Abstract
Novel imaging of active transcription sites in interphase nuclei of intestinal epithelial cells in situ demonstrated that key genes associated with Wnt and Notch signaling were dynamically regulated as the cells underwent normal maturation during their migration along the mouse crypt-villus axis (CVA). However, oscillating patterns of activation of these genes were displaced along this axis in the histologically normal intestinal mucosa of Apc1638N/+ mice before tumor development. Gene expression profiling then showed that the normal reprogramming of cells along the crypt-villus axis (CVA) was dampened in the Apc1638N/+ mice, with an overrepresentation of c-myc target genes among those loci affected in the mutant mice. Moreover, in the Apc1638N/+ mice, there was a perturbed pattern of expression of lineage specific markers along the CVA consistent with transcription site repression of the Math1 gene, and genes encoding enzymes of every step of the tricarboxylic acid cycle were down-regulated in the crypt of Apc1638N/+ mice compared to wild-type, but not in the villus. These changes may alter energy metabolism and generate a pseudohypoxic state, suggested by elevated expression of Hif1α and its target genes. Thus, although intestinal tumors develop in Apc1638N/+ mice upon focal loss or inactivation of the wild-type allele, our results show that in the Apc1638N/+ mouse, inheritance of only a single wild-type Apc allele perturbs the dynamic and complex reprogramming underlying normal cell maturation which links epithelial function and homeostasis with architectural organization of the intestine.
Introduction
Reprogramming of Intestinal epithelial cells as they leave the progenitor cell compartment at the base of the crypt and mature along the villi towards the lumen generates the multiple cell lineages necessary for normal functioning of the tissue. Homeostasis is established by the correct allocation of cells to these different lineages. The reprogramming during this cell maturation involves altered expression of genes that drive proliferation and markers of differentiation lineages (1, 2). Disruptions in this reprogramming – either persistent expression of underlying drivers of proliferation or failure of proper differentiation – cause tumor development.
In ApcMin/+, ApcΔ716 and Apc1638N/+ mice, intestinal tumors develop when the inherited mutant Apc allele is complemented by somatic focal loss, mutation or silencing of the wild-type allele (3–7), conforming to the hypothesis that for tumorigenesis, both alleles of a tumor suppressor gene must be inactivated (8, 9). Unclear, however, is how the inherited mutation affects the intestinal mucosa, and probability of tumor formation, before reduction of Apc mutation to homozygosity. For example, an ~85% decrease of APC protein is necessary for generation of ~1 tumor per mouse (10), but mice that inherit one mutant Apc allele, or at tumor risk for other reasons, exhibit an expanded proliferative compartment (11).
We therefore compared the intestinal mucosa of C57Bl6 wild-type (WT) mice to histologically normal mucosa of congenic Apc1638N/+ littermates, before tumors develop due to focal loss of the WT allele. Unlike ApcMin/+ or ApcΔ716 mice, in which large numbers of tumors develop within months of birth, only ~3 tumors develop from 6–9 months in Apc1638N/+ mice (6), thus permitting analysis of effects of the inherited mutant allele before loss of the WT allele and development of mucosal pathology.
A novel method of active transcription site imaging in single cells in situ revealed that during normal maturation of intestinal cells along the crypt-villus axis (CVA), regulation of genes associated with Wnt and Notch signaling was much more dynamic than apparent from analysis of steady state RNA or protein levels. Moreover, there was significant displacement in the Apc1638N/+ mice of oscillating patterns of these active transcriptional units responsible for cell reprogramming. These pathways cooperate to maintain crypt cells in a progenitor cell phenotype and in lineage specific allocation (12), and we also found that the inherited mutation dampened cell reprogramming and perturbed expression pattern of lineage specific markers in the villus. Moreover, crypt cells exhibited altered expression of genes that encode enzymes of the tricarboxylic acid (TCA) cycle, and perturbed expression of Hif1α and its targets. Thus, the single wild-type Apc allele in the Apc1638N/+ mouse is insufficient to maintain normal pathways and patterns of cell maturation along the crypt-villus axis.
Materials and Methods
Mice
Generation, maintenance, genotyping and pattern of tumor formation of Apc1638N/+ mice are described (4, 6, 13). Experiments were approved by the IACUC of Montefiore Medical Center and the Albert Einstein College of Medicine. Apc1638N/+ mice and Apc+/+ littermates were fed a completely defined diet (AIN76A) (13). Upon sacrifice, the intestine was rapidly dissected, portions of each region fixed in formalin and then embedded in paraffin, or were used for isolation of cells from along the crypt-villus axis.
Transcription site detection
Active transcript sites were detected based on methods described (14, 15). Formalin fixed, paraffin embedded sections (4uM) were heated in a 55°C dry oven for 1 hour, placed in decloaking buffer for deparaffinization, cooled, and treated with ammonia/70% ethanol (20 minutes) and sodium borohydride (50 minutes, 4°C) to reduce autofluorescence. Pre-hybridization with 50% formamide/2XSSC was at room temperature, 30 minutes; slides were hybridized overnight with 20ng of probe at 37°C protected from light in a humidified chamber. Probes for FISH were designed using OLIGO-6.0 software, with specificity verified using the NCI GeneBank BLAST program. For each target nascent transcript, three to eight 50-mer DNA probes were synthesized with 4–5 modified thymidine bases conjugated to succinimidyl ester fluorescent Cy3 or Cy5 dyes (GE Healthcare). Fluorescence of the multiple probes for each target mRNA localized to a specific site. Following hybridization, slides were washed in prewarmed buffers on a shaker protected from light: 50% formamide/2XSSC, 20 minutes; 2XSSC, 1XSSC, 0.5XSSC for 15 minutes each; then in PBSM and nuclei stained with DAPI (4′,6-Diamidino-2phenylindole) before mounting with Prolong gold antifade (Molecular Probes). In transcription site detection, fluorescence intensity of hybridized probes is amplified by the many nascent transcripts within a small volume. Fluorescent signals were detected with an Olympus AX70 microscope, UApo 40X, 1.35NA and PlanApo60X, 1.4NA objectives, and a CoolSNAP-HQ CCD camera (Photometrics) using filters for DAPI (#SP100), FITC (#SP101), Cy3 (#SP-102v2), and Cy5 (#SP104v2) (Chroma Technology). Three-dimensional images acquired with a 200nm Z step size were analyzed using IPLab software version 3.61 (BD Biosciences). Fluorescent spots were identified as transcription sites on the basis of location within the DAPI-stained nuclei, fluorescence intensity, volume, shape, and absence of autofluorescence detected in the FITC channel. Active transcription sites for each locus were counted in a minimum of 50 well-oriented crypt-villi from 3 mice per genotype. The percentage of transcription sites for each gene was calculated from the total number of transcription sites and nuclei detected.
