Abstract
Colonization of a host by an active transposon can increase mutation rates or cause sterility, a phenotype termed hybrid dysgenesis. As an example, intercrosses of certain Drosophila virilis strains can produce dysgenic progeny. The Penelope element is present only in a subset of laboratory strains and has been implicated as a causative agent of the dysgenic phenotype. We have also introduced Penelope into Drosophila melanogaster, which are otherwise naive to the element. We have taken advantage of these natural and experimentally induced colonization processes to probe the evolution of small RNA pathways in response to transposon challenge. In both species, Penelope was predominantly targeted by endo-small-interfering RNAs (siRNAs) rather than by piwi-interacting RNAs (piRNAs). Although we do observe correlations between Penelope transcription and dysgenesis, we could not correlate differences in maternally deposited Penelope piRNAs with the sterility of progeny. Instead, we found that strains that produced dysgenic progeny differed in their production of piRNAs from clusters in subtelomeric regions, possibly indicating that changes in the overall piRNA repertoire underlie dysgenesis. Considered together, our data reveal unexpected plasticity in small RNA pathways in germ cells, both in the character of their responses to invading transposons and in the piRNA clusters that define their ability to respond to mobile elements.
Keywords: Drosophila virilis, piRNA, siRNA, transposon
INTRODUCTION
RNA interference (RNAi) and related pathways represent deeply conserved mechanisms to combat both exogenous pathogenic and endogenous parasitic nucleic acids (Ghildiyal and Zamore 2009; Kim et al. 2009; Malone and Hannon 2009). In both the plant and animal kingdoms, infection by viruses can be met by the production of small interfering RNAs (siRNAs) that direct destruction of viral RNAs (Hammond et al. 2000; Zamore et al. 2000; Molnar et al. 2005). These RNAs are most probably produced by Dicer-dependent cleavage of replication intermediates or by Dicing of complementary sense and antisense transcripts that arise during viral gene expression (Bernstein et al. 2001; Blevins et al. 2006). The resulting small RNAs, 21–22 nucleotides (nt) in length, join Argonaute proteins and guide these to target RNAs, which are cleaved by the innate nuclease activity of the Piwi domain (Liu et al. 2004; Okamura et al. 2004; Song et al. 2004). Evidence for a direct and critical role for RNAi in combating viral infection comes from the existence of viral proteins that both negate host RNAi pathways and are essential for productive infection (Lucy et al. 2000).
siRNAs have also been implicated in controlling the endogenous counterpart of viruses, mobile genetic elements. In plants, 24-nt siRNAs join a specific Argonaute protein, AGO4, to direct DNA methylation of repeated sequences in the genome (Zilberman et al. 2004; Qi et al. 2006). A burst of small RNA production during pollen development has been hypothesized to identify transposable elements in germ cell genomes and to transmit that information to the next generation (Slotkin et al. 2009). In animals, the first indication of the intersection of small RNA pathways and transposon control came from the observation in Caenorhabditis elegans that certain mutations simultaneously disrupted exogenously triggered RNA interference and permitted movement of an otherwise inert TC3 transposon (Ketting et al. 1999). In fact, several such “mutator” genes are now known to be essential for RNAi, although their precise biochemical roles in RNAi pathways have yet to be determined. siRNA pathways also act in transposon control in Drosophila, both in the germ line and in somatic cells (Chung et al. 2008; Czech et al. 2008; Ghildiyal et al. 2008; Okamura et al. 2008).
In the germ cells of most multicellular animals examined to date, another class of small RNAs either collaborates with siRNAs or predominates in repressing mobile elements (Aravin et al. 2001). These are dubbed piwi-interacting RNAs (piRNAs) based upon their association with an animal-specific clade of Argonaute proteins (Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006; Vagin et al. 2006), which had Drosophila Piwi as its founding member (Cox et al. 1998, 2000). piRNAs differ from siRNAs in several important respects. First and foremost, they are not produced via canonical biogenesis pathways from double-stranded precursors. Instead, they arise via one of two distinct processing mechanisms. The first produces “primary” piRNAs. These are generated from discrete genomic loci, termed piRNA clusters, that are often highly enriched for transposon fragments (Brennecke et al. 2007). piRNA clusters are transcribed into long, single-stranded, continuous precursors that are cleaved by an unknown processing machinery into discrete small RNAs. Secondary piRNAs are produced through the catalytic activity of the Piwi proteins themselves (Aravin et al. 2007; Brennecke et al. 2007; Gunawardane et al. 2007). Upon recognition of a substrate, in this case often a transposon mRNA or a piRNA cluster transcript, piRNAs direct target cleavage much as is seen for siRNAs in the canonical pathway. However, in this instance, the target RNA can itself give rise to a new small RNA with its 5′ end at the cleavage site. This type of biogenesis mechanism, termed the ping-pong cycle, has the potential to skew piRNA populations toward elements that are highly expressed at any given time.
The diversity of transposable elements and the degree to which they burden the genomes of even closely related species is extremely variable. A prior study comparing only a single piRNA cluster, flamenco, between three Drosophilid species that diverged ∼12 million years ago showed a very similar overall arrangement and even a conservation of general element types; however, not a single individual element within this locus was shared between melanogaster, yakuba, and erecta (Malone et al. 2009). These studies strongly support the value of more systematic comparisons between piRNA pathways among Drosophila species.
Drosophila virilis belongs to the virilis group of Drosophila subgenus. It has been separated from Drosophila melanogaster by ∼50–60 million years of divergent evolution (Spicer and Bell 2002) and may reflect characteristics of the ancestral species of the whole Drosophila clade. D. virilis displays a syndrome of hybrid dysgenesis in progeny of intercrosses between different strains of the species (Lozovskaya et al. 1990; Petrov et al. 1995). Similar sterility syndromes have been well studied in D. melanogaster, where they have been traced to the mobilization of single transposons, such as the I- or P-element (Picard 1976; Kidwell et al. 1977). Recent studies have defined the underlying molecular basis of transposon activation during dysgenesis as a lack of maternally deposited piRNAs targeting the subject element, leading ultimately to a loss of silencing of that specific transposon in the germ cells of progeny (Brennecke et al. 2007, 2008; Chambeyron et al. 2008). In virilis, dysgenesis differs fundamentally in that several unrelated elements are simultaneously derepressed. Previous studies suggested that a key driver in virilis dysgenesis syndromes is the Penelope retroelement, which was proposed not only to become mobile itself, but also to mobilize other elements present within the genome in dysgenic progeny (Petrov et al. 1995; Evgen'ev et al. 1997; Blumenstiel and Hartl 2005).
