Abstract
The 3′ untranslated regions (UTRs) of many plant viral RNAs contain cap-independent translation elements (3′ CITEs). Among the 3′ CITEs, the Barley yellow dwarf virus (BYDV)-like translation elements (BTEs) form a structurally variable and widely distributed group. Viruses in three genera were known to harbor 3′ BTEs, defined by the presence of a 17-nt consensus sequence. To understand BTE function, knowledge of phylogenetically conserved structure is essential, yet the secondary structure has been determined only for the BYDV BTE. Here we show that Rose spring dwarf-associated luteovirus, and two viruses in a fourth genus, Umbravirus, contain functional BTEs, despite deviating in the 17 nt consensus sequence. Structure probing by selective 2′-hydroxyl acylation and primer extension (SHAPE) revealed conserved and highly variable structures in BTEs in all four genera. We conclude that BTEs tolerate striking evolutionary plasticity in structure, while retaining the ability to stimulate cap-independent translation.
Introduction
Upon entry into the host cell, the genomic RNA of a positive-sense RNA virus must first be translated to generate the viral proteins necessary for RNA replication. The very small number of initially infecting RNAs must compete with actively translating host mRNAs for the host translation machinery. Thus, plant viral RNAs have evolved a variety of mechanisms to usurp host factors for their own translation. An increasing number of translation enhancer elements have been identified in the untranslated regions (UTRs) of plant positive-sense RNA viruses (Dreher and Miller, 2006; Kneller et al., 2006; Miller et al., 2010).
All nonviral eukaryotic mRNAs have a 5′-cap structure, and all plant and most animal mRNAs have a 3′-poly (A) tail. These modifications are essential for recruitment of translation initiation factors and the ribosome to the mRNA and for mRNA stability (Hentze et al., 2007; Jackson et al., 2010). In contrast to canonical mRNAs, mRNAs of many positive-sense RNA viruses lack a 5′ cap structure and instead contain a cap-independent translation element (CITE), that allows for efficient translation initiation. Among the CITEs located in the 5′ UTRs of animal and some plant viruses, are internal ribosome entry sites (IRESes) which recruit the 40S ribosomal subunit directly to the mRNA (Filbin and Kieft, 2009; Jan, 2006; Niepel and Gallie, 1999; Zeenko and Gallie, 2005; Karetnikov and Lehto, 2007). In contrast, many uncapped plant viral RNAs harbor a CITE in the 3′ UTR of the genome (Kneller et al., 2006; Fabian and White, 2004; Scheets and Redinbaugh, 2006; Stupina et al., 2008; Wang, et al., 2009). The 3′-CITEs identified so far fall into about seven or eight distinct classes based on their sequence and secondary structures (Miller, et al., 2007). These elements show no obvious sequence or structural similarity to each other, except that most harbor a stem–loop in which the loop sequence is known or predicted to base pair to the 5′-UTR (Guo et al., 2001; Fabian and White, 2004; Miller and White, 2006; Karetnikov and Lehto, 2008).
Barley yellow dwarf virus (BYDV) and viruses in four genera (below) harbor a BYDV-like CITE (BTE) in their 3′ UTRs. Phylogenetic comparisons, mutagenesis, and structural probing of the BYDV BTE revealed that the BTEs contain a 17 nt conserved sequence, (17 nt CS) GAUCCCUGGGAAACAGG that forms a stem-loop (SL-I) in which the underlined bases are paired (Guo, et al., 2000; Mizumoto et al., 2003; Shen and Miller, 2004; Meulewaeter et al., 2004). The BYDV BTE adopts a cruciform secondary structure with three stem-loops radiating from the central hub which is connected to the rest of the viral genome by a fourth basal helix (Guo et al., 2000). The loop of one of the stem-loops (SL-III in the BYDV BTE) must base pair to the 5′ UTR in a kissing stem-loop interaction to facilitate cap-independent translation (Guo, et al., 2001; Rakotondrafara et al., 2006). This interaction may ensure localization of initiation factor eIF4G, which binds the BTE (Treder et al., 2008), to the 5′ end where the 40S ribosomal subunit is recruited.
To understand how the BTE structure brings about its function, phylogenetic comparisons with other viral 3′ CITEs reveal which sequences and structures are essential and which are not. Indeed, 3′ CITEs resembling, but differing significantly from, the BYDV BTE have been demonstrated in other viruses, including Tobacco necrosis viruses A and D (TNV-A, TNV-D) (Meulewaeter et al., 2004; Shen and Miller, 2004) and Red clover necrotic mosaic virus (RCNMV) RNA 1 (Mizumoto et al., 2003). These were identified as BTEs, because they harbor a sequence that is identical, or nearly-identical, to the 17 nt CS. The BTEs from TNVs A and D (which, despite their similar names, are quite distinct viruses), are predicted to lack a structural homolog to SL-II of the BYDV BTE (Meulewaeter et al., 2004; Shen and Miller, 2004). In contrast, the BTE of RCNMV is predicted to have five stem-loops radiating from the central core, in addition to the basal helix (Mizumoto et al., 2003; Sarawaneeyaruk et al., 2009). However, no direct structural analyses have been performed on any of the BTEs except that of BYDV (Guo et al., 2000).
To understand how the BTEs function, it is necessary to determine the essential sequence and structural features they share. Sequences matching or resembling the 17 nt CS are present in all members of the Luteo-, Diantho- and Necrovirus genera (Mizumoto et al., 2003; Shen and Miller, 2004; Wang, et al., 1997) and, we report here, in two viruses of genus Umbravirus (Table 1). Here we determine the functions and structures of 3′ UTR elements which contain sequences resembling, but not perfectly matching, the 17 nt BTE consensus, or which contain secondary structures that differ from those characterized to date. We report: (i) the first known BTEs in genus Umbravirus, (ii) a functional BTE in the Rose spring dwarf-associated luteovirus (RSDaV) 3′UTR which differs substantially from other known BTEs by having large tracts of unpaired bases, (iii) less functional BTEs and non-functional BTE-like sequences that differ from consensus in the 17 nt CS, (iv) chemical determination of actual secondary structures of BTEs from four viral genera, and (v) conserved structural features common to all BTEs. These results reveal that the BTE structure shows substantial plasticity, both in phylogenetic terms, and possibly physically, compared to other known cap-independent translation elements or IRESes.
TABLE 1. 17 nt conserved sequence in all known or predicted BTEs.
| Virus | 17 nt conserved sequence | Accession no. | Reference |
|---|---|---|---|
| Genus Luteovirus | |||
| Barley yellow dwarf virus –PAV (BYDV-PAV) | GGAUCCUGGGAAACAGG | AF235167 | (Wang et al. 1997) |
| Barley yellow dwarf virus –PAS (BYDV-PAS) | .A............... | AF218798 | |
| Soybean dwarf virus (SbDV) | ................. | L24049 | |
| Bean leafroll virus (BLRV) | .A............... | NC_003369 | |
| Rose spring dwarf associated virus (RSDaV) | .........U....... | EU024678 | |
| Genus Dianthovirus | |||
| Red clover necrotic mosaic virus (RCNMV) | ...C............. | NC_003756 | (Mizumoto et al., 2003) |
| Carnation ringspot virus (CRSV) | ........A........ | L18870 | |
| Sweet clover necrotic mosaic virus (SCNMV) | ...C.....U....... | NC_003806 | |
| Genus Necrovirus | |||
| Olive latent-1 (OLV-1) | .........U.G..... | NC_001721 | |
| Olive mild mosaic virus (OMMV) | ................. | NC_006939 | |
| Tobacco necrosis A (TNV-A) | ................. | NC_001777 | (Meulewaeter et al., 2004) |
| Tobacco necrosis D (TNV-D) | ................. | U62546 | (Shen and Miller, 2004) |
| Leek white stripe virus (LWSV) | A.....C.-AG...G.. | NC_001822 | |
| Black beet scorch virus (BBSV) | .A.......U....... | AF452884 | |
| Genus Umbravirus | |||
| Tobacco bushy top virus (TBTV) | ................. | AF402620 | |
| Groundnut rosette virus (GRV) | ......G.......U.. | NC_003603 | |
Dot: identical to BYDV-PAV, underlined: paired bases in SL-I of BYDV BTE. GenBank® accession numbers of the viral genome sequence and references to publications that demonstrate a functional BTE are shown.
