Abstract
Heterogeneous nuclear ribonucleoproteins (hnRNPs) comprise a family of RNA-binding proteins. The complexity and diversity associated with the hnRNPs render them multifunctional, involved not only in processing heterogeneous nuclear RNAs (hnRNAs) into mature mRNAs, but also acting as trans-factors in regulating gene expression. Heterogeneous nuclear ribonucleoprotein E1 (hnRNP E1), a subgroup of hnRNPs, is a KH-triple repeat containing RNA-binding protein. It is encoded by an intronless gene arising from hnRNP E2 through a retrotransposition event. hnRNP E1 is ubiquitously expressed and functions in regulating major steps of gene expression, including pre-mRNA processing, mRNA stability, and translation. Given its wide-ranging functions in the nucleus and cytoplasm and interaction with multiple proteins, we propose a post-transcriptional regulon model that explains hnRNP E1's widespread functional diversity.
Keywords: hnRNP, hnRNP E1, PCBP1, pre-mRNA processing, mRNA stability, translation, post-transcriptional regulon
INTRODUCTION
Research over the last two decades has generated a growing appreciation of the importance of gene regulatory processes at the post-transcriptional level (mRNA processing, mRNA stability and turnover, and translation). Nearly all of the mechanistic pathways discovered for such regulation involve the binding of proteins to coding or untranslated regions (UTRs) of mRNAs. Major questions that are being actively addressed are how RNA-binding proteins recognize specific RNA sites, what effects they have on RNA structure and processing dynamics, and how they regulate different functions. Often, these RNA-binding proteins, as is the case with the heterogeneous nuclear ribonucleoproteins (hnRNPs), are multifunctional, involved not only in processing heterogeneous nuclear RNAs (hnRNAs) into mature mRNAs, but also acting as trans-factors in regulating gene expression (Dreyfuss et al. 1993).
Based on high-affinity and sequence-specific RNA interaction, the hnRNPs have been categorized into defined groups (Dreyfuss et al. 1993). The poly(C)-binding proteins (PCBPs) are one such subgroup. PCBPs comprise two subsets, the hnRNPs K/J and the hnRNP E (1-4) (also referred to as α-CPs or PCBP 1-4) (Matunis et al. 1992; Makeyev and Liebhaber 2002). PCBPs in general have been extensively reviewed (Bomsztyk et al. 1997; Ostareck-Lederer et al. 1998; Makeyev and Liebhaber 2002). Herein, we provide a brief overview of the hnRNP family of proteins and subsequently focus specifically on hnRNP E1. We discuss its origin, structure, functional diversity, targets, and protein partners and summarize its multifunctional roles in maintaining cellular homeostasis during differentiation and cancer progression.
hnRNP FAMILY
Definition
In eukaryotic cells, nascent transcripts that are produced by RNA polymerase II undergo extensive processing before a functional mRNA is produced. These mRNA precursors are referred to as heterogeneous nuclear ribonucleic acids (hnRNAs), a historical term that refers to their size, heterogeneity, and cellular location, and is often used interchangeably with pre-mRNA (Dreyfuss et al. 1993). It must be added, however, that only a subset of hnRNAs serves as actual precursors to mRNAs, and the functions of the remainder is still unclear (Dreyfuss et al. 1993). From the moment hnRNAs are produced by RNA polymerase II and until their maturation into mRNAs, the hnRNAs are associated with proteins in large complexes (Gall 1956). Collectively, these hnRNA-binding proteins are termed hnRNP proteins (Dreyfuss et al. 1993).
hnRNP proteins are multifunctional: Along with other RNP proteins, they participate in pre-mRNA processing such as splicing and are important determinants of mRNA export, localization, translation, and stability (Dreyfuss et al. 2002). Many of the hnRNP proteins remain associated with the mRNA as it is transported, through nuclear pores, to ribosomes, hence undergoing constant nuclear–cytoplasmic shuttling (Dreyfuss et al. 2002). The hnRNP complex is highly dynamic; it is successively remodeled through the loss or acquisition of hnRNP proteins and other proteins as a consequence of specific processes. From its initial production, the hnRNP complex is modified at every step along the pathway of mRNA biogenesis until it emerges into an mRNP complex, ready to engage the mRNA turnover and translation machinery. The interaction and processing of the hnRNA with hnRNP proteins, as well as other RNA-binding proteins, creates what has been classically termed the “mRNP code” (Singh and Valcarcel 2005), in which the protein composition of the complex reveals its specificity and cytoplasmic fate. The mRNP code is only a predicted model system and perhaps does not represent its actual existence within the cell.
Discovery
Evidence for the existence of an hnRNP complex began to emerge following the examination of lampbrush chromosomes in amphibian oocytes (Gall 1956). Electron micrographs of these chromosomes revealed loops, which consisted of particles connected by thin strands. The interconnecting strands were subsequently shown to be hnRNA, and the particles were shown to be RNPs (Sommerville and Scheer 1981). The use of improved chromosome spreading techniques revealed a “beads-on-a-string” conformation for hnRNP complexes that were similar to the model for the packaging of DNA by histones (Miller and Bakken 1972). Biochemical characterization of nuclear RNPs appeared to support the existence of an hnRNP complex in vivo (Samarina et al. 1966). They identified a nuclear hnRNP complex sedimenting at 30S that was composed of rapidly labeled RNA and protein. Due in part to the instability of the RNA component, it was hypothesized that the RNA species in this particle was pre-mRNA. The name “informofers” was given to these 30S particles. These structures were postulated to transmit information from the nucleus to the cytoplasm and were thought to be composed of a single protein migrating at ∼40 kDa bound to ∼500 nucleotides (nt) of RNA (Samarina et al. 1968). The unstable nature of the RNP complexes led to the proposal that these 30S particles were the remnants of a large complex. Treatment of nuclear extracts with ribonuclease inhibitors shifted the sedimentation value of the complex from a homogeneous 30S particle to a heterogeneous distribution of particles ranging from 30S to ∼200S (Samarina et al. 1968). The idea that the 200S complex was composed of multiple 30S particles was further supported by the conversion of large complexes to 30S by deliberate ribonuclease digestion. Other studies proposed a “fibril model” for hnRNP structure. This model proposed that hnRNPs form a fibril structure rather than a particle (Stevenin et al. 1982). The inability to differentiate proteins that may be adventitiously bound to hnRNP complexes from authentic hnRNPs severely hampered the acceptance of this model. While the existence of hnRNP complexes was generally accepted, it was unclear whether a single hnRNP complex or multiple forms existed. Evidence against a sole hnRNP complex, and for differential association of hnRNPs with pre-mRNAs, was obtained from both in vitro RNA-binding studies and immunohistochemical studies. The idea of transcript-specific packaging is further supported by the finding that the ratio of individual hnRNPs varies with different primary transcripts (Amero et al. 1992; Matunis et al. 1993).
The hnRNP protein family
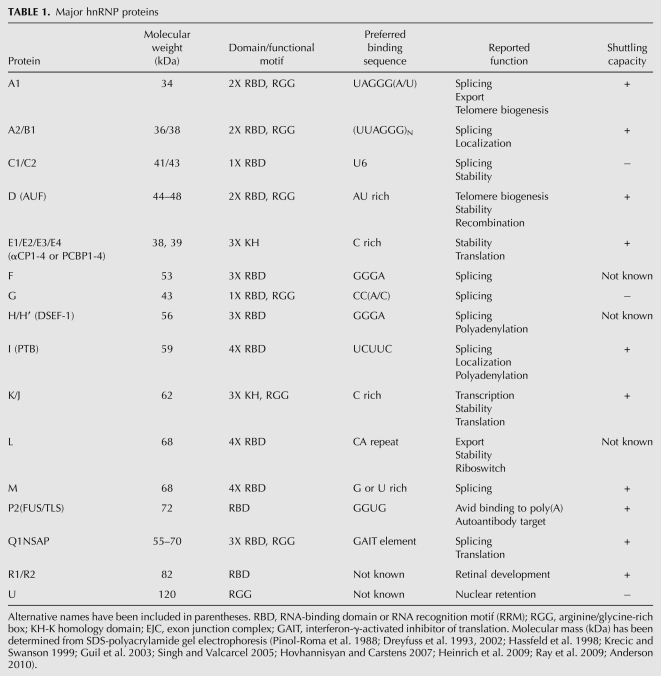
In human (HeLa) cells, a combination of immunopurification and two-dimensional gel electrophoresis has revealed the protein composition of hnRNP complexes: They are composed of at least 20 abundant, major hnRNP proteins and other less abundant, minor hnRNP proteins. The abundant proteins, designated hnRNP A1 through hnRNP U, have molecular weights ranging from 34 kDa to 120 kDa and form the core hnRNPs (Pinol-Roma et al. 1988; Dreyfuss et al. 1993, 2002) and are summarized in Table 1. Among these, hnRNP proteins A1, A2/B1, B2, C1, and C2 were the first identified based on their somewhat indiscriminate association with nascent transcripts and were subsequently termed the “core” hnRNP proteins (Beyer et al. 1977). Minor hnRNP proteins are ones that are not stably bound to hnRNAs during immunopurification or those that are preferentially associated with only a specific subset of hnRNAs (Dreyfuss et al. 1993, 2002; Krecic and Swanson 1999; Singh and Valcarcel 2005), and they are summarized in Table 2.
TABLE 1.
Major hnRNP proteins
TABLE 2.
Minor hnRNP proteins
hnRNP proteins are among the most abundant nuclear proteins, rivaling histones, and when complexed to hnRNAs, form major components of the nucleus. The mRNAs contained within certain hnRNP complexes have been estimated to be present at ∼100 million copies per nucleus, while others are present only in small amounts (Kamma et al. 1995; Dreyfuss et al. 2002). A unique combinatorial arrangement or “constellation” of hnRNP proteins is thought to form on each hnRNA dictated by the specific RNA sequence and relative expression of the hnRNPs (including abundance levels and post-translational modifications) (Habelhah et al. 2001; Mandal et al. 2001). However, as the “core” hnRNP proteins, such as A1, are extremely abundant, each protein is likely to be in vast excess over its respective binding sites. Therefore, in addition to their sequence-specific binding, nonspecific interactions are also prevalent among the hnRNP proteins. A systematic immunohistochemical survey revealed that hnRNP proteins are ubiquitously expressed in all tissue types to varying abundance levels and that their relative stoichiometry across cell types is not fixed (Kamma et al. 1995). Furthermore, hnRNP A1, D, E, F/H, and K were demonstrated to be present in both the nucleus and cytoplasm, suggestive of cytoplasmic roles for these hnRNPs (Dreyfuss et al. 2002).