Penelope does not belong to conventional long interspersed nuclear element or long terminal repeat (LTR) retroelement classes, but instead represents an active member of a little-studied element family termed, “Penelope-like elements” (PLEs) (Evgen'ev and Arkhipova 2005). These are distinguished by the presence of a GIY-YIG endonuclease domain and the ability to retain introns despite moving via an RNA intermediate. The reverse transcriptase encoded by Penelope's single open reading frame (ORF) shows the greatest similarity to telomerases (Arkhipova et al. 2003). PLEs are present in many animal genomes, and their reverse transcriptase moiety can be also found in several protists, fungi, and plants, indicating an ancient origin (Evgen'ev and Arkhipova 2005). These unusual elements are probably active only within the virilis group of Drosophila and perhaps also within a few fish species (Dalle Nogare et al. 2002).
As with the I- and P-elements, which can trigger dysgenesis syndromes in melanogaster, active Penelope seems to be in the process of colonizing D. virilis. Intact and potentially mobile copies of this element are absent from many of the older sequestered laboratory strains, but are present in some recently collected strains (Zelentsova et al. 1999). Like the D. melanogaster I-element, all D. virilis strains studied contain heterochromatic, highly diverged copies of Penelope, apparently representing remnants of previous invasions (Lyozin et al. 2001).
Recent reports have implicated RNA silencing in the repression of hybrid dysgenesis in virilis, since small RNAs homologous to Penelope were detected in nondysgenic, but not in dysgenic, embryos (Blumenstiel and Hartl 2005). Since these studies were done prior to our understanding of the role of piRNAs in transposon silencing and without the resolution possible with deep sequencing, we felt that a detailed study of virilis dysgenesis could provide an important evolutionary perspective on hybrid sterility syndromes.
RESULTS AND DISCUSSION
Small RNA populations in D. virilis
We profiled small RNAs from D. virilis ovaries, testes, and somatic tissues using deep sequencing (Fig. 1). We performed these studies using several strains, with a particular focus on strains 9 and 160. These were chosen because intercrosses between strain 9 females and strain 160 males produce progeny with a moderately penetrant dysgenic phenotype, with ∼40% of testes and 60% of ovaries showing uni- or bilateral gonadal atrophy (Lozovskaya et al. 1990). Reciprocal crosses produce the same abnormalities, but with at least a 10-fold lower frequency.
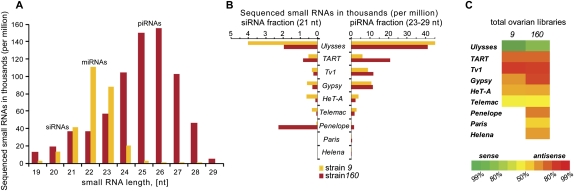
FIGURE 1.
Characteristics of transposon-derived small RNAs in strains 9 and 160. (A) Small RNA profiles obtained from total ovary small RNAs isolated from D. virilis strain 160. (B) Relative quantities of normalized siRNA (21 nt) and piRNA (23–29 nt) fractions homologous to various TEs in ovary libraries of D. virilis strains 9 and 160. (C) Strand asymmetry of piRNAs mapped to a set of characterized transposons in total ovary libraries of D. virilis.
Small RNA populations (19–29 nt) were gel isolated and cloned by procedures that require 5′ phosphate and 3′ hydroxyl termini. These were sequenced on the Illumina platform to yield 3–9 million reads per sample that could be successfully mapped to the D. virilis genome. In gonadal tissues, these had a size distribution similar to that seen in other Drosophilids (Fig. 1A).
Overall, the transposons resident in D. virilis are not as well annotated as those in D. melanogaster. We therefore focused most closely on a few well-characterized examples. Many of these were initially discovered based upon their causing insertional mutations in the progeny of dysgenic crosses. These include Ulysses, Penelope, Paris, Helena, Telemac, and Tv1 (Scheinker et al. 1990; Petrov et al. 1995; Evgen'ev et al. 1997). A number of additional elements, including HeT-A, TART, and Gypsy family members, were annotated based upon their high degree of homology with analogous elements in D. melanogaster (Mizrokhi and Mazo 1991; Casacuberta and Pardue 2005).
We observed a wide variation in the degree to which individual transposons were targeted in the ovaries or testes of a given strain (Supplemental Fig. S1). We noted several differences between strains. In particular, sequences homologous to Penelope were much more abundant in the 160 strain than they were in strain 9, and more prevalent in testis than in ovaries (Fig. 1B; Supplemental Fig. S1). This is in accord with the presence in strain 160 of full-length, active Penelope copies, whereas strain 9 harbors only divergent, heterchromatic, Penelope remnants. There were more subtle differences that could potentially be explained by copy number differences between strains; for example, TART is preferentially targeted in the ovaries of strain 160, while Ulysses is more strongly represented in the testis small RNAs of strain 9.
Penelope is selectively targeted by the siRNA pathway in both virilis and melanogaster
To define the nature of the response to each element in the tissues sampled, we annotated individual small RNA species. We removed degradation products of abundant cellular RNAs from each data set and then divided sequences into three classes. Annotated microRNAs were identified based upon comparisons to miRBase. piRNAs were then defined within the remainder based upon their size being between 23 and 29 nt. The rest of the sequences were classified as potential endo-siRNAs. Although nonannotated microRNAs could fall into the last group, they would not be predicted to match transposons and would therefore fail to impact our analysis.