Results
A functional BTE in umbravirus 3′ UTRs
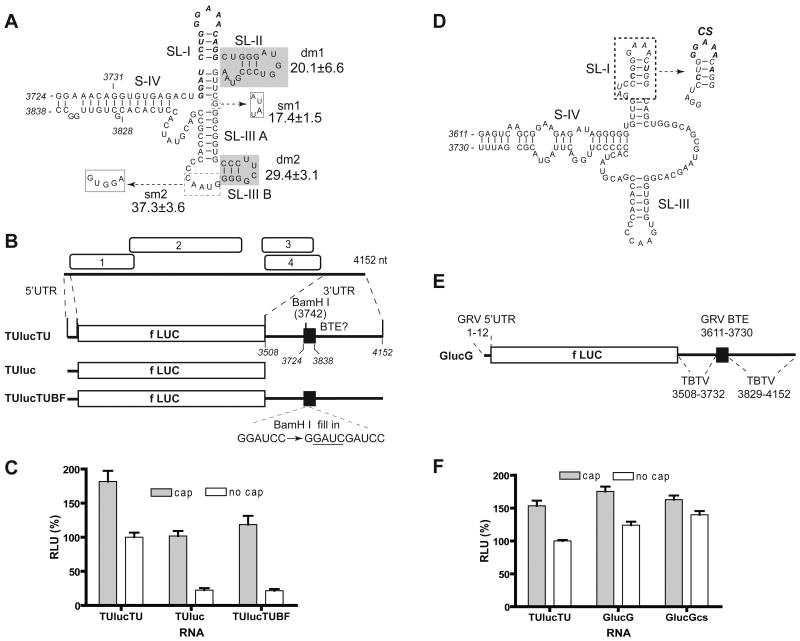
The tract spanning nts 3742-3758 in the 3′ UTR of the tobacco bushy top umbravirus (TBTV) genome (GenBank AF402620) matches the 17 nt CS consensus perfectly (Table 1). Moreover, the predicted secondary structure of the putative TBTV BTE resembles that of BYDV except that the first four bases of the conserved sequence may be weakly base paired to extend SL-I, and there is a small stem-loop expected to branch from the end of SL-III (Fig. 1A).
Fig. 1.
Umbravirus 3′ UTRs contain a BTE. (A) Representative predicted (Mfold) secondary structure of the BTE in the 3′ UTR of TBTV genomic RNA, and relative translation activities of selected mutants. 17 nt CS is in bold italics. The shaded bases were deleted in mutants dm1 and dm2 as indicated. In mutants sm1 and sm2, bases in dashed boxes were replaced by bases in solid boxes (dashed arrows). The luciferase activity generated by translation of uncapped mutant RNAs, as a percentage of wild type (100%) are indicated. (B) Genome organization of TBTV RNA and maps of translation reporter constructs containing TBTV UTRs. TUlucTU contains both complete UTRs of TBTV flanking the firefly luciferase coding sequence (fLUC). TUluc has only the TBTV 5′UTR. TUlucTUBF differs from TUlucTU by only a 4 nt insertion (underlined) in the BamH I site. Bases are numbered according to their position in the TBTV genome. (C) Relative translation activities of capped and uncapped reporter mRNAs in wheat germ extract. Luciferase activities obtained from the indicated RNAs are normalized to uncapped TUlucTU (defined as 100%) and shown as RLU (relative luciferase units). Error bars indicate standard error. (D) Predicted secondary structure of the GRV BTE. The 17 nt CS (italics) is in the dashed box, with bases that deviate from consensus indicated in bold italics. Mutant sequence (CS) which is identical to the consensus 17 nt CS is indicated at right by dashed arrow. (E) Map of reporter RNA GlucG. GlucG contains the GRV 5′UTR and a chimeric 3′UTR consisting of the TBTV 3′ UTR with the putative GRV BTE in place of the TBTV BTE. Positions of bases from each virus are indicated. (F) Relative translation activities of capped and uncapped reporter mRNAs in wheat germ extract.
To determine whether the predicted TBTV BTE confers cap-independent translation, a reporter construct was prepared which consists of the full-length 5′ and 3′ untranslated regions of TBTV genome flanking the firefly luciferase ORF (construct TUlucTU, Fig 1B). The mRNA was produced by in vitro transcription driven by a T7 RNA polymerase promoter immediately upstream of the TBTV 5′ UTR. The resulting TUlucTU transcripts were translated in wheat germ extract in amounts (4 nM) well below the saturating levels, so the levels of luciferase activity were proportional to the translation efficiency of the mRNA. Because the viral cap-independent translation element is defined as a sequence sufficient to functionally replace a 5′ cap, translation of both capped and uncapped TUlucTU RNAs was measured. Uncapped TUlucTU translated 57% as efficiently as the capped version (Fig 1C). Deletion of the 3′UTR, or a four base insertion (GAUC) in the natural BamH I site in the 17 nt CS (construct TUlucTUBF), reduced luciferase expression of uncapped RNA to 20% of the capped version. The GAUC insertion is known to knock out cap-independent translation activity of the BYDV and TNV BTEs (Guo, et al., 2000; Shen and Miller, 2004). Notice that the capped transcripts lacking the viral 3′ UTR (TUluc) or containing the GAUC insertion (TUlucTUBF) translated at levels similar to that of uncapped transcript harboring the intact TBTV 3′ UTR (Fig. 1C). Moreover, when these constructs were tested in protoplasts, TUlucTU translated about 30-fold more efficiently than TUlucTUBF. However, translation of both constructs was low in protoplasts, probably due to the very short 10 nt 5′ UTR which may be removed by 5′-3′ cellular exonucleases in the absence of a 5′ cap. Overall, these results resemble those observed previously with BYDV (Guo, et al., 2000; Wang, et al., 1997; Wang et al., 1999) and TNV BTEs (Meulewaeter et al., 2004; Shen and Miller, 2004), indicating that TBTV harbors a 3′ BTE.
To map the minimal length of TBTV BTE necessary for cap-independent translation, deletions were made in the 3′ UTR of TUlucTU and corresponding RNAs were tested in wheat germ extract for their ability to support translation. Translation levels with 3′ UTRs consisting of TBTV nts 3712-3872 or nt 3724-3838 were greater than 80% of the levels obtained from full-length viral 3′UTR (TUlucTU). In contrast, uncapped reporter mRNA containing only TBTV nts 3731-3828 (Fig. 1A) in the 3′ UTR translated less than 20% as efficiently as with the full 3′ UTR. Thus we define the element spanning nts 3724 to 3838 as the TBTV BTE.
We next tested the necessity of predicted structures of the TBTV BTE that are absent in all other BTEs (SL-IIIB) or absent in TNV BTEs (SL-II) (Fig. 1A). Deletion of SL-II (dm1), replacement of the G in the junction between SL-I and SL-III with AUAU (sm1) or deletion of SL-IIIB (dm2), reduced translation to approximately 20% of that obtained with uncapped, wild type TUlucTU (Fig. 1A). Replacement of the GUAAC sequence in loop III, which is predicted to kiss with a loop in the 5′ UTR, with the sequence GUGGA (sm2) retained just over one-third of wild type translation activity. However, structural probing data (below) showed that the sequences at the end of SL-III may not exist in the predicted structures. In summary, the TBTV BTE is sensitive to changes, even in regions that are not obviously conserved in other BTEs.
