Summary
Several approaches have been developed for screening combinatorial libraries or collections of synthetic molecules for agonists or antagonists of protein function, each with its own advantages and limitations. In this report, we describe an experimental platform that seamlessly couples massively parallel bead-based screening of one bead one compound combinatorial libraries with microarray-based quantitative comparisons of the binding affinities of the many hits isolated from the bead library. Combined with other technical improvements, this technique allows the rapid identification of the best protein ligands in combinatorial libraries containing millions of compounds without the need for labor-intensive re-synthesis of the hits.
Keywords: Combinatorial library, peptoid, peptide, split and pool synthesis, magnet, microarray, screening
Introduction
The discovery of synthetic molecules able to recognize proteins with high specificity and affinity is an issue of great current interest. Nowadays, most such molecules are discovered through screening efforts, of which there are two broad types. The first are functional screens, in which soluble small molecules are introduced into the wells of microtiter plates and assayed individually for their ability to alter the activity of an enzyme, elicit a certain phenotype in a cell, etc. Functional screens, while powerful, have several limitations. It is impractical to screen more than approximately 1,000,000 different compounds and even this is a major undertaking. Because of the necessity of handling a large number of individual compounds, an elaborate infrastructure of automated instrumentation is required and these screens are expensive.
Alternatively, one can employ binding assays. For libraries of synthetic molecules, the compounds of interest are generally displayed on a suitable solid support and exposed to a soluble, labeled protein under the desired conditions and retention of the label is monitored. This approach was developed first for bead-displayed peptide libraries created by split and pool synthesis (Lam et al., 1991), where each bead displays many copies of a single molecule. The identity of the “hits” in a bead-binding assay must be determined post-screening. For peptides and certain other oligomeric molecules (Alluri et al., 2003), sensitive analytical techniques are available that allow the structure of the “hits” to be determined directly from a single bead. If this is not the case, various encoding strategies can be employed to characterize the structure of hits indirectly. (Liu et al., 2002; Ohlmeyer et al., 1993) More recently, microarrays have been employed in binding screens. (Lam and Renil, 2002; MacBeath et al., 1999; Uttamchandani et al., 2005) In this format, thousands of different molecules are printed onto chemically modified glass slides so as to become attached covalently to the surface (Bradner et al., 2006; Kuruvilla et al., 2002).
Bead-based and microarray screening have complementary strengths and weaknesses. The major advantage of bead-based screens is that a large number of compounds can be screened easily and cheaply in a single experiment. This is because the binding screen is done as a batch assay and it is unnecessary to spatially segregate all of the beads prior to the screen. Microarray fabrication does require the physical separation of compounds into the wells of microtiter plates prior to spotting and thus requires some, but not all, of the infrastructure employed for functional screening. Moreover, the number of compounds that can be spotted onto a single slide is limited to a few tens of thousands. On the other hand, many microarrays can be made from small amounts of compounds, facilitating quantitative analysis via titration experiments. Moreover, in any one experiment, the relative binding characteristics of all of the compounds on the array can be compared. Such studies are difficult to do with bead libraries, because labor-intensive re-synthesis and detailed binding studies are usually required to identify the best ligands from the large number of hits that may result from a bead-based screen.
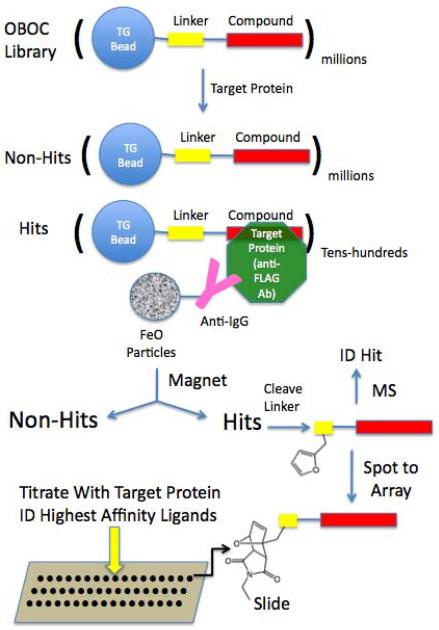
In this report, we describe a method for screening synthetic libraries and characterizing the resultant hits that combines many of the attractive features of bead library screening and microarray-based analysis in a seamless fashion. This allows very large libraries of millions of compounds to be screened rapidly and cheaply for the highest affinity protein ligands present. The key features of this method are the separation of hits from non-hits using magnetic capture and the ability to both identify the sequence of the hits and spot them onto microarrays for subsequent quantitative analysis without the need for hit re-synthesis (see Fig. 1). This approach allows millions of synthetic molecules to be analyzed quickly and easily for binding to a protein of interest and greatly facilitates the determination of which of these compounds exhibits the best affinity and specificity for the target.