The majority of elements were targeted by both siRNA and piRNA pathways as observed for transposons in melanogaster. For virtually all elements, for example, Ulysses, the piRNA pathway was dominant as expected. However, Penelope in strain 160 was unusual, showing a striking bias toward siRNAs (Fig. 1B).
Since active Penelope appears in the process of colonizing D. virilis, we wished to ascertain whether prominent siRNA responses were also provoked during Penelope colonization of another Drosophila species. The melanogaster genome contains no remnants of Penelope and is thus entirely naive to this element (Kapitonov and Jurka 2003). We previously developed several transgenic strains of D. melanogaster containing full-length copies of Penelope and the Penelope ORF under the control of the heat-shock (HS) promoter (Pyatkov et al. 2002). Penelope transposition occurred in all strains containing full-length copies, and the process of its amplification and colonization is still ongoing with current copy numbers in individual strains varying from three to 15 (data not shown).
We prepared small RNA libraries from two strains, A1 and A2, where Penelope amplification had been demonstrated. A1 is presently estimated to have four to five copies and A2 10–12 copies of intact, active Penelope. We selected and analyzed all small RNAs from these libraries corresponding to the Penelope consensus sequence. In both cases, we detected only siRNAs in ovary or testis (Fig. 2A; Supplemental Table S1). Moreover, we prepared Piwi, Aub, Ago3, and Ago2 RNP complexes from ovaries and determined their small RNA content. In accord with the size profiles of Penelope sequences in total small RNA libraries, we detected matches to this element only in Ago2 RISC (Supplemental Table S1). Previous studies in melanogaster have demonstrated the accumulation of endo-siRNAs that are bound to Ago2 and target a variety of mobile elements (Chung et al. 2008; Czech et al. 2008; Ghildiyal et al. 2008; Kawamura et al. 2008). However, in all cases examined to date, these elements were also clearly recognized by the piRNA pathway. A precedent for the behavior of Penelope might be found in the MT family in mouse ovary (Holt et al. 2006). These are relatively new invaders of mice and are predominantly and selectively under the control of the endo-siRNA pathway (Tam et al. 2008).
FIGURE 2.
Penelope-derived small RNAs. (A) Size distribution of Penelope-derived small RNAs in strains of D. virilis and transgenic D. melanogaster. (B,C) Penelope-derived siRNAs (21 nt) were plotted along the Penelope consensus sequence in transgenic strains A1 and A2 of D. melanogaster, respectively. (D) Penelope-derived siRNAs (21 nt) in an ovary small RNA library from strain 160. The structure of the consensus Penelope element, containing two “pseudo-LTRs” used in transformation experiments is shown at the bottom of the figure.
The mechanisms by which mobile elements become recognized by the siRNA pathway are unknown. However, rearranged Penelope copies containing long inverted repeats, which could give rise to double-stranded RNA (dsRNA), were previously isolated from the A1 and A2 strains. Penelope siRNAs precisely correspond to these regions (Fig. 2B,C). In contrast, Penelope siRNAs are distributed uniformly along the length of the consensus in virilis strain 160 (Fig. 2D), suggesting a different mechanism of recognition for the element. There appears to be no intrinsic propensity for Penelope sequences, per se, to be recognized as targets for siRNAs in melanogaster, since strains carrying only the ORF under the heat-shock promoter do not produce element-derived siRNAs even after Penelope ORF transcription was strongly induced by temperature elevation (Supplemental Table S1).
Observations from both melanogaster and virilis are consistent with a newly invading element coming at least in part under the control of the siRNA pathway during the process of adaptation. Whether this reflects an ancient role of the siRNA pathway in combating invading viruses remains to be seen. However, it is intriguing that even newly integrated green fluorescent protein transgenes can be recognized and selectively targeted by the siRNA system in Drosophila cells (Hartig et al. 2009).
Flexibility in the organization of the D. virilis piRNA pathway
The two best-studied models, D. melanogaster and mammals, differ in the operation of the ping-pong cycle, which may imply variation in the mechanisms by which each organism discriminates transposons from protein-encoding genes. In D. virilis, we noted transposon-specific patterns in the distribution of sense and antisense small RNAs. As has been observed in both germ-line and somatic tissues of D. melanogaster, siRNAs showed no enrichment for the sense or antisense strand of any of the elements examined (Vagin et al. 2006; Chung et al. 2008; Czech et al. 2008; Ghildiyal et al. 2008; data not shown). However, piRNAs corresponding to most elements were strongly skewed toward antisense species (Fig. 1C; Brennecke et al. 2007; Gunawardane et al. 2007; Lau et al. 2009). A notable exception was the Ulysses element, which showed the opposite character. In fact, in strain 9, 99% of Ulysses piRNAs corresponded to its coding strand.
piRNAs can be divided into primary and secondary species depending upon their biogenesis mechanism. Primary piRNAs have a strong bias for a 5′ U, and this characteristic is seen in the strain 9 Ulysses sense piRNA populations. piRNAs are uniformly distributed along the Ulysses consensus sequence, indicating that the elements that give rise to these species are likely near full length (Fig. 3A).
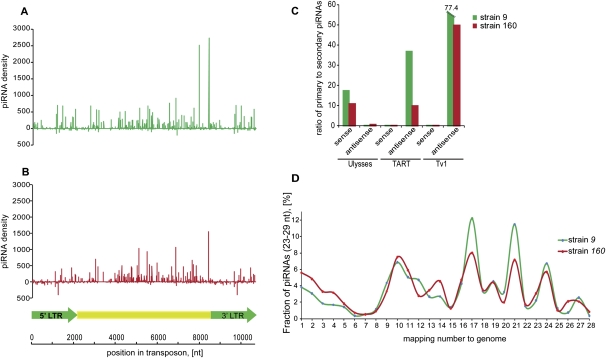
FIGURE 3.
Comparative analysis of Ulysses-piRNAs patterns in strains 9 and 160. Density of piRNAs from ovary libraries from strains 9 (A) and 160 (B) plotted over the Ulysses consensus sequence. (C) Processing categories of Ulysses, TART, and Tv1-piRNAs isolated from ovaries of both strains. (D) Frequency distribution of genomic mappings of Ulysses-piRNAs.