Expression array analysis
Epithelial cells were isolated progressively from the top of the villus (F1) to the bottom of the crypt (F10) as described (eg (1, 2). Expression profiling with RNA isolated from the F1 and F10 cells for each genotype was done on GeneChip Mouse Genome 430 2.0 Arrays (Affymetrix) for 4 mice for each cell position and genotype. Mean values by genotype and position for each sequence for were calculated, and the F1/F10 ratios calculated for each genotype, and then the absolute value of the ratio of these ratios calculated.
Western blots
Lysates of cells from fractions F10 (crypt bottom) to F2 (near villus top) were fractionated by electrophoresis, blotted and specific peptides detected as described (1). Antibodies: Hif1α (Cayman Chemical); VEGF (Santa Cruz); H2k (Cell Signaling). Cells from fraction 1 (villus tip) were not analyzed due to low yield. The experiment was repeated 3 times, using cells isolated from 3 different mice of each genotype.
Results
Dynamics of Intestinal cell reprogramming in wild-type and Apc1638N/+ mice
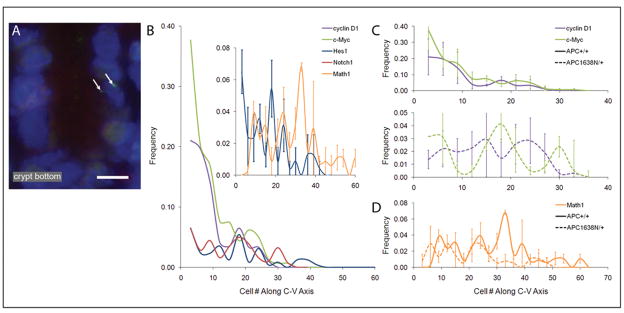
Analysis of intestinal cell reprogramming during maturation along the crypt-villus axis employed a validated method that detects active transcription of specific loci in single cells in situ (14–18). Since highest concentration of an RNA sequence is at its site of transcription, fluorescent oligonucleotide probes hybridizing to these transcripts produce a bright image at this site, indicating active transcription of the locus queried (figure 1A; additional examples in (14–20)). Assay of activation of gene transcription in single cells in situ, unlike measurement of steady-state levels of RNA or protein, assesses the state of the “rheostats” (ie functional state of the transcriptional machinery) that respond rapidly to internal and external signals. In WT mice, frequency of cyclin D1 and c-myc active transcription sites was higher in cells near the bottom of the crypt (Fig 1B), consistent with function of these genes in driving cell–cycling in this compartment, and with reports they are direct targets of Wnt signaling, more active at the bottom of the crypt (21–23). Similarly, active Notch1 or Hes1 transcription sites, the latter a direct target of activated Notch signaling, were also more frequent in cells in the lower half of the CVA, although localization was not as striking as for cyclin D1 and c-myc. Importantly, reprogramming of cells as they mature along the CVA was highly dynamic: for each gene, there was an oscillating pattern of cells with locus activation along the CVA, which may reflect compensatory regulation that overshoots at each position of stimulation and repression, or biological compartmentalization of activation resulting from lateral inhibition during differentiation. These oscillations contrast with monotonic decreases and increases in steady state expression levels of genes that characterize proliferation or differentiated functions, respectively, along the CVA axis that we reported (1, 2, 24, 25).
Fig 1.
1A: 2 active transcript sites (alleles) encoding Hes1 in an intestinal interphase nucleus.. The number of active transcription sites for each gene was scored in relation to cell position from the bottom of well-oriented crypts. Active transcription sites were scored in each of 50 well-oriented crypt/villi, 3 mice per genotype. Mean number of cells with active transcription sites were binned for each set of 3 consecutive positions (eg 1–3, 4–6), and plotted as a function of position along the crypt-villus axis. 1B frequency of cells with active transcription sites along the crypt-villus axis for cyclin D1, c-myc, Notch1 and Hes1 in wild-type Apc+/+ mice. Insert directly compares positions along the crypt-villus axis in Apc+/+ mice of cells with active transcription sites for Hes1 and Math1. 1C-top position of cells with active transcription sites for cyclin D1 and c-myc in the histologically normal intestinal mucosa of Apc+/+ mice; 1C-bottom, of Apc1638N/+ mutant mice; 1D directly compares distribution of cells with active transcription sites for Math1 in Apc+/+ and Apc1638N/+ mice.
Figure 1B (insert) also shows that oscillations of cells with active Hes1 or Math1 transcription sites, both components of Notch signaling, had different periodicities along the CVA, especially at cell positions 18 and 36. Consistent with repression of Math1 by Hes1, there was weak pair-wise correlation of Hes1 and Math1 active transcription sites (Spearmann coefficient of correlation, r=0.116, p=0.625) demonstrating these genes were not transcriptionally active in the same cells at the same time. Indeed, when assayed simultaneously, no cell was identified in which Hes1 and Math1 were simultaneously active, regardless of genotype. In contrast, pair-wise correlation of Notch1 and c-Myc was significant (r=0.618, p=0.035), demonstrating concordant transcriptional activation of these genes that drive proliferation.
The histologically normal mucosa of Apc1638N/+ and Apc+/+ littermates were compared, with all mice on a congenic C57Bl6 background and fed a chemically defined control diet (AIN76A). This defined, invariant, diet minimizes environmental variables (eg phytochemical content) that can markedly alter Apc initiated tumorigenesis and thus underlying mechanisms (eg. (26–29). Differences between the two genotypes were clear: preferential transcription of the cyclin D1 and c-myc genes at the bottom of the crypt of WT mice was shifted to higher positions along the CVA of the Apc1638N/+ mice (fig 1C). Moreover, a major peak of active transcription of Math1 in the wild-type mice at cell positions 30–36, and a smaller peak at 36–42, were absent in the Apc1638N/+ mice (p<0.002; fig 1D), with additional repression of active Math1 transcription sites at cell position 9 that approached significance (p=0.06).
