Abstract
Chronic in-vivo imaging studies of the brain require a labeling method that is fast, long-lasting, efficient, non-toxic and cell-type specific. Over the last decade, adeno-associated virus (AAV) has been used to stably express fluorescent proteins in neurons in vivo. However, AAV’s main limitation for many studies (such as those of neuronal development) is the necessity of second-strand DNA synthesis, which delays peak transgene expression. The development of double-stranded AAV (dsAAV) vectors has overcome this limitation allowing rapid transgene expression. Here we have injected different serotypes (1,2,6,7,8,9) of a dsAAV vector carrying the green fluorescent protein (GFP) gene into the developing and adult mouse visual cortex and characterized its expression. We observed labeling of both neurons and astrocytes with serotype-specific tropism. dsAAV-GFP labeling showed high levels of neuronal GFP expression as early as 2 days post-injection and as long as a month, surpassing conventional AAV’s onset of expression and matching its longevity. Neurons labeled with dsAAV-GFP appeared structurally and electrophysiologically identical to non-labeled neurons, suggesting that dsAAV-GFP is neither cytotoxic nor alters normal neuronal function. We also demonstrated that dsAAV-labeled cells can be imaged with subcellular resolution in vivo over multiple days. We conclude that dsAAV is an excellent vector for rapid labeling and long-term in vivo imaging studies of astrocytes and neurons on the single cell level within the developing and adult visual cortex.
Keywords: AAV, Labeling, Visual Cortex, Imaging, GFP
INTRODUCTION
The experience-dependent development of the visual system circuitry is a well studied model for neuronal development and plasticity. Recently much interest has centered around the rapid structural changes in cellular structures which may underlie functional plasticity in this system (Majewska, 2007). Understanding these changes requires the ability to image small structural elements (such as axons, dendrites and glial processes) dynamically over many days in the native milieu that is the intact visual cortex (Portera-Cailliau et al., 2005; Majewska et al., 2006). Moreover, the ability to perform electrophysiological recordings on cells undergoing small structural changes would allow for detailed structure/function studies to be conducted. The superficial location of the visual cortex makes it accessible for in vivo imaging. To conduct a fluorescence-based chronic in vivo imaging study, however, a labeling method that is fast, long-lasting, efficient, non-toxic and cell-type specific is necessary. Many of the current methods to label neurons in vivo fall short of meeting these requirements. In this study, we examine the ability of the adeno-associated viral (AAV) vector to achieve these necessary facets of in vivo imaging in the visual cortex.
Recombinant AAV has been successful in achieving stable, long-lasting transgene expression in the nervous system without significantly compromising cell viability (Chamberlin et al., 1998; Xiao et al., 1999; Tenenbaum et al., 2004). While AAV vectors primarily transduce neurons making studies of glia difficult, the identification of multiple serotypes of AAV has expanded the tropism of AAV vectors. Neuronal transduction by serotypes 1, 2 and 5–9 (Kaplitt et al., 1994; Bartlett et al., 1998; Davidson et al., 2000; Burger et al., 2004; Paterna et al., 2004; Cearley and Wolfe, 2006; Harding et al., 2006; Taymans et al., 2007) and glial transduction by serotypes 2, 5, 7 and 8 (Kaplitt et al., 1994; Davidson et al., 2000; Harding et al., 2006) has been described. The efficiency and cell type specificity of transduction, however, appears to be specific to the brain area studied (Taymans et al., 2007). To date, no studies characterizing the tropism of multiple serotypes of AAV vectors in the visual cortex have been reported. One of the aims of the current study is to identify an AAV serotype capable of transducing neurons and/or glia in the mouse visual cortex.
The use of AAV vectors for rapid labeling and imaging of cells can be problematic due to the long delay between cell entry and transgene production. AAV vectors have a single-stranded DNA genome that must be converted into a double-stranded genome before transgene expression can begin (Ferrari et al., 1996; Fisher et al., 1996). This process can take up to a month (Stettler et al., 2006) precluding many studies (such as those involving developmental phenomena). To overcome this limitation, a self-complementary, double-stranded (ds) DNA genome was created, allowing more rapid and robust transgene expression (McCarty et al., 2001; Wang et al., 2003). dsAAV vectors have been shown to successfully transduce neurons and glia in vitro (Howard et al., 2008) and in vivo (Fu et al., 2003; McCarty et al., 2003; Chen et al., 2007). However, the comparison of transgene expression between a dsAAV and ssAAV vector at various times after injection into mouse brain has not been reported.
One of the features that makes rAAV vectors a desired vector for gene delivery in the brain is the low toxicity and immunogenicity (McCown, 2005). To perform repeated in vivo imaging of AAV-labeled cells, it is necessary for minimal toxicity from transduction and the fluorescent label, GFP. Additionally, transduction and expression of GFP should not alter the electrophysiological properties of a neuron when compared to an equivalent, non-transduced neuron. To address this concern, we tested for the ability to perform multiday in vivo imaging of transduced neurons and we tested the electrophysiological properties of transduced neurons compared to non-transduced neurons.
In this study, we identify an optimal AAV vector for rapid and efficient labeling of neurons in the mouse visual cortex for in vivo imaging and electrophysiological recordings. Our results show that of the AAV serotypes tested, AAV1 most efficiently transduced neurons in the visual cortex. A double-stranded AAV vector more rapidly labeled cells in the visual cortex compared to a single-stranded AAV vector. Using a dsAAV1 vector expressing GFP, we were able to image labeled neuronal processes in vivo in the visual cortex over several days and GFP-labeled neurons exhibited similar electrophysiological properties compared to non-labeled neurons.
MATERIALS AND METHODS
Animals
C57Bl6 mice were obtained from Charles River Laboratories (Wilmington, MA). Post-natal day (P)35-P49 and P120 mice were used to compare dsAAV serotypes. P12, P21, and P84–P98 mice were used to determine the timeline of dsAAV-mediated expression. The animal and experimental protocols were approved by the University of Rochester University Committee on Animal Resources (UCAR) in accordance with the PHS Policy on Humane Care and Use of Laboratory Animals and conformed to the National Institute of Health guidelines.
Packaging, Purification and Titering of AAV vectors
The construction of dsAAV-GFP packaging plasmid is based on AAV serotype 2 genome and has been described previously (Wang et al., 2003). To generate pssAAV-GFP, the 5’ ITR of dsAAV-GFP was replaced with the 5’ITR of pAAV-LacZ (Stratagene, La Jolla, CA). Both plasmids contain the cytomegalovirus immediate-early enhancer/promoter to drive expression of GFP. Viral stocks were prepared using the triple-transfection method (Xiao et al., 1998; Howard et al., 2008). Briefly, twenty 15 cm dishes containing HEK293 cells at 85–95% confluency were transfected by CaCl2 method with pHelper (Stratagene, La Jolla, CA), pdsAAV-GFP (Wang et al., 2003) or pssAAV-GFP (Xiao et al., 1998) and a plasmid containing rep/cap genes for serotype1 pXR1 (aka pXX12; (Rabinowitz, 2002 #170)), pXX2 (Xiao et al., 1998), pXR5 (Rabinowitz et al., 2002), pAAV2/6 (Rutledge et al., 1998), pAAV7 (Gao et al., 2002), pAAV8 (Gao et al., 2002), and pAAV9 (Gao et al., 2004). Plasmids used for packaging AAV were generously provided by Dr. Xiao Xiao (UNC, Chapel Hill, NC). Approximately 48 hours post-transfection, cells were harvested, lysed by freeze/thaw, and purified by centrifugation on CsCl gradient. Final samples were dialyzed in PBS to 1013vg/ml, aliquoted and stored at −80°C until use. All vectors were titered by quantitative PCR using GFP as the target sequence. Viral titers are recorded as viral genome/ml.
Virus Microinjection
Mice were anesthetized with Avertin (200mg/kg, IP, Sigma), administered Bupronex (0.1 mg/kg, SC, Bedford Labs, Bedford, OH), secured in a stereotaxic frame with ear cups, and their skulls exposed. For mice P21 and older, the skull was thinned over visual cortex in both hemispheres and a small portion of skull removed. Injections were made bilaterally. For mice P12 and younger, a micropipette was inserted directly through the skull. A glass micropipette with a diameter of 20 µm was used to inject dsAAV-GFP or ssAAV-GFP 500 µm below the surface of the cortex in P21 and older mice, or 400 um below the surface of the cortex in P12 and younger mice. One microliter of virus (titer 1013 vg/ml) was delivered over a 10 minute period via a mechanical plunger controlled by a micropump injector (World Precision Instruments, Sarasota, FL). Following virus injection the pipette was withdrawn and the scalp sutured. Animals were allowed to recover under a heat lamp before being returned to the animal facility. Seven days after injection, mice were perfused transcardially with 0.1M phosphate buffered saline (PBS) followed by 4% paraformaldehyde (PFA) in 0.1M PBS and brains were harvested. In order to determine the timeline of dsAAV expression, mice were also perfused 2 days or 4 weeks following dsAAV injection. In the text, n denotes the number of individual injections for each serotype.