Strain 160 harbored an antisense population of Ulyssess piRNAs in addition to its abundant sense species (Figs. 1B, 3B). In this case, antisense species showed a particular bias toward LTRs within the consensus element. Although the sense and antisense species within those regions showed a signature of the ping-pong amplification loop, it is the antisense species that showed more enrichment for secondary piRNAs (Fig. 3C). In both strains, highly abundant Ulysses piRNAs generally mapped many times within the genome, generally reflecting the known copy numbers of the element (Fig. 3D; Zelentsova et al. 1999).
Two explanations can be envisioned for the striking differences between Ulysses and other virilis elements. The first reflects the remarkable similarity between the Ulysses piRNA pattern and the construction of the piRNA pathway in mouse gonocytes and juvenile testes (Aravin et al. 2008). In gonocytes, individual insertions of full-length elements are somehow identified and used for the production of primary piRNAs, producing predominantly sense species. Secondary species are enriched in antisense piRNAs and appear to come from transcripts generated from more conventional piRNA clusters. Thus, Ulysses could behave differently from other virilis elements, with dispersed, full-length elements serving as the source of primary species in strains 9 and 160. In strain 160, antisense content could be supplied by the strain-specific insertions of LTRs into piRNA clusters, which serve as a basis for ping-pong, but in the inverse of the normal orientation. An alternative hypothesis posits the insertion of one or more Ulysses elements in a highly expressed, single-stranded piRNA cluster in an orientation that would give rise to sense, primary piRNAs. Antisense transcripts, which act as fodder for ping-pong and the production of secondary piRNAs, could arise from fragments inserted into other piRNA clusters or from dispersed copies that, by chance, adopt a cellular promoter in an antisense configuration.
The unusual behavior of Ulysses prompted an examination of remaining elements. As examples, Tart and Tv1 are well targeted by the piRNA pathway, with a bias toward antisense piRNAs (Fig. 1C). In these cases and in all others where both a strong piRNA response and ample ping-pong partners could be detected, secondary piRNAs were enriched for sense species (Fig. 3C). Thus, the majority of elements in virilis engage the piRNA pathway in the melanogaster orientation, with piRNA clusters serving as the source for both primary piRNAs and antisense species within the cycle.
Maternal inheritance of virilis small RNAs and impacts on hybrid dysgenesis
It has been proposed that the Penelope transposon acts as an inducer of hybrid dysgenesis in intercrosses of virilis strains that differ in this element. Notably, dysgenesis correlates not only with an increase in Penelope expression but also with a derepression of other transposons, suggesting that Penelope might somehow interfere with silencing of unrelated elements by the RNAi machinery (Blumenstiel and Hartl 2005). Previous studies of hybrid dysgenesis syndromes in melanogaster demonstrated a critical role for maternally inherited piRNAs in controlling the activity of certain mobile elements (Brennecke et al. 2008), and this was consistent with a prior hypothesis for the control of Penelope (Blumenstiel and Hartl 2005).
If embryos are isolated prior to the onset of zygotic transcription, their small RNA populations reflect those of the maternal germ cells. In accord with this hypothesis, and with previous studies in melanogaster, endo-siRNA and piRNA populations corresponding to many different transposons were faithfully transmitted from strain 160 mothers to their progeny (Fig. 4A). Though the minority population of Penelope piRNAs was maternally inherited, Penelope siRNAs were not (Fig. 4A). This indicated that Penelope siRNAs might be present in the somatic rather than the germ cell compartment within the ovary (Chung et al. 2008; Czech et al. 2008; Ghildiyal et al. 2008). A comparison with total small RNA populations in carcass versus gonadal tissues (Fig. 4B) supported this hypothesis. Since siRNAs that target Penelope are not transmitted to the embryo, it is unlikely that failure of endo-siRNA pathway to repress Penelope can be responsible for its activation in dysgenic progeny.
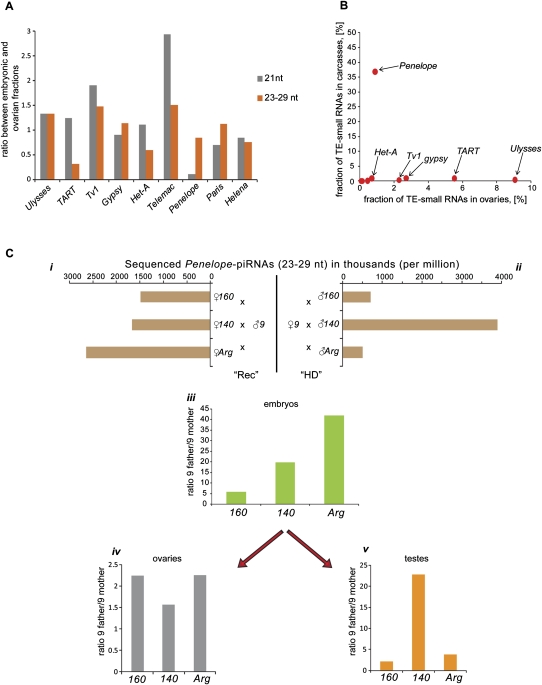
FIGURE 4.
Maternal deposition and levels of TE-derived small RNAs in D. virilis strains and hybrids. (A) Maternal deposition of TE-derived small RNAs in strain 160. The values were normalized to the levels of small RNA measured in ovaries of each strain. (B) Relative proportion (%) of various TE-derived small RNAs in somatic tissues (carcasses) and germ line (ovaries). (C) Maternal effect of piRNAs deposited into embryos on the levels of piRNAs in the gonads of F1 hybrids between strain 9 and strains containing multiple Penelope copies (160, 140, and Argentina). (i) Levels of Penelope-derived piRNAs in the ovaries of the three strains; (ii) levels of Penelope-derived piRNAs in the testes of the three strains; (iii) relative levels of Penelope-derived piRNA deposited into embryos from crosses involving strain 9, which lacks Penelope and strains carrying multiple Penelope copies (160, Argentina, and 140). Ratios of piRNA levels are shown comparing crosses with strain 9 fathers to crosses with strain 9 mothers. (iv, v) Relative levels of Penelope-derived piRNAs in F1 gonads of crosses involving strains 160, 140, and Argentina with the indicated strain 9 parent.