Figure 1. Overview of the integrated magnetic screening and testing of hits on microarrays.
Millions of 75 μm TentaGel beads from a one bead one compound (OBOC) library are incubated with target protein (anti-FLAG antibody in this study), washed, then incubated with anti-target protein antibodies linked covalently to iron oxide containing particles (Dynabeads). Beads that bind the target protein and therefore also attract Dynabeads, are retained on the side of the tube using a powerful magnet and non-magnetic beads are removed. Each of the putative “hit” beads is separated into the well of a microtiter plate and the compounds are removed from the beads by cleavage of a linker. The compounds are then spotted onto a maleimide-activated glass slide via a Diels-Alder reaction involving a conserved furan-containing monomer incorporated into each sequence. The structure of each putative hit is deduced by tandem mass spectrometry (MS). The compound microarrays are then probed with different concentrations of the target protein to determine the intrinsic affinity of each of the hit compounds for the target. In this way, no re-synthesis of the hits is necessary until the best binders are identified.
Results
The central goal of this study was to establish a screening strategy that would allow millions of bead-displayed compounds to be screened on resin rapidly and cheaply, followed by transfer of the hits to a microarray where their binding to the target of interest could be quantified (Fig. 1). Our previous work has shown that TentaGel, comprised of a polystyrene core coated with very long amine-terminated polyethylene glycol (PEG) chains is a superior bead surface for protein-binding screens due to its low non-specific protein binding capacity (Alluri et al., 2003). However, there is no simple way to release molecules built off of the terminal amine group from the resin. Therefore, we first focused on the development of a suitable linker arm that would support both efficient cleavage of hits from the beads and subsequent spotting onto maleimide-modified glass slides (Reddy and Kodadek, 2005).
Two linker types were explored, both based on well-known protocols for the specific cleavage of proteins, the cyanogen bromide-mediated cleavage C-terminal of Methionine (which has also been used by others recently (Thakkar et al., 2009)) and hydrolysis of the Asp-Pro peptide bond with dilute trifluoroacetic acid (TFA) (Crimmins et al., 2005). In the case of the Asp-Pro linker, a Cys residue was included to facilitate Michael addition of the cleaved molecule to the maleimide-terminated slides. In the Met-containing linkers, we found that a Cys residue led to side-reactions that decreased the purity of cleaved compounds, and rendered identification of the hit compounds difficult (data not shown). Therefore, we incorporated a furan-containing peptoid residue (Nffa; see Fig. 2). This supports linkage to the array via Diels-Alder reaction (Houseman et al., 2002). As is described in the Supplementary Material, FLAG peptide or Myc peptide were synthesized on 75 μm TentaGel beads with either the Cys-Asp-Pro or Nffa-Met linker (written in the N to C direction). We demonstrated that enough compound is produced from cleavage of a single bead with CNBr or dilute TFA to sequence the peptide using tandem MALDI mass spectrometry (MS). Moreover, when the compound was spotted onto an array and probed with either anti-Myc or anti-FLAG antibody, enough antibody was captured to easily detect a signal upon subsequent incubation with fluorescently labeled secondary antibody. More extensive work with small libraries showed that the Nffa-Met linker produced somewhat cleaner results when the molecules were sequenced by MS, but that about two-fold less compound was spotted onto the slides when compared to the Cys-Asp-Pro-linked compounds. While both linkers are suitable for use, we employed the Nffa-Met for the remainder of this study.