Immunofluorescent Staining
After overnight fixation in 4% PFA at 4°C, brains were cryoprotected for 24-hours each in 10, 20, 30% sucrose at 4°C. Brains were sectioned coronally on a freezing, sliding microtome (Microme; Global Medical Instrumentation, Inc., Ramsey, MN) at 50 µm thickness into 0.1M PBS. For immunofluorescent labeling, sections were washed several times in 0.1M PBS. To block endogenous peroxidase activity, sections were placed in a solution containing 3% H2O2 in 0.1M PBS for 20 minutes at room temperature. Sections were washed briefly in 0.1M PBS and blocked for 1 hour at room temperature in a solution containing 0.3% Triton-X, 5% normal donkey serum (NDS) in 0.1M PBS. Sections were washed in 0.1M PBS and transferred into an antibody dilution buffer containing 0.3% Triton-X, 3% NDS in 0.1M PBS along with one of the following primary antibodies: mouse anti-neuronal nuclei (NeuN, 1:250, IgG, Millipore), rabbit anti-Iba1 (1:500, IgG, Wako Pure Chemical Industries, Ltd., Richmond, VA). rabbit anti-glial fibrillary acidic protein (GFAP,1:1,500,IgG, DakoCytomation, Glostrup, Denmark) or mouse anti-GAD67 (1:1000, Millipore) at 4°C in a humidified chamber for 48-hours. Tissue was washed with 0.1M PBS and incubated for 4-hours at room temperature with Alexa Fluor 594 donkey anti-rabbit IgG or Alexa Fluor 594 donkey anti-mouse IgG (1:250, Molecular Probes, Carlsbad, CA) in a solution containing 0.5% bovine serum albumin (BSA), 3% NDS in 0.1M PBS. Sections were washed in 0.1M PBS. Tissue sections were mounted in 0.1M PBS and cover-slipped with Prolong Gold anti-fade reagent (Molecular Probes, Carlsbad, CA), slide edges sealed and protected from light until analysis.
Image acquisition and analysis
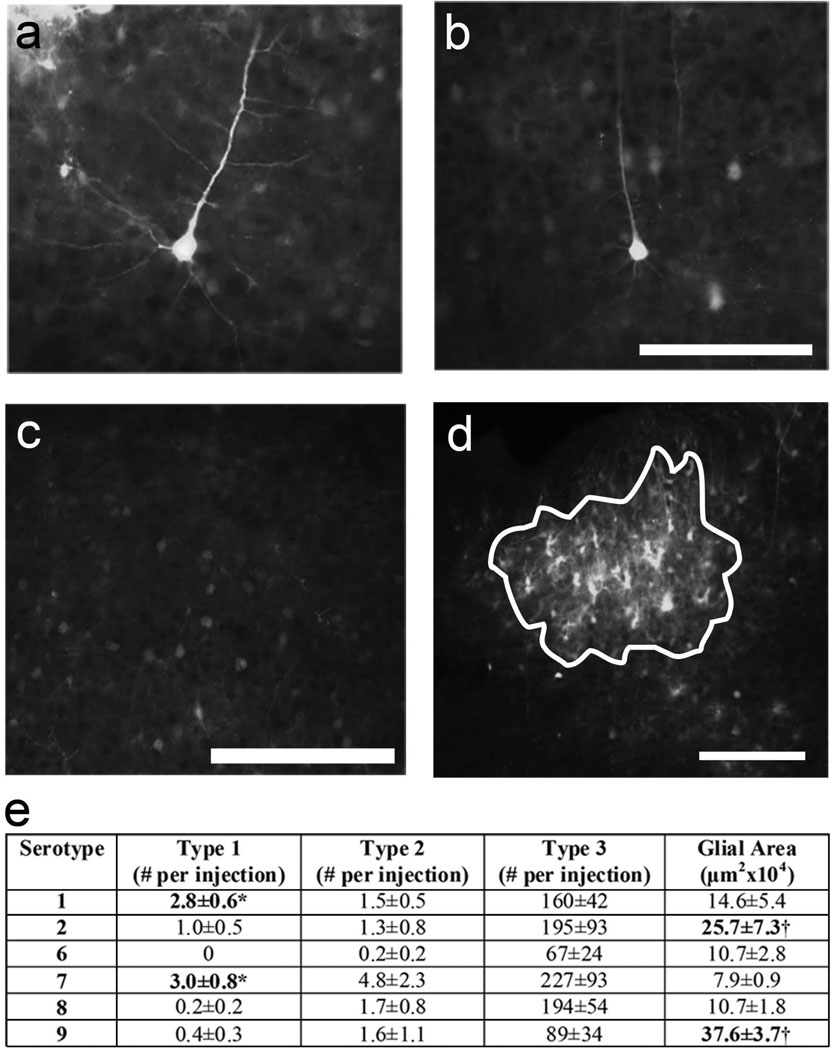
Injection sites were viewed on an AX70 Microscope (Olympus, Center Valley, PA) using epifluorescence. Digital images were obtained using a MicroFire camera (Optronics, Muskogee, OK) and Image Pro software (Media Cybernetics, Bethesda, MD). Images were analyzed off-line using free-ware NIH Image-J. Labeled neurons were manually counted in all sections taken from each injection site and divided into one of three categories (see Figure 4). Type 1 cells were defined as thoroughly-labeled neurons where proximal and distal dendrites and dendritic spines were clearly visible. Type 2 cells were defined by having clearly labeled cell bodies and proximal dendrites, while in Type 3 cells only the cell bodies were apparent. Injections were typically characterized by very strong, dense labeling at the center where individual cells were difficult to identify surrounded by a less dense “halo” of individual cells. Neuronal counts were made in the less dense “halo” region surrounding the bright injection core. While this suggests that our numbers underestimate the number of neurons labeled by the dsAAV, the counts are relevant for imaging purposes as the dense core labeling precludes the visualization of individual cells. Additionally, using immunohistochemistry we determined that the majority of cells in the dense core were GFAP+ suggesting that glial labeling is most pronounced in this region. Indeed while individual labeled glia were observed, they were generally very near the edges of the dense core unlike labeled neurons which could be found quite far from the site of injection. To compare the amount of glial labeling in injections of different serotypes, we identified the section in which the area of the core was largest, outlined the bright core labeling and measured its area in that section (Figure 4d).
Figure 4. Serotype comparison of dsAAV-GFP transduction of cells in mouse visual cortex.
One week after injection in P35–P49 mice, mouse brain sections were analyzed for GFP fluorescence to identify transduction of neurons and glia. Neurons were classified as Type 1 (a), Type 2 (b) and Type 3 (c; see text for type descriptions). dsAAV1-GFP and dsAAV7-GFP exhibited the most neurons with Type 1 labeling. dsAAV2-GFP and dsAAV9-GFP produced the largest area of transduced glia (d; white lines delineate the glial area - see text for description) compared to other serotypes tested. Bolded numbers represent values that are statistically significantly different using a one-way ANOVA. Numbers labeled with an asterisk represent values that are statistically significantly different compared to dsAAV6,8,9 (p<0.05; Tukey’s post-hoc test) while those labeled with a cross are statistically significant compared to dsAAV1,6,7,8 (p<0.01; Tukey’s post-hoc test). Scale bars: 100 µm (a,b), 200 µm (c), 50 µm (d).
To verify cell identification, a subset of injections was labeled immunocytochemically for either astrocytes (GFAP), microglia (Iba1), or neurons (NeuN) and imaged on a confocal microscope (Zeiss, Thornwood, NY). To determine whether inhibitory cells were labeled in addition to morphologically identifiable pyramidal cells, a subset of injections was labeled immunocytochemically with the inhibitory cell marker GAD67. Sections were imaged on an AX70 microscope (Olympus) using epifluorescence and images were analyzed using ImageJ freeware to determine the extent of colocalization of marker label and GFP. Statistical comparisons of different serotypes and timeline of dsAAV1 expression were made using a one-way ANOVA analysis with Tukey's post-hoc test. Statistical comparisons of dsAAV and ssAAV expression were made using a paired t-test.
Two-photon imaging
A custom-made two-photon laser scanning microscope (Majewska et al., 2000) was used for in vivo imaging. The microscope consists of a modified Fluoview confocal scanhead (Olympus) and a Ti:S laser providing 100 fs pulses at 80 MHz at a wavelength of 920 nm (Mai-Tai, Spectra-physics). Fluorescence was detected using photomultiplier tubes (HC125-02, Hamamatsu, Bridgewater, NJ) in whole field detection mode. For in vivo imaging, mice were anesthetized with Avertin (200mg/kg, IP). The skull over the injection site was thinned or removed. The craniotomy was initially identified under whole field fluorescence illumination and areas with superficial dendrites and glia were identified using a 20×, 0.9 5 NA lens (IR2, Olympus). Image acquisition was accomplished using Fluoview software. In cases of repeat imaging the animal’s scalp was sutured between imaging sessions. Axon terminals were identified based on their morphology (Majewska et al., 2006) and their locations were marked on different days. Stable, lost and new terminals are expressed as a percentage of total terminals observed on the first day of imaging. For acute slice imaging, two photomultiplier tubes were used for the GFP and Alexa-594 channels. Slices were placed in a recording chamber under the two-photon microscope and were imaged after a whole cell recording was obtained. Fluorescence from the slices was directed through a dichroic (Chroma, Rockingham, VT) to the photomultipler detectors. Analysis of images was carried out offline in ImageJ.