Structure of hnRNP proteins
cDNA sequence analysis of the hnRNP proteins has revealed a modular structure consisting of one or more RNA-binding motifs and at least one auxiliary domain that regulates protein–protein interactions and subcellular localization (see Table 1; Dreyfuss et al. 2002). Three types of RNA-binding motifs have been identified: (1) the RNP-CS-RBD motif (Bandziulis et al. 1989; Query et al. 1989); (2) the RGG (Arg-Gly-Gly) boxes (Kiledjian and Dreyfuss 1992); and (3) the K-homology (KH) domains (Burd and Dreyfuss 1994).
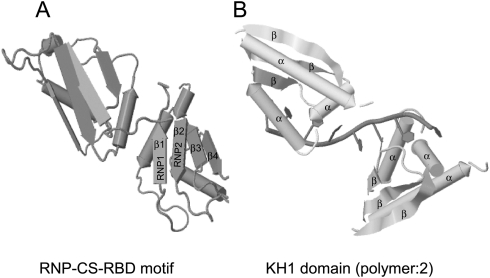
The best characterized and the most common RNA-binding motif among hnRNPs is the RNP consensus sequence–RNA-binding domain (RNP-CS-RBD), also referred to as the RNA recognition motif, RRM, RNP 80, or RNP motif (Dreyfuss et al. 1988). The hallmarks of this motif are two highly conserved RNP1 octameric and RNP2 hexameric sequences (Dreyfuss et al. 1988). These are located about 30 residues apart in these motifs of ∼90 amino acids (Dreyfuss et al. 1988). RNP2 is more conserved than RNP1 and was found based on sequence similarity between hnRNP A1 and the poly(A)-binding protein (PABP). The three-dimensional structure of this motif has been resolved (Fig. 1A). The N-terminal RBD folds into a βαββαβ structure. The RNP1 and RNP2 are juxtaposed on the centrally located β3 and β1, respectively, and make direct contact with the RNA. The β-sheet surface of the RBD constitutes an exposed surface that can serve as a platform for RNA binding (Gorlach et al. 1992). Important determinants of RNA-binding specificity of the hnRNP proteins were found to reside within the variable regions of the loops connecting the β-strands and the terminal regions of the RBD (Gorlach et al. 1992). These regions lie adjacent to the highly conserved sequences RNP1 and RNP2 and differ in amino acid sequence between several hnRNP proteins. Hence, these residues are the source of RNA-binding specificity differences between family members (Gorlach et al. 1992).
FIGURE 1.
Structure of hnRNP proteins. (A) The RNP-CS-RBD motif has a βαββαβ structure. The octameric RNP1 and hexameric RNP2 sequences are juxtaposed on β3 and β1, respectively. The structure shown is of multiple isomorphous replacement (MIR) of two RBDs of human hnRNP A1 at 1.75 Å (PDB ID: 1HA1) (Shamoo et al. 1997). (B) The KH domain comprises a triple β-sheet platform supporting three α-helical segments. Structure shown is of X-ray crystallography (resolution 3 Å) of KH1 domain (2-polymer) of human hnRNP E1 (PBD ID: 1ZTG) (Sidiqi et al. 2005). Both structures have been derived by the JMol algorithm.
The RGG box, originally described in hnRNP U and found to be responsible for the sole RNA-binding activity of this protein, is characterized by closely spaced clusters of Arg-Gly-Gly tripeptide repeats with interspersed aromatic (Phe, Tyr) residues. Both the number and spacing of the RGG repeats in different proteins are variable. In hnRNP U, the RGG box is both necessary and sufficient for RNA-binding activity (Kiledjian and Dreyfuss 1992); however, in other cases such as with hnRNP A1 and G, it is found in combination with other RNA-binding elements like the RBD. This suggests that the RGG box possibly plays another role besides contributing to RNA binding. Many of the Arg residues within the RGG box are sites for methylation and may act to modulate RNA binding.
The KH domain was first identified in hnRNP K as a triple repeat and was later found in other RNA-binding proteins from evolutionary distant organisms (Siomi et al. 1993a; Burd and Dreyfuss 1994). Initially the KH domain was believed to be a 45-aminoacid-long stretch (Gibson et al. 1993), but has been now revised to be an ∼70-amino acid domain, comprising a triple β-sheet platform supporting three α-helical segments (βααββα fold) (Fig. 1B; Dejgaard and Leffers 1996; Musco et al. 1996, 1997; Lewis et al. 1999, 2000; Chkheidze and Liebhaber 2003). Resolution of the crystal structure indicates that a loop (GKXG tetrapeptide) between the first two helices (α1 and α2) of the KH domain specifically interacts with a 4–5-nt stretch in the RNA (Musco et al. 1996; Buckanovich and Darnell 1997; Jensen et al. 2000). KH domains are divided into two subgroups based on the N- or C-terminal extension that they have; Type I has a C-terminal βα extension, whereas Type II has an N-terminal αβ extension (Grishin 2001).
Among eukaryotes, the KH domain is present in the PCBPs (hnRNP K/J and hnRNP E1-E4) (Siomi et al. 1993a; Kiledjian et al. 1995; Leffers et al. 1995); the product of the fragile X mental retardation gene (FMR1) (Siomi et al. 1993b); splicing modulators Mer1 in yeast (Nandabalan et al. 1993) and PS1 in Drosophila (Siebel et al. 1995); vigilin (Schmidt et al. 1992); RNA-associated Src substrate Sam68 (Courtneidge and Fumagalli 1994); signal transduction and activation of RNA (STAR) family of RNA-binding proteins (Musco et al. 1996; Vernet and Artzt 1997); and Nova-1, an auto-antigen in paraneoplastic opsoclonus myoclonus ataxia (Buckanovich et al. 1993). These proteins have different numbers of KH repeats as well as varying specificity, with a few exceptions like Nova, for poly(C) stretches in RNA (Makeyev and Liebhaber 2002). KH domains are also prevalent among some prokaryotic proteins like ribonuclease PNP (Regnier et al. 1987), the transcription elongation factor NusA (Liu and Hanna 1995), and the ribosomal protein S3 (Urlaub et al. 1995).
hnRNP proteins also contain other domains that mediate important functional specificity. These domains are termed auxiliary domains. Contrary to RNA-binding domains such as the RBD and RGG boxes, auxiliary domains are divergent in protein sequence and are unstructured. The most frequently found and best-characterized auxiliary domains are the glycine-rich domains found in the hnRNP A/B proteins (Weighardt et al. 1996). Both hnRNP A1 and A2/B1 have similar structures: They contain two RBDs at their N termini and a glycine-rich auxiliary domain at their C termini; thus, they are referred to as 2xRBD-Gly proteins (Matunis et al. 1992).
Nucleo-cytoplasmic shuttling of hnRNP proteins
Within the auxiliary domain of hnRNP A1 is a glycine-rich, 38-amino acid (YNDFGNYNNQSSNFGPMKGGNFGGRSSGPY) nucleo-cytoplasmic shuttling (NS) domain called M9 (Siomi and Dreyfuss 1995). The M9 domain is both necessary and sufficient to confer nuclear localization; attachment of this region to other proteins results in nuclear localization of the resulting chimeras. Neither the RBDs nor the RGG box is required for nuclear import of hnRNP A1. The M9 domain is a nuclear localization domain and does not bear resemblance to the classical NLS. Interestingly, the M9 domain also acts as a nuclear export signal, allowing export of hnRNP A1 in a temperature-dependent manner (Michael et al. 1995; Weighardt et al. 1996). Like many of the hnRNP proteins, hnRNP A1 is predominantly nuclear at steady state; and it, along with select members from the A, B, E, groups and hnRNP D, I, and K, shuttles rapidly between the nucleus and cytoplasm, whereas others such as hnRNP C and U are strictly nuclear. The hnRNPs C and U contain classical nuclear localization signal (NLS) sequences and are exclusively sequestered to the nucleus, as compared to noncanonical NS domains present in hnRNP A1. hnRNP K contains both a classical NLS and an NS domain (Michael et al. 1997), and MAPK/ERK-dependent phosphorylation of Ser 284 and 358 residues within the NS domain induces it to shuttle to the cytoplasm, where it subsequently inhibits translation (Habelhah et al. 2001). hnRNP E2 possesses two functionally independent NLS, a nanomeric segment between KH2 and KH3 (NLS I) and a dodecameric segment (NLS II) within the KH3 domain, that allow for its nucleo-cytoplasmic shuttling. hnRNP E3 and E4 are exclusively cytosolic and lack either the NLS I or II present in hnRNP E2 (Chkheidze and Liebhaber 2003).
Post-translation modifications regulating hnRNPs
As previously mentioned, hnRNP proteins undergo several post-translational modifications, and such changes regulate their subcellular localization. Modifications reported on hnRNPs include phosphorylation, sumoylation, ubiquitination, and methylation. For example, hnRNP proteins belonging to the A, B, and C groups and hnRNP G, K, and U are all phosphorylated in vivo (Dreyfuss et al. 1993). As mentioned above, ERK-mediated phosphorylation of hnRNP K mediates its shuttling to the cytosol, where it subsequently silences translation of target transcripts (Habelhah et al. 2001).
hnRNP P2 (FUS) is required for oncogenic transformation in BCR/ABL-transformed myeloid progenitor cells. BCR/ABL induces protein kinase CβII (PKCβII), which, in turn, phosphorylates hnRNP P2 (FUS) and stabilizes it (Iervolino et al. 2002). Conversely, protein kinase C-zeta (PKCζ) phosphorylates hnRNP A1 and causes its ubiquitination (Dreyfuss et al. 1993). Ubiquitinated hnRNP A1 causes proteolysis of hnRNP P2 (FUS), thus acting to prevent oncogenic transformation induced by BCR/ABL (Perrotti et al. 2000).