We next wished to probe the question of whether differences in the small population maternally deposited Penelope piRNAs were consistent with the suppression of hybrid dysgenesis. In melanogaster crosses in which dysgenesis is induced by an imbalance in maternal and paternal I- or P-elements, embryos destined to develop into sterile progeny have much lower levels of element-targeting piRNAs than do embryos that will develop into fertile progeny (Brennecke et al. 2008). This difference is maintained in adult gonads, where a deficit in piRNAs permits element activity, eventually leading to the secondary phenotype of sterility. We therefore examined the progeny of intercrosses between the “M-like” strain 9 and the “P-like” strain 160, where progeny of strain 9 mothers show the dysgenic phenotype. We also used two additional strains. These are deemed “neutral” because they do not produce dysgenic progeny when crossed with either strain 9 or strain 160, irrespective of the identity of the maternal parent. Notably, each of these neutral strains contains many intact euchromatic copies of Penelope, much like the P-like 160 strain.
We began by assessing levels of piRNAs in parental gonads (Fig. 4C i, ii). Overall, the ovaries of the P-strain, 160, and the neutral strains, 140 and Argentina (Arg), had very similar Penelope piRNA levels. The same was true of testes for 160 and Arg, but 140 showed substantially higher levels. In accord with these observations, all crosses in which the maternal parent was strain 160, Arg, or 140 showed greater numbers of deposited Penelope piRNAs than did crosses in which the maternal parent was strain 9 (Fig. 4C, iii).
We next examined the adult progeny of these crosses. In accord with prior observations, we observed induction of Penelope transcription in the ovaries of hybrids between strain 160 males and strain 9 females, but we failed to detect any Penelope transcription in the hybrids between both neutral strains used in the study and strain 9 (data not shown). Daughters of strain 9 fathers (with 160, 140, or Arg mothers) had relatively more Penelope piRNAs than daughters of strain 9 mothers (Fig. 4C, iv). However, differences in relative Penelope piRNA abundance did not correlate with the dysgenic phenotype. For example, dysgenic daughters of strain 9 mothers intercrossed with 160 had levels of adult piRNAs that were nearly identical to fertile daughters of intercrosses with 140. Similarly, in testes (Fig. 4C, v), dysgenic progeny in which strain 9 females were crossed to strain 160 males showed relative levels of Penelope piRNAs that were very similar to those observed in crosses between 9 and Argentina. There were variations in Penelope piRNAs in testes depending upon the nature of the parents in crosses between 9 and 140; however, these did not correlate with any observable phenotype.
Considered together, our data support maternal transmission of piRNA populations in D. virilis as had been previously observed in melanogaster. They also are consistent with the hypothesis that maternally inherited piRNA populations promote effective responses in progeny by priming the ping-pong cycle, since all progeny of strain 9 mothers have fewer piRNAs in daughter ovaries than daughters of strain 9 fathers (Fig. 4C, iv). However, our studies raise doubts about a critical role of maternally deposited Penelope small RNAs in the control of this element, though they do not exclude a possible role for Penelope, per se, in hybrid dysgenesis.
Differential expression of piRNA clusters in D. virilis strains
piRNA clusters represent an evolutionary record of transposon exposure and acquisition of control (Brennecke et al. 2007, 2008). Our previous studies in D. melanogaster are consistent with a model in which the content of piRNA clusters evolves rapidly. Even piRNA clusters of closely related strains can differ because of the insertion of new elements into these loci (Brennecke et al. 2008). However, piRNA loci themselves are broadly conserved between strains. For example, the flamenco cluster, located on X, and the 42AB cluster on chromosome 2 cannot only be found in all melanogaster strains examined to date, but are also functionally conserved at syntenic positions through ∼12 million years of divergent evolution in Drosophila yakuba and Drosophila erecta (Malone et al. 2009).
Despite its overall conservation, the flamenco piRNA locus exists as a variety of permissive and restrictive alleles that have been experimentally induced or are naturally occurring (Prud'homme et al. 1995; Brennecke et al. 2007; Mevel-Ninio et al. 2007; Malone et al. 2009). One of the persistent mysteries prior to the identification of flamenco as a piRNA locus was that these alleles simultaneously impacted multiple transposons that were only distantly related (Prud'homme et al. 1995; Mevel-Ninio et al. 2007). This seemed strongly reminiscent of the derepression of multiple element classes in crosses between strain 9 females and strain 160 males. Thus, we envisioned the possibility that variation in a maternally deposited cluster might induce the types of impacts observed in dysgenic virilis crosses.
To investigate this possibility, we identified piRNA clusters within the D. virilis genome by focusing on only the ∼20% of piRNAs that map uniquely because of polymorphisms (Brennecke et al. 2007; Lau et al. 2009). We focused on the 20 loci that contributed the greatest number of piRNAs, since we reasoned that those would be most likely to impact the phenotype of progeny (Supplemental Table S2). We noted clusters that, like flamenco, produce piRNAs from only one genomic strand, and clusters that produce piRNAs from both genomic strands. Like flamenco itself, many of the single-strand clusters mapped to pericentromeric heterochromatin and were not maternally deposited, indicating their expression in the somatic compartment of the gonad.
We also identified a number of clusters that produced piRNAs from both genomic strands. These were clearly active in the germ-line compartment, since the piRNAs that they produced were efficiently deposited in early embryos. We used a combination of in situ hybridization and genomic PCR with cluster-specific oligonucleotides to confirm the presence of cluster loci in all four strains and to verify their genomic locations as predicted from the genome assembly (for example, see Fig. 5A; Supplemental Table S2).
FIGURE 5.