Electrophysiology
Mice (P16–18) were decapitated 1 week after dsAAV1-GFP was injected bilaterally in visual cortex. The brains were rapidly removed and immersed in ice-cold artificial cerebral spinal fluid (ACSF) saturated with 95%O2 and 5%CO2. The ACSF contained (mM): 126 NaCl, 3 KCl, 2.5 CaCl2.2H2O, 1.3 MgCl2.6H2O, 1.1 NaH 2PO4, 10 Glucose, 26 NaHCO3. Coronal slices (400 µm) were cut on a Vibratome (1000 plus, TPI, St. Louis, MO) and incubated for at least 1 hour in a recovery chamber prior to recording. Slices were transferred to a recording chamber on the two-photon microscope, and perfused continuously with ACSF saturated with 95% O2 and 5%CO2 at a flow of 2–3 ml/min. Injection sites in visual cortex were identified under epifluorescence. Recording patch pipettes (5–8 MΩ) were filled with (mM) 135 k-gluconate, 10 KCl, 10 HEPES, 8 NaCl, 4 MgATP, 0.3Na-ATP, 0.1 Alexa594 (Invitrogen, Carlsbad, CA), (pH7.25, 290mOsm). Whole-cell current clamp recordings were made using a patch clamp amplifier (MultiClamp 700A, Axon Instruments, Toronto, Canada). Data acquisition and analysis were performed using a digitizer (DigiData 1322A, Axon Instruments) and pClamp 9.2 (Axon Instruments). Signals were filtered at 2 kHz, and sampled at 10 kHz. The membrane potential was determined from the value recorded immediately after achieving the whole-cell configuration. All chemicals for electrophysiology were purchased from Sigma Aldrich unless otherwise stated. Statistical comparisons were made using a one-tail Student’s t-test.
RESULTS
dsAAV-GFP targets neurons and astrocytes in visual cortex in a serotype specific manner
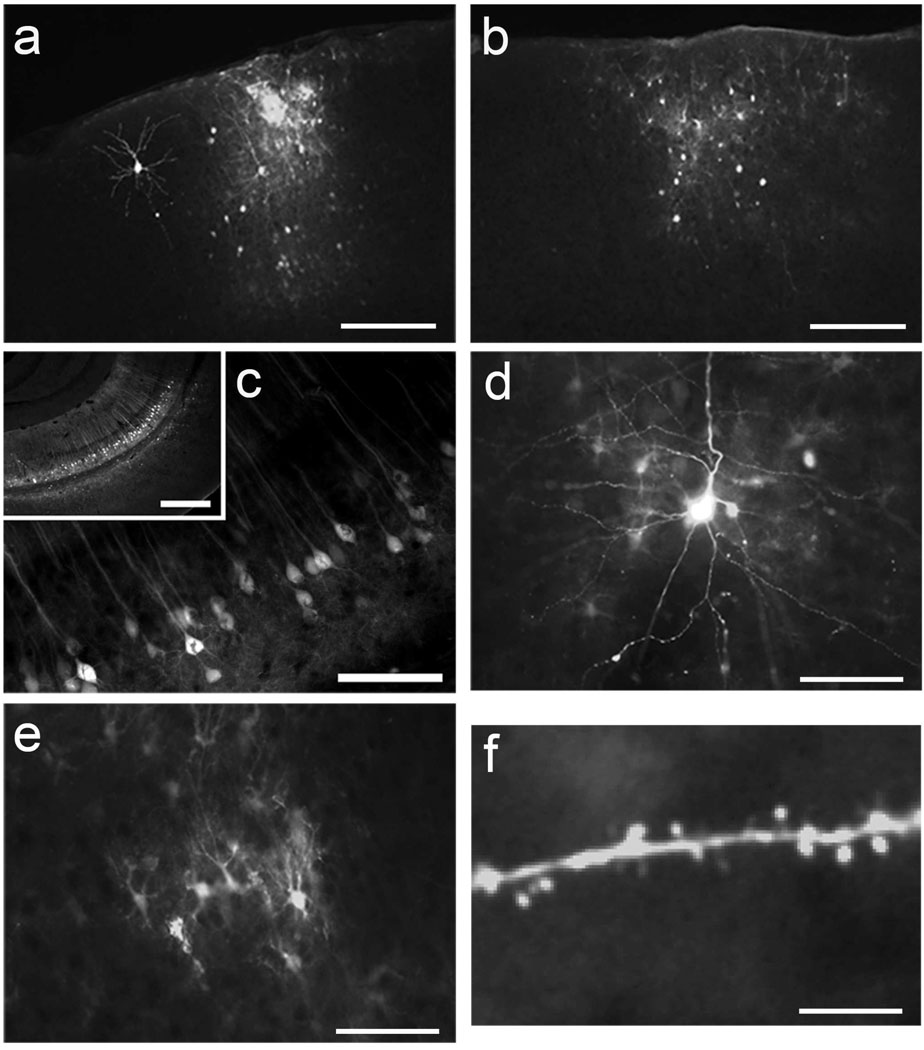
In order to determine the serotype(s) of AAV that most efficiently transduce cells in the mouse visual cortex, we injected serotypes 1, 2, 6, 7, 8 and 9 of dsAAV-GFP into the visual cortex of young adult mice [n=13 (7), 6 (3), 6 (3), 6 (3), 6 (3), 8 (4), injection sites (animals) respectively]. One microliter of virus (1013 vg/ml) was injected over a period of 10 minutes at a depth of 500 µm below the level of the pia. After 7 days animals were perfused and their brains harvested. GFP labeling in visual cortex was observed in fixed sections under a fluorescent microscope. All injections of dsAAV-GFP resulted in both neuronal and non-neuronal labeling. Injection sites were typically characterized by strong non-neuronal labeling centered in a bright core around the injection site with neuronal labeling around the perimeter (Figure 1a,b). The spread of total labeling (all neurons and glia) laterally from the point of injection ranged from 100 to 600 µm in diameter (Figure 1 a,b).
Figure 1. Transduction of mouse visual cortex by dsAAV-GFP.
P35–P49 mice were injected intracranially with dsAAV-GFP and perfused after one week. Both neuronal and non-neuronal labeling is apparent in dsAAV1-GFP (a) and dsAAV7-GFP (b) injection sites. In the hippocampus dsAAV1-GFP injection resulted in clear labeling of a large number of neurons without apparent glial labeling (c; inset shows lower resolution view of hippocampal CA1 layer). The level of GFP expression in neurons and astrocytes transduced by dsAAV1-GFP in the visual cortex make fine structures such as dendrites (d) glial processes (e), and dendritic spines (f) clearly visible. Scale bars: 250 µm (a, b, c-inset), 200 µm (c), 50 µm (d, e), 20 µm (f).
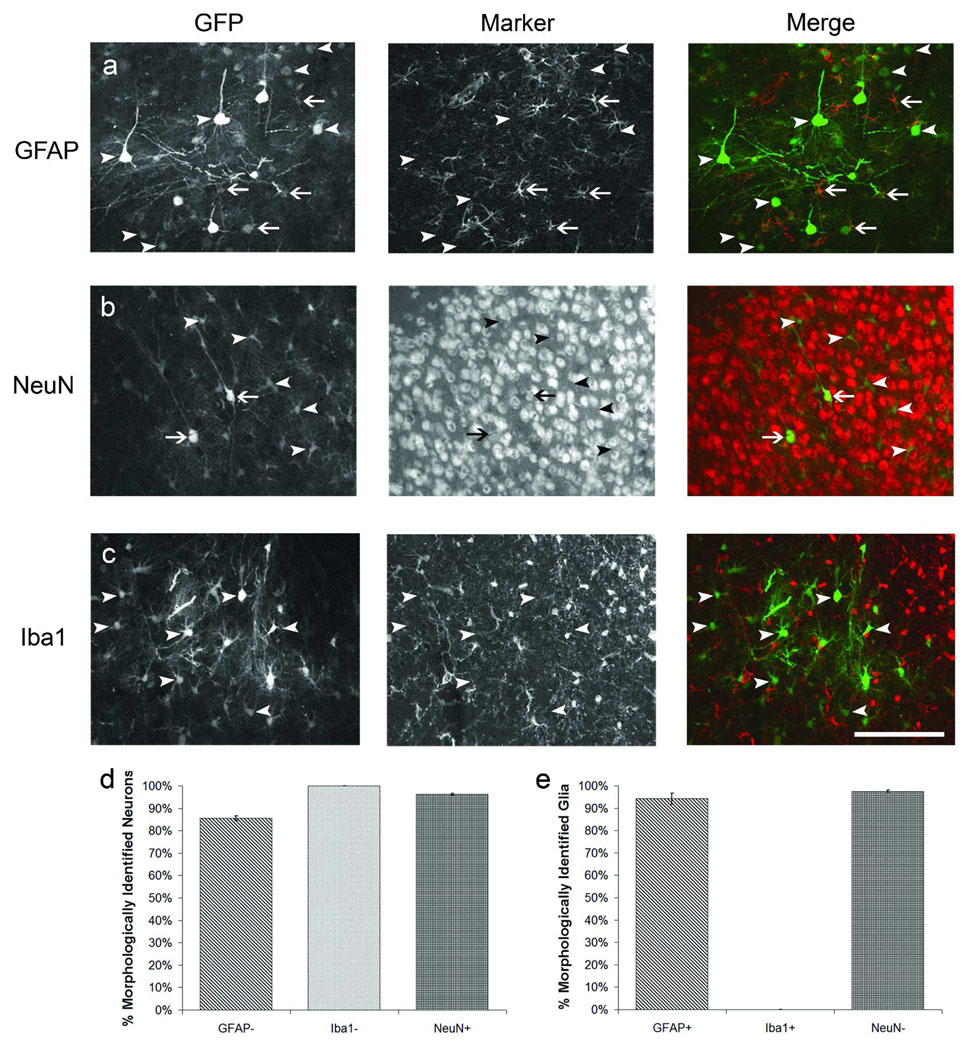
To ascertain the cell-specificity of staining, brain sections from dsAAV1-GFP injected animals were immunofluorescently labeled with either GFAP for astrocytes, Iba1 for microglia, or NeuN for neurons. The double-labeling revealed that astrocytes and neurons were the primary labeled cell types in AAV1 injections as GFP-labeled microglia were not observed (Figure 2). We identified neurons and glia morphologically using GFP fluorescence and then verified phenotype by co-localization with GFAP and NeuN staining. Cells were correctly identified as neurons or astrocytes on average 95% of the time (Figure 2d,e). For the remainder of our analysis we used the GFP fluorescence morphology to assess neuronal vs. astrocytic labeling.