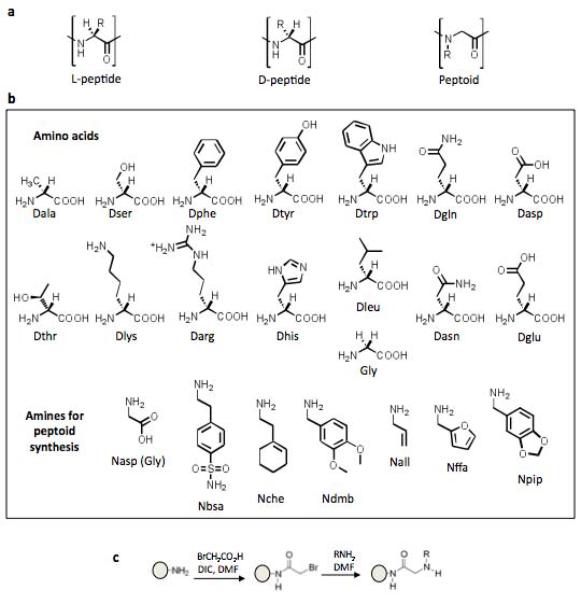
Figure 2. Composition of the combinatorial library employed in this study (X-X-X-X-X-X-Nffa-Met, where X = any of the peptide or peptoid monomers shown).
A) Structures of an L-peptide, D-peptide, and peptoid. B) Structures of the monomers used for library synthesis. C) The sub-monomer synthesis approach14, which illustrates how the amines shown in part b were incorporated into the library. The amino acids were incorporated using standard peptide couplings.
A combinatorial library was made by split and pool synthesis with the composition NH2-X6-Nffa-Met-TentaGel, where X = Nall, Nbsa, Nche, Ndmb, Npip, Gly, Dala, Darg, Dasn, Dasp, Dgln, Dglu, Dhis, Dleu, Dlys, Dphe, Dser, Dthr, Dtrp, or Dtyr (Fig. 2). Peptide couplings were done in the usual way, whereas the peptoid residues were inserted using the sub-monomer method of Zuckermann and co-workrs (Figliozzi et al., 1996) (Fig. 2c). The theoretical diversity of the library was 206 = 64 million compounds. Approximately one gram of 75 μm TentaGel resin, consisting of about four million beads, was employed for the synthesis, so most of the beads should display a unique D-peptide or D-peptide-peptoid hybrid. To carry out the screen, approximately two million beads were incubated with anti-FLAG antibody (67 nM in 5% milk blocking buffer) as a model target protein. This antibody recognizes the octapeptide N-Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Lys-C with high affinity. In previous studies, we had employed biotinylated proteins as targets and identified beads displaying protein-binding molecules by examination of the entire population under a fluorescent microscope after incubation with streptavidin (SA)-coated quantum dots. However, this is impractical with millions of beads, so we developed a more facile procedure to enrich “hits” from the library. After incubation of the antibody with the beads, secondary antibody-coated iron oxide particles (Invitrogen/Dynal) were added to the tube and the suspension was mixed. A strong magnet was then placed on the side of the tube, which was then made vertical. We anticipated that TentaGel beads that had bound the anti-FLAG antibody would be retained by the magnet through a peptide/peptoid•anti-FLAG antibody•secondary antibody-Dynabead bridging interaction (Fig. 1), while beads that do not bind to the anti-FLAG antibody should settle to the bottom of the tube. To ensure that no potential hits were left behind, after removing the beads that did not bind to the magnet with a pipette, we reintroduced new Dynabeads to this population and repeated the magnetic isolation procedure. We found that two rounds of this picked up several beads that were not retained by the magnet in the first round, but additional rounds did not yield more hits. A detailed procedure is provided in the Supplementary Material.
63 beads were retained by the magnet and separated manually into individual wells of a microtiter plate. We also included in other wells several beads that were not retained by the magnet as negative controls. Beads displaying FLAG peptide-Nffa-Met and Myc peptide-Nffa-Met were also included as further controls. The compounds were released into solution by treatment with 30 mg/ml CNBr in a 5:4:1 acetonitrile:acetic acid:water overnight. After transferring the resultant solution to a new plate, the solvent was evaporated and the compounds were processed as described in the Methods section such that some of the sample was used to spot onto maleimide-activated, PEGylated glass slides and some was employed for MALDI mass spectrometry-based sequencing. About 60% of the hits could be sequenced unambiguously (see Table 1)
16 copies of each array of 100 compounds (the hits and various controls) were spotted onto each microscope slide. Each array, isolated by applying a Whatman Fast Frame to the slide, was then incubated for two hours with either anti-Myc antibody or various concentrations of anti-FLAG antibody. After washing, the amount of antibody captured at each spot was quantified by subsequent hybridization with fluorescently-labeled secondary antibody, another wash, drying and scanning. Some of the results are shown in Fig. 3. From these data, quantitative binding curves for each compound spotted on the array could be derived (see Fig. 3 and Table 1). No binding of the anti-FLAG antibody to the Myc peptide was observed, nor was binding of the anti-Myc antibody to any of the hits or the FLAG peptide.