A potential BTE in the 3′ UTR of Groundnut rosette umbravirus (GRV) contains the sequence 3639-GGAUCCCGGGAAACUGG-3655 (GenBank NC_003603), which differs from the 17 nt CS at two positions (bold, italic). This would be expected to weaken or disrupt the SL-I base pairing in the consensus (underlined). However, Mfold predicts a four base-pair SL-I, as in the canonical BTE, but with the two strands of the helix shifted, giving a GAAA tetraloop (Fig. 1D). Also, there is no structural equivalent to SL-II, instead there is a tract of 17 unpaired bases upstream of SL-III (Fig. 1D). The 17 nt CS tracts of both umbraviruses are located near the middle of the 645 nt (TBTV) or 535 nt (GRV) 3′ UTR (Fig. 1). This differs from the BYDV and TNV BTEs which are located at the 5′ end of the 3′ UTR, immediately downstream of the stop codon of the upstream ORF.
We were unable to obtain clones of the GRV sequences because the virus is a serious pathogen limited to Africa. Thus, to test the GRV sequence for BTE function, the 12 nt GRV 5′ UTR and the 120 nt predicted GRV BTE sequence were synthesized, and subcloned into the TUlucTU clone in place of the TBTV 5′UTR and BTE, respectively, giving rise to construct GlucG (Fig 1E). Uncapped GlucG mRNA translated slightly more efficiently than uncapped TUlucTU mRNA and 70% as efficiently as capped GlucG mRNA (Fig. 1F). Thus, functional replacement of the TBTV BTE with the GRV sequence indicates that the GRV sequence also functions as a cap-independent translation element. To further analyze the significance of the deviations from the 17 nt CS, we mutated the GRV 17 nt CS to match the BYDV consensus sequence. The uncapped transcript translated slightly more efficiently than wild type and nearly as well as the capped version (GlucGcs, Fig. 1F). Thus, while the perfect consensus may provide slightly more efficient cap-independent translation than the natural GRV 17 nt CS, it appears that the BTE may tolerate the base differences in the wild type 17 nt CS of GRV with little effect on function, at least in uncompetitive conditions in wheat germ extract.
The RSDaV genome contains a functional BTE element in the 3′UTR
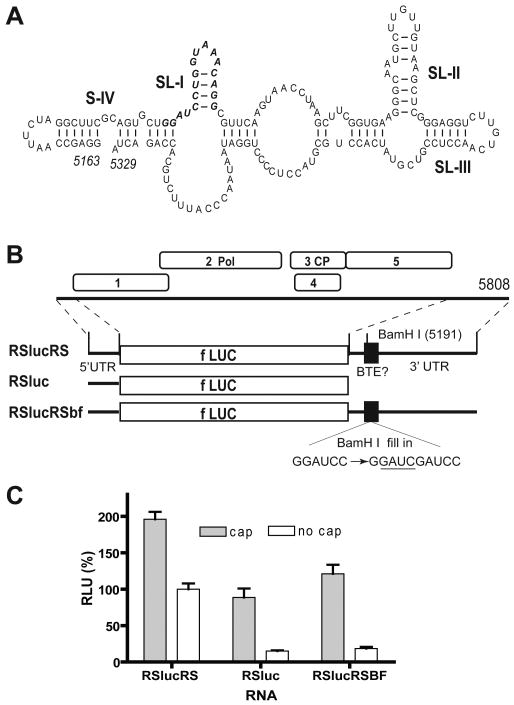
The genome organization and sequence of recently-discovered Rose spring dwarf-associated virus (RSDaV) are very similar to those of luteoviruses including BYDV (Salem et al., 2008), indicating that RSDaV belongs in genus Luteovirus. The 3′ UTR of RSDaV also harbors a 17 nt CS (bases 5190-5206) that differs from the BTE consensus at one base (a G-U transversion at position 10, Table 1). Downstream of this sequence is a predicted stem-loop capable of forming a kissing-loop interaction with the 5′ UTR, like the BYDV BTE. As in the case of BYDV, this stem-loop contains five predicted loop bases (UUGUC in RSDaV, UGUCA in BYDV) complementary to loop sequences in the predicted 5′ end of subgenomic RNA1 and the 5′ end of genomic RNA (Salem et al., 2008). However, the predicted RSDaV BTE secondary structure differs strikingly from other known and predicted (below) BTEs (Fig. 2A). All known BTEs are predicted to contain stem loops that radiate from a central hub, while the putative RSDaV BTE is predicted to have large tracks of unpaired or weakly paired sequence between predicted stem-loops SL-I, SL-II and SL-III and these putative stem-loops don't all project from a central hub. Many suboptimal structures with very similar minimum free energies were predicted by Mfold (Salem et al., 2008), indicating a relatively unstructured RNA.
Fig. 2.
RSDaV 3′UTR contains a functional BTE. (A) A representative predicted secondary structure of the RSDaV BTE (Salem et al., 2008). 17 nt CS is shown in bold italic. (B) Genome organization of RSDaV RNA and maps of translation reporter constructs. RSlucRS has both UTRs of RSDaV flanking the luciferase coding sequence (fLUC). RSluc has only the RSDaV 5′UTR. RSlucRSBF differs from RSlucRS only by a 4 nt GAUC insertion (underlined) in the BamH I site. Bases are numbered according to their position in the RSDaV genome. (C) Relative translation activities of capped and uncapped reporter mRNAs in wheat germ extract. Luciferase activities are normalized to uncapped RSlucRS (defined as 100%). Error bars indicate standard error.
To test whether the predicted RSDaV BTE confers cap-independent translation, it was tested by the same type of construct that was used for TBTV: an mRNA encoding the firefly luciferase gene flanked by the complete 5′ and 3′ UTRs of RSDaV genomic RNA (construct RSlucRS, Fig. 2B). The uncapped transcript yielded 50% as much luciferase as the capped transcript (Fig. 2C). The ratio of luciferase obtained from uncapped to capped transcripts dropped to 15-17% when reporter RNAs lacking the viral 3′ UTR, or containing the GAUC insertion in the BamH I site of the 17 nt CS were translated. Importantly, the capped versions of constructs with the mutant or deleted 3′ UTR translated about as efficiently as uncapped mRNA containing wild type UTRs, as was observed for other BTEs (e.g. Fig. 1C). Thus, we conclude that the RSDaV 3′ UTR contains a functional 3′ BTE.
Structure solution probing of BTE elements
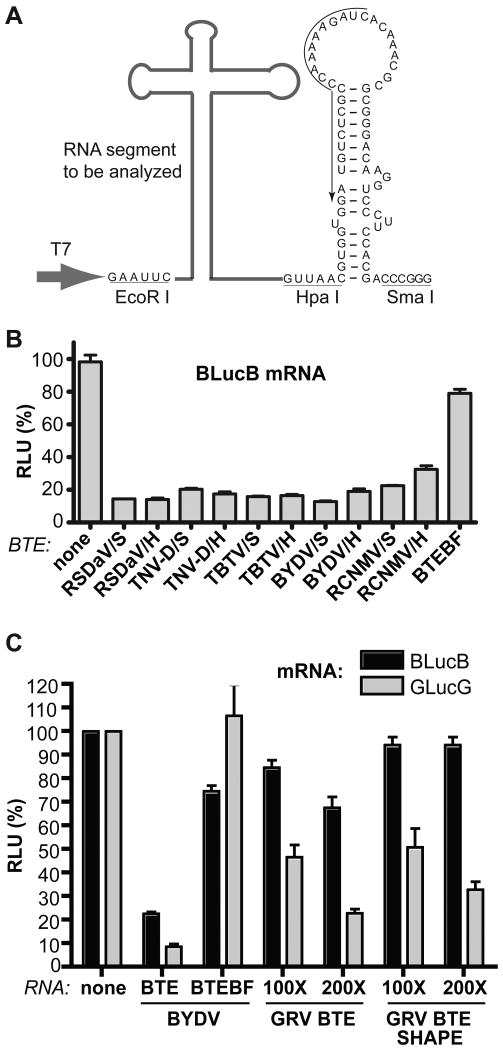
Although the secondary structures of many BTEs have been predicted, only that of BYDV has been determined directly. Because the predicted structures are so diverse, we set out to directly probe the secondary structures of diverse BTEs by Selective 2′-Hydroxyl Acylation analyzed by Primer Extension (SHAPE) (Mortimer and Weeks, 2007; Wang, et al., 2009; Wilkinson, et al., 2006). SHAPE chemistry reveals the position of unpaired or otherwise conformationally unconstrained nucleotides, whose 2′-hydroxyl group is able to form a 2′-O-ester product with SHAPE reagents N-methylisatoic anhydride (NMIA) or 1-methyl-7-nitroisatoic anhydride (1M7) (Mortimer and Weeks, 2007; Wilkinson, et al., 2006). The modified sites block reverse transcriptase so they can be identified by primer extension followed by polyacrylamide gel electrophoresis. Based on the previous solution structure probing of Pea enation mosaic virus (PEMV) RNA2 (Wang, et al., 2009), we designed a cassette which contains the T7 promoter sequence, and EcoR I and Hpa I sites upstream of PEMV RNA2 bases 3924-3981 (which are not part of a translation enhancer), which are followed by a Sma I site (Fig. 3A). The PEMV RNA2 sequence between the Hpa I and Sma I sites forms a stem-loop that serves as an excellent primer binding site (Wang, et al., 2009). To investigate the BTE structures, we took advantage of this sequence as a universal primer binding site, by placing the BTE of interest between the T7 promoter and the Hpa I site (Fig. 3A). Selected BTEs, were amplified and inserted between the EcoR I and Hpa I sites, and the resulting constructs were linearized with Sma I to allow transcription of RNAs for structure probing.