In vivo methylation of specific Arg residues within the RGG box of hnRNP A1 influences the RNA-binding properties of the RGG repeats (Kim et al. 1997). Similarly, methylation of hnRNP K at Arg residues promotes its binding to p53, which, in turn, stimulates the transcriptional activity of p53 (Chen et al. 2008). Additionally, hnRNPs A1, A3, F, H, and K have also been shown to undergo modification by sumoylation (Li et al. 2004). The sumoylation site for hnRNP K was mapped to its KH domain, whereas in other hnRNPs, it was mapped to their RBD. Whether the sumoylation of the hnRNPs affects their RNA-binding affinity is not known.
Functions
hnRNP proteins influence the structure and interaction of hnRNAs with other components required for processing of pre-mRNAs, thus intrinsically regulating their fate (Dreyfuss et al. 1993). They are multifunctional proteins participating in a variety of cellular processes. Broadly speaking, hnRNP proteins are principally involved in RNA metabolism, either in the nucleus or the cytoplasm. Within the nucleus, hnRNP proteins participate in RNA splicing (Caceres et al. 1994; Chou et al. 1999; Mourelatos et al. 2001), 3′-end processing (Kessler et al. 1997), transcriptional regulation (Miau et al. 1998), and immunoglobulin gene recombination (Dempsey et al. 1999). hnRNP A1 functions as an antagonist to the splicing regulator Ser-Arg (SR) protein SF2/ASF. This enables hnRNP A1 to function as an exon splicing repressor (Mayeda and Krainer 1992). Similarly, in neuronal cells, alternative splicing of c-src N1 exon is regulated by hnRNPs F and H (Krecic and Swanson 1999). hnRNP proteins are also involved in nucleo-cytoplasmic transport of mRNAs (Gallouzi and Steitz 2001), mRNA localization (Carson et al. 2001), translation (Habelhah et al. 2001), and mRNA stability (Xu et al. 2001). Classical examples of hnRNPs' role in mRNA localization are provided by hnRNP A2 and the Drosophila hnRNP Squid. hnRNP A2 selectively binds the cytoplasmic transport sequence of myelin basic protein and determines its localization to oligodendrocytes (Hoek et al. 1998), and Squid helps in the localization of the Gurken protein during Drosophila oogenesis (Norvell et al. 1999). While it is currently believed that hnRNPs are involved in pre-mRNA splicing, hnRNPs may also participate in RNA polymerase II transcription. For example, hnRNP K has been shown to bind to the RNA polymerase II transcriptional machinery and increase the transcription of the c-myc gene (Lynch et al. 2005). hnRNP Q (NSAP1) has been shown to mediate translational repression of pro-inflammatory genes, thus ensuring resolution of inflammation (Mukhopadhyay et al. 2009). hnRNP L functions as a “riboswitch” determining expression of pro-inflammatory or anti-inflammatory proteins. It causes translational up-regulation of VEGF-A during hypoxia by inhibiting translation silencing of pro-inflammatory genes mediated by the hnRNP Q-containing interferon-gamma-activated inhibitor of translation (GAIT) complex (Ray et al. 2009).
Predicted mechanisms for hnRNP functions
hnRNPs have been implicated in different steps of pre-mRNA processing and mRNA metabolism. The role of the hnRNPs during these steps are likely reflective of some or all of their biological functions (see Tables 1, 2 for the various functions of the different hnRNPs). These properties may allow hnRNPs to bind and manipulate RNA·RNA base-pairing in pre-mRNAs and thus modulate or influence RNA structure, which is essential for many of the steps in pre-mRNA processing. The assembly of RNA duplexes may also require that hnRNPs act as “matchmakers.” These are RNA-binding proteins that may interact with each other when bound to their respective RNA substrates and thus facilitate RNA·RNA base-pairing (Conaway and Conaway 1988). These activities have been used to develop several different models to explain how hnRNPs function (Conaway and Conaway 1988). The first model developed from the observation that metazoan hnRNPs are found in large complexes with pre-mRNAs when examined by electron microscopy (Borissova et al. 1981). These nucleosome-like structures have been referred to as ribonucleosomes, indicating that their main function was in the packaging of pre-mRNAs. This model predicts that hnRNPs bind nonspecifically to pre-mRNAs to form ribonucleosomes, which serves to protect the pre-mRNA from degradation. The second model integrates the transcript-specific binding properties of hnRNPs with their proposed function. Through their association with each other and with the pre-mRNA, hnRNPs mold each transcript into a structure that is recognizable by other trans-acting RNA processing factors. The third model proposes that hnRNPs function at individual steps in the pre-mRNA processing pathway by directly mediating the binding of other factors through protein–protein interactions.
hnRNP E1: UBIQUITOUSLY EXPRESSED YET DISCRETELY FUNCTIONAL
Origin
Human hnRNP E1 was initially defined as clone sub2.3, having similarity to hnRNP E2 and containing poly(C)-binding activity (Aasheim et al. 1994). hnRNP E1 maps to 2p12-p13 and hnRNP E2 to 12q13.12-q13.13 (Tommerup and Leffers 1996). hnRNP E1 and E2 are highly homologous proteins with 82% amino acid identity. The KH domains in the two are 93% identical. Through a series of unique and detailed bioinformatic analyses (Makeyev et al. 1999), hnRNP E1 was identified as an intronless gene coexpressing with its intron-containing hnRNP E2 paralog. Based on three discrete observations, it was proposed that hnRNP E1 originated via a retrotransposal event that gave rise to a processed pseudogene that was surprisingly functional. The evidence for this is that: (1) hnRNP E1 is intronless (expressed as one exon) but is identical and colinear with the hnRNP E2 mRNA (containing multiple exons); (2) hnRNP E1 has only one locus in the human and mouse genomes; and (3) hnRNP E1 lacks alternatively spliced forms (Makeyev et al. 1999). It was predicted that hnRNP E1 originated ∼400 million years ago, well before the mammalian radiation (Makeyev et al. 1999), and that it probably acquired a distinct cellular function from hnRNP E2, allowing for its evolutionary selection. It contains a 31-amino acid auxiliary domain between KH II and III (Makeyev et al. 1999), which may explain the nonredundant functions and binding specificities of hnRNP E1 and E2. hnRNP E1 has been identified in several murine tissues including bone marrow, liver, heart, kidney, brain, lung, and placenta (Chkheidze et al. 1999; Makeyev et al. 1999) and is considered to be ubiquitously expressed.
Structure
hnRNP E1 has three KH domains; two are sequestered to the N-terminus, followed by a nonconserved variable region and a C-terminal KH3 domain. As mentioned above, KH domains are divided into two subgroups; Type I has a C-terminal βα extension, whereas Type II has an N-terminal αβ extension (Grishin 2001). hnRNP E1 belongs to the Type I subset (Makeyev and Liebhaber 2002). The KH1 and KH3 domains have been predicted to bind RNA, whereas the precise function of the KH2 domain is unknown (Leffers et al. 1995; Dejgaard and Leffers 1996). Specificity for poly(C)-rich binding is postulated to be due to the narrow binding cleft formed within the KH-domain structure that is accessible only to smaller pyrimidine bases. Binding is stabilized by formation of hydrogen bonds between sugar hydroxyl groups of cytosine with the binding cleft in the KH domain. Since uracil residues lack these hydroxyl groups, hydrogen bonding with the KH domain does not occur. This explains the comparatively decreased affinity of the KH domain to poly(U) with respect to poly(C) stretches (Sidiqi et al. 2005).
Localization of hnRNP E1
Immunofluorescence studies have revealed a predominantly nuclear localization for hnRNP E1 (Chkheidze and Liebhaber 2003). Confocal microscopy showed that hnRNP E1 is sequestered to nuclear speckles, sites of accumulation of splicing factors prior to being loaded on the nascent transcripts. Hence, hnRNP E1 might have a very prominent role in either loading splice factors onto nascent transcripts or functioning as a splicing factor itself (Mintz et al. 1999; Chkheidze and Liebhaber 2003). Nuclear localization can be attributed to a nonameric nuclear localization signal (NLS I), putatively located between the KH2 and KH3 domains, and lacks the NLS II present in hnRNP E2 (Chkheidze and Liebhaber 2003). In addition to its nuclear localization, hnRNP E1 has been observed in the cytoplasm (Gamarnik and Andino 1997), suggesting that it has both nuclear and cytoplasmic functions.
hnRNP E1 functions
Transcriptional activator
hnRNP E1, along with other hnRNP E proteins, functions as a transcriptional activator of the mouse μ-opioid receptor (MOR) gene by binding to a 26-nt cis-acting polypyrimidine stretch within the proximal promoter (Kim et al. 2005). hnRNP E1 induces MOR mRNA expression by ∼4×-folds, and siRNA-mediated silencing of hnRNP E1 significantly inhibits MOR mRNA expression (Kim et al. 2005). hnRNP K has been shown to interact with RNA polymerase II and cause transcriptional up-regulation of the c-myc gene (Lynch et al. 2005). It will be of interest to determine whether hnRNP E1 also interacts with RNA polymerase II or is mediating its transcriptional effects more indirectly through binding other transcriptional regulators.
Regulator attenuating alternative splicing
hnRNP E1, in combination with U1 small nuclear RNP, abrogates splicing of a growth hormone receptor (GHR) pseudo-exon implicated in Laron syndrome. Using RNAi experiments, hnRNP E1 was demonstrated to be a silencer of GHR pseudoexon expression (Akker et al. 2007). These results corroborated earlier studies wherein hnRNP E1 was shown to colocalize with the splice factor SC35 in nuclear speckle (Chkheidze and Liebhaber 2003). In addition, mitogenic stimulation has recently been reported to promote colocalization of hnRNP E1 with SC35 and alternative splicing and exon inclusion from a CD44 minigene (Meng et al. 2007).
mRNA stability
hnRNP E1 regulates stability of a broad spectrum of mRNAs, including collagens I and III (Thiele et al. 2004), α-globin (Holcik and Liebhaber 1997), β-globin (Yu and Russell 2001), tyrosine hydroxylase (Czyzyk-Krzeska and Beresh 1996), erythropoietin (Czyzyk-Krzeska and Bendixen 1999), androgen receptor (Yeap et al. 2002), neurofilament-M (NF-M) (Thyagarajan and Szaro 2004), and renin (Skalweit et al. 2003).