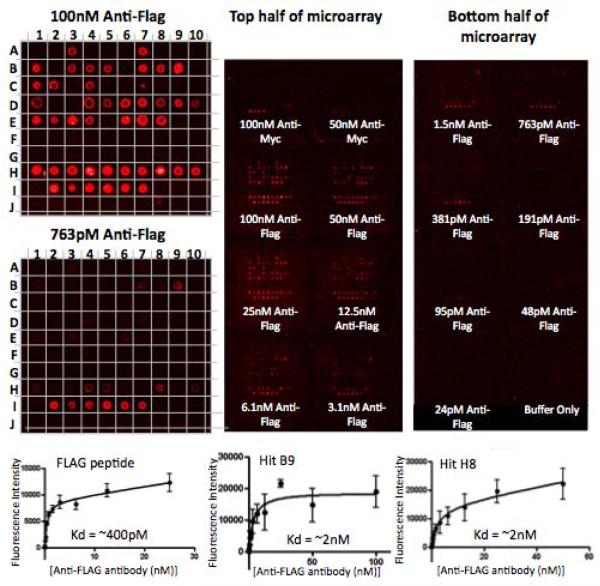
Figure 3. Microarray-based analysis of the hits isolated in the magnet-assisted screening procedure.
16 replicate arrays of hit compounds as well as positive and negative controls were spotted onto each of three microarray slides and hybridized with anti-Myc antibody or decreasing concentrations of anti-FLAG antibody, followed by red fluorescently labeled secondary antibodies. Displayed is the image of one of the three slides (right), with the 100 nM and 763 pM anti-FLAG antibody hybridized portions of the slide magnified (left). Anti-Myc antibody only binds Myc peptide, while anti-FLAG antibody binds FLAG peptide as well as many of the hits, but not the negative controls. The binding curves for FLAG peptide and two of the best hits are shown on the bottom. A1–E8, G10–I1 = Hits from X-X-X-X-X-X-Nffa-Met library screen, E9–G9 = Negatives from the screen, I2–I7 = FLAG peptide, I9–J4 = Myc peptide, I8, J5–J10 = blank. See Table 1 for sequences and binding affinities.
The data show that the compounds separate into two distinct classes: high affinity anti-FLAG ligands and those that do not bind the antibody detectably (false positives from the bead screen). The best of the hits had apparent KDs only about 5-fold higher than the native FLAG peptide antigen (see Fig. 3 and Table 1), while many displayed 10-to 100-fold lower affinity.
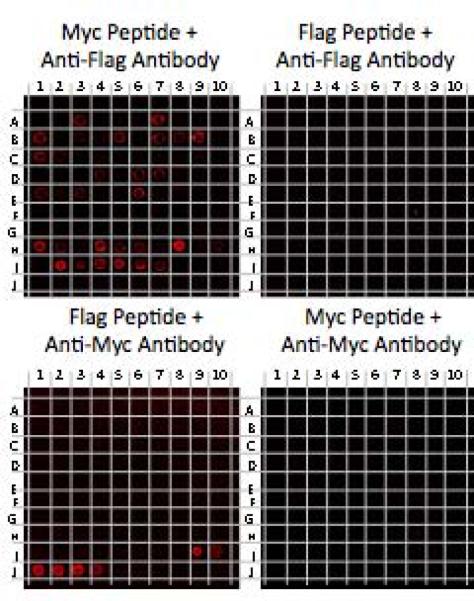
To address if the higher affinity hits bind to anti-FLAG antibody in the antigen binding site, we carried out a competition experiment in which the anti-FLAG antibody was first incubated with an excess of FLAG peptide or, as a control, the Myc peptide, before hybridization to the array. As shown in Fig. 4, the soluble FLAG peptide abrogated binding of the antibody to all of the molecules on the microarray, whereas the Myc peptide had little or no effect. While we cannot absolutely rule out allosteric competition, these data argue that all of the ligands derived from this screen bind to the peptide-binding site of the antibody.