Fig. 3.
Cassette for structure probing of BTEs, and test of function by trans-inhibition of translation. (A) RNA cassette for structure probing and reverse transcription. The cDNA of the RNA element of interest (bold gray line) was inserted between EcoR I and Hpa I sites of the structure probing cassette upstream of nts 3924-3981 from PEMV RNA2 which form the stem-loop between the Hpa I and Sma I sites. Sma I-linearized plasmids were used as templates to transcribe the RNA elements for trans-inhibition assays and structure probing. The black arrow along the sequence indicates sequence to which the 32P-labeled oligomer was annealed for primer extension. Large gray arrow indicates start site of transcription by T7 RNA polymerase. (B) Relative translation levels of uncapped 4 nM BlucB (reporter mRNA with BYDV UTRs flanking firefly luciferase coding sequence) in wheat germ extract containing 400 nM of the indicated viral BTE RNAs from the cassette in panel A (sequences are shown in Table 2). DNA was linearized with Sma I or Hpa I prior to transcription, as indicated. (C) Relative translation levels of 4 nM BlucB (black bars) or GlucG (gray bars) in the presence of BTE RNAs lacking (BTE) or containing (BTE SHAPE) the PEMV sequence used for the SHAPE primer.
To ensure that the each BTE folded into a functional conformation in the context of the structure probing cassette, we tested the efficiency with which each BTE inhibited translation in trans, comparing the BTE RNA lacking the extraneous sequences to the version containing the stem-loop from the expression cassette, i.e. the same RNA that will be used in structure probing assays. Previously it was established that the trans-inhibition efficiencies of three unrelated CITEs: the STNV TED, BYDV BTE and PEMV RNA2 PTE, correlated well with their translation enhancing activity in cis (Gazo et al., 2004; Guo, et al., 2000; Wang, et al., 2009). This is because they function by binding translation initiation factor complex eIF4F, and trans-inhibition is likely due to competition by the trans-inhibiting CITE with the mRNA for eIF4F. BTEs representing all four genera, BYDV and RSDaV (Luteovirus), TBTV (Umbravirus), TNV-D (Necrovirus), and RCNMV (Dianthovirus), were added to a translation reaction containing the BYDV reporter mRNA BlucB. Except for the RCNMV BTE, all BTEs inhibited translation by 75% to 85% when added in 100-fold excess over the mRNA (Fig 4B). The RCNMV BTE reduced BlucB translation by 60-75%. Importantly, the negative control, BTEBF, reduced translation of BlucB by less than 20% when present at the same concentrations as the trans-inhibiting RNAs. The presence of the 3′ stem-loop in the structure probing cassette (Sma I-linearized transcripts) had no deleterious effect on trans-inhibition (Fig. 3B). In some cases presence of the primer binding site enhanced trans-inhibition (compare RCNMV transcripts from Sma I- vs. Hpa I-linearized templates, Fig. 3B).
Fig. 4.
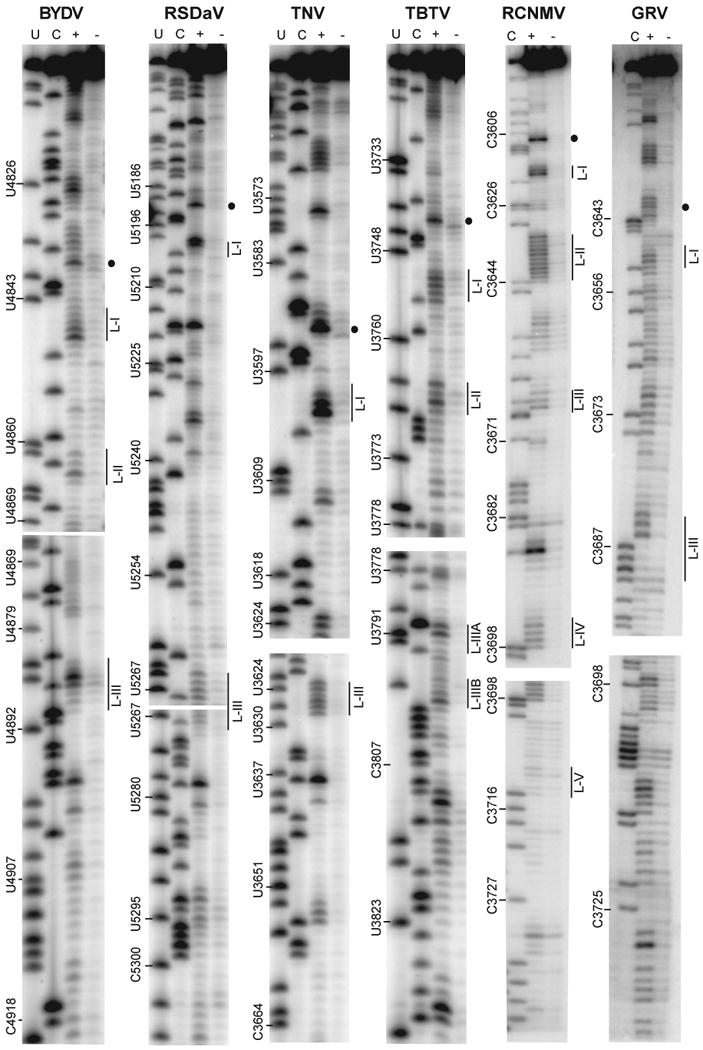
SHAPE analysis of BTEs from six viral genomes. Primer extension products from RNA treated (+) or not treated (-) with 5 mM 1M7 (See Materials and methods for details). The sequencing ladders showing positions of U and C residues in the BTEs (lanes U, C) were generated by reverse transcribing unmodified RNA in the presence of dideoxyATP (U lane) or dideoxyGTP (C lane) using the same 5′-labeled primer that was used for SHAPE. Numbers to the left of each gel indicate genomic positions of indicated bases. Position of the hypermodified G residue at the beginning of the 17 nt CS is indicated by the dot to the right of each gel. Regions corresponding to selected loops (Fig. 5) are indicated by L-[loop number].
In contrast to the above BTEs, the GRV BTE only slightly inhibited BlucB translation in trans (Fig. 3C). We speculated that because its 17 nt CS deviates most from consensus, the BTE of GRV may not compete well with the BYDV BTE in BlucB. To test this, we observed ability of the GRV BTE to trans-inhibit mRNA containing the GRV BTE in cis, i.e. GlucG. Indeed, the 100-fold excess of GRV BTE (with or without the SHAPE primer sequence from PEMV RNA 2) trans-inhibited translation of GlucG by 50%, and a 200-fold excess inhibited even more (Fig. 3C). We conclude that the RNAs used for structure probing are highly likely to form the conformations in which they function to stimulate translation in cis., and that the GRV BTE does not compete as well for translation machinery as the other BTEs.