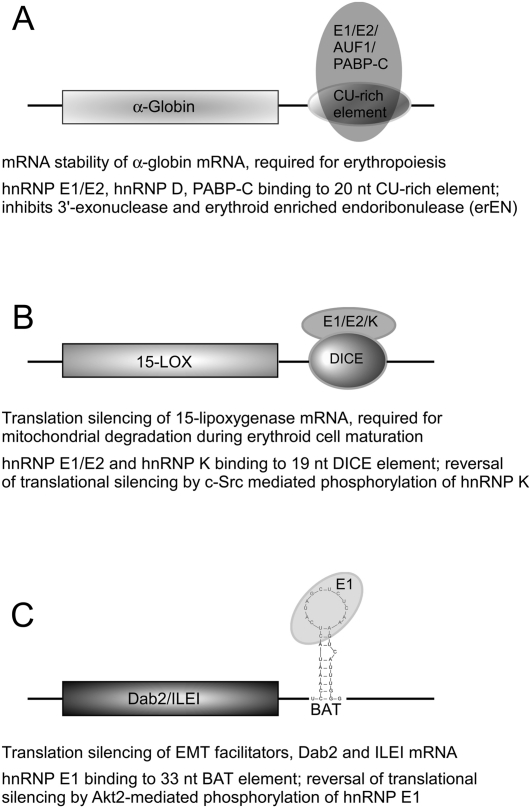
The continuous presence of α-globin mRNA and its translated product is a prerequisite during erythropoiesis. Interestingly, α-globin message stability is regulated by an α-complex binding to a 20-nt CU-rich region in the 3′-UTR of α-globin mRNA (Weiss and Liebhaber 1994; Holcik and Liebhaber 1997). The α-complex is comprised of hnRNP E1, hnRNP E2, hnRNP D (AUF1), and cytoplasmic poly(A)-binding protein (PABP-C) (Kiledjian et al. 1995, 1997; Chkheidze et al. 1999; Wang et al. 1999; Henics 2000). Two possible mechanisms have been proposed by which the α-complex modulates mRNA stability. PABP-C binds to hnRNP E1/E2 and the poly(A) tail of the α-globin mRNA and either attenuates deadenylation by inhibiting the deadenylating 3′-exonuclease or prevents endoribonucleolytic cleavage by the erythroid-enriched endoribonuclease (ErEN) (Fig. 2A; Wang et al. 1999; Wang and Kiledjian 2000a,b; Waggoner and Liebhaber 2003; Ostareck-Lederer and Ostareck 2004). It will be of interest to determine which of the two pathways is involved in regulating α-globin mRNA stability and how it is switched off in mature erythrocytes, when α-globin is not required.
FIGURE 2.
Post-transcriptional regulation by hnRNP E1 during development and differentiation. hnRNP E1 alone, or in combination with other hnRNPs, modulates mRNA stability of α-globin (A) or the translation of 15-LOX (B) or Dab2 (C) mRNAs. The regulatory mechanism involves binding to putative cis elements in the 3′-UTRs, which in most cases is disrupted by phosphorylation of one or more of the bound hnRNP proteins. A brief description of each regulatory mechanism is given. (15-LOX) 15-lipoxygenase; (DICE) differentiation control element; (BAT) TGFβ-activated translation element; (Dab2) disabled-2; (ILEI) interleukin-like EMT inducer.
Translational regulation
hnRNP E1 has been shown to function as both a translational coactivator and co-repressor. It functions as an IRES trans-activating factor (ITAF), binding a stem–loop IV structure in the viral internal ribosome entry site (IRES) element, and stimulates translation of poliovirus RNA (Gamarnik and Andino 1997). Similarly, it stimulates IRES-mediated translation of the Bag-1 (Bcl-2 associated gene) mRNA (Pickering et al. 2003). On the other hand, hnRNP E1 causes translational repression of 15-lipoxygenase (15-LOX) during erythrocyte maturation (Ostareck et al. 1997), human papillomavirus type 16 (HPV-16) L2 mRNA (Collier et al. 1998), disabled-2 (Dab2), and interleukin-like EMT inducer (ILEI) mRNAs during transforming growth factor-beta (TGFβ)-induced epithelial–mesenchymal transition (EMT) (Chaudhury et al. 2010), and A2 response element containing mRNAs in dendrites within neurons during neuronal development (Kosturko et al. 2006). In most of the above cases, hnRNP E1 functions with other proteins to induce translational stimulation or repression.
15-lipoxygenase (15-LOX) mRNA (Fig. 2B).
During the process of erythroid differentiation, the nucleus is compacted and subsequently extruded from the terminally differentiating erythroblast, and thus transcriptional activities are halted (Najfeld et al. 1988). Thus, subsequent developmental stages are regulated post-transcriptionally through sequestered mRNP complexes. During the final stage of erythrocyte maturation, the activation of 15-LOX is required for mediating mitochondrial degradation. This timely activation of 15-LOX is achieved by reversal of translational silencing of the 15-LOX mRNA (Ostareck-Lederer and Ostareck 2004). It has been shown that hnRNP E1 and hnRNP K bind to a 19-nt CU-rich repeat, the differentiation control element (DICE) in the 3′-UTR of the 15-LOX mRNA, and inhibit its translation until its activation is required for completion of differentiation (Ostareck-Lederer et al. 1994, 1998; Ostareck et al. 1997). Translational silencing occurs at the initiation step by preventing 80S complex formation. Specifically, the 60S large ribosomal subunit is prevented from binding to the 43S pre-initiation complex (consisting of the 40S small ribosomal subunit, eIF2-GTP, Met-tRNAi, eIF1/1A, and eIF3) at the initiating codon (Ostareck et al. 2001). Activation or reversal of translational silencing of 15-LOX mRNA is achieved by c-Src-mediated phosphorylation of hnRNP K at Tyr 72, 225, 230, 234, 236, and 380. Phosphorylation disrupts the hnRNP K/E1 mRNP complex from binding to the DICE elements and thereby relieves translational silencing (Ostareck-Lederer et al. 2002). Interestingly, hnRNP K activates its kinase, c-Src, through a positive feedback loop involving translational regulation through its 3′-UTR (Ostareck-Lederer et al. 2002). No modification of hnRNP E1 has been reported that attenuates its binding to the DICE. Of note, there has been one contrasting report in which translational silencing of DICE-containing reporter constructs was not observed in HeLa cells (Kong et al. 2006), even though the cellular context was not well defined in the reported studies.
HPV-16 L2 mRNA.
The capsid proteins, viral latent L1 and L2, of HPV-16 are detected only in terminally differentiated human cells, until which time the expression of L1 and L2 is silenced. hnRNP E1/2 and hnRNP K mediate this translational inhibition by binding to putative cis-acting inhibitory sequences in the 3′-end of the coding region (Collier et al. 1998). Translational inhibition of L2 mRNAs follows the same recourse as 15-LOX, even though hnRNP E1, hnRNP K, or hnRNP E2 can individually attenuate translation of L2 mRNAs in vitro. Interestingly, the L2 sequence has only 22% cytosines with no well-defined poly(C) repeats, suggesting that hnRNP E1 can preferentially bind to nonpoly(C) regions in mRNA. Earlier work has shown that hnRNP E1 can bind to poly(U) regions (Leffers et al. 1995), albeit with different affinity.
Dab2 and ILEI mRNAs (Fig. 2C).
Disabled-2 (Dab2) and interleukin-like inducer of EMT (ILEI) are two genes required for transforming growth factor β (TGFβ)-mediated EMT (Prunier and Howe 2005; Waerner et al. 2006). Recently, we have elucidated a novel post-transcriptional regulatory mechanism by which hnRNP E1 silences translation of Dab2 and ILEI mRNAs (Chaudhury et al. 2010). We identified a novel, 33-nt structural element in the 3′-UTRs of Dab2 and ILEI that inhibited translation of their respective mRNAs, an inhibition that was relieved following TGFβ treatment of cells. The 33-nt element, called the “BAT element” for TGFβ activated translational element, was sufficient to mediate translational inhibition in vitro and in vivo. Using the BAT element, the mRNP complex (i.e., BAT complex) was affinity-purified, and hnRNP E1 was identified as an integral and functional component of this complex. TGFβ activates the protein kinase Akt2, which subsequently phosphorylates hnRNP E1 at Ser43 residue, culminating in the disruption of the BAT complex and concurrent reversal of translational silencing of Dab2 and ILEI mRNAs. Ectopic overexpression of hnRNP E1 in normal mouse mammary gland (NMuMG) cells prevented the translation of Dab2 and ILEI mRNAs and the induction of the mesenchymal phenotype in response to TGFβ. Conversely, shRNA-mediated silencing of hnRNP E1 resulted in the constitutive translation of Dab2 and ILEI and of the mesenchymal phenotype irrespective of TGFβ treatment. Knockin of a phosphomutant hnRNP E1 (S43A) into the hnRNP E1 knockdown cells rescued the epithelial phenotype, but not the sensitivity to TGFβ-induced EMT seen in parental cells, unequivocally showing the importance of hnRNP E1 phosphorylation for EMT (Chaudhury et al. 2010).