Attenuated cell maturation in Apc1638N/+ mice
Since canonical Wnt and Notch signaling cooperate in regulating intestinal cell maturation, we determined whether alterations in transcription site patterns of Apc1638N/+ mice perturbed maturation by assaying expression profiles of villus and crypt cells. We have documented that the cell fractions isolated by sequential elution exhibit gradients of proliferation and differentiation markers that decrease and increase monotonically along the CVA (1, 2, 24, 25). Here we determined gene expression profiles of the extremes of these gradients: top of the villus (F1 fraction) and bottom of the crypt (F10 fraction) of each of 4 Apc+/+ and 4 Apc1638N/+ mice.
As expected from different functions of cells in crypts and villi, unsupervised clustering using all 31,213 probe sets unambiguously distinguished cells from these two compartments, regardless of genotype (branch 1 and 2, supplemental fig. 1). These data also suggested genotypes were distinguishable, with 3 of 4 mice of each genotype separating between branches 1a and 1b, and 2c and 2d (supp. fig 1), leading to more in-depth analyses.
Four sets of differentially regulated sequences were distinguished: by crypt or villus position and by genotype (supplemental figure 2; Supplemental Tables S1-S4). The 3457 sequences differentially expressed between villus (F1) and crypt (F10) for WT mice were consistent with those we previously reported for intestinal epithelial maturation along the CVA (eg enriched in genes contributing to cell cycle regulation, enterocyte differentiation, cytoskeleton assembly, and lipid metabolism (1). Genotype comparisons identified 73 sequences differentially expressed between the WT and mutant genotypes in the villi (criteria: change >2 fold, and p<0.05) and 259 in crypt cells. Thus, changes were ~4 fold more frequent between genotypes in crypts, where Wnt signaling is normally active, than in the villi, where Wnt signaling may be shut down and therefore not readily compromised.
To address whether the wild-type Apc allele in the Apc1638N+ mice was haploinsufficient for regulating normal reprogramming along the CVA, the villus/crypt ratios (F1/F10) for the mean of each sequence were compared. Using combined criteria (>2 fold change, p<0.05), there was no sequence for which F1/F10 was elevated or repressed for either wild-type or Apc1638N/+ mice, but which showed reciprocal change in the other genotype (supplemental figure 3), consistent with the apparent normal histology and functioning of the intestinal mucosa in Apc1638N/+ mice. However, while there was overlap between sequence subsets for which F1/F10 was altered in wild-type or mutant mice (supplemental figure 3), there were also distinct differences in extent of change, illustrated by the following. Sequences were identified differentially expressed between the crypt and villi in the wild type mice (F1/F10 >2 fold change, p<0.05), and also altered in the same direction in the mutant mice, but with no criteria imposed for magnitude of change or p value. For the 1720 sequences down-regulated, 82% were down-regulated less in the Apc1638N/+ mice (figure 2A – area under red-dotted line); for the 1187 sequences up-regulated, 68% were up-regulated less in Apc1638N/+ mice (figure 2B). A Z-test for binomial proportions demonstrated that the probability these results were due to chance was very small (p<0.0001). Inserts (figure 2A,B) show p values for differences in expression in villus(F1)/crypt(F10) for each sequence in the Apc1638N/+ mice, with sequences in the same order on the abscissas of both the figures and the inserts. As predicted, sequences that changed less in Apc1638N/+ than in wild-type mice (ie, those to the left of the x-axis, figures 2A, B), show higher p values in mutant mice, and hence less significant change. Importantly, for the mutant mice (inserts), most p values remain above the significance criteria (eg p<0.05), Thus, general patterns of cell reprogramming along the crypt-villus axis in Apc1638N/+ mice are maintained, consistent with the normal histology of the tissue, but are dampened in comparison to reprogramming in the wild-type mice.
Fig 2.
A: 1720 sequences down-regulated in the F1 fraction relative to F10 for both genotypes B. 1187 sequences up-regulated in F1 fraction relative to F10 for both genotypes. Inserts in A and B are the p values for F1 relative to F10 in Apc1638N/+ mice of the same sequences on the ordinates in each panel in the same order. C: Overlap between c-myc targets and the 2217 sequences with dampened F1/F10 values (ie sequences for which values fall below the dotted red line in A and B) in the mutant mice compared to F1/F10 in wild-type. A binomial Z-test determined that the intersection of 255 genes was in excess of chance. The underlying probability of selecting a c-myc gene was p=0.0374 (=1160/31000); the null hypothesis was p=0.0374 vs. p not equal 0.0374 and the test was based on randomly selecting 2217 genes from among the 31,000. The data yielded a z = 19.3, corresponding to P<0.0001: i.e., the 255 genes among the 2217 identified are in excess of that expected by chance.
Of the 2217 sequences attenuated in change along the CVA in mutant compared to WT mice (ie below the lines in fig. 2A,B), 255 overlapped with a c-myc target data base of 1160 sequences (30) a highly significant enrichment of c-myc target genes among sequences modulated in alteration in the Apc1638N/+ mice (p<0.0001, fig 2C). This is consistent with displaced distribution of c-myc active transcription sites in the mutant mice (Fig 1), and is important in that c-myc expression drives normal maturation of intestinal epithelial cells along the CVA (1), and its derepression is necessary for Apc initiated tumor formation (31, 32).
Perturbed lineage specific markers in Apc1638N/+ mice
Wnt and Notch signaling are determinants of lineage specific allocation of intestinal progenitor cells (33): Math1 expression drives secretory cell differentiation (34, 35) while in its absence, the default is enterocyte differentiation (35). We therefore determined whether alterations in transcription site distribution in the mutant mice, and especially repression of Math1 transcription site activation (Fig 1), was reflected in the expression pattern of lineage specific markers.