Figure 4. Competition assays on microarrays.
100 nM of anti-FLAG or anti-Myc antibodies were pre-incubated with 100 μM FLAG or Myc peptides and hybridized to microarrays that were replicates of the one shown in Fig. 3. Shown are representative images of the scanned slides after hybridization with red fluorescently labeled secondary antibodies. Anti-FLAG antibody binds to FLAG peptide and hit compounds on the arrays when pre-incubated with Myc peptide, but all of these binding events are blocked when anti-FLAG antibody is pre-incubated with FLAG peptide. Anti-Myc binds to Myc peptide displaying spots after pre-incubation with FLAG peptide, but not Myc peptide. A1–E8, G10–I1 = Hits from X-X-X-X-X-X-Nffa-Met library screen, E9–G9 = Negatives from the screen, I2–I7 = FLAG peptide, I9–J4 = Myc peptide, I8, J5–J10 = blank.
Discussion
In this report, we demonstrate a powerful protocol that allows libraries comprised of millions of compounds to be screened rapidly for the highest affinity protein ligands. This method employs on-bead screening of one bead one compound (OBOC) libraries against a target protein, where the putative hits are enriched by magnetic isolation. The beads are then distributed into the wells of microtiter plates, the compounds released by cleavage of a special linker, then spotted onto maleimide-activated glass slides to which they attach covalently. The arrays are then titrated with different concentrations of the target protein, allowing the affinity of each compound on the array to be determined. Importantly, this procedure eliminates the requirement for re-synthesis of any of the hits prior to these binding assays, which would be impractical for the number of hits obtained. Central to the execution of this strategy was the development of two linkers, Nffa-Met and Cys-Asp-Pro, that allow hydrophilic TentaGel beads to be employed as the screening platform, but also support efficient post-screening cleavage of the molecule from the beads followed by covalent linkage to the maleimide-activated slides.
In addition to assessing the relative affinities of the hits for the target protein, the arrays can also be employed to monitor other binding parameters. For example, as shown in Fig. 4, a competition binding experiment using excess soluble FLAG peptide suggested that all of the molecules on the array that bound anti-FLAG antibody recognize the antigen-binding region of the antibody since the soluble peptide abrogated binding of the antibody to the immobilized compounds. This is consistent with the lack of binding of the anti-Myc antibody to any of the anti-FLAG antibody ligands (Fig. 4). This type of determination of whether a certain ligand binds to the target protein competitively with another is of interest in the construction of bivalent ligands (Erlanson et al., 2004; Maly et al., 2000; Shuker et al., 1996), where one links two molecules that bind different surfaces of the protein together with a suitable tether to create a higher affinity species. For example, one may have in hand a modest affinity ligand for a protein of interest, screen a library for new ligands, then use this technique to rapidly determine which of these new ligands competes with the one in hand.
As evidenced by their failure to bind anti-FLAG antibody on the array, several of the compounds isolated in the magnet-assisted bead screen were clearly false positives. These molecules do not resemble the consensus sequence of the high affinity hits (see Table 1). The isolation of these false positives could be due to a variety of factors. Some TentaGel beads that do not display target protein ligands may simply have been trapped physically with the true hits during incubation with the Dynabeads. Alternatively, it is possible that some of the false positives are secondary antibody ligands, since these were not pre-cleared from the library prior to the introduction of the anti-FLAG antibody. A variety of technical glitches could also explain these results, for example, poor coupling of a particular molecule to the array due to low solubility. Again, this was not checked specifically because we are not interested in what might be missed in a very high throughput protocol such as this, but rather are focused on identifying the highest affinity and best behaved protein ligands amongst the hits obtained. Indeed, the important point relevant to the false positives that emerge from the bead screen is that they are easily identified in the microarray step and thus are not a matter of concern.