Figure 2. Immunocytochemical identification of dsAAV-GFP labeled cells in visual cortex.
P35–P49 mice were injected with dsAAV1-GFP and perfused after one week. Brain sections were immunocytochemically labeled to distinguish astrocytes (GFAP, a), neurons (NeuN, b), or microglia (Iba1, c). Virally transduced GFP expression is shown in the first column, the phenotype marker in the second, and a merged representation of the first two panels is shown in the third column. Arrows represent cells which label with both GFP and the marker while arrowheads represent cells which label with GFP but not the marker. Graphs show the percentage of neurons (d) or glia (e) that were correctly identified by blinded observer based on their morphology in the GFP channel. Scale bar: 200 µm.
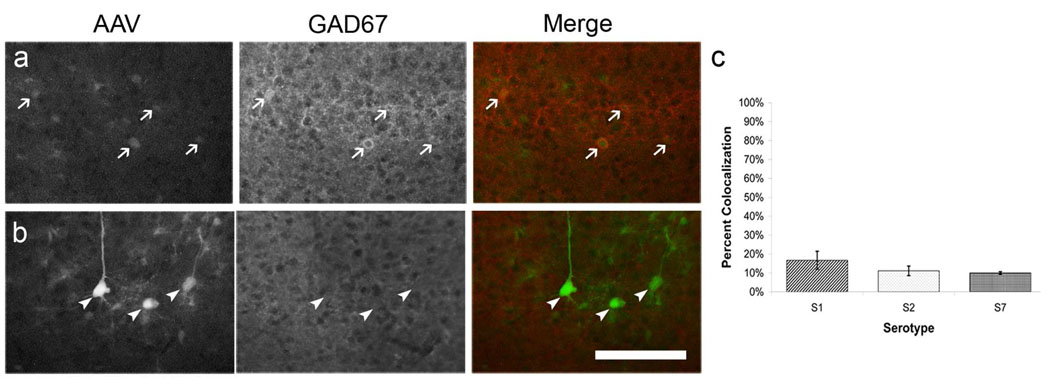
Most injections resulted in dense labeling of neurons. The majority of these neurons, however, were weakly fluorescent whereby only the cell body was clearly delineated while processes were not visible. Of these weakly labeled cells, 10–20% colocalized with the inhibitory cell marker GAD67 (Figure 3a,c). Well labeled neuronal cells were primarily layer II/III and layer V pyramidal cells and did not colocalize with the inhibitory marker (Figure 1d, Figure3b). While labeling in the center of the injection site was intense and fine structures could not be discerned over the background fluorescence, labeled astrocytes with clear processes could be imaged at the edges of the injection site (Figure 1e). Interestingly, this pattern of labeling appears to be specific to visual cortex as injections of dsAAV1 into the hippocampus resulted in clear labeling of CA1 neurons with no observable glial labeling (Figure 1c).
Figure 3. dsAAV-GFP labels excitatory and inhibitory neurons in visual cortex.
P120 mice were injected with serotype 1,2 or 7 of dsAAV-GFP and perfused after one week. Brain sections were immunocytochemically labeled with GAD67 to distinguish inhibitory cells. Virally transduced GFP expression is shown in the first column, the phenotype marker in the second, and a merged representation of the first two panels is shown in the third column. Arrows represent cells which label with both GFP and the marker (inhibitory neurons; a) while arrowheads represent cells which label with GFP but not the marker (excitatory neurons; b). Graph shows the percentage of all GFP-labeled neurons that colocalized with GAD67 by serotype (c). Scale bar: 100 µm.
For more detailed comparisons among serotypes, neurons were analyzed as three types: Type 1 – extensively labeled neurons with visible proximal and distal dendrites and dendritic spines, Type 2 – labeled neurons with visible proximal dendrites only and Type 3 – only cell bodies visible (Figure 4 a,b,c). Neuronal counts were made outside the bright core where individual cells could be easily identified. While this suggests that our numbers underestimate the number of neurons labeled by the dsAAV, the counts are relevant for imaging purposes as the dense core labeling precludes the clear visualization of individual cells. Based on these criteria, dsAAV1, 2, 7 and 8 were most efficient at labeling large numbers of neurons. dsAAV1 (n=13) and dsAAV7 (n=6) produced significantly more Type 1 transduced neurons where fine processes including dendritic spines could be discerned (Figure 4e; p<0.05 versus dsAAV2,6,8,9; one-way ANOVA analysis with Tukey's post-hoc test). Transduction was also compared by the area of the bright core around the injections site which comprised mostly labeled glia (Figure 4d,e; see methods for full description of analysis). dsAAV9 and dsAAV2 (n=8, n=6, respectively) injection resulted in the largest core of glial labeling (p<0.01; one-way ANOVA analysis with Tukey's post-hoc test compared to serotypes 1,6,7,8 (dsAAV9) and compared to serotypes 6,7,8 (dsAAV2)). Overall, dsAAV1-GFP and dsAAV7-GFP produced the highest numbers of Type 1 neurons that allowed visualization of dendritic processes and dendritic spines and would be most appropriate for imaging small compartments in the visual cortex in vivo.
dsAAV-GFP allows rapid, efficient and long-lasting labeling of neurons and astrocytes in developing visual cortex compared to ssAAV-GFP
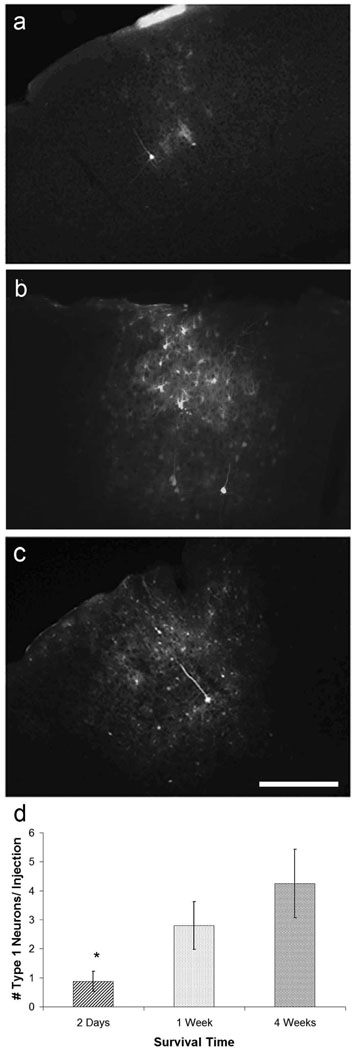
Initial characterization of transduction was conducted in adult visual cortex. Based on our results in the adult cortex, we chose serotype 1 to conduct subsequent labeling studies. We next determined whether dsAAV could be used to label neurons in the developing visual cortex in both a rapid and sustained manner. We injected mice (n=5 injections (n=3 mice) per timepoint) at the beginning of the visual critical period (~P21) with dsAAV1-GFP and analyzed the extent of labeling at 2 days, 1 week, and 4 weeks post-injection. Type 1 and Type 2 labeled neurons with visible processes were observed as early as 2 days after virus injection, and the numbers of Type 1, 2 and 3 neurons remained stable between 1 and 4 weeks post-injection, the last time point studied (Figure 5; p<0.05 one-way ANOVA). This demonstrates that dsAAV can label neurons rapidly (<2 days) and maintain high levels of expression for an extended period (at least 1 month) in developing visual cortex. dsAAV1 appears to be effective at labeling neurons at any age as our experiments using dsAAV1-GFP injections in mice of different ages (P12, P21, P35–P49, P84–P98) yielded qualitatively similar results (data not shown).
Figure 5. dsAAV-GFP labeling is rapid and long lasting.
Neuronal and non-neuronal labeling is detected as early as two days after dsAAV1-GFP injection (a). Labeling of both neurons and glia increases at one week (b) and stays relatively stable between one and four weeks (c). Well labeled (type1) neurons appear as early as two days after injection, but their numbers are smaller than at 1 and 4 weeks post-injection (one-way ANOVA, p<0.05; Tukey’s post-hoc test). Scale bar: 200 µm.
