (red arrow), but are anchored such that they generate a sliding force (arrowheads) that finally acts on the nucleus. Black arrows depict the force vectors that produce a resulting force (plain arrow) pointing in the observed directions of nuclear movement. A ‘pulling/pushing model’ (b) relies rather on microtubule dynamics. Microtubules are captured by anchor protein complexes involving OsKCH1 on both the nuclear envelop and the cell cortex. Depending on the accessory protein complexes, microtubule dynamics are differentially regulated, resulting either in growth or shrinkage. Black arrows again depict the force vectors. (This figure is available in colour at JXB online.)
In plant cells that prepare for division, typically a migration of the nucleus occurs towards the site where the prospective cell plate will form, as recently reviewed (Nick, 2008). Such migration is similarly observed in tobacco BY-2 cells and was found to be highly sensitive to inhibitors of microtubules and microfilaments (Katsuta and Shibaoka, 1988; Katsuta et al., 1990) indicating a tight interplay between both types of cytoskeletal elements. Katsuta et al. (1990) proposed that the premitotic network of microtubules might serve as a scaffold for the positioning of actin filaments that establish and maintain the position of the nucleus and the mitotic apparatus. KCH binds to both microtubules and microfilaments and cross-links actin and microtubules in vitro (Xu et al., 2009). Moreover they are predominantly localized at contact points between actin filaments and microtubules in vivo (Frey et al., 2009). It was observed that in the BY-2 KCH1 overexpressor the premitotic movement of the nucleus into the cell centre in BY-2 is clearly delayed, correlating with delayed mitotic activity during the culture cycle and elevated cell expansion. This is accompanied by the accumulation of GFP-tagged KCH1 along perinuclear filaments surrounding the nucleus as long as the nuclear envelope is contiguous, but repartitioned from the nucleus as soon as the nuclear envelope disintegrates. The most straightforward model to explain these observations would assume that KCH1 proteins could participate in the control of premitotic nuclear positioning (Fig. 8B).
In classical studies, Murata and Wada analysed the functions of the nucleus and the preprophase band for the formation of the ensuing cell plate and demonstrated that the position of the cell plate is determined by the nucleus (Miyao et al., 2003). The preprophase band in turn has a guiding function for the correct orientation of the cell plate (Traas et al., 1987; McClinton and Sung, 1997; Miyao et al., 2003). A delayed movement of the nucleus into the cell centre would thus lead to a delayed onset of division, as observed in the KCH1 overexpressor. The orientation of the ensuing cell plate would, in these cells, however, not be structurally altered, as the preprophase bands only starts to develop when the nucleus reaches its position. As cell division and cell growth are coupled via antagonistic signalling mechanisms that compete for auxin as a common trigger (cell division is controlled auxin-dependently via G-protein signalling while cell expansion is stimulated by auxin via the receptor ABP1), a delay in cell division would concomitantly result in a stimulation of cell expansion (Ullah et al., 2001; Campanoni and Nick, 2005). The observed effect of KCH1 on the overall cell size could in this context be seen as a secondary effect due to the delayed onset of mitosis.
Figure 8B shows two principal models that are not necessarily mutually exclusive and display how KCH1 possibly might contribute to nuclear movement. Microtubules and actin filaments might transmit forces that are generated by KCH1 at the perinuclear contact sites to the cortex such that the nucleus is either pulled or pushed or both. Alternatively, KCH1 might simply anchor the perinuclear network at the cell cortex and move the nucleus by mutual sliding of actin filaments and/or microtubules in the cortical cytoplasm. Future studies on the specific motor properties of KCH1, as well as the identification of putative regulatory factors, might contribute to a better understanding of a possible involvement of this interesting class of kinesins in nuclear positioning in plant cells.
Nuclear positioning has been studied in S. cerevisiae, S. pombe, filamentous fungi, and a variety of animal cells and tissues and the systems that orient and move nuclei were found to be moderately conserved and involve as key players dynein, dynactin, and other proteins at the plus ends of astral microtubules, mediating interaction with the cell cortex and the actin cytoskeleton (Morris, 2003; Yamamoto and Hiraoka, 2003). Both repulsive and attractive forces are generated by a combination of microtubule polymerization and depolymerization events, complemented by a tuneable, dynein-mediated sliding of microtubules along the cell cortex (Adames and Cooper, 2000). In plants, which lack dyneins and its associated proteins (Lawrence et al., 2001), the mechanisms for moving nuclei must involve a variety of fundamentally different players, interacting with both premitotic microtubules and the actin cytoskeleton. Our knowledge of the molecular details has, however, remained incomplete. Could KCH proteins be the functional homologues of dyneins as cortical anchors with minus-end directed motor activity? In order to find answers to these questions, future research will aim to identify binding partners of KCH and the regulatory circuits modulating its motor activity to get further insight into the putative cellular functions of this fascinating class of unconventional molecular motors.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Colocalization of OsKCH1 with microtubules and actin filaments in tobacco BY-2 cells.