Characterization of a dual-strand D. virilis piRNA cluster uniquely expressed in strain 160. (A) Example of in situ hybridization with cluster DNA. Arrows indicate hybridization at the telomere of chromosome 2 and at the 7C region of the X-chromosome of strain 9. Virtually all probes hybridized with variable efficiency to the heterochromatic chromocenter but also to other discrete genomic locations, which agreed with sequence-based assignments. The consistent hybridization with the 7C region on the X-chromosome of strain 9 was absent in strain 160. (B) Uniquely mapped piRNAs were plotted over cluster 1 (see Supplemental Table S1), which contains TART elements and protein-coding regions. These are differentially processed into piRNAs in strains 160 (red) and 9 (green).
Despite being present in all strains, the four clusters that produced the greatest numbers of piRNAs in strain 160 were completely inactive in the M-like and neutral strains studied (Fig. 5B). Notably, all four polymorphic virilis clusters were present in subtelomeric locations. One highly peculiar feature of these clusters is that they contain protein-coding genes that generate abundant piRNAs (Fig. 5B). Since we lack genetic mutations within piRNA pathway genes in virilis, it is impossible to determine whether these abundant piRNAs have any effect on the expression of cluster-resident genes in the germ line. However, it should be noted that similar, abundant, gene-derived piRNAs do not seem to affect their generative loci, for example, TJ, in melanogaster (Saito et al. 2009).
Considered together, these observations demonstrate that the activity of piRNA clusters can vary substantially between strains. Despite their apparent presence, a number of telomeric clusters that are the most active producers of piRNAs in strain 160 fail to produce piRNAs in strains 9, 140, and Argentina. The HP1-family member Rhino has recently been linked to piRNA production in melanogaster (Klattenhoff et al. 2009). This protein is thought to be targeted to piRNA clusters based, at least in part, upon specific histone modifications. It is tempting to speculate that changes in telomeric heterochromatin, which are well correlated with telomere length in other organisms (Schoeftner and Blasco 2009), might modulate the binding of a Rhino-like protein to the subtelomeric virilis clusters that function in a strain-specific fashion. Alternatively, the subtelomeric clusters may have become activated in strain 160 through the accumulation of mutations that somehow redefine these regions as piRNA clusters in germ cells.
Our studies, which probed the response to invading Penelope elements in two species, have revealed an unexpected flexibility in small RNA responses to transposons in germ cells. Both in melanogaster and in virilis strain 160, Penelope appears to be predominantly controlled by the siRNA rather than by the piRNA pathway. In both cases the siRNA pathway is unable to completely silence Penelope, which remains capable of occasional transposition in D. melanogaster transgenic strains and in D. virilis strain 160.
It was shown previously that in strain 160, an X-linked locus is responsible for producing the majority of Penelope-derived small RNAs (Blumenstiel and Hartl 2005). Our data now identify these as siRNAs, but suggest that they are restricted to the soma. piRNAs, which are implicated in Penelope silencing in the germ line, are apparently not X-chromosome derived, and we observe even more Penelope piRNAs in dysgenic males than in those arising from the reciprocal cross. The data presented herein explain the apparent discrepancy between the location of the Penelope small RNA “master-locus” on the X-chromosome and the inability of prior studies to correlate this locus with HD-associated gonadal sterility (Blumenstiel and Hartl 2005).
In melanogaster, it is unclear how Penelope siRNAs are produced; however, their distribution within the element consensus is consistent with the presence of documented rearranged copies that can produce dsRNA. It is tempting to speculate that the presence of such copies might even have been selected during the process of transgenesis as an accommodation to the presence of the new element. In either case, the Penelope element, which remains weakly active in the melanogaster germ line, does not apparently present a strong challenge to the species. In this sense, it may be behaving as a highly evolved parasite, which can propagate without substantial harm to its host. This, combined with its low level of activity, may simply not yet have allowed sufficient generations or applied sufficient selective pressure for Penelope to come under the control of the melanogaster piRNA system.
Strain 160 appears to differ from the other virilis strains analyzed in many respects. Most relevant to this study is its production of piRNAs from a number of subtelomeric loci that are not used for this purpose in other strains. The sterility and other abnormalities observed in the progeny of crosses involving strain 160 in a way resemble phenomena often seen in the progeny of interspecies crosses where mobilization of transposons sometimes occurs (Zelentsova et al. 1986; O'Neill et al. 1998; Labrador et al. 1999). It is noteworthy that in the progeny of dysgenic crosses involving strain 160 we previously isolated multiple chromosomal aberrations, which often coincide with species-specific inversions described within the virilis group. Consequently, we imagine that crosses involving strain 160 may lead to a burst of inversion polymorphism, providing a catalyst for chromosomal aberrations that create reproductive isolation (Evgen'ev et al. 2000). Thus, our observations of the divergence of piRNA pathways in the recently separated virilis strains examined herein may be providing a glimpse into how changes in the piRNA pathway, the acquisition of new piRNA clusters, and transposon content could ultimately lead to reproductive isolation and serve as one of many catalysts for speciation.
MATERIALS AND METHODS
Drosophila strains
Drosophila virilis strains 160 (b, gp, cd, pe, gl), 9 (wild type, Batumi, Caucasus), Argentina, and 140 (eb, va) were obtained from the Stock Center of the Institute of Developmental Biology, Moscow.
Drosophila melanogaster transgenic strains: A1 contains four to five copies of Penelope; strain A2 contains 12–14 copies of Penelope. The development and characteristics of these strains have been described previously (Pyatkov et al. 2002). Strain “hs” represents a strain transformed with Penelope ORF under the D. melanogaster Hsp70 heat-shock promoter. The constructs and transformation procedures were previously described (Pyatkov et al. 2002). The parental strain y,w67c23(2), lacking Penelope, served as a control. All flies were reared at 25°C on standard resin-sugar-yeast-agar medium containing propionic acid and methylparaben as mold inhibitors. Embryos (0–2 h) were collected using agar plates at 25°C.