Role of hnRNP E1 in tumorigenesis and cancer progression
It has been suggested that hnRNP proteins, and specifically hnRNP E1, play a critical and well-defined role in carcinogenesis. The consensus binding site for hnRNP A1 is UAGGG(A/U), which closely mimics telomeric single-stranded repeats, (TTAGGG)n. In fact, hnRNP E1, A1, and D have been shown to form a heteromeric complex on telomeric repeats in vitro (Ishikawa et al. 1993), suggesting that they may be involved in the maintenance of telomeric length. This notion is reinforced by the observation that deficient expression of hnRNPs can cause significant telomere shortening and oncogenic transformation (LaBranche et al. 1998). Reduced hnRNP E1 expression is also a prerequisite for human papillomavirus (HPV) proliferation and subsequent incidence of cervical carcinoma from cervical dysplasia (Pillai et al. 2003). Recently, miR328 has been shown to function as a “decoy” for hnRNP E2. hnRNP E2 binds to the cis element in the 5′-UTR of C/EBPα and inhibits its translation and subsequent myeloid cell differentiation (Beitzinger and Meister 2010; Eiring et al. 2010). miR328 sequesters away hnRNP E2, allowing translation of C/EBPα, which promotes chronic myelogenous leukemia (CML). Given the sequence similarity between hnRNP E2 and hnRNP E1, it will be of interest to determine whether miR328, or a similar miRNA, similarly functions as a decoy for hnRNP E1.
Spreading initiation centers (SICs) are defined as ribonucleoprotein macromolecular complexes, distinct from focal adhesions, and are involved in initiation of cell spreading during cancer metastasis (de Hoog et al. 2004). Using stable isotope labeling by amino acids in cell culture (SILAC), hnRNP E1, hnRNP K, and the Sm family protein FUS/TLS were identified as intricate constituents of SICs (de Hoog et al. 2004). Down-regulation of hnRNP E1, using neutralizing antibodies, stimulated cell spreading, consistent with our observation that shRNA-mediated silencing of hnRNP E1 induces constitutive EMT, another prerequisite for metastatic progression (de Hoog et al. 2004; Chaudhury et al. 2010). Finally, two recent reports suggest that hnRNP E1 may be involved in controlling embryonic and cancer stem cell proliferation and differentiation. In nasopharyngeal carcinoma CNE-2 cells (Cao et al. 2010) and in mouse embryonic stem cells (Baharvand et al. 2008), cellular reprogramming was associated with a decrease in the expression of hnRNP E1.
How does hnRNP E1 achieve functional diversity?
At the mechanistic level, an interesting question that arises regarding hnRNP E1 is how are its multifaceted functions mediated. How does it both regulate translation and stabilize specific mRNAs during differentiation of hematopoietic cells? In the case of hnRNP K, it has been proposed that its auxiliary domain acts as a docking platform on the RNA that allows it to recruit and interact with molecules from various signaling pathways (Bomsztyk et al. 2004). A similar situation can be envisaged for hnRNP E1; however, due to the nonconserved nature of these auxiliary domains, it is difficult to explain how they could be mediating conserved or similar functions across different species.
Another model that has been proposed to explain hnRNP E1's functional diversity is that it serves as a bridging molecule or “chaperone” on the RNA that facilitates the association of proteins in multicomponent regulatory systems, which, in turn, render stabilization to secondary nucleic acid structures (Ostareck-Lederer et al. 1998). However, it was subsequently demonstrated that hnRNP E (hnRNP E2, to be precise) causes mRNA stabilization irrespective of its RNA binding function, thus questioning the universality of the chaperone model (Makeyev and Liebhaber 2002).
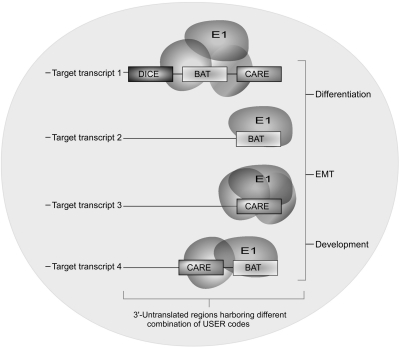
Given its wide-ranging functions in the nucleus and cytoplasm and numerous interacting partners, it is perhaps appropriate to envisage a post-transcriptional regulon model for hnRNP E1's functional diversity (Keene and Tenenbaum 2002). Post-transcriptional regulons involve untranslated sequence elements for regulation (USER) codes (the 5′- and 3′-UTR cis elements) bound by well-defined protein complexes that mediate regulatory function (Keene and Tenenbaum 2002). Post-transcriptional regulons, operating at different levels of gene expression, may have evolved as mechanisms to discretely, rapidly, and coordinately suppress multiple genes involved in a regulatory pathway (Fig. 3). These regulons can potentially coordinate overlapping functions temporally in different cellular locations (Anderson 2010). Coordinated translational regulation of Dab2 and ILEI mRNAs by TGFβ indicates that this may constitute a post-transcriptional BAT regulon inhibiting the expression of related EMT genes (Chaudhury et al. 2010). Similarly, hnRNP E1 along with hnRNP E2 and hnRNP K associates with the neurofilament-M (NF-M) mRNA during postnatal development based on the relative abundance of either the hnRNPs or the NF-M mRNA, which is suggestive of a dynamic post-transcriptional regulon (Thyagarajan and Szaro 2008). The regulon assembly possibly happens during mRNA processing and subsequent export to the cytoplasm, in turn pre-deciding the fate of the transcripts (Moore et al. 2005). Overall, such a model provides a rationale and energetically favorable framework for integrating functional diversity of hnRNP E1.
FIGURE 3.
Post-transcriptional regulons potentiate functional diversity of hnRNP E1. Putative cis elements in the 3′-UTRs of target transcripts, untranslated sequence elements for regulation (USER) codes, determine the content of the mRNP complex that will form. Many USER codes, such as DICE, BAT, and CARE, have been defined in mediating post-transcriptional regulation. and one transcript can harbor more than one USER code. hnRNP E1 and other hnRNPs bind alone or as a multifactorial mRNP complex to USER codes and give rise to post-transcriptional regulons. This model takes into account the multifunctional, yet conserved, diversity of hnRNP E1. As shown here, hnRNP E1 binding to other proteins and specific USER codes, BAT, DICE, or CARE elements, dictates their subsequent function during differentiation, EMT, and development. (BAT) TGFβ-activated translation element; (DICE) differentiation control element; (CARE) CACA-rich elements that bind hnRNP L.
Regulating the regulator
Considering the cytoplasmic and nuclear functions of hnRNP E1, it can be assumed that factors controlling its subcellular localization will likely affect its diverse functions. Earlier studies suggested that the phosphorylation of hnRNP E1 affects its RNA binding affinity (Leffers et al. 1995); however, only recently has this been confirmed. p21-activated kinase 1 (Pak1) phosphorylates hnRNP E1 at Thr 60 and 127 residues, causing its release from a DICE-minigene and reversal of translational silencing (Meng et al. 2007). Our recent studies also clearly demonstrate that phosphorylation of hnRNP E1 by Akt2 at the Ser 43 residue causes loss of binding to the BAT RNA element (Chaudhury et al. 2010). Resolution of the crystal structure of the phosphorylated forms will definitely lead to a clearer understanding of how post-translational modification attenuates hnRNP E1's RNA binding potential. It will also be of interest to determine whether phosphorylation affects the subcellular localization or half-life of hnRNP E1. Possibly, RNA binding masks the NLS1 site and prevents nuclear localization of hnRNP E1, and its phosphorylation and release from the RNA allow the NLS site to be exposed, thereby promoting nuclear import. Further investigation is essential to unravel the signaling pathways regulating hnRNP E1 expression, turnover, and function.
Concluding remarks and future implications
Considerable progress has been achieved in determining the multifunctional roles of hnRNP E1 in regulating cellular homeostasis. However, many important questions need to be addressed. For example, how does hnRNP E1 silence translation of 15-LOX mRNA, yet promote stability and, in turn, translation of stabilized α-globin mRNA? Are these different functions specified by the topology of hnRNP E1 and/or its binding partners to a specific mRNA? How many distinct hnRNP E1 complexes exist in vivo, and what is their exact composition? Are the interacting partners of hnRNP E1 compatible or mutually exclusive, and when and where do they assemble? Cells may also regulate hnRNP E1 through a variety of protein kinases and signaling pathways. How these impinge on hnRNP E1 is unclear, but could provide important clues regarding the regulation of hnRNP E1 functions. Answers to these questions await a comprehensive integration of genetic, biochemical, molecular biology, and structural analyses focused on hnRNP E1 protein–protein and protein–RNA complexes.
ACKNOWLEDGMENTS
This work was supported by grants CA55536 and CA80095 from the National Cancer Institute to P.H.H. A.C. is supported by American Heart Association (Ohio Valley affiliate) Pre-doctoral Fellowship 075080B.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2254110.