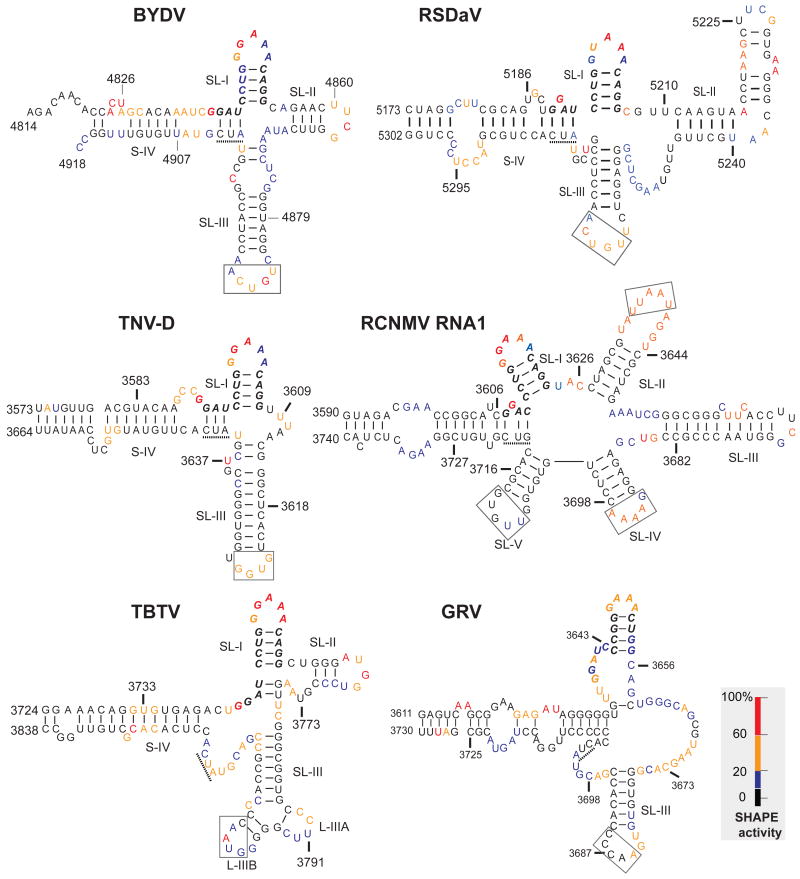
Results of primer extension following exposure of the BTE transcripts to SHAPE reagents reveal that the predicted loops are modified more heavily than flanking paired bases (Fig. 4). The SHAPE reactivities for each nucleotide in the BTE are superimposed on the best fitting RNA secondary structure in Fig. 5. SHAPE probing revealed the presence of SL-I topped by the GNRNA (N = any base, R = purine) pentaloop sequence in all tested BTEs. The first four bases of the 17 nt CS, GGAU (GGAC in RCNMV RNA1) showed some intriguing common features. One of the first two G's of the 17 nt CS was highly modified by SHAPE. In BYDV and TNV-D, the first G of the 17 nt CS is highly modified, while the adjacent GAU sequence has potential to base pair to the AUC sequence (underlined, Fig. 5) adjacent to the opposite strand of stem IV. In the RCNMV BTE, the second G of the 17 nt CS is modified and the flanking G and AC may base pair to a GUC in the same relative position as the AUC in the above two BTEs. Thus there appears to be natural covariation to allow the C of the GGAC in RCNMV RNA1 to base pair in a similar fashion to stem-IV as the U in the GGAU sequence of the other BTEs. The TBTV and RSDaV BTEs also have potential base pairing between the GAU and an AUC at the end of stem-IV, but the SHAPE probing of the TBTV BTE favors different interactions, because in both cases the AUC is moderately accessible to SHAPE reagent, while the pairing most parsimonious with the data is between the AU doublet at positions 3 and 4 of the CS and a UG on the opposite side of stem-I (TBTV, Fig. 5). The only BTE lacking a highly modified G at the beginning of the 17 nt CS is the weaker, non-consensus GRV BTE in which G's at that position were modified only moderately.
Fig. 5.
Superposition of SHAPE reactivity of each nucleotide on the best-fitting secondary structures of BTEs. Bases are color coded based on the intensity of the bands in Fig. 4 which reflects the level of modification by 1M7. Nucleotides are numbered according to their positions in the viral genome. 17 nt CS is in bold italics. The three base sequence (AUC or GUC) complementary to the 5′ end of the 17 nt CS (see text) is underlined. Boxed bases are known (BYDV and TNV-D) or predicted (other BTEs) to base pair to the 5′ UTR.
Another characteristic structural element predicted by Mfold and verified by SHAPE in all BTEs is the highly stable SL-III. Although it is not always the third stem-loop downstream of SL-I, with the exception of the RCNMV BTE, we define SL-III as the GC-rich stem-loop containing a loop capable of base pairing to the 5′ UTR, as is the case for BYDV. The terminal loop of SL-III in BYDV and TNV-D BTEs was shown experimentally to base pair to a loop region in the 5′ UTR of the genomic RNA (Guo, et al., 2001; Miller and White, 2006; Shen and Miller, 2004). What we call SL-III in the RSDaV BTE is predicted to base pair to a loop in the genomic 5′ UTR (Salem et al., 2008). In contrast, the TBTV BTE interaction with the 5′ UTR remains ambiguous because the genomic 5′ UTR has only 10 nucleotides. Moreover, SHAPE analysis is consistent with a different conformation at the distal end of SL-III than predicted by Mfold as in Fig. 1. The end branches into two very short stem-loops instead of one three base-pair stem-loop protruding from a large asymmetrical bulge (compare TBTV Fig. 5 with Fig 1A).
The SHAPE results also differ from the three most stable structures of the RSDaV BTE predicted using Mfold (Salem et al., 2008) (Fig. 2A). The predicted stem-loop II does not exist, instead these bases participate in a long, bulged stem-loop that we call SL-II (Fig. 5). We were unable to identify a structure that reconciled all of the structure probing data, hence some unmodified nucleotides are shown as single-stranded and some modified ones are base paired. SHAPE probing revealed that SL-III is in a different position than predicted and, in fact, is located near SL-I, which more closely resembles its position in the other BTEs. However RSDaV SL-III is flanked by a very large number of unpaired bases. The basal helix of the RSDaV BTE also resembles the other BTEs more closely than predicted because SHAPE reveals that it connects to the viral genome at the end of the helix, rather than at the side of a stem-loop as predicted (compare Fig.6 with Fig. 2A).
At 150 nt long, the RCNMV RNA1 BTE (known as 3′TE-DR1, Mizumoto et al., 2003) is the longest and most structurally complicated of the BTEs. The SHAPE data are in good agreement with the structure predicted by Okuno's group (Mizumoto et al., 2003; Sarawaneeyaruk et al., 2009) (Fig. 5). However, the SL-II loop is larger than predicted and predicted loop V was poorly modified by 1M7 indicating that this region is inaccessible, base-paired or constrained in some other way. The UU bases in the SL-III loop were not reactive to 1M7, perhaps because they are constrained in a highly stable UNCG tetraloop flanked by a C-G base pair (Molinaro and Tinoco, 1995). The loops of SL-II, SL-IV and SL-V each have potential to base pair to predicted single stranded tracts in the 5′ UTR (boxed Fig. 5), but Sarawaneeyaruk et al. (2009) showed that no single loop in the RCNMV BTE is required to base pair to the 5′ UTR for cap-independent translation. However the authors did not quite rule out the possibility that any of the three loops may be sufficient for base pairing to the 5′ UTR.