Supplementary Video S1: Cell stably expressing GFP KCH1-800 during early stages of mitosis.
Supplementary Video S2: Cells stably expressing GFP KCH1-800 during cytokinesis.
Supplementary Video S3: Cells stably expressing GFP KCH1-800 subsequent cytokinesis.
Supplementary Video S4. Cells stably expressing GFP KCH1-800 during interphase.
Supplementary Material
Acknowledgments
The authors are grateful to Dr Michael Riemann (University of Karlsruhe) for providing us with the primers Rubg237F/Rubg304R for Ubiquitin amplification. We thank Franziska Bühler, Juliane Draksler, and Kerstin Schwarz for their contributions to Figs 3 and 6. Angelika Piernitzki is acknowledged for her assistance with rice cultivation. This work was financially supported by the Centre for Functional Nanostructures of the University of Karlsruhe and the German Research Council (Project Ni 324/12-1).
Glossary
Abbreviations
- OsKCH1
Oryza sativa kinesin with calponin-homology domain 1
References
- Adames NR, Cooper JA. Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. Journal of Cell Biology. 2000;149:863–874. doi: 10.1083/jcb.149.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin TI, Bivens NJ. Stimulation of radial expansion in arabidopsis roots by inhibitors of actomyosin and vesicle secretion but not by various inhibitors of metabolism. Planta. 1995;197:514–521. doi: 10.1007/BF00196673. [DOI] [PubMed] [Google Scholar]
- Blancaflor EB. Cortical actin filaments potentially interact with cortical microtubules in regulating polarity of cell expansion in primary roots of maize (Zea mays L.) Journal of Plant Growth Regulation. 2000;19:406–414. doi: 10.1007/s003440000044. [DOI] [PubMed] [Google Scholar]
- Breitling F, Little M. Carboxy-terminal regions on the surface of tubulin and microtubules. Epitope locations of YOL1/34, DM1A and DM1B. Journal of Molecular Biology. 1986;189:367–370. doi: 10.1016/0022-2836(86)90517-6. [DOI] [PubMed] [Google Scholar]
- Campanoni P, Nick P. Auxin-dependent cell division and cell elongation. 1-Naphthaleneacetic acid and 2,4-dichlorophenoxyacetic acid activate different pathways. Plant Physiology. 2005;137:939–948. doi: 10.1104/pp.104.053843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Shimomura S, Sitbon F, Sandberg G, Jones AM. The role of auxin-binding protein 1 in the expansion of tobacco leaf cells. The Plant Journal. 2001a;28:607–617. doi: 10.1046/j.1365-313x.2001.01152.x. [DOI] [PubMed] [Google Scholar]
- Chen JG, Ullah H, Young JC, Sussman MR, Jones AM. ABP1 is required for organized cell elongation and division in Arabidopsis embryogenesis. Genes and Development. 2001b;15:902–911. doi: 10.1101/gad.866201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collings DA. Crossed-wires: interactions and cross-talk between the microtubule and microfilament networks in plants. In: Nick P, editor. Plant microtubules. Berlin/Heidelberg: Springer Verlag; 2008. pp. 47–79. [Google Scholar]
- Collings DA, Lill AW, Himmelspach R, Wasteneys GO. Hypersensitivity to cytoskeletal antagonists demonstrates microtubule-microfilament cross-talk in the control of root elongation in Arabidopsis thaliana. New Phytologist. 2006;170:275–290. doi: 10.1111/j.1469-8137.2006.01671.x. [DOI] [PubMed] [Google Scholar]
- Deeks MJ, Fendrych M, Smertenko A, Bell KS, Oparka K, Cvrckova F, Zarsky V, Hussey PJ. The plant formin AtFH4 interacts with both actin and microtubules, and contains a newly identified microtubule-binding domain. Journal of Cell Science. 2010;123:1209–1215. doi: 10.1242/jcs.065557. [DOI] [PubMed] [Google Scholar]
- Frey N, Klotz J, Nick P. Dynamic bridges: a calponin-domain kinesin from rice links actin filaments and microtubules in both cycling and non-cycling cells. Plant and Cell Physiology. 2009;50:1493–1506. doi: 10.1093/pcp/pcp094. [DOI] [PubMed] [Google Scholar]