REFERENCES
- Aasheim HC, Loukianova T, Deggerdal A, Smeland EB 1994. Tissue specific expression and cDNA structure of a human transcript encoding a nucleic acid binding [oligo(dC)] protein related to the pre-mRNA binding protein K. Nucleic Acids Res 22: 959–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akker SA, Misra S, Aslam S, Morgan EL, Smith PJ, Khoo B, Chew SL 2007. Pre-spliceosomal binding of U1 small nuclear ribonucleoprotein (RNP) and heterogenous nuclear RNP E1 is associated with suppression of a growth hormone receptor pseudoexon. Mol Endocrinol 21: 2529–2540 [DOI] [PubMed] [Google Scholar]
- Amero SA, Raychaudhuri G, Cass CL, van Venrooij WJ, Habets WJ, Krainer AR, Beyer AL 1992. Independent deposition of heterogeneous nuclear ribonucleoproteins and small nuclear ribonucleoprotein particles at sites of transcription. Proc Natl Acad Sci 89: 8409–8413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P 2010. Post-transcriptional regulons coordinate the initiation and resolution of inflammation. Nat Rev Immunol 10: 24–35 [DOI] [PubMed] [Google Scholar]
- Baharvand H, Fathi A, Gourabi H, Mollamohammadi S, Salekdeh GH 2008. Identification of mouse embryonic stem cell-associated proteins. J Proteome Res 7: 412–423 [DOI] [PubMed] [Google Scholar]
- Bandziulis RJ, Swanson MS, Dreyfuss G 1989. RNA-binding proteins as developmental regulators. Genes Dev 3: 431–437 [DOI] [PubMed] [Google Scholar]
- Beitzinger M, Meister G 2010. MicroRNAs: From decay to decoy. Cell 140: 612–614 [DOI] [PubMed] [Google Scholar]
- Beyer AL, Christensen ME, Walker BW, LeStourgeon WM 1977. Identification and characterization of the packaging proteins of core 40S hnRNP particles. Cell 11: 127–138 [DOI] [PubMed] [Google Scholar]
- Bomsztyk K, Van Seuningen I, Suzuki H, Denisenko O, Ostrowski J 1997. Diverse molecular interactions of the hnRNP K protein. FEBS Lett 403: 113–115 [DOI] [PubMed] [Google Scholar]
- Bomsztyk K, Denisenko O, Ostrowski J 2004. hnRNP K: One protein multiple processes. Bioessays 26: 629–638 [DOI] [PubMed] [Google Scholar]
- Borissova OF, Krichevskaya AA, Samarina OP 1981. Structural investigation of nuclear RNP particles containing pre-mRNA by different fluorescence techniques. Nucleic Acids Res 9: 663–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckanovich RJ, Darnell RB 1997. The neuronal RNA binding protein Nova-1 recognizes specific RNA targets in vitro and in vivo. Mol Cell Biol 17: 3194–3201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckanovich RJ, Posner JB, Darnell RB 1993. Nova, the paraneoplastic Ri antigen, is homologous to an RNA-binding protein and is specifically expressed in the developing motor system. Neuron 11: 657–672 [DOI] [PubMed] [Google Scholar]
- Burd CG, Dreyfuss G 1994. Conserved structures and diversity of functions of RNA-binding proteins. Science 265: 615–621 [DOI] [PubMed] [Google Scholar]
- Caceres JF, Stamm S, Helfman DM, Krainer AR 1994. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science 265: 1706–1709 [DOI] [PubMed] [Google Scholar]
- Cao JX, Cui YX, Long ZJ, Dai ZM, Lin JY, Liang Y, Zheng FM, Zeng YX, Liu Q 2010. Pluripotency-associated genes in human nasopharyngeal carcinoma CNE-2 cells are reactivated by a unique epigenetic sub-microenvironment. BMC Cancer 10: 68 doi: 10.1186/1471-2407-10-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson JH, Cui H, Krueger W, Schlepchenko B, Brumwell C, Barbarese E 2001. RNA trafficking in oligodendrocytes. Results Probl Cell Differ 34: 69–81 [DOI] [PubMed] [Google Scholar]
- Chaudhury A, Hussey GS, Ray PS, Jin G, Fox PL, Howe PH 2010. TGF-β-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nat Cell Biol 12: 286–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhou X, Liu N, Wang C, Zhang L, Mo W, Hu G 2008. Arginine methylation of hnRNP K enhances p53 transcriptional activity. FEBS Lett 582: 1761–1765 [DOI] [PubMed] [Google Scholar]
- Chkheidze AN, Liebhaber SA 2003. A novel set of nuclear localization signals determine distributions of the αCP RNA-binding proteins. Mol Cell Biol 23: 8405–8415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chkheidze AN, Lyakhov DL, Makeyev AV, Morales J, Kong J, Liebhaber SA 1999. Assembly of the α-globin mRNA stability complex reflects binary interaction between the pyrimidine-rich 3′ untranslated region determinant and poly(C) binding protein αCP. Mol Cell Biol 19: 4572–4581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou MY, Rooke N, Turck CW, Black DL 1999. hnRNP H is a component of a splicing enhancer complex that activates a c-src alternative exon in neuronal cells. Mol Cell Biol 19: 69–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier B, Goobar-Larsson L, Sokolowski M, Schwartz S 1998. Translational inhibition in vitro of human papillomavirus type 16 L2 mRNA mediated through interaction with heterogenous ribonucleoprotein K and poly(rC)-binding proteins 1 and 2. J Biol Chem 273: 22648–22656 [DOI] [PubMed] [Google Scholar]
- Conaway RC, Conaway JW 1988. ATP activates transcription initiation from promoters by RNA polymerase II in a reversible step prior to RNA synthesis. J Biol Chem 263: 2962–2968 [PubMed] [Google Scholar]
- Courtneidge SA, Fumagalli S 1994. A mitotic function for Src? Trends Cell Biol 4: 345–347 [DOI] [PubMed] [Google Scholar]
- Czyzyk-Krzeska MF, Bendixen AC 1999. Identification of the poly(C) binding protein in the complex associated with the 3′ untranslated region of erythropoietin messenger RNA. Blood 93: 2111–2120 [PubMed] [Google Scholar]
- Czyzyk-Krzeska MF, Beresh JE 1996. Characterization of the hypoxia-inducible protein binding site within the pyrimidine-rich tract in the 3′-untranslated region of the tyrosine hydroxylase mRNA. J Biol Chem 271: 3293–3299 [DOI] [PubMed] [Google Scholar]
- de Hoog CL, Foster LJ, Mann M 2004. RNA and RNA binding proteins participate in early stages of cell spreading through spreading initiation centers. Cell 117: 649–662 [DOI] [PubMed] [Google Scholar]
- Dejgaard K, Leffers H 1996. Characterization of the nucleic-acid-binding activity of KH domains. Different properties of different domains. Eur J Biochem 241: 425–431 [DOI] [PubMed] [Google Scholar]
- Dempsey LA, Sun H, Hanakahi LA, Maizels N 1999. G4 DNA binding by LR1 and its subunits, nucleolin and hnRNP D: A role for G–G pairing in immunoglobulin switch recombination. J Biol Chem 274: 1066–1071 [DOI] [PubMed] [Google Scholar]
- Dreyfuss G, Philipson L, Mattaj IW 1988. Ribonucleoprotein particles in cellular processes. J Cell Biol 106: 1419–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G, Matunis MJ, Pinol-Roma S, Burd CG 1993. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem 62: 289–321 [DOI] [PubMed] [Google Scholar]
- Dreyfuss G, Kim VN, Kataoka N 2002. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol 3: 195–205 [DOI] [PubMed] [Google Scholar]
- Eiring AM, Harb JG, Neviani P, Garton C, Oaks JJ, Spizzo R, Liu S, Schwind S, Santhanam R, Hickey CJ, et al. 2010. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell 140: 652–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG 1956. Small granules in the amphibian oocyte nucleus and their relationship to RNA. J Biophys Biochem Cytol 2: 393–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallouzi IE, Steitz JA 2001. Delineation of mRNA export pathways by the use of cell-permeable peptides. Science 294: 1895–1901 [DOI] [PubMed] [Google Scholar]
- Gamarnik AV, Andino R 1997. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA 3: 882–892 [PMC free article] [PubMed] [Google Scholar]
- Gibson TJ, Thompson JD, Heringa J 1993. The KH domain occurs in a diverse set of RNA-binding proteins that include the antiterminator NusA and is probably involved in binding to nucleic acid. FEBS Lett 324: 361–366 [DOI] [PubMed] [Google Scholar]
- Gorlach M, Wittekind M, Beckman RA, Mueller L, Dreyfuss G 1992. Interaction of the RNA-binding domain of the hnRNP C proteins with RNA. EMBO J 11: 3289–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishin NV 2001. KH domain: One motif, two folds. Nucleic Acids Res 29: 638–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guil S, Gattoni R, Carrascal M, Abián J, Stévenin J, Bach-Elias M 2003. Roles of hnRNP A1, SR proteins, and p68 Helicase in c-H-ras alternative splicing regulation. Mol Cell Biol 23: 2927–2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habelhah H, Shah K, Huang L, Ostareck-Lederer A, Burlingame AL, Shokat KM, Hentze MW, Ronai Z 2001. ERK phosphorylation drives cytoplasmic accumulation of hnRNP-K and inhibition of mRNA translation. Nat Cell Biol 3: 325–330 [DOI] [PubMed] [Google Scholar]
- Hassfeld W, Chan KLE, Mathison DA, Portman D, Dreyfuss G, Steiner G, Tan EM 1998. Molecular definition of heterogeneous nuclear ribonucleoprotein R (hnRNP R) using autoimuune antibody: Immunological relationship with hnRNP P. Nucleic Acids Res 26: 439–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich B, Zhang Z, Raitskin O, Hiller M, Benderska N, Hartmann AM, Bracco L, Elliott D, Ben-Ari S, Soreq H, et al. 2009. Heterogeneous nuclear ribonucleoprotein G regulates splice site selection by binding to CC(A/C)-rich regions in pre-mRNA. J Biol Chem 284: 14303–14315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henics T 2000. Differentiation-dependent cytoplasmic distribution and in vivo RNA association of proteins recognized by the 3′-UTR stability element of α-globin mRNA in erythroleukemic cells. Biochem Biophys Res Commun 279: 40–46 [DOI] [PubMed] [Google Scholar]