Because sample size was limited, and expected differences in expression modest, we present the individual data points for each gene in each of 4 Apc1638N/+ mice compared to 4 WT mice, along with the median value for each gene, which minimizes the contribution of outliers. While changes by genotype for each gene were not statistically significant, the overall differences in pattern of expression in the mutant compared to the WT mice was striking. The median of 9 of 10 secretory cell markers was lower in the Apc1638N/+ mice than in WT (Fig 3A). This included two markers of the secretion of mucus by goblet cells (Muc2 and Tff3) and 7 of 8 markers of the heterogenous population of enteroendocrine cells, which secrete a variety of hormones, growth, and neuroendocrine factors. The exception was glucagon, normally a product of pancreatic alpha cells, but processed by post-translational cleavage to produce GLP-1 in intestinal enteroendocrine cells. In contrast to this decreased pattern of secretory cell markers, all 6 enterocyte cell markers were elevated in villus cells of mutant mice compared to WT (Fig 3A). Thus, while perturbation of expression of each gene is modest in the histologically normal intestinal mucosa of Apc1638N/+ mice, the pattern of decreases of secretory, and increases in enterocyte, markers is consistent with the decrease in active Math1 transcription sites along the CVA.
Fig. 3.
Ratio of lineage specific marker expression in the villi (F1 cell fraction) of 4 Apc1638N/+ mice relative to 4 Apc+/+ mice. Individual data points, and the median for each gene (horizontal line) are shown. Data are plotted on a log2 Y axis for markers of goblet cells, enteroendocrine cells and enterocytes.
Deregulated metabolic pathways in Apc1638N/+ mice
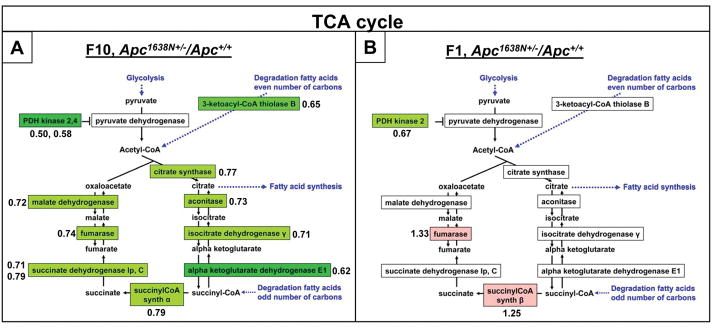
We reported that dietary risk for colon and intestinal tumors in mice was characterized by decreased expression in the histologically normal intestinal and colonic mucosa of sequences encoding enzymes of the TCA cycle, and suggested that elevated Wnt signaling was associated with shift towards glycolytic metabolism – a metabolic state well documented in colon tumors, but not known to exist in the normal tissue at risk (27). We therefore analyzed sequences encoding TCA cycle enzymes in relationship to genetic risk. Expression of the sequence encoding at least one enzyme of every step in the TCA cycle, as well as of 3-ketoacyl-CoA thiolase, which generates acetyl-CoA from metabolism of lipids, was decreased by 20–50% in the crypts of Apc1638N/+ mice compared to WT (Fig. 4A), similar to changes in the mucosa for dietary induced risk (27). In contrast, these genotype associated changes were not seen in villus cells (Fig 4B). Thus, the overall pattern of expression suggests a functional shift in metabolism specifically localized to the bottom of the crypt.
Fig 4.
Gene expression profiles were determined utilizing Affymetrix expression arrays (figure 2). Ratios of sequence mean expression compared data from F1 cell fractions (top of villus) for 4 different Apc1638N/+ mice to the same fraction for 4 different Apc+/+ mice. The same was done for F10 cell fractions (crypt bottom). Green color depicts decreased expression, red increased expression, with extent of change depicted by color intensity. For those sequences for which change is indicated, the ratio of expression in Apc1638N/+ to WT mice is shown next to the sequence. Where there are two numbers, they refer to the 2 different isoforms of the gene indicated.
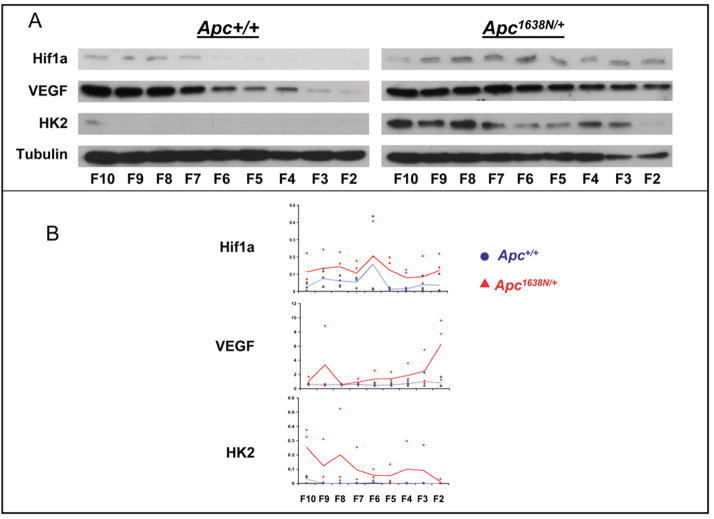
Oncogenic mutations in enzymes of the TCA cycle can generate a pseudohypoxic response and a shift of metabolism towards glycolysis, characterized by increased expression of Hif1α, a transcription factor regulated post-translationally that coordinates hypoxic response (36, 37). Hif1α protein levels from each of the fractions along the CVA axis of Apc1638N/+ mice were elevated in a representative experiment (fig 5A), and the mean Hif1α protein levels were elevated in each fraction in the mutant mice (3 experiments, 3 different mice of each genotype (fig 5B)). The elevation increased in fractions 6 to 2 that encompass villus cells expressing differentiated functions (1), although this was not statistically significant for individual cell positions or when the curves were modeled by a repeated measures analysis of patterns of expression across CVA levels and genotypes (fig 5, figure legend). However, VEGF protein, an angiogenic factor regulated by Hif1α, was progressively increased in expression in the same fractions 6 to 2 in the villus in Apc1638N/+ mice relative to wild-type (fig 5A,B). A quadratic model showed this to be a significant genotype by CVA level interaction (p<0.015), suggesting a curvilinear increasing trend from crypt bottom to villus top (legend, fig 5). Finally, hexokinase 2, another Hif1α target, was at background levels in every fraction in wild-type mice, but was elevated in every fraction in Apc1638N/+ mice (fig 5A, B). Here, a linear model (legend, fig 5) revealed a significant genotype by CVA level interaction (p<0.029) suggesting a decreasing trend from crypt bottom to villus top.