Discussion
We define BTEs as RNA sequence elements, located naturally in the 3′ UTR, that confer efficient cap-independent translation, and that harbor a sequence resembling the 17 nt CS. We showed here that BTEs exist in viruses in four genera representing portions of two virus families. One genus, Umbravirus, has not been assigned to a family. To our knowledge, this is the first report of BTEs in the Umbravirus genus. Interestingly, a different umbravirus, PEMV RNA 2, contains a completely different CITE (the PTE) (Wang et al., 2009), and a fourth umbravirus, Carrot mottle mimic virus, seems to have neither a BTE nor a PTE in its 3′ UTR. This lack of relationship between type of CITE, and classification of the virus, extends to the other genera and their families. Of the three genera in the Luteoviridae, only genus Luteovirus harbors a BTE. The CITEs of the other Luteoviridae genera, if any, are unknown. Of the eight genera in the Tombusviridae, only two, Necrovirus and Dianthovirus harbor BTEs. The other Tombusviridae genera contain a variety of diverse, and apparently unrelated CITES (Meulewaeter et al., 1998; Gazo et al., 2004; Fabian and White, 2006; Scheets and Redinbaugh, 2006; Truniger et al., 2008; Stupina et al., 2008; Miller, et al., 2007; Miller et al., 2010), including one genus, Panicovirus, which harbors the same type of CITE (PTE) as the umbravirus PEMV RNA2 (Jeffrey et al., 2006; Wang et al., 2009).
Based on the results presented here and elsewhere (Guo, et al., 2000; Mizumoto et al., 2003; Sarawaneeyaruk et al., 2009), the consensus of known functional 17 nt conserved sequences is: GgAuCCuGGgAAACaGG (bases in lower case can be altered) and it must be able to form a four base pair helix. The consensus of the 17 nt CS of all known and predicted BTEs is ggAuCCuGggaaACaGG (Table 1). While it is not known whether all the non-consensus 17 nt conserved sequences are functional in their natural context, it is likely that the 17 nt CS tolerates different types of deviations, but not a large number in the same sequence.
The situation is more ambiguous with regard to secondary structure. All functional and structure-probed BTEs have the following features. (i) A long, bulged, basal connecting helix from which a number of helices radiate at the distal end. (ii) The 17 nt CS which begins at the distal end of the basal helix. (iii) Three of the 5′-proximal four bases of the 17 nt CS have potential to base pair to a complementary sequence on the opposite side of the basal helix, but if this base pairing exists it is not essential, and in TBTV these bases may instead pair to bases downstream of SL-I (Fig. 5). (iv) The first or second guanidylate of the 17 nt CS is modified by SHAPE reagent (dots, Fig. 4) indicating it is exposed and unlikely to be base paired. The lesser modification of the G in the 17 nt CS of the GRV BTE may explain the weak activity of the GRV BTE as measured in the trans-inhibition assay (Fig. 3C). (v) Bases 5-17 of the 17 nt CS form a four base pair helix connected by a five base loop that fits the consensus GNRNA pentaloop motif (or in rare cases a GNRA tetraloop). The GNRNA pentaloop is highly stable, resembles a GNRA tetraloop, and is known to be a protein binding site in bacteriophage lambda RNA (Legault et al., 1998). (vi) A stable stem-loop we call SL-III has at least six uninterrupted base pairs, of which at least four are G-C or C-G pairs. Its loop is predicted to base pair to the 5′ UTR, except for RCNMV in which the kissing stem-loop, if any, may be one of three different loops. (vii) Unpaired bases link together many of the helices radiating from the central hub, suggesting a relatively floppy structure or structure determined by non-Watson-Crick interactions. (viii) The orientation of SL-III relative to the hub is expected to be variable or flexible as it is linked to the hub by unpaired, SHAPE-modifiable bases.
We speculate that the basal helix functions as a platform for proper folding of the rest of the BTE. In all of the most stable structures predicted by Mfold analysis of the entire BYDV genome, the BTE folds correctly and protrudes distinctly (data not shown). We propose that this makes the BTE accessible to translational machinery and also aids in allowing the kissing stem-loop III access to the 5′ end of the genome to which it base pairs. One suggested model for the BTE mechanism was that the GGAUC at the 5′ end of the 17 nt CS may base pair to the 3′ end of the 18S rRNA, analogous to the Shine-Dalgarno interaction in bacteria (Wang, et al., 1997). However, the presence of natural deviations from consensus, such as the GGACC in RCNMV, and the lack of SHAPE accessibility of all but one highly modified G in this sequence render that model unlikely.
Previous data indicate that the BTE binds the eIF4G subunit of eIF4F, leading to our model that the BTE delivers eIF4F to the 5′ UTR by long-distance base pairing and that this, in turn, recruits the 40S ribosomal subunit to the 5′ end (Treder et al., 2008). The 40S subunit then binds the 5′ end of the RNA and scans to the start codon by the same mechanism as on normal capped mRNAs. (Rakotondrafara et al., 2006).
One candidate for a protein binding site in the BTE is the 17 nt CS. In all BTEs (except for the weak GRV BTE, which contains a GNRA tetraloop), the 17 nt CS contains a consensus GNRNA pentaloop motif. High resolution NMR structural analysis of bacteriophage lambda box b RNA revealed a similar stem-loop containing the GNRNA loop. In this loop, the G and A are paired and all bases except the protruding fourth base are stacked in a helix, folding as in a GNRA tetraloop (Legault et al., 1998). A GNRA or GNRNA loop is sufficient for binding of the lambda N protein, while the 4th protruding base of the GNRNA loop is required for subsequent binding of the E. coli transcription elongation factor NusA. We speculate that the 17 nt CS forms a similar structure which is bound specifically by eIF4F (directly by the eIF4G subunit and indirectly by the eIF4E subunit) and other as-yet unidentified BTE-binding proteins (Treder et al., 2008). The data presented here will guide future research into the RNA structural requirements for high affinity binding to these factors.
Materials and Methods
Plasmids
A full-length clone of the Tobacco bushy top virus (TBTV) genome was assembled from cDNA segments as described (Mo et al., 2003). TUlucTU is a firefly luciferase (luc2, Promega) reporter construct with firefly luciferase gene flanked by the 5′ and 3′ UTRs of TBTV (Fig. 1). The 10 nt 5′ UTR was fused directly between the T7 promoter and the luciferase start codon by PCR using primer T7-TBTV5′Luc2 5′ and Luc2-r1. The PCR product was digested by EcoR I and Xba I and inserted into EcoR I and Xba I-digested pKF19K-2 vector (TaKaRa). The 3′UTR of TBTV was amplified using primer TBTV 3′UTRf and TBTV 3′UTRr, digested with Pst I and Xba I and inserted into Pst I and Xba I-digested pKF19K-2 vector containing the TBTV 5′UTR and luc2 as described above. The resulting construct was designated as pTUlucTU. pTUlucTUBF is a mutant vector generated from pTUlucTU with a duplication of the GATC sequence in the 17 CS of the BTE. It was generated by digesting with BamH I and then filling in the sticky ends with DNA polymerase Pfx (Invitrogen) followed by blunt-end re-ligation.
pGlucG was derived by inserting the PCR-amplified 12 nt 5′UTR of GRV into EcoR I/Sca I-digested pTUlucTU, in place of the TBTV 5′ UTR. The resulting intermediary construct was cut with XbaI/XhoI in the 3′ UTR to replace the TBTV BTE sequence with the PCR fragment containing the GRV BTE sequence to give rise to the finished pGlucG construct (Fig. 1E).
pRSlucRS is a firefly reporter construct with the RSDaV 5′ and 3′ UTRs. The RSDaV 5′ UTR was amplified by overlapping PCR using primer RSDaV 5′UTRf/RSDaV OL1, RSDaV OL1/ luc72-53r to amplify two segments and then fused together using primer RSDaV 5′UTRf / luc72-53r. The resulting PCR product was digested with Xba I and Hind III and then ligated into Xba I and Hind III-digested pGEM®-luc (Promega). The 3′UTR of RSDaV was amplified using primers RSDaV 3′UTRf / RSDaV OL3, RSDaV OL4/ RSDaV 3′UTRr from two cDNA segments and then fused to generate the full RSDaV 3′UTR by PCR using RSDaV 3′UTRf /RSDaV 3′UTRr. The PCR product was digested with Stu I and Sac I and then ligated into Stu I and Sac I-digested pGEM®-luc vector with RSDaV 5′ UTR as described above. The resulting construct was designated pRSlucRS.