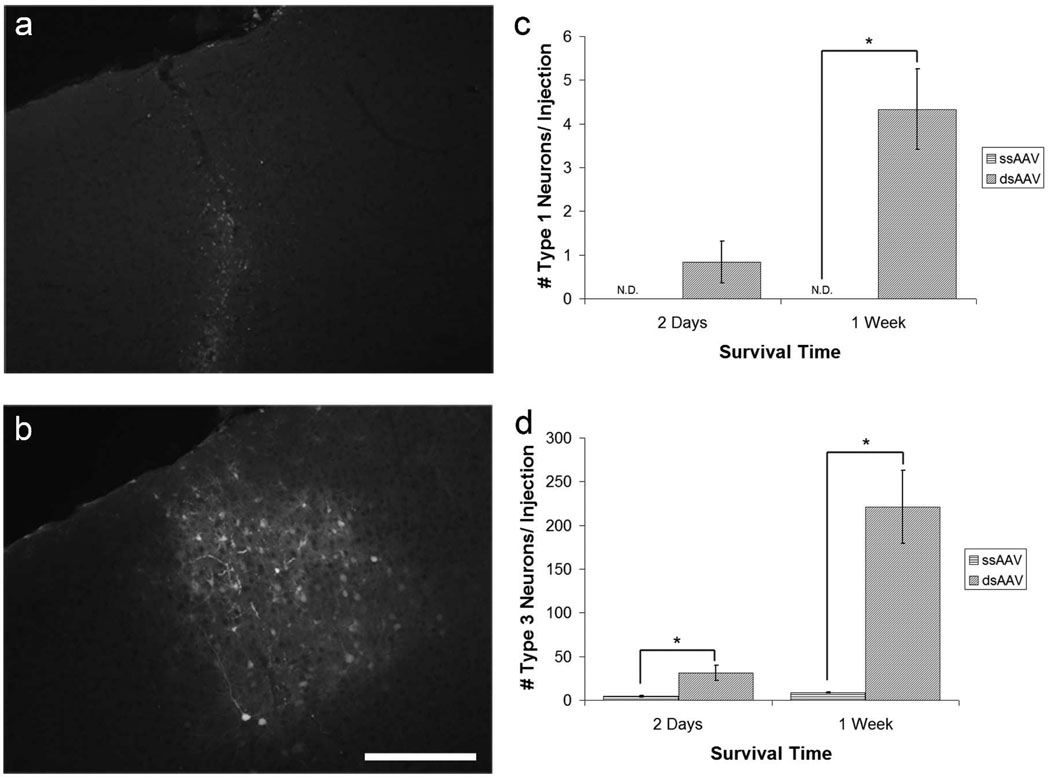
To determine whether dsAAV can label cells in the visual cortex more efficiently than the more commonly used ssAAV, we injected 6 animals at age P21, the beginning of the visual critical period, with a single stranded version of the dsAAV virus (ssAAV1-GFP) at an equivalent titer. We compared labeling at 2 days (n=6 injections, 3 animals per group) and 1 week (n=6 injections, 3 animals per group) after injection with either dsAAV1-GFP or ssAAV1-GFP. Brains labeled with ssAAV1-GFP showed weak, diffuse labeling and neuronal processes were not observed (Figure 6a). ssAAV1-GFP transduced brains were also devoid of labeled glia. By comparison, dsAAV1-GFP injection resulted in more intense labeling and labeled a larger number of neurons and astrocytes (Figure 6b). Brains labeled with dsAAV1-GFP had significantly more labeling of Type 3 neurons (neuronal cell bodies; see above for details) than ssAAV1-GFP at both 2 days and 1 week post-injection (Figure 6d; p<0.01; Student’s t-test). dsAAV1-GFP labeled brains also had significantly more labeling of Type 1 neurons (extensively labeled neurons; see above for details) at 1 week post-injection than those labeled with ssAAV1-GFP (Figure 6c; p<0.05; Student’s t-test). These data suggest that dsAAV1-GFP more rapidly and efficiently labels neurons for imaging compared to ssAAV1-GFP.
Figure 6. dsAAV-GFP results in more efficient rapid neuronal labeling than ssAAV.
Mice injected with ssAAV1-GFP at P21 (a) show much less extensive and intense labeling than those injected with dsAAV1-GFP (b) at one week post-injection. dsAAV1-GFP injections had significantly more Type 1 neurons compared to ssAAV1-GFP injections after 1 week (c, p<0.05; paired t-test; ND= Not Detected) and significantly more Type 3 neurons both 2 days and 1 week post-injection (d, p<0.01; paired t-test). Labeled glia were not observed in ssAAV1-GFP labeled brains. Scale bar: 200 µm.
dsAAV-GFP labeled neurons and glia can be imaged in an alive anesthetized animal
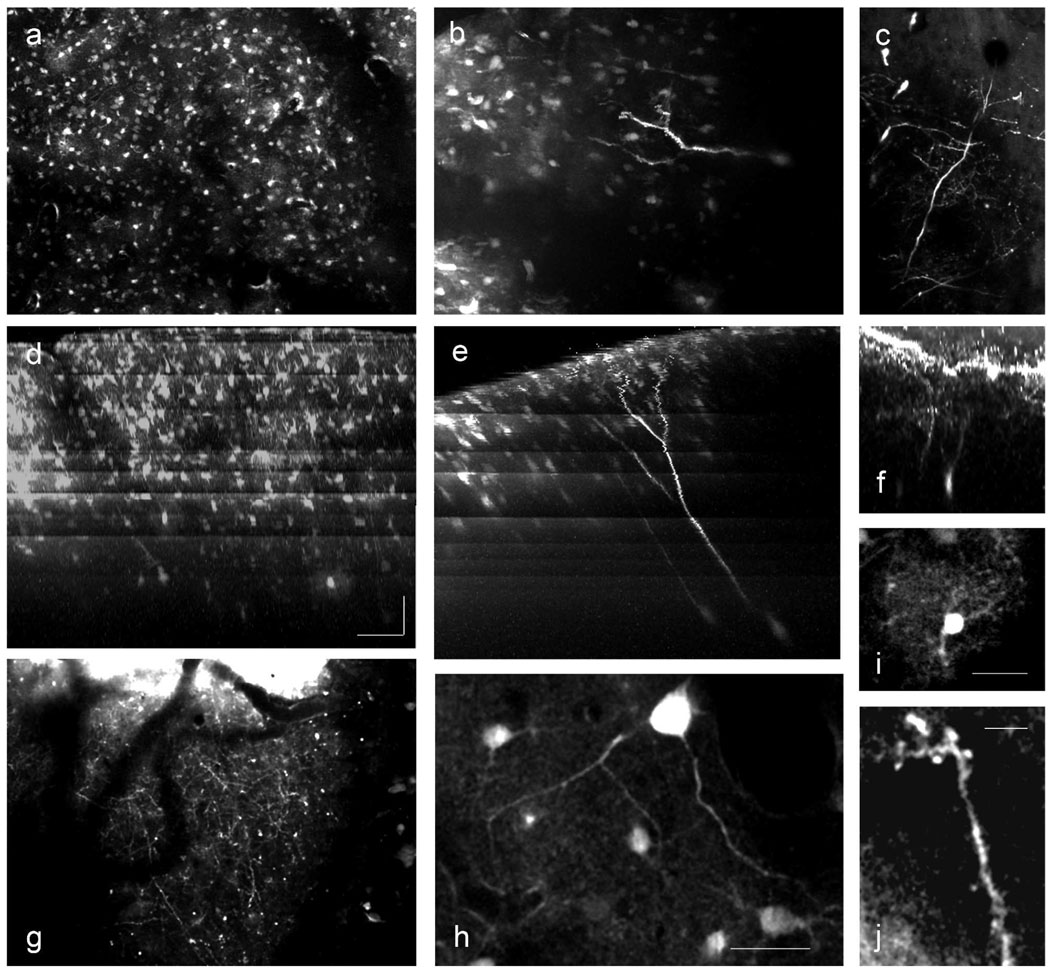
To determine whether dsAAV1-GFP-labeled neurons and glia could be imaged in vivo, we injected dsAAV1-GFP into the visual cortex and performed in vivo two-photon microscopy (Figure 7 and Movie S1). Labeled neurons and glia could be imaged in the center of the injection site despite the dense labeling (Figure 7a,b). Fine processes, however, were not visible likely due to the high background caused by labeled neuropil. On the edges of the injection sites, labeled neurons and glia were easily discernible over the background staining and dendrites (including dendritic spines), axons and fine glial processes could be imaged (Figure 7c,g,i,j).
Figure 7. In vivo imaging of dsAAV1-GFP labeled neurons.
dsAAV1-transduced neurons and astrocytes could be readily imaged in vivo. Images shown are projections of three dimensional stacks imaged using two-photon microscopy (a–c), or coronal reconstructions of those same images (d–f). The banding pattern in the coronal reconstructions (d–f) is a result of increasing the laser power during imaging to compensate for signal loss deeper within the cortex. At the injection site neuronal and glial cell bodies can be imaged, but fine structure is lost because of the high level of background label (a,d), unless very bright labeling is present (b,e). At the edge of the injection site, fully labeled neurons (c,f) and dense axonal labeling (g) is apparent. Higher magnification images show dendritic structure (h), glial arborization (i) and dendritic spines (j). Scale bars: 80 µm x,y; 100 µm z (a–g); 25 µm (h,i), 5 µm (j).