- Hoek KS, Kidd GJ, Carson JH, Smith R 1998. hnRNP A2 selectively binds the cytoplasmic transport sequence of myelin basic protein mRNA. Biochemistry 37: 7021–7029 [DOI] [PubMed] [Google Scholar]
- Holcik M, Liebhaber SA 1997. Four highly stable eukaryotic mRNAs assemble 3′ untranslated region RNA–protein complexes sharing cis and trans components. Proc Natl Acad Sci 94: 2410–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovhannisyan RH, Carstens RP 2007. Heterogeneous ribonucleoprotein M is a splicing regulatory protein that can enhance or silence splicing of alternatively spliced exons. J Biol Chem 282: 36265–36274 [DOI] [PubMed] [Google Scholar]
- Iervolino A, Santilli G, Trotta R, Guerzoni C, Cesi V, Bergamaschi A, Gambacorti-Passerini C, Calabretta B, Perrotti D 2002. hnRNP A1 nucleocytoplasmic shuttling activity is required for normal myelopoiesis and BCR/ABL leukemogenesis. Mol Cell Biol 22: 2255–2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa F, Matunis MJ, Dreyfuss G, Cech TR 1993. Nuclear proteins that bind the pre-mRNA 3′ splice site sequence r(UUAG/G) and the human telomeric DNA sequence d(TTAGGG)n. Mol Cell Biol 13: 4301–4310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KB, Musunuru K, Lewis HA, Burley SK, Darnell RB 2000. The tetranucleotide UCAY directs the specific recognition of RNA by the Nova K-homology 3 domain. Proc Natl Acad Sci 97: 5740–5745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamma H, Portman DS, Dreyfuss G 1995. Cell type-specific expression of hnRNP proteins. Exp Cell Res 221: 187–196 [DOI] [PubMed] [Google Scholar]
- Keene JD, Tenenbaum SA 2002. Eukaryotic mRNPs may represent posttranscriptional operons. Mol Cell 9: 1161–1167 [DOI] [PubMed] [Google Scholar]
- Kessler MM, Henry MF, Shen E, Zhao J, Gross S, Silver PA, Moore CL 1997. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes Dev 11: 2545–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiledjian M, Dreyfuss G 1992. Primary structure and binding activity of the hnRNP U protein: Binding RNA through RGG box. EMBO J 11: 2655–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiledjian M, Wang X, Liebhaber SA 1995. Identification of two KH domain proteins in the α-globin mRNP stability complex. EMBO J 14: 4357–4364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiledjian M, DeMaria CT, Brewer G, Novick K 1997. Identification of AUF1 (heterogeneous nuclear ribonucleoprotein D) as a component of the α-globin mRNA stability complex. Mol Cell Biol 17: 4870–4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Merrill BM, Rajpurohit R, Kumar A, Stone KL, Papov VV, Schneiders JM, Szer W, Wilson SH, Paik WK, et al. 1997. Identification of NG-methylarginine residues in human heterogeneous RNP protein A1: Phe/Gly-Gly-Gly-Arg-Gly-Gly-Gly/Phe is a preferred recognition motif. Biochemistry 36: 5185–5192 [DOI] [PubMed] [Google Scholar]
- Kim SS, Pandey KK, Choi HS, Kim SY, Law PY, Wei LN, Loh HH 2005. Poly(C) binding protein family is a transcription factor in μ-opioid receptor gene expression. Mol Pharmacol 68: 729–736 [DOI] [PubMed] [Google Scholar]
- Kong J, Sumaroka M, Eastmond DL, Liebhaber SA 2006. Shared stabilization functions of pyrimidine-rich determinants in the erythroid 15-lipoxygenase and α-globin mRNAs. Mol Cell Biol 26: 5603–5614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosturko LD, Maggipinto MJ, Korza G, Lee JW, Carson JH, Barbarese E 2006. Heterogeneous nuclear ribonucleoprotein (hnRNP) E1 binds to hnRNP A2 and inhibits translation of A2 response element mRNAs. Mol Biol Cell 17: 3521–3533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krecic AM, Swanson MS 1999. hnRNP complexes: Composition, structure, and function. Curr Opin Cell Biol 11: 363–371 [DOI] [PubMed] [Google Scholar]
- LaBranche H, Dupuis S, Ben-David Y, Bani MR, Wellinger RJ, Chabot B 1998. Telomere elongation by hnRNP A1 and a derivative that interacts with telomeric repeats and telomerase. Nat Genet 19: 199–202 [DOI] [PubMed] [Google Scholar]
- Leffers H, Dejgaard K, Celis JE 1995. Characterisation of two major cellular poly(rC)-binding human proteins, each containing three K-homologous (KH) domains. Eur J Biochem 230: 447–453 [PubMed] [Google Scholar]
- Lewis HA, Chen H, Edo C, Buckanovich RJ, Yang YY, Musunuru K, Zhong R, Darnell RB, Burley SK 1999. Crystal structures of Nova-1 and Nova-2 K-homology RNA-binding domains. Structure 7: 191–203 [DOI] [PubMed] [Google Scholar]
- Lewis HA, Musunuru K, Jensen KB, Edo C, Chen H, Darnell RB, Burley SK 2000. Sequence-specific RNA binding by a Nova KH domain: Implications for paraneoplastic disease and the fragile X syndrome. Cell 100: 323–332 [DOI] [PubMed] [Google Scholar]
- Li T, Evdokimov E, Shen RF, Chao CC, Tekle E, Wang T, Stadtman ER, Yang DC, Chock PB 2004. Sumoylation of heterogeneous nuclear ribonucleoproteins, zinc finger proteins, and nuclear pore complex proteins: A proteomic analysis. Proc Natl Acad Sci 101: 8551–8556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Hanna MM 1995. NusA interferes with interactions between the nascent RNA and the C-terminal domain of the α subunit of RNA polymerase in Escherichia coli transcription complexes. Proc Natl Acad Sci 92: 5012–5016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Chen L, Ravitz MJ, Mehtani S, Korenblat K, Pazin MJ, Schmidt EV 2005. hnRNP K binds a core polypyrimidine element in the eukaryotic translation initiation factor 4E (eIF4E) promoter, and its regulation of eIF4E contributes to neoplastic transformation. Mol Cell Biol 25: 6436–6453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev AV, Liebhaber SA 2002. The poly(C)-binding proteins: A multiplicity of functions and a search for mechanisms. RNA 8: 265–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev AV, Chkheidze AN, Liebhaber SA 1999. A set of highly conserved RNA-binding proteins, αCP-1 and αCP-2, implicated in mRNA stabilization, are coexpressed from an intronless gene and its intron-containing paralog. J Biol Chem 274: 24849–24857 [DOI] [PubMed] [Google Scholar]
- Mandal M, Vadlamudi R, Nguyen D, Wang RA, Costa L, Bagheri-Yarmand R, Mendelsohn J, Kumar R 2001. Growth factors regulate heterogeneous nuclear ribonucleoprotein K expression and function. J Biol Chem 276: 9699–9704 [DOI] [PubMed] [Google Scholar]
- Matunis MJ, Michael WM, Dreyfuss G 1992. Characterization and primary structure of the poly(C)-binding heterogeneous nuclear ribonucleoprotein complex K protein. Mol Cell Biol 12: 164–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis EL, Matunis MJ, Dreyfuss G 1993. Association of individual hnRNP proteins and snRNPs with nascent transcripts. J Cell Biol 121: 219–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A, Krainer AR 1992. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell 68: 365–375 [DOI] [PubMed] [Google Scholar]
- Meng Q, Rayala SK, Gururaj AE, Talukder AH, O'Malley BW, Kumar R 2007. Signaling-dependent and coordinated regulation of transcription, splicing, and translation resides in a single coregulator, PCBP1. Proc Natl Acad Sci 104: 5866–5871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miau LH, Chang CJ, Shen BJ, Tsai WH, Lee SC 1998. Identification of heterogeneous nuclear ribonucleoprotein K (hnRNP K) as a repressor of C/EBPβ-mediated gene activation. J Biol Chem 273: 10784–10791 [DOI] [PubMed] [Google Scholar]
- Michael WM, Choi M, Dreyfuss G 1995. A nuclear export signal in hnRNP A1: A signal-mediated, temperature-dependent nuclear protein export pathway. Cell 83: 415–422 [DOI] [PubMed] [Google Scholar]
- Michael WM, Eder PS, Dreyfuss G 1997. The K nuclear shuttling domain: A novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO J 16: 3587–3598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller OL Jr, Bakken AH 1972. Morphological studies of transcription. Acta Endocrinol Suppl (Copenh) 168: 155–177 [DOI] [PubMed] [Google Scholar]
- Mintz PJ, Patterson SD, Neuwald AF, Spahr CS, Spector DL 1999. Purification and biochemical characterization of interchromatin granule clusters. EMBO J 18: 4308–4320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore CM, Gaballa A, Hui M, Ye RW, Helmann JD 2005. Genetic and physiological responses of Bacillus subtilis to metal ion stress. Mol Microbiol 57: 27–40 [DOI] [PubMed] [Google Scholar]
- Mourelatos Z, Abel L, Yong J, Kataoka N, Dreyfuss G 2001. SMN interacts with a novel family of hnRNP and spliceosomal proteins. EMBO J 20: 5443–5452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay R, Jia J, Arif A, Ray PS, Fox PL 2009. The GAIT system: A gatekeeper of inflammatory gene expression. Trends Biochem Sci 34: 324–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musco G, Stier G, Joseph C, Castiglione Morelli MA, Nilges M, Gibson TJ, Pastore A 1996. Three-dimensional structure and stability of the KH domain: Molecular insights into the fragile X syndrome. Cell 85: 237–245 [DOI] [PubMed] [Google Scholar]
- Musco G, Kharrat A, Stier G, Fraternali F, Gibson TJ, Nilges M, Pastore A 1997. The solution structure of the first KH domain of FMR1, the protein responsible for the fragile X syndrome. Nat Struct Biol 4: 712–716 [DOI] [PubMed] [Google Scholar]
- Najfeld V, Zucker-Franklin D, Adamson J, Singer J, Troy K, Fialkow PJ 1988. Evidence for clonal development and stem cell origin of M7 megakaryocytic leukemia. Leukemia 2: 351–357 [PubMed] [Google Scholar]
- Nandabalan K, Price L, Roeder GS 1993. Mutations in U1 snRNA bypass the requirement for a cell type-specific RNA splicing factor. Cell 73: 407–415 [DOI] [PubMed] [Google Scholar]