Fig 5.
Panel A immunoblot results from 1 of the 3 experiments; panel B quantitative analysis of the data from the 3 different experiments. Mixed models repeated measures analysis (MMRMA) was used as an exploratory tool to discover patterns of expression across crypt levels and between genotypes. The models were constructed first with crypt level as a dummy variable and, in cases where there appeared to be a linear or curvilinear trend, as a linear or a quadratic model, respectively. The interaction terms for genotype x crypt level were included in an appropriate way in each of the models, using p<0.05 as a guide to whether a particular model was significant. For Hif1α protein, the data suggest pattern of expression across crypts-villi is the same for both genotypes but that expression levels for Hif1α are uniformly higher for Apc1638N/+ mice (although the MMRMA could not verify this observation). For VEGF protein, the quadratic model revealed significant genotype x CVA interaction (quadratic p<0.015, linear p<0.006), suggesting curvilinear increasing trend in expression from F10 to F2 for Apc1638N/+ mice, while the expression level is relatively constant across crypt-villi for Apc+/+ mice. For Hk2 protein, the linear model revealed a significant genotype x CVA interaction (p<0.029), suggesting a linear decreasing trend in expression from F10 to F2 for Apc1638N/+ mice, while expression level is relatively constant across crypts-villi for genotype Apc+/+.
Discussion
These data demonstrate that although the intestinal mucosa of the Apc1638N/+ mouse exhibits normal histology and function, the complex reprogramming of intestinal epithelial cells as they migrate from the progenitor cell compartment in the crypt is perturbed. This was determined by altered patterns of transcriptional activation along the CVA of key genes in the Notch pathway (Hes1 and Math1), and two genes (cyclin D1 and c-myc) that drive proliferation in the progenitor cell compartment regulated by Wnt and Notch signaling, fundamental developmental pathways that cooperate in maintaining the crypt progenitor cell compartment. Expression profiling then showed that although general patterns of reprogramming were maintained in villus compared to crypt cells in the mucosa of Apc1638N/+ mice, there was a significant compromise in the reprogramming in the mutant mice compared to their WT littermates. These changes in the mutant mice encompassed perturbed profiles of lineage specific markers and altered expression of sequences that govern metabolic patterns. Changes are modest, consistent with the fact that the tissue continues to appear morphologically and functionally normal, but overall patterns of change indicate that it is significantly perturbed in comparison with the mucosa in wild-type mice. It has been reported that enterocyte migration along the crypt-villus axis is decreased in ApcMin+ mice (38), consistent with the dampened maturation of cells in the villi. Although this was not detected in the Apc1638N/+ mouse (39), this may be related to the much more modest tumor phenotype in Apc1638N/+ compared to ApcMin/+.
In the Apc1638N/+ mouse, loss of the mutant allele is not detected until development of frank tumors (40), an observation we confirmed ((41), and additional data not shown). Moreover, we determined that the expression of the wild-type allele Apc is reduced in the histologically normal mucosa by 40–60%, coupled with a 25% reduction in APC protein of 25% (not shown). In addition, the Apc1638N/+ mice do not accumulate significant levels of a truncated APC protein encoded by the mutant allele. Therefore, we conclude that the changes in the underlying molecular biology of the mucosa are due to haploinsufficiency of the wild-type allele.
The shift of cells exhibiting active cyclinD1 and c-myc transcription, and Notch signaling, along the CVA in the mucosa is important because these genes and pathways likely drive the expanded proliferative compartment that characterizes the mucosa at genetic and/or nutritional risk for tumor development (11). Continued proliferation of cells with a progenitor cell phenotype into zones where cell cycling is normally repressed can contribute to hyperplastic growth and expand the stem-like cell compartment which must be targeted by the loss of the second Apc allele for tumors to form (42). Affects on apoptosis may be less important: rates in the intestinal mucosa are very low and a mutation affecting short-chain fatty acid metabolism that reduce this >90% does not cause tumor development (43). Further, targeted inactivation of Tcf4 in the mouse, which, in complex with β-catenin, is a major effector of intestinal Wnt signaling regulated by Apc, leads to post-partum lethality as the mucosa deteriorates and cannot be regenerated due to premature differentiation, but not apoptosis, of intestinal progenitor cells (44).
Math1 drives secretory cell differentiation of intestinal epithelial cells, and in its absence there is default to the enterocyte lineage (35). Thus, the decrease in 9 of 10 secretory cell markers and the complementary increase in 6 of 6 enterocyte markers was consistent with that predicted by the repression of Math1 transcription sites in the mutant mice. However, altered expression of these markers likely reflects perturbed coordination of differentiation programs, rather than significant shifts in overall lineage allocation, since the mucosa appears normal until focal loss of the second Apc allele and tumor initiation.
A novel finding was decreased expression of genes that encode enzymes of every step of the TCA cycle in crypt cells in Apc1638N/+ mice compared to Apc+/+ mice, in contrast to the lack of such changes in the villus cells of the same mice. This is similar to alterations we reported in the mucosa of both the small and large intestine of mice at nutritional risk for tumor formation (27), which we recently found is also enriched in the crypt (not shown). Here we have shown association of these changes with perturbed expression of Hif1α, and its targets, VEGF and hexokinase 2. We hypothesize that these data reflect a shift in the tissue towards glycolytic metabolism, and generation of a (pseudo)hypoxic state that promotes tumorigenesis. It has been suggested that a shift towards glycolysis favors proliferation, higher in the crypt, by providing biochemical intermediates for synthesis of macromolecules and increase in biomass (45). In regards to the potentially greater shift in the crypts of mutant compared to wild-type mice, it is important that hypoxia and Hif1α expression are characteristics of stem cell niches (46–50), and that genes encoding enzymes of the TCA cycle are bona fide proto-oncogenes that, when mutated, lead to accumulation of TCA cycle intermediates (51–53) that can trigger Hif1α expression either by succinate inhibition of prolyl hydroxylase activity, and/or by generating increases in reactive oxygen species (54). Moreover, elevated Hif1α expression has been shown to be a direct cause of intestinal polyp formation in Peutz-Jaeghers syndrome, mediating a metabolic shift that drives tumorigenesis (55). Down regulation of the TCA cycle, a shift towards glycolytic metabolism, and a hypoxic response contributing to higher probability of tumor development in the intestinal mucosa by either genetic or environmental influences can be important in both screening strategies for early detection and as targets for chemoprevention.