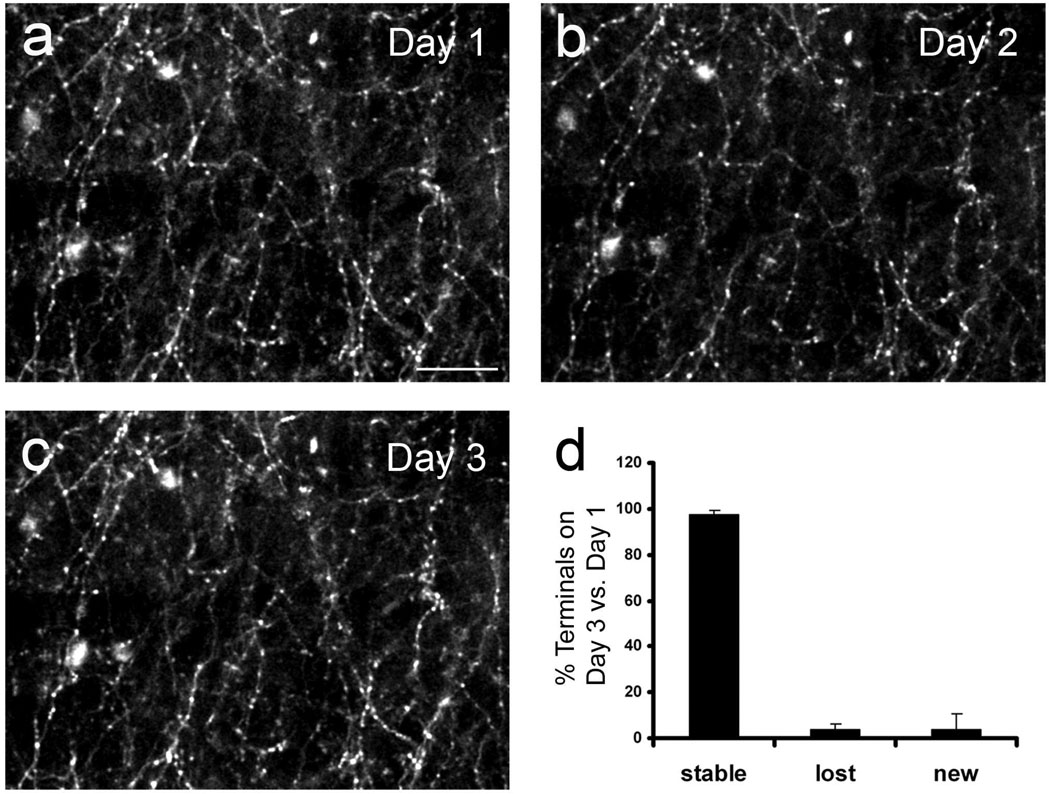
We also carried out chronic imaging of axonal structure in vivo. Over three days, the same axonal field could be identified and re-imaged (Figure 8). In agreement with our previous imaging studies in transgenic mice expressing GFP (Majewska et al., 2006), axonal boutons remained highly stable and the vast majority (>95%) could be re-identified on the subsequent imaging session. These experiments demonstrate that dsAAV1-GFP labeling can be used for high-resolution acute and chronic in vivo imaging of subcellular neuronal structures, including the stability of axon terminals.
Figure 8. Chronic imaging of dsAAV1-GFP labeled axons.
A field of dsAAV1-GFP-labeled axons was imaged in vivo through a thin skull over a period of 3 days (a–c). The same area was identified based on the blood vessel pattern during repeat imaging sessions. Axon terminals, identified by their morphological characteristics (Majewska et al., 2006), were remarkably stable with very few being lost (3.8±2.5) or gained (4±6.6) during the three day period (3 cortical areas, 2 animals (d)). Scale bar = 20 µm.
dsAAV-GFP labeled neurons do not show signs of virus-induced damage
dsAAV-transduced neurons appeared structurally intact without overt signs of virally-induced damage. Over-expression of GFP did not cause increased axonal dynamics or altered dendritic morphology such as fragmentation or blebbing which is characteristic of degeneration. To determine whether transduction by dsAAV altered the electrophysiological activity of labeled neurons, we injected dsAAV1-GFP into the visual cortex of developing mice (P9–P11) and prepared acute slices from these animals 7 days post-injection. Slices were imaged using two-photon microscopy and had a high density of labeled neurons and glia. Blind whole cell recordings were obtained from neurons in the visual cortex of transduced slices. Alexa 594 was included in the recording pipette to determine the identity of recorded neurons which were often too deep in the slice to be observed using transmitted and epifluorescence microscopy. Labeled neurons were identified as those that showed signal in both red (Alexa594) and green (GFP) detection channels when imaged with the two-photon microscope, while non-transduced control neurons only had signal in the red detection channel. Transduced and non-transduced neurons were not significantly different with respect to their resting and active electrophysiological properties such as resting potential, threshold, action potential amplitude or input resistance (p>0.05; t-test; Figure 9 and Table 1). These electrophysiological data, together with the lack of structural defects observed upon in vivo imaging, suggest the dsAAV is not overtly cytotoxic to neurons at least up to 7 days post-injection.
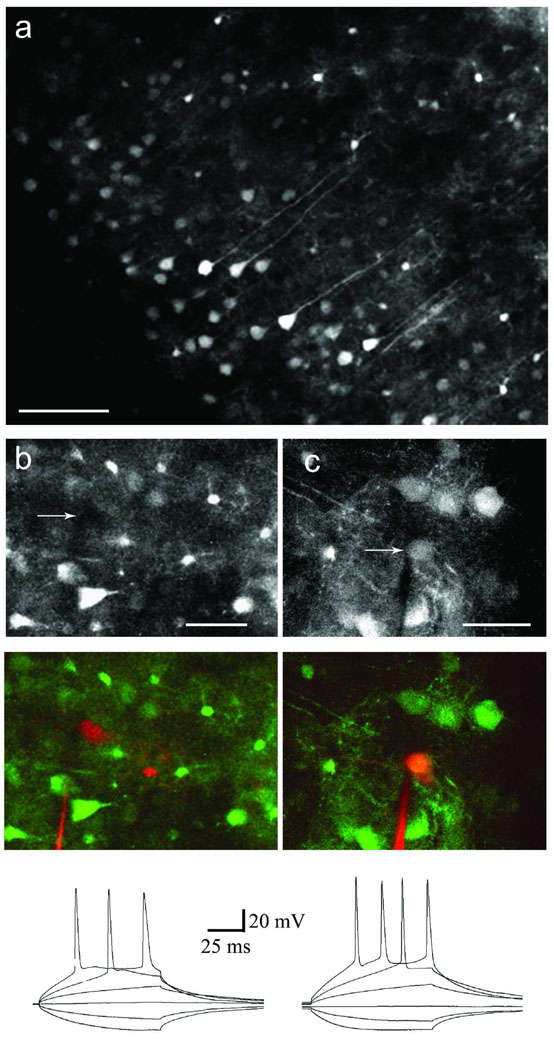
Figure 9. Intrinsic and active properties of dsAAV1-GFP labeled neurons.
Two-photon imaging of an acute slice made from an animal injected with dsAAV1-GFP (A). Notice that many neurons and glia are visible with varying intensities of labeling. Non-labeled (B) and labeled (C) neuronal properties were assayed using whole cell recording. Alexa594 was included in the recording pipette to allow visualization of recorded neurons. Top panels represent images collected in the GFP detector channel (arrows indicate location of recorded neuron), while middle panels show the overlay of both Alexa 594 and GFP detector channels. Notice the lack of signal in the GFP channel of the non-labeled neuron in B. Bottom panels show recordings obtained from the neurons featured in the top panels. A family of step currents was injected at the soma to determine the input resistance and action potential amplitude and threshold. The intrinsic and active properties of transduced neurons were not significantly different than those of their non-transduced neighbors (see Table S1; p>0.05; Student’s t-test). Scale bars: 80 µm (A), 20 µm (B), 30 µm (C).
Table 1.
Intrinsic and firing properties of dsAAV-labeled neurons.
| dsAAV-labeled neurons |
Non-dsAAV labeled neurons |
P value (t-test) |
|
|---|---|---|---|
| N (cells) | 9 | 5 | |
| Resting potential (mV) | −65.56±2.02 | −66.72±0.94 | 0.5588 |
| Threshold (mV) | −44.45±2.83 | −39.78±1.30 | 0.1078 |
| Action potential amplitude (mV) |
77.03±1.55 | 77.67±1.65 | 0.8177 |
| Input resistance (MΩ) | 188.00±25.22 | 166.22±21.32 | 0.5618 |
Whole cell recordings were obtained from dsAAV1-labeled neurons and neighboring non-labeled neurons. There were no statistically significant differences (p>0.05; Student’s t-test) in the intrinsic and firing properties between the two neuronal sets. Data is reported as mean±SEM.