While we have employed a library of oligomers comprised of mixed D-peptide and peptoid residues in this study, the screening method described here could be used with any type of library whose sequence can be determined directly from the amount of compound on a single bead or encoded on that bead (Liu et al., 2002). Thus, while D-peptide/peptoid hybrid libraries are good sources of protein ligands for in vitro applications and perhaps the development of injectable pharmaceuticals, this technique should be applicable to the discovery of orally bioavailable molecules as well. This study employed an antibody as a model target protein but this technique could be employed with almost any soluble protein target. The protein could be chemically biotinylated and hit beads isolated using streptavidin-coated Dynabeads.
Significance
We have developed a convenient and efficient method for the screening of very large one bead one compound libraries by combining several different technical advances. This protocol was demonstrated here for mixed peptide/peptoid libraries, but could be applied to any compound class where the structure of the molecule can be obtained from a single bead. The salient feature of the technique is to carry out the screen on hydrophilic beads and then to transfer candidate hits to a microarray for more detailed analysis. This allows the best hits to be identified without the need for tedious re-synthesis of many different compounds.
Experimental Procedures
Library hybridization and magnetic screening
TBST swelled beads were washed with TBST, then blocked with 50 mg/ml dried skim milk (Carnation) in 1:1 TBST:StartingBlock (Sigma) for 1 hour at room temperature (RT) in a 5 ml or 10 ml disposable reaction column. M2 monoclonal anti-flag antibody (Sigma) was diluted in 50 mg/ml milk in 1:1 TBST:StartingBlock at a concentration of 10 μg/ml and hybridized to beads for 1hr at RT. Beads were washed with TBST 8 times, resuspended in StartingBlock, and transferred to a 15 ml conical tube. 10 μl of 10 mg/ml sheep anti-mouse IgG antibody conjugated M280 Dynabeads was added per ml of StartingBlock. Typically, 3ml of solution was used per ~500,000 beads screened at each of the hybridization steps. For the library screen with biotinylated beads spiked in, 6ml of solution was used for ~2 million beads. The Dynabeads were hybridized with the library beads anywhere from 20 minutes to 2 hours. TBST was added to the tubes up to 14 mls, then the 15 ml conicals were placed in a DynaMag-15. Tubes were inverted slowly for 2 minutes, then left upright until the beads settled to the bottom. Solution and the beads at the bottom of the tube were transferred to a new 15 ml conical with a 5 ml pipette. Two more washes were performed, where 14 ml TBST was added, the tubes inverted, and placed back into the DynaMag-15 and solution drained as before (hit beads should be stuck on the sides of the tubes while in the DynaMag-15). After the last wash, 1 ml TBST was added to the tube and all beads and Dynabeads were collected to the TBST by inversion and rotation of the tube. The beads and TBST were transferred to a 1.5 ml eppendorf tube and placed under a dissecting microscope. A hand-held rectangular rare-earth metal magnet was very carefully placed next to the tube and the tube rotated while visualizing the beads under the microscope. Hit beads should follow the magnet while any negatives should stay at the bottom of the tube. Any negative beads were removed from the bottom with a 200 μl pipetteman while the hits were kept on the side of the tube next to the magnet. The tube was inverted until all Dynabeads were in suspension and the tube centrifuged briefly to let the hits settle to the bottom while the Dynabeads stayed in suspension. This was accomplished by pressing “short spin” until the speed reached 2,500 rpm, or pressing start and then stop as soon as the speed reached 2,500 rpm. While visualizing the clump of hit beads on the bottom of the tube, most of the dynabeads and TBST was drained from the top using a 1000 μl pipetteman. 1 ml HPLC water was added to the tube, the tube inverted, and spun down as before. Again, most of the solution was drained while taking great care not to suck up the beads from the bottom of the tube. This washing step was repeated 6 times. For hit beads from libraries containing the methionine linker, most of the water was drained, and 1 ml acetonitrile added. Beads were then transferred to a 96-well plate and sorted one bead per well under a dissecting microscope. This can be quite tedious or simple, depending on technique (>100 hits can be sorted in less than an hour using the technique described in supplementary materials). 20 μl of 30 mg/ml CNBr in 5:4:1 acetonitrile:AcOH:water was added per well, the plate covered with sticky foil and placed on a shaker at room temperature overnight. The next day, the foil was removed and the 96-well plate left to air dry in a chemical hood for several hours. 20 μl HPLC grade water was added, the plate covered and left on a shaker for 1hr at RT. 10 μl from each well was transferred to a 384-well plate containing 10 ul/well DMSO, and the plate sealed and set aside for microarray spotting. 10 μl acetonitrile was added to each of the wells in the 96-well plate containing hits beads. This plate was sealed and set aside for MS sequencing. For hit beads containing the Asp-Pro linker, after the water washing of hit beads to get rid of most of the Dynabeads and TBST, beads were re-suspended in HPLC grade water and transferred to a small Petri dish under a dissecting microscope. A 10 μl pipetteman was set at 1μl and beads were transferred one bead at a time to thin-walled PCR tubes. 20 μl per tube of 0.1% TFA in water was added, and the tubes heated to 95°C in a PCR machine with heated lid for 40 minutes. 10 μl per well were transferred to a 384-well plate containing 10 μl DMSO for microarray spotting. 10 μl per well of acetonitrile was added to the 96-well plate containing hit beads for subsequent MS sequencing.