Cytological analysis
Larvae were grown at 18°C on medium supplemented with live yeast solution for 2 d before dissection. Salivary glands from third instar larvae were dissected in 45% acetic acid and squashed (Lim 1993). Procedures and labeling of DNA probes for in situ hybridization were as described (Lim 1993).
Immunoprecipitation
Immunoprecipitation experiments using either monoclonal anti-FLAG antibodies (for tagged strains) or rabbit polyclonal antisera against Ago2 and the N-termini of Piwi, Aub, and Ago3 have been described previously (Brennecke et al. 2007; Czech et al. 2008).
Small RNA libraries
Cloning of small RNAs from total RNA and immunopurified complexes was performed as described previously (Brennecke et al. 2007).
Small RNA sequences were mapped to the latest releases of the D. melanogaster and D. virilis genomes. Cluster analysis was performed on libraries after subtraction of small RNA reads matching to all rRNAs, tRNAs, snRNAs, and miRNA. Extraction of piRNA clusters was performed as described (Brennecke et al. 2007). Transposon-derived small RNAs with up to three mismatches were mapped onto Repbase. For the Penelope analysis, only small RNAs matching 1–3394 nt of U49102.2 GI:1555193 were considered as the true Penelope-derived sequences.
Small RNA counts were normalized to 1 million small RNAs after subtracting abundant noncoding RNAs such as rRNAs, tRNAs, and snoRNAs.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank M. Rooks, Laura Cardone, and Melissa Kramer for help with Illumina sequencing; G. Assaf and O. Tam for help with bioinformatics analysis; and E. Rozhkova for fly husbandry. We also thank members of the Hannon laboratory for helpful discussions. G.J.H. is an investigator of the Howard Hughes Medical institute, and this work was supported by grants from the NIH and by a kind gift from Kathryn W. Davis. Work at the IMB was supported by the Program of Molecular and Cellular Biology RAN. Sequences reported in this study can be accessed using GEO accession number: GSE22067.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2217810.
REFERENCES
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA 2001. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol 11: 1017–1027 [DOI] [PubMed] [Google Scholar]
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. 2006. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442: 203–207 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ, Brennecke J 2007. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 318: 761–764 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ 2008. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31: 785–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkhipova IR, Pyatkov KI, Meselson M, Evgen'ev MB 2003. Retroelements containing introns in diverse invertebrate taxa. Nat Genet 33: 123–124 [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366 [DOI] [PubMed] [Google Scholar]
- Blevins T, Rajeswaran R, Shivaprasad PV, Beknazariants D, Si-Ammour A, Park HS, Vazquez F, Robertson D, Meins F Jr, Hohn T, et al. 2006. Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res 34: 6233–6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenstiel JP, Hartl DL 2005. Evidence for maternally transmitted small interfering RNA in the repression of transposition in Drosophila virilis. Proc Natl Acad Sci 102: 15965–15970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103 [DOI] [PubMed] [Google Scholar]
- Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ 2008. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 322: 1387–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casacuberta E, Pardue ML 2005. HeT-A and TART, two Drosophila retrotransposons with a bona fide role in chromosome structure for more than 60 million years. Cytogenet Genome Res 110: 152–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambeyron S, Popkova A, Payen-Groschene G, Brun C, Laouini D, Pelisson A, Bucheton A 2008. piRNA-mediated nuclear accumulation of retrotransposon transcripts in the Drosophila female germline. Proc Natl Acad Sci 105: 14964–14969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WJ, Okamura K, Martin R, Lai EC 2008. Endogenous RNA interference provides a somatic defense against Drosophila transposons. Curr Biol 18: 795–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H 1998. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev 12: 3715–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Lin H 2000. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development 127: 503–514 [DOI] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, et al. 2008. An endogenous small interfering RNA pathway in Drosophila. Nature 453: 798–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalle Nogare DE, Clark MS, Elgar G, Frame IG, Poulter RT 2002. Xena, a full-length basal retroelement from tetraodontid fish. Mol Biol Evol 19: 247–255 [DOI] [PubMed] [Google Scholar]
- Evgen'ev MB, Arkhipova IR 2005. Penelope-like elements—a new class of retroelements: Distribution, function and possible evolutionary significance. Cytogenet Genome Res 110: 510–521 [DOI] [PubMed] [Google Scholar]
- Evgen'ev MB, Zelentsova H, Shostak N, Kozitsina M, Barskyi V, Lankenau DH, Corces VG 1997. Penelope, a new family of transposable elements and its possible role in hybrid dysgenesis in Drosophila virilis. Proc Natl Acad Sci 94: 196–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evgen'ev MB, Zelentsova H, Poluectova H, Lyozin GT, Veleikodvorskaja V, Pyatkov KI, Zhivotovsky LA, Kidwell MG 2000. Mobile elements and chromosomal evolution in the virilis group of Drosophila. Proc Natl Acad Sci 97: 11337–11342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD 2009. Small silencing RNAs: An expanding universe. Nat Rev Genet 10: 94–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, et al. 2008. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 320: 1077–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA 2006. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442: 199–202 [DOI] [PubMed] [Google Scholar]
- Grivna ST, Pyhtila B, Lin H 2006. MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc Natl Acad Sci 103: 13415–13420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC 2007. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315: 1587–1590 [DOI] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404: 293–296 [DOI] [PubMed] [Google Scholar]
- Hartig JV, Esslinger S, Bottcher R, Saito K, Forstemann K 2009. Endo-siRNAs depend on a new isoform of loquacious and target artificially introduced, high-copy sequences. EMBO J 28: 2932–2944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt JE, Roman SD, Aitken RJ, McLaughlin EA 2006. Identification and characterization of a novel Mt-retrotransposon highly represented in the female mouse germline. Genomics 87: 490–499 [DOI] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J 2003. Molecular paleontology of transposable elements in the Drosophila melanogaster genome. Proc Natl Acad Sci 100: 6569–6574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC, Siomi H 2008. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature 453: 793–797 [DOI] [PubMed] [Google Scholar]