- Giani S, Qin X, Faoro F, Breviario D. In rice, oryzalin and abscisic acid differentially affect tubulin mRNA and protein levels. Planta. 1998;205:334–341. doi: 10.1007/s004250050328. [DOI] [PubMed] [Google Scholar]
- Goode BL, Drubin DG, Barnes G. Functional cooperation between the microtubule and actin cytoskeletons. Current Opinion in Cell Biology. 2000;12:63–71. doi: 10.1016/s0955-0674(99)00058-7. [DOI] [PubMed] [Google Scholar]
- Guindon S, Lethiec F, Duroux P, Gascuel O. PHYML Online: a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Research. 2005;33:W557–W559. doi: 10.1093/nar/gki352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holweg C, Susslin C, Nick P. Capturing in vivo dynamics of the actin cytoskeleton stimulated by auxin or light. Plant and Cell Physiology. 2004;45:855–863. doi: 10.1093/pcp/pch102. [DOI] [PubMed] [Google Scholar]
- Katsuta J, Hashiguchi Y, Shibaoka H. The role of the cytoskeleton in positioning of the nucleus in premitotic tobacco BY-2 cells. Journal of Cell Science. 1990;95:413–422. [Google Scholar]
- Katsuta J, Shibaoka H. The roles of the cytoskeleton and the cell wall in nuclear positioning in tobacco BY-2 Cells. Plant and Cell Physiology. 1988;29:403–413. [Google Scholar]
- Kim HJ, Triplett BA. Cotton fiber growth in planta and in vitro. Models for plant cell elongation and cell wall biogenesis. Plant Physiology. 2001;127:1361–1366. [PMC free article] [PubMed] [Google Scholar]
- Lawrence CJ, Morris NR, Meagher RB, Dawe RK. Dyneins have run their course in plant lineage. Traffic. 2001;2:362–363. doi: 10.1034/j.1600-0854.2001.25020508.x. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive tree of life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- Liu B, Cyr RJ, Palevitz BA. A kinesin-like protein, KatAp, in the cells of arabidopsis and other plants. The Plant Cell. 1996;8:119–132. doi: 10.1105/tpc.8.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisch J, Fiserova J, Fischer L, Nick P. Tobacco Arp3 is localized to actin-nucleation sites in vivo. Journal of Experimental Botany. 2009;60:603–614. doi: 10.1093/jxb/ern307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClinton R, Sung Z. Organization of cortical microtubules at the plasma membrane in Arabidopsis. Planta. 1997;201:252–260. doi: 10.1007/s004250050064. [DOI] [PubMed] [Google Scholar]
- Mitsui H, Yamaguchi-Shinozaki K, Shinozaki K, Nishikawa K, Takahashi H. Identification of a gene family (kat) encoding kinesin-like proteins in Arabidopsis thaliana and the characterization of secondary structure of KatA. Molecular and General Genetics. 1993;238:362–368. doi: 10.1007/BF00291995. [DOI] [PubMed] [Google Scholar]
- Miyao A, Tanaka K, Murata K, Sawaki H, Takeda S, Abe K, Shinozuka Y, Onosato K, Hirochika H. Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. The Plant Cell. 2003;15:1771–1780. doi: 10.1105/tpc.012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris NR. Nuclear positioning: the means is at the ends. Current Opinion in Cell Biology. 2003;15:54–59. doi: 10.1016/s0955-0674(02)00004-2. [DOI] [PubMed] [Google Scholar]
- Nagata T, Kumagai F. Plant cell biology through the window of the highly synchronized tobacco BY-2 cell line. Methods in Cell Science. 1999;21:123–127. doi: 10.1023/a:1009832822096. [DOI] [PubMed] [Google Scholar]
- Nagata T, Nemoto Y, Hasezawa S. Tobacco BY-2 cell line as the ‘HeLa’ cell in the cell biology of higher plants. International Review of Cytology. 1992;132:1–30. [Google Scholar]
- Nick P. Control of cell axis. In: Nick P, editor. Plant microtubules. Berlin/Heidelberg: Springer Verlag; 2008. pp. 23–46. [Google Scholar]