Microarray spotting, hybridization, and data analysis
Contents of the 384-well plates were printed onto maleimide coated glass slides using a NanoPrint LM 360 (TeleChem International Inc., Sunnyvale CA.) with MP946 Micro Spotting Pins. A 10% Ethanol (Midwest Grain Products, EM-AX007309) and water mixture was used to wash the pins before printing and after spotting of each compound. Multiple wash/sonicate/dry cycles were used between each sample pick-up and print cycle. Spots were printed on the slide to fit within the wells of a 16-well Whatman Fast Frame (Whatman, 10486003), which enables 16 isolated hybridization events on a single slide. Slides were left in 50% humidity for 12 hours before printing. After printing, the humidifier was turned off and the slides were left for at least 12 hours before free maleimide groups were blocked with 2% β-mercaptoethanol in DMF for 1 hr by placing the slides in glass slide-holders inside of glass containers on a shaker in the chemical hood. Slides were washed sequentially with DMF 30 min, Tetrahydrofuran (TFH) 30 min, DMF 30 min, Acetonitrile 3 × 20min, Isopropanol 3 × 20min, 1×TBST once for 20 min then 0.1×TBST once for 20 min. Washed slides were spun dry for 5 minutes at 2,000 rpm.
Dry slides were placed inside the Whatman Fast Frame following the provided instructions and each of the 16 wells per slide was blocked with 100 μl of StartingBlock (Fisher) for 1 hr at room temperature using a multichannel pipetteman. Wells were drained and washed once with 120 μl TBST. TBST was drained one well at a time before adding 100μl of appropriate concentrations of protein(s) diluted in 1:1 TBST:StartingBlock. The FastFrame was placed on wet paper towels inside of a glass cake-pan which was sealed with Glad Press'n Seal and placed on an orbital shaker for 2–4 hours at room temperature. Each well was washed with TBST 6 times before adding 4 μg/ml Alexa647-Goat anti-mouse IgG secondary antibodies diluted in 1:1 TBST:StartingBlock for 1hr at room temperature. Slides were washed 5 × 3min with 1×TBST then once with 0.1×TBST, spun dry at 2,000 rpm, then scanned using a GenePix Autoloader 4200AL Scanner (Molecular Devices, Sunnyvale CA.). Slides were scanned with a power of 100 and PMT of 500–600. Gal files were created and used to determine fluorescence intensity of each of the spots using GenePixPro6.0. Gal files were aligned manually, then automatic spotfinding followed by manual correction of spots performed for each of the scanned slides. GPR files were created and Median fluorescence-Background fluorescence values for each of the spots were cut and pasted in Excel and arranged (using simple macros) to simplify transferring results to GraphPad Prism 5.0 software for binding curve analyses.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health Director's Pioneer Award (DP1OD000663).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Data Further experimental details and supplementary experimental information are available online.