- Ketting RF, Haverkamp TH, van Luenen HG, Plasterk RH 1999. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell 99: 133–141 [DOI] [PubMed] [Google Scholar]
- Kidwell MG, Kidwell JF, Sved JA 1977. Hybrid dysgenesis in Drosophila melanogaster: A syndrome of aberrant traits including mutation, sterility and male recombination. Genetics 86: 813–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC 2009. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10: 126–139 [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Xi H, Li C, Lee S, Xu J, Khurana JS, Zhang F, Schultz N, Koppetsch BS, Nowosielska A, et al. 2009. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell 138: 1137–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrador M, Farre M, Utzet F, Fontdevila A 1999. Interspecific hybridization increases transposition rates of Osvaldo. Mol Biol Evol 16: 931–937 [DOI] [PubMed] [Google Scholar]
- Lau NC, Robine N, Martin R, Chung WJ, Niki Y, Berezikov E, Lai EC 2009. Abundant primary piRNAs, endo-siRNAs, and microRNAs in a Drosophila ovary cell line. Genome Res 19: 1776–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JK 1993. In situ hybridization with biotinylated DNA. Drosoph Inf Serv 72: 73–77 [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441 [DOI] [PubMed] [Google Scholar]
- Lozovskaya ER, Scheinker VS, Evgen'ev MB 1990. A hybrid dysgenesis syndrome in Drosophila virilis. Genetics 126: 619–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucy AP, Guo HS, Li WX, Ding SW 2000. Suppression of post-transcriptional gene silencing by a plant viral protein localized in the nucleus. EMBO J 19: 1672–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyozin GT, Makarova KS, Velikodvorskaja VV, Zelentsova HS, Khechumian RR, Kidwell MG, Koonin EV, Evgen'ev MB 2001. The structure and evolution of Penelope in the virilis species group of Drosophila: An ancient lineage of retroelements. J Mol Evol 52: 445–456 [DOI] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ 2009. Small RNAs as guardians of the genome. Cell 136: 656–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ 2009. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137: 522–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevel-Ninio M, Pelisson A, Kinder J, Campos AR, Bucheton A 2007. The flamenco locus controls the gypsy and ZAM retroviruses and is required for Drosophila oogenesis. Genetics 175: 1615–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizrokhi LJ, Mazo AM 1991. Cloning and analysis of the mobile element gypsy from D. virilis. Nucleic Acids Res 19: 913–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar A, Csorba T, Lakatos L, Varallyay E, Lacomme C, Burgyan J 2005. Plant virus-derived small interfering RNAs originate predominantly from highly structured single-stranded viral RNAs. J Virol 79: 7812–7818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill RJ, O'Neill MJ, Graves JA 1998. Undermethylation associated with retroelement activation and chromosome remodelling in an interspecific mammalian hybrid. Nature 393: 68–72 [DOI] [PubMed] [Google Scholar]
- Okamura K, Ishizuka A, Siomi H, Siomi MC 2004. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev 18: 1655–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Chung WJ, Ruby JG, Guo H, Bartel DP, Lai EC 2008. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature 453: 803–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov DA, Schutzman JL, Hartl DL, Lozovskaya ER 1995. Diverse transposable elements are mobilized in hybrid dysgenesis in Drosophila virilis. Proc Natl Acad Sci 92: 8050–8054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard G 1976. Non-mendelian female sterility in Drosophila melanogaster: Hereditary transmission of I factor. Genetics 83: 107–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud'homme N, Gans M, Masson M, Terzian C, Bucheton A 1995. Flamenco, a gene controlling the gypsy retrovirus of Drosophila melanogaster. Genetics 139: 697–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyatkov KI, Shostak NG, Zelentsova ES, Lyozin GT, Melekhin MI, Finnegan DJ, Kidwell MG, Evgen'ev MB 2002. Penelope retroelements from Drosophila virilis are active after transformation of Drosophila melanogaster. Proc Natl Acad Sci 99: 16150–16155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, He X, Wang XJ, Kohany O, Jurka J, Hannon GJ 2006. Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature 443: 1008–1012 [DOI] [PubMed] [Google Scholar]
- Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC 2009. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 461: 1296–1299 [DOI] [PubMed] [Google Scholar]
- Scheinker VS, Lozovskaya ER, Bishop JG, Corces VG, Evgen'ev MB 1990. A long terminal repeat-containing retrotransposon is mobilized during hybrid dysgenesis in Drosophila virilis. Proc Natl Acad Sci 87: 9615–9619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S, Blasco MA 2009. A ‘higher order’ of telomere regulation: Telomere heterochromatin and telomeric RNAs. EMBO J 28: 2323–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, Vaughn M, Borges F, Tanurdzic M, Becker JD, Feijo JA, Martienssen RA 2009. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136: 461–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L 2004. Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305: 1434–1437 [DOI] [PubMed] [Google Scholar]
- Spicer GS, Bell CD 2002. Molecular phylogeny of the Drosophila virilis species group (Diptera: Drosophilidae) inferred from mitochondrial 12S and 16S ribosomal RNA genes. Ann Entomol Soc Am 95: 156–161 [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al. 2008. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453: 534–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD 2006. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313: 320–324 [DOI] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP 2000. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101: 25–33 [DOI] [PubMed] [Google Scholar]
- Zelentsova ES, Vashakidze RP, Krayev AS, Evgenev MB 1986. Dispersed repeats in Drosophila-virilis - elements mobilized by interspecific hybridization. Chromosoma 93: 469–476 [Google Scholar]
- Zelentsova H, Poluectova H, Mnjoian L, Lyozin G, Veleikodvorskaja V, Zhivotovsky L, Kidwell MG, Evgen'ev MB 1999. Distribution and evolution of mobile elements in the virilis species group of Drosophila. Chromosoma 108: 443–456 [DOI] [PubMed] [Google Scholar]
- Zilberman D, Cao X, Johansen LK, Xie Z, Carrington JC, Jacobsen SE 2004. Role of Arabidopsis ARGONAUTE4 in RNA-directed DNA methylation triggered by inverted repeats. Curr Biol 14: 1214–1220 [DOI] [PubMed] [Google Scholar]