To generate RNAs from plasmids for trans-inhibition assays and solution structure probing, a universal cassette was generated and inserted into the PUC19 vector. The cassette contains the T7 promoter sequence, EcoR I and Hpa I sites, a stem-loop segment from PEMV RNA2 (nt 3924-3981) and a Sma I site sequence (Fig. 3). Individual BTE segments (Table 2) were amplified and inserted between the EcoR I and Hpa I sites.
TABLE 2. Sequences of the selected BTE elements for structure probing.
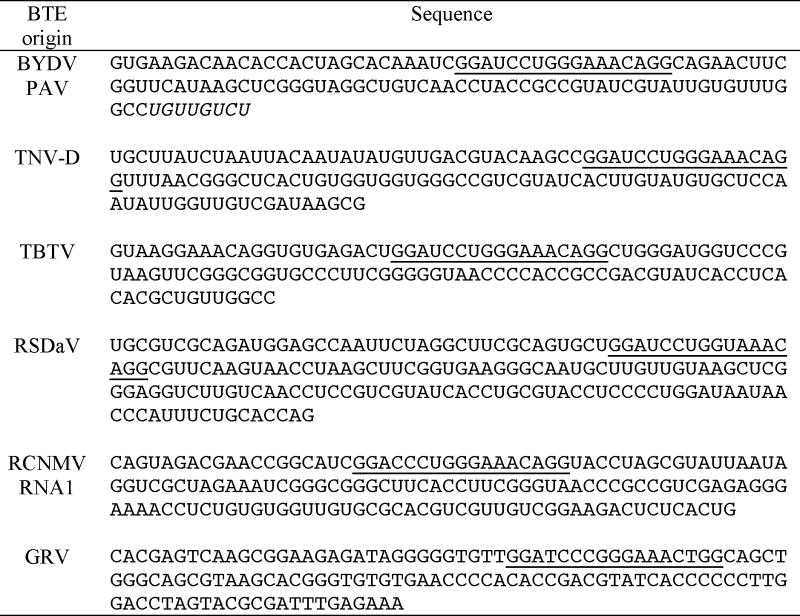 |
The cDNAs were amplified and inserted between EcoR I and Hpa I sites of the universal SHAPE vector (Fig. 3A). The 17 nt CS is underlined. Extra sequence introduced on the 3′ end of BYDV PAV BTE for extension of SL III was indicated in italic.
RNA preparation
To generate templates for transcription of mRNAs, plasmids pTUlucTU, pGlucG, pBlucB, and pRSlucRS were linearized with Hind III, Hind III, Sma I and Sac I, respectively. Capped and uncapped RNAs were synthesized by using Message Machine® and MEGAscript® (Ambion), respectively, according to the manufacturer's instructions. Small RNA segments for trans-inhibition assays and solution structure probing were transcribed from Hpa I or Sma I linearized plasmids using MEGAshortscript™ (Ambion) except for the GLucG derived segments. The GLucG BTE with or without the PEMV primer sequence were transcribed from Kpn I-linearized plasmid to include the SHAPE cassette for structure probing or from PCR-amplified GRV BTE segment designed to exclude the cassette for trans-inhibition studies. For trans-inhibition and structure probing, RNA integrity was estimated by 0.8% agarose gel electrophoresis. Final concentration and purity was determined by spectrophotometry.
In vitro translation
In vitro translation reactions were performed in wheat germ extract (Promega) as described (Wang, et al., 2009). Nonsaturating amounts of RNAs (0.2 pmol) were translated in wheat germ extract in a total volume of 25 μl with amino acids mixture, 93 mM potassium acetate and 2.1 mM MgCl2 according to the manufacturer's instructions. In trans-inhibition experiments, the mRNA and competitor RNA were mixed prior to addition to the translation reaction. After 1 hour incubation at room temperature, 2 μl of the translation reaction product was mixed into 50 μl of Luciferase assay reagent (Promega), and measured immediately on a GloMax™20/20 Luminometer (Promega).
In vivo translation
Uncapped TUlucTU or TUlucTUBF RNAs were co-electroporated into oat protoplasts with capped mRNA encoding Renilla luciferase as an internal control. Protoplasts were prepared and assays performed as described previously (Rakotondrafara et al., 2006). Four hours following the electroporation, the luciferase activities were measured with Dual Luciferase Reporter Assay System™ of Promega and Firefly relative light units (RLUs) were normalized against the Renilla relative light units. RLUs measured in the absence of added luciferase mRNA were subtracted from the values obtained with TUlucTU or TUlucTUBF mRNAs. All samples were tested in triplicate.
Structure probing
Plasmids with individual BTE segments in the PEMV RNA2-derived universal cassette were linearized with Sma I to be used as template for generation of RNA transcript using Megashortscript™ Kit (Ambion). Selective 2′-Hydroxyl Acylation analyzed by Primer Extension (SHAPE) (Wilkinson, et al., 2006) was applied to probe selected BTE elements following the procedure described previously (Wang, et al., 2009; Wilkinson, et al., 2006). Briefly, 500 ng of RNA was heat denatured and renatured in SHAPE buffer (50 mM HEPES-KOH, pH 7.2, 100 mM KCl and 8 mM MgCl2) at room temperature. 1-methyl-7-nitroisatoic anhydride (1M7) in 50 mM in anhydrous DMSO (Mortimer and Weeks, 2007) was mixed into the renatured RNA aliquot at a 1/10 (v/v) ratio. After 2 min at room temperature, the RNA was mixed with four-fold excess tRNA and precipitated in 3 volumes of ethanol and 1/10 volume 3M sodium acetate. Control RNA was treated with same amount of DMSO without 1M7. The primer (GATCTTTTTGGGCGAGACATC (Wang, et al., 2009), shown here in Fig. 3, was 5′ end-labeled with γ-[32P] ATP and used for the extension reaction. Primer extension, gel electrophoresis and visualization by phosphorimager were performed as described previously (Wang, et al., 2009; Wilkinson, et al., 2006). RNA secondary structures were deduced from solution probing data and the best fitting Mfold prediction (Zuker, 2003).