Wnt signaling may contribute to intestinal tumorigenesis as a continuum of effects related to extent of altered signaling (56), increases in Wnt signaling beyond those sufficient for initiation are necessary for intestinal tumor progression, (57), and embryonic stem cell differentiation is modulated as a function of extent of β-catenin signaling levels (58). Thus, it is tempting to speculate that the inherited Apc1638 mutation causes modest changes in Wnt signaling that drive the altered transcriptional and expression patterns. While steady state levels of expression of several Wnt target genes (c-myc, cyclin D1, Sox9, Lgr5 and jagged1) were, as expected, higher at the bottom of the crypt of both normal and mutant mice, these steady state levels were not significantly different at any position along the crypt-villus axis of Apc1638N/+ compared to WT mice (data not shown). However, whether Wnt signaling is functionally altered in the mucosa of Apc1638N/+ mice is not easily resolved. For example, if inactivation of one Apc allele decreases Apc expression, modestly increasing Wnt activity, this would greatly increase the probability that stochastic variations in expression of the wild-type allele could transiently exceed a threshold sufficient to significantly alter steady state levels of direct Wnt targets (59). This also applies to the variation in VEGF and HEK2 levels that are seen (figure 5), although these changes in Apc1638N/+ compared to Apc+/+ mice reach statistical significance. While such focal and transient changes might not be detected in cell populations isolated from the mucosa, the important effect of these stochastic variations in tumor suppressor gene expression has been discussed in detail (59). Alternatively, the alterations in the mucosa of Apc1638N/+ mice may depend on perturbation of one of the many other functions that have been reported for APC, rather than changes in Wnt signaling.
In summary, in Apc1638N/+ mice few tumors develop over an extended period and the histologically normal intestinal mucosa can be readily investigated. We have found that in this histologically normal mucosa, there are significant alterations in the dynamics of cell reprogramming along the crypt-villus axis and of markers of normal cell maturation. We have previously shown that some of these changes are present in the mucosa at dietary risk (27). Thus, just as alterations at distant tissue sites contribute to tumor metastasis by generating receptive environments, probability of tumor development at the primary site may be modulated by alterations that precede the reduction of the inherited mutation to homozygosity, or the generation of initiating mutations. Therefore these findings have important implications for understanding the mechanism of risk and tumor formation in this tissue, and for clinical approaches to early detection and prevention.
Supplementary Material
Acknowledgments
Supported by grants U54CA100926, RO1CA114265, RO1CA135561, R33CA-083208, EB2060, GM80247 and Cancer Center Support Grant P013330 from the National Cancer Institute.
References
- 1.Mariadason JM, Nicholas C, L’Italien KE, Zhuang M, Smartt HJ, Heerdt BG, et al. Gene expression profiling of intestinal epithelial cell maturation along the crypt-villus axis. Gastroenterology. 2005;128:1081–8. doi: 10.1053/j.gastro.2005.01.054. [DOI] [PubMed] [Google Scholar]
- 2.Smartt H, Guilmeau S, Nasser S, Nicholas C, Bancroft L, Simpson S, et al. p27kip1 regulates cdk2 activity in the proliferating zone of the mouse intestinal epithelium: potential role in neoplasia. Gastroenterology. 2007;207:232–43. doi: 10.1053/j.gastro.2007.04.043. [DOI] [PubMed] [Google Scholar]
- 3.Su L-K, Kinzler KW, Vogelstein B, Preisinger AC, Moser AR, Luongo C, et al. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science. 1992;256:668–70. doi: 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- 4.Fodde R, Edelmann W, Yang K, van Leeuwen C, Carlson C, Renault B, et al. A targeted chain-termination mutation in the mouse Apc gene results in multiple intestinal tumors. Proc Nat Acad Sci USA. 1994;91:8969–73. doi: 10.1073/pnas.91.19.8969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levy DB, Smith KJ, Beazer-Barclay Y, Hamilton SR, Vogelstein B, Kinzler KW. Inactivation of Both APC alleles in human and mouse tumors. Cancer Res. 1994;54:5953–8. [PubMed] [Google Scholar]
- 6.Yang K, Edelmann W, Fan K, Lau K, Kolli VR, Fodde R, et al. A mouse model of human familial adenomatous polyposis. J Exp Zool. 1997;277:245–54. [PubMed] [Google Scholar]
- 7.Oshima M, Oshima H, Kitagawa K, Kobayashi M, Itakura C, Taketo M. Loss of Apc heterozygosity and abnormal tissue building in nascent intestinal polyps in mice carrying a truncated Apc gene. Proc Nat Acad Sci USA. 1995;92:4482–6. doi: 10.1073/pnas.92.10.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knudson AG., Jr Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A. 1971;68:820–3. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knudson AG. Hereditary cancer, oncogenes and antioncogenes. Cancer Res. 1985;45:1437–43. [PubMed] [Google Scholar]
- 10.Li Q, Ishikawa TO, Oshima M, Taketo MM. The threshold level of adenomatous polyposis coli protein for mouse intestinal tumorigenesis. Cancer Res. 2005;65:8622–7. doi: 10.1158/0008-5472.CAN-05-2145. [DOI] [PubMed] [Google Scholar]
- 11.Lipkin M, Blattner WE, Fraumeni JF, Lynch HT, Deschner E, Winawer S. Tritiated thymidine (0p,0h)labeling distribution as a marker for hereditary predisposition to colon cancer. Cancer Res. 1983;43:1899–904. [PubMed] [Google Scholar]