- Nick P, Furuya M. Phytochrome dependent decrease of gibberellin-sensitivity. Plant Growth Regulation. 1993;12:195–206. [Google Scholar]
- Nick P, Heuing A, Ehmann B. Plant chaperonins: a role in microtubule-dependent wall formation? Protoplasma. 2000;211:234–244. [Google Scholar]
- Nick P, Yatou O, Furuya M, Lambert A–M. Auxin-dependent microtubule responses and seedling development are affected in a rice mutant resistant to EPC. The Plant Journal. 1994;6:651–663. [Google Scholar]
- Perfus-Barbeoch L, Jones AM, Assmann SM. Plant heterotrimeric G protein function: insights from Arabidopsis and rice mutants. Current Opinion in Plant Biology. 2004;7:719–731. doi: 10.1016/j.pbi.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Petrasek J, Schwarzerova K. Actin and microtubule cytoskeleton interactions. Current Opinion in Plant Biology. 2009;12:728–734. doi: 10.1016/j.pbi.2009.09.010. [DOI] [PubMed] [Google Scholar]
- Preuss ML, Kovar DR, Lee YR, Staiger CJ, Delmer DP, Liu B. A plant-specific kinesin binds to actin microfilaments and interacts with cortical microtubules in cotton fibers. Plant Physiology. 2004;136:3945–3955. doi: 10.1104/pp.104.052340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson D, Simmons M, Reddy A. Comprehensive comparative analysis of kinesins in photosynthetic eukaryotes. BMC Genomics. 2006;7:18. doi: 10.1186/1471-2164-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemann M, Riemann M, Takano M. Rice JASMONATE RESISTANT 1 is involved in phytochrome and jasmonate signalling. Plant, Cell and Environment. 2008;31:783–792. doi: 10.1111/j.1365-3040.2008.01790.x. [DOI] [PubMed] [Google Scholar]
- Rodriguez OC, Schaefer AW, Mandato CA, Forscher P, Bement WM, Waterman-Storer CM. Conserved microtubule-actin interactions in cell movement and morphogenesis. Nature Cell Biology. 2003;5:599–609. doi: 10.1038/ncb0703-599. [DOI] [PubMed] [Google Scholar]
- Seagull R. The effects of microtubule and microfilament disrupting agents on cytoskeletal arrays and wall deposition in developing cotton fibers. Protoplasma. 1990;159:44–59. [Google Scholar]
- Tamura K, Nakatani K, Mitsui H, Ohashi Y, Takahashi H. Characterization of katD, a kinesin-like protein gene specifically expressed in floral tissues of Arabidopsis thaliana. Gene. 1999;230:23–32. doi: 10.1016/s0378-1119(99)00070-0. [DOI] [PubMed] [Google Scholar]
- Traas JA, Doonan JH, Rawlins DJ, Shaw PJ, Watts J, Lloyd CW. An actin network is present in the cytoplasm throughout the cell cycle of carrot cells and associates with the dividing nucleus. Journal of Cell Biology. 1987;105:387–395. doi: 10.1083/jcb.105.1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah H, Chen JG, Young JC, Im KH, Sussman MR, Jones AM. Modulation of cell proliferation by heterotrimeric G protein in Arabidopsis. Science. 2001;292:2066–2069. doi: 10.1126/science.1059040. [DOI] [PubMed] [Google Scholar]
- Wang QY, Nick P. The auxin response of actin is altered in the rice mutant Yin-Yang. Protoplasma. 1998;204:22–33. doi: 10.1007/BF01282290. [DOI] [PubMed] [Google Scholar]
- Wasteneys GO, Galway ME. Remodeling the cytoskeleton for growth and form: an overview with some new views. Annual Review of Plant Biology. 2003;54:691–722. doi: 10.1146/annurev.arplant.54.031902.134818. [DOI] [PubMed] [Google Scholar]
- Xu T, Qu Z, Yang X, Qin X, Xiong J, Wang Y, Ren D, Liu G. A cotton kinesin GhKCH2 interacts with both microtubules and microfilaments. Biochemistry Journal. 2009;421:171–180. doi: 10.1042/BJ20082020. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Hiraoka Y. Cytoplasmic dynein in fungi: insights from nuclear migration. Journal of Cell Science. 2003;116:4501–4512. doi: 10.1242/jcs.00835. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.