References
- Alluri PG, Reddy MM, Bacchawat-Sikder K, Olivos HJ, Kodadek T. Isolation of protein ligands from large peptoid libraries. J Amer Chem Soc. 2003;125:13995–14004. doi: 10.1021/ja036417x. [DOI] [PubMed] [Google Scholar]
- Bradner JE, McPherson OM, Mazischek R, Barnes-Seeman D, Shen JP, Dhaliwal J, Stevenson KE, Duffner JL, Park SB, Neuberg DS, et al. A robust small-molecule microarray platform for screening cell lysates. Chem & Biol. 2006;13:493–504. doi: 10.1016/j.chembiol.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Crimmins DL, Mische SM, Denslow ND. Chemical cleavage of proteins in solution. In: Coligan John E, et al., editors. Current protocols in protein science. 2005. Chapter 11, Unit 11 14. [DOI] [PubMed] [Google Scholar]
- Erlanson DA, Wells JA, Braisted AC. Tethering: fragment-based drug discovery. Ann Rev Biophys Biomol Structure. 2004;33:199–223. doi: 10.1146/annurev.biophys.33.110502.140409. [DOI] [PubMed] [Google Scholar]
- Figliozzi GM, Goldsmith R, Ng SC, Banville SC, Zuckermann RN. Synthesis of N-substituted glycine peptoid libraries. Methods Enzymol. 1996;267:437–447. doi: 10.1016/s0076-6879(96)67027-x. [DOI] [PubMed] [Google Scholar]
- Houseman BT, Huh JH, Kron SJ, Mrksich M. Peptide chips for the quantitative evaluation of protein kinase activity. Nature biotechnology. 2002;20:270–274. doi: 10.1038/nbt0302-270. [DOI] [PubMed] [Google Scholar]
- Kuruvilla FG, Shamji AF, Sternson SM, Hergenrother PJ, Schreiber SL. Dissecting glucose signaling with diversity-oriented synthesis and small-molecule microarrays. Nature. 2002;416:653–657. doi: 10.1038/416653a. [DOI] [PubMed] [Google Scholar]
- Lam KS, Renil M. From combinatorial chemistry to chemical microarray. Curr, Opin In Chem Biol. 2002;6:353–358. doi: 10.1016/s1367-5931(02)00326-5. [DOI] [PubMed] [Google Scholar]
- Lam KS, Salmon SE, Hersh EM, Hruby VJ, Kazmierski WM, Knapp RJ. A new type of synthetic peptide library for identifying ligand-binding activity. Nature. 1991;354:82–84. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- Liu R, Marik J, Lam KS. A novel peptide-based encoding system for “one-bead one-compound” peptidomimetic and small molecule combinatorial libraries. J Amer Chem Soc. 2002;124:7678–7680. doi: 10.1021/ja026421t. [DOI] [PubMed] [Google Scholar]
- MacBeath G, Koehler AN, Schreiber SL. Printing small molecules as microarrays and detecting protein-ligand interactions en masse. J Amer Chem Soc. 1999;121:7967–7968. [Google Scholar]
- Maly DJ, Choong IC, Ellman JA. Combinatorial target-guided ligand assembly: identification of potent subtype-selective c-Src inhibitors. Proc Natl Acad Sci USA. 2000;97:2419–2424. doi: 10.1073/pnas.97.6.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlmeyer MH, Swanson RN, Dillard LW, Reader JC, Asouline G, Kobayashi R, Wigler M, Still WC. Complex synthetic chemical libraries indexed with molecular tags. Proc Natl Acad Sci USA. 1993;90:10922–10926. doi: 10.1073/pnas.90.23.10922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy MM, Kodadek T. Protein “fingerprinting” in complex mixtures with peptoid microarrays. Proc Natl Acad Sci U S A. 2005;102:12672–12677. doi: 10.1073/pnas.0501208102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuker SB, Hajduk PJ, Meadows RP, Fesik SW. Discovering high-affinity ligands for proteins: SAR by NMR. Science. 1996;274:1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- Thakkar A, Cohen AS, Connolly MD, Zuckermann RN, Pei D. High-Throughput Sequencing of Peptoids and Peptide-Peptoid Hybrids by Partial Edman Degradation and Mass Spectrometry. Journal of combinatorial chemistry. 2009 doi: 10.1021/cc8001734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uttamchandani M, Walsh DP, Yao SQ, Chang Y-T. Small molecule microarrays: recent advances and applications. Curr Op Chem Biol. 2005;9:4–13. doi: 10.1016/j.cbpa.2004.12.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.