DISCUSSION
Most studies of the adeno-associated viral vector transduction of the brain examine a single-stranded version of the virus (Tenenbaum et al., 2004; Broekman et al., 2006; Cearley and Wolfe, 2006; Taymans et al., 2007). The development of a double-stranded DNA genome in an AAV vector was accomplished by mutating the 5’ ITR (Wang et al., 2003) and allowed for more efficient transduction of target cells. The double-stranded DNA genome negates the need for second-strand synthesis prior to transcription of transgene which makes it potentially more useful than ssAAV for studies requiring fast expression. In the current study we have characterized a double-stranded AAV vector as a labeling method for in vivo developmental studies of visual cortex. We find that dsAAV can label neurons and glia both faster and more robustly than ssAAV, but like ssAAV, the labeling is long-lasting and results in minimal cytotoxicity. Additionally, dsAAV-labeled neurons and glia, as well as their processes, can be imaged in living acute slices as well as in the intact brain. While our studies have concentrated on labeling in the visual cortex, injections that targeted other structures such as other cortical areas and the hippocampus suggest that dsAAV can also be used to label neurons and glia in other brain areas, although the cell types targeted for transduction are dependent on the specific brain area being transduced.
While studies of AAV have found primarily neuronal labeling, the majority of these studies used ssAAV (Kaplitt et al., 1994; Bartlett et al., 1998; Davidson et al., 2000; Burger et al., 2004; Paterna et al., 2004; Cearley and Wolfe, 2006; Harding et al., 2006; Taymans et al., 2007). In contrast, a recent study found that one serotype of a self-complimentary AAV vector produced both neuronal and non-neuronal labeling (Fu et al., 2003). We find the density and efficiency of this non-neuronal labeling is serotype-dependent, and serotypes 2 and 9 were particularly efficient at labeling glia (Figure 4e). These glial-labeling capabilities make dsAAV a viable labeling technique for studying and manipulating astrocytes in healthy and diseased tissue in the adult and developing visual cortex.
Similar to previous studies using ssAAV, we found that dsAAV efficiently labeled neurons in mice of different ages. In particular, injections of dsAAV1 and 7 resulted in large numbers (more than 200) of infected Type 3 neurons per injection in P35–P49 mice. Only 3–4 neurons per injection exhibited Type 1 labeling whereby small structures such as dendrites and axons could be easily discerned over the background fluorescence. However, a limited number of well-labeled neurons is desirable in imaging studies (Deisseroth et al., 2006), as too many labeled neurons in a small area make it difficult to discern the origin of individual processes, and fine structures can be obscured by nearby bright objects. Indeed we demonstrate that fine processes of neurons and glia labeled with dsAAV can be imaged with high resolution both in fixed brain slices and in the intact brain using two-photon microscopy.
ssAAVs have previously been used to label neuronal populations for in vivo imaging in the visual cortex of the primate (Stettler et al., 2006). In these experiments, imaging was delayed for 3 weeks or more to allow for complete labeling of neurons by ssAAV so that axons and axon terminals could be clearly visualized in vivo. In comparison, we characterize the efficiency of different dsAAV serotypes at targeting specific cell types in visual cortex and demonstrate high level expression of GFP as early as 2 days post-injection of dsAAV1-GFP. This timescale rivals that of vectors such as Sindbis virus, whose use is limited by excessive toxicity (Jeromin et al., 2003; Kim et al., 2004), while surpassing that of conventional ssAAV vectors. The difference in labeling between dsAAV and ssAAV-injected brains may be due to availability of the cellular enzymes needed to catalyze second strand DNA synthesis of ssAAV (Ferrari et al., 1996; Fisher et al., 1996). More recently, a study found that the instability of the newly synthesized double-stranded DNA of a ssAAV genome is the primary contributing factor for the low transduction efficiency of ssAAV (Wang et al., 2007). Consistent with this study, we observed lower genome recovery from rat primary cortical cultures transduced with ssAAV1 versus dsAAV1 at 2 days post-transduction even though comparable numbers of viral genomes were injected (based on qPCR analysis and standard DNA gel analysis of purified viral genomes from ssAAV and dsAAV; data not shown). Further studies and characterization of ssAAV and dsAAV serotype 1 are needed to identify the mechanism(s) underlying the differences in transgene expression.
dsAAV-mediated expression was maintained for at least 4 weeks, possibly due to the low level of toxicity of dsAAV. This possibility is supported by our observation that transduced neurons appeared structurally similar to non-transduced neurons at 4 weeks post-injection, and the electrophysiological parameters of transduced neurons were identical to those of non-transduced neighboring neurons at 7 days post-injection. Immunocytochemical studies showed very limited inflammatory response around the injection site and the majority of labeled and non-labeled glia retained non-reactive morphologies. Additionally, chronic imaging revealed high stability of axon terminals in dsAAV-transduced neurons. These observations suggest that the cortex suffered limited damage from the viral transduction and the physical injection of virus.
Previous studies have shown that AAV transgene expression mediated by the cytomegalovirus immediate-early enhancer/promoter (CMV) decreases over time and this decrease is dependent on the brain region transduced (McCown et al., 1996; Klein et al., 1998; Lo et al., 1999). We found that expression of GFP from the CMV promoter is sufficient for in vivo imaging of cells in the visual cortex from 2 days to 4 weeks post-transduction. Future studies examining longer time points may identify the need to use alternative promoters capable of transgene expression at time points longer than 4 weeks.
In conclusion, dsAAV retains the advantages of ssAAV of neuronal transduction and prolonged high levels of expression, while providing the additional advantages of glial labeling and rapid expression. All of these properties facilitate imaging of rapid structural changes in cellular structures while the minimal cytopathic effects characteristic of AAV vectors allow long-term imaging and electrophysiological study of labeled neurons. As such, dsAAV is an excellent vector for in vivo labeling of the visual cortex and is appropriate for in vitro and in vivo studies of structure and function in both the adult and the developing brain.
Supplementary Material
Supplementary movie S1. In vivo imaging of dsAAV1-GFP labeled neurons.
This movie shows in vivo imaging of visual cortex in a mouse injected with dsAAV1-GFP. Each frame in the movie shows an image taken deeper in the cortex at increments of 5 µm, starting at the pia and ending ~750 µm below the surface. The laser power was increased several times (resulting in sudden increases in brightness) to compensate for signal loss deeper within the brain. This movie corresponds to the reconstructions shown in Figure 6A,D. Notice that astrocytes and neurons can be discerned even in the center of the injection and at depths of ~700 µm below the level of the pia.
ACKNOWLEDGMENTS
This work was supported in part by the Burroughs Wellcome Fund, Whitehall Foundation, and Sloan Foundation (A.K.M.), NIH vision training grant (E.A.K.) and by the Intramural Research Program at the National Institute on Drug Abuse, NIH (B.K.H.). We would like to thank John Olschowka and Lee Trojanczyk for help with immunocytochemical experiments and Douglas Howard and Priscila Castillo for their technical assistance with viral preparation and characterization. We thank Dr. Yun Wang for his review and comments on the manuscript.