Acknowledgments
The authors thank Xiaohan Mo (Yunnan Academy of Tobacco Science, Kunming, P. R. China) for generously providing the TBTV cDNA clones, and Bryce W. Falk (University of California, Davis) for generously providing RSDaV cDNA clones. We thank Min Zeng for contributing to construction of pRSlucRS and preliminary translation assays. This work was supported by National Institutes of Health Grant R01 GM067104. This journal paper of the Iowa Agriculture and Home Economics Experiment Station, Ames, IA, Project No. 3608 was also supported by Hatch Act and State of Iowa funds.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Dreher TW, Miller WA. Translational control in positive strand RNA plant viruses. Virology. 2006;344:185–197. doi: 10.1016/j.virol.2005.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, White KA. 5′-3′ RNA-RNA interaction facilitates cap- and poly(A) tail-independent translation of tomato bushy stunt virus mRNA: a potential common mechanism for Tombusviridae. J Biol Chem. 2004;279:28862–28872. doi: 10.1074/jbc.M401272200. [DOI] [PubMed] [Google Scholar]
- Fabian MR, White KA. Analysis of a 3′-translation enhancer in a tombusvirus: a dynamic model for RNA-RNA interactions of mRNA termini. RNA. 2006;12:1304–1314. doi: 10.1261/rna.69506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filbin ME, Kieft JS. Toward a structural understanding of IRES RNA function. Current Opinion in Structural Biology. 2009;19:267–276. doi: 10.1016/j.sbi.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazo BM, Murphy P, Gatchel JR, Browning KS. A novel interaction of Cap-binding protein complexes eukaryotic initiation factor (eIF) 4F and eIF(iso)4F with a region in the 3′-untranslated region of satellite tobacco necrosis virus. J Biol Chem. 2004;279:13584–13592. doi: 10.1074/jbc.M311361200. [DOI] [PubMed] [Google Scholar]
- Guo L, Allen E, Miller WA. Structure and function of a cap-independent translation element that functions in either the 3′ or the 5′ untranslated region. RNA. 2000;6:1808–1820. doi: 10.1017/s1355838200001539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Allen EM, Miller WA. Base-pairing between untranslated regions facilitates translation of uncapped, nonpolyadenylated viral RNA. Mol Cell. 2001;7:1103–1109. doi: 10.1016/s1097-2765(01)00252-0. [DOI] [PubMed] [Google Scholar]
- Hentze MW, Gebauer F, Preiss T. cis-Regulatory Sequences and trans-Acting Factors in Translational Control. In: Mathews MB, Sonenberg N, Hershey J, editors. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 269–295. [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan E. Divergent IRES elements in invertebrates. Virus Res. 2006;119:16–28. doi: 10.1016/j.virusres.2005.10.011. [DOI] [PubMed] [Google Scholar]
- Jeffrey SB, Desvoyes B, Yamamura Y, Scholthof KB. A translational enhancer element on the 3′ -proximal end of the Panicum mosaic virus genome. FEBS letters. 2006;580:2591–2597. doi: 10.1016/j.febslet.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Karetnikov A, Lehto K. The RNA2 5′ leader of Blackcurrant reversion virus mediates efficient in vivo translation through an internal ribosomal entry site mechanism. J Gen Virol. 2007;88:286–297. doi: 10.1099/vir.0.82307-0. [DOI] [PubMed] [Google Scholar]
- Karetnikov A, Lehto K. Translation mechanisms involving long-distance base pairing interactions between the 5′ and 3′ non-translated regions and internal ribosomal entry are conserved for both genomic RNAs of Blackcurrant reversion nepovirus. Virology. 2008;371:292–308. doi: 10.1016/j.virol.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Kneller ELP, Rakotondrafara AM, Miller WA. Cap-independent translation of plant viral RNAs. Virus Res. 2006;119:63–75. doi: 10.1016/j.virusres.2005.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legault P, Li J, Mogridge J, Kay LE, Greenblatt J. NMR structure of the bacteriophage lambda N peptide/boxB RNA complex: recognition of a GNRA fold by an arginine-rich motif. Cell. 1998;93:289–299. doi: 10.1016/s0092-8674(00)81579-2. [DOI] [PubMed] [Google Scholar]
- Meulewaeter F, van Montagu M, Cornelissen M. Features of the autonomous function of the translational enhancer domain of satellite tobacco necrosis virus. RNA. 1998;4:1347–1356. doi: 10.1017/s135583829898092x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulewaeter F, van Lipzig R, Gultyaev AP, Pleij CW, Van Damme D, Cornelissen M, van Eldik G. Conservation of RNA structures enables TNV and BYDV 5′ and 3′ elements to cooperate synergistically in cap-independent translation. Nucleic Acids Res. 2004;32:1721–1730. doi: 10.1093/nar/gkh338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WA, Wang Z, Treder K. The amazing diversity of cap-independent translation elements in the 3′-untranslated regions of plant viral RNAs. Biochem Soc Trans. 2007;35:1629–33. doi: 10.1042/BST0351629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WA, White KA. Long-Distance RNA-RNA Interactions in Plant Virus Gene Expression and Replication. Annu Rev Phytopathol. 2006;44:447–467. doi: 10.1146/annurev.phyto.44.070505.143353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WA, Kraft JJ, Wang Z, Fan Q. Roles of cis-acting elements in translation of viral RNAs. In: Caranta C, Lopez-Moya JJ, Aranda M, Tepfer M, editors. Plant Viruses: Molecular Biology. Horizon Press; 2010. In press. [Google Scholar]
- Mizumoto H, Tatsuta M, Kaido M, Mise K, Okuno T. Cap-independent translational enhancement by the 3′ untranslated region of red clover necrotic mosaic virus RNA1. J Virol. 2003;77:12113–12121. doi: 10.1128/JVI.77.22.12113-12121.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo XH, Qin XY, Wu J, Yang C, Wu JY, Duan YQ, Li TF, Chen HR. Complete nucleotide sequence and genome organization of a Chinese isolate of tobacco bushy top virus. Arch Virol. 2003;148:389–397. doi: 10.1007/s00705-002-0919-y. [DOI] [PubMed] [Google Scholar]
- Molinaro M, Tinoco I., Jr Use of ultra stable UNCG tetraloop hairpins to fold RNA structures: thermodynamic and spectroscopic applications. Nucl Acids Res. 1995;23:3056–3063. doi: 10.1093/nar/23.15.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer SA, Weeks KM. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J Am Chem Soc. 2007;129:4144–4145. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- Niepel M, Gallie DR. Identification and characterization of the functional elements within the tobacco etch virus 5′ leader required for cap-independent translation. J Virol. 1999;73:9080–9088. doi: 10.1128/jvi.73.11.9080-9088.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakotondrafara AM, Polacek C, Harris E, Miller WA. Oscillating kissing stem-loop interactions mediate 5′ scanning-dependent translation by a viral 3′-cap-independent translation element. RNA. 2006;12:1893–906. doi: 10.1261/rna.115606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem NM, Miller WA, Rowhani A, Golino DA, Moyne AL, Falk BW. Rose spring dwarf-associated virus has RNA structural and gene-expression features like those of Barley yellow dwarf virus. Virology. 2008;375:354–360. doi: 10.1016/j.virol.2008.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarawaneeyaruk S, Iwakawa Ho, Mizumoto H, Murakami H, Kaido M, Mise K, Okuno T. Host-dependent roles of the viral 5′ untranslated region (UTR) in RNA stabilization and cap-independent translational enhancement mediated by the 3′ UTR of Red clover necrotic mosaic virus RNA1. Virology. 2009;391:107–118. doi: 10.1016/j.virol.2009.05.037. [DOI] [PubMed] [Google Scholar]
- Scheets K, Redinbaugh MG. Infectious cDNA transcripts of Maize necrotic streak virus: infectivity and translational characteristics. Virology. 2006;350:171–183. doi: 10.1016/j.virol.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Shen R, Miller WA. The 3′ untranslated region of tobacco necrosis virus RNA contains a barley yellow dwarf virus-like cap-independent translation element. J Virol. 2004;78:4655–4664. doi: 10.1128/JVI.78.9.4655-4664.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupina VA, Meskauskas A, McCormack JC, Yingling YG, Shapiro BA, Dinman JD, Simon AE. The 3′ proximal translational enhancer of Turnip crinkle virus binds to 60S ribosomal subunits. RNA. 2008;14:2379–2393. doi: 10.1261/rna.1227808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treder K, Pettit Kneller EL, Allen EM, Wang Z, Browning KS, Miller WA. The 3′ cap-independent translation element of Barley yellow dwarf virus binds eIF4F via the eIF4G subunit to initiate translation. RNA. 2008;14:134–147. doi: 10.1261/rna.777308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truniger V, Nieto C, Gonzalez-Ibeas D, Aranda M. Mechanism of plant eIF4E-mediated resistance against a Carmovirus (Tombusviridae): cap-independent translation of a viral RNA controlled in cis by an (a)virulence determinant. Plant J. 2008;58:716–727. doi: 10.1111/j.1365-313X.2008.03630.x. [DOI] [PubMed] [Google Scholar]
- Wang S, Browning KS, Miller WA. A viral sequence in the 3′-untranslated region mimics a 5′ cap in facilitating translation of uncapped mRNA. EMBO J. 1997;16:4107–4116. doi: 10.1093/emboj/16.13.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Guo L, Allen E, Miller WA. A potential mechanism for selective control of cap-independent translation by a viral RNA sequence in cis and in trans. RNA. 1999;5:728–738. doi: 10.1017/s1355838299981979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Treder K, Miller WA. Structure of a viral cap-independent translation element that functions via high affinity binding to the eIF4E subunit of eIF4F. J Biol Chem. 2009;284:14189–14202. doi: 10.1074/jbc.M808841200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- Zeenko V, Gallie DR. Cap-independent translation of tobacco etch virus is conferred by an RNA pseudoknot in the 5′-leader. J Biol Chem. 2005;280:26813–26824. doi: 10.1074/jbc.M503576200. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucl Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]