- 12.Sancho E, Batlle E, Clevers H. Live and let die in the intestinal epithelium. Curr Op Cell Biol. 2003;15:763–70. doi: 10.1016/j.ceb.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 13.Yang WC, Mathew J, Velcich A, Edelmann W, Kucherlapati R, Lipkin M, et al. Targeted inactivation of the p21 WAF1/cip1 gene enhances Apc initiated tumor formation and the tumor promoting activity of a Western-style high risk diet by altering cell maturation in the intestinal mucosa. Cancer Res. 2001;61:565–9. [PubMed] [Google Scholar]
- 14.Capodieci P, Donovan M, Buchinsky H, Jeffers Y, Cordon-Cardo C, Gerald W, et al. Gene expression profiling in single cells within tissue. Nat Methods. 2005;2:663–5. doi: 10.1038/NMETH786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levsky JM, Shenoy SM, Pezo RC, Singer RH. Single-cell gene expression profiling. Science. 2002;297:836–40. doi: 10.1126/science.1072241. [DOI] [PubMed] [Google Scholar]
- 16.Wilson AJ, Velcich A, Arango D, Kurland AR, Shenoy SM, Pezo RC, et al. Novel detection and differential utilization of a c-myc transcriptional block in colon cancer chemoprevention. Cancer Res. 2002;62:6006–10. [PubMed] [Google Scholar]
- 17.Pezo R, Gandhi S, Shirley L, Pestell R, Augenlicht L, Singer R. Single-cell profiling of transcription site activation predicts chemotherapeutic response of human colorectal tumor cells. Cancer Res. 2008;68:4977–82. doi: 10.1158/0008-5472.CAN-07-6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maier S, Daroqui CM, Scherer S, Roepcke S, Velcich A, Shenoy SM, et al. Butyrate and vitamin D3 induce transcriptional attenuation at the cyclin D1 locusin colonic carcinoma cells. J Cell Physiol. 2009;218:638–42. doi: 10.1002/jcp.21642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larson DR, Singer RH, Zenklusen D. A single molecule view of gene expression. Trends Cell Biol. 2009;19:630–7. doi: 10.1016/j.tcb.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Darzacq X, Yao J, Larson DR, Causse SZ, Bosanac L, de Turris V, et al. Imaging transcription in living cells. Annu Rev Biophys. 2009;38:173–96. doi: 10.1146/annurev.biophys.050708.133728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tetsu O, McCormick F. B-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–6. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 22.Shtutman M, Zhurinsky J, Simcha I, Albanese C, D’Amico M, Pestell R, et al. The cyclin D1 gene is a target of the B-catenin/LEF-1 pathway. Proc Nat Acad Sci USA. 1999;96:5522–7. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He T-C, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, et al. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–12. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 24.Guilmeau S, Flandez M, Bancroft L, Sellers R, Tear B, Stanley P, et al. Intestinal deletion of protein O-fucosyltransferase in the mouse inhibits Notch signaling and causes entero-colitis. Gastroenterology. 2008;135:849–60. doi: 10.1053/j.gastro.2008.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flandez M, Guilmeau S, Blache P, Augenlicht LH. KLF4 regulation in intestinal epithelial cell maturation. Exp Cell Res. 2008;314:3712–23. doi: 10.1016/j.yexcr.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang K, Edelmann W, Fan K, Lau K, Leung D, Newmark H, et al. Dietary modulation of carcinoma development in a mouse model for human familial polyposis. Cancer Res. 1998;58:5713–7. [PubMed] [Google Scholar]
- 27.Yang K, Kurihara N, Fan K, Newmark H, Rigas B, Bancroft L, et al. Dietary induction of colonic tumors in a mouse model of sporadic colon cancer. Cancer Res. 2008;68:7803–10. doi: 10.1158/0008-5472.CAN-08-1209. [DOI] [PubMed] [Google Scholar]
- 28.Yang W, Bancroft L, Nicholas C, Lozonschi I, Augenlicht LH. Targeted inactivation of p27kip1 is sufficient for large and small intestinal tumorigenesis in the mouse, which can be augmented by a western-style high-risk diet. Cancer Res. 2003;63:4990–6. [PubMed] [Google Scholar]
- 29.Yang W, Velcich A, Lozonschi I, Liang J, Nicholas C, Zhuang M, et al. Inactivation of p21WAF1/cip1 enhances intestinal tumor formation in Muc2−/− mice. Am J Pathol. 2005;166:1239–46. doi: 10.1016/S0002-9440(10)62342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.website m. http://www.myccancergene.org/site/mycTargetDB.asp.
- 31.Ignatenko NA, Holubec H, Besselsen DG, Blohm-Mangone KA, Padilla-Torres JL, Nagle RB, et al. Role of c-Myc in intestinal tumorigenesis of the ApcMin/+ mouse. Cancer Biol Ther. 2006;5:1658–64. doi: 10.4161/cbt.5.12.3376. [DOI] [PubMed] [Google Scholar]
- 32.Sansom OJ, Meniel VS, Muncan V, Phesse TJ, Wilkins JA, Reed KR, et al. Myc deletion rescues Apc deficiency in the small intestine. Nature. 2007;446:676–9. doi: 10.1038/nature05674. [DOI] [PubMed] [Google Scholar]
- 33.Fre S, Huyghe M, Mourikis P, Robine S, Louvard D, Artavanis-Tsakonas S. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005;435:964–8. doi: 10.1038/nature03589. [DOI] [PubMed] [Google Scholar]
- 34.Yang Q, Bermingham NA, Finegold MJ, Zoghbi HY. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 2001;294:2155–8. doi: 10.1126/science.1065718. [DOI] [PubMed] [Google Scholar]
- 35.Jensen J, Pedersen EE, Galante P, Hald J, Heller RS, Ishibashi M, et al. Control of endodermal endocrine development by Hes-1. Nat Genet. 2000;24:36–44. doi: 10.1038/71657. [DOI] [PubMed] [Google Scholar]