REFERENCES
- Bartlett JS, Samulski RJ, McCown TJ. Selective and rapid uptake of adeno-associated virus type 2 in brain. Hum Gene Ther. 1998;9:1181–1186. doi: 10.1089/hum.1998.9.8-1181. [DOI] [PubMed] [Google Scholar]
- Broekman ML, Comer LA, Hyman BT, Sena-Esteves M. Adeno-associated virus vectors serotyped with AAV8 capsid are more efficient than AAV-1 or -2 serotypes for widespread gene delivery to the neonatal mouse brain. Neuroscience. 2006;138:501–510. doi: 10.1016/j.neuroscience.2005.11.057. [DOI] [PubMed] [Google Scholar]
- Burger C, Gorbatyuk OS, Velardo MJ, Peden CS, Williams P, Zolotukhin S, Reier PJ, Mandel RJ, Muzyczka N. Recombinant AAV viral vectors pseudotyped with viral capsids from serotypes 1, 2, and 5 display differential efficiency and cell tropism after delivery to different regions of the central nervous system. Mol Ther. 2004;10:302–317. doi: 10.1016/j.ymthe.2004.05.024. [DOI] [PubMed] [Google Scholar]
- Cearley CN, Wolfe JH. Transduction characteristics of adeno-associated virus vectors expressing cap serotypes 7, 8, 9, and Rh10 in the mouse brain. Mol Ther. 2006;13:528–537. doi: 10.1016/j.ymthe.2005.11.015. [DOI] [PubMed] [Google Scholar]
- Chamberlin NL, Du B, de Lacalle S, Saper CB. Recombinant adeno-associated virus vector: use for transgene expression and anterograde tract tracing in the CNS. Brain Res. 1998;793:169–175. doi: 10.1016/s0006-8993(98)00169-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SL, Ma HI, Han JM, Tao PL, Law PY, Loh HH. dsAAV type 2-mediated gene transfer of MORS196A-EGFP into spinal cord as a pain management paradigm. Proc Natl Acad Sci U S A. 2007;104:20096–20101. doi: 10.1073/pnas.0703409104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson BL, Stein CS, Heth JA, Martins I, Kotin RM, Derksen TA, Zabner J, Ghodsi A, Chiorini JA. Recombinant adeno-associated virus type 2, 4, and 5 vectors: transduction of variant cell types and regions in the mammalian central nervous system. Proc Natl Acad Sci U S A. 2000;97:3428–3432. doi: 10.1073/pnas.050581197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisseroth K, Feng G, Majewska AK, Miesenbock G, Ting A, Schnitzer MJ. Next-generation optical technologies for illuminating genetically targeted brain circuits. J Neurosci. 2006;26:10380–10386. doi: 10.1523/JNEUROSCI.3863-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari FK, Samulski T, Shenk T, Samulski RJ. Second-strand synthesis is a rate-limiting step for efficient transduction by recombinant adeno-associated virus vectors. J Virol. 1996;70:3227–3234. doi: 10.1128/jvi.70.5.3227-3234.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher KJ, Gao GP, Weitzman MD, DeMatteo R, Burda JF, Wilson JM. Transduction with recombinant adeno-associated virus for gene therapy is limited by leading-strand synthesis. J Virol. 1996;70:520–532. doi: 10.1128/jvi.70.1.520-532.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Muenzer J, Samulski RJ, Breese G, Sifford J, Zeng X, McCarty DM. Self-complementary adeno-associated virus serotype 2 vector: global distribution and broad dispersion of AAV-mediated transgene expression in mouse brain. Mol Ther. 2003;8:911–917. doi: 10.1016/j.ymthe.2003.08.021. [DOI] [PubMed] [Google Scholar]
- Gao G, Vandenberghe LH, Alvira MR, Lu Y, Calcedo R, Zhou X, Wilson JM. Clades of Adeno-associated viruses are widely disseminated in human tissues. J Virol. 2004;78:6381–6388. doi: 10.1128/JVI.78.12.6381-6388.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao GP, Alvira MR, Wang L, Calcedo R, Johnston J, Wilson JM. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc Natl Acad Sci U S A. 2002;99:11854–11859. doi: 10.1073/pnas.182412299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding TC, Dickinson PJ, Roberts BN, Yendluri S, Gonzalez-Edick M, Lecouteur RA, Jooss KU. Enhanced gene transfer efficiency in the murine striatum and an orthotopic glioblastoma tumor model, using AAV-7- and AAV-8-pseudotyped vectors. Hum Gene Ther. 2006;17:807–820. doi: 10.1089/hum.2006.17.807. [DOI] [PubMed] [Google Scholar]
- Howard DB, Powers K, Wang Y, Harvey BK. Tropism and toxicity of adeno-associated viral vector serotypes 1, 2, 5, 6, 7, 8, and 9 in rat neurons and glia in vitro. Virology. 2008;372:24–34. doi: 10.1016/j.virol.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeromin A, Yuan LL, Frick A, Pfaffinger P, Johnston D. A modified Sindbis vector for prolonged gene expression in neurons. J Neurophysiol. 2003;90:2741–2745. doi: 10.1152/jn.00464.2003. [DOI] [PubMed] [Google Scholar]
- Kaplitt MG, Leone P, Samulski RJ, Xiao X, Pfaff DW, O'Malley KL, During MJ. Long-term gene expression and phenotypic correction using adeno-associated virus vectors in the mammalian brain. Nat Genet. 1994;8:148–154. doi: 10.1038/ng1094-148. [DOI] [PubMed] [Google Scholar]
- Kim J, Dittgen T, Nimmerjahn A, Waters J, Pawlak V, Helmchen F, Schlesinger S, Seeburg PH, Osten P. Sindbis vector SINrep(nsP2S726): a tool for rapid heterologous expression with attenuated cytotoxicity in neurons. J Neurosci Methods. 2004;133:81–90. doi: 10.1016/j.jneumeth.2003.09.029. [DOI] [PubMed] [Google Scholar]
- Klein RL, Meyer EM, Peel AL, Zolotukhin S, Meyers C, Muzyczka N, King MA. Neuron-specific transduction in the rat septohippocampal or nigrostriatal pathway by recombinant adeno-associated virus vectors. Exp Neurol. 1998;150:183–194. doi: 10.1006/exnr.1997.6736. [DOI] [PubMed] [Google Scholar]
- Lo WD, Qu G, Sferra TJ, Clark R, Chen R, Johnson PR. Adeno-associated virus-mediated gene transfer to the brain: duration and modulation of expression. Hum Gene Ther. 1999;10:201–213. doi: 10.1089/10430349950018995. [DOI] [PubMed] [Google Scholar]
- Majewska A. Dynamic remodeling of dendritic spines in developmental visual plasticity. Cell Science Reviews. 2007;3:85–98. [Google Scholar]
- Majewska A, Yiu G, Yuste R. A custom-made two-photon microscope and deconvolution system. Pflugers Arch. 2000;441:398–408. doi: 10.1007/s004240000435. [DOI] [PubMed] [Google Scholar]
- Majewska AK, Newton JR, Sur M. Remodeling of synaptic structure in sensory cortical areas in vivo. J Neurosci. 2006;26:3021–3029. doi: 10.1523/JNEUROSCI.4454-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty DM, Fu H, Monahan PE, Toulson CE, Naik P, Samulski RJ. Adeno-associated virus terminal repeat (TR) mutant generates self-complementary vectors to overcome the rate-limiting step to transduction in vivo. Gene Ther. 2003;10:2112–2118. doi: 10.1038/sj.gt.3302134. [DOI] [PubMed] [Google Scholar]
- McCarty DM, Monahan PE, Samulski RJ. Self-complementary recombinant adeno-associated virus (scAAV) vectors promote efficient transduction independently of DNA synthesis. Gene Ther. 2001;8:1248–1254. doi: 10.1038/sj.gt.3301514. [DOI] [PubMed] [Google Scholar]
- McCown TJ. Adeno-associated virus (AAV) vectors in the CNS. Curr Gene Ther. 2005;5:333–338. doi: 10.2174/1566523054064995. [DOI] [PubMed] [Google Scholar]
- McCown TJ, Xiao X, Li J, Breese GR, Samulski RJ. Differential and persistent expression patterns of CNS gene transfer by an adeno-associated virus (AAV) vector. Brain Res. 1996;713:99–107. doi: 10.1016/0006-8993(95)01488-8. [DOI] [PubMed] [Google Scholar]
- Paterna JC, Feldon J, Bueler H. Transduction profiles of recombinant adeno-associated virus vectors derived from serotypes 2 and 5 in the nigrostriatal system of rats. J Virol. 2004;78:6808–6817. doi: 10.1128/JVI.78.13.6808-6817.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portera-Cailliau C, Weimer RM, De Paola V, Caroni P, Svoboda K. Diverse modes of axon elaboration in the developing neocortex. PLoS Biol. 2005;3:e272. doi: 10.1371/journal.pbio.0030272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz JE, Rolling F, Li C, Conrath H, Xiao W, Xiao X, Samulski RJ. Cross-packaging of a single adeno-associated virus (AAV) type 2 vector genome into multiple AAV serotypes enables transduction with broad specificity. J Virol. 2002;76:791–801. doi: 10.1128/JVI.76.2.791-801.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutledge EA, Halbert CL, Russell DW. Infectious clones and vectors derived from adeno-associated virus (AAV) serotypes other than AAV type 2. J Virol. 1998;72:309–319. doi: 10.1128/jvi.72.1.309-319.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stettler DD, Yamahachi H, Li W, Denk W, Gilbert CD. Axons and synaptic boutons are highly dynamic in adult visual cortex. Neuron. 2006;49:877–887. doi: 10.1016/j.neuron.2006.02.018. [DOI] [PubMed] [Google Scholar]
- Taymans JM, Vandenberghe LH, Haute CV, Thiry I, Deroose CM, Mortelmans L, Wilson JM, Debyser Z, Baekelandt V. Comparative analysis of adeno-associated viral vector serotypes 1, 2, 5, 7, and 8 in mouse brain. Hum Gene Ther. 2007;18:195–206. doi: 10.1089/hum.2006.178. [DOI] [PubMed] [Google Scholar]
- Tenenbaum L, Chtarto A, Lehtonen E, Velu T, Brotchi J, Levivier M. Recombinant AAV-mediated gene delivery to the central nervous system. J Gene Med. 2004;6 Suppl 1:S212–S222. doi: 10.1002/jgm.506. [DOI] [PubMed] [Google Scholar]
- Wang J, Xie J, Lu H, Chen L, Hauck B, Samulski RJ, Xiao W. Existence of transient functional double-stranded DNA intermediates during recombinant AAV transduction. Proc Natl Acad Sci U S A. 2007;104:13104–13109. doi: 10.1073/pnas.0702778104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Ma HI, Li J, Sun L, Zhang J, Xiao X. Rapid and highly efficient transduction by double-stranded adeno-associated virus vectors in vitro and in vivo. Gene Ther. 2003;10:2105–2111. doi: 10.1038/sj.gt.3302133. [DOI] [PubMed] [Google Scholar]
- Xiao W, Chirmule N, Berta SC, McCullough B, Gao G, Wilson JM. Gene therapy vectors based on adeno-associated virus type 1. J Virol. 1999;73:3994–4003. doi: 10.1128/jvi.73.5.3994-4003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X, Li J, Samulski RJ. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J Virol. 1998;72:2224–2232. doi: 10.1128/jvi.72.3.2224-2232.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary movie S1. In vivo imaging of dsAAV1-GFP labeled neurons.
This movie shows in vivo imaging of visual cortex in a mouse injected with dsAAV1-GFP. Each frame in the movie shows an image taken deeper in the cortex at increments of 5 µm, starting at the pia and ending ~750 µm below the surface. The laser power was increased several times (resulting in sudden increases in brightness) to compensate for signal loss deeper within the brain. This movie corresponds to the reconstructions shown in Figure 6A,D. Notice that astrocytes and neurons can be discerned even in the center of the injection and at depths of ~700 µm below the level of the pia.