- Norvell A, Kelley RL, Wehr K, Schupbach T 1999. Specific isoforms of squid, a Drosophila hnRNP, perform distinct roles in Gurken localization during oogenesis. Genes Dev 13: 864–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostareck DH, Ostareck-Lederer A, Wilm M, Thiele BJ, Mann M, Hentze MW 1997. mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3′ end. Cell 89: 597–606 [DOI] [PubMed] [Google Scholar]
- Ostareck DH, Ostareck-Lederer A, Shatsky IN, Hentze MW 2001. Lipoxygenase mRNA silencing in erythroid differentiation: The 3′UTR regulatory complex controls 60S ribosomal subunit joining. Cell 104: 281–290 [DOI] [PubMed] [Google Scholar]
- Ostareck-Lederer A, Ostareck DH 2004. Control of mRNA translation and stability in haematopoietic cells: The function of hnRNPs K and E1/E2. Biol Cell 96: 407–411 [DOI] [PubMed] [Google Scholar]
- Ostareck-Lederer A, Ostareck DH, Standart N, Thiele BJ 1994. Translation of 15-lipoxygenase mRNA is inhibited by a protein that binds to a repeated sequence in the 3′ untranslated region. EMBO J 13: 1476–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostareck-Lederer A, Ostareck DH, Hentze MW 1998. Cytoplasmic regulatory functions of the KH-domain proteins hnRNPs K and E1/E2. Trends Biochem Sci 23: 409–411 [DOI] [PubMed] [Google Scholar]
- Ostareck-Lederer A, Ostareck DH, Cans C, Neubauer G, Bomsztyk K, Superti-Furga G, Hentze MW 2002. c-Src-mediated phosphorylation of hnRNP K drives translational activation of specifically silenced mRNAs. Mol Cell Biol 22: 4535–4543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrotti D, Iervolino A, Cesi V, Cirinna M, Lombardini S, Grassilli E, Bonatti S, Claudio PP, Calabretta B 2000. BCR-ABL prevents c-jun-mediated and proteasome-dependent FUS (TLS) proteolysis through a protein kinase CβII-dependent pathway. Mol Cell Biol 20: 6159–6169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering BM, Mitchell SA, Evans JR, Willis AE 2003. Polypyrimidine tract binding protein and poly r(C) binding protein 1 interact with the BAG-1 IRES and stimulate its activity in vitro and in vivo. Nucleic Acids Res 31: 639–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai MR, Chacko P, Kesari LA, Jayaprakash PG, Jayaram HN, Antony AC 2003. Expression of folate receptors and heterogeneous nuclear ribonucleoprotein E1 in women with human papillomavirus mediated transformation of cervical tissue to cancer. J Clin Pathol 56: 569–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinol-Roma S, Choi YD, Matunis MJ, Dreyfuss G 1988. Immunopurification of heterogeneous nuclear ribonucleoprotein particles reveals an assortment of RNA-binding proteins. Genes Dev 2: 215–227 [DOI] [PubMed] [Google Scholar]
- Prunier C, Howe PH 2005. Disabled-2 (Dab2) is required for transforming growth factor β-induced epithelial to mesenchymal transition (EMT). J Biol Chem 280: 17540–17548 [DOI] [PubMed] [Google Scholar]
- Query CC, Bentley RC, Keene JD 1989. A common RNA recognition motif identified within a defined U1 RNA binding domain of the 70K U1 snRNP protein. Cell 57: 89–101 [DOI] [PubMed] [Google Scholar]
- Ray PS, Jia J, Yao P, Majumder M, Hatzoglou M, Fox PL 2009. A stress-responsive RNA switch regulates VEGFA expression. Nature 457: 915–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regnier P, Grunberg-Manago M, Portier C 1987. Nucleotide sequence of the pnp gene of Escherichia coli encoding polynucleotide phosphorylase. Homology of the primary structure of the protein with the RNA-binding domain of ribosomal protein S1. J Biol Chem 262: 63–68 [PubMed] [Google Scholar]
- Samarina OP, Krichevskaya AA, Georgiev GP 1966. Nuclear ribonucleoprotein particles containing messenger ribonucleic acid. Nature 210: 1319–1322 [DOI] [PubMed] [Google Scholar]
- Samarina OP, Lukanidin EM, Molnar J, Georgiev GP 1968. Structural organization of nuclear complexes containing DNA-like RNA. J Mol Biol 33: 251–263 [DOI] [PubMed] [Google Scholar]
- Schmidt C, Henkel B, Poschl E, Zorbas H, Purschke WG, Gloe TR, Muller PK 1992. Complete cDNA sequence of chicken vigilin, a novel protein with amplified and evolutionary conserved domains. Eur J Biochem 206: 625–634 [DOI] [PubMed] [Google Scholar]
- Shamoo Y, Krueger U, Rice LM, Williams KR, Steitz TA 1997. Crystal structure of the two RNA binding domains of human hnRNP A1 at 1.75 Å resolution. Nat Struct Biol 4: 215–222 [DOI] [PubMed] [Google Scholar]
- Sidiqi M, Wilce JA, Vivian JP, Porter CJ, Barker A, Leedman PJ, Wilce MC 2005. Structure and RNA binding of the third KH domain of poly(C)-binding protein 1. Nucleic Acids Res 33: 1213–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebel CW, Admon A, Rio DC 1995. Soma-specific expression and cloning of PSI, a negative regulator of P element pre-mRNA splicing. Genes Dev 9: 269–283 [DOI] [PubMed] [Google Scholar]
- Singh R, Valcarcel J 2005. Building specificity with nonspecific RNA-binding proteins. Nat Struct Mol Biol 12: 645–653 [DOI] [PubMed] [Google Scholar]
- Siomi H, Dreyfuss G 1995. A nuclear localization domain in the hnRNP A1 protein. J Cell Biol 129: 551–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Matunis MJ, Michael WM, Dreyfuss G 1993a. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res 21: 1193–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Siomi MC, Nussbaum RL, Dreyfuss G 1993b. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell 74: 291–298 [DOI] [PubMed] [Google Scholar]
- Skalweit A, Doller A, Huth A, Kahne T, Persson PB, Thiele BJ 2003. Post-transcriptional control of renin synthesis: Identification of proteins interacting with renin mRNA 3′-untranslated region. Circ Res 92: 419–427 [DOI] [PubMed] [Google Scholar]
- Sommerville J, Scheer U 1981. Structural organization of nascent transcripts and hnRNA molecules in amphibian oocytes. Mol Biol Rep 7: 53–56 [DOI] [PubMed] [Google Scholar]
- Stevenin J, Gattoni R, Keohavong P, Jacob M 1982. Mild nuclease treatment as a probe for a nonrandom distribution of adenovirus-specific RNA sequences and of cellular RNA in nuclear ribonucleoprotein fibrils. J Mol Biol 155: 185–205 [DOI] [PubMed] [Google Scholar]
- Thiele BJ, Doller A, Kahne T, Pregla R, Hetzer R, Regitz-Zagrosek V 2004. RNA-binding proteins heterogeneous nuclear ribonucleoprotein A1, E1, and K are involved in post-transcriptional control of collagen I and III synthesis. Circ Res 95: 1058–1066 [DOI] [PubMed] [Google Scholar]
- Thyagarajan A, Szaro BG 2004. Phylogenetically conserved binding of specific K homology domain proteins to the 3′-untranslated region of the vertebrate middle neurofilament mRNA. J Biol Chem 279: 49680–49688 [DOI] [PubMed] [Google Scholar]
- Thyagarajan A, Szaro BG 2008. Dynamic endogenous association of neurofilament mRNAs with K-homology domain ribonucleoproteins in developing cerebral cortex. Brain Res 1189: 33–42 [DOI] [PubMed] [Google Scholar]
- Tommerup N, Leffers H 1996. Assignment of human KH-box-containing genes by in situ hybridization: HNRNPK maps to 9q21.32-q21.33, PCBP1 to 2p12-p13, and PCBP2 to 12q13.12-q13.13, distal to FRA12A. Genomics 32: 297–298 [DOI] [PubMed] [Google Scholar]
- Urlaub H, Kruft V, Bischof O, Muller EC, Wittmann-Liebold B 1995. Protein–rRNA binding features and their structural and functional implications in ribosomes as determined by cross-linking studies. EMBO J 14: 4578–4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernet C, Artzt K 1997. STAR, a gene family involved in signal transduction and activation of RNA. Trends Genet 13: 479–484 [DOI] [PubMed] [Google Scholar]
- Waerner T, Alacakaptan M, Tamir I, Oberauer R, Gal A, Brabletz T, Schreiber M, Jechlinger M, Beug H 2006. ILEI: A cytokine essential for EMT, tumor formation, and late events in metastasis in epithelial cells. Cancer Cell 10: 227–239 [DOI] [PubMed] [Google Scholar]
- Waggoner SA, Liebhaber SA 2003. Regulation of α-globin mRNA stability. Exp Biol Med (Maywood) 228: 387–395 [DOI] [PubMed] [Google Scholar]
- Wang Z, Kiledjian M 2000a. Identification of an erythroid-enriched endoribonuclease activity involved in specific mRNA cleavage. EMBO J 19: 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Kiledjian M 2000b. The poly(A)-binding protein and an mRNA stability protein jointly regulate an endoribonuclease activity. Mol Cell Biol 20: 6334–6341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Day N, Trifillis P, Kiledjian M 1999. An mRNA stability complex functions with poly(A)-binding protein to stabilize mRNA in vitro. Mol Cell Biol 19: 4552–4560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weighardt F, Biamonti G, Riva S 1996. The roles of heterogeneous nuclear ribonucleoproteins (hnRNP) in RNA metabolism. Bioessays 18: 747–756 [DOI] [PubMed] [Google Scholar]
- Weiss IM, Liebhaber SA 1994. Erythroid cell-specific determinants of α-globin mRNA stability. Mol Cell Biol 14: 8123–8132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Yen T, Geczy CL 2001. Il-10 up-regulates macrophage expression of the S100 protein S100A8. J Immunol 166: 6358–6366 [DOI] [PubMed] [Google Scholar]
- Yeap BB, Voon DC, Vivian JP, McCulloch RK, Thomson AM, Giles KM, Czyzyk-Krzeska MF, Furneaux H, Wilce MC, Wilce JA, et al. 2002. Novel binding of HuR and poly(C)-binding protein to a conserved UC-rich motif within the 3′-untranslated region of the androgen receptor messenger RNA. J Biol Chem 277: 27183–27192 [DOI] [PubMed] [Google Scholar]
- Yu J, Russell JE 2001. Structural and functional analysis of an mRNP complex that mediates the high stability of human β-globin mRNA. Mol Cell Biol 21: 5879–5888 [DOI] [PMC free article] [PubMed] [Google Scholar]