- 36.Guzy RD, Sharma B, Bell E, Chandel NS, Schumacker PT. Loss of the SdhB, but Not the SdhA, subunit of complex II triggers reactive oxygen species-dependent hypoxia-inducible factor activation and tumorigenesis. Mol Cell Biol. 2008;28:718–31. doi: 10.1128/MCB.01338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keith B, Simon MC. Hypoxia-inducible factors, stem cells, and cancer. Cell. 2007;129:465–72. doi: 10.1016/j.cell.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahmoud NN, Boolbol SK, Bilinski RT, Martucci C, Chadburn A, Bertagnolli MM. Apc gene mutation is associated with a dominant-negative effect upon intestinal cell migration. Cancer Res. 1997;57:5045–50. [PubMed] [Google Scholar]
- 39.Mahmoud NN, Bilinski RT, Churchill MR, Edelmann W, Kucherlapati R, Bertagnolli MM. Genotype-phenotype correlation in murine Apc mutation: differences in enterocyte migration and response to sulindac. Cancer Res. 1999;59:353–9. [PubMed] [Google Scholar]
- 40.Smits R, Kartheuser A, Jagmohan-Changur S, Leblanc V, Breukel C, de Vries A, et al. Loss of Apc and the entire chromosome 18 but absence of mutations at the Ras and Tp53 genes in intestinal tumors from Apc1638N, a mouse model for Apc-driven carcinogenesis. Carcinogenesis. 1997;18:321–7. doi: 10.1093/carcin/18.2.321. [DOI] [PubMed] [Google Scholar]
- 41.Yang K, Popova N, Yang W, Lozonschi I, Tadesse S, Kent S, et al. Interaction of Muc2 and Apc on Wnt signaling and in intestinal tumorigenesis: potential role of chronic inflammation. Cancer Res. 2008;68:7313–22. doi: 10.1158/0008-5472.CAN-08-0598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–11. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 43.Augenlicht LH, Anthony GM, Chruch TL, Edelmann W, Kucherlapati R, Yang KY, et al. Short chain fatty acid metabolism, apoptosis and Apc initiated tumorigenesis in the mouse gastrointestinal mucosa. Cancer Res. 1999;59:6005–9. [PubMed] [Google Scholar]
- 44.Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, et al. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nature Genetics. 1998;19:379–83. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- 45.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cipolleschi MG, Dello Sbarba P, Olivotto M. The role of hypoxia in the maintenance of hematopoietic stem cells. Blood. 1993;82:2031–7. [PubMed] [Google Scholar]
- 47.Danet GH, Pan Y, Luongo JL, Bonnet DA, Simon MC. Expansion of human SCID-repopulating cells under hypoxic conditions. J Clin Invest. 2003;112:126–35. doi: 10.1172/JCI17669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morrison SJ, Csete M, Groves AK, Melega W, Wold B, Anderson DJ. Culture in reduced levels of oxygen promotes clonogenic sympathoadrenal differentiation by isolated neural crest stem cells. J Neurosci. 2000;20:7370–6. doi: 10.1523/JNEUROSCI.20-19-07370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramirez-Bergeron DL, Simon MC. Hypoxia-inducible factor and the development of stem cells of the cardiovascular system. Stem Cells. 2001;19:279–86. doi: 10.1634/stemcells.19-4-279. [DOI] [PubMed] [Google Scholar]
- 50.Studer L, Csete M, Lee SH, Kabbani N, Walikonis J, Wold B, et al. Enhanced proliferation, survival, and dopaminergic differentiation of CNS precursors in lowered oxygen. J Neurosci. 2000;20:7377–83. doi: 10.1523/JNEUROSCI.20-19-07377.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pollard PJ, Briere JJ, Alam NA, Barwell J, Barclay E, Wortham NC, et al. Accumulation of Krebs cycle intermediates and over-expression of HIF1alpha in tumours which result from germline FH and SDH mutations. Hum Mol Genet. 2005;14:2231–9. doi: 10.1093/hmg/ddi227. [DOI] [PubMed] [Google Scholar]
- 52.King A, Selak MA, Gottlieb E. Succinate dehydrogenase and fumarate hydratase: linking mitochondrial dysfunction and cancer. Oncogene. 2006;25:4675–82. doi: 10.1038/sj.onc.1209594. [DOI] [PubMed] [Google Scholar]
- 53.Koivunen P, Hirsila M, Remes AM, Hassinen IE, Kivirikko KI, Myllyharju J. Inhibition of hypoxia-inducible factor (HIF) hydroxylases by citric acid cycle intermediates: possible links between cell metabolism and stabilization of HIF. J Biol Chem. 2007;282:4524–32. doi: 10.1074/jbc.M610415200. [DOI] [PubMed] [Google Scholar]
- 54.Chandel NS, Maltepe E, Goldwasser E, Mathieu CE, Simon MC, Schumacker PT. Mitochondrial reactive oxygen species trigger hypoxia-induced transcription. Proc Natl Acad Sci U S A. 1998;95:11715–20. doi: 10.1073/pnas.95.20.11715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shackelford DB, Vasquez DS, Corbeil J, Wu S, Leblanc M, Wu CL, et al. mTOR and HIF-1alpha-mediated tumor metabolism in an LKB1 mouse model of Peutz-Jeghers syndrome. Proc Natl Acad Sci U S A. 2009;106:11137–42. doi: 10.1073/pnas.0900465106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Albuquerque C, Breukel C, van der Luijt R, Fidalgo P, Lage P, Slors FJ, et al. The ‘just-right’ signaling model: APC somatic mutations are selected based on a specific level of activation of the beta-catenin signaling cascade. Hum Mol Genet. 2002;11:1549–60. doi: 10.1093/hmg/11.13.1549. [DOI] [PubMed] [Google Scholar]
- 57.Oyama T, Yamada Y, Hata K, Tomita H, Hirata A, Sheng H, et al. Further upregulation of {beta}-catenin/Tcf transcription is involved in the development of macroscopic tumors in the colon of Apc Min/+ mice. Carcinogenesis. 2008;29:666–72. doi: 10.1093/carcin/bgn001. [DOI] [PubMed] [Google Scholar]
- 58.Kielman MF, Rindapaa M, Gaspar C, van Poppel N, Breukel C, van Leeuwen S, et al. Apc modulates embryonic stem-cell differentiation by controlling the dosage of beta-catenin signaling. Nat Genet. 2002;32:594–605. doi: 10.1038/ng1045. [DOI] [PubMed] [Google Scholar]
- 59.Cook DL, Gerber AN, Tapscott SJ. Modeling stochastic gene expression: implications for haploinsufficiency. Proc Natl Acad Sci U S A. 1998;95:15641–6. doi: 10.1073/pnas.95.26.15641. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.